Abstract
Identifying novel therapeutics for the treatment of substance use disorder (SUD) is an area of intensive investigation. Prior strategies that have attempted to modify one or a few neurotransmitter receptors have had limited success, and currently there are no FDA-approved medications for the treatment of cocaine, methamphetamine, and marijuana use disorders. Because drugs of abuse are known to alter the expression of numerous genes in reward-related brain regions, epigenetic-based therapies have emerged as intriguing targets for therapeutic innovation. Here, I evaluate potential therapeutic approaches and challenges in targeting epigenetic factors for the treatment of SUD and highlight examples of promising strategies and future directions.
Emerging support for epigenetic pharmacotherapies in SUD
Initially coined by Conrad Waddington in 1942, contemporary use of the term epigenetics generally refers to the broad spectrum of molecular mechanisms that regulate chromatin dynamics and gene expression, independent of changes in DNA sequence (For reviews on epigenetics see [1–5]). Biochemically, DNA methylation and histone modifications (e.g., acetylation, methylation and phosphorylation) are the primary epigenetic mechanisms that have been investigated. However, hundreds of additional modifications to DNA, histones, and RNAs, with yet to be determined functions, have been recently identified [6–8]. Acting as a guide or anchor for chromatin remodeling complexes, long non-coding RNAs are also considered to be part of the epigenetic repertoire by facilitating gene- and cell type-specific activity of ubiquitous histone and DNA-modifying proteins [9, 10]. In the central nervous system, recent evidence indicates that modifications to DNA and histones play an essential role in the development and maintenance of neuronal networks, learning and memories processes, and behavioral output in response to environmental stimuli [11]. In some cases, epigenetic alterations are also associated with enduring, pathophysiological changes in neural function and disease susceptibility [12].
In multifactorial, polygenic disorders such as SUD, epigenetics is a particularly intriguing area of research. Indeed, repeated exposure to drugs of abuse alters the expression of hundreds of genes in reward-related brain regions, triggering maladaptive changes at the molecular, cellular, and circuit levels that promote drug-seeking and -taking behaviors [13–15]. Concomitant deviations in histone- and DNA-modifying enzyme activity and gene-specific epigenetic modifications are also observed following drug use. For example, in preclinical studies, cocaine [16–22], methamphetamine [23, 24], amphetamine [25], nicotine [26, 27], 3,4-methylenedioxy-methamphetamine (MDMA) [28], opiates [29, 30], ethanol [31, 32], cannabis [33, 34], and inhalants [35] alter DNA and/or histone modification the in nucleus accumbens, a key brain region involved in reward processing. Similar to pre-clinical studies, variations in epigenetic modifications, non-coding RNAs, and histone-modifying enzymes have also been observed in post-mortem brain samples from human drug abusers [14, 36–38].
In animal models for SUD, pharmacological manipulation of some epigenetic-related proteins is sufficient to ameliorate drug-induced behavioral, transcriptional, and physiological changes [39]. For example, non-selective histone deacetylase (HDAC) inhibitors trichostatin A and phenylbutyrate dose-dependently reduced cocaine self-administration [40], and the HDAC3-selective inhibitor, RGFP-966, enhanced extinction of cocaine conditioned place preference (CPP) [41], an animal model that measures contextual reward processing. Sirtinol, a class III histone deacetylase (sirtuin) inhibitor, reduced cocaine conditioned place preference, whereas the sirtuin agonist, resveratrol, had the opposite effect [42]. Following repeated cocaine administration, histone acetylation reader protein, BRD4, is elevated in the nucleus accumbens and recruited to promoter regions of addiction-related genes, and pharmacological inhibition of BRD4 attenuated transcriptional and behavioral responses to cocaine and heroin [37, 43]. Additionally, pharmacological manipulation of other DNA- and histone-modifying enzymes such as, DNMT [44–53], KDM6b [54], G9a [20], and PRMT1 [55] alter behavioral responses to drugs of abuse (Table 1). Outside of small molecule inhibitors, viral-mediated manipulation of DNA- and/or histone-modifying genes has also been found to alter drug-seeking behaviors [56–61]. Thus, with the potential to reverse or normalize the extensive transcriptional dysregulation and maladaptive behaviors caused by repeated drug use, epigenetic pharmacotherapy is a promising area of drug discovery for SUD.
Table 1:
Effects of epigenetic pharmacotherapies on drug-seeking behaviors.
| Drug of abuse | Epigenetic inhibitor | Target(s) | Route of delivery | Behavioral effect | Reference(s) |
|---|---|---|---|---|---|
| Cocaine | TSA | HDACs | systemic | ↓ SA | [40] |
| Phenylbutyrate | HDACs | systemic | ↓ SA | [40] | |
| Garcinol | HATs | systemic | ↓ reinstatement of SA | [113] | |
| RGFP-966 | HDAC3 | systemic | ↑ extinction of SA | [68] | |
| RGFP-966 | HDAC3 | systemic | ↑ extinction of CPP | [41] | |
| RG108 | DNMTs | intra-NAc | ↓ incubation of craving | [44] | |
| TSA | HDACs | intra-NAcS | ↑ SA | [114] | |
| JQ1 | BETs | systemic and intra-NAc | ↓ acquisition of CPP | [43] | |
| SKLB-639 | PRMT1 | systemic | ↓ acquisition of CPP | [55] | |
| GSK-J4 | KDM6B | systemic | ↓ reinstatement of CPP | [54] | |
| methionine | DNA methylation | systemic | ↓ acquisition of CPP | [52] | |
| BIX01294 | GLP/G9a | intra-NAc | ↑ acquisition of CPP | [20] | |
| Sirtinol | Sirtuins | intra-NAc | ↓ acquisition of CPP | [42] | |
| Nicotine | phenylbutyrate | HDACs | systemic | ↓ acquisition of CPP | [115] |
| NaB | HDACs | systemic | ↓ reinstatement of SA | [116] | |
| Morphine | NaB | HDACs | systemic | ↑ extinction of CPP | [117] |
| NaB | HDACs | systemic | ↑ acquisition of CPP | [118] | |
| Heroin | JQ1 | BETs | intra-DS | ↓ SA | [37] |
| NaB | HDACs | systemic | ↑ reinstatement of SA | [119] | |
| Ethanol | NaB and MS-275 | HDACs | systemic | ↓ SA | [120] |
| RG108 | DNMTs | intra-mPFC | ↓ SA | [48] | |
| 5-Aza-dc | DNMTs | intra-mPFC | ↓ SA | [53] |
5-Aza-dc, 5-aza-2′-deoxycytidine; BETs, bromodomain and extra terminal domain; CPP, conditioned place preference; DNMTs, DNA methyltransferases; DS, dorsal striatum; GLP, G9a-like protein; HATs, histone acetyltransferases; KDM6B, lysine demethylase 6B; mPFC, medial prefrontal cortex; NaB, sodium butyrate; PRMT1, protein arginine methyltransferase 1; NAc, nucleus accumbens; NAcS, nucleus accumbens shell; SA, self-administration; TSA, Trichostatin A.
Epigenetic therapeutic innovation and potential treatments for SUD
Despite the thousands of lives lost during the current opioid epidemic and the high rates of cocaine-linked deaths in African American and Hispanic populations over the past 3 decades [62], a dearth of new and effective clinical treatments for SUD remains. In fact, almost all current FDA-approved pharmacological treatments for SUD and other psychiatric disorders are based on neurotransmitter receptor agonist/antagonist that were identified over 40 years ago [63] (Figure 1). It is clear, however, that SUD is more complex than changes in neurotransmission, and new multifaceted treatment options are urgently needed. Although preclinical studies have revealed a plethora of potential therapeutic targets [39, 64, 65], few pharmaceutical companies have shown interest in pursuing novel treatments for SUD [66]. This is particularly troubling considering that some companies contributed to and profited from the current opioid epidemic.
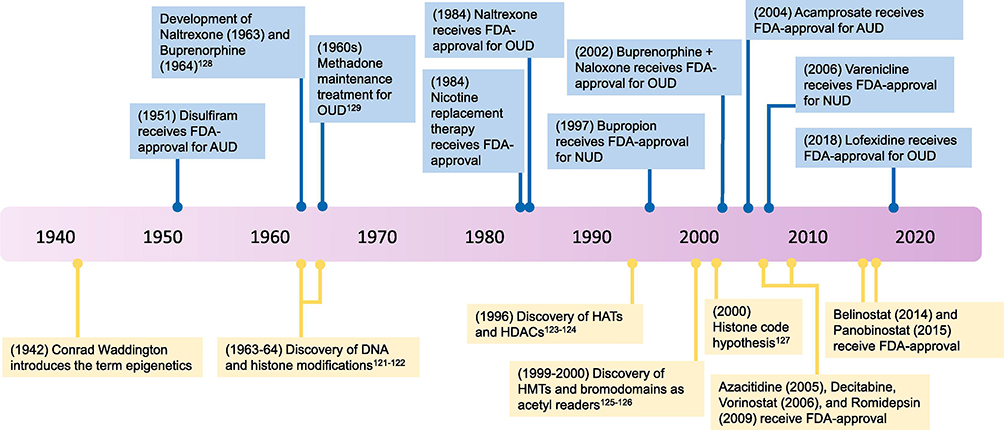
Figure 1: Timeline of milestones in SUD treatments and epigenetics.
(Top) Relatively few FDA-approved treatments for SUD are currently available, and most treatments target neurotransmitter receptors. There are no FDA-approved medications for cocaine, methamphetamine, and cannabis use disorders. (Bottom) The majority of histone and DNA-modifying proteins were identified in the last few decades. Multiple epigenetic-based therapies have received FDA-approval in recent years for the treatment of cancer, but there are currently no FDA-approved, epigenetic-related treatments for SUD. AUD, alcohol use disorder; FDA, Food and Drug Administration; HATs, histone acetyltransferases; HDACs, histone deacetylases; HMTs, histone methyltransferases; NUD, nicotine use disorder; OUD, opioid use disorder.
While many pharmaceutical companies have abandoned their psychiatry and neuroscience research programs, drug discovery and development in the field of epigenetics has recently flourished. Illustrating the recent surge of interest, over 200 epigenetic-related clinical trials are currently recruiting patients, compared to only 12 recruiting studies from 2000–2010 (clinicaltrials.gov). Seven epigenetic-related drugs have been FDA-approved for various types of cancers (Azacitidine, Decitabine, Vorinostat, Romidepsin, Panobinostat, and Belinostat) and epilepsy/bipolar disorder (valproic acid), and over 200,000 compounds targeting histone- and DNA-modifying enzymes are published on ChEMBL, a chemical database of bioactive molecules [67]. Though the vast majority epigenetic-based treatments are aimed at treating cancer, many of the same enzymes are dysregulated in the brain following chronic drug use. Several pre-clinical studies have found that epigenetic inhibitors, with similar mechanisms as FDA-approved epigenetic drugs, reduce drug-seeking behaviors [40, 53], indicating that related therapies could be tested in patients with SUD.
Decreasing drug intake and craving, alleviating withdrawal symptoms, and preventing relapse are the primary treatment goals for patients with SUD. In preclinical studies, the HDAC3-selective inhibitor, RGFP-966, enhanced memory processes involved in extinction of cocaine-seeking behavior and attenuated relapse-like behavior for cocaine [41, 68]. Based on these results, HDAC3-selective inhibitors might be useful as a co-therapy to facilitate cognitive-behavioral treatments aimed at reducing drug consumption and relapse in SUD patients. Increased drug craving during abstinence is mediated by numerous molecular neuroadaptations that drive drug-seeking behaviors [69, 70]. The BET inhibitor, JQ1, has been shown to decrease the expression of multiple genes and/or proteins (e.g., Bdnf, GluA1) [43, 71, 72] that are elevated during abstinence. Therefore, with the potential to reverse multiple drug-induced molecular factors that drive craving and relapse, BET inhibitors may promote abstinence in SUD patients. Persistent drug use to alleviate withdrawal symptoms is another aspect that contributes to the cycle of addiction. The HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) has been shown to reduce alcohol withdrawal-induced hyperalgesia in rodents [73], and perhaps similar epigenetic treatments may be an effective way to mitigate withdrawal symptoms in SUD patients. To facilitate clinical studies, the recent development of PET ligands for multiple histone-modifying proteins [74] could also be used to measure target engagement and brain biomarkers in SUD patients. Thus, epigenetic-based therapies for SUD and other psychiatric disorders may help fill a void where traditional medications have failed or have had limited success.
Epigenetic biomarkers for SUD
In patients with SUD, epigenetic differences are potentially detectable in easily accessible tissues such as blood, saliva, and cerebral spinal fluid [75]. Thus far, most studies have compared DNA methylation of specific genes or levels of non-coding RNAs in blood samples of control vs. SUD patients [76–79]. Some epigenetic changes have been correlated with drug history and/or propensity to relapse [80], an indication that the epigenome may offer a new source of biomarkers to identify patient subpopulations and treatment opportunities for personalized medicine. Given that that vast majority of drugs entering clinical trials for neuropsychiatric diseases do not produce marketable compounds [81], adding epigenetic biomarkers to clinical studies may diminish drug failure rates. Blood-derived epigenetic biomarkers, however, do have limitations, as epigenetic modifications are known to vary widely across tissues [82]. To overcome this challenge, PET ligands are now being utilized to measure histone-modifying proteins in the human brain. Radioligands for HDACs and bromodomain and extra terminal domain (BET) inhibitors have been developed and tested in animals and/or humans [74, 83, 84]. Age- and disease-related changes in HDAC activity are presently being studied in Alzheimer’s patients [85] and similar strategies are likely to be employed in future SUD studies. Therefore, the integration of epigenetic biomarkers with existing diagnostic tools (e.g., physiologic measurements and genetic variables) will conceivably improve therapeutic decision making and successful clinical outcomes in SUD patients.
Future directions and limitations of epigenetic-based therapies for SUD
With the staggering loss of life from the ongoing opioid crisis [86, 87], there has never been a more important time to identify effective treatments for SUD. So why are there so few pharmacological treatments options? SUD and other psychiatric disorders have long been stigmatized and perhaps this is one reason why many pharmaceutical companies have not pursued new therapeutic avenues. Another reason may be related to the complexity of SUD. Indeed, chronic drug use evokes a torrent of maladaptive neuroadaptations–intricate changes that may be difficult to reverse using traditional, single neurotransmitter receptor-targeted therapies. With the ability to regulate multiple transcriptional networks altered by drugs of abuse, epigenetic pharmacotherapy has emerged as a new treatment tactic. However, some may contend that epigenetic-based treatments are too risky because of their pleiotropic activities and potential side-effects. While adverse side-effects are always a threat for new therapies, one could argue that epigenetic-based medication is a tractable approach based on several factors. First, multiple epigenetic drugs have been FDA-approved and many more have completed phase I and II clinical trials, indicating that these treatments can be tolerated in humans. Furthermore, the potential side effects may be reduced in SUD, as pre-clinical SUD experiments typically require much lower effective doses and treatment frequency compared to cancer studies [43, 88]. Second, several safe and commonly-prescribed medications such as statins, antiepileptics and others exhibit direct epigenetic effects in addition to their commonly understood mechanisms of action [89–91]. Third, humans are exposed to endogenous and exogenous epigenetic inhibitors on a daily basis. The foods that we eat contain health-promoting compounds that modify epigenetic enzyme activity (e.g, catechins, curcumin, lycopene, resveratrol) [92]. Additionally, our gut bacteria and liver produce molecules with epigenetic effects (e.g., beta hydroxybutyrate) that enter the CNS and promote brain health [93, 94]. Though clearly more studies are needed to understand epigenetic-based treatment regimen for SUD (e.g., dose and duration), such medications are a potentially safe option.
In order for epigenetic pharmacotherapy to become a viable clinical option for SUD, further work needs to be done to verify the effectiveness of epigenetic inhibitors in advanced animal models that more accurately resemble specific aspects of compulsive drug seeking observed in human drug users. Thus far, the majority of studies have used the conditioned place preference procedure in male rodents to study the effects of epigenetic pharmacotherapies on drug-seeking behaviors. While this procedure is sufficient at measuring contextual reward processing, animals are only acutely exposed to drugs and the drugs are administered by the experimenter [95]. In self-administration procedures, the gold standard of SUD models, animals learn to press a lever (or nose poke) to receive a drug infusion [96]. Over the years, several variations of the self-administration procedure have been developed to model specific aspects of addiction, yet few studies have examined epigenetic mechanisms in these advanced models— a major impediment in epigenetic drug development for SUD (For more information on animal models of SUD, see [97–104]).
Testing epigenetic compounds that are FDA-approved or in phase II/III clinical trials and able to penetrate the blood brain barrier should be a priority, as these treatment options are more likely to be quickly approved for clinical testing in SUD patients. Likewise, natural or endogenous epigenetic inhibitors that are known to be relatively safe is another promising treatment route. Though epigenetic pharmacotherapy is still in its infancy, isoform-specific epigenetic inhibitors are already being developed to address potential side-effects of first-generation, non-selective epigenetic compounds. Some of these isoform-specific HDAC inhibitors have been shown to reduce drug-seeking behaviors in animals [68] and may be considered for clinical testing. Though selective epigenetic inhibitors will likely reduce potential side effects, a major limiting factor in epigenetic pharmacotherapy for SUD is that many epigenetic targets are expressed in all cells and/or tissues. Moreover, some drug-induced epigenetic modifications are regulated in a cell type-specific manner within the brain [105]. New gene- and cell type-specific epigenetic techniques have been utilized in pre-clinical studies to address these issues [106, 107]. Viral-mediated gene delivery, which has recently been utilized in humans (e.g., Clinicaltrial.gov identifier NCT00749957), could potentially be employed alone or in combination with other techniques (e.g, ultrasound) [108, 109] to achieve brain region- and/or cell type-specific epigenetic manipulations in SUD patients. Additionally, certain non-coding RNAs are selectively expressed in the brain and in specific cell types within the brain [110, 111] and may be targeted in patients using chemically modified, single-stranded oligonucleotides [112], though more research is needed in this area. Therefore, as advances in epigenetic pharmacotherapies and viral-mediated approaches unfold, the prospect of clinically effective treatments for multifactorial brain diseases, such as SUD, may be realized in the near future.
Acknowledgments
Funding: NIH/NIDA grant R00DA040744
Footnotes
The author declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Allis CD, Jenuwein T, The molecular hallmarks of epigenetic control, Nat Rev Genet 17(8) (2016) 487–500. [DOI] [PubMed] [Google Scholar]
- [2].Patel DJ, Wang Z, Readout of epigenetic modifications, Annu Rev Biochem 82 (2013) 81–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Marmorstein R, Zhou MM, Writers and readers of histone acetylation: structure, mechanism, and inhibition, Cold Spring Harb Perspect Biol 6(7) (2014) a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hyun K, Jeon J, Park K, Kim J, Writing, erasing and reading histone lysine methylations, Exp Mol Med 49(4) (2017) e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kumar S, Chinnusamy V, Mohapatra T, Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond, Front Genet 9 (2018) 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Farrelly LA, Thompson RE, Zhao S, Lepack AE, Lyu Y, Bhanu NV, Zhang B, Loh YE, Ramakrishnan A, Vadodaria KC, Heard KJ, Erikson G, Nakadai T, Bastle RM, Lukasak BJ, Zebroski H 3rd, Alenina N, Bader M, Berton O, Roeder RG, Molina H, Gage FH, Shen L, Garcia BA, Li H, Muir TW, Maze I, Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3, Nature (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Satterlee JS, Basanta-Sanchez M, Blanco S, Li JB, Meyer K, Pollock J, Sadri-Vakili G, Rybak-Wolf A, Novel RNA modifications in the nervous system: form and function, J Neurosci 34(46) (2014) 15170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao Y, Garcia BA, Comprehensive Catalog of Currently Documented Histone Modifications, Cold Spring Harb Perspect Biol 7(9) (2015) a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sartor GC, St Laurent G 3rd, Wahlestedt C, The Emerging Role of Non-Coding RNAs in Drug Addiction, Front Genet 3 (2012) 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang KC, Chang HY, Molecular mechanisms of long noncoding RNAs, Mol Cell 43(6) (2011) 904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Houston I, Peter CJ, Mitchell A, Straubhaar J, Rogaev E, Akbarian S, Epigenetics in the human brain, Neuropsychopharmacology 38(1) (2013) 183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Narayan P, Dragunow M, Pharmacology of epigenetics in brain disorders, Br J Pharmacol 159(2) (2010) 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Walker DM, Cates HM, Loh YE, Purushothaman I, Ramakrishnan A, Cahill KM, Lardner CK, Godino A, Kronman HG, Rabkin J, Lorsch ZS, Mews P, Doyle MA, Feng J, Labonte B, Koo JW, Bagot RC, Logan RW, Seney ML, Calipari ES, Shen L, Nestler EJ, Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry, Biol Psychiatry 84(12) (2018) 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou Z, Yuan Q, Mash DC, Goldman D, Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol, Proc Natl Acad Sci U S A 108(16) (2011) 6626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C, Laitman B, Stockman V, LaPlant Q, Cahill ME, Nestler EJ, Shen L, Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens, Genome Biol 15(4) (2014) R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ, Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum, Neuron 48(2) (2005) 303–14. [DOI] [PubMed] [Google Scholar]
- [17].Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA, CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors, J Neurosci 31(47) (2011) 16941–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rogge GA, Wood MA, The role of histone acetylation in cocaine-induced neural plasticity and behavior, Neuropsychopharmacology 38(1) (2013) 94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ferguson D, Koo JW, Feng J, Heller E, Rabkin J, Heshmati M, Renthal W, Neve R, Liu X, Shao N, Sartorelli V, Shen L, Nestler EJ, Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action, J Neurosci 33(41) (2013) 16088–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Maze I, Covington HE 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ, Essential role of the histone methyltransferase G9a in cocaine-induced plasticity, Science 327(5962) (2010) 213–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Damez-Werno DM, Sun H, Scobie KN, Shao N, Rabkin J, Dias C, Calipari ES, Maze I, Pena CJ, Walker DM, Cahill ME, Chandra R, Gancarz A, Mouzon E, Landry JA, Cates H, Lobo MK, Dietz D, Allis CD, Guccione E, Turecki G, Defilippi P, Neve RL, Hurd YL, Shen L, Nestler EJ, Histone arginine methylation in cocaine action in the nucleus accumbens, Proc Natl Acad Sci U S A 113(34) (2016) 9623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].LaPlant Q, Nestler EJ, CRACKing the histone code: cocaine’s effects on chromatin structure and function, Horm Behav 59(3) (2011) 321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Aguilar-Valles A, Vaissiere T, Griggs EM, Mikaelsson MA, Takacs IF, Young EJ, Rumbaugh G, Miller CA, Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation, Biol Psychiatry 76(1) (2014) 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martin TA, Jayanthi S, McCoy MT, Brannock C, Ladenheim B, Garrett T, Lehrmann E, Becker KG, Cadet JL, Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens, PLoS One 7(3) (2012) e34236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mychasiuk R, Muhammad A, Ilnytskyy S, Kolb B, Persistent gene expression changes in NAc, mPFC, and OFC associated with previous nicotine or amphetamine exposure, Behav Brain Res 256 (2013) 655–61. [DOI] [PubMed] [Google Scholar]
- [26].Chase KA, Gavin DP, Guidotti A, Sharma RP, Histone methylation at H3K9: evidence for a restrictive epigenome in schizophrenia, Schizophr Res 149(1–3) (2013) 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Levine A, Huang Y, Drisaldi B, Griffin EA Jr., Pollak DD, Xu S, Yin D, Schaffran C, Kandel DB, Kandel ER, Molecular mechanism for a gateway drug: epigenetic changes initiated by nicotine prime gene expression by cocaine, Sci Transl Med 3(107) (2011) 107ra109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Caputi FF, Palmisano M, Carboni L, Candeletti S, Romualdi P, Opioid gene expression changes and post-translational histone modifications at promoter regions in the rat nucleus accumbens after acute and repeated 3,4-methylenedioxy-methamphetamine (MDMA) exposure, Pharmacol Res 114 (2016) 209–218. [DOI] [PubMed] [Google Scholar]
- [29].Sheng J, Lv Z, Wang L, Zhou Y, Hui B, Histone H3 phosphoacetylation is critical for heroin-induced place preference, Neuroreport 22(12) (2011) 575–80. [DOI] [PubMed] [Google Scholar]
- [30].Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, Neve RL, Zachariou V, Shen L, Nestler EJ, Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens, J Neurosci 32(48) (2012) 17454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Botia B, Legastelois R, Alaux-Cantin S, Naassila M, Expression of ethanol-induced behavioral sensitization is associated with alteration of chromatin remodeling in mice, PLoS One 7(10) (2012) e47527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Subbanna S, Nagre NN, Shivakumar M, Umapathy NS, Psychoyos D, Basavarajappa BS, Ethanol induced acetylation of histone at G9a exon1 and G9a-mediated histone H3 dimethylation leads to neurodegeneration in neonatal mice, Neuroscience 258 (2014) 422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].DiNieri JA, Wang X, Szutorisz H, Spano SM, Kaur J, Casaccia P, Dow-Edwards D, Hurd YL, Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring, Biol Psychiatry 70(8) (2011) 763–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, Hurd YL, Genome-Wide DNA Methylation Profiling Reveals Epigenetic Changes in the Rat Nucleus Accumbens Associated With Cross-Generational Effects of Adolescent THC Exposure, Neuropsychopharmacology 40(13) (2015) 2993–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sanchez-Serrano SL, Cruz SL, Lamas M, Repeated toluene exposure modifies the acetylation pattern of histones H3 and H4 in the rat brain, Neurosci Lett 489(3) (2011) 142–7. [DOI] [PubMed] [Google Scholar]
- [36].Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ, Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers, J Neurochem 116(3) (2011) 459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Egervari G, Landry J, Callens J, Fullard JF, Roussos P, Keller E, Hurd YL, Striatal H3K27 Acetylation Linked to Glutamatergic Gene Dysregulation in Human Heroin Abusers Holds Promise as Therapeutic Target, Biol Psychiatry 81(7) (2017) 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kozlenkov A, Jaffe AE, Timashpolsky A, Apontes P, Rudchenko S, Barbu M, Byne W, Hurd YL, Horvath S, Dracheva S, DNA Methylation Profiling of Human Prefrontal Cortex Neurons in Heroin Users Shows Significant Difference between Genomic Contexts of Hyper- and Hypomethylation and a Younger Epigenetic Age, Genes (Basel) 8(6) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].De Sa Nogueira D, Merienne K, Befort K, Neuroepigenetics and addictive behaviors: Where do we stand?, Neurosci Biobehav Rev (in press) (2018). [DOI] [PubMed] [Google Scholar]
- [40].Romieu P, Host L, Gobaille S, Sandner G, Aunis D, Zwiller J, Histone deacetylase inhibitors decrease cocaine but not sucrose self-administration in rats, J Neurosci 28(38) (2008) 9342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA, HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner, Proc Natl Acad Sci U S A 110(7) (2013) 2647–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE 3rd, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ, Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins, Neuron 62(3) (2009) 335–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sartor GC, Powell SK, Brothers SP, Wahlestedt C, Epigenetic Readers of Lysine Acetylation Regulate Cocaine-Induced Plasticity, J Neurosci 35(45) (2015) 15062–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Massart R, Barnea R, Dikshtein Y, Suderman M, Meir O, Hallett M, Kennedy P, Nestler EJ, Szyf M, Yadid G, Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving, J Neurosci 35(21) (2015) 8042–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Anier K, Zharkovsky A, Kalda A, S-adenosylmethionine modifies cocaine-induced DNA methylation and increases locomotor sensitization in mice, Int J Neuropsychopharmacol 16(9) (2013) 2053–66. [DOI] [PubMed] [Google Scholar]
- [46].Urb M, Niinep K, Matsalu T, Kipper K, Herodes K, Zharkovsky A, Timmusk T, Anier K, Kalda A, The role of DNA methyltransferase activity in cocaine treatment and withdrawal in the nucleus accumbens of mice, Addict Biol (2019). [DOI] [PubMed] [Google Scholar]
- [47].Fonteneau M, Filliol D, Anglard P, Befort K, Romieu P, Zwiller J, Inhibition of DNA methyltransferases regulates cocaine self-administration by rats: a genome-wide DNA methylation study, Genes Brain Behav 16(3) (2017) 313–327. [DOI] [PubMed] [Google Scholar]
- [48].Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, Sun H, Schuebel K, Zhou Z, Yuan Q, Vendruscolo LF, Goldman D, Heilig M, DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity, J Neurosci 35(15) (2015) 6153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, Tahir A, Sweatt JD, DNA methylation regulates associative reward learning, Nat Neurosci 16(10) (2013) 1445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].LaPlant Q, Vialou V, Covington HE 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ, Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens, Nat Neurosci 13(9) (2010) 1137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Han J, Li Y, Wang D, Wei C, Yang X, Sui N, Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice, Eur J Pharmacol 642(1–3) (2010) 93–8. [DOI] [PubMed] [Google Scholar]
- [52].Tian W, Zhao M, Li M, Song T, Zhang M, Quan L, Li S, Sun ZS, Reversal of cocaine-conditioned place preference through methyl supplementation in mice: altering global DNA methylation in the prefrontal cortex, PLoS One 7(3) (2012) e33435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Qiao X, Yin F, Ji Y, Li Y, Yan P, Lai J, 5-Aza-2’-deoxycytidine in the medial prefrontal cortex regulates alcohol-related behavior and Ntf3-TrkC expression in rats, PLoS One 12(6) (2017) e0179469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang YX, Akumuo RC, Espana RA, Yan CX, Gao WJ, Li YC, The histone demethylase KDM6B in the medial prefrontal cortex epigenetically regulates cocaine reward memory, Neuropharmacology 141 (2018) 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li Y, Zhu R, Wang W, Fu D, Hou J, Ji S, Chen B, Hu Z, Shao X, Yu X, Zhao Q, Zhang B, Du C, Bu Q, Hu C, Tang Y, Zhong L, Yang S, Zhao Y, Cen X, Arginine Methyltransferase 1 in the Nucleus Accumbens Regulates Behavioral Effects of Cocaine, J Neurosci 35(37) (2015) 12890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lopez AJ, Hemstedt TJ, Jia Y, Hwang PH, Campbell RR, Kwapis JL, White AO, Chitnis O, Scarfone VM, Matheos DP, Lynch G, Wood MA, Epigenetic regulation of immediate-early gene Nr4a2/Nurr1 in the medial habenula during reinstatement of cocaine-associated behavior, Neuropharmacology 153 (2019) 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Alaghband Y, Kwapis JL, Lopez AJ, White AO, Aimiuwu OV, Al-Kachak A, Bodinayake KK, Oparaugo NC, Dang R, Astarabadi M, Matheos DP, Wood MA, Distinct roles for the deacetylase domain of HDAC3 in the hippocampus and medial prefrontal cortex in the formation and extinction of memory, Neurobiol Learn Mem 145 (2017) 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Heller EA, Hamilton PJ, Burek DD, Lombroso SI, Pena CJ, Neve RL, Nestler EJ, Targeted Epigenetic Remodeling of the Cdk5 Gene in Nucleus Accumbens Regulates Cocaine- and Stress-Evoked Behavior, J Neurosci 36(17) (2016) 4690–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Taniguchi M, Carreira MB, Cooper YA, Bobadilla AC, Heinsbroek JA, Koike N, Larson EB, Balmuth EA, Hughes BW, Penrod RD, Kumar J, Smith LN, Guzman D, Takahashi JS, Kim TK, Kalivas PW, Self DW, Lin Y, Cowan CW, HDAC5 and Its Target Gene, Npas4, Function in the Nucleus Accumbens to Regulate Cocaine-Conditioned Behaviors, Neuron 96(1) (2017) 130–144 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cannella N, Oliveira AMM, Hemstedt T, Lissek T, Buechler E, Bading H, Spanagel R, Dnmt3a2 in the Nucleus Accumbens Shell Is Required for Reinstatement of Cocaine Seeking, J Neurosci 38(34) (2018) 7516–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Anderson EM, Larson EB, Guzman D, Wissman AM, Neve RL, Nestler EJ, Self DW, Overexpression of the Histone Dimethyltransferase G9a in Nucleus Accumbens Shell Increases Cocaine Self-Administration, Stress-Induced Reinstatement, and Anxiety, J Neurosci 38(4) (2018) 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shiels MS, Freedman ND, Thomas D, Berrington de Gonzalez A, Trends in U.S. Drug Overdose Deaths in Non-Hispanic Black, Hispanic, and Non-Hispanic White Persons, 2000–2015, Ann Intern Med 168(6) (2018) 453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].van Gerven J, Cohen A, Vanishing clinical psychopharmacology, Br J Clin Pharmacol 72(1) (2011) 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chiamulera C, Padovani L, Corsi M, Drug discovery for the treatment of substance use disorders: novel targets, repurposing, and the need for new paradigms, Curr Opin Pharmacol 35 (2017) 120–124. [DOI] [PubMed] [Google Scholar]
- [65].Rasmussen K, White DA, Acri JB, NIDA’s medication development priorities in response to the Opioid Crisis: ten most wanted, Neuropsychopharmacology 44(4) (2019) 657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fulco CE, Liverman CT, Laurence E, Development of Medications for the Treatment of Opiate and Cocaine Addictions: Issues for the Government and Private Sector, National Academies Press (US), Washington (DC), 1995. [PubMed] [Google Scholar]
- [67].Prachayasittikul V, Prathipati P, Pratiwi R, Phanus-Umporn C, Malik AA, Schaduangrat N, Seenprachawong K, Wongchitrat P, Supokawej A, Prachayasittikul V, Wikberg JE, Nantasenamat C, Exploring the epigenetic drug discovery landscape, Expert Opin Drug Discov 12(4) (2017) 345–362. [DOI] [PubMed] [Google Scholar]
- [68].Hitchcock LN, Raybuck JD, Wood MA, Lattal KM, Effects of a histone deacetylase 3 inhibitor on extinction and reinstatement of cocaine self-administration in rats, Psychopharmacology (Berl) 236(1) (2019) 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Loweth JA, Tseng KY, Wolf ME, Adaptations in AMPA receptor transmission in the nucleus accumbens contributing to incubation of cocaine craving, Neuropharmacology 76 Pt B (2014) 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y, Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving, J Neurosci 23(3) (2003) 742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD, BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice, Nat Neurosci 18(10) (2015) 1464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sartor GC, Malvezzi AM, Kumar A, Andrade NS, Wiedner HJ, Vilca SJ, Janczura KJ, Bagheri A, Al-Ali H, Powell SK, Brown PT, Volmar CH, Foster TC, Zeier Z, Wahlestedt C, Enhancement of BDNF Expression and Memory by HDAC Inhibition Requires BET Bromodomain Reader Proteins, J Neurosci 39(4) (2019) 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pradhan AA, Tipton AF, Zhang H, Akbari A, Pandey SC, Effect of Histone Deacetylase Inhibitor on Ethanol Withdrawal-Induced Hyperalgesia in Rats, Int J Neuropsychopharmacol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tago T, Toyohara J, Advances in the Development of PET Ligands Targeting Histone Deacetylases for the Assessment of Neurodegenerative Diseases, Molecules 23(2) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Andersen AM, Dogan MV, Beach SR, Philibert RA, Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders, Genes (Basel) 6(4) (2015) 991–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Xu H, Wang F, Kranzler HR, Gelernter J, Zhang H, Alcohol and nicotine codependence-associated DNA methylation changes in promoter regions of addiction-related genes, Sci Rep 7 (2017) 41816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kuhnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JBJ, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D, A DNA methylation biomarker of alcohol consumption, Mol Psychiatry 23(2) (2018) 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lax E, Warhaftig G, Ohana D, Maayan R, Delayahu Y, Roska P, Ponizovsky AM, Weizman A, Yadid G, Szyf M, A DNA Methylation Signature of Addiction in T Cells and Its Reversal With DHEA Intervention, Front Mol Neurosci 11 (2018) 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Toyama K, Kiyosawa N, Watanabe K, Ishizuka H, Identification of Circulating miRNAs Differentially Regulated by Opioid Treatment, Int J Mol Sci 18(9) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bruckmann C, Di Santo A, Karle KN, Batra A, Nieratschker V, Validation of differential GDAP1 DNA methylation in alcohol dependence and its potential function as a biomarker for disease severity and therapy outcome, Epigenetics 11(6) (2016) 456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Craven R, The risky business of drug development in neurology, Lancet Neurol 10(2) (2011) 116–7. [DOI] [PubMed] [Google Scholar]
- [82].Bakulski KM, Halladay A, Hu VW, Mill J, Fallin MD, Epigenetic Research in Neuropsychiatric Disorders: the “Tissue Issue”, Curr Behav Neurosci Rep 3(3) (2016) 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang C, Schroeder FA, Hooker JM, Development of New Positron Emission Tomography Radiotracer for BET Imaging, ACS Chem Neurosci 8(1) (2017) 17–21. [DOI] [PubMed] [Google Scholar]
- [84].Wey HY, Gilbert TM, Zurcher NR, She A, Bhanot A, Taillon BD, Schroeder FA, Wang C, Haggarty SJ, Hooker JM, Insights into neuroepigenetics through human histone deacetylase PET imaging, Sci Transl Med 8(351) (2016) 351ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Couto PJ, Millis RM, PET Imaging of Epigenetic Influences on Alzheimer’s Disease, Int J Alzheimers Dis 2015 (2015) 575078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].J K, Drug Deaths in America Are Rising Faster Than Ever, The New York Times, 2017. [Google Scholar]
- [87].Scholl L, Seth P, Kariisa M, Wilson N, Baldwin G, Drug and Opioid-Involved Overdose Deaths — United States, 2013. –2017, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, Heslin MJ, Arnoletti JP, Richardson JH, Chen D, Hanna CA, Cramer SL, Yang ES, Qi J, Bradner JE, Yoon KJ, The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models, Oncogene 35(7) (2016) 833–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Raynal NJ, Da Costa EM, Lee JT, Gharibyan V, Ahmed S, Zhang H, Sato T, Malouf GG, Issa JJ, Repositioning FDA-Approved Drugs in Combination with Epigenetic Drugs to Reprogram Colon Cancer Epigenome, Mol Cancer Ther 16(2) (2017) 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Singh RS, Chaudhary DK, Mohan A, Kumar P, Chaturvedi CP, Ecelbarger CM, Godbole MM, Tiwari S, Greater efficacy of atorvastatin versus a non-statin lipid-lowering agent against renal injury: potential role as a histone deacetylase inhibitor, Sci Rep 6 (2016) 38034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lin YC, Lin JH, Chou CW, Chang YF, Yeh SH, Chen CC, Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC½, Cancer Res 68(7) (2008) 2375–83. [DOI] [PubMed] [Google Scholar]
- [92].Taormina G, Mirisola MG, Longevity: epigenetic and biomolecular aspects, Biomol Concepts 6(2) (2015) 105–17. [DOI] [PubMed] [Google Scholar]
- [93].Sleiman SF, Henry J, Al-Haddad R, El Hayek L, Abou Haidar E, Stringer T, Ulja D, Karuppagounder SS, Holson EB, Ratan RR, Ninan I, Chao MV, Exercise promotes the expression of brain derived neurotrophic factor (BDNF) through the action of the ketone body beta-hydroxybutyrate, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J, The neuroactive potential of the human gut microbiota in quality of life and depression, Nat Microbiol 4(4) (2019) 623–632. [DOI] [PubMed] [Google Scholar]
- [95].Tzschentke TM, Measuring reward with the conditioned place preference (CPP) paradigm: update of the last decade, Addict Biol 12(3–4) (2007) 227–462. [DOI] [PubMed] [Google Scholar]
- [96].O’Connor EC, Chapman K, Butler P, Mead AN, The predictive validity of the rat self-administration model for abuse liability, Neurosci Biobehav Rev 35(3) (2011) 912–38. [DOI] [PubMed] [Google Scholar]
- [97].Kawa AB, Allain F, Robinson TE, Samaha AN, The transition to cocaine addiction: the importance of pharmacokinetics for preclinical models, Psychopharmacology (Berl) 236(4) (2019) 1145–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Smith RJ, Laiks LS, Behavioral and neural mechanisms underlying habitual and compulsive drug seeking, Prog Neuropsychopharmacol Biol Psychiatry 87(Pt A) (2018) 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Muller CP, Animal models of psychoactive drug use and addiction - Present problems and future needs for translational approaches, Behav Brain Res 352 (2018) 109–115. [DOI] [PubMed] [Google Scholar]
- [100].Bickel WK, Koffarnus MN, Moody L, Wilson AG, The behavioral- and neuro-economic process of temporal discounting: A candidate behavioral marker of addiction, Neuropharmacology 76 Pt B (2014) 518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Allain F, Samaha AN, Revisiting long-access versus short-access cocaine self-administration in rats: intermittent intake promotes addiction symptoms independent of session length, Addict Biol 24(4) (2019) 641–651. [DOI] [PubMed] [Google Scholar]
- [102].Goltseker K, Hopf FW, Barak S, Advances in behavioral animal models of alcohol use disorder, Alcohol 74 (2019) 73–82. [DOI] [PubMed] [Google Scholar]
- [103].Augier E, Barbier E, Dulman RS, Licheri V, Augier G, Domi E, Barchiesi R, Farris S, Natt D, Mayfield RD, Adermark L, Heilig M, A molecular mechanism for choosing alcohol over an alternative reward, Science 360(6395) (2018) 1321–1326. [DOI] [PubMed] [Google Scholar]
- [104].Riley AL, Hempel BJ, Clasen MM, Sex as a biological variable: Drug use and abuse, Physiol Behav 187 (2018) 79–96. [DOI] [PubMed] [Google Scholar]
- [105].Jordi E, Heiman M, Marion-Poll L, Guermonprez P, Cheng SK, Nairn AC, Greengard P, Girault JA, Differential effects of cocaine on histone posttranslational modifications in identified populations of striatal neurons, Proc Natl Acad Sci U S A 110(23) (2013) 9511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H, Hu X, Li C, Yao X, Shen X, Sun Y, Wei Y, Liu F, Ying W, Zhang J, Tang C, Zhang X, Xu H, Shi L, Cheng L, Huang P, Yang H, In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice, Nat Neurosci 21(3) (2018) 440–446. [DOI] [PubMed] [Google Scholar]
- [107].Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y, Yamauchi T, Sakurai M, O’Keefe DD, Nunez-Delicado E, Guillen P, Campistol JM, Wu CJ, Lu LF, Esteban CR, Izpisua Belmonte JC, In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation, Cell 171(7) (2017) 1495–1507 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Stavarache MA, Petersen N, Jurgens EM, Milstein ER, Rosenfeld ZB, Ballon DJ, Kaplitt MG, Safe and stable noninvasive focal gene delivery to the mammalian brain following focused ultrasound, J Neurosurg (2018) 1–10. [DOI] [PubMed] [Google Scholar]
- [109].Dallapiazza RF, Timbie KF, Holmberg S, Gatesman J, Lopes MB, Price RJ, Miller GW, Elias WJ, Noninvasive neuromodulation and thalamic mapping with low-intensity focused ultrasound, J Neurosurg 128(3) (2018) 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gendron J, Colace-Sauty C, Beaume N, Cartonnet H, Guegan J, Ulveling D, Pardanaud-Glavieux C, Moszer I, Cheval H, Ravassard P, Long non-coding RNA repertoire and open chromatin regions constitute midbrain dopaminergic neuron - specific molecular signatures, Sci Rep 9(1) (2019) 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bannon MJ, Savonen CL, Jia H, Dachet F, Halter SD, Schmidt CJ, Lipovich L, Kapatos G, Identification of long noncoding RNAs dysregulated in the midbrain of human cocaine abusers, J Neurochem 135(1) (2015) 50–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C, Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation, Nat Biotechnol 30(5) (2012) 453–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Dunbar AB, Taylor JR, Garcinol Blocks the Reconsolidation of Multiple Cocaine-Paired Cues after a Single Cocaine-Reactivation Session, Neuropsychopharmacology 42(9) (2017) 1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wang L, Lv Z, Hu Z, Sheng J, Hui B, Sun J, Ma L, Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement, Neuropsychopharmacology 35(4) (2010) 913–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pastor V, Host L, Zwiller J, Bernabeu R, Histone deacetylase inhibition decreases preference without affecting aversion for nicotine, J Neurochem 116(4) (2011) 636–45. [DOI] [PubMed] [Google Scholar]
- [116].Castino MR, Cornish JL, Clemens KJ, Inhibition of histone deacetylases facilitates extinction and attenuates reinstatement of nicotine self-administration in rats, PLoS One 10(4) (2015) e0124796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wang R, Zhang Y, Qing H, Liu M, Yang P, The extinction of morphine-induced conditioned place preference by histone deacetylase inhibition, Neurosci Lett 483(2) (2010) 137–42. [DOI] [PubMed] [Google Scholar]
- [118].Sanchis-Segura C, Lopez-Atalaya JP, Barco A, Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition, Neuropsychopharmacology 34(13) (2009) 2642–54. [DOI] [PubMed] [Google Scholar]
- [119].Chen WS, Xu WJ, Zhu HQ, Gao L, Lai MJ, Zhang FQ, Zhou WH, Liu HF, Effects of histone deacetylase inhibitor sodium butyrate on heroin seeking behavior in the nucleus accumbens in rats, Brain Res 1652 (2016) 151–157. [DOI] [PubMed] [Google Scholar]
- [120].Simon-O’Brien E, Alaux-Cantin S, Warnault V, Buttolo R, Naassila M, Vilpoux C, The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals, Addict Biol 20(4) (2015) 676–89. [DOI] [PubMed] [Google Scholar]
- [121].Friedman OM, Mahapatra GN, Stevenson R, The methylation of deoxyribonucleosides by diazomethane, Biochim Biophys Acta 68 (1963) 144–6. [DOI] [PubMed] [Google Scholar]
- [122].Allfrey VG, Faulkner R, Mirsky AE, Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis, Proc Natl Acad Sci U S A 51 (1964) 786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD, Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation, Cell 84(6) (1996) 843–51. [DOI] [PubMed] [Google Scholar]
- [124].Taunton J, Hassig CA, Schreiber SL, A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p, Science 272(5260) (1996) 408–11. [DOI] [PubMed] [Google Scholar]
- [125].Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM, Structure and ligand of a histone acetyltransferase bromodomain, Nature 399(6735) (1999) 491–6. [DOI] [PubMed] [Google Scholar]
- [126].Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T, Regulation of chromatin structure by site-specific histone H3 methyltransferases, Nature 406(6796) (2000) 593–9. [DOI] [PubMed] [Google Scholar]
- [127].Strahl BD, Allis CD, The language of covalent histone modifications, Nature 403(6765) (2000) 41–5. [DOI] [PubMed] [Google Scholar]
- [128].Campbell ND, Lovell AM, The history of the development of buprenorphine as an addiction therapeutic, Ann N Y Acad Sci 1248 (2012) 124–39. [DOI] [PubMed] [Google Scholar]
- [129].Dole VP, Nyswander M, A Medical Treatment for Diacetylmorphine (Heroin) Addiction. A Clinical Trial with Methadone Hydrochloride, JAMA 193 (1965) 646–50. [DOI] [PubMed] [Google Scholar]