Abstract
Next-generation sequencing (NGS) has been applied in the field of infectious diseases. Bronchoalveolar lavage fluid (BALF) is considered a sterile type of specimen that is suitable for detecting pathogens of respiratory infections. The aim of this study was to comprehensively identify causative pathogens using NGS in BALF samples from immunocompetent pediatric patients with respiratory failure. Ten patients hospitalized with respiratory failure were included. BALF samples obtained in the acute phase were used to prepare DNA- and RNA-sequencing libraries. The libraries were sequenced on MiSeq, and the sequence data were analyzed using metagenome analysis tools. A mean of 2,041,216 total reads were sequenced for each library. Significant bacterial or viral sequencing reads were detected in eight of the 10 patients. Furthermore, candidate pathogens were detected in three patients in whom etiologic agents were not identified by conventional methods. The complete genome of enterovirus D68 was identified in two patients, and phylogenetic analysis suggested that both strains belong to subclade B3, which is an epidemic strain that has spread worldwide in recent years. Our results suggest that NGS can be applied for comprehensive molecular diagnostics as well as surveillance of pathogens in BALF from patients with respiratory infection.
Subject terms: Clinical microbiology, Infectious-disease diagnostics, Viral infection, Paediatric research
Introduction
In the field of infectious diseases, identification of etiologic microorganisms is essential for definitive diagnosis and decisions regarding appropriate management. Establishment of bacterial and fungal cultures is the gold standard method for identification of causative microorganisms. However, about 2–3 days are generally required to obtain quantitative culture test results, and this method is not suitable for identification of unculturable bacteria. For identification of viral pathogens, PCR and antigen tests are commonly used, but only a defined set of candidate microorganisms can be examined. Furthermore, virus isolation is a reliable method to determine the causative pathogen, but it is time-consuming, and its sensitivity may be insufficient. Although combinations of these procedures are performed, no significant pathogens are identified in 34–57% of pediatric patients1 and 13–62% of adult patients with pneumonia2,3.
Microbiological diagnosis of respiratory infection can be determined when a specific pathogen is isolated from sterile materials such as bronchoalveolar lavage fluid (BALF), transtracheal aspiration, percutaneous lung aspiration, and pleural effusion; when a significant quantity of bacteria is cultured from sputum; or when the presence of pathogens that do not usually colonize the upper respiratory tract is proven1,4,5. Sputum is generally used for detecting pathogens that cause lower respiratory tract infections. However, obtaining sputum is difficult, especially in pediatric patients, and distinguishing pathogens from resident bacteria in the oral cavity is sometimes difficult as well6. Therefore, BALF is more suitable than sputum for identifying pathogens of respiratory diseases because it can be collected from the locus of infection with less contamination with oral bacteria. BALF is often obtained for PCR of Pneumocystis jirovecii or cytomegalovirus (CMV) in patients with severe pneumonia7,8. BALF can be obtained with a relatively safe procedure and with low morbidity and mortality9. The diagnostic yield of causative pathogens from BALF was 28–68% in lower respiratory tract infections in immunosuppressed children including those with hematological malignancy and organ transplantation10. Furthermore, positive or negative results following microbiological examination of BALF lead to alteration in the management of the infection in 38.7–72.7% of patients9–11.
Next-generation sequencing (NGS) has been applied for comprehensive detection of causative pathogens in various infectious diseases12–16. We have demonstrated the utility of NGS for identification of causative or potentially causative pathogens of encephalitis, fulminant hepatitis, bloodstream infection, and acute myocarditis17–20. In several previous studies, NGS was applied to detect pathogens from BALF samples. Recently, Miao et al. have shown that significant reads of candidate pathogens were detected in 34% of BALF samples obtained from patients with infectious and noninfectious diseases21. However, BALF samples examined in previous studies were obtained mainly from adult patients, and no study was performed to simultaneously detect both bacteria and viruses22–24. In this study, we conducted comprehensive detection of pathogens from BALF samples from immunocompetent pediatric patients with severe respiratory failure using NGS.
Results
Comparison of DNA extraction methods for NGS
A comparison of DNA extraction methods was performed using three BALF samples in which results of bacterial cultures and CMV PCR were available. BALF-1 was obtained from a 3-month-old boy with interstitial pneumonia. Staphylococcus aureus was isolated by culturing, and CMV DNA was detected with real-time PCR (2,101,786 IU/ml). BALF-2 was obtained from a 6-month-old patient with extremely low birth weight and chronic lung disease, and Serratia marcescens was isolated from a transtracheal aspiration sample. BALF-3 was obtained from a 7-year-old patient with pulmonary alveolar proteinosis, and no bacteria were isolated. We compared the detection efficiency of pathogen-derived reads by NGS between two types of DNA extraction kits: QIAamp DNA Microbiome kit (MB kit) and the QIAamp UCP Pathogen Mini kit (Path kit). Classifications of total reads from each library are shown in Fig. 1A. Proportions of bacterial reads were higher in libraries prepared with the MB kit than the Path kit in BALF-1 and BALF-2 (14.8% vs. 0.1% and 52.6% vs. 1.2%, respectively). Numbers of sequencing reads assigned to each bacterial species or CMV are expressed as reads per million (RPM) and shown in Fig. 1B. In BALF-1, the total number of bacterial reads obtained from the library prepared with the MB kit was more than 100 times higher than that with the Path kit. Most bacterial reads were aligned with the S. aureus genome in both libraries. CMV-derived reads were detectable in libraries obtained with both extraction kits. In BALF-2, approximately 80 times more total bacterial reads were obtained from the library with the MB kit compared to the Path kit. Among bacterial reads, Streptococcus mitis-derived reads were dominant in both libraries. Other than S. mitis, more than 200 RPM of four bacterial species were obtained from the library with the MB kit, whereas reads of Staphylococcus epidermidis and Enterococcus faecalis were not obtained from the library with the Path kit. S. epidermidis and E. faecalis were detected with real-time PCR using DNA extracted with the MB kit (1.6 × 105 and 3.5 × 103 copies/ml, respectively), whereas they were not detected in DNA extracted using the Path kit. In BALF-3 with a negative culture test, a small number of reads of Cutibacterium acnes, which is considered a member of the normal oral cavity flora, was detected in both libraries.
Figure 1.

Comparison of DNA extraction kits for preparing DNA libraries to detect pathogen-derived reads. DNA libraries were prepared using DNA extracted with the QIAamp DNA Microbiome kit (MB kit) or the QIAamp UCP Pathogen Mini kit (Path kit) from bronchoalveolar lavage fluid (BALF) samples. The composition of sequence data in each DNA library was compared. Each bar represents the composition of sequence reads including adapter, trimmed reads, human genome, bacteria, virus, eukaryotes, and others (A). Numbers of sequencing reads assigned to each bacterial species or cytomegalovirus (CMV) are shown (B). RPM: reads per million.
The comparison of the read mapping results is shown in Fig. 2. The average coverage (depth) and the fraction of reference covered (coverage) of sequenced reads mapped to the reference genome of S. aureus (NC_007795) or S. mitis (NC_013853) were much higher in libraries prepared with the MB kit than those with the Path kit. Taken together, these results indicate that the MB kit is more suitable than the Path kit for preparing a DNA library to detect bacterial reads in BALF samples. Considering that the difference in CMV reads was small in BALF-1, the MB kit was used in further experiments.
Figure 2.
Comparison of the coverage plots of bacterial genomes. Sequencing reads detected in each bronchoalveolar lavage fluid (BALF) sample were mapped against the reference genome of S. aureus (NC_007795) in BALF-1 and S. mitis (NC_013853) in BALF-2. DNA for library preparation was extracted using the QIAamp DNA Microbiome kit (MB kit) or the QIAamp UCP Pathogen Mini kit (Path kit). Blue and dark blue colors in the bacterial genome alignment represent average and maximal coverage in the aggregated 1-kbp region, respectively.
Comprehensive detection of pathogen-derived sequences from BALF samples from pediatric patients with severe respiratory failure
We investigated 10 BALF samples from pediatric patients with severe respiratory failure (patients 1–10) using NGS to identify etiologic agents. In addition, one patient in the chronic phase of idiopathic interstitial pneumonia was included as a control (patient 11). Patient characteristics are listed in Table 1. A mean of 2,041,216 total reads was sequenced for each library. The classification of the sequence data is shown in Supplementary Table 1. To avoid making calls based on spurious alignments or cross-contamination, we considered bacterial or viral reads as significant when the number of sequences aligned to each reference genome was above 10 RPM, and the sequenced reads covered wide ranges of the reference genome with the read mapping approach. Composition of bacterial reads at the genus level of each DNA-sequencing library is shown in Fig. 3, and a summary of the pathogens with significant bacterial and viral reads is listed in Table 2. A significant number of four types of bacterial reads was detected in three BALF samples with DNA-sequencing (patients 1, 2, and 4), and NGS results for three bacteria were consistent with the results of transtracheal aspiration culture. Sequencing reads of bacteria identified with DNA-sequencing in these three patients were also detected with RNA-sequencing (data not shown). Substantial bacterial reads of Cutibacterium and Rastonia, which are considered contamination, were detected in libraries from distilled water (Fig. 3 and Supplementary Table 1). Sequence coverage and depth of each reference bacterial genome are shown in Fig. 4. In patient 1, Stenotrophomonas maltophilia and Pseudomonas aeruginosa reads were detected with NGS, whereas only the latter was isolated in a small amount with culturing. Conversely, no significant bacterial read was identified in patient 5 or 6, in which Streptococcus pneumoniae and Haemophilus influenzae, respectively, were isolated with culturing of transtracheal aspiration.
Table 1.
Patient characteristics.
| Pt No. | Age | Sex | Underlying disease | Respiratory failure | Circulatory failure | X-ray findings of pneumonia | ICU LOS (days) | Mechanical ventilation (days) | ECMO | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5y 9m | F | Cerebral palsya | + | + | + | 12 | 7 | − | Recovered |
| 2 | 28d | M | none | + | + | − | 14 | 5 | − | Recovered |
| 3 | 0y 11m | F | TFO, Bronchomalacia | + | + | − | 10 | 6 | + | Recovered |
| 4 | 0y 4m | F | 21 trisomy | + | − | − | 8 | 4 | − | Recovered |
| 5 | 16d | M | none | + | + | − | 9 | 4 | − | Recovered |
| 6 | 0y 3m | F | none | + | + | − | 13 | 9 | − | Recovered |
| 7 | 4y 11m | M | none | + | − | − | 7 | 6 | − | Recovered |
| 8 | 8y 10m | F | none | + | − | + | 8 | 6 | − | Recovered |
| 9 | 0y 1m | F | none | + | + | + | 59 | 52 | + | Recovered |
| 10 | 7y 0m | F | none | + | + | − | 15 | 12 | + | Recovered |
| 11 | 2y 5m | F | IIPsb | − | − | − | − | − | − | − |
Abbreviation: ECMO, extracorporeal membrane oxygenation; ICU, intensive care unit; IIPs, idiopathic interstitial pneumonias; LOS, length of stay. TFO, Tetralogy for Fallot. aA patient with cerebral palsy after bacterial meningitis. She had undergone ventriculoperitoneal shunt placement procedure for hydrocephalus. bA patient with chronic phase of IIPs used for negative control.
Figure 3.
Genus composition of BALF samples from patients with severe respiratory failure. Each bar represents taxa at the genus level of taxonomic hierarchy. Bacterial reads with fewer than 10 reads or without annotations at the genus level were classified as “Others”. Pt 1-10 and NTC 1-3 indicate libraries prepared from bronchoalveolar lavage fluid (BALF) and distilled water, respectively.
Table 2.
Sequence results using NGS for detection of pathogens from BALF samples.
| Pt No. | NGS results | Conventional method results | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA-sequencing | RNA-sequencing | Bacterial culture | Viral antigen | PCR | |||||||
| Total reads | Detected pathogen | Number of reads | RPM | Total reads | Detected pathogen | Number of reads | RPM | ||||
| 1 | 2,603,238 | S. maltophilia | 100,083 | 38,446 | 2,303,752 | HMPV | 322 | 140 | P. aeruginosa (±) | HMPV | — |
| P. aerginosa | 16,947 | 6,510 | |||||||||
| 2 | 2,631,210 | H. influenzae | 103,738 | 39,426 | 2,937,810 | HRSV-A | 342 | 116 | H. influenzae (2+) | HRSV | HRSV-A |
| 3 | 2,071,836 | — | — | — | 2,676,318 | HRV-B | 50,138 | 18,734 | — | — | HRV |
| 4 | 2,754,852 | M. catarrhalis | 2,428 | 881 | 2,946,090 | — | — | — | M. catarrhalis (2+) | — | — |
| 5 | 3,608,962 | — | — | — | 2,402,562 | HRSV-A | 1,612 | 671 | S. pneumoniae (±) | — | HRSV-A |
| 6 | 1,489,176 | — | — | — | 1,685,054 | HRSV-B | 83,212 | 49,382 | H. influenzae (2+) | HRSV | HRSV-B |
| 7 | 1,660,692 | — | — | — | 1,531,292 | EV-D68 | 115,343 | 75,324 | — | — | — |
| 8 | 1,419,054 | — | — | — | 1,549,278 | EV-D68 | 784,380 | 506,287 | — | — | — |
| 9 | 1,373,122 | — | — | — | 1,478,618 | — | — | — | — | — | CMV |
| 10 | 1,581,982 | — | — | — | 1,517,564 | — | — | — | — | HMPV | — |
| 11a | 1,252,872 | — | — | — | 1,431,422 | — | — | — | — | — | — |
Abbreviation: CMV, cytomegalovirus; EV-D68, enterovirus D-68; HMPV, human metapneumovirus; HRSV, human respiratory syncytial virus; HRV, human rhinovirus; Pt, patient; RPM, read per million. aA patient with chronic phase of idiopathic interstitial pneumonias used for negative control.
Figure 4.
Coverage plots of bacterial genomes detected in patients with severe respiratory failure. Sequencing reads detected in bronchoalveolar lavage fluids of each patient were mapped to the reference genome of S. maltophilia, P. aeruginosa, H. influenzae, and M. catarrhalis. Blue and dark blue colors in the bacterial genome alignment represent average and maximal coverage in the aggregated 1-kbp region, respectively.
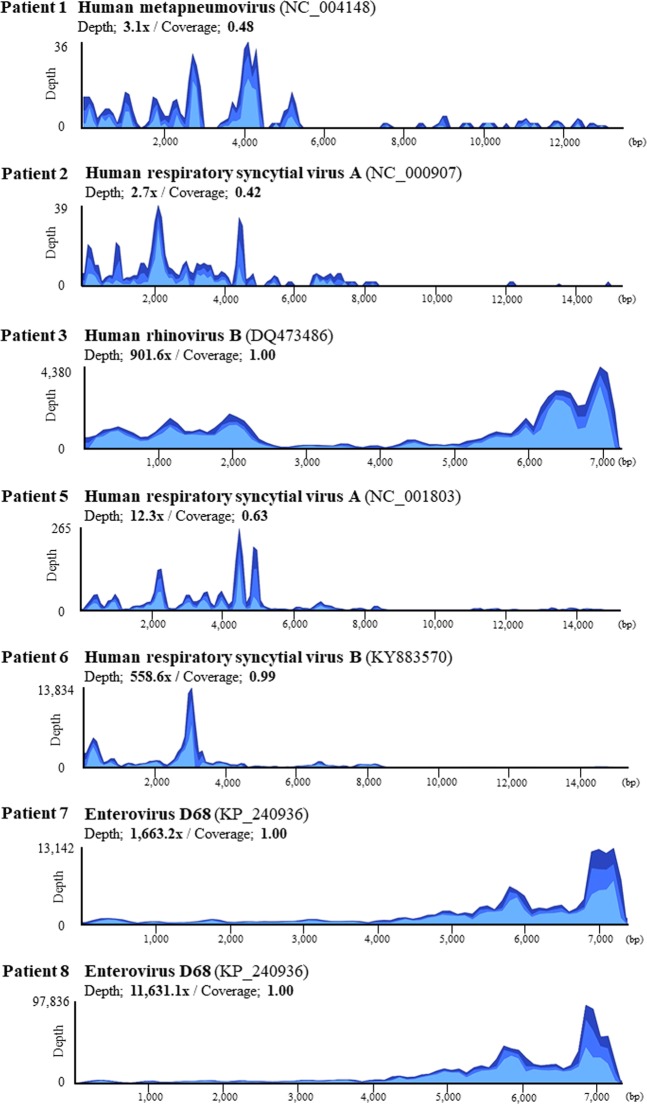
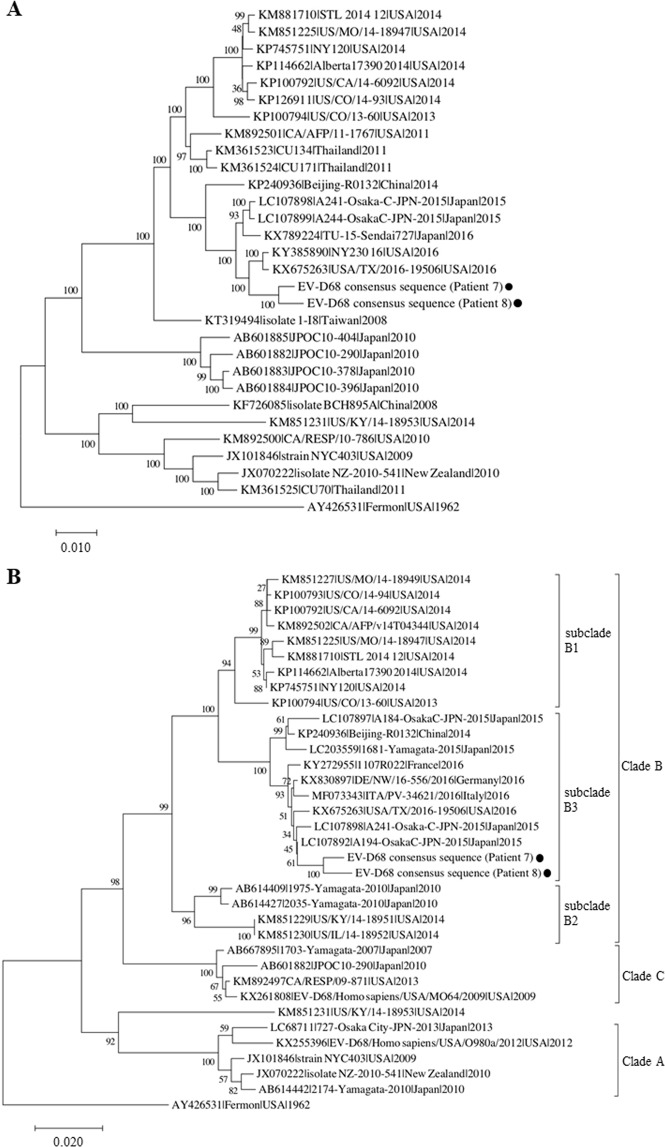
Candidate pathogenic respiratory viruses were detected in seven of 10 patients with RNA-sequencing: human respiratory syncytial virus (HRSV) from three patients, enterovirus D68 (EV-D68) from two patients, and human metapneumovirus (HMPV) and human rhinovirus B (HRV-B) from one patient each. Viral antigen test results were confirmed by retrospective review of medical records, and most were consistent with NGS results. Additionally, we performed multiplex RT-PCR to confirm the presence of the viral genome that was identified with NGS, except EV-D68. As a result, the NGS-based approach for detection of the causative virus was consistent with the results of multiplex virus PCR except in patient 1 in whom HMPV was not confirmed with PCR. Sequence coverage and depth of each reference viral genome are shown in Fig. 5. In patients 3, 6, 7, and 8, read mapping to each reference genome was achieved with high coverage and depth, and almost the complete viral genome was obtained. Phylogenetic analysis of the EV-D68 full genome suggested that the EV-D68 sequences derived from patients 7 and 8 were genetically related to the EV-D68 viruses circulating worldwide in 2016 (Fig. 6A). Based on phylogenetic analysis of the VP1 region, the EV-D68 strains detected in this study belong to subclade B3 (Fig. 6B). Among the two NGS-negative cases in which neither bacteria nor viruses were detected, CMV and HMPV were detected with PCR and an antigen test, respectively (patients 9 and 10). No significant bacterial or viral reads were identified from the negative control BALF sample (Patient 11). Consensus sequences of detected viruses with high coverage (HRV-B from patient 3, HRSV-B from patient 6, and EV-D68 from patients 7 and 8) are deposited in the DNA Data bank of Japan (Accession numbers: LC495296, LC495297, LC495298, and LC495299).
Figure 5.
Coverage plots of viral genomes detected in patients with severe respiratory failure. Sequencing reads detected in bronchoalveolar lavage fluids of each patient were mapped to the reference genome of human metapneumovirus (HMPV), human respiratory syncytial virus (HRSV)-A, human rhinovirus (HRV)-B, HRSV-B, and enterovirus (EV)-D68. Light blue, blue, and dark blue colors in the viral genome alignment represent minimal, average, and maximal coverage in the aggregated 10-bp region, respectively.
Figure 6.
Phylogenetic analysis of enterovirus D68. Phylogenetic trees of full genomes (A) and VP1 sequences (B) of enterovirus D68 (EV-D68) consensus sequences obtained from patients 7 and 8 are shown. The phylogenetic relationships of the registered sequences of EV-D68 were estimated using the neighbor-joining method with 1,000 replicates using MEGA743. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in units of the number of base differences per site. Clades were defined previously based on VP1 sequences37,50,51. GenBank accession number, strain name, country of origin, and year of detection are shown for each strain. Scale bars show the genetic distance.
Discussion
In this study, we used NGS to perform comprehensive detection of pathogenic microorganisms from BALF samples from 10 pediatric patients with severe respiratory failure. We expected that all microorganisms including viruses could be detected by conducting both DNA- and RNA-sequencing; indeed, a significant number of bacterial or viral sequencing reads was detected in eight of 10 patients. Furthermore, candidate causative pathogens of respiratory failure were detected in three cases (patients 3, 7, 8) in which pathogenic microorganisms were not identified by conventional methods. To the best of our knowledge, this is the first study to use NGS to comprehensively investigate both bacteria and viruses from BALF specimens in pediatric patients with respiratory failure.
In the present study, we first verified two DNA extraction kits to improve efficiency in bacteria detection from BALF samples, and showed that the QIAamp DNA Microbiome kit was more suitable. The QIAamp DNA Microbiome kit is a “microbial enrichment” DNA extraction kit. After the human cell lysis step, exposed nucleic acids are degraded with Benzonase nuclease, whereas the bacterial cells theoretically remain intact. In this experiment, this microbial enrichment procedure resulted in an approximately 100-fold increase in the number of bacterial reads compared with the other DNA extraction kit.
A discrepancy in the results between NGS and the culture test was observed in some cases. In patient 1, the number of reads annotated to S. maltophilia was much higher than that of P. aeruginosa, which was the only bacterial strain isolated from aspirated sputum by culturing. In contrast, no significant bacterial read of isolated bacteria was identified with NGS in patient 5 or 6. Bacterial culturing with samples from these patients was performed using transtracheal aspirated sputum obtained on the same day as BALF. Therefore, the discrepancy between NGS and the culture test results may reflect the difference in the specimens, and some of the isolated bacteria from aspirated sputum could be colonizing bacteria in the upper respiratory tract. Unfortunately, because the culture test using BALF was not performed in some patients, a comparison of the results of NGS and culturing was difficult. Furthermore, sufficient amounts of BALF were not available in some cases, which may have resulted in a false negative NGS-based result. On the other hand, a discrepancy between NGS and the bacterial culture results of BALF or other specimens was observed in a previous study21.
In this study, a significant number of viral reads was detected in seven of 10 patients. Among them, reads of more than 10,000 RPM were annotated to each virus reference genome in four patients, suggesting that BALF contains abundant reads of pathogenic viruses for respiratory infections. Because NGS does not require virus-specific primers, the NGS-based approach is especially useful for detection of RNA viruses, which have much higher mutation rates than DNA viruses16. The pathogenicity of the detected viruses has already been established in immunocompetent pediatric patients25–30. In general, HRV is considered to have relatively weak virulence, although it sometimes causes severe respiratory disease, especially in patients with immunodeficiency or anatomical abnormalities of the airway30. HRV is classified into three subtypes, HRV-A, -B, and -C, based on gene sequence analysis. The detection rate of HRV-B is lower than the other subtypes. Whether the severity of clinical symptoms is related to HRV subtypes remains inconclusive30–32. Patient 3, in whom HRV-B was detected, had congenital tracheomalacia as an underlying disease, suggesting that HRV-B may have been a causative pathogen or trigger of respiratory failure.
EV-D68 was identified in two patients without underlying diseases (patients 7 and 8). EV-D68 can cause severe lower respiratory illness and asthma exacerbation, mostly in children29,33. EV-D68 infections are more likely to be associated with severe and life-threating respiratory diseases than other enterovirus genotypes29. In 2014, a large-scale outbreak of severe respiratory infection caused by EV-D68 was reported in the US and other countries29,33. In Japan, EV-D68 outbreaks coincided with spikes in acute asthma exacerbations and acute flaccid myelitis in 201534, and another epidemic of EV-D68 was reported in 201835. Phylogenetic analysis suggested that the EV-D68 sequences identified in this experiment belong to subclade B3 and are related to epidemic strains that spread worldwide in 201636,37. These results indicate that the NGS-based approach enables us not only to comprehensively detect the causative virus, but also to specify genotypes of the detected virus in a single test, which would contribute to molecular epidemiological studies. In patient 1, HMPV was detected with NGS and the antigen test, but it was not detected with PCR. The primer sequences may not have been appropriate for detection of the HMPV strain in this patient.
Co-infection with bacteria and viruses was observed in four of 10 patients. Using NGS, S. maltophilia, P. aeruginosa, and HMPV were detected in patient 1, and H. influenzae and HRSV were detected in patient 2. HRSV was identified with NGS in patients 5 and 6, whereas S. pneumoniae and H. influenzae were isolated in patient 5 and patient 6, respectively, by culturing. Although concurrent bacterial infection in infants with HRSV or HMPV infection is relatively uncommon, co-infection may worsen the clinical symptoms of virus infection28,38. Therefore, respiratory failure may have been induced by co-infection with bacteria and the respiratory viruses in these patients. Conversely, because viral shedding of HRSV may continue for as long as 3 to 4 weeks, especially in young children39, detection of a small number of HRSV reads may reflect its presence as a bystander.
In our study, neither a significant number of bacterial reads nor viral reads were detected in two patients by NGS. Patient 10 was diagnosed with fulminant myocarditis with respiratory failure. In this patient, an antigen test using a nasopharyngeal swab sample was HMPV positive, whereas HMPV was not detected in BALF, neither with NGS nor PCR. Although HMPV may be a trigger of development of fulminant myocarditis, a false-positive result of the HMPV antigen test should be considered. In patient 9, a discrepancy between NGS and CMV PCR results was observed. Insufficient host genome digestion may have decreased the sensitivity of detection of CMV DNA because the concentration of extracted DNA was relatively higher than that of other specimens. Another reason for the discrepancy may be that CMV particles that are latent in human cells were simultaneously digested with Benzonase nuclease at the time of DNA extraction. Considering the above, more sequence depth may be required to detect DNA viruses with NGS.
Some limitations of our NGS-based approach for detecting pathogens should be discussed. First, the microbial enrichment procedure increased the number of bacterial reads, but still, 92.9–99.6% of trimmed NGS reads were derived from human DNA. More efficient methods to remove host nucleic acids are desirable to improve the sensitivity of detecting pathogen-derived genomes and to reduce the cost of the NGS procedure. Second, NGS can identify fungi in clinical samples21, but we could not validate whether our NGS procedure could detect fungal DNA. Third, we considered more than 10 RPM of bacterial or viral reads as significant in this study. However, determining a threshold read count was difficult. Furthermore, detection of viral or bacterial reads with NGS does not always indicate the presence of viable or pathogenic microorganisms. Finally, we utilized MePIC v2.0 (National Institute of Infectious Disease, Tokyo, Japan)40 as a metagenomic pathogen identification tool in this study. This pipeline incorporates the MEGABLAST program for a homology search against known nucleotide sequences registered in the NCBI nt database. Therefore, microorganisms whose sequences are unknown or not registered in the database cannot be detected. In addition, reproducibility issues in bioinformatics may occur due to several reasons such as the short half-life of the bioinformatics software, the complexity of the pipelines, and the incompleteness in workflow description41.
In conclusion, we demonstrated the utility of the NGS-based approach for detection of pathogens in BALF from pediatric patients with severe respiratory failure. Although further improvement of NGS workflow may be required, NGS can be applied for molecular diagnostics as well as surveillance of pathogens in the field of infectious diseases.
Methods
Ethics statement
This study was performed in compliance with relevant laws and institutional guidelines and was approved by the Institutional Review Board of Nagoya University Graduate School of Medicine and Aichi Children’s Health and Medical Center. Written informed consent was obtained from all patients or their legal guardians.
Patients and samples
BALF samples were obtained from 10 pediatric patients hospitalized in the pediatric intensive care unit with respiratory failure and investigated with NGS for comprehensive detection of pathogens. No patients had a history of an immunocompromised state. Patient characteristics are listed in Table 1. BALF was obtained during the acute phase of the illness and cryopreserved at −80 °C until use. Until the NGS analysis was completed, the researchers who conducted experiments and analyses were not notified of the medical information of each patient or of the results of conventional microorganism tests including cultures, viral antigen tests, and PCR.
Library preparation and sequencing
First, we compared two DNA extraction kits: the QIAamp DNA Microbiome kit (Qiagen, Hilden, Germany) and the QIAamp UCP Pathogen Mini kit (Qiagen) for efficacy of detection of bacterial reads. Based on the results, the QIAamp DNA Microbiome kit was used in further experiments. RNA extraction was performed using the NucleoSpin RNA Blood kit (MACHEREY-NAGEL, Düren, Germany). The extracted RNA was immediately converted to cDNA and amplified with the REPLI-g WTA Single Cell kit (Qiagen) in accordance with the manufacturer’s instructions. DNA and RNA concentrations were measured using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Waltham, MA, USA) and a Qubit RNA HS assay kit (Thermo Fisher Scientific), respectively.
The Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) was used to prepare all the libraries from extracted DNA or generated cDNA, as described above. Libraries were also prepared from distilled water as a preparation control (NTC 1-3). Library quality was analyzed using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA) or Agilent 2100 Bioanalyzer (Agilent Technologies). The library concentration was quantified using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Then, libraries were sequenced on MiSeq (Illumina) with the 2 × 150 bp paired-end protocol.
Processing of sequence data
For analysis of the sequence data, the FASTQ files were uploaded to and processed with MePIC v2.0 (National Institute of Infectious Disease)40. First, unnecessary adapter sequences and low-quality bases (Q-score cutoff, 20) were trimmed off in the pipeline. Then, human-derived reads were detected using the BWA program and removed from the downstream analysis. For the remaining reads, the MEGABLAST program (E-value cutoff, 1e–30) was used to search similar sequences of known nucleotide sequences registered in the NCBI nt database. Finally, the search result was downloaded and summarized regarding taxonomic information using MEGAN6 (University of Tübingen, Tübingen, Germany) (bit score >250 (bacteria)/>50 (virus))42. The raw sequence data in FASTQ format was also used for alignment with the reference genome with CLC Genomics Workbench 9.5 (CLC bio; Qiagen) (length fraction = 0.9 (bacteria)/0.8 (virus); similarity fraction = 0.9 (bacteria)/0.8 (virus); nonspecific reads were ignored).
Full-length consensus sequences of EV-D68 were obtained by assigning the most common nucleotide sequence gained in this study to each nucleotide position of the reference genome (KP240936). Phylogenetic analysis of the full genome and the VP1 region of EV-D68 was conducted using the neighbor joining method by MEGA743.
Real-time PCR
Real-time PCR for detection of CMV was performed with the QuantiTect Multiplex PCR kit (Qiagen), and amplification was conducted using the QuantStudioTM3 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) as previously described44. To confirm the presence of viruses detected by NGS, reverse transcription PCR (RT-PCR) was performed with the Cycleave PCR respiratory virus detection kit (Takara Bio, Kusatsu, Japan), and amplification was conducted using the QuantStudioTM3 Real-Time PCR System in accordance with the manufacturer’s instructions. Real-time PCR for S. epidermidis and E. faecalis was performed by a commercial laboratory (TechnoSuruga Laboratory, Shizuoka, Japan) using Rotor-Gene (Qiagen). Information about the primers and probes used for PCR is shown in Supplementary Table 244–49.
Supplementary information
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan [17K10107 to J.K.].
Author Contributions
S.T., J.K., Y.O. and Y.I. conceived and designed the experiments. S.K., S.W. and T.I. collected clinical samples and data. S.T., J.K., K.H., Y.O., T.O., T.S. and Y.T. performed the experiments. S.T., J.K., K.H., Y.O. and Y.I. analyzed and interpreted the data. S.T. and J.K. wrote the manuscript. All authors read and approved the final manuscript.
Data Availability
Consensus sequences of detected viruses are available in the DNA Data bank of Japan (Accession numbers: LC495296, LC495297, LC495298, and LC495299). All other data generated or analyzed during this study are included in this article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49372-x.
References
- 1.Mani, C. S. & Murray, D. L. Acute pneumonia and its complications. In Principles and Practice of Pediatric Infectious Diseases, 4th ed. (eds Long, S. S., Pickering, L. K. & Prober, C. G.) 235–245 (Elsevier Saunders, Edinburgh, 2012).
- 2.Jain S, et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gadsby NJ, et al. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis. 2016;62:817–823. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Thoracic Society; Infectious Disease Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Cre Med171, 388–416 (2005). [DOI] [PubMed]
- 5.Mandell LA, et al. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin Infect Dis. 2003;37:1405–1433. doi: 10.1086/380488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lode H, Schaberg T, Raffenberg M, Mauch H. Diagnostic problems in lower respiratory tract infections. J Antimicrob Chemother. 1993;32(Suppl A):29–37. doi: 10.1093/jac/32.suppl_A.29. [DOI] [PubMed] [Google Scholar]
- 7.Hohenthal U, et al. Bronchoalveolar lavage in immunocompromised patients with haematological malignancy-value of new microbiological methods. Eur J Haematol. 2005;74:203–211. doi: 10.1111/j.1600-0609.2004.00373.x. [DOI] [PubMed] [Google Scholar]
- 8.Bewig B, et al. Detection of CMV pneumonitis after lung transplantation using PCR of DNA from bronchoalveolar lavage cells. Respiration. 2000;67:166–172. doi: 10.1159/000029481. [DOI] [PubMed] [Google Scholar]
- 9.Rao U, et al. Utility of bronchoalveolar lavage in the diagnosis of pulmonary infection in children with haematological malignancies. Ir J Med Sci. 2013;182:177–183. doi: 10.1007/s11845-012-0852-3. [DOI] [PubMed] [Google Scholar]
- 10.Nadimpalli S, et al. Diagnostic yield of bronchoalveolar lavage in immunocompromised children with malignant and non-malignant disorders. Pediatr Pulmonol. 2017;52:820–826. doi: 10.1002/ppul.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efrati O, et al. Fiberoptic bronchoscopy and bronchoalveolar lavage for the evaluation of pulmonary disease in children with primary immunodeficiency and cancer. Pediatr Blood Cancer. 2007;48:324–329. doi: 10.1002/pbc.20784. [DOI] [PubMed] [Google Scholar]
- 12.Schlaberg R, et al. Viral pathogen detection by metagenomics and pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J Infect Dis. 2017;215:1407–1415. doi: 10.1093/infdis/jix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houldctoft CJ, Beale MA, Breuer J. Clinical and biological insights from viral genome sequencing. Nat Rev Microbiol. 2017;15:183–192. doi: 10.1038/nrmicro.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MR, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf EH, et al. Unbiased detection of respiratory viruses by use of RNA sequencing-based metagenomics: a systematic comparison to a commercial PCR panel. J Clin Microbiol. 2016;54:1000–1007. doi: 10.1128/JCM.03060-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorburn F, et al. The use of next generation sequencing in the diagnosis and typing of respiratory infections. J Clin Virol. 2015;69:96–100. doi: 10.1016/j.jcv.2015.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawada J, et al. Identification of viruses in cases of pediatric acute encephalitis and encephalopathy using next-generation sequencing. Sci Rep. 2016;6:33452. doi: 10.1038/srep33452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki T, et al. Comprehensive detection of viruses in pediatric patients with acute liver failure using next-generation sequencing. J Clin Virol. 2017;96:67–72. doi: 10.1016/j.jcv.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Horiba K, et al. Comprehensive detection of pathogens in immunocompromised children with bloodstream infections by next-generation sequencing. Sci Rep. 2018;8:3784. doi: 10.1038/s41598-018-22133-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi S, et al. Identification of potential pathogenic viruses in patients with acute myocarditis using next-generation sequencing. J Med Virol. 2018;90:1814–1821. doi: 10.1002/jmv.25263. [DOI] [PubMed] [Google Scholar]
- 21.Miao Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(S2):S231–S240. doi: 10.1093/cid/ciy693. [DOI] [PubMed] [Google Scholar]
- 22.Seo S, et al. Idiopathic pneumonia syndrome after hematopoietic cell transplantation: evidence of occult infectious disease. Blood. 2015;125:3789–3797. doi: 10.1182/blood-2014-12-617035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewandowska DW, et al. Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection. PLoS ONE. 2017;12:e0177349. doi: 10.1371/journal.pone.0177340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young JC, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant. 2015;15:200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain S, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372:835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krilov L, et al. The association of rhinoviruses with lower respiratory tract disease in hospitalized patients. J Med Virol. 1986;19:345–352. doi: 10.1002/jmv.1890190407. [DOI] [PubMed] [Google Scholar]
- 27.Choi SH, et al. Viral infection in patients with severe pneumonia requiring intensive care unit admission. Am J Respir Crit Care Med. 2012;186:325–332. doi: 10.1164/rccm.201112-2240OC. [DOI] [PubMed] [Google Scholar]
- 28.Nolan VG, et al. Etiology and impact of coinfections in children hospitalized with community-acquired pneumonia. J Infect Dis. 2018;218:179–188. doi: 10.1093/infdis/jix641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esposito S, Bosis S, Niesters H, Principi N. Enterovirus D68 infection. Viruses. 2015;7:6043–6050. doi: 10.3390/v7112925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esposito S, et al. Impact of rhinoviruses on pediatric community-acquired pneumonia. Eur J Clin Microbiol Infect Dis. 2012;31:1637–1645. doi: 10.1007/s10096-011-1487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin ET, et al. Heterotypic infection and spread of rhinovirus A, B, and C among childcare attendees. J Infect Dis. 2018;218:848–855. doi: 10.1093/infdis/jiy232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn JG, Kim DS, Kim KH. Clinical characteristics and cytokine profiles of children with acute lower respiratory tract infections caused by human rhinovirus. PLoS One. 2018;13:e0198624. doi: 10.1371/journal.pone.0198624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm-Hansen CC, Midgley SE, Fischer TK. Global emergence of enterovirus D68: a systematic review. Lancet Infect Dis. 2016;16:e64–e75. doi: 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- 34.Korematsu S, et al. “Spike” in acute asthma exacerbations during enterovirus D68 epidemic in Japan: A nation-wide survey. Allergol Int. 2018;67:55–60. doi: 10.1016/j.alit.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 35.National Institute of Infectious Diseases. Infectious Agents Surveillance Report. (2018) Available at: https://www.niid.go.jp/niid/images/iasr/rapid/topics/ev68/151006/ev68mon_181129.gif. (Accessed: 7th December 2018).
- 36.Majumdar M, et al. Environmental surveillance reveals complex enterovirus circulation patterns in human population. Open Forum Infect Dis. 2018;10:ofy250. doi: 10.1093/ofid/ofy250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, et al. Enterovirus D68 subclade B3 strain circulating and causing an outbreak in the United States in 2016. Sci Rep. 2017;7:1242. doi: 10.1038/s41598-017-01349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruuskane O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet. 2011;377:1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.American Academy of Pediatrics. Respiratory Syncytial Virus. In Red book: 2015 Report of the Committee on Infectious Diseases. 30th ed. (eds Kimberlin, D. W., Brady, M. T., Jackson, M. A. & Long, S. S.) 667–675 (American Academy of Pediatrics, Elk Grove Village, IL, 2015).
- 40.Takeuchi F, et al. MePIC, metagenomic pathogen identification for clinical specimens. Jpn J Infect Dis. 2014;67:62–65. doi: 10.7883/yoken.67.62. [DOI] [PubMed] [Google Scholar]
- 41.Kulkarni N, et al. Reproducible bioinformatics project: a community for reproducible bioinformatics analysis pipelines. BMC Bioinformatics. 2018;19(10):349. doi: 10.1186/s12859-018-2296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huson DH, et al. MEGAN community edition -interactive exploration and analysis of large-scale microbe sequencing data. PLoS Comut. Biol. 2016;12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wada K, et al. Simultaneous quantification of Epstein-Barr virus, cytomegalovirus, and herpesvirus 6 DNA in samples from transplant recipients by multiplex real-time PCR assay. J Clin Microbiol. 2007;45:1426–1432. doi: 10.1128/JCM.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mentel R, Wegner U, Bruns R, Gurtler L. Real-time PCR to improve the diagnosis of respiratory syncytial virus infection. J Med Microbiol. 2003;52:893–896. doi: 10.1099/jmm.0.05290-0. [DOI] [PubMed] [Google Scholar]
- 46.Hamano-Hasegawa K, et al. Comprehensive detection of causative pathogens using real-time PCR to diagnose pediatric community-acquired pneumonia. J Infect Chemother. 2008;14:424–432. doi: 10.1007/s10156-008-0648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mackay IM, et al. Molecular assays for detection of human metapneumovirus. J Clin Microbiol. 2003;41:100–105. doi: 10.1128/JCM.41.1.100-105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheltinga SA, Templeton KE, Beersma MF, Claas EC. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J Clin Virol. 2005;33:306–311. doi: 10.1016/j.jcv.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tokarz R, et al. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;95:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaida A, et al. Distinct genetic clades of enterovirus D68 detected in 2010, 2013, and 2015 in Osaka City, Japan. PLoS One. 2017;12:e0184335. doi: 10.1371/journal.pone.0184335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Consensus sequences of detected viruses are available in the DNA Data bank of Japan (Accession numbers: LC495296, LC495297, LC495298, and LC495299). All other data generated or analyzed during this study are included in this article.