Abstract
Primary cardiac fibroblasts are notoriously difficult to maintain for extended periods of time in cell culture, due to the plasticity of their phenotype and sensitivity to mechanical input. In order to study cardiac fibroblast activation in vitro, we have developed cell culture conditions which promote the quiescent fibroblast phenotype in primary cells. Using elastic silicone substrata, both rat and mouse primary cardiac fibroblasts could be maintained in a quiescent state for more than 3 days after isolation and these cells showed low expression of myofibroblast markers, including fibronectin extracellular domain A, non-muscle myosin IIB, platelet-derived growth factor receptor-alpha and alpha-smooth muscle actin. Gene expression was also more fibroblast-like vs. that of myofibroblasts, as Tcf21 was significantly upregulated, while Fn1-EDA, Col1A1 and Col1A2 were markedly downregulated. Cell culture conditions (eg. serum, nutrient concentration) are critical for the control of temporal fibroblast proliferation. We propose that eliminating mechanical stimulus and limiting the nutrient content of cell culture media can extend the quiescent nature of primary cardiac fibroblasts for physiological analyses in vitro.
Subject terms: Biological techniques, Cell biology, Cell growth
Introduction
The term “fibroblast” is assigned to a heterogeneous group of highly-motile stromal cells found in the interstitium, whose function and phenotype is tissue-dependent1–3. In the heart, cardiac fibroblasts serve to maintain tissue homeostasis and regulate extracellular matrix turnover. While there are no molecular markers that are entirely specific to cardiac fibroblasts, they are typically positive for transcription factor 21 (TCF21)4–6, vimentin7–9, and CD90 (or Thy-1)10–12. When subject to stress or injury, fibroblasts lose their normal phenotype and activate into myofibroblasts. Unlike quiescent fibroblasts, myofibroblasts are highly contractile and are characterized by a radically organized cytoskeleton featuring alpha-smooth muscle actin (αSMA) -positive stress fibers13–16, mature focal adhesions17–19, and increased production of periostin20,21 and fibrillar collagens22,23. In addition, the activation of cardiac myofibroblasts is also associated with alternative splicing of fibronectin, specifically denoted by the inclusion of the cell-associated extracellular domain A (ED-A)24,25.
Myofibroblast activation is a hallmark of cardiovascular disease, as these cells are responsible for the excessive deposition of extracellular matrix (ECM) proteins and are the primary drivers of fibrosis and its related pathologies26,27. Induction of the myofibroblast phenotype has been associated with a multitude of stimuli and the most common effector in this process is transforming growth factor-beta (TGF-β)28,29, which is associated with the initial inflammatory response after vascular or myocardial injury. Moreover, cardiac myofibroblasts are further driven to promote fibrogenesis in response to the autocrine and paracrine effects of other pro-inflammatory cytokines, such as platelet-derived growth factor (PDGF)30. Along with this response, cardiac myofibroblasts also exhibit a pronounced increase in PDGF receptor-alpha (PDGFRα) expression in disease states31–33. Hypertrophic agents such as growth factors and Angiotensin-II34,35, hyperglycemia36,37, and the presence of reactive oxygen species (ROS)38,39 have also been shown to contribute to this transition in phenotype. Finally, biomechanical input has also been implicated in myofibroblast activation. Isometric tension promotes the release of latent TGF-β present in the ECM40,41, and the formation of stress fibers with the incorporation of αSMA42 and this could be likened in vitro to seeding cells on stiff plastic surfaces. Similarly, mechanical loading and stretching modulates fibroblast function and phenotype, promoting the deposition of ED-A fibronectin25 and enhancing TGF-β signaling43. While it has traditionally been viewed as a permanent event, the activation of myofibroblasts has recently been observed as a reversible process in resident fibroblasts in vivo5,44.
Despite these findings, the mechanisms which govern the cardiac fibroblast and myofibroblast phenotypes are largely uncharacterized, as common cell culture techniques are not commensurate to physiologically-relevant conditions, and in vivo transgenic models are difficult to generate without affecting other stromal cells. Even when isolated from healthy myocardium, the spontaneous phenotype of primary cardiac fibroblasts in conventional cell culture is of pro-fibrotic, activated myofibroblasts within hours of plating45. However, it has been shown that culturing primary fibroblasts on elastic surfaces which are biomimetic to their native tissues can help to alleviate myofibroblast activation46. In the case of myocardium, culture surfaces with a compressibility, or elastic modulus (E), of ~7 kPa are representative of healthy tissue, while surfaces with E > 10 kPa are considered fibrotic47,48. This presents a considerable hurdle in that conventional polystyrene tissue culture plates are significantly stiffer, often upwards of E = 3 GPa49,50. In addition, while it is common practice to passage primary cells to promote homogeneity in the culture population, passaging further drives myofibroblast activation and prevents physiologically- pertinent studies45. This phenotypic plasticity presents a unique problem in that fibroblast physiology that is representative of healthy myocardium cannot be readily observed and manipulated in vitro.
In spite of the unstable nature of the cardiac fibroblast phenotype, the capacity to maintain these cells in a quiescent state in two-dimensional cell culture would enable much more accurate and reproducible means by which to study their physiology, and their response to genetic manipulation and pharmacological treatment. In this study, we present conditions in which unpassaged (P0) primary cardiac fibroblasts can be maintained for more than 72 hours in vitro without significant activation of the myofibroblast phenotype. These data support an alternative means by which to investigate the molecular and cellular physiology of primary cardiac fibroblasts in vivo studies.
Results
Myofibroblast markers are downregulated in low nutrient conditions with restricted biomechanical input
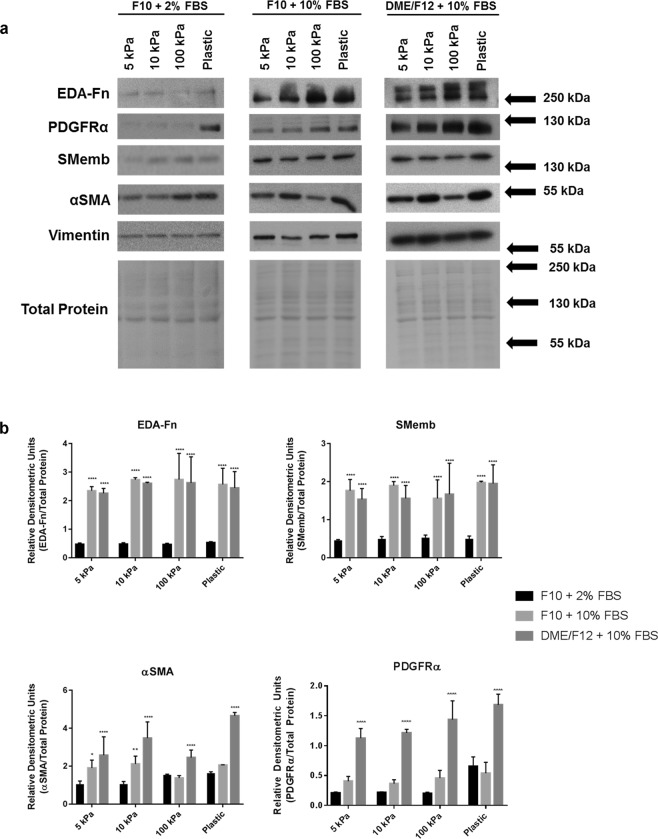
Mechanobiological properties (stiffness) of the ECM govern myofibroblast activation and function, and do so in the absence of input from TGFβ1/Smad signalling50. Moreover, myofibroblasts are contractile and sense and modulate stiffness within the ECM through focal adhesions via integrin binding23. In order to determine the physiological effects of two-dimensional cell culture on primary cardiac fibroblasts, we not only examined the influence of cell culture medium and serum, but also whether the compressibility of the culture surface was a greater factor in the spontaneous phenotype of the cells in vitro. After three days in culture, variable expression of myofibroblast markers ED-A fibronectin, non-muscle myosin heavy chain (SMemb or myosin IIB), and αSMA was observed in conditions of either low nutrient media (F10 with 2% FBS) or high nutrient media (DMEM/F12 with 10% FBS), plated on substrates that mimic the compressibility of healthy myocardium (5 kPa), or fibrosis-stiff substrate (100 kPa) (Fig. 1). On stiff substrate in combination with high serum, the preponderance of expression of myofibroblast markers is evident and significantly greater the other conditions tested. This increase was markedly evident with the expression of PDGRFα, which had a strong response to both substrate stiffness and medium composition. In mouse primary cardiac fibroblasts, we observed a significant increase in EDA-Fn, and PDGFRα, which was exhibited in concert with an increase in substrate stiffness (Supplemental Fig. 2). While conditions which favored the fibroblast phenotype were of F10 medium with low serum, it was evident that lower plate compressibility also imparted greater control of cell phenotype. The data supports the hypothesis that tuning the fibroblast substrate for reduced biomechanical input (ie. cells plated on elastic substrate versus stiff plastic) in combination with low nutrient conditions reduces the activation of fibroblasts cultured for extended periods.
Figure 1.
Myofibroblast marker expression in rat primary cardiac fibroblasts after >72 hours in culture. (a) Rat cardiac fibroblasts were plated on elastic plates with characteristic elastic moduli of 5 kPa, 10 kPa, and 100 kPa, with the indicated culture medium. Cells plated on conventional polystyrene tissue culture plates (rigid substrate – ranging from 30 to 100 MPa) were used as a comparative control. Protein from unpassaged primary cardiac fibroblasts was harvested ~3 days after plating. Vimentin expression was used as a pan-phenotypic control, and protein expression was normalized to total protein loading. (b) Graphical representation of data in (A). Data shown as the mean ± SD and is representative of n = 3–6 biological replicates. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001 when compared to cells cultured in F10 medium with 2% FBS on the same substrate.
αSMA is excluded from the cytoskeleton in quiescent cardiac fibroblasts
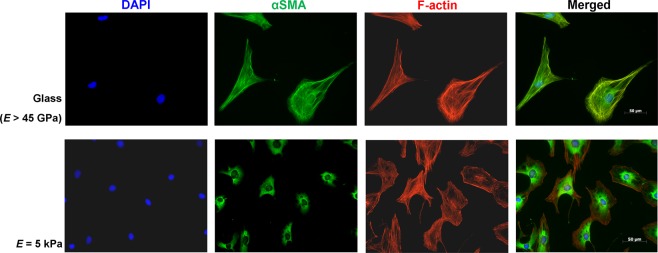
As αSMA is often viewed as the gold-standard marker for tissue fibrosis, and was evidently present in all samples we studied, we sought to compare its subcellular localization in our proposed in vitro model. Unpassaged primary rat cardiac fibroblasts were maintained in vitro for >72 hours; Fig. 2 provides comparative fields of cardiac fibroblasts plated on glass (rigid substrate) and fibroblasts plated on “cardiac soft” 5 kPa plates. F-actin and αSMA are double-stained in these fields, and the relative size of the cells in each set is remarkably different. αSMA is not incorporated into the cytoskeleton of the inactivated fibroblasts plated on the elastic substrata, indicating that these cells have not formed stress fibers, and are not actively contractile51. The merged fields of cells plated on glass show complete incorporation of αSMA into stress fibers, which reflects their phenotype of activated myofibroblasts.
Figure 2.
αSMA is excluded from F-actin stress fibers when cultured on 5 kPa culture surfaces. P0 rat cardiac fibroblasts were seeded at low confluency (<10%) on either glass (rigid substrate) or elastic coverslips and probed for αSMA (green) and F-actin (red) 3 days after plating. Images are representative of n = 3 independent biological replicates. Scale bar = 50 μm.
Cardiac fibroblast gene expression is further affected by biomechanical input, in vitro
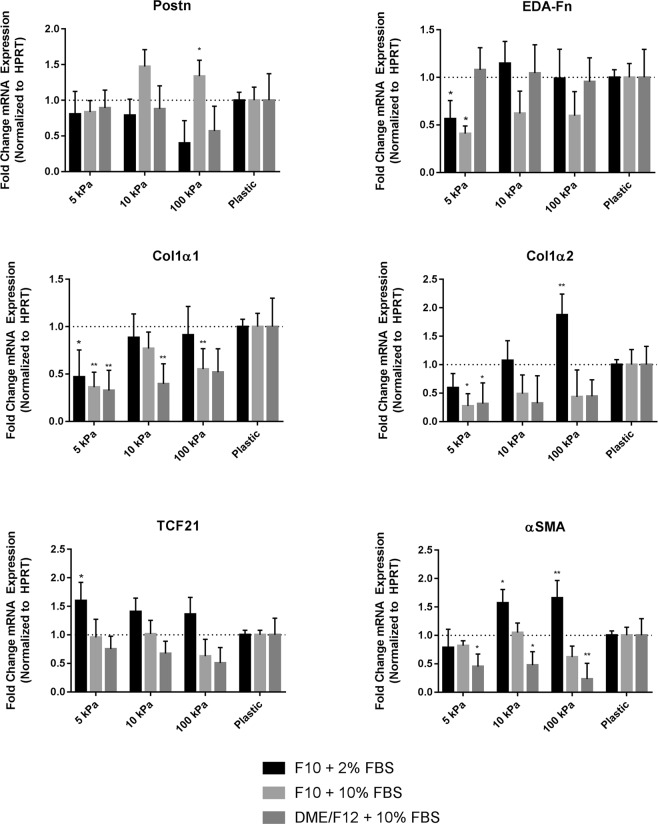
In addition to protein expression, we sought to determine the effects of culture medium and/or substrate stiffness on gene expression in cultured primary cardiac fibroblasts. After three days in vitro, relative mRNA abundance of fibroblast- and myofibroblast-expressed genes including collagen type I monomers, αSMA, ED-A fibronectin, periostin (a secreted, matricellular protein), and Tcf21 were examined. Tcf21 is required for fibroblast formation, and is a nucleus-localized protein expressed in adult fibroblasts52, whereas periostin is a marker for activated fibroblasts53,54. We found Tcf21 expression to be significantly elevated in the fibroblasts harvested from 5kPa substrate with F10 medium and 2% FBS, versus expression on plastic plates, whereas Postn was elevated in the 100 kPa plates relative to its expression on plastic plates in similar conditions (Fig. 3). ED-A fibronectin (Fn1-EDA) gene expression was significantly lower on 5 kPa plates versus the non-elastic plastic controls, whereas αSMA was elevated in 10 and 100 kPa plates versus plastic in low nutrient and low serum conditions. Collagen monomer (Col1a1 and Col1a2) expression also shows similar responsiveness to plate stiffness, with expression increasing as substrate stiffness increases. While our results show some heterogeneity in the variable expression of marker mRNAs, which appears to be gene-dependent, the myofibroblast markers are generally upregulated in response to plating of fibroblasts on stiff substrates. When considering the effects of the cell culture medium, the conditions which resulted in the most inhibition of myofibroblast gene expression were those in F10 medium with 2% serum, although this effect can apparently be overridden by the compressibility of the tissue culture plate.
Figure 3.
Fibroblast and myofibroblast gene expression in primary cardiac fibroblasts after >72 hours in culture. RNA was harvested from P0 rat cardiac fibroblasts 3 days after plating and used for qPCR. Samples from cells cultured on conventional plastic tissue (rigid substrate) culture surfaces were used as comparative controls. All reactions were performed in technical triplicates and were normalized to HPRT. Data shown as the mean ± SD and is representative of n = 3 biological replicates. *P < 0.05, **P < 0.01, when compared to cells cultured on plastic in the same culture medium.
Cardiac fibroblast proliferation can be limited in extended cell culture
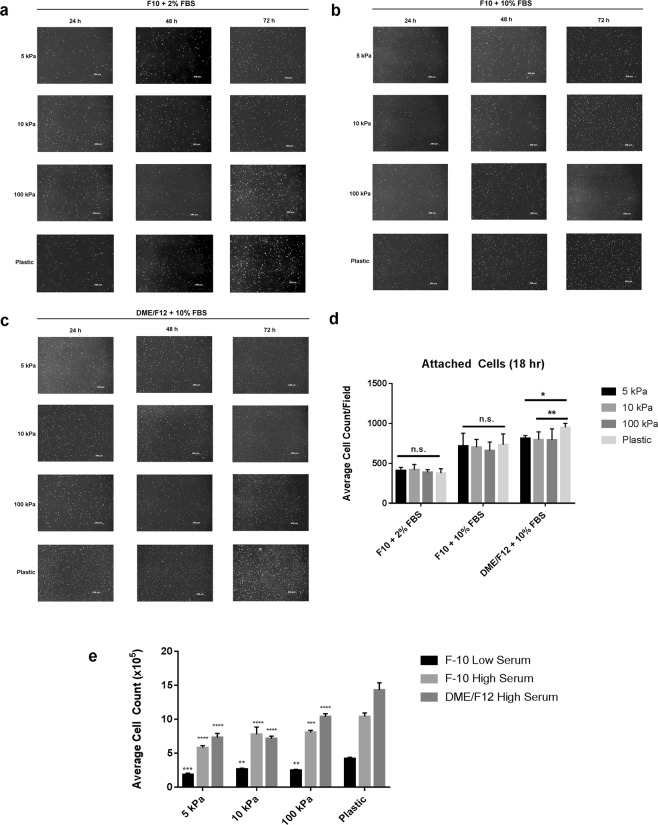
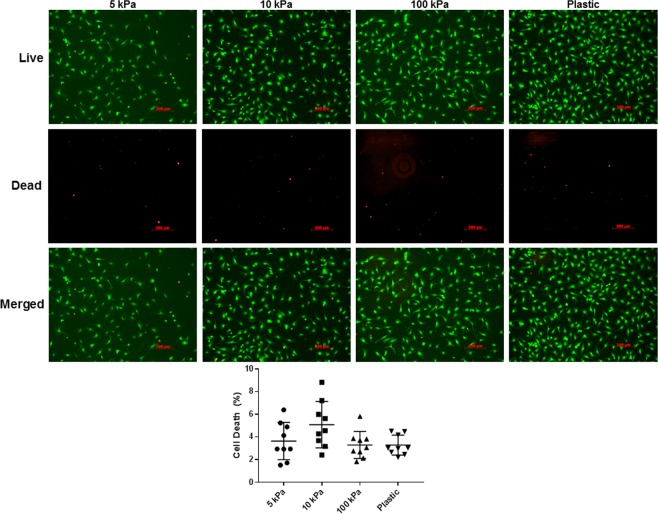
To further characterize isolated rat primary cardiac fibroblasts in vitro, we examined their proliferative behavior under various conditions. Specifically, we sought to investigate the responsiveness of fibroblast and myofibroblast proliferation in low and high serum conditions using media with low nutrient (F10) and high nutrient (DMEM/F12) content. In low serum conditions, cardiac fibroblast proliferation is suppressed by plating on 5 kPa, 10 kPa and 100 kPa plates with F10 medium, versus plastic controls (Fig. 4). A similar trend was observed using high serum conditions with the same medium, as well as with DMEM/F12; however the expansion of the cell population in conditions with DMEM/F12 was much more pronounced, especially on stiff plastic. To determine whether this observation was due to a difference in initial cell attachment between culture conditions, we counted the number of attached cells at 18 hours post-plating, after washing off any debris. We did not observe a significant difference in cell attachment when cells were cultured in low nutrient conditions however there was preferential attachment on stiffer culture surfaces when the cells were maintained in DMEM/F12 + 10% FBS. Additionally, to confirm that the observed lack of proliferation in F10 culture medium was not due to excessive cell death, at 96 hours post-plating the cells were stained with Calcein-AM (stains viable cells) and ethidium homodimer (stains nucleic acid/dead cells). We observed no significant difference in cell death between the various elastic moduli of the cell culture plates, suggesting that the compressibility of the culture substrata does indeed affect primary cardiac fibroblast activation and proliferation in vitro (Fig. 5). These results support the suggestion that proliferation of activated myofibroblasts is higher than inactive fibroblasts in culture, and that the state of activation can be controlled by both substrate stiffness and the nutrient content of the culture medium. Thus, we reveal an interaction between substrate stiffness, activation state of cardiac fibroblasts, and the proliferative capacity of primary cardiac fibroblasts in vitro.
Figure 4.
Nutrient restriction and decreased biomechanical input inhibits proliferation in primary cardiac fibroblasts. (a–c) P0 rat cardiac fibroblasts were plated on elastic plates with various elastic moduli, with the indicated culture medium, and stained with proliferation staining agent 24 hours after seeding. Images were captured immediately after staining and every 24 hours subsequently. Cells plated on conventional plastic tissue culture plates (rigid substrate) were used as a comparative control. Images are representative of n = 3 biological replicates. (d) Histrographical representation of cell counts 18 hours after plating and attachment (e) Histrographical representation of cell counts obtained after 3 days in culture (t > 72 hours). Data displayed as mean ± SD and is representative of n = 6 biological replicates. **P < 0.01, ***P < 0.005, ****P < 0.001 when compared to cells cultured on plastic in the same medium.
Figure 5.
Assessment of cell viability in restricted nutrient conditions after >72 hours in culture. P0 rat cardiac fibroblasts were seeded onto substrata of various elastic moduli and cultured in F10 medium supplemented with 2% FBS for 4 days. Cell viability was assessed with Calcein-AM (green) staining, and cell death by ethidium homodimer (red). Each biological replicate was assessed by capturing 3 random fields at 10X magnification. Images are representative of n = 3 biological replicates, with 9 technical replicates each.
Discussion
Cardiac fibroblasts and activated myofibroblasts are component cells of the myocardium that contribute to the maintenance of the ECM in homeostatic hearts and to cardiac fibrosis after injury, respectively3. A novel interpretation of cardiac fibrosis (and other tissue fibrosis) is that rather than simply described as “over-active” wound healing, the organism may be attempting to utilize developmental programs that are typically active when generating functional muscle23. In the heart, fibroblasts are derived from different embryonic sources, with the majority from the epicardium (EMT contributing 80% of cells), and most of the remaining fibroblasts from the endocardium (EndoMT contributing 18%)55. Thus, defining these cells in a molecular context is difficult due to the lack of identification of a specific marker common to all fibroblasts. The mixed origin of cardiac fibroblasts notwithstanding, the common role of the activated myofibroblast is to generate and remodel the ECM23. Furthermore, the recent focus on cardiac fibrosis and the involvement of myofibroblasts and specific markers for them, including periostin, ED-A fibronectin, SMemb, and αSMA provides new impetus to direct fibrosis research to focus on activated myofibroblasts, which require refinements in approaches for their practical study4,17,45.
Accordingly, the estimation of the impact of fibroblast activation to myofibroblasts in the damaged and failing heart have become a much sought-after topic. Nonetheless, while the burgeoning number of scientific reports published during the past ten years reflects the overall acceptance of cardiac fibrosis as a player in the evolution of heart failure, methods to distinguish the activated from non-activated form of cardiac fibroblasts have not kept pace, with few exceptions. We, and other groups, have previously published studies including both unpassaged and passaged primary cardiac fibroblasts plated on stiff plastic substrata in an attempt to procure a baseline phenotype in vitro56–58. Despite these efforts, observing primary cardiac fibroblasts in conventional cell culture is limited to the first 12 to 16 hours before seeing overt activation of the myofibroblast phenotype45. As a result, the maintenance of the quiescent phenotype in culture has become a pressing issue, and is regularly overlooked in experimental design.
Herein we have provided detailed results to contextualize the importance of nutrients in culture media, culturing cells to eliminate biomechanical input, and minimize the impact of serum on fibroblast activation to allow for reliable culture of inactive cardiac fibroblasts. Upon examination of protein expression in rat primary cardiac fibroblasts maintained in culture for three days, it was evident that ED-A fibronectin, αSMA, and SMemb are differentially expressed among all conditions tested. The most differentially-expressed pro-fibrotic marker was PDGFRα, which had increased expression on stiff plastic substrata, and was even more highly-expressed in conditions of high nutrient and serum concentrations. A similar expression pattern for myofibroblast markers was observed in mouse cardiac fibroblasts, which were maintained in culture for a total of 10 days post-isolation (Supplemental Fig. 2). Recent evidence indicates that these markers may also serve to drive fibroblast activation, as myosin II may mediate myofibroblast activation in stiffened fibrotic lungs59, and that in the setting of a stiff matrix, αSMA incorporation into contractile stress fibers facilitates mesenchymal stromal cell fate by controlling YAP release60. In addition, PDGFRα has recently garnered interest as a marker of fibroblast activation and ECM remodeling, as its expression is upregulated in models of fibrosis an heart failure30,31. Along with the data presented here, the addition of PDGFRα to the gamut of markers often used to describe cardiac fibroblasts and myofibroblasts will provide a clearer understanding of a cell’s position on the spectrum spanning the two phenotypes. Very recently, the fibronectin ED-A domain has been shown to promote binding to latent TGF-β-binding protein-1 (LTBP-1), and thus enhances fibronectin-associated storage of TGF-β, which may then increase its availability for extracellular activation and stimulation of fibrosis25. The appearance of fibronectin ED-A domain remains a useful marker heralding the activation of fibroblasts to myofibroblasts, and their subsequent production and local accumulation of disordered ECM. In addition, we also observed more severe upregulation of all three markers, along with Hippo and EMT markers, when the elastic substrata were coated with soluble fibronectin (Supplementary Fig. 1). These findings corroborate current work which postulates that fibronectin is an ECM component drives fibrogenesis and heart failure6. Taken together, these results underscore the utility of ED-A fibronectin as a marker of phenotype plasticity in response to biomechanical and nutritive inputs in culture, and strengthen the case to use it as such in concert with αSMA incorporation into myofibroblasts’ stress fibers.
Beyond the expression of αSMA, its subcellular localization is paramount to its effects on cell phenotype. We observed not only that αSMA is indeed present in unpassaged fibroblasts on elastic substrates, but also that its incorporation into stress fibers is a greater indicator of the myofibroblast phenotype. Similar findings have been demonstrated in other stromal cell types, including subcutaneous61, dermal59, and hepatic portal fibroblasts60. Furthermore, αSMA can itself be a driving force in myofibroblast activation, as ectopic overexpression in stromal cells has been shown activate the myofibroblast phenotype, and generate a feed-forward loop in fibrogenesis62. It should also be noted that in the myocardium, αSMA is not only expressed by cells of mesenchymal origin, but can also be expressed by stressed or injured cardiomyocytes expressing fetal gene programs during active remodelling and dedifferentiation53–55. Collectively, these results suggest that αSMA cannot be entirely relied upon for accurate characterization of cardiac fibroblasts, and that its subcellular localization should also be considered when conducting in vitro cardiovascular studies.
Based on the findings of this study, we propose that relatively inactive primary cardiac fibroblasts can indeed be maintained in vitro for a period of time that is adequate for most molecular assays, provided that certain parameters (ie. passage number, culture medium, plate compressibility) are addressed (Fig. 6). The majority of published literature which implements primary cardiac cell culture, including those previously published by our lab, do not take into account the biomechanical, nutritional, and hormonal input to which the cells are subject in two-dimensional culture. Likewise, primary cardiac fibroblasts which are subject to passaging are also often used in order to maximize cell numbers and decrease the number of animals or tissue specimens for a given experiment. This is often seen as reasonable as it is simple, convenient, and offers some flexibility to the type of molecular assays employed in fibroblast-centric research. Using the methods described here, fewer cells are required to seed plates and passaging is not used; this can significantly decrease the material requirements for a given experiment. While diverging from traditional cell culture methods is not easily accomplished from a technical perspective, it is essential to consider all factors when designing experiments and interpreting data, especially as current in vitro studies are still using what would be considered as activated myofibroblasts. It is known that cardiac fibroblasts exist on a delicate phenotypic spectrum that is highly reactive to the extracellular environment and methods should be adapted to generate accurate and reproducible results. This will facilitate the generation of data which is relevant to physiological conditions, but will also promote rigor and consistency in the literature. The ability to control myofibroblast activation greatly improves the sensitivity of genetic and pharmacological assays. Future exploration into medium composition (eg. serum-free media) would certainly enable further refinements on this method. Thus, to better understand the pathogenesis of cardiac fibrosis and the effects of potential therapeutic interventions, the consequences of in vitro conditions on cell physiology should be carefully considered.
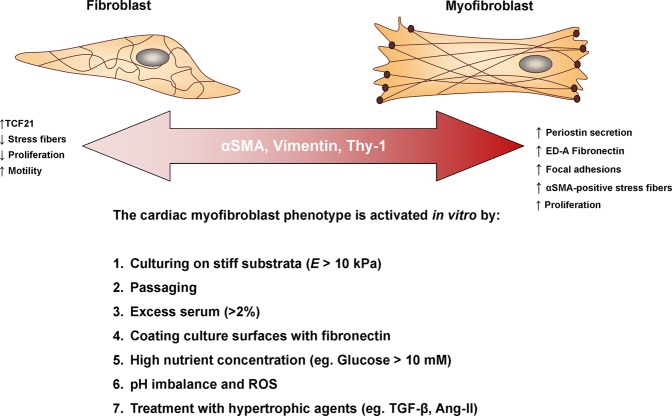
Figure 6.
A schematic depicting the various factors affecting the fibroblast phenotype in vitro. Herein we summarize our main findings along with other known elements which influence primary cardiac fibroblast activation in vitro. We suggest that α-SMA is an ever-present marker on the cardiac fibroblast-myofibroblast spectrum, and that its incorporation in myofibroblast stress fibers is a key component of defining the activated phenotype. Previous studies on conventional plastic tissue culture plates have confirmed that passaging45, hyperglycemic-like conditions67, and treatment with ROS and hypertrophic agents indeed activate the myofibroblast phenotype and we conclude that excess serum also contributes to this activation.
Materials and Methods
Animal ethics
All experimental protocols involving live animals were reviewed and approved by the University of Manitoba Animal Care Committee, and were generated in accordance with the standards of the Canadian Council of Animal Care.
Preparation of elastic tissue culture surfaces
PrimeCoat silicone elastic tissue culture plates (10 cm) and coverslips (ExCellness Biotech SA, Lausanne, Switzerland) were coated with 10 μg/mL porcine gelatin type A (0.2 mL/cm2) in sterile water overnight at 37 °C, 5% CO2. Prior to plating cells, the supernatant was removed and replaced with complete culture medium. Polystyrene (non-elastic plastic) tissue culture dishes and glass coverslips were also treated in a similar manner and used as comparative controls when evaluating various elastic moduli.
Isolation of rat primary cardiac fibroblasts
Rat primary cardiac fibroblasts were isolated, as previously described56,57,63 with some modifications. Male Sprague-Dawley rats weighing 101–125 g were anaesthetized with a ketamine-xylazine cocktail (100 mg/kg ketamine; 10 mg/kg xylazine) via intraperitoneal injection. Upon loss of limb reflexes, heparin (6 mg/kg) was administered intravenously via the femoral artery. Hearts were excised and briefly placed in Dulbecco’s Modified Eagle’s medium/Ham’s F12 nutrient mixture (DMEM/F12) prior to cannulating via the aorta on a Langendorff apparatus. The hearts were then subject to retrograde perfusion with DMEM/F12, followed by Minimum Essential Medium, Spinner’s Modification (S-MEM) to cease cardiac contraction and promote cell dissociation. Finally, the hearts were perfused with S-MEM supplemented with 640 U/mL collagenase type II (Worthington Biochemical Corporation, Lakewood, NJ) with recirculation for 25 minutes. Once digested, the tissue was incubated at 37 °C, 5% CO2 for 10 minutes before neutralizing the collagenase with 10 mL of DMEM/F12 supplemented with 2% fetal bovine serum (FBS) and further dissociated by trituration with a serological pipette. The resulting cell suspension was then passed through a 40 μm sterile cell strainer (Thermo Fisher Scientific, Waltham, MA) to remove any undigested tissue and debris. The cells were pelleted by centrifugation at 200 × g for 7 minutes, and re-suspended in 45 mL of complete cell culture medium. Three different types of media were used for comparison: F10 with 2% FBS, F10 with 10% FBS, and DMEM/F12 with 1 μM ascorbic acid and 10% FBS. All media was supplemented with 100 U/mL penicillin-streptomycin. For each 10 dish, 3 mL of cell suspension was added to a total of 10 mL of medium at plating. For coverslips in 35 mm or 6-well dishes, 0.5 mL of cell suspension was added to a total of 2 mL medium per dish or well. Fibroblasts were allowed to adhere for 2.5 hours at 37 °C, 5% CO2; cultures were then briefly washed twice with phosphate buffered saline (PBS; pH 7.4) supplemented with penicillin-streptomycin and then fresh complete culture medium was added. The following day, the cultures were once again washed twice with PBS, and the growth medium was replaced. The culture medium was subsequently replaced once per day until harvesting.
Isolation of mouse primary cardiac fibroblasts
Primary mouse cardiac fibroblasts were isolated using a modified version of a previously-published protocol64. In short, 8- to 12-week old male C57BL/6 mice were anaesthetized with 3% isofluorane until loss of limb reflexes. The chest was opened to excise the heart with a portion of the ascending aorta remaining attached. The heart was promptly flushed with 10 mL of EDTA buffer (5 mM EDTA, 130 mM NaCl, 5 mM KCl, 500 nM NaH2PO4, 10 mM HEPES, 10 mM Glucose, 10 mM 2,3-Butanedione 2-monoxime, and 10 mM Taurine, pH 7.8), followed by 3 mL of perfusion buffer (1 mM MgCl2, 130 mM NaCl, 5 mM KCl, 500 nM NaH2PO4, 10 mM HEPES, 10 mM Glucose, 10 mM 2,3-Butanedione 2-monoxime, and 10 mM Taurine, pH 7.8) using a 27-gauge needle inserted into the ventricles via the apex. While still injecting through the apex, the heart was perfused twice with 25 mL S-MEM supplemented with collagenase type II (330 U/mL), while collecting the solution in a 10 cm tissue culture dish. Once the heart was sufficiently digested, it was gently pulled apart with forceps and allowed to digest in 20 mL of the collagenase solution at 37 °C, 5% CO2 for another 10 minutes. After triturating the tissue, the resulting suspension was neutralized with 10 mL of complete culture medium and passed through a 40 μm cell strainer and pelleted by centrifugation, as mentioned above. The resulting pellet was resuspended in 40 mL of DMEM/F12 supplemented with 10% FBS, 1 μM ascorbic acid, and 100 U/mL penicillin-streptomycin and the cells were then evenly distributed among 4–10 cm dishes and allowed to adhere for 3 hours. Adherent cells were then gently washed once with pre-warmed PBS (pH 7.4) supplemented with antibiotics, and the DMEM/F12 growth medium was replaced. The culture medium was replaced in a similar fashion every 24 hours until they adopted a spindle-shaped morphology (~3–4 days), after which the medium was switched to F10 supplemented with 2% FBS and Insulin-Transferrin-Selenium-Sodium Pyruvate (ITS-A; Thermo Fisher). The cells were harvested once they reached 40–50% confluency, at approximately 10 days post-isolation.
Cell proliferation imaging and counting
Approximately 18 hours after plating, unpassaged (P0) fibroblast cultures were washed twice with PBS. Pre-warmed, serum-free and antibiotic-free medium was then supplemented with Cytopainter Green Cell Proliferation Agent (Abcam, Cambridge, UK). Cells were treated with the dye solution at 37 °C, 5% CO2 for 30 minutes, protected from light. The dye solution was removed, and the cells were briefly washed twice with PBS before replacing with complete culture medium. The cells were imaged every 24 hours post-plating with a Zeiss LSM 5 Pascal microscope using 4X and 10X objectives and an excitation wavelength of 488 nm. Images were processed using AxioVision Microscopy software (Zeiss, rel. 4.8).
Initial cell counts at 18 hours post-plating were performed by using phase contrast microscopy images and ImageJ software65, using three randomly-selected fields for each biological replicate, totaling 9 technical replicates for each cell culture condition. Manual cell counting was accomplished using a Moxi Z automated cell counter (Orflo Technologies, Ketchum, ID). Cells were trypsinized and re-suspended in an excess volume of complete culture medium, and a 1:10 dilution of the cell suspension was used for each count. Two counts were taken for each biological replicate to ensure accuracy for each measurement. Manual cell counting was performed in triplicate for each time point and cell proliferation imaging was accomplished by selecting three randomly-chosen fields for each cell culture condition.
Protein isolation
After approximately 80 hours after plating, cells were trypsinized and pelleted by centrifugation at 200 × g for 5 minutes. Cell pellets were then washed with PBS and re-pelleted by centrifugation for 3 minutes. The supernatant was removed and the pellets were lysed with RIPA lysis buffer supplemented with protease inhibitor cocktail (P8340; Sigma-Aldrich Canada Co., Oakville, ON) and phosphatase inhibitors (10 mM NaF, 1 mM Na3VO4, and 10 mM EGTA). The resulting lysates were then vortexed and incubated on ice for 30 minutes. Following incubation, the lysates were briefly sonicated for 5 seconds, and then centrifuged at 16 000 × g for 15 minutes at 4 °C. Supernatants were transferred to new microcentrifuge tubes and protein concentrations were determined using a bicinchoninic acid (BCA) assay.
Immunoblotting
SDS-PAGE of 25 μg of protein was performed on 8% reducing gels. Proteins were transferred at 4 °C onto PVDF membranes in tris-glycine buffer containing 20% methanol. Total protein loading was measured prior to blotting using Ponceau S staining and densitometric analysis. Non-specific binding sites were blocked with 5% skim milk in tris-buffered saline supplemented with 0.1% Tween-20 (TBS-T) at room temperature. The blots were thoroughly washed in TBS-T before applying primary antibodies overnight at 4 °C with shaking. Primary antibodies were used at the following dilutions: ED-A (cellular) fibronectin (1:1000; MAB1940; MilliporeSigma, Burlington, MA; or NBP1-91258; Novus), SMemb (1:1000; ab684; Abcam), αSMA (1:5000; A2547; Sigma). Because the primary fibroblasts originated from a heterogenous population of cells, vimentin (1:2000; ab8069; Abcam) and platelet-derived growth factor receptor alpha (PDGFRα; 1:1000; ab134123; Abcam) were used as a phenotype controls on each blot. Appropriate HRP-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were applied at a 1:5000 dilution for 1 hour at room temperature. Protein detection was done using ECL substrate, and protein bands were visualized on blue X-ray film. Protein expression was measured by relative densitometry using Quantity One® analysis software (version 4.6.9; Bio-Rad).
RNA isolation and quantitative PCR
Primary cardiac fibroblasts were isolated by trypsinization and centrifugation at 200 × g for 5 minutes. Column-based RNA isolation was performed using the PureLink® RNA Mini kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Approximate RNA concentration and purity was assessed by measuring the absorbance at 260 and 280 nm using a NanoDrop™ Lite Spectrophotometer (Thermo Scientific).
Two-step qPCR was performed first by synthesizing cDNA from 100 ng of RNA, using the Maxima™ First Strand cDNA synthesis for RT-qPCR (Thermo Scientific), and included initial treatment with dsDNase. Amplification reactions were prepared according to the Luna® Universal qPCR Master Mix (New England Biolabs, Ipswich, MA) protocol, using 1 μL of of cDNA template and 200 nM of forward and reverse primers in a final volume of 10 μL. PCR amplification was performed in triplicate for each reaction on a QuantStudio 3 Real-Time PCR System (Applied Biosystems, Foster City, CA) using the fast cycling mode. The following cycling program was used: initial denaturation at 95 °C (60 seconds), followed by 40 cycles of denaturation at 95 °C (15 seconds) and extension at 60 °C (30 seconds). After amplification, a continuous melt curve was generated from 60 °C to 95 °C. Relative gene expression was calculated using the 2−ΔΔCt method66, using the samples from cells plated on plastic as controls for each sample set, and normalized to HPRT. Primer pairs and their corresponding targets are illustrated in Table 1.
Table 1.
List of primer pairs used in quantitative PCR.
| Gene | Accession | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| Acta2 | NM_031004.2 | AGATCGTCCGTGACATCAAGG | TCATTCCCGATGGTGATCAC |
| Col1a1 | NM_053304.1 | TGCTCCTCTTAGGGGCCA | CGTCTCACCATTAGGGACCCT |
| Col1a2 | NM_053356.1 | TGACCAGCCTCGCTCACAG | CAATCCAGTAGTAATCGCTCTTCCA |
| Fn1 | NM_019143.2 | ACTGCAGTGACCAACATTGACC | CACCCTGTACCTGGAAACTTGC |
| Hprt1 | NM_012583.2 | CTCATGGACTGATTATGGACAGGAC | GCAGGTCAGCAAAGAACTTATAGCC |
| Postn | NM_001108550.1 | GCTTCAGAAGCCACTTTGTC | CGCCAACTACATCGACAAGG |
| Tcf21 | NM_001032397.1 | CATTCACCCAGTCAACCTGA | CCACTTCCTTTAGGTCACTCTC |
Fluorescence immunocytochemistry
Primary cardiac fibroblasts were seeded at a low confluency onto either glass coverslips in 6-well dishes, or elastic (E = 5 kPa) silicone coverslips (ExCellness) in 35 mm dishes, coated with porcine gelatin type A, and maintained in culture with F10 medium with 2% FBS for 72 hours. The cells were briefly washed in PBS and fixed in 4% paraformaldehyde for 15 minutes at room temperature. After another brief wash in PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS for 15 minutes, and non-specific binding sites were blocked for 1 hour in 5% normal goat serum (Invitrogen) in PBS. The blocking agent was removed by another set of washes before applying primary antibodies diluted in 1% bovine serum albumin (BSA) in PBS. αSMA was probed using a 1:50 dilution (A2547; Sigma) and incubated overnight at 4 °C in a humidified chamber. The following day, the cells were thoroughly washed three times in PBS and incubated with Alexa Fluor 488-conjugated secondary antibody (1:500; A27023; Invitrogen) for 1 hour at room temperature. After a brief wash in PBS, F-actin was stained using a 1:500 dilution of rhodamine-phalloidin (R415; Invitrogen) in PBS. After several washes over a period of 30 minutes, the coverslips were thoroughly dried using gentle suction and mounted on glass slides using Fluoroshield™ mounting medium with DAPI (Abcam) and allowed to cure at room temperature for 24 hours. Cells were imaged using a Zeiss LSM 5 Pascal microscope as described above, using DAPI, FITC and Texas Red detection channels.
Cell viability assay
The viability of cells in each culture condition was assessed after 96 hours in culture. Cells were washed twice with pre-warmed PBS, then treated with 2 μM Calcein-AM (C3100, Thermo Fisher) and 2.5 μM ethidium homodimer (E1169, Thermo Fisher) in PBS for 30 minutes at 37 °C, 5% CO2. The stains were then gently removed by aspiration and replaced with fresh PBS. Cells were imaged immediately, using FITC and Texas Red detection channels.
Data analysis and statistics
Statistical analyses and graphs were generated using Graph Pad Prism 7. All data are presented as the mean ± standard deviation, unless otherwise indicated in figure legends. Individual biological replicates are counted as one experiment involving cells from only one animal. Grouped data analyses were performed using one-way or two-way ANOVA followed by Tukey’s post-hoc test, with significance recorded if P < 0.05.
Supplementary information
Acknowledgements
N.M.L.’s work is funded by studentships from the Canadian Institutes of Health Research (CIHR), the Institute of Cardiovascular Sciences in the Albrechtsen Research Centre and Research Manitoba. I.M.C.D. is grateful to the Heart & Stroke Foundation of Canada and the Canadian Institutes for Health Research for a grant-in-aid, and an operating grant, respectively, to support this research. Operating patronage is also provided by the St. Boniface Hospital and Research Foundation, as well as the St. Boniface Hospital Albrechtsen Research Centre. We are grateful to Dr. Mark Hnatowich for his assistance with qPCR primer design, as well as for his review of the draft manuscript.
Author Contributions
N.M.L. contributed to experimental design and execution, data interpretation, statistical analyses, as well as manuscript and figure preparation. S.G. Rattan contributed to experiment execution and optimization, and statistical analyses. I.M.C. Dixon contributed to experimental design, data interpretation, and manuscript writing. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49285-9.
References
- 1.Pinto AR, et al. Revisiting Cardiac Cellular Composition. Circulation research. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldsmith EC, et al. Organization of fibroblasts in the heart. Developmental dynamics: an official publication of the American Association of Anatomists. 2004;230:787–794. doi: 10.1002/dvdy.20095. [DOI] [PubMed] [Google Scholar]
- 3.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac Fibrosis: The Fibroblast Awakens. Circulation research. 2016;118:1021–1040. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanisicak O, et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nature communications. 2016;7:12260. doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molkentin JD, et al. Fibroblast-Specific Genetic Manipulation of p38 Mitogen-Activated Protein Kinase In Vivo Reveals Its Central Regulatory Role in Fibrosis. Circulation. 2017;136:549–561. doi: 10.1161/CIRCULATIONAHA.116.026238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valiente-Alandi Iñigo, Potter Sarah J., Salvador Ane M., Schafer Allison E., Schips Tobias, Carrillo-Salinas Francisco, Gibson Aaron M., Nieman Michelle L., Perkins Charles, Sargent Michelle A., Huo Jiuzhou, Lorenz John N., DeFalco Tony, Molkentin Jeffery D., Alcaide Pilar, Blaxall Burns C. Inhibiting Fibronectin Attenuates Fibrosis and Improves Cardiac Function in a Model of Heart Failure. Circulation. 2018;138(12):1236–1252. doi: 10.1161/CIRCULATIONAHA.118.034609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng F, et al. Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-beta-Slug signaling. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E4320–4327. doi: 10.1073/pnas.1519197113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circulation research. 2009;105:1164–1176. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lane EB, Hogan BL, Kurkinen M, Garrels JI. Co-expression of vimentin and cytokeratins in parietal endoderm cells of early mouse embryo. Nature. 1983;303:701–704. doi: 10.1038/303701a0. [DOI] [PubMed] [Google Scholar]
- 10.Hudon-David F, Bouzeghrane F, Couture P, Thibault G. Thy-1 expression by cardiac fibroblasts: lack of association with myofibroblast contractile markers. Journal of molecular and cellular cardiology. 2007;42:991–1000. doi: 10.1016/j.yjmcc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Gago-Lopez N, et al. THY-1 receptor expression differentiates cardiosphere-derived cells with divergent cardiogenic differentiation potential. Stem cell reports. 2014;2:576–591. doi: 10.1016/j.stemcr.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang Y, et al. CD90(+) cardiac fibroblasts reduce fibrosis of acute myocardial injury in rats. The international journal of biochemistry & cell biology. 2018;96:20–28. doi: 10.1016/j.biocel.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Darby I, Skalli O, Gabbiani G. Alpha-smooth muscle actin is transiently expressed by myofibroblasts during experimental wound healing. Laboratory investigation; a journal of technical methods and pathology. 1990;63:21–29. [PubMed] [Google Scholar]
- 14.Black FM, et al. The vascular smooth muscle alpha-actin gene is reactivated during cardiac hypertrophy provoked by load. The Journal of clinical investigation. 1991;88:1581–1588. doi: 10.1172/JCI115470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie KO, Taatjes DJ, Schwarz J. vonTurkovich, M. & Low, R. B. Cardiac myofibroblasts express alpha smooth muscle actin during right ventricular pressure overload in the rabbit. The American journal of pathology. 1991;139:207–216. [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nature reviews. Molecular cell biology. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 17.Hinz B, Dugina V, Ballestrem C, Wehrle-Haller B, Chaponnier C. Alpha-smooth muscle actin is crucial for focal adhesion maturation in myofibroblasts. Molecular biology of the cell. 2003;14:2508–2519. doi: 10.1091/mbc.e02-11-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dugina V, Fontao L, Chaponnier C, Vasiliev J, Gabbiani G. Focal adhesion features during myofibroblastic differentiation are controlled by intracellular and extracellular factors. Journal of cell science. 2001;114:3285–3296. doi: 10.1242/jcs.114.18.3285. [DOI] [PubMed] [Google Scholar]
- 19.Thannickal VJ, et al. Myofibroblast differentiation by transforming growth factor-beta1 is dependent on cell adhesion and integrin signaling via focal adhesion kinase. The Journal of biological chemistry. 2003;278:12384–12389. doi: 10.1074/jbc.M208544200. [DOI] [PubMed] [Google Scholar]
- 20.Zhao S, et al. Periostin expression is upregulated and associated with myocardial fibrosis in human failing hearts. Journal of cardiology. 2014;63:373–378. doi: 10.1016/j.jjcc.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Snider P, et al. Origin of cardiac fibroblasts and the role of periostin. Circulation research. 2009;105:934–947. doi: 10.1161/CIRCRESAHA.109.201400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K, Rekhter MD, Gordon D, Phan SH. Myofibroblasts and their role in lung collagen gene expression during pulmonary fibrosis. A combined immunohistochemical and in situ hybridization study. The American journal of pathology. 1994;145:114–125. [PMC free article] [PubMed] [Google Scholar]
- 23.Klingberg F, Hinz B, White ES. The myofibroblast matrix: implications for tissue repair and fibrosis. The Journal of pathology. 2013;229:298–309. doi: 10.1002/path.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serini G, et al. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. The Journal of cell biology. 1998;142:873–881. doi: 10.1083/jcb.142.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klingberg Franco, Chau Grace, Walraven Marielle, Boo Stellar, Koehler Anne, Chow Melissa L., Olsen Abby L., Im Michelle, Lodyga Monika, Wells Rebecca G., White Eric S., Hinz Boris. The fibronectin ED-A domain enhances recruitment of latent TGF-β-binding protein-1 to the fibroblast matrix. Journal of Cell Science. 2018;131(5):jcs201293. doi: 10.1242/jcs.201293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schelbert, E. B. et al. Myocardial Fibrosis Quantified by Extracellular Volume Is Associated With Subsequent Hospitalization for Heart Failure, Death, or Both Across the Spectrum of Ejection Fraction and Heart Failure Stage. Journal of the American Heart Association4, 10.1161/JAHA.115.002613 (2015). [DOI] [PMC free article] [PubMed]
- 27.Ahmad T, et al. Biomarkers of myocardial stress and fibrosis as predictors of mode of death in patients with chronic heart failure. JACC. Heart failure. 2014;2:260–268. doi: 10.1016/j.jchf.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letterio JJ, Roberts AB. Regulation of immune responses by TGF-beta. Annual review of immunology. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 29.Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (TGF)-beta signaling in cardiac remodeling. Journal of molecular and cellular cardiology. 2011;51:600–606. doi: 10.1016/j.yjmcc.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore-Morris T, Guimaraes-Camboa N, Yutzey KE, Puceat M, Evans SM. Cardiac fibroblasts: from development to heart failure. J Mol Med (Berl) 2015;93:823–830. doi: 10.1007/s00109-015-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zymek P, et al. The role of platelet-derived growth factor signaling in healing myocardial infarcts. Journal of the American College of Cardiology. 2006;48:2315–2323. doi: 10.1016/j.jacc.2006.07.060. [DOI] [PubMed] [Google Scholar]
- 32.Bonner JC. Regulation of PDGF and its receptors in fibrotic diseases. Cytokine & growth factor reviews. 2004;15:255–273. doi: 10.1016/j.cytogfr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Ali SR, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res. 2014;115:625–635. doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- 34.Lee AA, Dillmann WH, McCulloch AD, Villarreal FJ. Angiotensin II stimulates the autocrine production of transforming growth factor-beta 1 in adult rat cardiac fibroblasts. Journal of molecular and cellular cardiology. 1995;27:2347–2357. doi: 10.1016/S0022-2828(95)91983-X. [DOI] [PubMed] [Google Scholar]
- 35.Bai J, et al. Metformin inhibits angiotensin II-induced differentiation of cardiac fibroblasts into myofibroblasts. PloS one. 2013;8:e72120. doi: 10.1371/journal.pone.0072120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutchinson KR, Lord CK, West TA, Stewart JA., Jr. Cardiac fibroblast-dependent extracellular matrix accumulation is associated with diastolic stiffness in type 2 diabetes. PloS one. 2013;8:e72080. doi: 10.1371/journal.pone.0072080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh VP, Baker KM, Kumar R. Activation of the intracellular renin-angiotensin system in cardiac fibroblasts by high glucose: role in extracellular matrix production. American journal of physiology. Heart and circulatory physiology. 2008;294:H1675–1684. doi: 10.1152/ajpheart.91493.2007. [DOI] [PubMed] [Google Scholar]
- 38.Xie Z, Singh M, Singh K. ERK1/2 and JNKs, but not p38 kinase, are involved in reactive oxygen species-mediated induction of osteopontin gene expression by angiotensin II and interleukin-1beta in adult rat cardiac fibroblasts. Journal of cellular physiology. 2004;198:399–407. doi: 10.1002/jcp.10419. [DOI] [PubMed] [Google Scholar]
- 39.Sano M, et al. ERK and p38 MAPK, but not NF-kappaB, are critically involved in reactive oxygen species-mediated induction of IL-6 by angiotensin II in cardiac fibroblasts. Circulation research. 2001;89:661–669. doi: 10.1161/hh2001.098873. [DOI] [PubMed] [Google Scholar]
- 40.Sarrazy V, et al. Integrins alphavbeta5 and alphavbeta3 promote latent TGF-beta1 activation by human cardiac fibroblast contraction. Cardiovascular research. 2014;102:407–417. doi: 10.1093/cvr/cvu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wipff PJ, Rifkin DB, Meister JJ, Hinz B. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. The Journal of cell biology. 2007;179:1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goffin JM, et al. Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibers. The Journal of cell biology. 2006;172:259–268. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinz B. Tissue stiffness, latent TGF-beta1 activation, and mechanical signal transduction: implications for the pathogenesis and treatment of fibrosis. Current rheumatology reports. 2009;11:120–126. doi: 10.1007/s11926-009-0017-1. [DOI] [PubMed] [Google Scholar]
- 44.Fu X, et al. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. The Journal of clinical investigation. 2018;128:2127–2143. doi: 10.1172/JCI98215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santiago JJ, et al. Cardiac fibroblast to myofibroblast differentiation in vivo and in vitro: expression of focal adhesion components in neonatal and adult rat ventricular myofibroblasts. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239:1573–1584. doi: 10.1002/dvdy.22280. [DOI] [PubMed] [Google Scholar]
- 46.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophysical journal. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKee CT, Last JA, Russell P, Murphy CJ. Indentation versus tensile measurements of Young’s modulus for soft biological tissues. Tissue engineering. Part B, Reviews. 2011;17:155–164. doi: 10.1089/ten.TEB.2010.0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wells RG. Tissue mechanics and fibrosis. Biochimica et biophysica acta. 2013;1832:884–890. doi: 10.1016/j.bbadis.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Putten S, Shafieyan Y, Hinz B. Mechanical control of cardiac myofibroblasts. J Mol Cell Cardiol. 2016;93:133–142. doi: 10.1016/j.yjmcc.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 50.Hinz B. The myofibroblast: paradigm for a mechanically active cell. Journal of biomechanics. 2010;43:146–155. doi: 10.1016/j.jbiomech.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 51.Hinz B. Masters and servants of the force: the role of matrix adhesions in myofibroblast force perception and transmission. European journal of cell biology. 2006;85:175–181. doi: 10.1016/j.ejcb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 52.Clement S, et al. Expression and function of alpha-smooth muscle actin during embryonic-stem-cell-derived cardiomyocyte differentiation. J Cell Sci. 2007;120:229–238. doi: 10.1242/jcs.03340. [DOI] [PubMed] [Google Scholar]
- 53.Jones C, Ehrlich HP. Fibroblast expression of alpha-smooth muscle actin, alpha2beta1 integrin and alphavbeta3 integrin: influence of surface rigidity. Experimental and molecular pathology. 2011;91:394–399. doi: 10.1016/j.yexmp.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, et al. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- 55.Yu J, et al. Topological Arrangement of Cardiac Fibroblasts Regulates Cellular Plasticity. Circ Res. 2018;123:73–85. doi: 10.1161/CIRCRESAHA.118.312589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunnington RH, et al. Antifibrotic properties of c-Ski and its regulation of cardiac myofibroblast phenotype and contractility. American journal of physiology. Cell physiology. 2011;300:C176–186. doi: 10.1152/ajpcell.00050.2010. [DOI] [PubMed] [Google Scholar]
- 57.Gupta SS, et al. Inhibition of autophagy inhibits the conversion of cardiac fibroblasts to cardiac myofibroblasts. Oncotarget. 2016;7:78516–78531. doi: 10.18632/oncotarget.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagalingam RS, et al. Regulation of cardiac fibroblast MMP2 gene expression by scleraxis. J Mol Cell Cardiol. 2018;120:64–73. doi: 10.1016/j.yjmcc.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 59.Eid H, Chen JH, de Bold AJ. Regulation of alpha-smooth muscle actin expression in adult cardiomyocytes through a tyrosine kinase signal transduction pathway. Annals of the New York Academy of Sciences. 1995;752:192–201. doi: 10.1111/j.1749-6632.1995.tb17422.x. [DOI] [PubMed] [Google Scholar]
- 60.Chen MC, et al. Dedifferentiation of atrial cardiomyocytes in cardiac valve disease: unrelated to atrial fibrillation. Cardiovascular pathology: the official journal of the Society for Cardiovascular Pathology. 2008;17:156–165. doi: 10.1016/j.carpath.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 61.Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol Biol Cell. 2001;12:2730–2741. doi: 10.1091/mbc.12.9.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talele NP, Fradette J, Davies JE, Kapus A, Hinz B. Expression of alpha-Smooth Muscle Actin Determines the Fate of Mesenchymal Stromal Cells. Stem cell reports. 2015;4:1016–1030. doi: 10.1016/j.stemcr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hao J, Wang B, Jones SC, Jassal DS, Dixon IM. Interaction between angiotensin II and Smad proteins in fibroblasts in failing heart and in vitro. American journal of physiology. Heart and circulatory physiology. 2000;279:H3020–3030. doi: 10.1152/ajpheart.2000.279.6.H3020. [DOI] [PubMed] [Google Scholar]
- 64.Ackers-Johnson M, et al. A Simplified, Langendorff-Free Method for Concomitant Isolation of Viable Cardiac Myocytes and Nonmyocytes From the Adult Mouse Heart. Circ Res. 2016;119:909–920. doi: 10.1161/CIRCRESAHA.116.309202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 67.Fowlkes V, et al. Type II diabetes promotes a myofibroblast phenotype in cardiac fibroblasts. Life sciences. 2013;92:669–676. doi: 10.1016/j.lfs.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.