Abstract
We report the synthesis and applications of a novel N-doped graphene quantum dots (GQDs) using hydrothermal reaction between citric acid and p-aminophenol. The synthesized N-doped GQDs have been characterized physico-chemically and evaluated its antioxidant, antimicrobial, DNA binding and cleavage activities. siRNA loading studies were performed and their effects on cells were evaluated. Obtained results indicate that monodisperse solution of N-doped GQDs has been obtained with particles size ca. ∼10.9 ± 1.3 nm. UV–Vis spectroscopy studies of the interactions between the N-doped GQDs and calf thymus DNA (CT-DNA) showed that the compound interact with CT-DNA via both intercalative and electrostatic binding. The DNA cleavage study showed that the N-doped GQDs cleaved DNA without any external agents. The antioxidant activity of N-doped GQDS was very active when compared to BHT. As the concentration of the compound increased, the antioxidant activity also increased. Cell viability assay demonstrated that the Ndoped GQDs showed cell viability (70%) when the concentration reached 200 μg/mL for A549 and also MDA-MB-231, 150 μg/mL for NIH-3T3 cell lines at 24 h incubation. N-doped GQDs were coated with Eudragit RS 100 and EphA2-siRNA was loaded. As a result of the studies on these formulations, it was concluded that there may be significant effects on A549 cells. The microscopy results revealed that N-doped GQDs was quickly internalized into the cell. Our novel N-doped-GQDs with siRNA are candidate for in situ tumor suppression via DNA and mRNA breakage.
Keywords: N-doped Graphene quantum dots, DNA binding, DNA cleavage, DNA damage, Cell viability, siRNA uptake
1. Introduction
Quantum dots (QDs), which are light-emitting nanoparticles, have been applied in in vitro and in vivo transfection experiments. QDs are known to be spherical semiconductor particles that have a core shell structure. The semiconductor structure of these particles and their fluorescence properties are dependent on their size, and they are important tools for diagnosing disease (Biju et al., 2010, Şenel and Büyükköroğlu, 2018).
Graphene quantum dots (GQDs) are a novel, zero-dimensional nanomaterial made from graphene. GQDs are small graphene fragments, with particle sizes in the range of 3–20 nm, and favourable surface grafting that involves π–π conjugations (Benítez-Martínez and Valcárcel, 2015, Kuo et al., 2016). GQDs have been widely studied in recent years due to their optoelectronic properties and their potential applications (Zhu et al., 2015, Lai, 2017, Safardoust-Hojaghan et al., 2017). Recently, they have been utilized for bioimaging and biosensing (Giovannelli et al., 2002, Sun et al., 2013, Lai, 2017) due to their desirable properties for biomedical applications, such as photoluminescence, a large surface area, good water solubility, good biocompatibility and low cytotoxicity (Nurunnabi et al., 2013, Zhou et al., 2016).
The rapid development of GQDs has also shown that these nanomaterials may have antioxidant (Zheng et al., 2015), antimicrobial (Szunerits and Boukherroub, 2016) and cytotoxic effects (Lalwani et al., 2016); therefore, they could be used in areas other than biosensing and bioimaging. For example, the chemical composition of some GQDs in relation to their antioxidant and free radical scavenging activities has been investigated and compared (Ruiz et al., 2017, Wang et al., 2017). The inhibitory effective concentration of the best performing GQDs was lower than that of ascorbic acid and other carbon nanomaterials (Ruiz et al., 2017). To the best of our knowledge, there have been very few antioxidant studies on N-doped GQDS and GQDs.
Due to their nanoscale size and biocompatibility, GQDs can be used as effective carriers for drug delivery (De Jong and Borm, 2008). They may also be useful in chemotherapeutics for cancer treatment and stem cell differentiation and imaging (Lee et al., 2011). Additionally, several studies have shown that some GQDs are biocompatible with A549 or HeLa cells up to 200 µg/mL (Yuan et al., 2014, Jiang et al., 2015).
The conjugation of graphene quantum dot nanoparticles into synthetic (Kuo et al., 2016) and natural (Nigam et al., 2014) polymer molecules has recently attracted considerable research attention. To investigate this concept, we used a hydrothermal method to synthesize N-doped GQDs from citric acid and p-aminophenol. The N-doped GQDs were purified by column chromatography and characterised. The antimicrobial and antioxidant activities and the DNA interactions of the N-doped GQDs were screened. Furthermore, the potential of the GQDs to incorporate into Eudragit RS 100 polymeric nanoparticles for bioimaging and targeted drug delivery was explored.
2. Materials
Citric acid (CA), 4-aminophenol (4-APhOH), 2,2-diphenyl-1-picrylhydrazyl (DPPH), butylated hydroxytoluene (BHT), silica gel 60 (0.063–0.200 mm), ethidium bromide (EB), calf thymus DNA (CT-DNA) and pBR322 DNA were obtained from commercial sources and used without further purification. Spectrophotometry-grade dimethyl sulfoxide (DMSO), Dulbecco's modified Eagle’s medium (DMEM), RPMI-1640 medium and MTT dye were purchased from Sigma–Aldrich (Germany). Eudragit® RS100 was purchased from Evonik Industries (Germany). siRNA EphA2 was purchased from Santa Cruz Biotechnology (Germany).
3. Methods
3.1. Synthesis of N-doped GQDs
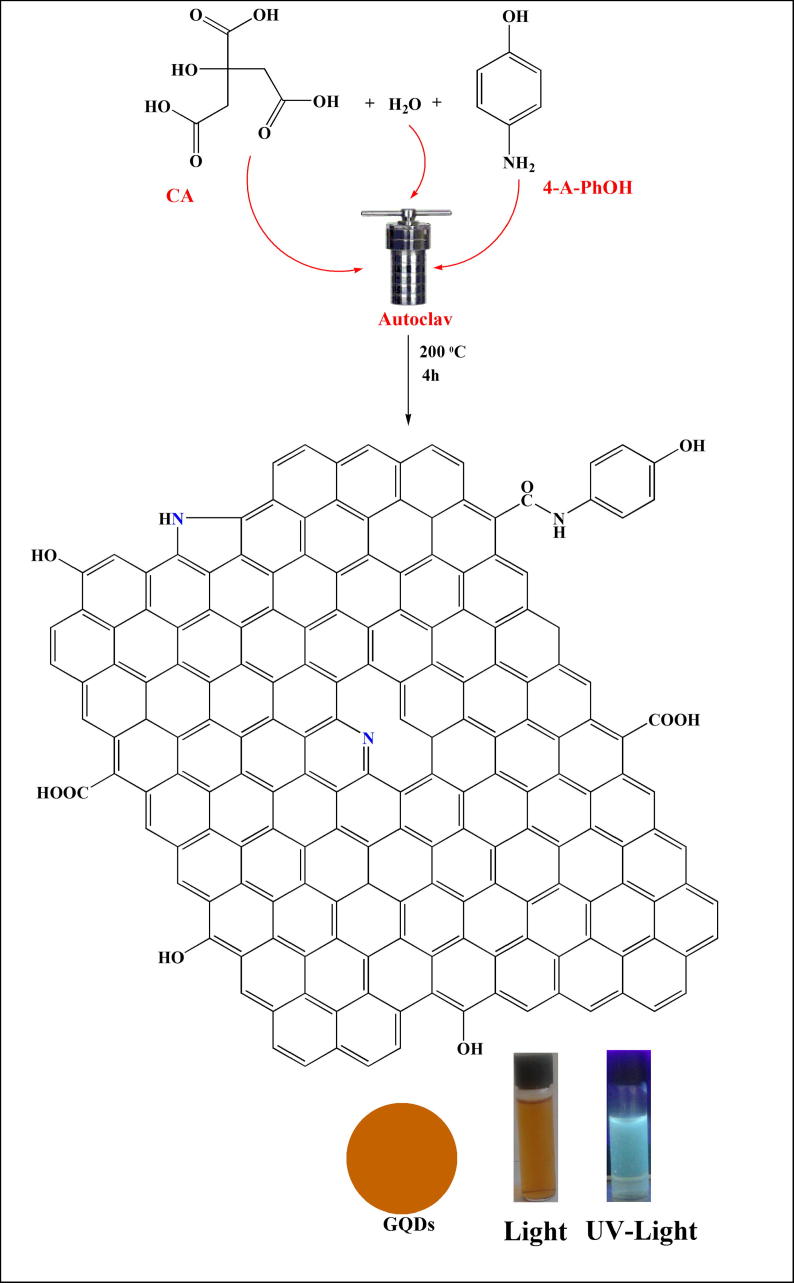
Citric acid (1.20 g, 6.25 mmol) and p-aminophenol (0.68 g, 6.25 mmol) were dissolved in 50 mL of deionized distilled water incubated in an autoclave at 200 °C for 4 h. The mixture was then evaporated and purified by column chromatography (silica gel 60 g, eluent; etanol-CHCl3, 2:3) to separate the N-doped GQDs. The N-doped GQDs was obtained from the supernatant and were collected upon evaporation of the solution.
3.2. Characterisation studies
The characterization studies investigated the following: Fourier transform infrared spectroscopy (FTIR) absorption spectra using a Perkin Elmer BX II spectrometer, UV–Vis (ultraviolet–visible) spectra using a PG Instruments T + 80 UV/Vis spectrophotometer, TEM-EDX analysis using a JEOL TEM-1400-EDX instrument and particle size using dynamic light scattering (DLS).
3.3. Screening for antimicrobial activity
The microorganisms listed in Table 1 were used to assess antimicrobial activity. Ampicillin and fluconazole are well-known broad-spectrum antibiotics that have different mechanisms of activity and were used as controls (Li et al., 2012). The minimum inhibitory concentration was determined via the broth microdilution test (CLSI, 2015). Briefly, the N-doped GQDs were dissolved in a minimal volume of H2O and serially diluted (1–500 µg/µL) in Mueller-Hinton broth. The plates were inoculated with the N-doped GQDs and incubated at 37 °C. The test results were obtained for bacteria and fungi after 24 and 48 h, respectively.
Table 1.
Antimicrobial activities of N-doped GQDs (µg/mL).
| Compound | Antibiotic |
||
|---|---|---|---|
| Microorganism | N-doped GQDs | Ampicillin | Fluconasol |
| S. aureus ATCC 25923 | 16 | 1 | – |
| E. faecalis ATCC 29212 | 8 | 2 | – |
| B. cereus NRRL B-3711 | 64 | 2 | – |
| B. subtilis ATCC 6633 | 64 | 1 | – |
| E. coli ATCC 25922 | 64 | 2 | – |
| E. coli ATCC 35218 | 64 | 2 | – |
| P. aeruginosa ATCC 27853 | 125 | 2 | – |
| P. vulgaris ATCC 13315 | 64 | 2 | – |
| C. albicans ATCC 60193 | 8 | – | 1 |
| C. tropicalis ATCC 13803 | 16 | – | 1 |
3.4. DNA binding and cleavage experiments
UV–Vis spectra titrations were carried out to investigate the binding affinity between CT-DNA and the N-doped GQDs. These analyses were performed according to Zeyrek et al. (2018). Briefly, 3 mL of Tris–HCl/NaCl buffer (5 µM Tris–HCl, 50 µM NaCl, pH 7.2) and the solutions of N-doped GQDs of buffered CT-DNA were added to each cuvette together and incubated at room temperature for 5 min. The UV–Vis absorbance of CT-DNA solution in Tris buffer was recorded at 260 and 280 nm wavelengths. An absorbance value of 1.8–1.9 indicates that the DNA is sufficiently free of protein (Marmur, 1961).
In cleavage experiments using H2O2 as oxidizing agent, pBR322 was incubated with N-doped GQDs for 3 h at 37 °C. The samples were then loaded onto a 1% agarose gel in TAE buffer (Tris-Acetate-EDTA, pH 8.3), and electrophoresis was applied for 1 h at 60 V. The bands were observed and imaged under UV light.
3.5. Antioxidant activity (DPPH: UV–Vis-based assay)
The DPPH-based assay is a popular method for analysing the antioxidant activity because of it is simple and sensitive (Kedare and Singh, 2011, Sachdev and Gopinath, 2015). Butylated hydroxytoluene (BHT) is a lipophilic organic compound most commonly used as an antioxidant in food safety (Yehye et al., 2015). Briefly, a 20 µg/mL ethanol solution was first prepared for each measurement of DPPH. N-doped GQDs with different concentrations (10, 20, 40, 60 and 80 µg/mL) were then added into the DPPH solutions and allowed to incubate in the dark for 1 h. After the incubation, the absorbance change of each sample was monitored at 517 nm and used to calculate the antioxidant activity of the N-doped GQDs against DPPH (Garcia et al., 2012).
3.6. Cell culture studies
Mouse fibroblast cells (NIH-3T3), human lung carcinoma cells (A549) and human breast adenocarcinoma cells (MDA-MB-231) were used to evaluate the cell viability when exposed to N-doped GQDs. RPMI-1640 and DMEM media were supplemented with 10% FBS and 1% antibiotics (penicillin/streptomycin). The NIH-3T3 cells were incubated in RPMI-1640 medium, and the A549 and MDA-MB-231 cell lines were incubated in DMEM, at 37 °C for 24 h in a humidified atmosphere containing 5% CO2 (Şenel et al., 2015).
3.6.1. Cell viability analysis
The thiazoyl blue colorimetric (MTT) assay was used to determine cell viability. Briefly, 2 × 104 cells were seeded into a 96-well plate (Greiner CellStar, Sigma-Aldrich, Germany) and incubated for 24 h. When the desired cell density was reached, N-doped GQDs were applied to the cells and incubated for 24 and 48 h at concentrations of 1, 5, 10, 15, 20 and 50 µg/well. After incubation, the culture medium was aspirated, and 20 µL of MTT dye (5 mg/mL in PBS) was added. At the end of an additional incubation period of 4 h, 200 µL of DMSO was added to each well, and the absorbance was measured at 570 nm after 30 min using a multi-mode microplate reader (Cytation 5, BioTek Instruments, Germany). All concentrations were investigated in 8 wells, and all analyses were repeated in triplicate. Cells that were exposed only to the culture medium were used as a control. The results were calculated as the percentage of inhibition compared to untreated control cells, where cell viability was presumed to be 100% (Gencer et al., 2010, Şenel et al., 2015).
3.6.2. Determination of genotoxicity by comet assay
NIH-3T3, A549 and MDA-MB-231 cells were treated with N-doped GQDs as described in the MTT assay. Before the trypsinisation of the cells with trypsin/EDTA (0.25%), the cells were assessed with a confocal microscope to verify whether the QDs had passed into the cells. The treated cells were then harvested from the flask, and the comet assay was applied. Briefly, 80 µL of removed cells (2 × 105) were resuspended in low-melting agarose (1% in Milli-Q distilled water) and placed on precoated normal-melting agarose (1%). The slides were maintained at 4 °C for 10 min. After incubation, the slides were placed in lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris-base, 1% v/v Triton-X, 1% sodium N-lauroyl sarcosinate, pH 10) for 1 h at 4 °C. After lysis, the slides were washed with electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13) and placed in a horizontal electrophoresis tank filled with electrophoresis buffer for 30 min at 25 V and 300 mA. After the electrophoretic run, the slides were placed in a neutralizing buffer (1 M Tris, pH 7.5) 3 times for 3 min, followed by incubation in 98% alcohol for 5 min. The dried microscopic slides were stained with 20 µL of EtBr (20 µg/mL). One hundred randomly selected cells from each tested concentration were analysed under a fluorescence microscope (Leica DM1000) at 400× magnification and scored using Open Comet v1.3 open access software. The degree of DNA damage in the cells was presented as the percentage of tail DNA. To visually determine the type of comets, they are categorized according to the DNA tail intensity in 5 different classes: class 0 (no tail or 0–6%), class 1 (6–17%), class 2 (17–35%), class 3 (35–60%) and class 4 (from 60% to nearly all DNA in the tail). The positive control was 20 µL of H2O2. The experiment was performed in triplicate (Giovannelli et al., 2002, Kim et al., 2016, Mitkovska et al., 2017).
3.7. Preparation of GQDs/Eudragit formulations (Eu-GQDs)
To achieve the formulation of Eu-GQDs, Eudragit-coated GQDs were prepared by a simple solvent evaporation method. First, 50 mg of Eudragit RS 100 (Eu) was dissolved in 5 mL of ethanol by mixing on magnetic stirrer at 500 rpm for 1 h. The 5 mL GQD (4 mg/mL) sample was then added dropwise into 5 mL of Eu solution and stirred at 500 rpm for 1 h. After mixing, the dispersions were evaporated to remove the ethanol with a Rotavapor (Büchi, Switzerland), forming the Eu-GQDs. The formed dispersion was centrifuged at 10,000 rpm for 20 min to remove non-encapsulated GQDs. The pellet was resuspended in distilled water from a Milli-Q® ultrapure system.
3.7.1. Eu-GQDs characterisation studies
For the characterization analysis, the particle size, particle size distribution and zeta potential were characterized by a Zetasizer NanoZS (Malvern Instruments, UK). The measurements were made by taking the average of three repeated measurements.
3.7.2. Binding studies with EpHA2 siRNA of Eu-GQDs
To prepare the siRNA-loaded formulation, EphA2 siRNA was mixed with Eu-GQD dispersions at ratios of 0.5/1, 1/1, 2/1 and 3/1 (N/P). The dispersions were then incubated at 37 °C for 20 min to form electrostatic interactions between the negatively charged siRNA and positively charged Eudragit particles. To determine the siRNA binding ratio of the formulations and to evaluate possible siRNA fragmentation, gel retardation analysis was performed using 1.5% agarose gels for 2 h at 50 V. The resulting images were examined with a gel imaging system (Uvitec Alliance 4.7, Cambridge, UK) (Şenel et al., 2015).
3.7.3. Transfection studies
To analyse the transfection of the formulations, transfection assays were performed that were similar to those previously carried out in our laboratory (Şenel et al., 2015). FITC-labelled siRNA was used as genetic material, and images were evaluated after 24 h.
3.7.4. Determination of genotoxicity and cytotoxicity of Eu-GQDs
In this experiment, Eu-GQDs and siRNA-loaded Eu-GQDSs were evaluated for their cytotoxicity and genotoxicity in A549 cells. These cells were selected for study because EpHA2 protein is highly expressed in them. The genotoxicity and cytotoxicity studies were performed as described in the Determination of genotoxicity by comet assay and Cell viability assay sections.
3.8. Statistical analyses
The MTT and comet assay data are presented as the mean ± standard deviation on the graphic. The differences between the treated groups and the control groups were analysed from the concentration-effect-time curves for MTT and from the tail intensity or foci intensity for the comet assay using linear regression analysis, with Minitab 18, and means of analysis of variance and Student’s t-test, respectively. A p-value < 0.05 was considered to be statistically significant.
4. Results and discussion
We used a hydrothermal method to synthesize the N-doped GQDs from citric acid and p-aminophenol (Fig. 1). The N-doped GQDs were purified by column chromatography and characterized with regard to morphology, elemental content and optical properties.
Fig. 1.
Synthesis of N-doped GQDs.
4.1. UV–Vis, FTIR and TGA (Thermogravimetric analysis) studies
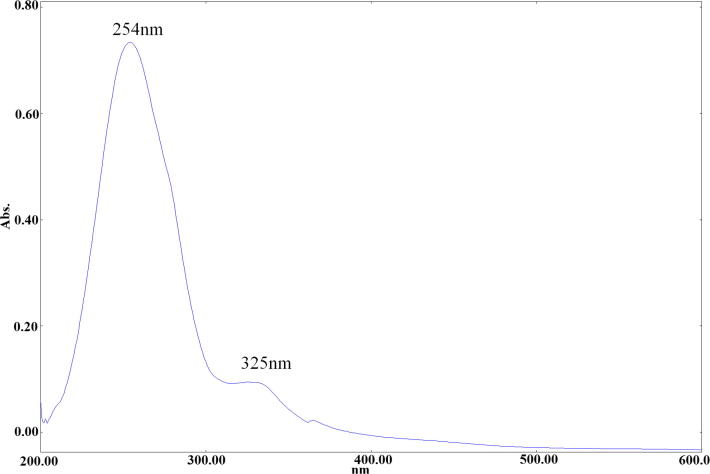
The UV–Vis spectrum of the N-doped GQDs was studied in H2O. The N-doped GQDs showed two bands, at 269.12 and 310.92 nm. They were assigned to the π–π* and n–π* transition of C C, C—N and C—O (Fig. 2).
Fig. 2.
The UV–Vis spectrum of N-doped GQDs.
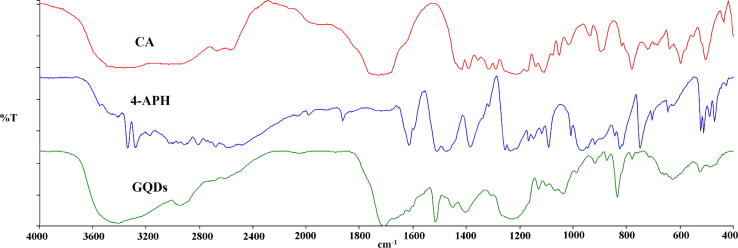
The FTIR spectra of the N-doped GQDs exhibited characteristic changes in the functional group frequencies when compared with the spectrum of the starting materials 4-aminophenol (4-APhOH) and citric acid (CA) (Fig. 3). The OH, NH2, COOH, C—H, C O, C C, C—N and C—O vibration bands were observed at 3416, 3339–3283, 3373, 2952, 1731, 1509, 1615–1475 and 1386 cm−1, respectively, in the starting materials. The OH, NH, COOH, C—H, C O, C C, C—N and C—O vibration band were observed at 3582, 3415, 3226, 2931, 1706, 1518, 1614–1445 and 1345 cm−1, respectively, in the N-doped GQDs (Fig. 3). The frequency of the OH and NH bonds shifted to higher values in the N-doped GQDs, while the COOH, C O and C—N bonds shifted to lower values in the N-doped GQDs.
Fig. 3.
The FTIR spectrum of N-doped GQDs.
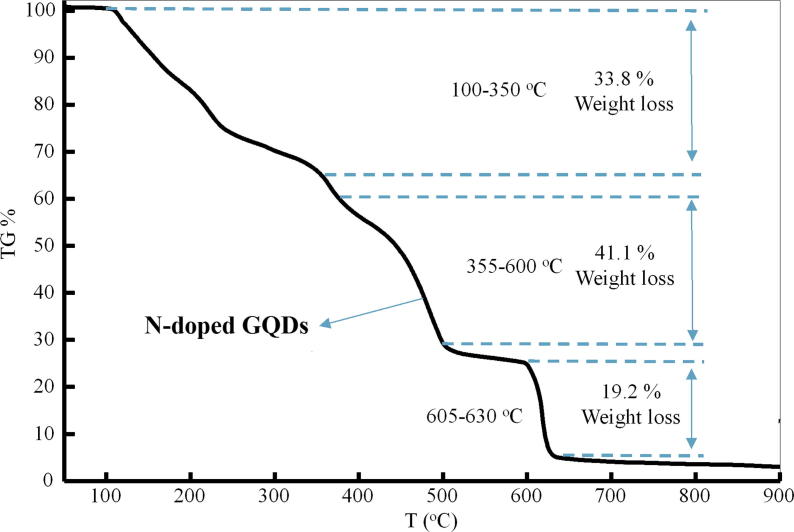
Thermal degradation of N-doped GQDs is shown in Fig. 4. In this illustration, the degradation temperature and total weight loss are given. The N-doped GQDs decomposed in three steps. The first step in the decomposition occurred at 100–350 °C, incurring a 33.80% weight loss, which could have been due to the removal of hydroxyl, carboxyl and amide groups and solvent/water. The second decomposition step occurred at 355–600 °C, which could have been due to the elimination of phenyl groups, with a total weight loss of 41.10%. The final decomposition step occurred at 600–635 °C, likely due to the elimination of pyridine and pyrolidine groups, with a total weight loss of 19.20%. A plateau was obtained when heated above 650 °C. No degradation was observed after 650 °C (Fig. 4).
Fig. 4.
The TGA curve of N-doped GQDs.
4.2. TEM, EDX and DLS studies
The structure and morphology of the N-doped GQDs were further confirmed by transmission electron microscopy (Fig. 5). The sample solution was treated in an ultrasonic bath before deposition to decrease aggregation. The TEM analysis confirmed the presence of the spherical shape morphology of the N-doped GQDs nanoparticle, with particle sizes of approximately 7–11 nm (Fig. 5).
Fig. 5.
TEM images of N-doped GQDs.
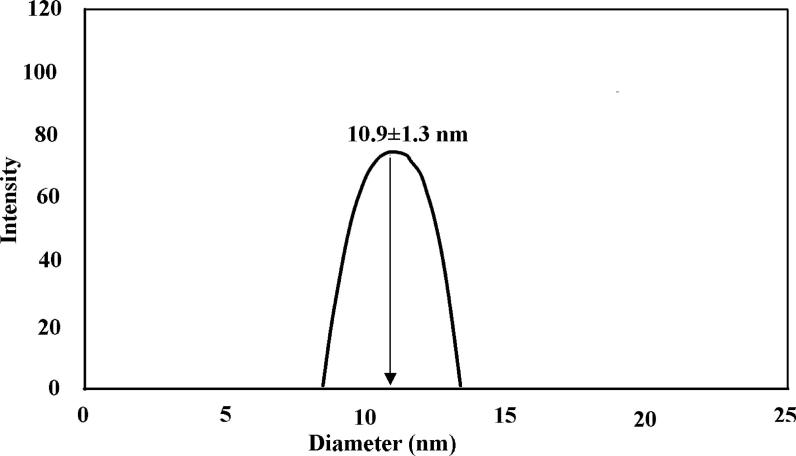
The DLS results are presented as the average values of ten consecutive measurements with an integration time of 2 s. Each measurement was repeated at least three times. In brief, the prepared N-doped GQDs were suspended in DI water, and the measurements were performed using a DLS analyser. The particle size of the N-doped GQDs was 10.9 ± 1.3 nm (Fig. 6), which was supported by the TEM analysis. In a non-clumping suspension, the particle size measured by DLS will be similar or slightly larger than that found with TEM. The determined dimensions indicate that the emission characteristics of the GQDs are the result of the quantum size effect in the quantum-sized nanoparticles and/or the recombination of holes and electrons. The DLS results showed that the particle size was compatible with TEM.
Fig. 6.
DLS analysis of N-doped GQDs.
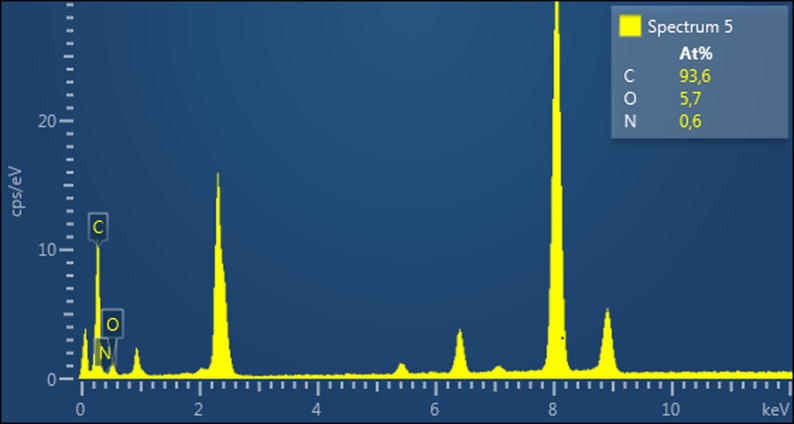
The presence of elements in the analysed structures was also confirmed by the application of EDX measurements. The amount of each element detected by EDX is shown in Fig. 7. Carbon, nitrogen and oxygen were detected on the surface of the nanoparticles, and the presence of nitrogen on the surface was noteworthy. In EDX, only those elements on the surface can be detected. Therefore, this method cannot provide a complete elemental analysis. Nevertheless, the EDX data confirmed the presence of nitrogen in the N-doped GQDs, and accordingly, N-doped GQDs can be said to have formed.
Fig. 7.
EDX spectrum of N-doped GQDs.
4.3. Antimicrobial activity of N-doped GQDs
The minimal inhibitory concentrations (MICs) of the N-doped GQDs are presented in Table 1. These data are the average of three broth microdilution tests. The antimicrobial activity spectrum of the N-doped GQDs varied greatly. The N-doped GQDs showed a strong antibacterial and antifungal effect against E. faecalis ATCC 29212 and C. albicans ATCC 60193, while it had a weak effect against P. aeruginosa ATCC 27853. They had a stronger antibacterial effect against S. aureus ATCC 25923 compared to B. cereus NRRL B-3711, B. subtilis ATCC 6633, E. coli ATCC 25922, E. coli ATCC 35218 and P. vulgaris ATCC 13315.
The N-doped GQDs were shown to have different effects on different microorganisms. It is thought that the cell walls of the tested organisms, i.e., single-layered in Gram (+) bacteria, multilayered in Gram (−) bacteria, and more complex in yeast cells, leads to these differences in activity. In the study, N-doped GQDs were compared with ampicillin and fluconazole standards and showed low activity against the tested organisms compared to the reference drugs. The low lipophilicity of N-doped GQDs decreases their effectiveness by reducing their ability to penetrate the lipid membrane, making them unable to inhibit and block the growth of the microorganism. In a study by Ristic et al. (2014), photodynamic antibacterial activities of electrochemically produced GQDs were evaluated against methicillin-resistant Staphylococcus aureus (MRSA) and Escherichia coli. Similar to our results, neither the GQDs nor light exposure alone had an impact on bacterial viability. The authors stated that the GQDs were not more effective than the currently available antibiotics, but would be potentially valuable when antibiotic resistance is encountered (Ristic et al., 2014).
While there are many detailed studies on the antibacterial activity of GO (graphene oxide) (Palmieri et al., 2017, Papi et al., 2016), there are insufficient studies on N-doped GQDs. The impact of GO on the cell wall of bacteria has been reported. Separate from the inner membrane, Gram-negative bacteria have a fine peptidoglycan layer, with an outer membrane, while Gram-positive bacteria lack an outer membrane but have a thicker peptidoglycan layer. The lack of an external membrane makes Gram-positive bacteria more susceptible to penetration by many antibiotics and GO. The thickness of the peptidoglycan layer was shown to play a role in the low MIC for Gram-negative E. coli and S. typhimurium compared to Gram-positive E. faecalis and B. subtilis. It has been shown that GO has no effect against the fungi C. albicans and C. tropicalis. However, GO was found to be effective against two fungal phytopathogens, F. graminearum and F. oxysporum (Palmieri et al., 2017).
Some studies have found Gram-positive S. aureus to be more sensitive to GO due to the lack of an outer membrane, external to the peptidoglycan layer, which was a protective for Gram-negative E. coli. In addition, the effect of GO was less apparent in the fungus C. albicans, with a thicker and more complex cell wall structure compared to bacteria (Papi et al., 2016). In our study, the N-doped GQDs were quite effective against C. albicans and C. tropicalis, and they showed similar effects against the bacteria E. faecalis and S. aureus.
The environmental activities of GO in different solutions (ultra-pure water, PBS, NaCl, MgCl2 and CaCl2) have been tested against bacteria (Palmieri et al., 2017). It has been observed that surface properties are important factors and affect surface bacterial colonization and can improve adhesion. In the same study, the effect of GO on the viability of two model microorganisms, Gram-negative E. coli and Gram-positive S. aureus, was analysed. According to this study, in water, as the GO concentration increases, the GO killing efficacy increases, while in other solutions, GO does not impact microbial growth because of aggregation that shields the GO edges. However, when the GO cluster size becomes much larger than the bacterial diameter, GO aggregates wrap around the bacteria and impede their growth (Palmieri et al., 2017). In our study, no aggregation was observed in the studied concentration range (8–125 µg/µL).
4.4. Interaction of N-doped GQDs with DNA
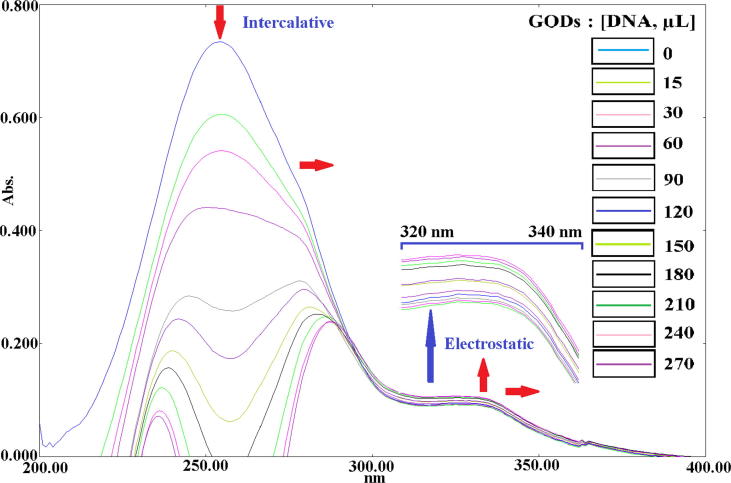
The potential CT-DNA binding ability of the N-doped GQDs was characterized by UV–Vis spectroscopy in the presence and absence of different CT-DNA concentrations (Fig. 8). UV–Vis spectroscopy is one of the methods used to investigate the effect of a material on DNA. If a hypochromic effect is observed in the spectrum, this can indicate intercalation with the DNA (Yıldız et al., 2015, Ünver et al., 2016). However, if the interaction of the material with the DNA is electrostatically or partially intercalative, a hyper-chromic effect is observed in the spectrum. In addition, a red and/or blue shift of the maximum absorption indicates that the difference between the HOMO and LUMO energy levels is reduced, and that the material interacts with the DNA (Yıldız et al., 2015). In the UV–Vis spectrum, two intense bands were observed (at 270 and 310 nm) for N-doped GQDs. When the concentration of the CT-DNA increased, hyperchromism and higher wavelengths of 1–10 nm were observed at 310 nm, while hypochromism of 1.0–96.0% and a red shift of 1 nm were observed at 270 nm. Moreover, when the maximum absorption shifted to red, the reduced energy between HOMO and LUMO indicated that the DNA interacted with the GQDs through both intercalation and electrostatic binding.
Fig. 8.
Absorption spectra of the N-doped GQDs in the presence and absence of increasing amounts of CT-DNA at room temperature in Tris–HCl/NaCl buffer (pH 7.2).
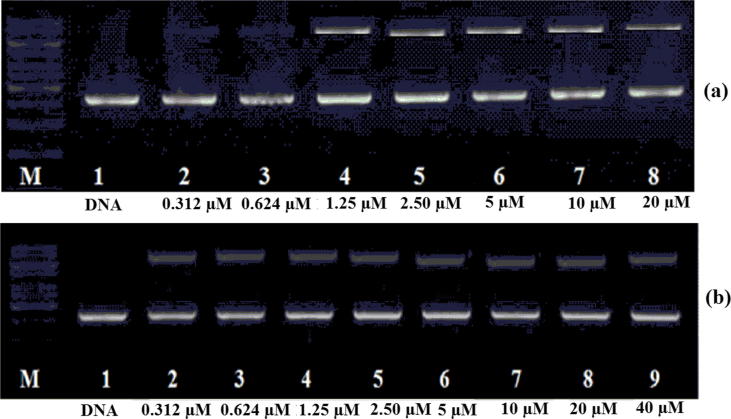
The cleavage activity assays were assessed by agarose gel electrophoresis using super helix (SC) plasmid pBR322 DNA in Tris-acetic acid-EDTA. When the DNA was incubated with increasing concentrations of N-doped GQDs (from 0.312 to 40 μM), super helix DNA (Form I) was degraded to an open circular form (Form II). The catalytic activity of N-doped GQDs is depicted in Fig. 9 (A: hydrolytic B: oxidative). According to the test results, N-doped GQDs showed DNA oxidative reaction at 0.312 μM and DNA hydrolytic reaction at 1.25 μM.
Fig. 9.
Agarose gel electrophoresis patterns for the hydrolytic cleavage of pBR322 DNA by N-doped GQDs (a) and oxidative cleavage of pBR322 DNA by N-doped GQDs (b).
4.5. DPPH assay
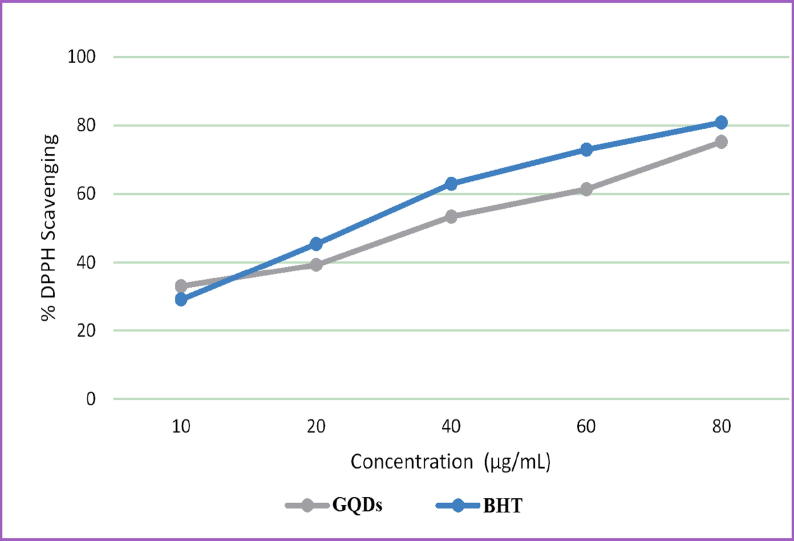
The antioxidant activity of the N-doped GQDs was assessed by the DPPH assay. UV–Vis spectroscopy of DPPH was used to evaluate the antioxidant activity of the N-doped GQDs. According to the average of three experiments, N-doped GQDs concentrations of 10, 20, 40, 60, and 80 µg/mL showed 33%, 39.0%, 53.0%, 61.4%, and 75% antioxidant activities (Fig. 10), respectively. As the concentration of the N-doped GQDs increased from 10 to 80 µg/mL, the antioxidant activity also increased. The concentration dependence of N-doped GQDs is consistent with that of graphene quantum dots and carbon nanodots (Chong et al., 2016, Zhang et al., 2017). The results of this study are noteworthy because there are few antioxidant studies on N-doped GQDs. The activity of the N-doped GQDS was high when compared to that of BHT.
Fig. 10.
DPPH free radical scavenging assay of N-doped GQDs.
4.6. Cell viability of N-doped GQDs
Cell-based assays are one of the important components of drug development studies. The synthesised N-doped GQDs were used to assess their direct cytotoxic effects, leading to cell viability, cell proliferation or cell death. Assessing cell viability is essential for daily cell manipulation and for subsequent processing and analysis. The characterization of chemicals that promote or inhibit cell proliferation is an important area of cell biology and drug discovery research (Eastman, 2017).
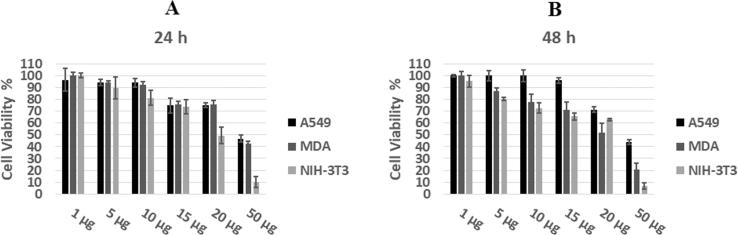
In this study, MTT assays were performed to assess the in vitro cell viability of N-doped GQDs against MDA-MB-231, NIH-3T3 and A549 cell lines. The cell growth inhibitory effect of the N-doped GQDs on each cell line was evaluated by calculating the IC50 from the cell growth curves; the results are presented in Fig. 11A, B and Table 2.
Fig. 11.
Cell viability results of N-doped GQDs after 24 (A) and 48 h (B) of incubation.
Table 2.
IC50 values of N-doped GQDs in the tested cell lines.
| IC50 Values µg/mL |
||
|---|---|---|
| 24 h | 48 h | |
| A549 | 385 ± 0.5 | 355 ± 1.8 |
| MDA-MB-231 | 425 ± 1.2 | 230 ± 0.3 |
| NIH-3T3 | 235 ± 0.7 | 315 ± 2.2 |
±SE: Standart Error.
The effect of the N-doped GQDs on the cell proliferation of all cell lines showed a concentration-dependent decrease. When compared to the control group (Fig. 11), the cell viability was found to be as high (70%) at 200 µg/mL for A549 and MDA, and 120 µg/mL for NIH-3T3, after a 24 h incubation time. After 48 h, a slight decrease in these values (more than 70% of live cells) was observed, resulting in 195 µg/mL for A549, 150 µg/mL for MDA and 100 µg/mL for NIH-3T3. The influence of N-doped GQDs on MDA-231 and NIH 3T3 cell proliferation was statistically significant (p < 0.05). No statistically significant effect of incubation time on cell viability was observed (p > 0.05). It is thought that the high A549 cell viability seen at low concentrations (i.e., 10–150 µg/mL) after 48 h may be due to antioxidant effects. Similarly, Qu et al. (2015) showed no cytotoxic effect against A549 cells up to 500 mg after 24 h. The cell proliferation showed statistical effects above concentrations of 150 μg/mL (p < 0.05).
GQDs are good candidates for molecular biology and biotechnological applications because of their excellent physicochemical properties. Due to the cytoplasm-nucleus shuttle system, graphene-containing nanomaterials might be used as carriers for drugs and genes (Zhang et al., 2016). Synthesized GQDs can enter the cell and pass through to the nucleus, thus interacting with both the proteins in the cell and the genetic material. It has been reported that these interactions can disrupt the cytoplasm and nucleus morphology and change gene expression to decrease cell viability (Jiang et al., 2015). Wang et al. (2013) reported that DOX (doxorubicin)-GQD conjugates were efficiently delivered to the nucleus to increase DOX-induced cytotoxicity in drug-resistant cancer cells. A major side effect seen in the study by Wang et al. was that normal cells were also affected by this toxicity. Therefore, it was hypothesised that GQDs could be put into a carrier system and conjugated with cancer cell-specific antibodies. In the Wang study, N-doped GQDs were loaded into a carrier system; the antibody binding process will be investigated in a future study.
4.7. Determination of genotoxicity by comet assay
GQD-based systems are an alternative route for nuclear cell staining because of their good distribution within the cell and nucleus. In our study, the N-doped GQDs were internalized in both the cytoplasm and the nucleus as a result of their small particle size. Our results were consistent with previous reports of GQDs (Kumawat et al., 2017).
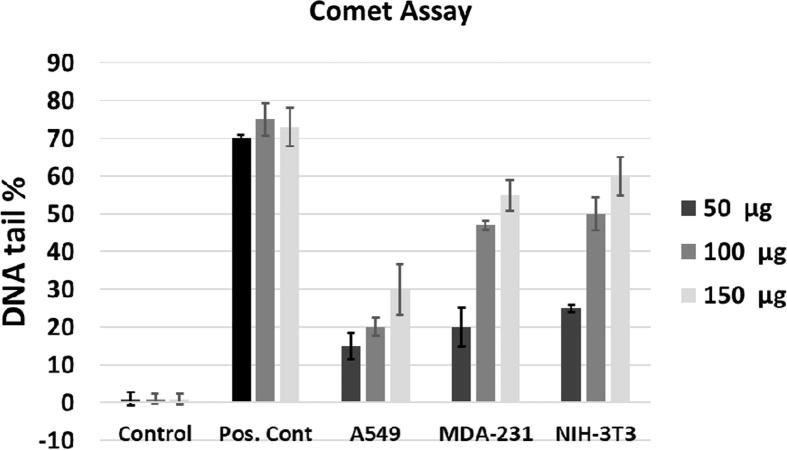
The MTT assay measures the metabolic activity of cells. Low viability in the MTT assay might be due to anti-proliferative, apoptotic or necrotic effects of the studied nanomaterials. A cytotoxic substance does not necessarily have to affect the genome, but it may be harmful to other parts of the cell. Therefore, genotoxicity is also an important parameter to test when studying the biological effects of nanomaterials. According to international recommendations, doses of 50, 100 and 150 µg/mL with cell viabilities above 70% were chosen for the comet assay (Kim et al., 2016). In the comet assay, undamaged DNA does not lose its integrity and does not form a comet, whereas damaged DNA forms a tail-shaped image due to its fragmentation caused by different molecular weights and electrical charges, which induces movement at differing speeds in an electrical field. Scoring is performed by measuring the percentage of DNA in the head and tail with various computer-based microscopy systems (Mitkovska et al., 2017). The results shown in Fig. 12 demonstrate the DNA damage induced at the applied doses.
Fig. 12.
DNA damage induced after treatment with N-doped GQDs and H2O2 (positive control) for A549, MDA-231 and 3T3 cell lines.
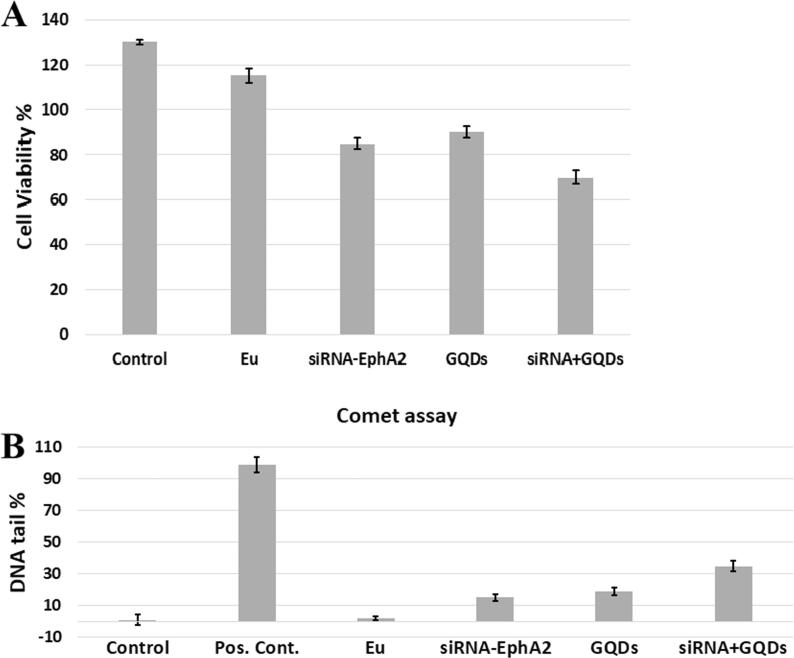
The percent of DNA in the tail showed statistically significant genotoxic damage at the 100 and 150 µg/mL doses compared to controls p < 0.005 (Fig. 12). A visual classification of the comets showed that class 1 and 2 comets predominated in the A549 cells, while class 2, 3 and 4 comets predominated in the MDA-231 and NIH-3T3 cells. Class 4 comets were particularly observed at the 150 mg doses for the MDA-231 and NIH-3T3 cells. Class 3 and 4 comets predominated in the positive control, which showed few class1 and 2 comets. Class 0 comets predominated in the control group, with few class 1comets. An increased percent of DNA in the tail was dependent on the concentration in all studied cells.
The presence of genotoxicity in the NIH3T3 and MDA-MB-231 cells demonstrated that the GQDs affected their genomes to a greater extent than in the A549 cells. A number of publications have suggested that the use of molecules with antioxidant activity together with chemotherapeutic agents in the treatment of cancer increases chemotherapeutic efficacy (Singh et al., 2018). Therefore, we surmise that using N-doped GQDs at doses that do not have a cytotoxic effect but do have antioxidant activity against A549 cells, together with a chemotherapeutic agent, could be useful for cancer cell cytotoxicity.
4.8. Cytotoxicity, genotoxicity and transfection of Eu-GQDs with EpHA2 siRNA
GQDs are known as zero-dimensional quantum dots with small fragments, 3–20 nm in size, and specific electronic and luminous properties (Benítez-Martínez and Valcárcel, 2015). They are widely used in various scientific fields due to their mechanical, optical, electrical and thermal properties, as well as their biocompatibility and transparency. Likewise, nanomaterials containing GQDs are promising materials in biomedical applications, especially in bioimaging, drug delivery and photothermal treatments (Chen et al., 2017). Eudragit® RS 100 is a polymer composed from esters of acrylic or methacrylic acid, with quaternary ammonium groups (Leyva-Gómez et al., 2018). Eudragit polymers have been reported to have less cytotoxicity to cells compared to cationic polymers such as polyethyleneimine (PEI) and poly(l-lysine) (PLL), which are known to have both in vitro and in vivo toxicity (Jain et al., 2015). Studies have shown that Eudragit polymers containing antisense oligodeoxynucleotides can be prepared and efficiently delivered to cells (Jain et al., 2015, Singh et al., 2015).
Membrane-associated receptor tyrosine kinase class A2 (EphA2) is overexpressed in many types of cancers, such as breast, prostate, bladder, skin, lung, ovarian and brain cancers. Particularly in the advanced stages of NSLC cancers, previous studies have shown that EphA2 is highly increased in the cytoplasm and membrane. This increase can be associated with a poor prognosis and survival rate (Tandon et al., 2011).
Antisense siRNA, which has recently attracted intense interest, has been investigated for their effect on cancer cells. Therefore, in this study, Eudragit® RS 100-coated GQDs (Eu-GQDs) were prepared and allowed to electrostatically interact with EphA2 siRNA.
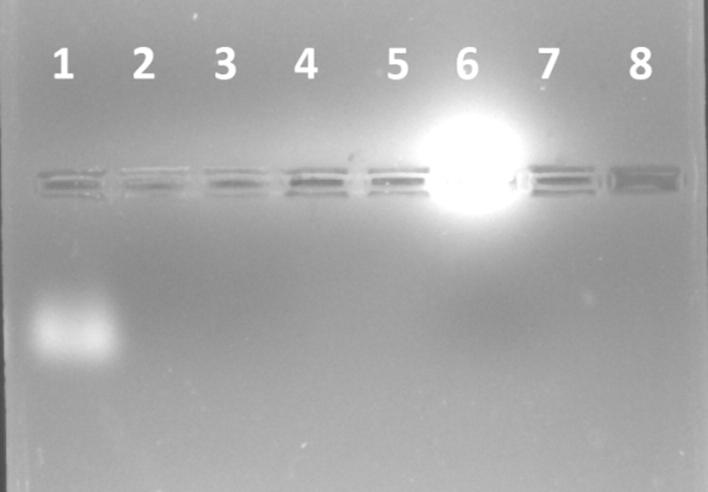
According to the characterization results, Eu-GQDs have an appropriate particle size and zeta potential for transfection assays. In general, the PDIs varied between 0.109 and 0.178, demonstrating uniformity of the nanoparticles, and the results are presented in Table 3. The particle size is a very important parameter for the cellular internalization of graphene-related nanomaterials (Zhang et al., 2016). It is thought that the relatively small particle size of the siRNA-loaded or unloaded nanoparticles facilitates their passage into the cell. However, the high zeta potential helps both the binding of siRNA and the successful cell transmission of the particles. According to the gel retardation studies (Fig. 13), the well for GQDs alone had high radiance, while the well for the Eu-GQDs did not show any radiance. It was shown that Eudragit successfully covered the GQDs, and the prepared nanoparticles could effectively bind to siRNA and did not cleave the siRNA at the studied ratios.
Table 3.
Particle size and zeta potential results of GQDs, Eu-GQDs and siRNA-loaded Eu-GQD formulations.
| GQDs | Eu-GQDs | siRNA/Eu-GQDs | |
|---|---|---|---|
| Particle Size (nm) | 10.9 ± 1.3 | 173.2 ± 0.2 | 198.4 ± 4.2 |
| Polydispersity Index (PDI) | 0.139 ± 0.1 | 0.109 ± 0.2 | 0.178 ± 0.6 |
| Zeta Potential (mV) | −2.86 ± 1.8 | +47.9 ± 2.2 | +35.2 ± 1.3 |
±SE: Standart Error.
Fig. 13.
Gel retardation assay of siRNA/Eu- GQDs, Lane 1: 1.5 µg naked siRNA; Lane 2–5 ratio of Eu-GQDs/siRNA, 0.5/1, 1/1, 2/1, 3/1, respectively; Lane 6: only N-doped GQDs; Lane 7: only Eu-GQDs; Lane 8: Free well.
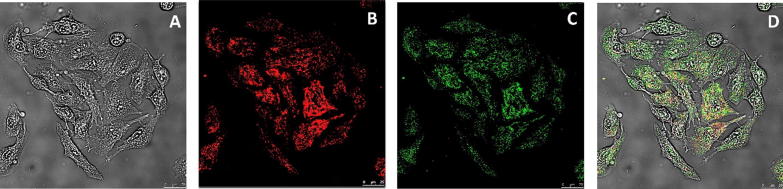
Studies have shown that quantum dots can be successfully used for imaging because they exhibit a high bioimaging potential (Wu et al., 2013, Joshi et al., 2016). In our study, the transfer of GQDs into the cell cytoplasm was also demonstrated. The transfection study was carried out to determine whether the prepared formulations could pass into the cell. The 0.5/1 (N/P) ratio was used for transfections. The transfection studies showed (Fig. 14) that FITC-siRNA and GQD-loaded formulations could pass into the cells with a 24 h incubation. FITC-siRNA and GQD-loaded formulations were observed by confocal microscopy at 20× and 40× magnification with extra zoom. According to the results, the siRNA and GQD formulations were easily transferred.
Fig. 14.
Confocal microscopy images at different wavelengths of the delivery of GQDs (578 nm) and siRNA (518 nm) to A549 cells. Bright field (A), 578 nm (B), 518 nm (C) and overlay (D).
Non-small cell lung cancer (NSCLC) is one of the leading causes of cancer-related deaths. Despite significant advances in diagnosis and treatment, little progress has been made regarding the mortality of NSCLC. Recently, the intratumoral application of chemotherapeutic drugs has shown promise in clinical trials. However, the high toxicity of the chemotherapeutic agents used and the need for high doses are limitations (Lee et al., 2017). Eph expression levels are associated with cancer progression, metastatic spread and patient survival (Ireton and Chen, 2005). Ephrin receptor-A2 (EphA2) has been identified as a new biomarker of tumour cells. For example, in all NSCLC cases, EphA2 has is overexpressed, while this protein is not expressed in normal epithelial cells and tissues in adults. Thus, the targeting and downregulation of EphA2 is important for tumour growth and migration (Ogawa et al., 2000, Brannan et al., 2009, Lee et al., 2017).
In the current study, the cytotoxicity and genotoxicity effects of the prepared formulations were investigated in the A549 cell line 72 h after the formulation was applied. The results are presented in Fig. 15A and B.
Fig. 15.
Cytotoxicity (A) and comet assay (B) results of EphA2 siRNA-loaded and N-doped GQD formulations.
A study of EphA2 siRNA by Landen et al. showed that paclitaxel-containing liposomal formulations produced a remarkable reduction in tumour growth in vivo. The effect of siRNA in this observed reduction is noteworthy (Landen et al., 2005). In another study performed on two orthotopic ovarian tumour models, continuous EphA2 silencing resulted in a decrease in tumour volume, vascular density and cell proliferation (Tanaka et al., 2010). Compared to the results of that study, our EphA2 siRNA and QGD-loaded formulations did not induce any statistically significant decrease in cell viability (p > 0.05). However, the comet analyses showed that the cells were killed at a higher rate. When only GQDs were applied, class 1 and 2 comets were predominant in the comet analyses. But in this study, which applied the same concentrations, class 3 comets was predominant (p < 0.05). In our previous studies, we reported that Eudragit® RS 100 up to 1 mg/mL produced a cell viability of greater than 70% in various cells (Wang et al., 2013, Öztürk et al., 2018). The reason for the decreased cell viability is thought to be due to the co-administration of siRNA and GQD. It is expected that siRNA will be more effective with longer incubation times, and further decreases in cell viability can be obtained.
5. Conclusion
Graphene quantum dots (GQDs) are very similar to traditional quantum dots, but their most distinctive features are that they do not contain heavy metals and have very small dimensions. They are currently used in many applications, such as drug transport, DANN-sensing, bioimaging and tissue engineering. In addition, the use of these materials with drugs has produced positive results for their use with drug carriers, particular regarding their ability to reduce cell viability in diseases such as cancer (Zhang et al., 2017, Islam et al., 2017, Lee et al., 2018).
In our study, the synthesized N-doped GQDs were confirmed by TEM, EDX, DLS, TG, FTIR and UV–Vis analyses. These green synthesized N-doped GQDs were tested for DNA binding, DNA cleavage, cell viability, antimicrobial and antioxidant activities. The N-doped GQDs showed dose–dependent cell viability, antimicrobial and antioxidant activities. The results show that the N-doped GQDs could bind to DNA via intercalation and electrostatic routes.
We believe that siRNA-containing formulations prepared with graphene quantum dot can be used to minimize the systemic toxicities of anticancer drugs and the undesirable side effects associated with conventional chemotherapy. Our findings showed that due to the luminescence properties of quantum dots, the formulations containing GQDs could enter into cells, and a decrease in cancer cells could be observed with a decrease in EpHA2, which is highly expressed in A549 cells. This reduction is thought to arise from the shearing effect of the graphene quantum dots on the DNA and from the effect of the siRNA. Therefore, at high doses, quantum dots can lower the cell viability by breaking the DNA in cancer cells; they can also provide bioimaging capabilities at low doses. Moreover, siRNA-GQD complexes may be effective even at low concentrations against A549 cells. To the best of our knowledge, this is the first report that has coupled Eudragit RS 100, siRNA and N-doped GQD nanoformulations to demonstrate lung cancer-specific effects. This study has shown that N-doped GQDs may be safe and cost-effective, with potential cell growth inhibition, DNA interaction, antioxidant and antimicrobial activities. These findings will encourage further exploration of N-doped GQDs as potential agents for future drug research and development.
Declaration of Competing Interest
The author reports no conflict of interest.
Acknowledgments
Acknowledgements
The authors are grateful to the Çanakkale Onsekiz Mart University, The Scientific Research Commission (COMU-BAP), Turkey for the financial support of this work, grant number 2018-1291. The authors would like to special thank to Şennur Görgülü and AUBIBAM for confocal microscopy analyses and to Koray Şenel for statistical analyses.
All authors agree to publish the manuscript. All the authors worked in the design of this study, the evaluation and writing of the experiments.
Footnotes
Peer review under responsibility of King Saud University.
References
- Benítez-Martínez S., Valcárcel M. Graphene quantum dots in analytical science. TrAC Trends Anal. Chem. 2015;72:93–113. doi: 10.1016/j.trac.2015.03.020. [DOI] [Google Scholar]
- Biju V., Mundayoor S., Omkumar R.V., Anas A., Ishikawa M. Bioconjugated quantum dots for cancer research: present status, prospects and remaining issues. Biotechnol. Adv. 2010;28(2):199–213. doi: 10.1016/j.biotechadv.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Brannan J.M., Sen B., Saigal B., Prudkin L., Behrens C., Solis L., Dong W., Bekele B.N., Wistuba I., Johnson F.M. EphA2 in the early pathogenesis and progression of non-small cell lung cancer. Cancer Prev. Res. (Phila) 2009;2(12):1039–1049. doi: 10.1158/1940-6207.CAPR-09-0212. [DOI] [PubMed] [Google Scholar]
- Chen F., Gao W., Qiu X., Zhang H., Liu L., Liao P., Fu W., Luo Y. Graphene quantum dots in biomedical applications: recent advances and future challenges. Front. Lab. Med. 2017;1(4):192–199. doi: 10.1016/j.flm.2017.12.006. [DOI] [Google Scholar]
- Chong Y., Ge C., Fang G., Tian X., Ma X., Wen T., Wamer W.G., Chen C., Chai Z., Yin J.J. Crossover between anti- and pro-oxidant activities of graphene quantum dots in the absence or presence of light. ACS Nano. 2016;10:8690–8699. doi: 10.1021/acsnano.6b04061. [DOI] [PubMed] [Google Scholar]
- CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2015. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard-tenth Edition. CLSI Document M07–A10. [Google Scholar]
- De Jong W.H., Borm P.J.A. Drug delivery and nanoparticles: applications and hazards. Int. J. Nanomed. 2008;3(2):133–149. doi: 10.2147/ijn.s596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman A. Improving anticancer drug development begins with cell culture: misinformation perpetrated by the misuse of cytotoxicity assays. Oncotarget. 2017;8(5):8854–8866. doi: 10.18632/oncotarget.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E.J., Oldoni T.L.C., Alencar S.M.D., Reis A., Loguercio A.D., Grande R.H.M. Antioxidant activity by DPPH assay of potential solutions to be applied on bleached teeth. Braz. Dent. J. 2012;23:22–27. doi: 10.1590/S0103-64402012000100004. [DOI] [PubMed] [Google Scholar]
- Gencer S., Cebeci A., Irmak-Yazicioglu M.B. Silencing of the MMP-3 gene by siRNA transfection in gastric cancer AGS cells. J. Gastrointestin. Liver Dis. 2010;20:19–26. [PubMed] [Google Scholar]
- Giovannelli L., Cozzi A., Guarnieri I., Dolara P., Moroni F. Comet assay as a novel approach for studying DNA damage in focal cerebral ischemia: differential effects of NMDA receptor antagonists and poly(ADP-ribose) polymerase inhibitors. J. Cereb. Blood Flow Metab. 2002;22(6):697–704. doi: 10.1097/00004647-200206000-00008. [DOI] [PubMed] [Google Scholar]
- Ireton R.C., Chen J. EphA2 receptor tyrosine kinase as a promising target for cancer therapeutics. Curr. Cancer Drug Targets. 2005;5:149–157. doi: 10.2174/1568009053765780. [DOI] [PubMed] [Google Scholar]
- Islam M.S., Tong L., Hasan M., Minett A.I., Gomes V.G., Deng Y., Roy A.K. In-situ direct grafting of graphene quantum dots onto carbon fibre by low temperature chemical synthesis for high performance flexible fabric supercapacitor. Mater. Today Commun. 2017;10:12–119. doi: 10.1016/j.mtcomm.2016.11.002. [DOI] [Google Scholar]
- Jain R., Dandekar P., Loretz B., Kochdand M., Lehr C.M. Dimethylaminoethyl methacrylate copolymer-siRNA nanoparticles for silencing a therapeutically relevant gene in macrophages. Med. Chem. Comm. 2015;6(4):691–701. doi: 10.1039/C4MD00490F. [DOI] [Google Scholar]
- Jiang D., Chen Y., Li N., Li W., Wang Z., Zhu J., Zhang H., Liu B., Xu S. Synthesis of luminescent graphene quantum dots with high quantum yield and their toxicity study. PLoS One. 2015;10(12):1–15. doi: 10.1371/journal.pone.0144906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P.N., Kundu S., Sanghi S.K., Sarkar D. In: Smart Drug Delivery Systems. Sezer A.D., editor. Crotia; Intech Open: 2016. Chapter 7: graphene quantum dots – from emergence to nanotheranostic applications; pp. 159–195. [Google Scholar]
- Kedare S.B., Singh R.P. Genesis and development of DPPH method of antioxidant assay. J. Food Sci. Technol. 2011;48(4):412–422. doi: 10.1007/s13197-011-0251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Koedrith P., Kim H.S., Yu W.J., Kim J.C., Seo Y.R.J. Comparative genotoxicity investigation using comet and gammaH2AX assays for screening of genotoxicants in HepG2 human hepatoma cells. Toxicol. Environ. Health Sci. 2016;8(1):68–78. doi: 10.1007/s13530-016-0263-3. [DOI] [Google Scholar]
- Kumawat M.K., Thakur M., Gurung R.B., Srivastava R. Graphene quantum dots for cell proliferation, nucleus imaging, and photoluminescent sensing applications. Sci. Rep. 2017;7(15858):1–16. doi: 10.1038/s41598-017-16025-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo W.S., Hsu C.L.L., Chen H.H., Chang C.Y., Kao H.F., Chou L.C.S., Chen Y.C., Chen S.J., Chang W.T., Tseng S.W., Wang J.Y., Pu Y.C. Graphene quantum dots conjugated with polymers for two-photon properties under two-photon excitation. Nanoscale. 2016;8:16874–16880. doi: 10.1039/c6nr02614a. [DOI] [PubMed] [Google Scholar]
- Landen C.N., Jr., Chavez-Reyes A., Bucana C., Schmandt R., Deavers M.T., Lopez-Berestein G., Sood A.K. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- Lai E.P.C. Graphene quantum dots for bioanalytical sensors and biochemical imaging applications. J. Res. Anal. 2017;3(3):88–90. [Google Scholar]
- Lalwani G., D'Agati M., Khan A.M., Sitharaman B. Toxicology of graphene-based nanomaterials. Adv. Drug Deliv. Rev. 2016;105:109–144. doi: 10.1016/j.addr.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.C., Haley C., Lim Y.X., Shi H., Lena A., Tang L., Wang Y., Lim C.T., Loh K.P. Origin of enhanced stem cell growth and differentiation on graphene and graphene oxide. ACS. Nano. 2011;5(9):7334–7341. doi: 10.1021/nn202190c. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Mohammed K.A., Kaye F., Moudgil B.M., Nasreen N. EphA2 targeted intratumoral therapy for non-small cell lung cancer using albumin mesospheres. Am. J. Transl. Res. 2017;9(7):3293–3303. [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Youn Y.H., Kwon K., Ko N.R. Recent advances in quantum dots for biomedical applications. J. Pharm. Investig. 2018;48:209–214. doi: 10.1007/s40005-018-0387-3. [DOI] [Google Scholar]
- Leyva-Gómez G., Piñón-Segundo E., Mendoza-Muñoz N., Zambrano-Zaragoza M.L., Mendoza-Elvira S., Quintanar-Guerrero D. Approaches in polymeric nanoparticles for vaginal drug delivery: a review of the state of the art. Int. J. Mol. Sci. 2018;19(6):1–19. doi: 10.3390/ijms19061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Feterl M., Mulyana Y., Warner J.M., Collins J.G., Keene F.R. In vitro susceptibility and cellular uptake for a new class of antimicrobial agents: dinuclear ruthenium(II) complexes. J. Antimicrob. Chemother. 2012;67:2686–2695. doi: 10.1093/jac/dks291. [DOI] [PubMed] [Google Scholar]
- Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961;3:208–218. doi: 10.1016/S0022-2836(61)80047-8. [DOI] [Google Scholar]
- Mitkovska W.I., Dimitrov H.A., Chassovnikarova T.G. In vivo genotoxicity and cytotoxicity assessment of allowable concentrations of nickel and lead: comet assay and nuclear abnormalities in acridine orange stained erythrocytes of common carp (Cyprinus carpio L.) Acta Zool. Bulg. Suppl. 2017;8:47–56. [Google Scholar]
- Nigam P., Waghmode S., Louis M., Wangnoo S., Chavan P., Sarkar D. Graphene quantum dots conjugated albumin nanoparticles for targeted drug delivery and imaging of pancreatic cancer. J. Mater. Chem. B. 2014;2:3190–3195. doi: 10.1039/C4TB00015C. [DOI] [PubMed] [Google Scholar]
- Nurunnabi M., Khatun Z., Huh K.M., Park S.Y., Lee D.Y., Cho K.J., Lee Y.K. In vivo biodistribution and toxicology of carboxylated graphene quantum dots. ACS Nano. 2013;7:6858–6867. doi: 10.1021/nn402043c. [DOI] [PubMed] [Google Scholar]
- Ogawa K., Pasqualini R., Lindberg R.A., Kain R., Freeman A.L., Pasquale E.B. The ephrin-A1 ligand and its receptor, EphA2, are expressed during tumor neovascularization. Oncogene. 2000;19:6043–6052. doi: 10.1038/sj.onc.1204004. [DOI] [PubMed] [Google Scholar]
- Öztürk A.A., Güven U.M., Yenilmez E., Şenel B. Effects of different derivatives of eudragit polymer on entrapment efficiency, in vitro dissolution, release kinetics and cell viability results on extended release flurbiprofen loaded nanomedicines. Lat. Am. J. Pharm. 2018;37(10):1981–1992. [Google Scholar]
- Qu D., Zheng M., Li J., Xie Z., Sun Z. Tailoring color emissions from N-doped graphene quantum dots for bioimaging applications. Light-Sci. App. 2015;4(e364):1–8. doi: 10.1038/lsa.2015.13. [DOI] [Google Scholar]
- Palmieri V., Carmela Lauriola M., Ciasca G., Conti C., De Spirito M., Papi M. The graphene oxide contradictory effects against human pathogens. Nanotechnology. 2017;28(15):1–18. doi: 10.1088/1361-6528/aa6150. [DOI] [PubMed] [Google Scholar]
- Papi M., Palmieri V., Bugli F., De Spirito M., Sanguinetti M., Ciancico C., Braidotti M.C., Gentilini S., Angelani L., Conti C. Biomimetic antimicrobial cloak by graphene-oxide agar hydrogel. Sci. Rep. 2016;7(12):1–7. doi: 10.1038/s41598-016-0010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic B.Z., Milenkovic M.M., Dakic I.R., Todorovic-Markovic B.M., Milosavljevic M.S., Budimir M.D., Paunovic V.G., Dramicanin M.D., Markovic Z.M., Trajkovic V.S. Photodynamic antibacterial effect of graphene quantum dots. Biomaterials. 2014;35(15):4428–4435. doi: 10.1016/j.biomaterials.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Ruiz V., Yate L., García I., Cabanero G., Grande H.J. Tuning the antioxidant activity of graphene quantum dots: protective nanomaterials against dye decoloration. Carbon. 2017;116:366–374. doi: 10.1016/j.carbon.2017.01.090. [DOI] [Google Scholar]
- Sachdev A., Gopinath P. Green synthesis of multifunctional carbon dots from coriander leaves and their potential application as antioxidants, sensors and bioimaging agents. Analyst. 2015;140:4260–4269. doi: 10.1039/C5AN00454C. [DOI] [PubMed] [Google Scholar]
- Safardoust-Hojaghan H., Salavati-Niasari M., Amiri O., Hassanpour M. Preparation of highly luminescent nitrogen doped graphene quantum dots and their application as a probe for detection of Staphylococcus aureus and E. coli. J. Mol. Liquids. 2017;241:1114–1119. doi: 10.1016/j.molliq.2017.06.106. [DOI] [Google Scholar]
- Singh S., Neelam, Arora S., Singla Y.P. An overview of multifaceted significance of eudragit polymers in drug delivery systems. Asian J. Pharm. Clin. Res. 2015;8(5):1–6. [Google Scholar]
- Singh K., Bhori M., Kasu Y.A., Bhat G., Marar T. Antioxidants as precision weapons in war against cancer chemotherapy induced toxicity – exploring the armoury of obscurity author links open overlay panel. Saudi Pharm. J. 2018;26(2):177–190. doi: 10.1016/j.jsps.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Wu L., Wei W., Qu X. Recent advances in graphene quantum dots for sensing. Mater. Today. 2013;16(11):433–442. doi: 10.1016/j.mattod.2013.10.020. [DOI] [Google Scholar]
- Szunerits S., Boukherroub R. Antibacterial activity of graphene-based materials. J. Mater. Chem. B. 2016;4:6892–6912. doi: 10.1039/C6TB01647B. [DOI] [PubMed] [Google Scholar]
- Şenel B., Büyükköroğlu G., Yazan Y. Solid lipid and chitosan particulate systems for delivery of siRNA. Pharmazie. 2015;70:698–705. doi: 10.1691/ph.2015.5026. [DOI] [PubMed] [Google Scholar]
- Şenel B., Büyükköroğlu G. In: Multifunctional Nanocarriers for Contemporary Healthcare Applications. Barkat A., Harshita A.B., Beg S., Ahmad F.J., editors. IGI Global; 2018. Chapter 14: nanocarriers for vaccine and gene delivery application; pp. 381–414. [Google Scholar]
- Tanaka T., Mangala L.S., Vivas-Mejia P.E., Nieves-Alicea R., Mann A.P., Mora E., Han H.D., Shahzad M.M., Liu X., Bhavane R., Gu J., Fakhoury J.R., Chiappini C., Lu C., Matsuo K., Godin B., Stone R.L., Nick A.M., Lopez-Berestein G., Sood A.K., Ferrari M. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res. 2010;70(9):3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon M., Vemula S.V., Mittal S.K. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert Opin Ther. Targets. 2011;15(1):31–51. doi: 10.1517/14728222.2011.538682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünver H., Boyacıoglu B., Zeyrek C.T., Yıldız M., Demir N., Yıldırım N., Karaosmanoglu O., Sivas H., Elmalı A. Synthesis, spectral and quantum chemical studies and use of (E)-3-[(3,5-bis(trifluoromethyl)phenylimino)methyl]benzene-1,2-diol and its Ni(II) and Cu(II) complexes as an anion sensor, DNA binding, DNA cleavage, anti-microbial, anti-mutagenic and anti-cancer agent. J. Mol. Struct. 2016;1125:162–176. doi: 10.1016/j.molstruc.2016.06.058. [DOI] [Google Scholar]
- Yehye W.A., Rahman N.A., Ariffin A., Abd Hamid S.B., Alhadi A.A., Kadir F.A., Yaeghoobi M. Understanding the chemistry behind the antioxidant activities of butylated hydroxytoluene (BHT): a review. Eur. J. Med. Chem. 2015;28(101):295–312. doi: 10.1016/j.ejmech.2015.06.026. [DOI] [PubMed] [Google Scholar]
- Yıldız M., Karpuz O., Zeyrek C.T., Boyacıoglu B., Dal H., Demir N., Yıldırım N., Unver H. Synthesis, biological activity, DNA binding and anion sensors, molecular structure and quantum chemical studies of a novel bidentate Schiff base derived from 3,5-bis(triflouromethyl)aniline and salicylaldehyde. J. Mol. Struct. 2015;1094:148–160. doi: 10.1016/j.molstruc.2015.03.047. [DOI] [Google Scholar]
- Yuan X.C., Liu Z.M., Guo Z.Y., Ji Y.H., Jin M., Wang X.P. Cellular distribution and cytotoxicity of graphene quantum dots with different functional groups. Nanoscale Res. Lett. 2014;9(1): 108:1–9. doi: 10.1186/1556-276X-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wu C.Y., Zhou X.J., Han T., Xin X.Z., Wu J.Y., Zhang J.Y., Guo S.W. Enhancing cell nucleus accumulation and DNA cleavage activity of anti-cancer drug via graphene quantum dots. Sci. Rep. 2013;3(2852):1–8. doi: 10.1038/srep02852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Chen J., Liu C., Qiu J., Wang X., Chen P., Xu C. A graphene quantum dots-hypochlorite hybrid system for the quantitative fluorescent determination of total antioxidant capacity. Small. 2017;13(30):1700–1709. doi: 10.1002/smll.201700709. [DOI] [PubMed] [Google Scholar]
- Wu X., Tian F., Wang W., Chen J., Wub M., Zha J.X. Fabrication of highly fluorescent graphene quantum dots using L-glutamic acid for in vitro/in vivo imaging and sensing. J. Mat. Chem. C. 2013;1:4676–4684. doi: 10.1039/C3TC30820K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyrek C.T., Ünver H., Boyacıoğlu B., Demir N., Yapar G., Dal H., Yıldız M. Synthesis, quantum chemical calculations and molecular docking studies, biological and anion sensor properties of (E)-4-[(4-ethoxy-phenylimino)methyl]-2-methoxyphenol. Croat. Chem. Acta. 2018;91(3):1–15. doi: 10.5562/cca3316. [DOI] [Google Scholar]
- Zhang B., Wei P., Zhou Z., Wei T. Interactions of graphene with mammalian cells: molecular mechanisms and biomedical insights. Adv. Drug Deliv. Rev. 2016;105(1):145–162. doi: 10.1016/j.addr.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Zhang W., Zeng Z., Wei J. Electrochemical study of DPPH radical scavenging for evaluating the antioxidant capacity of carbon nanodots. J. Phys. Chem. C. 2017;121:18635–18642. doi: 10.1021/acs.jpcc.7b05353. [DOI] [Google Scholar]
- Zheng X.T., Ananthanarayanan A., Luo K.Q., Chen P. Glowing graphene quantum dots and carbon dots: properties, syntheses, and biological applications. Small. 2015;11(14):1620–1636. doi: 10.1002/smll.201402648. [DOI] [PubMed] [Google Scholar]
- Zhou S., Xu H., Gan W., Yuan Q. Graphene quantum dots: recent progress in preparation and fluorescence sensing applications. RSC Adv. 2016;6:110775–110788. doi: 10.1039/C6RA24349E. [DOI] [Google Scholar]
- Zhu S., Song Y., Zhao X., Shao J., Zhang J., Yang B. The photoluminescence mechanism in carbon dots (graphene quantum dots, carbon nanodots, and polymer dots): current state and future perspective. Nano Res. 2015;8:355–381. doi: 10.1007/s12274-014-0644-3. [DOI] [Google Scholar]