Abstract
Although genetically modified mice can be generated with high efficiency by using CRISPR/Cas9-mediated genome editing in mouse zygotes, only the loci with a protospacer-adjacent motif (PAM) sequence are targetable. The present study investigated the usability of engineered Streptococcus pyogenes Cas9 (SpCas9-NG) in mouse zygotes. In addition to the 5′-NGG sequence, SpCas9-NG recognized the 5′-NGA, 5′-NGC and 5′-NGT sequences in mouse zygotes as PAMs that were appropriate for the generation of knockout mice. Moreover, SpCas9-NG-mediated genome editing enabled the generation of knock-in mice untargetable by the conventional SpCas9 in mouse zygotes. These results suggest that SpCas9-NG-mediated genome editing in zygotes is available for the generation of knockout and knock-in mice at the locus corresponding to NGN-PAM.
Subject terms: Genetic engineering, Transgenic organisms
Introduction
The Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Cas9 system, which consists of Cas9 endonuclease and a single guide-RNA (gRNA)1,2, is a widely utilized engineered endonuclease, and CRISPR/Cas9-mediated genome editing in mammalian zygotes is utilized for the generation of genetically modified animals2–8. The Cas9-gRNA complex recognizes a specific DNA sequence which consists of a base-pairing sequence to the spacer region of the gRNA and a protospacer adjacent motif (PAM), then induces a DNA double-strand break (DSB) in the target site. The repair of a DSB by error-prone non-homologous end-joining (NHEJ) sometimes forms insertion/deletion (indel) mutations, thereby disrupting the target gene or genome information7,8. Besides, DSB stimulates homology-directed repair (HDR), and thus knock-in via HDR is achieved by introducing exogenous donor DNA as a repair template together with the Cas9 and gRNA.
Streptococcus pyogenes (Sp)-derived Cas9 (SpCas9) is commonly used as a conventional CRISPR/Cas9 system. SpCas9 requires 5′-NGG as its PAM sequence, and consequently, the targetable locus is restricted9,10. Alternatively, other prokaryote-derived orthologous Cas9 endonucleases and Cpf1 (also known as Cas12a), which recognize different sequences as PAMs, are available for genome editing in mammalian cells including zygotes11–19. Although these systems contribute to the expansion of targetable loci, the requirement of specific polynucleotide sequences as PAMs still restricts the designable target loci for genome editing. It was reported that an orthologous Cas9 from Streptococcus canis can recognize and cut NNG-PAM-bearing target site in mammalian culture cells, but the availability to mammalian zygotes is not investigated20.
It has been reported that the protein engineering of Cas9 endonuclease enhances functions such as the requirement of a PAM sequence21–23, the accuracy of target recognition24–28 or endonuclease activities29–31. The xCas922 and SpCas9-NG23 are engineered Cas9 containing independent 7 amino acid substitutions of the wildtype SpCas9 and require a 5′-NGN sequence as the PAM. It was reported that SpCas9-NG more efficiently recognize and cleavage the target site bearing NGH-PAM compared with xCas9 in in vitro assay23 and SpCas9-NG induced NHEJ-mediated indels or nucleotide substitution via a fused-deaminase domain at the target loci corresponding to NGN-PAM in mammalian culture cell and plants23,32,33. It was reported that SpCas9-NG increases the targeting range of SpCas9 in the human coding sequence23, therefore the utilization of SpCas9-NG in mammalian zygotes is expected to expand the versatility of target designs for the generation of genetically modified animals. However, previous studies have suggested that SpCas9-NG reduces the efficiency of target mutagenesis compared with wildtype SpCas9 at NGG-PAM23. Thus, it is unclear whether SpCas9-NG could be used in place of the conventional SpCas9 for genome editing in zygotes.
In the present study, we evaluated the efficiency of SpCas9-NG-mediated genome-editing at endogenous target sites bearing NGN-PAM in mouse zygotes. Moreover, we attempted to generate knockout and knock-in mice using SpCas9-NG.
Results
We previously established a Cas9 expression construct optimized for mammalian zygotes. This construct-derived Cas9 mRNA has shown highly efficient target mutagenesis at various loci in mouse zygotes8,18,19,31,34,35, and we therefore used this plasmid vector as a template when reconstructing SpCas9-NG in the present study. Western blot analysis showed that SpCas9-NG expressed as well as the wildtype Cas9 in HEK293 cells (Supplementary Fig. 2).
By using this construct, we evaluated the efficiency of SpCas9-NG-mediated target mutagenesis in mouse embryos. We designed 9 gRNAs at a tyrosinase locus; they corresponded to the 5′-NGG, 5′-NGA, 5′-NGT and 5′-NGC sequences as PAMs (Supplementary Fig. 3A,B). Each gRNA was microinjected with SpCas9-NG mRNA into C57BL/6NCr-derived zygotes, and then blastocyst-stage embryos were subjected to the PCR-directed Sanger-sequencing, and each of the obtained chromatogram data was observed. As a result, the target sequences of NGA-, NGC- and NGT-PAM contained mutagenized sequence in almost all of the blastocyst (97.7%, 94.2% and 93.2%, respectively) in addition to the target sequence of NGG-PAM (98.1%) by using SpCas9-NG in contrast to the wildtype SpCas9, which generated only 16.7% mutants in NGA-PAM and 6.3% mutant in NGT-PAM (Supplementary Figs 3B,C and 4). The TIDE analysis35 also suggested that SpCas9-NG showed highly efficient scores in the target sequence of NGN-PAM in each blastocyst while the rates of mutagenic efficiencies in NGA-PAM and NGT-PAM by wildtype-SpCas9 were limited (2.2 and 2.6%, respectively) (Supplementary Fig. 3B,D). These results suggested that SpCas9-NG could recognize the 5′-NGN sequences as a PAM and functioned efficiently as an engineered endonuclease in mouse zygotes.
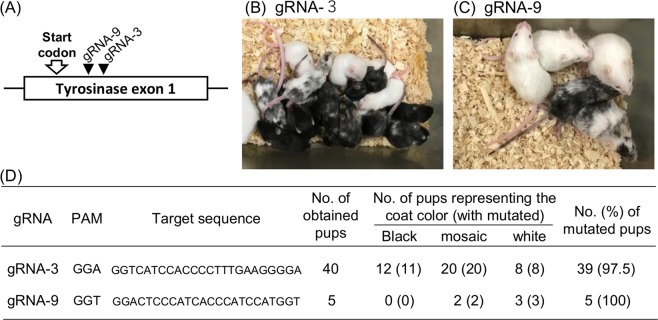
Next, we attempted to generate knockout mice using SpCas9-NG. Cas9 mRNA and gRNA-3 (5′-NGA as PAM; Fig. 1A and Supplementary Fig. 3) were injected into the C57BL/6NCr zygotes and the embryos were transferred to recipients, successfully yielding 40 offspring. Tail-tip-derived genomic DNA indicated that 39 of 40 F0 pups showed induced mutations at the target loci (Fig. 1D and Supplementary Fig. 5). The coat of 28 of 40 pups consisted of completely-white or black-white mosaic hair, suggesting tyrosinase deficiency (Fig. 1B,D). With the same efficiency as gRNA-3, gRNA-9 (5′-NGT as PAM) could induce the mutation efficiently at the target locus (5/5 pups), and the pups exhibited the tyrosinase deficiency (Fig. 1C,D and Supplementary Fig. 6). These results suggested that SpCas9-NG is available for the efficient generation of knockout mice via zygotes.
Figure 1.
Generation of Tyrosinase knockout mice using SpCas9-NG. (A) Schematics of the target loci for generation of tyrosinase knockout mice. (B,C) The coat colors of obtained pups generated by using gRNA-3 (B) and gRNA-9 (C). (D) The incidence of the represented coat color and mutagenic efficiencies of the obtained pups.
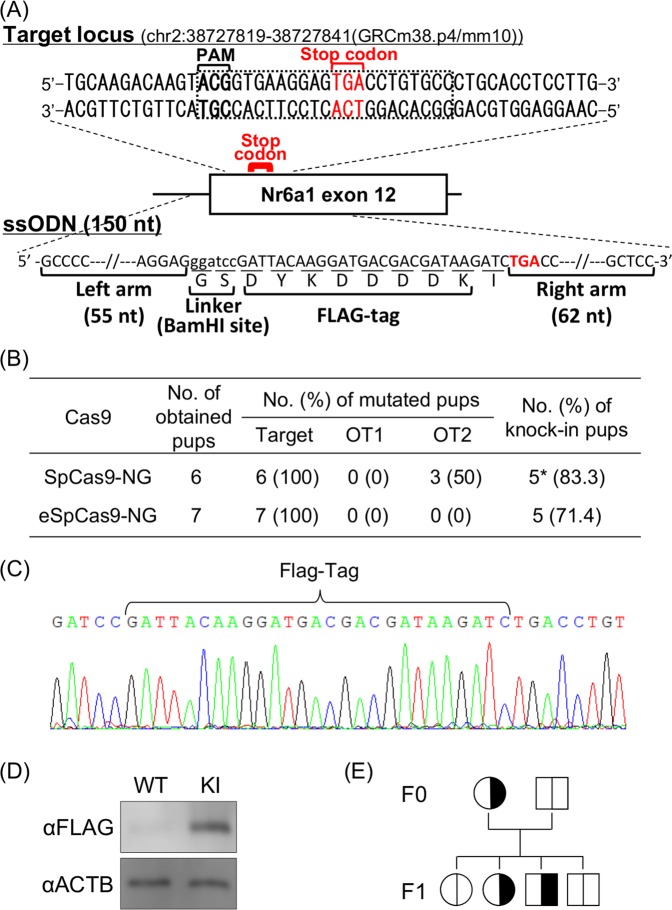
We next examined whether SpCas9-NG-mediated genome editing in zygotes could be utilized for the generation of knock-in mice. In the case of genome editing-mediated knock-in, it is preferable to design gRNA overlapping with the knock-in site in order to avoid retargeting of the knock-in allele36. Nuclear receptor subfamily 6, group A, member 1 (Nr6a1) is known as an orphan nuclear receptor which is expressed in a variety of tissues, including testis tissue, in adult mice37,38. Knock-in of the epitope-tag into the NR6A1 c-terminal is a suitable experiment for proof-of-principle because there is no canonical PAM (5′-NGG) at the 3′ sides of the stop codon in the plus- and minus-strand sequence (Fig. 2A), which is required for the design of a gRNA overlapping with the knock-in site. Thus, a gRNA (5′-NGT as PAM) was designed, and was microinjected together with SpCas9-NG mRNA and a single strand oligodeoxynucleotide (ssODN)-encoding Flag-tag sequence and homology arms (Fig. 2A). After embryo transfer, 6 pups were successfully obtained. The sequencing of the tail-tip derived-genome DNA revealed that mutagenesis took place in all of the pups (Supplementary Fig. 8A) and 5 of the 6 had the Flag-tag knock-in allele (Fig. 2B,C and Supplementary Figs 7A and 9A). Flag-tagged allele-derived Nr6a1 mRNA was expressed in a knock-in male testis, and Flag-tagged Nr6a1 protein expression was detected by western blot analysis (Fig. 2D and Supplementary Fig. 11). A knock-in female pup was mated with a wildtype male and the obtained offspring revealed that the knock-in allele was inherited normally in the next generation (Fig. 2E and Supplementary Fig. 12). These results suggested that SpCas9-NG-mediated genome editing in zygotes could be utilized for knock-in mouse generation. Meanwhile, 3 pups had unexpected mutations at one of the two examined predicted off-target locus, OT2 (Fig. 2B and Supplementary Fig. 10). Accordingly, we investigated in mouse zygotes the efficacy of the eSpCas9-NG variant—which is SpCas9-NG with the high-fidelity eSpCas9 variant—thereby reducing the off-target risk in somatic culture cells23. Although all 7 of the resulting pups had the target modification (Supplementary Fig. 8B) and 5 of them had the knock-in allele (Supplementary Figs 7B and 9B), no mutation was found in OT2 locus of any of the pups (Fig. 2B). These results suggested that eSpCas9-NG would have the potential to be available for the efficient and accurate genome editing in mouse zygotes as well as the previous study by somatic culture cells23.
Figure 2.
Generation of Nr6a1-Flag knock-in mice using SpCas9-NG and eSpCas9-NG. (A) Schematic illustration of the Nr6a1 gene structures, sequences around the target locus (upper), and ssODN template (lower). Black-dot box on the sequence around the target locus indicates the PAM and the sequence recognized by the Cas9-gRNA complex. The target sequence contains the stop codon of Nr6a1. The DNA sequence of ssODN is shown in Supplementary Table 1. (B) Mutagenic efficiencies at on-target and off-targets (as shown OT1 and OT2), and knock-in efficiencies of the obtained pups. The locus information is shown in Supplementary Table 2. *Two pups have a nucleotide substitution between Flag-tag encoding sequence and stop codon (shown in Supplementary Fig. 9A). (C) Waveform sequence data of the target locus of an obtained pup (see also Supplementary Figs 8 and 9). (D) Immunoblotting of Flag-tag (FLAG) and β-Actin (ACTB) using the protein extracted from the testes of wildtype and the knock-in males. (E) A pedigree obtained from Nr6a1-Flag knock-in F0 female generated by using SpCas9-NG. The knock-in alleles are shown by the black color, transmitting to the male and female pups (see also Supplementary Fig. 12).
Discussion
The present study investigated whether SpCas9-NG could be used for genome editing of NG-PAM loci via zygotes, and the results showed that both knockout and knock-in mice could be successfully generated using this approach. To date, the target design for genome editing in zygotes has been restricted to the locus where the specific polynucleotide sequence is present. Since SpCas9-NG requires only mononucleotide “G” as a PAM sequence, it expands the range of targetable loci to include loci that could not be achieved by wildtype SpCas9 and orthologous Cas9 in mammalian zygotes. Such expansion is particularly conducive to the production of knock-in animals. Because the Cas9-gRNA injected into the 1-cell-stage egg maintains its activity for a while during preimplantation development, it may be possible to induce unexpected NHEJ-mediated indels by retargeting of the Cas9-gRNA even if the knock-in is achieved at an early stage. Thus, it is necessary to design a gRNA to overlap the knock-in position so that the post-knock-in sequence is not a target for gRNA35. For precise modification of the restricted position, such as nucleotide substitution or insertion of a reporter gene or epitope-tag, a specific gRNA must be designed from limited alternatives. Thus, by increasing the designable loci of gRNA, the SpCas9-NG system is expected to expand the feasibility of such applications.
In contrast, it is concerned that the shortening of the PAM sequence also increases the number of potential off-target positions. In the previous study, off-target mutations by SpCas9-NG have been avoided by the use of the high fidelity eSpCas9 variant23, and the present study in mouse zygotes also supported it while it is preliminary. In addition to the eSpCas9 variant, it has been reported that the utilization of other high fidelity Cas9 variants24–28, shortening of the protospacer and engineering of a gRNA scaffold structure39,40 are all effective means of improving the target recognition of Cas9-gRNA. It is possible that these improvements would contribute to a reduction of the off-target risk of SpCas9-NG in zygotes.
In this study, we found the presence of mutated alleles was observed in almost all obtained offspring, and also observed the transmission of knock-in alleles to the next generation. In contrast, some pups exhibit a mosaic or black coat color as a phenotype while the mutant allele was detected by genotyping, suggesting that the wildtype allele remains as heterozygous mutation at a certain rate in mutated pups. As well, the results of TIDE assay showed various rates of the efficiencies because of containing an intact allele in some of the genetically-modified blastocysts (Supplementary Fig. 3D). In our previous studies on tyrosinase knockout using SpCas9, ST1-Cas9 or Cj-Cas9, almost all of the pups showed bi-allelic target mutagenesis and presented a completely white coat18,19,34. SpCas9-NG has been reported to have lower mutagenic activity compared to wildtype SpCas9 at NGG-PAM23, which could have affected the present results in zygotes. Additional improvements to the mutagenic activity of SpCas9-NG are expected, and should further expand the feasibility of efficient genome editing in zygotes.
Material and Methods
Ethics statement
All animal care and experiments conformed to the Guidelines for Animal Experiments of the University of Tokyo and were approved by the Animal Research Committee of the University of Tokyo (P18-093, 27 Sept 2018).
Construction of Cas9- and gRNA-plasmid DNA
According to the amino acid sequences shown in previous studies23,24, SpCas9-NG and its high-fidelity variant, eSpCas9-NG, were constructed by mutation PCR of pCAG-T3-hCAS9-pA plasmid vector (Addgene plasmid 48625)8. The vectors were sequenced using a commercial sequencing kit (Applied Biosystems, Foster City, CA, USA) and a DNA sequencer (Applied Biosystems) according to the manufacturer’s instructions. The gRNAs for tyrosinase and Nr6a1 were newly designed or designed according to a previous study34, and plasmid vectors coding gRNAs with a T3 promoter for each target were synthesized according to a previous study8. These vectors were sequenced as described above. All of the sequence information is provided in Supplementary Fig. 1.
In Vitro transcription of Cas9 mRNA and gRNAs
The in vitro synthesis of Cas9 mRNA and gRNA was performed as described previously8 using T3 RNA polymerase (New England Biolabs, MA, USA). The RNA transcripts were precipitated with absolute ethanol, washed, and resuspended in RNase-free water. The RNA solutions were stored at −80 °C until use.
Microinjection and embryo-transfer
The microinjection was performed using a microinjector (Narishige, Tokyo, Japan)-equipped microscope. Approximately 4 pL of RNA solution, containing 100 ng/μL of Cas9 mRNA and 20 ng/μL of gRNA (with 100 ng/μL ssODN as shown in Supplementary Table 1 only for the knock-in experiment), was injected into the cytoplasm of in vitro fertilization-derived C57BL/6NCr zygotes19. The injected zygotes were cultured in KSOMaa-BSA for 1–2 h, then transferred into the oviducts of 0.5 days post-coitum pseudopregnant ICR females; alternatively, the zygotes were cultured for 96 h and blastocysts were subjected to genotyping.
Detection of induced mutations at the on- and off-target sites
A search for a potential off-target locus for NR6A1 gRNA was performed using a CRISPOR Version 4.92 (http://crispor.tefor.net/)41. Briefly, an NR6A1 target sequence (5′-GGCACAGGTCACTCCTTCACCGT) was used for the search and the “20bp-NGN or GA(A/T)” was searched as PAM, then the resulting loci (off-targets for 0-1-2-3-4 mismatches were 0 - 0 - 2 - 52 - 518 loci) were adopted as potential off-target loci (Supplementary Table 2), and two of the resulting loci which has 2 mismatches were subjected to the analysis of mutation induction (Supplementary Table 3).
The genome DNA was extracted from the blastocysts or the tail tips of the pups according to our previous report19, then subjected to PCR using the primer sets shown in Supplementary Table 1. The PCR amplicons were purified by a FastGene Gel/PCR Extraction Kit (Nippon Genetics, Tokyo) and sequenced as described above. Mutation introduction in chromatogram data was detected as insertion or deletion of nucleotides into the wildtype sequence or as overlapping of multiple waveforms at the target site, as shown in a previous study8. Furthermore, each chromatogram data of blastocyst-derived Sanger sequencing was subjected to TIDE assay34 to measure the rates of mutant sequence in each blastocyst. TIDE-derived “overall efficiency” and its median of all blastocysts in each group were shown as the efficiency scores (Supplementary Fig. 3B,D).
Cell culture and DNA transfection
The HEK293 cell culture and DNA transfection were performed according to the previous report19,42. The constructed Cas9-expressing vector was transfected into the HEK293 cells by Lipofectamine LTX reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocols. The transfected cells were harvested 48 h after transfection and were subjected to western blot analysis.
RT-PCR
Total RNA was isolated from testicular cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and first-strand cDNA was produced using ReverTra Ace qPCR RT Master Mix (ToYoBo, Tokyo) according to the manufacturer’s protocol. PCR was performed using the primer sets shown in Supplementary Table 1.
PCR-restriction fragment-length polymorphism (RFLP)
The genomic PCR- or RT-PCR-produced amplicons were purified as described above and digested with BamHI (TaKaRa, Tokyo), and then the digested materials were measured by agarose-gel electrophoresis. In order to analyze the knock-in DNA sequence, the digested fragments of genomic PCR were extracted and sequenced by using the Nr6a1 reverse primer (Supplementary Table 2).
Immunoblotting
Western blot analysis was performed according to the process described in our previous report19. The harvested culture cells or testicular cells were suspended in Laemmli buffer. The antibodies used were anti-Flag M2 monoclonal antibody (F1804, Sigma, Carlsbad, CA, USA) and anti-β-actin polyclonal antibody (GTX109639, GeneTex, Inc., CA, USA). To visualize the protein-bound antibodies, horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) were used for the second layer, respectively. The signals were detected using an Immunostar LD Kit (Wako, Tokyo) and a C-DiGit scanner.
Supplementary information
Acknowledgements
We thank members of the Applied Genetics laboratory for their assistance with the study. The plasmid vectors encoding SpCas9-NG and eSpCas9-NG were deposited in Addgene (#131465 and #131466). This work was supported by a Grant in-Aid for Scientific Research (18K19261) from the Japan Society for the Promotion of Science.
Author Contributions
W.F. conceived and designed the experiments; W.F., H.I., T.K. and A.I. performed the experiments; K.S. and K.N. contributed reagents/materials/analysis tools; W.F. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49394-5.
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen B, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang D, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6:97–99. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 8.Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013;41:e187. doi: 10.1093/nar/gkt772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano H, et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada M, et al. Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems. Mol Cell. 2017;65:1109–1121.e1103. doi: 10.1016/j.molcel.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016;34:808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- 18.Fujii W, et al. Zygote-mediated generation of genome-modified mice using Streptococcus thermophilus 1-derived CRISPR/Cas system. Biochem Biophys Res Commun. 2016;477:473–476. doi: 10.1016/j.bbrc.2016.06.070. [DOI] [PubMed] [Google Scholar]
- 19.Fujii, W., Ikeda, A., Sugiura, K. & Naito, K. Efficient Generation of Genome-Modified Mice Using Campylobacter jejuni-Derived CRISPR/Cas. Int J Mol Sci, 18, 10.3390/ijms18112286 (2017). [DOI] [PMC free article] [PubMed]
- 20.Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv. 2018;4:eaau0766. doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu JH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimasu H, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JK, et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. 2018;9:3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casini A, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujii W, Onuma A, Sugiura K, Naito K. Efficient generation of genome-modified mice via offset-nicking by CRISPR/Cas system. Biochem Biophys Res Commun. 2014;445:791–794. doi: 10.1016/j.bbrc.2014.01.141. [DOI] [PubMed] [Google Scholar]
- 32.Hua K, Tao X, Han P, Wang R, Zhu JK. Genome Engineering in Rice Using Cas9 Variants that Recognize NG PAM Sequences. Mol Plant. 2019;12:1003–1014. doi: 10.1016/j.molp.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Endo M, et al. Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat Plants. 2019;5:14–17. doi: 10.1038/s41477-018-0321-8. [DOI] [PubMed] [Google Scholar]
- 34.Fujii W, Onuma A, Sugiura K, Naito K. One-step generation of phenotype-expressing triple-knockout mice with heritable mutated alleles by the CRISPR/Cas9 system. J Reprod Dev. 2014;60:324–327. doi: 10.1262/jrd.2013-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujii W. Generation of Knock-in Mouse by Genome Editing. Methods Mol Biol. 2017;1630:91–100. doi: 10.1007/978-1-4939-7128-2_8. [DOI] [PubMed] [Google Scholar]
- 37.Chen F, Cooney AJ, Wang Y, Law SW, O’Malley BW. Cloning of a novel orphan receptor (GCNF) expressed during germ cell development. Mol Endocrinol. 1994;8:1434–1444. doi: 10.1210/mend.8.10.7854358. [DOI] [PubMed] [Google Scholar]
- 38.Zechel C. The germ cell nuclear factor (GCNF) Mol Reprod Dev. 2005;72:550–556. doi: 10.1002/mrd.20377. [DOI] [PubMed] [Google Scholar]
- 39.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kocak DD, et al. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat Biotechnol. 2019;37:657–666. doi: 10.1038/s41587-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshioka S, Fujii W, Ogawa T, Sugiura K, Naito K. Development of a mono-promoter-driven CRISPR/Cas9 system in mammalian cells. Sci Rep. 2015;5:18341. doi: 10.1038/srep18341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.