Abstract
Lignin, an aromatic polymer found in plants, has been studied for years in many biological fields. Initially, when biofuel was produced from lignocellulosic biomass, lignin was regarded as waste generated by the biorefinery and had to be removed, because of its inhibitory effects on fermentative bacteria. Although it has since proven to be a natural resource for bio-products with considerable potential, its utilization is confined by its complex structure. Hence, the microbial degradation of lignin has attracted researchers' interest to overcome this problem. From this perspective, the studies have primarily focused on fungal systems, such as extracellular peroxidase and laccase from white- and brown-rot fungi. However, recent reports have suggested that bacteria play an increasing role in breaking down lignin. This paper, therefore, reviews the role of bacteria in lignin and lignin-related research. Several reports on bacterial species in soil that can degrade lignin and their enzymes are included. In addition, a cellulolytic anaerobic bacterium capable of solubilizing lignin and carbohydrate simultaneously has recently been identified, even though the enzyme involved has not been discovered yet. The assimilation of lignin-derived small molecules and their conversion to renewable chemicals by bacteria, such as muconic acid and polyhydroxyalkanoates, including genetic modification to enhance their capability was discussed. This review also covers the indirect use of bacteria for lignin degradation, which is concerned with whole-cell biosensors designed to detect the aromatic chemicals released from lignin transformation.
Keywords: lignin valorization, bacterial lignin degradation, enzymatic depolymerization, bacterial laccase, dye-decolorizing peroxidase, biosensor
Introduction
Lignin was considered a typical industrial by-product, available in pulp and paper waste, agricultural residue, and other hydrolytic industries (Gottlieb and Pelczar, 1951; Reddy and Yang, 2005; Santos et al., 2013). Specifically, large quantities of lignin were produced by biorefineries, as an alternative for industries that manufactured petroleum-based products. However, interest in lignin has dramatically increased in recent years as the demand for biomass-based products increased as a consequence of the energy crisis and the environmental problems humankind is facing.
Lignin is a complex aromatic polymer of high heterogeneity, and is attached to carbohydrate via an ester linkage within a plant to provide cell wall rigidity and protection. It comprises a considerable percentage of lignocellulose (as much as 30%) although it differs according to the type of source (Vanholme et al., 2010). Lignin includes phenolic compounds, of which there are mainly three groups, i.e., guaiacyl (G), syringyl (S), and hydroxyphenyl (H), which are polymerized via cross-linking mediated by an enzymatic radical reaction, giving rise to β-O-4 bonding dominantly and other C-C and C-O bonding (Ralph et al., 2004). Because of the intrinsic recalcitrance of lignin, it was simply regarded as waste to be removed from the pretreated biomass at the beginning of biofuel production, which is hampered by lignin-derived toxicity (Ezeji et al., 2007). To avoid this problem, even until recently, methods to recover the lignin from the pretreated liquid have mainly been reported, or strategies have been suggested to construct robust strains under lignin-derived toxic conditions (Cho et al., 2009; Lu et al., 2013; Lee et al., 2015). Nevertheless, lignin has vast potential to be used in biological systems as the second-most abundant source of aromatic compounds on the Earth. Thus, a process with a new perspective is required to not only decrease the recalcitrance of biomass but also enable lignin valorization (Ragauskas et al., 2014).
The first step in biological lignin valorization is the decomposition of the lignin polymer. The natural depolymerization of lignin by white- or brown-rot fungi, which produce extracellular oxidative enzymes, including laccase and peroxidases, has been intensively studied (Dashtban et al., 2010). Laccase can oxidize phenolic units with oxygen as an electron acceptor. The laccase from Trametes versicolor was shown to degrade the β-O-4 link within a non-phenolic lignin model (Kawai et al., 2002). In addition, treatment using the laccase within T. villosa removed lignin from wood (Gutiérrez et al., 2012). Other enzymes reactive to lignin are heme-containing peroxidases, such as lignin peroxidase (LiP), manganese peroxidase (MnP), and versatile peroxidase (VP), which needs hydrogen peroxide or a metal mediator. Phanerochaete chrysosporium has been found to be able to secrete several oxidases, and, among them, LiP has been used for a comparative analysis of other novel enzymes because of their high activity to lignin (Singh and Chen, 2008). In spite of the high catalytic efficiency of fungal enzymes, the challenges of expression are obstacles for their use in large-scale lignin treatment. In contrast, the lignin depolymerization activity of bacterial enzymes has been reported for limited cases. However, several species of bacteria capable of depolymerizing lignin have been isolated with lignin-modifying enzymes (Brown and Chang, 2014). Even more, the bacteria have a versatile metabolic pathway where lignin-derived compounds can be transformed into high-value chemicals (Jiménez et al., 2002). Reflecting this, the synthesis of polyhydroxyl-alkanoates (PHAs), for use as bioplastics, has been studied within soil bacteria, such as Pseudomas putida, using lignin-derived aromatic compounds (Linger et al., 2014).
In this review, therefore, we present research related to lignin valorization, but remain deeply focused on bacterial activity, which is covered in two sections, the first on lignin depolymerization and the second on the conversion of lignin to value-added products. It should be noted that the concept herein is to distinguish between lignin decomposition for the breakdown of polymers, and the degradation of lignin for the catabolism of lignin-derived phenolics. This includes the use of bacteria as an alternative approach to improve lignin valorization, such as the use of a bacterial sensor to screen enzymes and strains with exceptional catalytic properties.
Lignin Depolymerization to Produce Low-Molecular-Weight Phenolic Compounds
Lignin valorization by bacteria first requires lignin to be depolymerized to low-molecular-weight (LMW) phenolic compounds. Bacteria and enzymes in the bacterial strains capable of modifying lignin have been reported.
Bacterial Species Capable of Lignin Decomposition
Lignolytic microbes have been identified for years from various habitats, suggesting that they are widely distributed throughout nature, such as in soil, wood, plants, and even the gut. Among them the breakdown of lignin by bacterial strains has not been studied as widely as fungi, but several strains within the class of actinomycetes and proteobacteria are known. In general, Sterptomyces, Rhodococcus, Pseudomonas, and Bacillus strains have been reported to have lignin decomposition ability. These results are summarized in Table 1 and the details are presented below.
Table 1.
Lignin modifying bacterial strains.
| Genus | Species | Substrates | Evidences | References |
|---|---|---|---|---|
| Streptomyces | Streptomyces viridosporus T7A | Corn-stover lignocellulose | Lignin-derived APPL formation | Crawford et al., 1983 |
| Wheat straw biomass | Increased carbohydrate/lignin ratio | Zeng et al., 2013 | ||
| Streptomyces badius ATCC 39117 | 14C-labeled plant lignin | Detection of 14CO2 | McCarthy, 1987 | |
| Streptomyces coelicolor A3(2) | Grass lignocellulose | Lignin-derived APPL formation | Majumdar et al., 2014 | |
| Amycolatopsis sp. 75iv2 | Soft and hard wood | Weight loss in lignin | Antai and Crawford, 1981 | |
| Grass lignocellulose | Lignin-derived APPL formation | Brown et al., 2011 | ||
| Rhodococcus | Rhodococcus jostii RHA1 | Fluorescent lignin | Increase of fluorescence | Ahmad et al., 2010 |
| Kraft lignin | Sole carbon source for growth | Mycroft et al., 2015 | ||
| Rhodococcus erythropolis | Nitrated wheat lignin | Increase of absorbance at 430 nm | Taylor et al., 2012 | |
| Rhodococcus opacus DSM 1069 | Organosolv lignin | Sole carbon source for growth | Kosa and Ragauskas, 2013 | |
| Rhodococcus opacus PD630 | Organosolv lignin and corn stover lignin | Sole carbon source for growth | Kosa and Ragauskas, 2013; He et al., 2017 | |
| Enterobacter | Enterobacter lignolyticus SCF1 | Kraft lignin | Increased growth with lignin addition | DeAngelis et al., 2011 |
| Up-regulation of genes involved in lignin depolymerization | DeAngelis et al., 2013 | |||
| Enterobacter aerogenes | Kraft lignin | Sole carbon source for growth | Deschamps et al., 1980 | |
| Enterobacter soil sp. nov. | Kraft lignin | Sole carbon source for growth | Manter et al., 2011 | |
| Pseudomonas | Pseudomonas putida KT2440 | Alkali-pretreated liquor | Sole carbon source for growth | Salvachúa et al., 2015 |
| Pseudomonas putida A514 | Alkali insoluble lignin | Sole carbon source for growth | Lin et al., 2016 | |
| Pseudomonas putida NX-1 | Kraft lignin | Reduced particle size and guaiacyl unit decrease | Xu et al., 2018 | |
| Bacillus | Bacillus sp. | Kraft lignin | Sole carbon source for growth | Bandounas et al., 2011 |
| Bacillus pumilus | Kraft lignin | Decrease of molecular weight | Huang et al., 2013 | |
| Bacillus atrophaeus | Kraft lignin | Decrease of molecular weight | Huang et al., 2013 | |
| Bacillus sp. ITRC-S8 | Kraft lignin | Growth and decolorization | Raj et al., 2007 | |
| Bacillus ligninphilus | Alkaline lignin | Release of monomeric aromatic compounds | Zhu et al., 2017 | |
| Paenibacillus glucanolyticus | BioChoice lignin | Decreased average molecular weight of lignin | Mathews et al., 2016 | |
| Others | Thermobifida fusca | Switch grass and corn stover | Sole carbon source for growtha | Deng and Fong, 2011 |
| Clostridium thermocellum | Populus lignin | Decreased β-O-4 linkage and increased S/G index | Akinosho et al., 2017 | |
| Caldicellulosiruptor bescii | Switch grass | Release of lignin-derived aromatic compounds | Kataeva et al., 2013 |
Several Streptomyces strains are well-known for their ability to decompose lignin. Streptomyces viridosporus T7A is one of the best-studied strains, as a type of actinobacteria, resembling fungi in filamentous form and producing extracellular lignin-degrading enzymes. Crawford et al. (1983) reported that the intermediate formed from the culture of S. viridosporus T7A on corn stover lignocellulose was identified as a lignin-derived acid-precipitable polymeric lignin (APPL), enriched with phenolic hydroxyl groups. Recently, the bio-decomposition of lignin was demonstrated by more intensive chemical compositional analysis including Fourier-transform infrared spectroscopy (FT-IR), pyrolysis gas chromatography–mass spectrometry (Py-GCMS), and 13C cross polarization/magic angle spinning nuclear magnetic resonance (CP-MAS NMR) spectroscopy. The results revealed an increased carbohydrate/lignin ratio, and the deduction of the guaiacyl unit in lignin after S. viridosporus T7A cultivation on wheat straw biomass (Zeng et al., 2013). Other strains in this Streptomyces genus also showed activity toward lignin decomposition; for example, S. badius ATCC 39117 decomposed 14C-labeled plant lignin (McCarthy, 1987) and S. coelicolor A3(2) generated APPL in a medium with grass lignocellulose with the help of small laccase (Majumdar et al., 2014). Amycolatopsis sp. 75iv2, formerly S. setonii and S. griseus 75iv2, was grown on softwood, hardwood, and grass lignocellulose for 12 weeks, finally resulting in 34, 29, and 39% weight loss in lignin, respectively, which was comparable with the weight loss by S. viridosporus T7A (Antai and Crawford, 1981). The production of APPL, as shown in Streptomyces, was also reported for Amycolatopsis sp. 75iv2 on grass lignocellulose (Brown et al., 2011)
Rhodoccocus species are also an attractive strain for lignin breakdown. R. jostii RHA1, a soil bacterium, was initially identified as a polychlorinated biphenyl (PCB) degrader (Seto et al., 1995). Then, Ahmad et al. (2010) discovered its lignin-degrading activity by an assay using fluorescently modified lignin, and reported that it directly utilized kraft lignin and wheat straw lignocellulose as the sole carbon source within the medium to generate bio-products such as aromatic dicarboxylic acids and vanillin (Sainsbury et al., 2013; Mycroft et al., 2015). R. erythropolis, another PCB-degrading bacterium isolated from termite gut (Chung et al., 1994), also exhibited lignin metabolizing activity, which was detected by UV-vis assay using nitrated-lignin from wheat (Taylor et al., 2012). Moreover, the colony forming unit (CFU) of two oleaginous R. opacus DSM 1069 and PD630 on ethanol organosolv lignin as a sole carbon and energy source was increased over 300 times compared to the inoculum amount by 7 days, suggesting their capability of decomposing lignin (Kosa and Ragauskas, 2013). Although growth is slower than that of R. jostii RHA1, R. opacus PD630 was also reported to grow with alkali corn stover lignin as the only substrate (He et al., 2017).
Distinctive types of lignolytic bacteria, which are able to solubilize carbohydrate and lignin within lignocellulose simultaneously, are known. One example is Thermobifica fusca, which is an aerobic thermophile that harbors both enzymes in charge of lignin modification and cellulose hydrolysis (Wilson, 2004; Rahmanpour et al., 2016). This strain, with a heterologous expression of alcohol dehydrogenase, produced 1-propanol from untreated switch grass and corn stover biomass, implying that lignin was decomposed to enable cellulase to gain sufficient access to generate the carbon source (Deng and Fong, 2011). Clostridium thermocellum, which is an anaerobic and thermophilic organism, also have a similar function. Akinosho et al. (2017) characterized the plant cell wall after treatment with C. thermocellum and suggested that the β-O-4 linkage, which is dominantly found in lignin, was decreased and the syringyl/guaiacyl (S/G) index was increased by this strain. Caldicellulosiruptor bescii, originally isolated from a geothermally heated pool, has also attracted researchers' interest owing to its cellulytic and lignolytic activity. This bacteria was successfully grown on untreated switchgrass biomass at 78°C and released a number of lignin-derived aromatic compounds in the culture medium (Kataeva et al., 2013). Furthermore, engineered C. bescii, as T. fusca, utilized untreated biomass for ethanol production (Chung et al., 2014). These strains are noteworthy as strong lignin decomposers although they use sugars liberated from the biomass, rather than lignin, as their actual energy source.
DeAngelis et al. (2011) isolated a facultative anaerobic organism, Enterobacter lignolyticus SCF1, from rainforest soil by using a medium containing alkali-treated lignin as the sole carbon source. E. lignolyticus SCF1 was grown anaerobically on xylose, showing higher cell density by more than 2-fold with the addition of lignin, which infers the strain is capable of depolymerizing lignin. This was proven again in their succeeding research by transcriptomic and proteomic analyses, showing the up-regulation of four enzymes involved in the lignin depolymerization, such as dye-decolorizing peroxidase (DyP)-type peroxidase and glutathione S-transferase, in kraft lignin-amended sample (DeAngelis et al., 2013). Other strains E. aerogenes and E. soil sp. nov. were also isolated from soil using a culture medium containing kraft lignin as the sole carbon source. E. aerogenes is able to assimilate lignin and lignin-derived aromatics (Deschamps et al., 1980; Manter et al., 2011).
Pseudomonas strains are also attractive for use in lignin depolymerization with high potential for application in industrial biotechnology (Poblete-Castro et al., 2012; Nikel and de Lorenzo, 2018). Lignin decomposition by Pseudomonas species was first tested using a lake isolate by detecting the release of CO2 from 14C-labeled lignin (Haider et al., 1978). Several decades later, the results of many studies on lignolytic Pseudomonas strains were published, in which P. putida has been studied as an important example. The capability of P. putida KT2440 to depolymerize the lignin was indirectly determined by analyzing mcl-PHA produced from alkali-pretreated liquor (APL) with supplementation of 13C-labeled compound (Linger et al., 2014). P. putida KT2440, as well as its parental strain harboring the mega-plasmid, P. putida mt-2, was shown to decrease a significant amount of high molecular weight (HMW) component in APL by nearly 30% after seven-day culture, as measured by Klason lignin analysis and gel permeation chromatography (GPC) analysis (Salvachúa et al., 2015). Another P. putida A514, selected from among 15 P. putida strains for carbon source utilization, exhibited sufficient growth on the minimal medium supplemented with 1% alkali-insoluble lignin (Lin et al., 2016). A novel isolate, designated as P. putida NX-1, was also found to be capable of lignin modification (Xu et al., 2018). After the treatment of lignin with this strain, the morphology and chemical bonds were observed to change by using scanning electron microscopy (SEM) and FT-IR analysis, respectively, resulting in reduced particle size and decrease in the guaiacyl unit. In the case of P. fluorescens, although its lignolytic potential was reported with the identification and characterization of extracellular enzyme involved in the lignin deconstruction (Rahmanpour and Bugg, 2015), the actual decomposition of lignin has not been reported yet.
Many strains belonging to Bacillus genus are able to decompose lignin polymer. Especially, several isolates from soil, sediment, and sludge have recently been found and characterized for lignin treatment. A soil bacterium screened using kraft lignin was identified as Bacillus sp., showing 99% similarity to B. cereus and B. thuringiensis by 16S rRNA genotyping. This bacterium showed increased CFU after 72 h of culture including the HMW fraction of kraft lignin as the sole carbon source (Bandounas et al., 2011). Huang et al. (2013) also isolated several bacteria from rainforest soil in Peru, two of which, B. pumilus and B. atrophaeus, were chosen based on their high lignin-degrading activity. The GPC of kraft lignin products degraded by these two strains showed 50–70% removal of large lignin fragments that were extracted by dioxane. In the following evaluation using poplar biomass, a number of compounds were detected by LC-MS analysis, implying that lignin modification occurred to give LMW compounds. One isolate, ITRC-S8, from a sludge sample collected from the pulp industry showed efficient growth on kraft lignin and decolorization of the culture over time, and the reduction of color was well-correlated with that of kraft lignin within the medium (Raj et al., 2007). Zhu et al. (2017) published results, not only relating to the decolorization of the culture medium containing alkaline lignin but also concerning the formation of several compounds with a single phenol ring by B. ligniniphilus L1. In addition, Paenibacillus strains, formerly in the class of Bacillus, were studied to investigate the degradation of polymeric lignin (Chandra et al., 2008; Mathews et al., 2016). GPC analysis of Paenibacillus glucanolyticus, which was examined with various substrates including BioChoice lignin under anaerobic or aerobic conditions, showed that the average molecular weight of BioChoice lignin in the culture supernatant had decreased (Mathews et al., 2016).
Lignin deconstruction by Citrobacter and Klebsiella genus has been proposed, and a culture consisting of a mixture of these two was reported to improve the efficiency. Salvachúa et al. (2015) reported that C. freundii did not exhibit efficient bacterial growth on APL and lignin conversion in comparison with other strains examined, including P. putida KT2440. However, when C. freundii was co-cultured with another Citrobacter sp. isolated from sludge sample, effective decolorization of kraft lignin as much as 62% was found (Chandra and Bharagava, 2013). Moreover, high-performance liquid chromatography (HPLC) and GC-MS analyses indicated the formation of new metabolites, such as tri-, tetra-, and penta-chloro phenols. K. pneumoniae NX-1 was shown to be able to modify lignin with a 23.8% reduction in absorbance at 280 nm (A280) (Xu et al., 2018). In addition, the mixed culture of K. pneumonia and B. subtilis that was isolated from the sludge, was more efficient in terms of growth and lignin reduction (Yadav and Chandra, 2015).
Apart from this, bacterial lignin depolymerization was reported in Novosphingobium, Cupriavidus basilensis, Norcadia, Pandoraea species, among others (Haider et al., 1978; Chen et al., 2012; Shi et al., 2013; Si et al., 2018). Further, Pandoraea strains were used to assist with the pretreatment of biomass by facilitating lignin depolymerization by their lignolytic ability (Kumar et al., 2016; Liu et al., 2018).
Enzymes Involved in Lignin Depolymerization
In general, bacteria that grow in lignin secrete various oxidative enzymes that assist in lignin depolymerization or modification. Peroxidases such as fungal lignolytic peroxidases were rarely found in the bacteria. DyP-type peroxidases and laccases play an important role in bacterial lignin depolymerization (de Gonzalo et al., 2016). These are summarized in Table 2 and demonstrated below in detail.
Table 2.
Enzymes involved in lignin depolymerization.
| Enzyme | Protein | Origins | Substrates | References | ||
|---|---|---|---|---|---|---|
| Monomeric lignina | Dimeric ligninb | Polymeric ligninb | ||||
| Dye-decolorizing peroxidase | ||||||
| Subfamily A | DyP | Bacillus subtilis KCTC2023 | ABTS | VGE | – | Min et al., 2015 |
| Thermobifida fusca | ABTS | GGE | Kraft lignin | Rahmanpour et al., 2016 | ||
| Saccharomonospora viridis | 2,6-DMP | – | – | Yu et al., 2014 | ||
| Subfamily B | DyPB | Rhodococcus jostii RHA1 | ABTS | GGE | Nitrated lignin and kraft lignin | Ahmad et al., 2011 |
| DyP1B | Pseudomonas fluorescens Pf-5 | Guaiacol | – | Kraft lignin and wheat straw cellulose | Rahmanpour and Bugg, 2015 | |
| DyP | Pseudomonas putida MET94 | Syringaldehyde | GGE | Kraft lignin | Brissos et al., 2017 | |
| Klebsiella pneumoniaea | ABTS | – | – | Pfanzagl et al., 2018 | ||
| Enterobacter lignolyticus | ABTS | – | – | Shrestha et al., 2017 | ||
| Subfamily C | DyP2 | Amycolaptosis sp. 75iv2 | ABTS | GGE | – | Brown et al., 2012 |
| Streptomyces avermitilis | ABTS | – | – | Sugawara et al., 2017 | ||
| AnaPX | Anabaena sp. strain PCC7120 | Syringaldehyde | – | – | Ogola et al., 2009 | |
| Laccase | ||||||
| Laccase | Lac | Streptomyces coelicolor A3(2) | ABTS | GGE and VGE | Ethanosolv lignin | Majumdar et al., 2014 |
| Streptomyces lividans TK24 | ABTS | GGE and VGE | Ethanosolv lignin | |||
| Streptomyces vifidosporus T7A | ABTS | GGE and VGE | Ethanosolv lignin | |||
| Amycolaptosis sp. 75iv2 | ABTS | GGE and VGE | Ethanosolv lignin | |||
| Lac4 | Pantoea ananatis Sd-1 | Guaiacol | – | lignin | Shi et al., 2015 | |
| SilA | Streptomyces ipomoea | Sinapic acid | – | Kraft lignin | Moya et al., 2011 | |
| Multi-copper oxidase (Laccase-like multicopper oxidase, LMCO) | CueO | Escherichia coli | 2,6-DMP | – | PAHc | Grass and Rensing, 2001; Zeng et al., 2011 |
| Ochrobactrum sp. | ABTS | GGE and DDVA | Ca-lignosulfonate | Granja-Travez et al., 2018 | ||
| CotA | Bacillus subtilis | Sinapic acid | – | – | Ihssen et al., 2015 | |
| Bacillus licheniformis | Sinapic acid | – | – | Koschorreck et al., 2008 | ||
| Bacillus pumilus | Guaiacol | – | – | Ihssen et al., 2017 | ||
| CopA | Pseudomonas stutzeri | ABTS | – | HP-L | Strachan et al., 2014 | |
| Pseudomonas putida KT2440 | Guaiacol | GGE and DDVA | Ca-lignosulfonate | Granja-Travez and Bugg, 2018 | ||
| Pseudomonas fluorescens Pf-5 | Guaiacol | GGE and DDVA | Ca-lignosulfonate | Granja-Travez and Bugg, 2018 | ||
| Monocopper polyphenol oxidase | Tfu1114 | Thermobifida fusca | 2,6-DMP | – | Alkaline lignin and sugarcane bagasse | Chen et al., 2016 |
| Manganese-dependent superoxide dismutase | MnSOD | Sphingobacterium sp. T2 | – | – | Organosolv and kraft lignin | Rashid et al., 2015 |
Lignin derived aromatic compounds or conventional peroxidase substrates.
-, not examined in the paper.
PAH is a polymeric compound, but not lignin.
DyP-Type Peroxidases
DyP was first discovered in fungi as a unique heme-containing peroxidase, which is differentiated from plant peroxidase by the absence of a histidine residue involved in the heme-binding motif (Sugano et al., 2007). To date, more DyP-type peroxidases have been found in bacteria than fungi or other eukaryotes, and bacterial enzymes were shown to play a key role in lignin depolymerization, emphasizing its importance as bacterial peroxidase (Brown and Chang, 2014; Colpa et al., 2014). DyP-type peroxidase has been categorized into four subfamilies based on their amino acid sequence, A-D class, which is re-grouped as I, P, and V in a new classification considering structure based-sequence alignment (Yoshida and Sugano, 2015; Zamocky et al., 2015). Note that the A-C classes of DyP that are discussed in this review as DyPs in class D are primarily fungal enzymes.
DyPs in class A are reported to have a Tat-dependent secretion signal, which suggests they are involved in extracellular lignin depolymerization (Colpa et al., 2014). For example, a putative heme-containing DyP found from B. sutilis KCTC2023 (BsDyP) showed oxidative activity toward various substrates, including 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) and Reactive Blue dye (Min et al., 2015). In addition to using an oxidation indicator, BsDyP also exhibited efficient decomposition of veratrylglycerol-β-guaiacyl ether (VGE), a dimeric lignin model compound containing the β-O-4 bond. At pH 3 and 50°C, conversion of VGE to veratryl aldehyde by BsDyP was observed in the presence of hydrogen peroxide, giving rise to a final conversion rate of 53.5% of within 2 h of reaction. DyP from T. fusca (TfuDyP) is also included in class A showing periplasmic export by Tat-system (van Bloois et al., 2010). The Fraaije group evaluated the catalytic potential of TfuDyP using different classes of substrate, including flavors, lignin-derived monomers, and dyes (Lončar et al., 2016). The oxidative activity of TfuDyP toward kraft lignin and guaiacylglycerol-β-guaiacyl ether (GGE), dimeric lignin model compound, was demonstrated (Rahmanpour et al., 2016). Especially, the formation of new products, generated from the dimerization of GGE, was identified by HPLC, although TfuDyP encouraged oxidative coupling rather than cleavage of the linkage. The thermophilic actinomycete, Saccharomonospora viridis, also contains DyP-type peroxidase (SviDyP) in subfamily A (Yu et al., 2014). When tested with an azo dye, SviDyP was active in wide pH (5–10) and temperature (50–80°C) ranges, showing the highest activity at pH 7 and 70°C. With these beneficial properties, SviDyP was applied to pulp bleaching, which enhanced the brightness and changed the morphology, resulting in it being considered as a promising candidate for further industrial applications. Additional A-type DyPs were reported in Escherichia coli and Streptomyces (Sturm et al., 2006; Liu et al., 2011; de Gonzalo et al., 2016).
Unlike the A subfamily, DyPs in the B (as well as the C) class do not have disclosed signal peptides for secretion, inferring that they are in the cytoplasmic phase of the cell (Colpa et al., 2014). Although the possibility of export has been proposed with the assistance of encapsulin, which is co-transcribed with dypB, in Rhodococcus strains, it still remains unclear (Tamura et al., 2015; Putri et al., 2017). Nevertheless, B-type peroxidase has been prominently studied owing to its versatile and sensitive activity. Ahmad et al. (2011) identified significant reduction in lignin deconstruction by a R. jostii RHA1 mutant carrying dypB deletion in comparison to the wild type strain, which emphasizes the role of DyPB in lignin breakdown. Based on the finding, characterization of the RjDyPB mutant having N246A was conducted with several lignin-related substrates including nitrated lignin, kraft lignin, and GGE as β-aryl ether compound, showing high oxidation activity in the colorimetric assay. In addition to the colorimetric assay, HPLC and GC-MS analyses showed the release of syringaldehyde as a deconstructed product from kraft lignin. Furthermore, its capacity was significantly enhanced to provide a 5.4–23-fold increase in the presence of MnCl2 (Singh et al., 2013). P. fluorescens Pf-5 was found to contain three DyP-type peroxidases, i.e., two DyPB and one DyPA (Rahmanpour and Bugg, 2015). Among these three enzymes, the best action was reported with DyP1B, demonstrating activity against all the substrates that were used. In particular, the oxidation of Mn(II) and kraft lignin was observed only by DyP1B. Moreover, in the treatment of wheat straw lignocellulose with DyP1B, the release of a lignin fragment was identified by HPLC. MS data suggested that the fragment was consistent with a compound containing two-aryl C3 units, which were a G and H units, respectively. Another important B-type DyP was found in P. putida MET94. In Santos's report, PpDyP exhibited higher activities and a wider scope of substrates, including phenolic and non-phenolic lignin units, compared with BsDyP (Santos et al., 2014). Later, Marins's group conducted an extensive study on PpDyP to characterize its integrated redox and catalytic properties and the construction of mutants for increased activity (Mendes et al., 2015; Brissos et al., 2017). The directed evolution of PpDyP generated the 6E10 variant carrying three mutations, E188K, A142V, and H125Y. This mutant showed noticeably improved efficiency in the oxidation of a lignin-related source, such as syringyl-type phenolics, GGE and kraft lignin, and displayed 100-fold higher catalytic efficiency toward 2,6-dimethoxyphenol (DMP) (Brissos et al., 2017). Another meaningful feature in this mutation is that this variant was found to preferably react under alkaline conditions, pH 8.5, whereas the wild-type enzyme and typical B-type DyP prefer an acidic (pH 4–5) environment. Recently, it has been reported that Asp-143 and Arg-232 residues of B-type DyP from K. pneumoniaea (KpDyP) and E. lignolyticus (ElDyP) play an important role in the decomposition of hydrogen peroxide by the enzyme structure analysis (Shrestha et al., 2017; Pfanzagl et al., 2018). Analysis of enzymes based on the structure will be helpful in rational design of enzymes for lignin depolymerization.
A limited number of enzymes have been characterized for DyP in the C subgroup, of which representatives are known from the genomes of proteobacteria, actinobacteria and cyanobacteria (Zamocky et al., 2015). Nevertheless, a strong peroxidase, DyP2, was identified within Amycolatopsis sp. 75iv (Brown et al., 2012). The activity of DyP2 was determined with ABTS, two dyes, and a manganese cation. Here, DyP2 also showed high activity as a Mn-peroxidase, which was comparable with that of fungal Mn-peroxidase. The breakdown of GGE also occurred in the presence of DyP2 and hydrogen peroxide, proving its capability of lignin depolymerization. The effect of DyP2 on lignin was also verified by heterologous expression within the P. putida strain. The overexpression and secretion of DyP2 within P. putida A514 showed ~2-fold higher CFU than the wild-type strain when they were cultivated on lignin as the sole carbon source, suggesting DyP2 was involved in the depolymerization of the lignin substrate (Lin et al., 2016). In 2009, Ogola et al. (2009) published their study of a cyanobacterium mobilizing C-type peroxidase, which is in the Anabaena sp. strain PCC7120. This heme-dependent peroxidase, AnaPX, showed substantial activity on phenolic compounds, such as guaiacol and pyrogallol, and this was significantly increased 50-fold by the redox mediator syringaldehyde. Recently, another bacterial DyP within the C class was discovered from Streptomyces avermitilis, following AnaPX and DyP2 (Sugawara et al., 2017). Even though SaDyP2 was found to be active on a typical oxidation indicator, further research would be required to determine the actual activity of lignin modification.
Laccase and Laccase-Like Multicopper Oxidase (LMCO)
The use of laccase, multicopper-containing oxidase, has been proposed for a wide range of bio-industrial uses, including bioremediation, lignin modification, and bio-bleaching of pulp and even biofuel cells (Arias et al., 2003; Beneyton et al., 2011; Zheng et al., 2012; Chandra and Chowdhary, 2015; Wang et al., 2015). In fact, fewer types of bacterial laccase are known and the efficiency in their wild type is not as high as that of the fungal enzyme. However, laccases from bacterial species have some advantages over those from fungi, such as thermo- and halo-tolerance, resulting in their consideration as suitable catalyst for industrial applications (de Gonzalo et al., 2016).
As a representative of lignin depolymerizing bacteria, Streptomyces species have been known to possess laccase (Fernandes et al., 2014). Based on the discovery of laccase within the strain, some strategies have been suggested to produce laccase at high yield within the culture. One strategy suggested by Niladevi et al. (2008) was to use a laccase inducer. By examining several aromatic compounds, pyrogallol was chosen as the best aromatic inducer to produce laccase from S. psammoticu at the level of 116 U/g, which was an ~2-fold increase compared to the control. The use of 1 mM of pyrogallol, at the optimal condition, was able to enhance the production yield up to 3.9-fold higher level. Within the culture of two Streptomyces strains, S. lavendulae and S. cinnamomensis, the optimization of the medium formula was conducted for the simultaneous production of laccase and lignin peroxidase. The use of an optimal formulation was able to enhance the production and control the ratio of laccase:LiP activity as well (Jing, 2010; Jing and Wang, 2012).
Similar to the studies of DyP, in many studies laccase was characterized not only by an oxidation indicator, such as ABTS and a variety of dyes, which suggested their potential as lignin modification enzymes, but also by lignin-related compounds or lignin to show their capability of lignin depolymerization directly. From a genomic analysis of Pantoea ananatis Sd-1, four putative laccase were identified, designated as Lac1-Lac4. Among them, only Lac4 was found to be able to oxidize phenolic compounds at acidic pH 2.5 (Shi et al., 2015). The ability of Lac 4 to decompose lignin was also evaluated by an in vitro assay using lignin. With the addition of ABTS as a mediator, 38% of lignin was observed to undergo degradation, and GC-MS analysis revealed the generation of LMW aromatic compounds, including 1,4-benzenedicarboxaldehyde, benzenepropanoic acid and phenol, which are considered to be the basic phenolic units possibly found in natural lignin. Laccase from S. ipomoea was also characterized with lignin-related compounds, such as p-coumaric acid, ferulic acid, and kraft or organosolv lignin prepared from softwood and spruce (Moya et al., 2011). In this study, the polymerization of lignin and lignan was also observed to show the reactivity of this laccase. Small laccase (SLac) is differentiated from typical laccase by containing only two domains for Cu binding rather than three, resulting in a smaller size (Machczynski et al., 2004). Majumdar reported four SLac from S. doelicolor A3(2), S. lividans TK24, S. viridosporus T7A, and Amycolatopsis 75iv2 (Majumdar et al., 2014). Initially, a mutant strain deficient of the SLac encoding gene was used to investigate its role in lignin decomposition. The generation of APPL only in the wild-type strain confirmed that SLac governs the lignin process. Then, all four enzymes were assessed for their activity toward lignin by using lignin model compounds with a β-aryl ether structure and ethanosolve lignin. Analysis of the product showed that one compound was identified as vanillin at A340, providing evidence for the laccase-related lignin modification.
Several examples of laccase-like multicopper oxidase (LMCO) have been reported within the bacterial strains. In 2001, a multicopper oxidase, CueO, was found in Escherichia coli, which confers tolerance to copper-induced damage (Grass and Rensing, 2001). Then, its oxidative activity was investigated using 2,6-DMP, a typical substrate for laccase, and polycyclic aromatic hydrocarbon (PAH), but the activity toward the lignin-related compound was not determined (Grass and Rensing, 2001; Zeng et al., 2011). Instead, CueO from Ochrobactrum sp. showed activity toward β-aryl ether and biphenyl lignin as model compounds of lignin (Granja-Travez et al., 2018). In the assay of oxidizing lignosulfonate, the generation of vanillin was observed as lignin-deconstructed fragment. Structural analysis showed similarity with CueO from Escherichia coli. Another LMCO that is well-studied is CotA from Bacillus strains. CotA, which is the protein originally related with the outer spore coating, was found to have similarities with multicopper oxidase, especially in terms of structural features, suggesting that CotA is a laccase (Hullo et al., 2001). Based thereupon, the oxidation of CotA from B. subtilis was evaluated with syringaldazine and several dyes to demonstrate its laccase activity (Hullo et al., 2001; Wang et al., 2016). Sufficient activity was observed at pH 7.2, even exhibiting high tolerance to organic solvents, including acetone, ethyl acetate, and chloroform. Then, B. subtilis was also examined with 45 aromatic chemicals, including 13 aromatic carboxylic acids and aromatic aldehydes, which are all lignin-related compounds, and displayed considerably high activity surpassing that of enzymes from Streptomyces (Ihssen et al., 2015). CotA from B. licheniformis and B. pumilus was also demonstrated to be an enzyme suitable for lignin modification via the oxidation of lignin-derived phenolics, such as guaiacol and syringic acid (Koschorreck et al., 2008; Ihssen et al., 2017). In the case of a B. pumilus CotA, mutant carrying L386W/G417L/G57F (abbreviated as WLF) was constructed by site-directed mutagenesis to enhance the catalytic efficiency, and a method was established for the efficient expression of WLF within Escherichia coli, which renders it a suitable candidate for industrial applications (Chen et al., 2017; Luo et al., 2018).
Recently, multicopper oxidase was identified in Pseudomonas genus. CopA from P. stutzeri was found by functional screening using a biosensor developed to respond to monomeric phenolics (Strachan et al., 2014). Further evaluation showed that industrially purified lignin incubated with CopA in the presence of Cu(II) released newly formed compounds, including 2,6-dimethoxybenzen-1,4-diol and 4-hydroxy-3methoxybenzoic acid, detected by GC-MS. P. putida KT2440 and P. fluorescens Pf-5 have been elucidate to retain CopA-type multicopper oxidase (Granja-Travez and Bugg, 2018). These researchers conducted comprehensive studies to determine the CopA involvement in lignin oxidation. They oxidized the lignin model compounds, GGE and 5,5′-dehydrodivanillate (DDVA) to obtain the dimerized compound. The reaction of lignosulfonate led to the production of vanillic acid. In addition, P. putida KT2440 mutated by deleting copA genes showed reduced growth on aromatic compounds. All these results suggested that CopA is able to act as laccase.
A different type of oxidase was found in T. fusca and Sphingobacterium sp. T2, showing activity toward lignin modification similar to that of laccase. Chen et al. (2016) reported a monocopper polyphenol oxidase identified within T. fusca. In addition to an oxidation test with 2,6-DMP, oxidative activity was determined with alkaline lignin and bagasse. The treatment of alkaline lignin with this oxidase led to a decrease in the total phenolic content, as confirmed by spectral scanning and size exclusion chromatography analysis. Moreover, modification of the lignin structure in bagasse was observed by LC-MS. The other is a bacterial manganese-dependent superoxide dismutase within Sphingobacterium sp. T2, which was found in proteomic analysis (Rashid et al., 2015). Two enzymes, MnSOD1 and MnSOD2, were both shown to be active toward organosolv and kraft lignin as well as the lignin model compound. The reaction generated several oxidized products, some of which were found to be formed by the cleavage of aryl-Cα and Cα-Cβ linkange such as dihydroxybenzoic acid, 5-hydroxyvanillic acid, and 2-methoxyhydroquinone.
From Low-Molecular-Weight Lignin Product to Bioproducts
Catabolic Pathways for Aromatic Compounds Within Bacteria
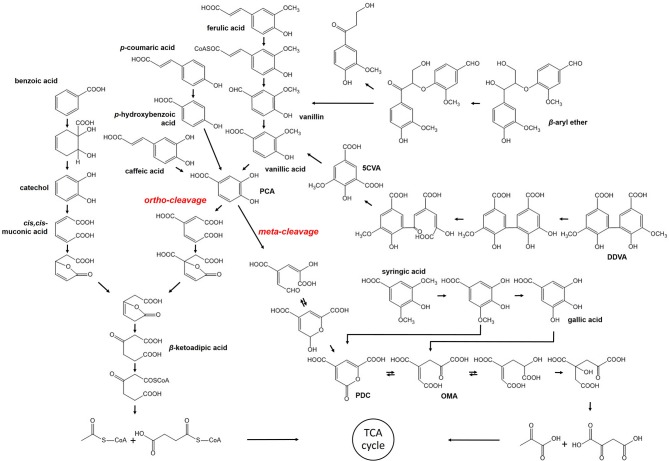
Most of the bacterial strains mentioned in the previous section have also been found capable of degrading lignin-derived oligomers or aromatic monomers, except for cellulolytic thermophiles. These strains converted high-molecular-weight polymers to compounds of lower molecular weight, followed by uptake to provide a source of energy. Several pathways within bacteria, in which LMW or monomeric compounds are digested via stepwise reaction to produce the important intermediate, such as protocatechuate (PCA) and catechol, have been discussed, even though they are not yet completely understood (Figure 1).
Figure 1.
Bacterial pathway for degradation of lignin-derived compounds.
β-aryl ether such as GGE is a dimeric compound that contains the β-O-4 linkage, which is dominantly found in the lignin structure up to ~50% (Masai et al., 1999; Zakzeski et al., 2010). Therefore, cleavage of the β-aryl ether is significant in the degradation of lignin-derived LMW compounds and observed in several lignolytic strains, including Sphingomonas paucimobilis, Delftia acidovorans, Rhodococcus equi, and so on (Masai et al., 2007). The metabolism has been well-characterized in S. paucimobilis SYK-6 (Masai et al., 2007). The β-aryl ether is catabolized to vanillic acid by NAD-dependent dehydrogenase, LigD, and glutathione-dependent β-esterase, LigEFG, which is coupled with the reduction of glutathione (Gall et al., 2014a,b). After oxidation of the compound, ether cleavage takes place as a stepwise reaction. In the first step, LigE or LigF are involved in the reduction of glutathione by nucleophilic attack, but their different chirality preferences lead them to attack the opposite enantiomers (Masai et al., 2003). Additionally, LigG was found to have no esterase activity. Nevertheless, LigG is essential in this cleavage reaction as it is solely able to remove glutathione from β-thioether intermediate. The product formed by β-esterase is then metabolized to vanillic acid, which is further degraded by another pathway described below.
The biphenyl structure is also included in polymeric lignin. Fortunately, the mineralization of biphenyl compounds has been extensively studied within bacterial strains capable of degrading PCBs (Pieper, 2005). The conversion of DDVA, a model of the different types of biphenyl, was also reported to take place via a catabolic pathway within S. paucimobilis SYK-6 (Peng et al., 2002). This reaction is initiated by O-demethylation with LigX, and followed by ring opening via meta-cleavage of dioxygenase, LigZ. Finally, the two units are split, producing 5-carboxyvanillate. Subsequently, a decarboxylation reaction occurs to convert 5-carboxyvanilliate to vanillic acid, which is catalyzed by LigW. Peng et al. (2005) reported the identification of LigW2 as the second decarboxylase. Demonstration of the growth of S. paucimobilis SYK-6 on DDVA led to the finding that ligW2 contributes to the conversion of 5-carboxyvanilliate as much as ligW. In addition to S. paucimobilis, the DDVA catabolic pathway was found in P. putida, P. fluorescens, S. viridosporus, and several soil bacteria (Crawford et al., 1981).
The degradation of lignin-derived monomeric compounds is known to occur via two different pathways and they are well-explained with S. paucimobilis SYK-6 and P. putida KT2440, respectively. In the case of ferulic acid, a precursor for lignin synthesis, the catalyzing enzymes are identical. Feruloyl-CoA synthetase starts the conversion of ferulic acid, encoded by ferA in S. paucimobilis and fcs in P. putida (Jiménez et al., 2002; Masai et al., 2007). Then, feruloyl-CoA is converted to vanillin with the help of FerB of S. paucimobilis or Ech of P. putida, which is further converted to vanillic acid via vanillin dehydrogenase. The feruloyl-CoA synthetase also catalyzes the transformation of p-coumaric acid and caffeic acid, but the following reactions produced p-hydroxybenzoic acid and PCA, respectively. The important intermediate along the degradation pathway is PCA, converged from most of the lignin-derived phenolics discussed in this section, including the conversion of vanillin produced by the cleavage of the dimeric compound. However, PCA proceeds through the different cleavage reactions within two strains. First, in S. paucimobilis SYK-6, PCA is converted by meta-cleavage instigated by 4, 5-dioxygenase, LigAB, and further degraded by LigCIJK to finally generate oxaloacetate and pyruvate (Masai et al., 2007). In contrast, ortho-cleavage occurs during the conversion of PCA to muconate within P. putida KT2440, which is catalyzed by the 3,4-dioxygenase, PcaGH (Jiménez et al., 2002). According to Bugg's report, more bacteria use the ortho-cleavage pathway (Bugg et al., 2011). P. putida KT2440 provides acetyl-CoA and succinyl-CoA as the final product via β-ketoadipate in which gene products from pcaBCDIJF are involved. Therefore, the pathway for aromatic compound catabolism in P. putida KT2440 is referred to as the upper β-ketoadipate pathway, which is observed in many other soil bacteria as well, such as Acinetobacter sp. ADP1, Burkholderia sp., and some other Pseudomonas strains (Jiménez et al., 2004; Fischer et al., 2008). This pathway covers the degradation of benzoic acid in another branch via catechol, the downstream degradation of which also forms β-ketoadipate.
Conversion of Lignin to Biochemicals
Various soil bacteria are able to degrade aromatic compounds via the β-ketoadipate pathway and produce renewable chemicals (Table 3). Within this pathway, several intermediates that are practically useful can be found, including vanillin, which is widely used in the food industry as flavor additive (da Silva et al., 2009). Sainsbury's report suggested the production of vanillin from lignin by engineered R. jostii RHA1 (Sainsbury et al., 2013). This strain was constructed by deleting vanillin dehydrogenase, catalyzing the conversion of vanillin to vanillic acid, to block the downstream reaction and accumulate vanillin. When the mutant was cultivated with the minimal media containing both glucose and wheat straw biomass, vanillin was produced with 96 mg/L of titer. The pathway within R. jostii RHA1 was also manipulated to generate the aromatic carboxylic acid, which could be a source for the production of bio-plastic (Mycroft et al., 2015). PCA is normally cleaved by 3,4-dioxygenase (ortho-cleavage) and converted to a muconic acid compound (Jiménez et al., 2002). However, the conversion was re-routed to a meta-cleavage by insertion of 2,3-dioxygenase from Paenibacillus sp. JJ-1b or 4,5-dioxygenase from S. paucimobilis, which finally gave rise to pyridine 2,5-dicarboxylic acid (2,5-PDCA) and 2,4-dicarboxylic acid (2,4-PDCA), respectively. One percent wheat straw lignocellulose within the culture was transformed to generate maximally 125 mg/L of PDCA. Re-routing the cleavage of PCA and catechol introducedinto P. putida KT2440 enabled the production of pyruvate to be accelerated (Johnson and Beckham, 2015). In this case, catechol was catabolized by the 2,3-meta-cleavage to replace succinate with pyruvate as the product. Furthermore, PCA cleavage by 4,5-dioxygenase from S. paucimobilis served to generate two molecules of pyruvate. Finally, two substrates, p-coumaric acid and benzoic acid, which are degraded via PCA and catechol, respectively, were utilized by this engineered strain, and converted to pyruvate to increase the yield 5-fold. Integration of the lactate dehydrogenase A encoding gene from B. Taurus, finally led to the production of L-lactate from pyruvate to obtain a yield of 41.1%.
Table 3.
Lignin-based chemical production.
| Strains | Genotype | Substrates | Titers | References | |
|---|---|---|---|---|---|
| Vanillin | Rhodococcus jostii RHA045 | R. jostii RHA1 derivative, Δvdh | Wheat straw cellulose | 96 mg/L | Sainsbury et al., 2013 |
| Pseudomonas putida KT2440 | P. putida KT2440 derivative, Δupp ΔPP_0166-0168 Δvdh ΔPP_3827-3832 ΔPP_2680 ΔPP_0545 ΔPP_1948 Ptac::ech:fcs | Ferulic acid | 1.3 g/L | Graf and Altenbuchner, 2014 | |
| Lipid | Rhodococcus opacus DSM1069 | Wild type | O2-treated kraft lignin | 67 mg/L | Wei et al., 2015 |
| Ultrasonicated EOL | 4 mg/ga | Kosa and Ragauskas, 2013 | |||
| Rhodococcus opacus PD630 and Rhodococcus jostii RHA VanA- | Wild type | Alkali-extracted lignin (from corn stover) | 330 mg/gb | He et al., 2017 | |
| Rhodococcus opacus PD630 | Wild type | Laccase-treated kraft lignin | 145 mg/L | Zhao et al., 2016 | |
| PHA | Pseudomonas putida KT2440 | Wild type | Alkaline pretreated liquor (APL) | 252 mg/L | Linger et al., 2014 |
| Pseudomonas putida ADVJ4C1 | P. putida A514 carrying pTDV and pGJ4C1 | Biorefinery waste | 161 mg/L | Lin et al., 2016 | |
| Pseudomonas putida Axyl_alkKphaGC1 | P. putida A514 carrying pTPxylAphaGC1 | Vanillic acid | 246 mg/L | Wang et al., 2018 | |
| Pandoraea sp. ISTKB | Wild type | 4-hydroxybenzoic acid | 409 mg/L | Kumar et al., 2017 | |
| Cupriavidus basilensis B-8 | Wild type | Kraft lignin | 128 mg/L | Shi et al., 2017 | |
| Alkaline pretreated liquor (APL) (from Rice straw) | 450 mg/L | Si et al., 2018 | |||
| Ralstonia eutropha H16 | Wild type | 4-hydroxybenzoic acid | 435 mg/L | Tomizawa et al., 2014 | |
| Muconic acid | Pseudomonas putida KT2440-JD1 | P. putida KT2440 derivative, randomly mutated on catR, R50C | Benzoic acid | n.m. | Van Duuren et al., 2011 |
| Pseudomonas putida KT2440-CJ103 | P. putida KT2440 derivative, ΔpcaHG::Ptac:aroY,ΔcatRBC::Ptac:catA:dmpKLMNOP | p-coumaric acid | 13.5 g/L | Vardon et al., 2015 | |
| Base-catalyzed depolymerized (BCD) lignin liquor | 0.5 g/L | Rodriguez et al., 2017 | |||
| Pseudomonas putida KT2440-CJ184 | P. putida KT2440 derivative, ΔpcaHG::Ptac:aroY:ecdB:ecdDΔcatRBC::Ptac:catA | p-coumaric acid | 15.6 g/L | Johnson et al., 2016 | |
| Pseudomonas putida MA-9 | P. putida KT2440 derivative, ΔcatBC ΔendA-1 ΔendA-2 Pcat::catA:catA2 Pgro-4::dmpKLMNOP | Hydrothermal depolymerized pine lignin | 13 g/L | Kohlstedt et al., 2018 | |
| Amycolaptosis sp ATCC 39116 MA-2 | Amycolaptosis sp ATCC 39116 derivative | Guaiacol | 3.1 g/L | Barton et al., 2018 |
mg lipid/g us-EOL.
mg lipid/g CDW.
n.m., not mentioned in the paper.
Polyhydroxyalkanoate (PHA) is another promising material applicable to the production of bioplastics in various ways. Therefore, the conversion of lignin to PHA has been regarded as a valuable reaction. Linger et al. (2014) evaluated the production of PHA by P. putida KT2440 by studying monomeric lignin model compounds and APL in which these compounds are possibly present. They confirmed that a model culture consisting of a heterogeneous mixture containing p-coumaric acid, ferulic acid, glucose, and acetic acid was able to assimilate all the substrates within 48 h, except for ferulic acid, and 34% of PHA was accumulated. Moreover, the use of actual APL as substrate led to the accumulation of 32% of PHA. In this study, mcl-PHA produced from APL was converted to hydrocarbon fuels, demonstrating the valorization of lignin. P. putida A514 was also reported to generate PHA from vanillic acid and kraft lignin (Lin et al., 2016; Wang et al., 2018). A combination of several strategies was employed within the strain to enhance the yield of mcl-PHA, which includes overexpression of heterologous DyP and vanillin dehydrogenase to facilitate the utilization of lignin and lignin-derived compounds (Lin et al., 2016). The final mutant strain was able to accumulate PHA using biorefinery waste as the sole substrate, achieving 160 mg/L under N-source limited conditions. When the transcription level was optimized within P. putida A514 by a strong promoter with its regulator protein induced by xylose, PHA production from vanillic acid reached 246 mg/L, which is the highest record among the reported titer (Wang et al., 2018). Apart from the aforementioned work, PHA production from lignin within other bacterial strains has also been investigated. For example, Tomizawa et al. (2014) screened a bacterial strain that has both the ability to accumulate PHA and to efficiently utilize the lignin-derived metabolite, which is Ralstonia eutropha H16. PHA production was determined within Pandoraea sp. ISTKB as well with the lignin model compounds and kraft lignin. The use of p-hydroxybenzoic acid as the substrate resulted in the highest PHA accumulation, which was consistent with the amount of biomass (Kumar et al., 2017). Moreover, genomic and proteomic analyses were implemented, and enabled the two important gene clusters involved in lignin degradation and PHA polymerization to be identified (Kumar et al., 2018). The production of PHA by Cupriavidus basilensis B-8 from kraft lignin and lignin fragmentation was demonstrated by SEM and GPC analyses (Shi et al., 2017).
The availability of P. putida KT2440 in the bio-production of value-added chemicals was one again demonstrated with the generation of muconic acid. The first study on the accumulation of cis, cis-muconic acid (MA) was published in 2011, where a mutant was generated by chemically induced mutation (Van Duuren et al., 2011). This variant strain, which was shown to grow on benzoate only in the presence of glucose, was able to produce MA from benzoate, which was derived from deficient CatR, the transcriptional regulator of the cat operon responsible for catechol degradation; in addition, the strain induced the expression of catA2, of which the gene product is the second catechol 1,2-dioxygenase, within the ben operon for the conversion of benzoate to catechol. Subsequently, an effort to produce MA was reported with the acceptance of a wide range of compounds (Vardon et al., 2015). In this report, the compound degraded via the PCA branch of the β-ketoadipate pathway was funneled to MA by eliminating PcaHG for the ring cleavage of PCA and by re-routing to catechol with insertion of the PCA-decarboxylate encoding gene. This engineered strain, CJ103, was used to accumulate MA from p-coumaric acid and depolymerized corn stover (Vardon et al., 2015; Rodriguez et al., 2017). Additionally, the MA synthesized within P. putida was purified and further converted to adipic acid and nylon by chemical hydrogenation and polymerization (Vardon et al., 2015; Kohlstedt et al., 2018). Recently, upgraded MA production was reported with a P. putida KT2440-derived mutant, where the co-expression of AroY and its cofactor EcdBD accelerated the conversion of PCA to catechol (Johnson et al., 2016). A different access method using the metabolically engineered Amycolatopsis sp. ATCC 39116 was reported to produce MA by the metabolism of guaiacol-based lignin monomer (Barton et al., 2018). Deletion of the genes producing putative muconate cycloisomerase led to the synthesis of as much as 3.1 g/L MA from guaiacol. MA was also obtained with this mutant from guaiacol-enriched lignin hydrolysate prepared by the hydrothermal conversion of pine, thereby demonstrating its potential to achieve valorization.
As a biofuel, lipids are in demand for bioconversion by employing a bacterial strain. The Rhodococcus strain harbors an aromatic compound catabolic pathway via β-ketoadipate and also shows preference for acetyl-CoA as a substrate for fatty acid synthesis, which can be derived via this catabolic pathway (Kosa and Ragauskas, 2012). Two oleaginous Rhodoccocus, R. opacus DSM 1069 and PD630, efficiently utilized a variety of lignin model compounds, pretreated lignin, and whole lignocellulosic biomass for lipid production (Kosa and Ragauskas, 2013; Wang et al., 2014; Wei et al., 2015). The co-fermentation of two strains, R. opacus PD630 and R. jostii RHA1 showed improved growth on lignin and lipid generation with a synergetic effect (He et al., 2017), as demonstrated for the enhanced depolymerization within the co-culture of Citrobacter strains (Chandra and Bharagava, 2013).
Lignin valorization to produce useful chemicals still has unlimited potential by the versatile pathways within bacteria. Several reports have described the chemical reactions for lignin-derived phenolic adhesive and resin, and the synthesis of polyurethane foam (Jin et al., 2010; Ramires et al., 2010; Pan and Saddler, 2013). The synthesis of divanillin-derived polyurethane by a chemical catalyst has also been reported, and the formation of divanillin with the bacterial enzyme, TfuDyP, has been identified (Lončar et al., 2016; Gang et al., 2017). The possible replacement of these chemical reactions by bacterial systems would enlarge the market for lignin conversion.
Bacterial Biosensors for Lignin Transformation
As mentioned before, the bioconversion of lignin to other chemicals has been achieved by using various cells or mutant strains. Although enzymes and cells related to lignin valorization are widely distributed in nature, only a fraction of them have been studied thus far. In addition, the low lignin transformation activity of these enzymes and strains is a major limitation for the industrial application of lignin. Transcriptional regulator-based whole cell biosensors are one of the most powerful tools to screen strains or enzymes capable of efficient lignin conversion, and would be a good choice to overcome this limitation. These microbial cell-based biosensors have been applied to numerous fields, including environmental analysis and medical diagnosis (Gu et al., 2004; Lee et al., 2013; Gui et al., 2017).
In recent years, bacterial biosensors used for the detection of lignin transformation were generally constructed by the fusion of fluorescence or luminescence reporters with promoters or transcription factors such that they were responsive to lignin-derived aromatic compounds (Figure 2). For example, two Escherichia coli biosensors were developed to exhibit a luminescent response to aromatic compounds. Here, the promoter regions of inaA and the aaeXAB operon encode the pH-inducible protein involved in stress responses and the p-hydroxybenzoic acid efflux system, respectively. These two regions, which were selected by transcriptional analysis, were fused with luciferase genes, luxCDABE (Lee et al., 2012). The luminescence increased when lignin-derived aromatic compounds, such as ferulic acid, p-coumaric acid, and vanillin, were added; however, sugar-derived furfural and HMF, which are also present within the pretreated lignocellulose, did not induce the luminescent signal (Figure 2A). These biosensors were used to determine the toxicity within pretreated lignocellulosic biomass with the aim of providing a non-inhibitory substrate for the biofuel production, as shown in the examples of uses (Lee and Mitchell, 2012; Monnappa et al., 2013). Another E. coli sensor was established by Hallam's group to detect mono-aromatic compounds resulting from lignin depolymerization. The sensing module “PemrR-GFP” was constructed by expressing GFP under the control of the EmrR promoter from the emrRAB multidrug efflux system operon (Figure 2B). This sensor was subsequently used to screen and identify bacterial strains containing the lignin transforming phenotype. The work ultimately led to the discovery of a novel multicopper oxidase within P. stutzeri (Strachan et al., 2014). Ho et al. (2017) modified this sensor, which enabled them to increase the sensitivity and selectivity of its fluorescent response by modeling analysis. Employing this sensor to screen the enzymes in a high-throughput system successfully screened 147 clones that were able to convert kraft lignin to vanillin and syringaldehyde, from 42,520 fosmid clones.
Figure 2.
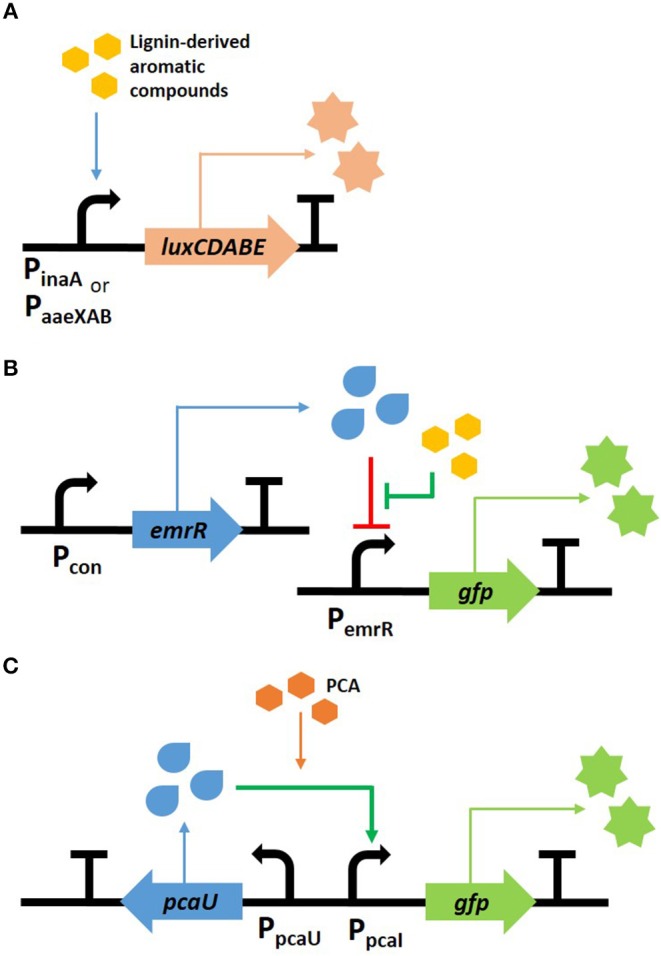
Scheme of biosensors for phenolic compound detection. Two E. coli sensors were developed to detect aromatic compounds using luminescent (A) and fluorescent (B) genes, and a fluorescent P. putida sensor was developed to respond to PCA (C).
Rhodococcus and Pseudomonas, which are lignin transforming strains, were used to construct a bioreporter strain, thereby expanding their uses for lignin conversion (DeLorenzo et al., 2017; Jha et al., 2018). Several biosensors derived from R. opacus PD630 were developed to detect the intracellular nitrogen level or aromatic metabolites. In particular, a nitrogen sensor, which was constructed using the promoter region of LPD03031 gene encoding a putative nitrite extrusion protein, was able to provide useful information for lipid production from lignin. This is because R. opacus prefers low nitrogen conditions for increased TAG synthesis (DeLorenzo et al., 2017). Unlike the other sensors discussed here, which respond to various aromatic compounds, the P. putida KT2440 sensor is unique in that it only reacts toward protocatechuate (PCA), the central intermediate found in the catabolic pathway of lignin-derived aromatic compounds (Jha et al., 2018). The transcription factor PcaU from A. baylyi ADP1 was fused with superfolder GFP (sfGFP) as a reporter protein (Figure 2C), and evolved by modulating the protein-DNA interaction within the promoter and the protein-protein interaction of the dimer. The optimal sensor, indicated as PcaU 1.2, showed a dose-dependent response to PCA. The biosensor module was introduced into mutant cells with various genetic backgrounds and the intracellular accumulation of PCA from 4-hydroxybenzoic acid was evaluated. The strain showing the highest fluorescence was deficient of PcaHG, in which case the accumulated PCA cannot be further metabolized; furthermore the other strain from which PobA was deleted resulted in the reaction with the lowest yield because 4-hydroxybenzoic acid is not converted to PCA.
Biosensors have several benefits over physico-chemical analysis, such as a fast and sensitive response, simple preparation, and in situ detection, which makes these sensors useful tools in the field of lignin valorization. Accordingly, the development and utilization of additional diverse biosensors for lignin-derived aromatic compounds are foreseen.
Challenges and Perspectives in Bacterial Lignin Valorization
Although lignin transformation in biorefinery has promised advanced process for the production of value-added chemicals as the alternative for a petroleum-based product, the structural complexity of lignin has made its utilization rate low. To enable the lignin valorization, the huge efforts have been made into each step, such as lignin characterization, biomass pretreatment, isolation of depolymerized compounds, and catalytic or thermochemical conversion, which are well-discussed in recent literature studies (Beckham et al., 2016; Cao et al., 2018; Ponnusamy et al., 2018; Wang et al., 2019).
In this review, we suggested that biological lignin valorization, in particular by bacterial microorganisms, could be a breakthrough for the efficient lignin utilization. As stated previously, bacteria show a wide spectrum of the aromatic substrate utilization or resistance, high adaptability in new environments, and easy genetic manipulation. Moreover, many researchers have discovered various lignolytic enzymes and catabolic pathway of aromatic compounds in bacterial cell, all which could facilitate the conversion of lignin to bio-chemicals in a single strain. The simultaneous lignin depolymerization and chemical production, called “consolidated bioprocessing,” has been proposed and the capability of bacterial strain has been proved (Linger et al., 2014). However, for the industrial feasibility of this bioprocessing of lignin, we still face several challenges needed to be solved, with largely two objectives: 1) efficient lignin depolymerization to supply considerable LMW lignin-related carbon substrate and 2) a significant increase in yield of value-added product from aromatic compounds.
Despite the discovery of various lignolytic enzymes, the enzyme research is still needed for the efficient extracellular decomposition of HMW to LMW lignin by bacterial strain. Especially, for the consolidated process, the enzyme is necessary to exhibit high catalytic efficiency in a mild condition, like as in the culture medium, although the most of the enzymes known are active in acidic condition (Janusz et al., 2017). Protein engineering will allow us to obtain improved lignin-decomposing enzyme. A variant of PpDyP could be an example, of which the activity toward kraft lignin at alkaline pH was enhanced by changing amino acids in the protein sequence (Brissos et al., 2017). Further discovery of novel efficient enzymes is also needed from the lignin-related environment with the aid of high-throughput sequencing and screening technology. Another issue on bacterial lignin depolymerization is that few secretion systems have been known. Several enzymes discussed in section Enzymes Involved in Lignin Depolymerization were reported as extracellular, but their secretion mechanisms involved were not fully studied, which makes it difficult to be engineered. Developing the enzyme secretion system in lignin-transforming strain will enable the active transport of enzyme, finally leading a considerable increase in lignin depolymerization.
For scaling up the lignin valorization, the high productivity of the target molecule should also be obtained by efficient uptake of substrate and their catabolism. Especially, as the high heterogeneity of lignin causes a diverse type of compounds in the depolymerized stream, the simultaneous uptake of those compounds is essential. Also, the lignin-depolymerized mixture possibly contains some compounds which are not preferred to consume by natural pathways, such as guaiacol (García-Hidalgo et al., 2019). In this sense, expanding the range of aromatic compounds as the substrate will be needed. Omics studies and synthetic biology will offer new genetic information and tools to identify and characterize the mechanism that can be further engineered. In addition, metabolic pathway could be tailored to funnel the substrate to the target product. Vardon et al. (2015) re-routed PCA to catechol pathway to maximize the production of cis, cis-muconate. Combining with computational studies would be more helpful for engineering, where metabolic flux could be possible to predict according to the design. Recently, the metabolic models of P. putida have been developed with single gene knockout by computational tool for the biochemical production (Tokic et al., 2019).
Here, we have several issues to be figured for significant lignin valorization. Nevertheless, several promising candidates have been suggested, including Pseudomonas species having DyP-type peroxidase (Salvachúa et al., 2015). In future work, engineering should be applied toward enzyme and bacterial strain to get an ideal bacterial system, where efficient lignin-degrading enzymes are sufficiently transported out and the aromatic compounds are efficiently assimilated, which are directly channeled into the product. Additionally, paired bacterial system or stepwise fermentation could also be possible for the efficient bacterial valorization.
Conclusions
The valorization of lignin is essential to increase the sustainability and economic feasibility of the bio-refinery process, although the decomposition of lignin is complicated by its structural rigidity. Lignin valorization has been studied using catalytic, thermochemical, and biological approaches for many decades. Among biological approaches, few studies related to bacterial lignin decomposition were reported as compared with those involving fungi. However, various studies are currently underway because of its ease genetic manipulation and its ability to produce value-added chemicals using lignin-related aromatics. Decomposition of the lignin polymer was performed by DyP-type peroxidases and laccase secreted from bacteria. And LMW compounds from lignin was converted into value-added products. The results of lignin valorization by bacteria are still insignificant. However, new approaches using a bacterial biosensor, synthetic biology, and various “omic” studies are expected to lead to the discovery of a number of more effective lignolytic bacteria and enzymes that could be investigated for lignin utilization. These bacteria and enzymes could be potential candidates to achieve future lignin valorization.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Intelligent Synthetic Biology Center of Global Frontier Project (NRF-2015M3A6A8065831) and the Bio & Medical Technology Development Program (NRF-2018M3A9H3024746) through the National Research Foundation of Korea, and the Research Initiative Program of KRIBB.
References
- Ahmad M., Roberts J. N., Hardiman E. M., Singh R., Eltis L. D., Bugg T. D. (2011). Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50, 5096–5107. 10.1021/bi101892z [DOI] [PubMed] [Google Scholar]
- Ahmad M., Taylor C. R., Pink D., Burton K., Eastwood D., Bending G. D., et al. (2010). Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol. Biosyst. 6, 815–821. 10.1039/b908966g [DOI] [PubMed] [Google Scholar]
- Akinosho H. O., Yoo C. G., Dumitrache A., Natzke J., Muchero W., Brown S. D., et al. (2017). Elucidating the structural changes to Populus lignin during consolidated bioprocessing with Clostridium thermocellum. ACS Sustain. Chem. Eng. 5, 7486–7491. 10.1021/acssuschemeng.7b01203 [DOI] [Google Scholar]
- Antai S. P., Crawford D. L. (1981). Degradation of softwood, hardwood, and grass lignocelluloses by two Streptomyces strains. Appl. Environ. Microbiol. 42, 378–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias M. E., Arenas M., Rodríguez J., Soliveri J., Ball A. S., Hernández M. (2003). Kraft pulp biobleaching and mediated oxidation of a nonphenolic substrate by laccase from Streptomyces cyaneus CECT 3335. Appl. Environ. Microbiol. 69, 1953–1958. 10.1128/AEM.69.4.1953-1958.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandounas L., Wierckx N. J., De Winde J. H., Ruijssenaars H. J. (2011). Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol. 11:94. 10.1186/1472-6750-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N., Horbal L., Starck S., Kohlstedt M., Luzhetskyy A., Wittmann C. (2018). Enabling the valorization of guaiacol-based lignin: integrated chemical and biochemical production of cis, cis-muconic acid using metabolically engineered Amycolatopsis sp ATCC 39116. Met. Eng. 45, 200–210. 10.1016/j.ymben.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Beckham G. T., Johnson C. W., Karp E. M., Salvachúa D., Vardon D. R. (2016). Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 42, 40–53. 10.1016/j.copbio.2016.02.030 [DOI] [PubMed] [Google Scholar]
- Beneyton T., Beyl Y., Guschin D. A., Griffiths A. D., Taly V., Schuhmann W. (2011). The thermophilic CotA laccase from Bacillus subtilis: bioelectrocatalytic evaluation of O2 reduction in the direct and mediated electron transfer regime. Electroanalysis 23, 1781–1789. 10.1002/elan.201100054 [DOI] [Google Scholar]
- Brissos V. N., Tavares D., Sousa A. C., Robalo M. P., Martins L. O. (2017). Engineering a bacterial DyP-type peroxidase for enhanced oxidation of lignin-related phenolics at alkaline pH. ACS Catal. 7, 3454–3465. 10.1021/acscatal.6b03331 [DOI] [Google Scholar]
- Brown M. E., Barros T., Chang M. C. (2012). Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem. Biol. 7, 2074–2081. 10.1021/cb300383y [DOI] [PubMed] [Google Scholar]
- Brown M. E., Chang M. C. (2014). Exploring bacterial lignin degradation. Curr. Opin. Chem. Biol. 19, 1–7. 10.1016/j.cbpa.2013.11.015 [DOI] [PubMed] [Google Scholar]
- Brown M. E., Walker M. C., Nakashige T. G., Iavarone A. T., Chang M. C. (2011). Discovery and characterization of heme enzymes from unsequenced bacteria: application to microbial lignin degradation. J. Am. Chem. Soc. 133, 18006–18009. 10.1021/ja203972q [DOI] [PubMed] [Google Scholar]
- Bugg T. D., Ahmad M., Hardiman E. M., Rahmanpour R. (2011). Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 28, 1883–1896. 10.1039/c1np00042j [DOI] [PubMed] [Google Scholar]
- Cao L., Iris K., Liu Y., Ruan X., Tsang D. C., Hunt A. J., et al. (2018). Lignin valorization for the production of renewable chemicals: state-of-the-art review and future prospects. Bioresource Technol. 269, 465–475. 10.1016/j.biortech.2018.08.065 [DOI] [PubMed] [Google Scholar]
- Chandra R., Bharagava R. N. (2013). Bacterial degradation of synthetic and kraft lignin by axenic and mixed culture and their metabolic products. J. Environ. Biol. 34, 991–999. [PubMed] [Google Scholar]
- Chandra R., Chowdhary P. (2015). Properties of bacterial laccases and their application in bioremediation of industrial wastes. Environ. Sci. Process. Impacts 17, 326–342. 10.1039/C4EM00627E [DOI] [PubMed] [Google Scholar]
- Chandra R., Singh S., Purohit H. J., Kapley A. (2008). Isolation and characterization of bacterial strains Paenibacillus sp. and Bacillus sp. for kraft lignin decolorization from pulp paper mill waste. J. Gen. Appl. Microbiol. Biol. 54, 399–407. 10.2323/jgam.54.399 [DOI] [PubMed] [Google Scholar]
- Chen C.-Y., Lee C.-C., Chen H.-S., Yang C.-H., Wang S.-P., Wu J.-H., et al. (2016). Modification of lignin in sugarcane bagasse by a monocopper hydrogen peroxide-generating oxidase from Thermobifida fusca. Process Biochem. 51, 1486–1495. 10.1016/j.procbio.2016.07.009 [DOI] [Google Scholar]
- Chen Y., Chai L., Tang C., Yang Z., Zheng Y., Shi Y., et al. (2012). Kraft lignin biodegradation by Novosphingobium sp. B-7 and analysis of the degradation process. Bioresour. Technol. 123, 682–685. 10.1016/j.biortech.2012.07.028 [DOI] [PubMed] [Google Scholar]
- Chen Y., Luo Q., Zhou W., Xie Z., Cai Y.-J., Liao X.-R., et al. (2017). Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis. Appl. Microbiol. Biotechnol. 101, 1935–1944. 10.1007/s00253-016-7962-1 [DOI] [PubMed] [Google Scholar]
- Cho D. H., Lee Y. J., Um Y., Sang B.-I., Kim Y. H. (2009). Detoxification of model phenolic compounds in lignocellulosic hydrolysates with peroxidase for butanol production from Clostridium beijerinckii. Appl. Microbiol. Biotechnol. 83, 1035–1043. 10.1007/s00253-009-1925-8 [DOI] [PubMed] [Google Scholar]
- Chung D., Cha M., Guss A. M., Westpheling J. (2014). Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc. Natl. Acad. Sci. U.S.A. 111, 8931–8936. 10.1073/pnas.1402210111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.-Y., Maeda M., Song E., Horikoshij K., Kudo T. (1994). A Gram-positive polychlorinated biphenyl-degrading bacterium, Rhodococcus erythropolis strain TA421, isolated from a termite ecosystem. Biosci. Biotechnol. Biochem. 58, 2111–2113. 10.1271/bbb.58.2111 [DOI] [Google Scholar]
- Colpa D. I., Fraaije M. W., van Bloois E. (2014). DyP-type peroxidases: a promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 41, 1–7. 10.1007/s10295-013-1371-6 [DOI] [PubMed] [Google Scholar]
- Crawford D. L., Pometto A. L., Crawford R. L. (1983). Lignin degradation by Streptomyces viridosporus: isolation and characterization of a new polymeric lignin degradation intermediate. Appl. Environ. Microbiol. 45,898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R. L., Crawford D. L., Dizikes G. J. (1981). Catabolism of the lignin substructure model compound dehydrodivanillin by a lignin-degrading Streptomyces. Arch. Microbiol. 129, 204–209. 10.1007/BF00425251 [DOI] [Google Scholar]
- da Silva E. B., Zabkova M., Araújo J., Cateto C., Barreiro M., Belgacem M., et al. (2009). An integrated process to produce vanillin and lignin-based polyurethanes from Kraft lignin. Chem. Eng. Res. Des. 87, 1276–1292. 10.1016/j.cherd.2009.05.008 [DOI] [Google Scholar]
- Dashtban M., Schraft H., Syed T. A., Qin W. (2010). Fungal biodegradation and enzymatic modification of lignin. Int. J. Biochem. Mol. Biol. 1, 36–50. [PMC free article] [PubMed] [Google Scholar]
- de Gonzalo G., Colpa D. I., Habib M. H., Fraaije M. W. (2016). Bacterial enzymes involved in lignin degradation. J. Biotechnol. 236, 110–119. 10.1016/j.jbiotec.2016.08.011 [DOI] [PubMed] [Google Scholar]
- DeAngelis K. M., D'haeseleer P., Chivian D., Fortney J. L., Khudyakov J., Simmons B., et al. (2011). Complete genome sequence of “Enterobacter lignolyticus” SCF1. Stand. Genomic Sci. 5:69. 10.4056/sigs.2104875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis K. M., Sharma D., Varney R., Simmons B. A., Isern N. G., Markillie L. M., et al. (2013). Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front. Microbiol. 4:280. 10.3389/fmicb.2013.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorenzo D. M., Henson W. R., Moon T. S. (2017). Development of chemical and metabolite sensors for Rhodococcus opacus PD630. ACS Synth. Biol. 6, 1973–1978. 10.1021/acssynbio.7b00192 [DOI] [PubMed] [Google Scholar]
- Deng Y., Fong S. S. (2011). Metabolic engineering of Thermobifida fusca for direct aerobic bioconversion of untreated lignocellulosic biomass to 1-propanol. Met. Eng. 13, 570–577. 10.1016/j.ymben.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Deschamps A., Mahoudeau G., Lebeault J. (1980). Fast degradation of kraft lignin by bacteria. Eur. J. Appl. Microbiol. Biotechnol. 9, 45–51. 10.1007/BF00500001 [DOI] [Google Scholar]
- Ezeji T., Qureshi N., Blaschek H. P. (2007). Butanol production from agricultural residues: impact of degradation products on Clostridium beijerinckii growth and butanol fermentation. Biotechnol. Bioeng. 97, 1460–1469. 10.1002/bit.21373 [DOI] [PubMed] [Google Scholar]
- Fernandes T. A. R., Da Silveira W. B., Passos F. M. L., Zucchi T. D. (2014). Laccases from Actinobacteria—what we have and what to expect. Adv. Microbiol. 4:285 10.4236/aim.2014.46035 [DOI] [Google Scholar]
- Fischer R., Bleichrodt F. S., Gerischer U. C. (2008). Aromatic degradative pathways in Acinetobacter baylyi underlie carbon catabolite repression. Microbiology 154, 3095–3103. 10.1099/mic.0.2008/016907-0 [DOI] [PubMed] [Google Scholar]
- Gall D. L., Kim H., Lu F., Donohue T. J., Noguera D. R., Ralph J. (2014a). Stereochemical features of glutathione-dependent enzymes in the Sphingobium sp. strain SYK-6 β-aryl etherase pathway. J. Biol. Chem. 289, 8656–8667. 10.1074/jbc.M113.536250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall D. L., Ralph J., Donohue T. J., Noguera D. R. (2014b). A group of sequence-related sphingomonad enzymes catalyzes cleavage of β-aryl ether linkages in lignin β-guaiacyl and β-syringyl ether dimers. Environ. Sci. Technol. 48, 12454–12463. 10.1021/es503886d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang H., Lee D., Choi K.-Y., Kim H.-N., Ryu H., Lee D.-S., et al. (2017). Development of high performance polyurethane elastomers using vanillin-based green polyol chain extender originating from lignocellulosic biomass. ACS Sustain. Chem. Eng. 5, 4582–4588. 10.1021/acssuschemeng.6b02960 [DOI] [Google Scholar]
- García-Hidalgo J., Ravi K., Kuré L.-L., Lidén G., Gorwa-Grauslund M. (2019). Identification of the two-component guaiacol demethylase system from Rhodococcus rhodochrous and expression in Pseudomonas putida EM42 for guaiacol assimilation. AMB Express 9:34. 10.1186/s13568-019-0759-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Pelczar M. J., Jr. (1951). Microbiological aspects of lignin degradation. Bacteriol. Rev. 15:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf N., Altenbuchner J. (2014). Genetic engineering of Pseudomonas putida KT2440 for rapid and high-yield production of vanillin from ferulic acid. Appl. Microbiol. Biotechnol. 98, 137–149. 10.1007/s00253-013-5303-1 [DOI] [PubMed] [Google Scholar]
- Granja-Travez R. S., Bugg T. D. (2018). Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: involvement in bacterial lignin oxidation. Arch. Biochem. Biophys. 660, 97–107. 10.1016/j.abb.2018.10.012 [DOI] [PubMed] [Google Scholar]
- Granja-Travez R. S., Wilkinson R. C., Persinoti G. F., Squina F. M., Fülöp V., Bugg T. D. (2018). Structural and functional characterisation of multi-copper oxidase CueO from lignin-degrading bacterium Ochrobactrum sp. reveal its activity towards lignin model compounds and lignosulfonate. FEBS J. 285, 1684–1700. 10.1111/febs.14437 [DOI] [PubMed] [Google Scholar]
- Grass G., Rensing C. (2001). CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Comm. 286, 902–908. 10.1006/bbrc.2001.5474 [DOI] [PubMed] [Google Scholar]
- Gu M. B., Mitchell R. J., Kim B. C. (2004). Whole-cell-based biosensors for environmental biomonitoring and application. Adv. Biochem. Eng. Biotechnol. 87, 269–305. 10.1007/b13533 [DOI] [PubMed] [Google Scholar]
- Gui Q., Lawson T., Shan S., Yan L., Liu Y. (2017). The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors 17:1623. 10.3390/s17071623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez A., Rencoret J., Cadena E. M., Rico A., Barth D., José C., et al. (2012). Demonstration of laccase-based removal of lignin from wood and non-wood plant feedstocks. Bioresour. Technol. 119, 114–122. 10.1016/j.biortech.2012.05.112 [DOI] [PubMed] [Google Scholar]
- Haider K., Trojanowski J., Sundman V. (1978). Screening for lignin degrading bacteria by means of 14C-labelled lignins. Arch. Microbiol. 119, 103–106. 10.1007/BF00407936 [DOI] [PubMed] [Google Scholar]
- He Y., Li X., Ben H., Xue X., Yang B. (2017). Lipid production from dilute alkali corn stover lignin by Rhodococcus strains. ACS Sustain. Chem. Eng. 5, 2302–2311. 10.1021/acssuschemeng.6b02627 [DOI] [Google Scholar]
- Ho J. C., Pawar S. V., Hallam S. J., Yadav V. G. (2017). An improved whole-cell biosensor for the discovery of lignin-transforming enzymes in functional metagenomic screens. ACS Synth. Biol. 7, 392–398. 10.1021/acssynbio.7b00412 [DOI] [PubMed] [Google Scholar]
- Huang X. F., Santhanam N., Badri D. V., Hunter W. J., Manter D. K., Decker S. R., et al. (2013). Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol. Bioeng. 110, 1616–1626. 10.1002/bit.24833 [DOI] [PubMed] [Google Scholar]
- Hullo M.-F., Moszer I., Danchin A., Martin-Verstraete I. (2001). CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183, 5426–5430. 10.1128/JB.183.18.5426-5430.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihssen J., Jankowska D., Ramsauer T., Reiss R., Luchsinger R., Wiesli L., et al. (2017). Engineered Bacillus pumilus laccase-like multi-copper oxidase for enhanced oxidation of the lignin model compound guaiacol. Protein Eng. Des. Sel. 30, 449–453. 10.1093/protein/gzx026 [DOI] [PubMed] [Google Scholar]
- Ihssen J., Reiss R., Luchsinger R., Thöny-Meyer L., Richter M. (2015). Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in Escherichia coli. Sci. Rep. 5:10465. 10.1038/srep10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janusz G., Pawlik A., Sulej J., Swiderska-Burek U., Jarosz-Wilkołazka A., Paszczynski A. (2017). Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 41, 941–962. 10.1093/femsre/fux049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha R. K., Bingen J. M., Johnson C. W., Kern T. L., Khanna P., Trettel D. S., et al. (2018). A protocatechuate biosensor for Pseudomonas putida KT2440 via promoter and protein evolution. Met. Eng. Comm. 6, 33–38. 10.1016/j.meteno.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Miñambres B., García J. L., Díaz E. (2002). Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4, 824–841. 10.1046/j.1462-2920.2002.00370.x [DOI] [PubMed] [Google Scholar]
- Jiménez J. I., Miñambres B., García J. L., Díaz E. (2004). Genomic insights in the metabolism of aromatic compounds in Pseudomonas. Pseudomonas 2004, 425–462. 10.1007/978-1-4419-9088-4_15 [DOI] [Google Scholar]
- Jin Y., Cheng X., Zheng Z. (2010). Preparation and characterization of phenol–formaldehyde adhesives modified with enzymatic hydrolysis lignin. Bioresour. Technol. 101, 2046–2048. 10.1016/j.biortech.2009.09.085 [DOI] [PubMed] [Google Scholar]
- Jing D. (2010). Improving the simultaneous production of laccase and lignin peroxidase from Streptomyces lavendulae by medium optimization. Bioresour. Technol. 101, 7592–7597. 10.1016/j.biortech.2010.04.087 [DOI] [PubMed] [Google Scholar]
- Jing D., Wang J. (2012). Controlling the simultaneous production of laccase and lignin peroxidase from Streptomyces cinnamomensis by medium formulation. Biotechnol. Biofuels 5:15. 10.1186/1754-6834-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. W., Beckham G. T. (2015). Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Met. Eng. 28, 240–247. 10.1016/j.ymben.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Johnson C. W., Salvachúa D., Khanna P., Smith H., Peterson D. J., Beckham G. T. (2016). Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Met. Eng. Comm. 3, 111–119. 10.1016/j.meteno.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataeva I., Foston M. B., Yang S.-J., Pattathil S., Biswal A. K., Poole Ii F. L., et al. (2013). Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energ. Environ. Sci. 6, 2186–2195. 10.1039/c3ee40932e [DOI] [Google Scholar]
- Kawai S., Nakagawa M., Ohashi H. (2002). Degradation mechanisms of a nonphenolic β-O-4 lignin model dimer by Trametes versicolor laccase in the presence of 1-hydroxybenzotriazole. Enzyme Microb. Technol. 30, 482–489. 10.1016/S0141-0229(01)00523-3 [DOI] [Google Scholar]
- Kohlstedt M., Starck S., Barton N., Stolzenberger J., Selzer M., Mehlmann K., et al. (2018). From lignin to nylon: cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Met. Eng. 47, 279–293. 10.1016/j.ymben.2018.03.003 [DOI] [PubMed] [Google Scholar]
- Kosa M., Ragauskas A. J. (2012). Bioconversion of lignin model compounds with oleaginous Rhodococci. Appl. Microbiol. Biotechnol. 93, 891–900. 10.1007/s00253-011-3743-z [DOI] [PubMed] [Google Scholar]
- Kosa M., Ragauskas A. J. (2013). Lignin to lipid bioconversion by oleaginous Rhodococci. Green Chem. 15, 2070–2074. 10.1039/c3gc40434j [DOI] [Google Scholar]
- Koschorreck K., Richter S. M., Ene A. B., Roduner E., Schmid R. D., Urlacher V. B. (2008). Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic acids. Appl. Microbiol. Biotechnol. 79, 217–224. 10.1007/s00253-008-1417-2 [DOI] [PubMed] [Google Scholar]
- Kumar M., Singhal A., Thakur I. S. (2016). Comparison of submerged and solid state pretreatment of sugarcane bagasse by Pandoraea sp. ISTKB: enzymatic and structural analysis. Bioresour. Technol. 203, 18–25. 10.1016/j.biortech.2015.12.034 [DOI] [PubMed] [Google Scholar]
- Kumar M., Singhal A., Verma P. K., Thakur I. S. (2017). Production and characterization of polyhydroxyalkanoate from lignin derivatives by Pandoraea sp. ISTKB. ACS Omega 2, 9156–9163. 10.1021/acsomega.7b01615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Verma S., Gazara R. K., Kumar M., Pandey A., Verma P. K., et al. (2018). Genomic and proteomic analysis of lignin degrading and polyhydroxyalkanoate accumulating β-proteobacterium Pandoraea sp. ISTKB. Biotechnol. Biofuels 11:154. 10.1186/s13068-018-1148-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Amasia M., Madou M., Mitchell R. J. (2013). Serum complement enhances the responses of genotoxin-and oxidative stress-sensitive Escherichia coli bioreporters. Biosens. Bioelectron. 46, 175–182. 10.1016/j.bios.2013.02.038 [DOI] [PubMed] [Google Scholar]
- Lee S., Lee J. H., Mitchell R. J. (2015). Analysis of Clostridium beijerinckii NCIMB 8052's transcriptional response to ferulic acid and its application to enhance the strain tolerance. Biotechnol. Biofuels 8:68. 10.1186/s13068-015-0252-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Mitchell R. J. (2012). Detection of toxic lignin hydrolysate-related compounds using an inaA:: luxCDABE fusion strain. J. Biotechnol. 157, 598–604. 10.1016/j.jbiotec.2011.06.018 [DOI] [PubMed] [Google Scholar]
- Lee S., Nam D., Jung J. Y., Oh M.-K., Sang B.-I., Mitchell R. J. (2012). Identification of Escherichia coli biomarkers responsive to various lignin-hydrolysate compounds. Bioresour. Technol. 114, 450–456. 10.1016/j.biortech.2012.02.085 [DOI] [PubMed] [Google Scholar]
- Lin L., Cheng Y., Pu Y., Sun S., Li X., Jin M., et al. (2016). Systems biology-guided biodesign of consolidated lignin conversion. Green Chem. 18, 5536–5547. 10.1039/C6GC01131D [DOI] [Google Scholar]
- Linger J. G., Vardon D. R., Guarnieri M. T., Karp E. M., Hunsinger G. B., Franden M. A., et al. (2014). Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. U.S.A. 111, 12013–12018. 10.1073/pnas.1410657111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Yan X., Zhuo S., Si M., Liu M., Wang S., et al. (2018). Pandoraea sp. B-6 assists the deep eutectic solvent pretreatment of rice straw via promoting lignin depolymerization. Bioresour. Technol. 257, 62–68. 10.1016/j.biortech.2018.02.029 [DOI] [PubMed] [Google Scholar]
- Liu X., Du Q., Wang Z., Zhu D., Huang Y., Li N., et al. (2011). Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes. J. Biol. Chem. 286, 14922–14931. 10.1074/jbc.M110.197780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lončar N., Colpa D. I., Fraaije M. W. (2016). Exploring the biocatalytic potential of a DyP-type peroxidase by profiling the substrate acceptance of Thermobifida fusca DyP peroxidase. Tetrahedron 72, 7276–7281. 10.1016/j.tet.2015.12.078 [DOI] [Google Scholar]
- Lu C., Dong J., Yang S.-T. (2013). Butanol production from wood pulping hydrolysate in an integrated fermentation–gas stripping process. Bioresour. Technol. 143, 467–475. 10.1016/j.biortech.2013.06.012 [DOI] [PubMed] [Google Scholar]
- Luo Q., Chen Y., Xia J., Wang K.-Q., Cai Y.-J., Liao X.-R., et al. (2018). Functional expression enhancement of Bacillus pumilus CotA-laccase mutant WLF through site-directed mutagenesis. Enzyme Microb. Technol. 109, 11–19. 10.1016/j.enzmictec.2017.07.013 [DOI] [PubMed] [Google Scholar]
- Machczynski M. C., Vijgenboom E., Samyn B., Canters G. W. (2004). Characterization of SLAC: a small laccase from Streptomyces coelicolor with unprecedented activity. Protein Sci. 13, 2388–2397. 10.1110/ps.04759104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S., Lukk T., Solbiati J. O., Bauer S., Nair S. K., Cronan J. E., et al. (2014). Roles of small laccases from Streptomyces in lignin degradation. Biochemistry 53, 4047–4058. 10.1021/bi500285t [DOI] [PubMed] [Google Scholar]
- Manter D. K., Hunter W. J., Vivanco J. M. (2011). Enterobacter soli sp. nov.: a lignin-degrading γ-proteobacteria isolated from soil. Curr. Microbiol. 62, 1044–1049. 10.1007/s00284-010-9809-9 [DOI] [PubMed] [Google Scholar]
- Masai E., Ichimura A., Sato Y., Miyauchi K., Katayama Y., Fukuda M. (2003). Roles of the enantioselective glutathione S-transferases in cleavage of β-aryl ether. J. Bacteriol. 185, 1768–1775. 10.1128/JB.185.6.1768-1775.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E., Katayama Y., Fukuda M. (2007). Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotech. Biochem. 71, 1–15. 10.1271/bbb.60437 [DOI] [PubMed] [Google Scholar]
- Masai E., Katayama Y., Nishikawa S., Fukuda M. (1999). Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J. Ind. Microbiol. Biotechnol. 23, 364–373. 10.1038/sj.jim.2900747 [DOI] [PubMed] [Google Scholar]
- Mathews S. L., Grunden A. M., Pawlak J. (2016). Degradation of lignocellulose and lignin by Paenibacillus glucanolyticus. Int. Biodeterior. Biodegrad. 110, 79–86. 10.1016/j.ibiod.2016.02.012 [DOI] [Google Scholar]
- McCarthy A. (1987). Lignocellulose-degrading actinomycetes. FEMS Microbiol. Lett. 46, 145–163. 10.1111/j.1574-6968.1987.tb02456.x [DOI] [Google Scholar]
- Mendes S., Brissos V., Gabriel A., Catarino T., Turner D. L., Todorovic S., et al. (2015). An integrated view of redox and catalytic properties of B-type PpDyP from Pseudomonas putida MET94 and its distal variants. Arch. Biochem. Biophys. 574, 99–107. 10.1016/j.abb.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Min K., Gong G., Woo H. M., Kim Y., Um Y. (2015). A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and β-ether lignin dimer. Sci. Rep. 5:8245. 10.1038/srep08245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnappa A. K., Lee S., Mitchell R. J. (2013). Sensing of plant hydrolysate-related phenolics with an aaeXAB:: luxCDABE bioreporter strain of Escherichia coli. Bioresour. Technol. 127, 429–434. 10.1016/j.biortech.2012.09.086 [DOI] [PubMed] [Google Scholar]
- Moya R., Saastamoinen P., Hernández M., Suurnäkki A., Arias E., Mattinen M.-L. (2011). Reactivity of bacterial and fungal laccases with lignin under alkaline conditions. Bioresour. Technol. 102, 10006–10012. 10.1016/j.biortech.2011.08.046 [DOI] [PubMed] [Google Scholar]
- Mycroft Z., Gomis M., Mines P., Law P., Bugg T. D. (2015). Biocatalytic conversion of lignin to aromatic dicarboxylic acids in Rhodococcus jostii RHA1 by re-routing aromatic degradation pathways. Green Chem. 17, 4974–4979. 10.1039/C5GC01347J [DOI] [Google Scholar]
- Nikel P. I., de Lorenzo V. (2018). Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Met. Eng. 50, 142–155. 10.1016/j.ymben.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Niladevi K., Sheejadevi P., Prema P. (2008). Strategies for enhancing laccase yield from Streptomyces psammoticus and its role in mediator-based decolorization of azo dyes. Appl. Biochem. Biotechnol. 151, 9–19. 10.1007/s12010-008-8175-6 [DOI] [PubMed] [Google Scholar]
- Ogola H. J. O., Kamiike T., Hashimoto N., Ashida H., Ishikawa T., Shibata H., et al. (2009). Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl. Environ. Microbiol. 75, 7509–7518. 10.1128/AEM.01121-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Saddler J. N. (2013). Effect of replacing polyol by organosolv and kraft lignin on the property and structure of rigid polyurethane foam. Biotechnol. Biofuels 6:12. 10.1186/1754-6834-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Masai E., Kasai D., Miyauchi K., Katayama Y., Fukuda M. (2005). A second 5-carboxyvanillate decarboxylase gene, ligW2, is important for lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 71, 5014–5021. 10.1128/AEM.71.9.5014-5021.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Masai E., Kitayama H., Harada K., Katayama Y., Fukuda M. (2002). Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 68, 4407–4415. 10.1128/AEM.68.9.4407-4415.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanzagl V., Nys K., Bellei M., Michlits H., Mlynek G., Battistuzzi G., et al. (2018). Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage. J. Biol. Chem. 293, 14823–14838. 10.1074/jbc.RA118.004773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper D. H. (2005). Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 67, 170–191. 10.1007/s00253-004-1810-4 [DOI] [PubMed] [Google Scholar]
- Poblete-Castro I., Becker J., Dohnt K., Dos Santos V. M., Wittmann C. (2012). Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 93, 2279–2290. 10.1007/s00253-012-3928-0 [DOI] [PubMed] [Google Scholar]
- Ponnusamy V. K., Nguyen D. D., Dharmaraja J., Shobana S., Banu R., Saratale R. G., et al. (2018). A review on lignin structure, pretreatments, fermentation reactions and biorefinery potential. Bioresource Technol. 271, 462–472. 10.1016/j.biortech.2018.09.070 [DOI] [PubMed] [Google Scholar]
- Putri R. M., Allende-Ballestero C., Luque D., Klem R., Rousou K.-A., Liu A., et al. (2017). Structural characterization of native and modified encapsulins as nanoplatforms for in vitro catalysis and cellular uptake. ACS Nano 11, 12796–12804. 10.1021/acsnano.7b07669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas A. J., Beckham G. T., Biddy M. J., Chandra R., Chen F., Davis M. F., et al. (2014). Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. 10.1126/science.1246843 [DOI] [PubMed] [Google Scholar]
- Rahmanpour R., Bugg T. D. (2015). Characterisation of Dyp-type peroxidases from Pseudomonas fluorescens Pf-5: oxidation of Mn (II) and polymeric lignin by Dyp1B. Arch. Biochem. Biophys. 574, 93–98. 10.1016/j.abb.2014.12.022 [DOI] [PubMed] [Google Scholar]
- Rahmanpour R., Rea D., Jamshidi S., Fülöp V., Bugg T. D. (2016). Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch. Biochem. Biophys. 594, 54–60. 10.1016/j.abb.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Raj A., Reddy M. K., Chandra R., Purohit H. J., Kapley A. (2007). Biodegradation of kraft-lignin by Bacillus sp. isolated from sludge of pulp and paper mill. Biodegradation 18, 783–792. 10.1007/s10532-007-9107-9 [DOI] [PubMed] [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P. F., et al. (2004). Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem. Rev. 3, 29–60. 10.1023/B:PHYT.0000047809.65444.a4 [DOI] [Google Scholar]
- Ramires E. C., Megiatto J. D., Jr., Gardrat C., Castellan A., Frollini E. (2010). Valorization of an industrial organosolv–sugarcane bagasse lignin: characterization and use as a matrix in biobased composites reinforced with sisal fibers. Biotechnol. Bioeng. 107, 612–621. 10.1002/bit.22847 [DOI] [PubMed] [Google Scholar]
- Rashid G. M., Taylor C. R., Liu Y., Zhang X., Rea D., FüLöP V., et al. (2015). Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. ACS Chem. Biol. 10, 2286–2294. 10.1021/acschembio.5b00298 [DOI] [PubMed] [Google Scholar]
- Reddy N., Yang Y. (2005). Biofibers from agricultural byproducts for industrial applications. Trends Biotechnol. 23, 22–27. 10.1016/j.tibtech.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Salvachúa D., Katahira R., Black B. A., Cleveland N. S., Reed M., et al. (2017). Base-catalyzed depolymerization of solid lignin-rich streams enables microbial conversion. ACS Sustain. Chem. Eng. 5, 8171–8180. 10.1021/acssuschemeng.7b01818 [DOI] [Google Scholar]
- Sainsbury P. D., Hardiman E. M., Ahmad M., Otani H., Seghezzi N., Eltis L. D., et al. (2013). Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. ACS Chem. Biol. 8, 2151–2156. 10.1021/cb400505a [DOI] [PubMed] [Google Scholar]
- Salvachúa D., Karp E. M., Nimlos C. T., Vardon D. R., Beckham G. T. (2015). Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 17, 4951–4967. 10.1039/C5GC01165E [DOI] [Google Scholar]
- Santos A., Mendes S., Brissos V., Martins L. O. (2014). New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl. Microbiol. Biotechnol. 98, 2053–2065. 10.1007/s00253-013-5041-4 [DOI] [PubMed] [Google Scholar]
- Santos R. B., Hart P., Jameel H., Chang H.-M. (2013). Wood based lignin reactions important to the biorefinery and pulp and paper industries. Bioresources 8, 1456–1477. 10.15376/biores.8.1.1456-1477 [DOI] [Google Scholar]
- Seto M., Kimbara K., Shimura M., Hatta T., Fukuda M., Yano K. (1995). A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61, 3353–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Liu Q., Ma J., Liao H., Xiong X., Zhang K., et al. (2015). An acid-stable bacterial laccase identified from the endophyte Pantoea ananatis Sd-1 genome exhibiting lignin degradation and dye decolorization abilities. Biotechnol. Lett. 37, 2279–2288. 10.1007/s10529-015-1914-1 [DOI] [PubMed] [Google Scholar]
- Shi Y., Chai L., Tang C., Yang Z., Zheng Y., Chen Y., et al. (2013). Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioproc. Biosyst. Eng. 36, 1957–1965. 10.1007/s00449-013-0972-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yan X., Li Q., Wang X., Xie S., Chai L., et al. (2017). Directed bioconversion of Kraft lignin to polyhydroxyalkanoate by Cupriavidus basilensis B-8 without any pretreatment. Process Biochem. 52, 238–242. 10.1016/j.procbio.2016.10.004 [DOI] [Google Scholar]
- Shrestha R., Huang G., Meekins D. A., Geisbrecht B. V., Li P. (2017). Mechanistic insights into dye-decolorizing peroxidase revealed by solvent isotope and viscosity effects. ACS Catal. 7, 6352–6364. 10.1021/acscatal.7b01861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M., Yan X., Liu M., Shi M., Wang Z., Wang S., et al. (2018). In situ lignin bioconversion promotes complete carbohydrate conversion of rice straw by Cupriavidus basilensis B-8. ACS Sustain. Chem. Eng. 6, 7969–7978. 10.1021/acssuschemeng.8b01336 [DOI] [Google Scholar]
- Singh D., Chen S. (2008). The white-rot fungus Phanerochaete chrysosporium: conditions for the production of lignin-degrading enzymes. Appl. Microbiol. Biotechnol. 81, 399–417. 10.1007/s00253-008-1706-9 [DOI] [PubMed] [Google Scholar]
- Singh R., Grigg J. C., Qin W., Kadla J. F., Murphy M. E., Eltis L. D. (2013). Improved manganese-oxidizing activity of DypB, a peroxidase from a lignolytic bacterium. ACS Chem. Biol. 8, 700–706. 10.1021/cb300608x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan C. R., Singh R., Vaninsberghe D., Ievdokymenko K., Budwill K., Mohn W. W., et al. (2014). Metagenomic scaffolds enable combinatorial lignin transformation. Proc. Natl. Acad. Sci. U.S.A. 111, 10143–10148. 10.1073/pnas.1401631111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A., Schierhorn A., Lindenstrauss U., Lilie H., Brüser T. (2006). YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. J. Biol. Chem. 281, 13972–13978. 10.1074/jbc.M511891200 [DOI] [PubMed] [Google Scholar]
- Sugano Y., Muramatsu R., Ichiyanagi A., Sato T., Shoda M. (2007). DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family ASP171 replaces the distal histidine of classical peroxidases. J. Biol. Chem. 282, 36652–36658. 10.1074/jbc.M706996200 [DOI] [PubMed] [Google Scholar]
- Sugawara K., Nishihashi Y., Narioka T., Yoshida T., Morita M., Sugano Y. (2017). Characterization of a novel DyP-type peroxidase from Streptomyces avermitilis. J. Biosci. Bioeng. 123, 425–430. 10.1016/j.jbiosc.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Tamura A., Fukutani Y., Takami T., Fujii M., Nakaguchi Y., Murakami Y., et al. (2015). Packaging guest proteins into the encapsulin nanocompartment from Rhodococcus erythropolis N771. Biotechnol. Bioeng. 112, 13–20. 10.1002/bit.25322 [DOI] [PubMed] [Google Scholar]
- Taylor C., Hardiman E., Ahmad M., Sainsbury P., Norris P., Bugg T. (2012). Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J. Appl. Microbiol. 113, 521–530. 10.1111/j.1365-2672.2012.05352.x [DOI] [PubMed] [Google Scholar]
- Tokic M., Miskovic L., Hatzimanikatis V. (2019). Large-scale kinetic metabolic models of Pseudomonas putida for a consistent design of metabolic engineering strategies. bioRxiv 2019:569152 10.1101/569152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S., Chuah J.-A., Matsumoto K., Doi Y., Numata K. (2014). Understanding the limitations in the biosynthesis of polyhydroxyalkanoate (PHA) from lignin derivatives. ACS Sustain. Chem. Eng. 2, 1106–1113. 10.1021/sc500066f [DOI] [Google Scholar]
- van Bloois E., Pazmiño D. E. T., Winter R. T., Fraaije M. W. (2010). A robust and extracellular heme-containing peroxidase from Thermobifida fusca as prototype of a bacterial peroxidase superfamily. Appl. Microbiol. Biotechnol. 86, 1419–1430. 10.1007/s00253-009-2369-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duuren J., Wijte D., Leprince A., Karge B., Puchałka J., Wery J., et al. (2011). Generation of a catR deficient mutant of P. putida KT2440 that produces cis, cis-muconate from benzoate at high rate and yield. J. Biotechnol. 156, 163–172. 10.1016/j.jbiotec.2011.08.030 [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. 10.1104/pp.110.155119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardon D. R., Franden M. A., Johnson C. W., Karp E. M., Guarnieri M. T., Linger J. G., et al. (2015). Adipic acid production from lignin. Energ. Environ. Sci. 8, 617–628. 10.1039/C4EE03230F [DOI] [Google Scholar]
- Wang B., Rezenom Y. H., Cho K.-C., Tran J. L., Lee D. G., Russell D. H., et al. (2014). Cultivation of lipid-producing bacteria with lignocellulosic biomass: effects of inhibitory compounds of lignocellulosic hydrolysates. Bioresour. Technol. 161, 162–170. 10.1016/j.biortech.2014.02.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Cui D., Lu L., Zhang N., Yang H., Zhao M., et al. (2016). Cloning and characterization of CotA laccase from Bacillus subtilis WD23 decoloring dyes. Ann. Microbiol. 66, 461–467. 10.1007/s13213-015-1128-8 [DOI] [Google Scholar]
- Wang H., Pu Y., Ragauskas A., Yang B. (2019). From lignin to valuable products–strategies, challenges, and prospects. Bioresour. Technol. 271, 449–461. 10.1016/j.biortech.2018.09.072 [DOI] [PubMed] [Google Scholar]
- Wang J., Feng J., Jia W., Chang S., Li S., Li Y. (2015). Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnol. Biofuels 8:145. 10.1186/s13068-015-0331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lin L., Dong J., Ling J., Wang W., Wang H., et al. (2018). Simultaneous improvements of Pseudomonas cell growth and polyhydroxyalkanoate production from a lignin derivative for lignin-consolidated bioprocessing. Appl. Environ. Microbiol. 84:18. 10.1128/AEM.01469-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Zeng G., Huang F., Kosa M., Huang D., Ragauskas A. J. (2015). Bioconversion of oxygen-pretreated Kraft lignin to microbial lipid with oleaginous Rhodococcus opacus DSM 1069. Green Chem. 17, 2784–2789. 10.1039/C5GC00422E [DOI] [Google Scholar]
- Wilson D. B. (2004). Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 4, 72–82. 10.1002/tcr.20002 [DOI] [PubMed] [Google Scholar]
- Xu Z., Qin L., Cai M., Hua W., Jin M. (2018). Biodegradation of kraft lignin by newly isolated Klebsiella pneumoniae, Pseudomonas putida, and Ochrobactrum tritici strains. Environ. Sci. Pollut. Res. 25, 14171–14181. 10.1007/s11356-018-1633-y [DOI] [PubMed] [Google Scholar]
- Yadav S., Chandra R. (2015). Syntrophic co-culture of Bacillus subtilis and Klebsiella pneumonia for degradation of kraft lignin discharged from rayon grade pulp industry. J. Environ. Sci. 33, 229–238. 10.1016/j.jes.2015.01.018 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Sugano Y. (2015). A structural and functional perspective of DyP-type peroxidase family. Arch. Biochem. Biophys. 574, 49–55. 10.1016/j.abb.2015.01.022 [DOI] [PubMed] [Google Scholar]
- Yu W., Liu W., Huang H., Zheng F., Wang X., Wu Y., et al. (2014). Application of a novel alkali-tolerant thermostable DyP-type peroxidase from Saccharomonospora viridis DSM 43017 in biobleaching of eucalyptus kraft pulp. PLoS ONE 9:e110319. 10.1371/journal.pone.0110319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakzeski J., Bruijnincx P. C., Jongerius A. L., Weckhuysen B. M. (2010). The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 110, 3552–3599. 10.1021/cr900354u [DOI] [PubMed] [Google Scholar]
- Zamocky M., Hofbauer S., Schaffner I., Gasselhuber B., Nicolussi A., Soudi M., et al. (2015). Independent evolution of four heme peroxidase superfamilies. Arch. Biochem. Biophys. 574, 108–119. 10.1016/j.abb.2014.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Lin X., Zhang J., Li X., Wong M. H. (2011). Oxidation of polycyclic aromatic hydrocarbons by the bacterial laccase CueO from E. coli. Appl. Microbiol. Biotechnol. 89, 1841–1849. 10.1007/s00253-010-3009-1 [DOI] [PubMed] [Google Scholar]
- Zeng J., Singh D., Laskar D., Chen S. (2013). Degradation of native wheat straw lignin by Streptomyces viridosporus T7A. Int. J. Environ. Sci. Technol. 10, 165–174. 10.1007/s13762-012-0085-z [DOI] [Google Scholar]
- Zhao C., Xie S., Pu Y., Zhang R., Huang F., Ragauskas A. J., et al. (2016). Synergistic enzymatic and microbial lignin conversion. Green Chem. 18, 1306–1312. 10.1039/C5GC01955A [DOI] [Google Scholar]
- Zheng Z., Li H., Li L., Shao W. (2012). Biobleaching of wheat straw pulp with recombinant laccase from the hyperthermophilic Thermus thermophilus. Biotechnol. Lett. 34, 541–547. 10.1007/s10529-011-0796-0 [DOI] [PubMed] [Google Scholar]
- Zhu D., Zhang P., Xie C., Zhang W., Sun J., Qian W.-J., et al. (2017). Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol. Biofuels 10:44. 10.1186/s13068-017-0735-y [DOI] [PMC free article] [PubMed] [Google Scholar]