Abstract
Chronic inflammation in the benign prostate has been associated with a higher risk of developing prostate cancer. While a range of immune lineages is found in the prostate including T cells, B cells and myeloid cells, the specific subsets of immune cells with each major lineage have not been well described. In this study, we use mass cytometry (CyTOF) to comprehensively and reproducibly profile immune cells in mouse and human prostate. Using 4 myeloid markers (CD11b, CD11c, F4/80, Ly6C) in the mouse, we identified 8 phenotypically-distinct myeloid populations, demonstrating considerable heterogeneity within the immune compartment of the mouse prostate. We then profiled the prostate immune microenvironment from 9 human patients. Unlike the mouse prostate which is myeloid-dominant, the immune compartment in the benign human prostate is consistently T-lymphocyte-dominant. Using the X-shift algorithm to identify individual immune subsets based on marker expression, we found 57 phenotypically-distinct immune cell types in the human prostate. Despite similar proportions of T, B and myeloid lineage cells in the benign human prostate of all patients evaluated, we observed considerable interpatient heterogeneity in the abundance of more specific immune subsets. These findings highlight the importance of studying the immune compartment in the prostate at a granular level and will lead to future studies addressing the functional role of specific immune subsets in prostate epithelial transformation.
Keywords: Prostate, immune, lymphocyte, myeloid, inflammation, mass cytometry
Introduction
The relationship between chronic inflammation and cancer has long been established. Many cancers, such as cervical cancer, colorectal cancer, and mesothelioma are associated with chronic inflammation [1]. Although the sources of inflammation are diverse—human papillomavirus infection for cervical cancer, inflammatory bowel disease for colorectal cancer, and asbestos for mesothelioma—chronic inflammation itself is thought to play an important role in carcinogenesis [2-4]. Inflammation is usually considered a defense mechanism of the body. Immune cells in areas of infection or tissue damage work to remove foreign agents and repair tissue through the secretion of cytokines and reactive oxygen species (ROS). During the normal inflammatory response, neutrophils secrete ROS to destroy damaged tissue, and other immune cells secrete cytokines to stimulate tissue repair [5]. However, during chronic inflammation, there is prolonged exposure to these signals. ROS can induce oxidative DNA damage and mutations in oncogenes and tumor suppressor genes [6]. Additionally, cytokines can stimulate receptors on epithelial cells and activate downstream oncogenic transcription factors like nuclear factor-κB (NF-κB) and signal transducer and activator of transcription 3 (STAT3), potentially leading to carcinogenesis [7].
Chronic inflammation of the prostate has been implicated as a risk factor for prostate cancer. A 2014 study by Gurel et al. found that in patients without prostate cancer those with inflammation in their prostate were 1.78 times more likely to develop prostate cancer and 2.24 times more likely to develop higher-grade prostate cancer [8]. While the mechanisms by which immune cells increase the risk of developing prostate cancer is poorly understood, there is evidence that inflammation is associated with prostate epithelial proliferation [9]. Proliferative inflammatory atrophy (PIA) lesions typically reside in close proximity to inflammatory cells and have been observed to transition into high-grade prostatic intraepithelial neoplasia, a precursor to prostate cancer [10-12]. Various immune cell subsets have also been shown to have a role in prostate carcinogenesis [13]. Of particular interest are B cells and myeloid-derived suppressor cells, which have been shown to promote prostate cancer in mouse models [14,15]. Additionally, inflammation induced by different mechanisms in the mouse prostate, such as bacterial and autoimmune inflammation, has been shown to promote proliferation of prostate progenitor cells and transformation [16-19].
While mice are useful models to define mechanisms regulating prostate development and carcinogenesis, there are important differences between the human and mouse immune system. Mice and humans differ in the types of immune cells in their bodies; while humans are dominant in neutrophils, mice are rich in lymphocytes [20]. Mouse and human immune cells also differ in the expression of certain cell surface antigens [21]. These species-specific differences are important to consider when studying the immune cells in the prostate of each species.
Recently, we have reported a population of human prostatic luminal cells defined by low expression of CD38 that is expanded near sites of chronic inflammation. These luminal cells express pro-inflammatory cytokines and chemokines, suggesting a two-way interaction between CD38lo luminal cells and immune cells in modulation of the prostate [22,23]. Importantly, CD38lo luminal cells are enriched for progenitor activity and can respond to oncogene expression to initiate human prostate cancer. Whether immune cells contribute to CD38lo progenitor cell expansion or tumor initiation has not been established.
While there have been many studies about the relationship between specific immune cells and prostate cancer, there is a lack of consensus about the effects of specific immune cell subsets on disease risk or patient outcome. Out of 10 studies between 2008 and 2014 exploring the effects of T cells on prostate cancer, half reported a positive correlation while the other half reported negative or no correlation with patient outcome [24]. One explanation is that many of these studies lack detail about immune cell subsets. For example, one of the 10 studies which found a positive correlation only used CD3 as a T cell marker [24,25]. In leaving out information about immune subsets, we run the risk of generalizing an immune cell compartment with a diverse range of functions [26]. In order to gain a better understanding of the immune cells in the prostate, it is important to know more about specific immune cell subsets and how they interact with tissue.
From a large, heterogenous tissue like the prostate, immune cells are traditionally profiled using flow cytometry or immunohistochemistry [27]. However, with the advent of mass cytometry (CyTOF) in the past decade, it is now possible to use many times the number of markers to gain a richer understanding of immune cell subsets isolated from mouse and human tissue [28]. Instead of fluorochromes conjugated to antibodies, CyTOF makes use of heavy metal isotopes conjugated to antibodies and measures them on a time-of-flight mass spectrometer, allowing for up to 40 simultaneous markers to be used [29]. Using this technique, we are able to analyze specific subsets of immune cell compartments and detect rare subsets [30]. In this study we use CyTOF to define the wild type prostate immune compartment, leading the way for future studies to address the functional role of specific immune subsets in epithelial progenitor activity and tumorigenesis.
Materials and methods
Immunohistochemistry
De-identified human prostate tissue was embedded in paraffin and sectioned at UCLA’s Translational Pathology Core Laboratory. Tissue sections were incubated in a vacuum oven at 60°C for 45-60 minutes, then transferred into xylene (Fisher Chemical) ×3, 100% alcohol (Decon Labs) ×2, 95% ethanol ×1, and 70% ethanol ×1, for 3 minutes each. Slides were washed in phosphate buffered saline (PBS) (Gibco) for 5 minutes prior to epitope unmasking using a heat antigen retrieval step. Staining of sections was performed using the manufacturer’s protocol for the following anti-human antibodies: CD45 (DakoCytomation), CD3 (DakoCytomation), CD4 (Thermo Scientific), CD8 (DakoCytomation), CD11c (Abcam), CD19 (DakoCytomation), CD20 (DakoCytomation), CD68 (DakoCytomation), and CD163 (Cell Marque).
Animal work
Immunocompetent male C57BL/6J and C57BL/6N (BL/6) mice from Jackson Laboratories and the UCLA Department of Radiation Oncology’s animal core facility were used in experiments. Mice were bred and maintained by the UCLA Division of Laboratory Animal Medicine (DLAM), and all animal protocols were approved by DLAM.
Mouse prostate dissociation to single cells
Male C57BL/6 mice were euthanized using CO2, and prostate tissue was isolated by dissection, diced with a razor blade, and dissociated in RPMI 1640 (Gibco) containing 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (Gibco), 1 mg/mL collagenase type I (Gibco), and 0.1 mg/mL DNase (Sigma) at 37°C on a nutating platform for 60-90 minutes. Cell pellets were washed in 1× DPBS (Gibco) and incubated in TrypLE Express Enzyme, no phenol red (Thermo Fisher Scientific) for 5 minutes at 37°C. Cells were drawn through an 18 G syringe and sequentially passed through a 100 µm and 70 µm cell strainer (Corning).
Human prostate cell preparation and sorting
De-identified human prostatectomy-derived prostate tissue samples were provided by the UCLA Translational Pathology Core Laboratory. Histologically benign regions were identified by a Genitourinary Pathologist and tissue was mechanically and enzymatically dissociated to single cells for fluorescence-activated cell sorting (FACS) as described previously [31]. Samples were run on a FACSAria II cell sorter (BD Biosciences).
Antibodies for mass cytometry
Pre-conjugated antibodies for mass cytometry were purchased from Fluidigm (South San Francisco, CA) or conjugated at UCLA using MaxPar X8® Polymer chemistry (Fluidigm, South San Francisco, CA) according to the manufacturer’s instructions. Antibodies used in mouse and human panels are shown in Tables 1, 2.
Table 1.
CyTOF antibodies for mouse prostate immune cells
| Species | Label | Target | Clone | Conjugation | Source |
|---|---|---|---|---|---|
| Mouse | 89Y | CD45 | 30-F11 | Pre-conjugated | DVS |
| Mouse | 139La | CD27 | LG.3A10 | Maxpar Kit | BioLegend |
| Mouse | 141Pr | CD138 | 281-2 | Maxpar Kit | BioLegend |
| Mouse | 144Nd | CD45R (B220) | RA3-6B2 | Pre-conjugated | DVS |
| Mouse | 145Nd | CD4 | RM4-5 | Pre-conjugated | DVS |
| Mouse | 146Nd | F4/80 | BM8 | Pre-conjugated | DVS |
| Mouse | 148Nd | CD11b | M1/70 | Pre-conjugated | DVS |
| Mouse | 152Sm | CD3e | 145-2C11 | Pre-conjugated | DVS |
| Mouse | 155Gd | CD25 | 3C7 | Maxpar Kit | BioLegend |
| Mouse | 162Dy | Ly6C | HK1.4 | Pre-conjugated | DVS |
| Mouse | 166Er | CD19 | 6D5 | Pre-conjugated | DVS |
| Mouse | 168Er | CD8a | 53-6.7 | Pre-conjugated | DVS |
| Mouse | 173Yb | CD117 | 2B8 | Maxpar Kit | BioLegend |
| Mouse | 176Yb | FcεR1a | MAR-1 | Pre-conjugated | DVS |
| Mouse | 209Bi | CD11c | N418 | Pre-conjugated | DVS |
Table 2.
CyTOF antibodies for human prostate immune cells
| Species | Label | Target | Clone | Conjugation | Source |
|---|---|---|---|---|---|
| Human | 89Y | CD45 | HI30 | Pre-conjugated | DVS |
| Human | 142Nd | CD19 | HIB19 | Pre-conjugated | DVS |
| Human | 143Nd | CD25 | M-A251 | Maxpar Kit | BD Biosciences |
| Human | 145Nd | CD4 | RPA-T4 | Maxpar Kit | BioLegend |
| Human | 146Nd | CD8a | RPA-T8 | Maxpar Kit | BioLegend |
| Human | 147Nd | CD11c | Bu15 | Pre-conjugated | DVS |
| Human | 149Sm | CD25 | 2A3 | Pre-conjugated | DVS |
| Human | 151Eu | CD103 | Ber-ACT8 | Pre-conjugated | DVS |
| Human | 153Eu | CD62L | DREG-56 | Pre-conjugated | DVS |
| Human | 156Gd | CD183 (CXCR3) | G025H7 | Pre-conjugated | DVS |
| Human | 158Gd | CD33 | WM53 | Pre-conjugated | DVS |
| Human | 159Tb | CD197 (CCR7) | G043H7 | Pre-conjugated | DVS |
| Human | 160Gd | CD14 | M5E2 | Pre-conjugated | DVS |
| Human | 163Dy | CD56 (NCAM) | NCAM16.2 | Pre-conjugated | DVS |
| Human | 164Dy | CD45RO | UCHL1 | Pre-conjugated | DVS |
| Human | 165Ho | CD16 | 3G8 | Pre-conjugated | DVS |
| Human | 169Tm | CD45RA | HI100 | Pre-conjugated | DVS |
| Human | 170Er | CD3 | UCHT1 | Pre-conjugated | DVS |
| Human | 209Bi | CD11b | ICRF44 | Pre-conjugated | DVS |
Cell surface staining for mass cytometry
Cell staining buffer was prepared with 1× DPBS (Gibco) containing 5% bovine serum albumin (BSA) protease-free (Sigma-Aldrich) and 0.2% NaN3 (Sigma-Aldrich). From single-cell suspension into tubes, 1×105-1.5×105 cells were aliquoted for the unstained control, and 3×105-1.8×106 cells were aliquoted for the stained samples. For Live/Dead staining with rhodium, samples were centrifuged and resuspended at 1×106 cells/mL in cell staining buffer containing 1 µM Cell-ID Intercalator-103Rh (Fluidigm) and incubated at 37°C for 15 minutes. For Live/Dead staining with cisplatin, samples were centrifuged and resuspended at 1×107 cells/mL in cell staining buffer. Stock Cell-ID Cisplatin (Fluidigm) was added to samples for a final concentration of 5 μM, and samples were incubated for 5 minutes at room temperature. Live/Dead stain was quenched with 2 mL cell staining buffer and centrifuged. After Live/Dead staining, the antibody cocktail was diluted to 1 µL of each antibody per 50 µL per sample. For mouse samples, cells were resuspended in 45 µL cell staining buffer and 5 µL of 5 µg/mL TruStain fcX (anti-mouse CD16/32) Antibody (BioLegend, 101319). Mouse samples were incubated at room temperature for 10 minutes, then 50 µL of the antibody cocktail was added. For human samples, cells were resuspended in 50 µL of cell staining buffer and 50 µL of the antibody cocktail was added. Samples were incubated with antibodies at room temperature for 30 minutes, washed twice with cell staining buffer and resuspended in 1 mL Maxpar Fix and Perm Buffer (Fluidigm) containing 125 nM Cell-ID Intercalator-Ir (Fluidigm). Samples were incubated for 12-48 hours at 4°C. Samples were then washed a total of 3 times with cell staining buffer, 1× DPBS (Gibco), then MilliQ Water (Millipore). Mouse prostate samples were passed through a 40 µm strainer (Corning) between the DPBS and MilliQ Water washes. After the final wash, cells were resuspended in a residual amount of MilliQ Water for mass cytometry.
Mass cytometry
Mass cytometry was performed at the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility. Pellets were washed 2× with Maxpar® cell staining buffer (Fluidigm, South San Francisco, CA), 2× with MilliQ water and resuspended in 10% EQ™ Four Element Calibration Beads (Fluidigm, South San Francisco, CA) containing natural abundance cerium (140/142Ce), europium (151/153Eu), holmium (165Ho), and lutetium (175/176Lu). Samples were run on a Helios® mass cytometer (Fluidigm, South San Francisco, CA) using an event rate of 300-500 events/second. Data was normalized using bead-based normalization in the CyTOF software.
CyTOF clustering
Manual gating for live CD45+ singlets in each sample was performed in FlowJo V10 (FlowJo LLC). Each sample was given a unique Sample ID, then all samples were concatenated into a single .fcs file. On this file T-Distributed Stochastic Neighbor Embedding (t-SNE) in FlowJo V10 was performed using all surface markers besides CD45 and the following settings: Iterations, 3000; Perplexity, 40; Eta (learning rate), 17594; Theta, 0.5; Force Recalculation, No; Limit Duration, No. Heatmaps of marker expression were generated using the Color Map Axis function. To cluster cells with Spanning-Tree Progression Analysis of Density-Normalized Events (SPADE), CD45+ cells were exported from FlowJo V10 as .fcs files. The .fcs files were uploaded to ImmPort Galaxy and run through the “Transform FCS data” function without transformation to generate a .fcs file compatible for SPADE clustering. SPADE V3.0, a stand-alone program from Dr. Qui’s lab at the Georgia Institute of Technology and Emory University for SPADE clustering not requiring MATLAB was used for SPADE clustering and generation of a minimum spanning tree. ImmPort Galaxy and SPADE V3.0 are freely available programs (Bhattacharya et al., 2014; Qiu et al., 2011). SPADE clustering was performed on T cells from human prostates with 200 target nodes. X-Shift clustering was performed in the VorteX clustering environment (Samusik et al., 2016). Manually gated live CD45+ singlets were gated in FlowJo V10, exported as a new .fcs file for each sample, and imported to VorteX. X-Shift clustering was performed using all shared surface markers using the following settings: arcsinh (x/f), f = 5; Noise Threshold, 1.0; Feature Rescaling, none; Normalization, none; Distance Measure, Angular Distance; Clustering Algorithm, X-shift algorithm; Density Estimate, N nearest neighbors; Define K Values, K values from 5 to 150 at 30 step intervals; Number of Neighbors, determine automatically (Kimball et al., 2018). The algorithm was run for the set K values, then VorteX was used to calculate the optimal K value at the elbow point of the graph of K value versus the number of clusters generated. These clusters were visualized on a minimum spanning tree (MST) generated in VorteX where each cluster was represented as a node on the MST.
Quantitative polymerase chain reaction
FACS was used to sort out CD3+ CD8- T cells and CD3+ CD8+ T cells from total dissociated human prostate. RNA was purified from sorted cells using the RNeasy micro kit (Qiagen) following the manufacturer’s protocol. Reverse transcriptase was used to convert RNA to cDNA, then qPCR was performed using the ViiA 7 Real-Time PCR System (Applied Biosystems) to quantify levels of CD4, CD8, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers used for CD4: CCACTCAGGGAAAGAAAGTGGT, GCGCGATCATTCAGCTTGGA. Primers used for CD8: AACCACAGGAACCGAAGACG, GGGTTAGACGTATCTCGCCG. GAPDH was measured using RT² qPCR Primer Assay for Human GAPDH (QIAGEN).
Statistical analysis
Statistical analysis was performed in Prism V7 (GraphPad). Unless otherwise stated, two-tailed Student’s T-test assuming unequal variance was performed to determine statistical significance. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Results
Range of immune cells are found in the human prostate
To study the prostate immune microenvironment, we first performed immunohistochemistry to detect immune cells in benign human prostate. Cells staining positive for the pan-leukocyte marker CD45 were detected in the prostate (Figure 1). To further investigate the immune cells in the prostate, we stained for markers of different immune lineages and found evidence of T cells (CD3, CD4, CD8), B cells (CD19, CD20), and various types of myeloid cells (CD11c, CD68, CD163) (Figure 1). Immune cells in the prostate were mainly localized to the stroma, while a smaller fraction of immune cells was found nestled between epithelial cells (Figure 1). The presence of many immune cell-types shows that the prostate has a diverse immune microenvironment that warrants a more comprehensive approach to characterize.
Figure 1.
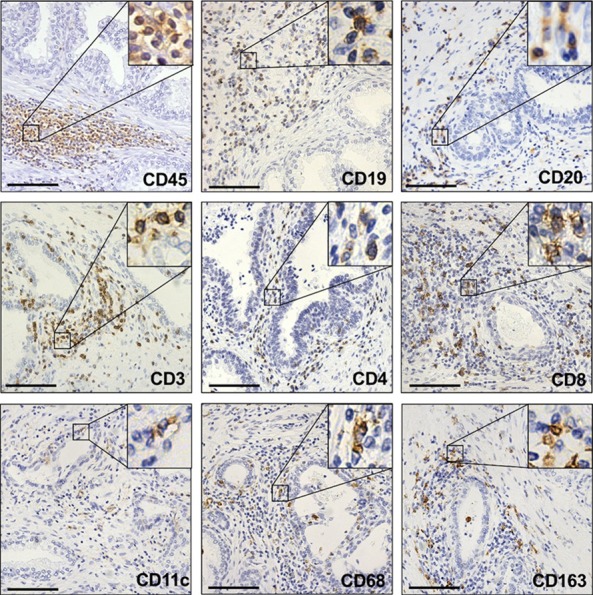
Detection of prostatic immune cells by immunohistochemistry. Representative images of benign human prostate stained for markers of immune cells. CD45 is a pan-leukocyte marker. CD19 and CD20 stain B cells. CD3, CD4, and CD8 stain T cells. CD11c, CD68, and CD163 stain myeloid cells. Insets show 4× magnification of positively-stained cells. Scale bars, 100 µm.
CyTOF allows reproducible detection of immune cells in the prostate
We used CyTOF to comprehensively phenotype immune cells in the prostate using a panel of established leukocyte surface markers. We first tested this assay in adult (12 week-old) wild type C57BL/6 mice, representing a consistent source of prostate tissue. Whole mouse prostate was mechanically and enzymatically dissociated to a single cell suspension which was then stained with a panel of leukocyte markers for CyTOF analysis, allowing for detection and characterization of immune cells present in the prostate. Myeloid cells were found to comprise the largest portion of the immune compartment of the mouse prostate (Figure 2A, 2B). We therefore designed our panel of 15 murine leukocyte markers with an emphasis on myeloid antigens (Table 1).
Figure 2.
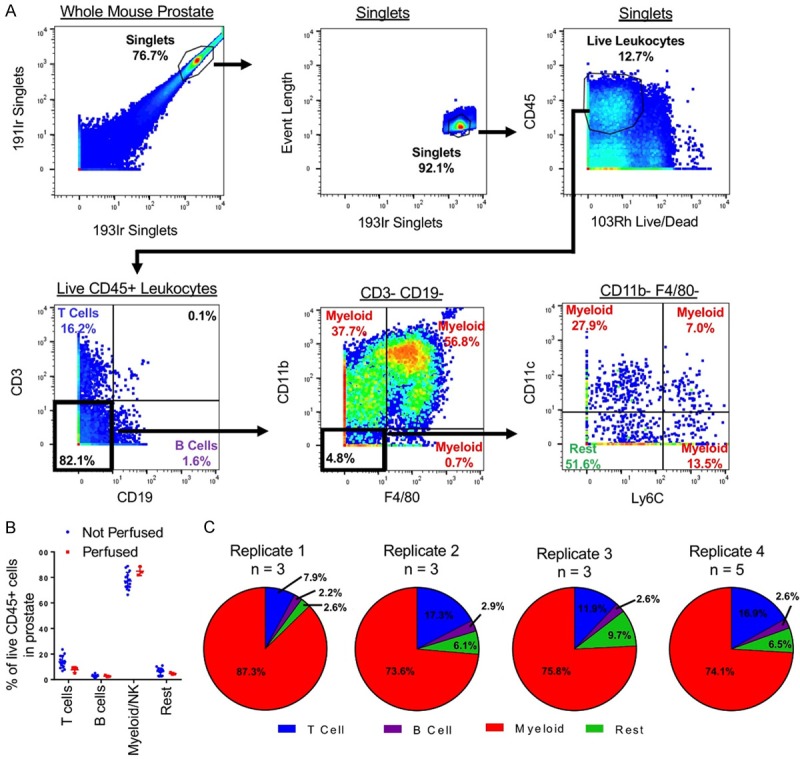
Characterization of the mouse prostate immune microenvironment with mass cytometry. A. Manual gating scheme for classifying mouse prostate immune cells into T cells, B cells, myeloid cells, and rest. B. Frequencies of major immune cell populations in the prostates of mice detected using CyTOF from mice that received no perfusion (n = 14) and those that were perfused with PBS (n = 3). P > 0.05. C. Pie charts showing frequencies of major immune cell populations in the prostates of un-perfused mice over 4 biological replicate CyTOF experiments.
To examine the gross immune cell composition of the mouse prostate, each immune cell in the prostate was classified into one of four major groups of immune cells. First, 191Ir and 193Ir were used to identify singlets. Then, live leukocytes were identified as expressing CD45 and lacking 103Rh signal. Next, cells expressing CD3 were classified as T cells and cells expressing CD19 were classified as B cells. Myeloid cells were classified with an absence of CD3 and CD19 and expression of at least one of the markers CD11b, CD11c, F4/80, and Ly6C. The remaining cells were classified as “Rest” (Figure 2A). The mouse prostate was found to be dominant in myeloid cells (M = 77.19%, SD = 6.79). Lymphocytes, consisting of T cells (M = 13.99%, SD = 4.88) and B cells (M = 2.57%, SD = 0.99), made up a minority of the immune cells in the mouse prostate (Figure 2B).
Myeloid dominance in the mouse prostate differs from immune cell composition of circulating blood, which is lymphoid dominant, and is consistent with myeloid dominance found in other non-mucosal organs in the mouse [20,32]. To further show that the immune cells detected using CyTOF correspond to immune cells in the tissue and are not overly influenced by cells passing through blood vessels, we perfused 3 mice with PBS to flush the circulatory system before harvesting the prostate. Perfused mice did not differ significantly in frequencies of T cells, B cells, or myeloid cells (Figure 2B), indicating that the assay is not altered by the presence or absence of leukocytes in prostatic blood vessels.
Next, we assessed the reproducibility of CyTOF phenotyping, comparing immune cell frequencies in the prostates of 14 mice over 4 replicate experiments. Between 4 experiments conducted on different days, we detected a very similar make up of prostatic immune cells when evaluating T cells, B cells, and myeloid cells (Figure 2C). These results indicate that CyTOF is a reproducible approach to define immune cells in the prostate.
CyTOF reveals heterogeneity of myeloid cells in the mouse prostate
An advantage of CyTOF is that it allows for comprehensive phenotypic characterization of individual cells. Using the immune markers in our panel we were able to further subdivide the major immune cell lineages into more specific immune subsets. Using just 4 markers—CD11b, CD11c, F4/80, and Ly6C—we identified 8 phenotypically-distinct myeloid subsets in the mouse prostate (Figure 3A, 3B). The most abundant myeloid population in the mouse prostate is CD11c+ CD11b- Ly6C- F4/80+, comprising nearly 40% of myeloid cells (Figure 3B, 3C). The other 7 myeloid populations range from 28.1% of total myeloid cells to less than 2.0% (Figure 3B, 3C). These results demonstrate that the myeloid compartment of the prostate is highly heterogeneous, containing many subpopulations which may each have a unique role in prostate biology. Subpopulations 1 and 2 can be further subdivided using the markers CD25 and CD117 (Figure 3A), showing greater immune cell heterogeneity when additional markers are used.
Figure 3.
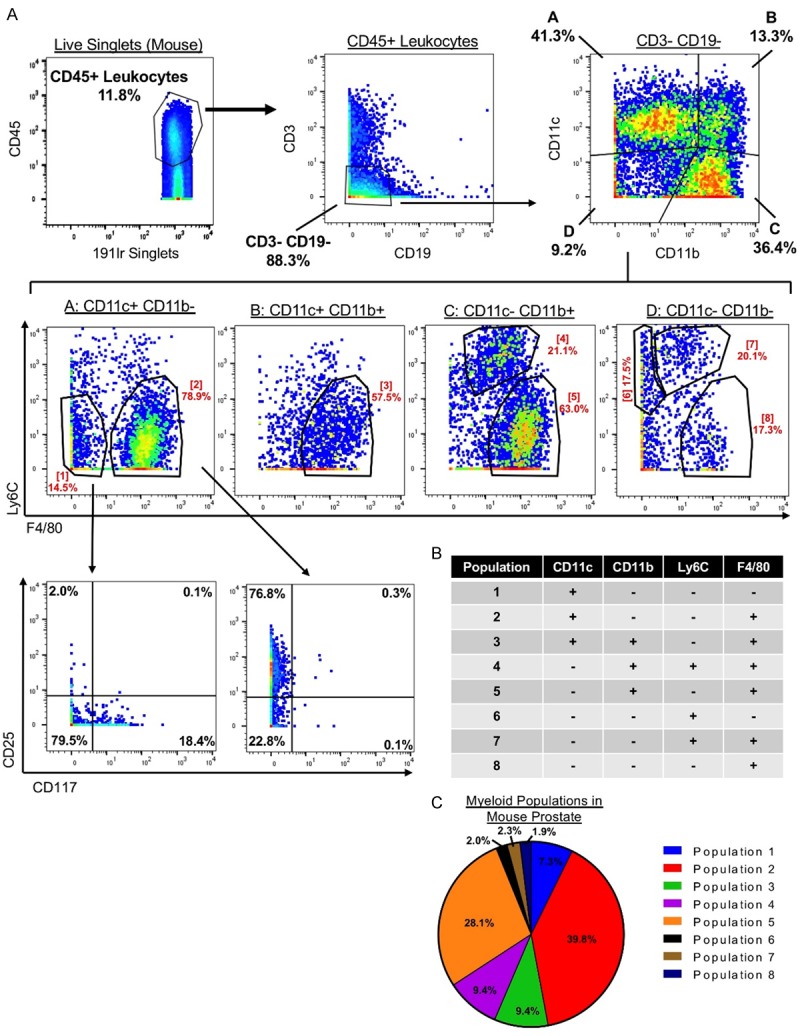
The mouse prostate contains a heterogeneous myeloid compartment. A. Gating scheme showing identification of 8 phenotypically distinct myeloid subsets in the mouse prostate using 4 myeloid markers CD11c, CD11b, Ly6C, and F4/80. Top: CD45+ CD3- CD19- cells are gated to study myeloid cells. CD11c and CD11b separate 4 populations (A-D). Middle: Ly6C and F4/80 further subdivide populations A-D into 8 myeloid populations (1-8). Bottom: Populations 1 and 2 are further subdivided using the markers CD25 and CD117. B. Expression of 4 myeloid markers in 8 identified myeloid population. C. Pie chart showing contribution of 8 myeloid populations to the total pool of myeloid cells in the mouse prostate.
Human prostate contains a unique immune microenvironment rich in T cells
Next, we set out to phenotype the immune compartment of the benign human prostate. Tissue samples used in these experiments came from benign regions of the prostate, confirmed by our genitourinary pathologist to be histologically benign, from patients undergoing radical prostatectomy. Tissues defined were dissociated to single cells, and CD45+ leukocytes were sorted using fluorescence-activated cell sorting (FACS). Sorted cells were stained with a panel of 19 human leukocyte markers for CyTOF analysis (Table 2).
We used CyTOF to profile the immune cell composition of benign human prostate samples from 9 patients (Figure 4A). Due to differences in lifestyle, age, diseases, and genetics when using human samples, we hypothesized that each patient would differ in the composition of prostatic immune cells. Interestingly, all 9 benign prostates were T cell dominant (M = 75.94%, SD = 11.30). B cells were the second largest fraction of prostatic immune cells (M = 16.27%, SD = 10.28), while myeloid cells made up a minority of immune cells in the benign human prostate (M = 4.92%, SD = 1.99) (Figure 4B, 4C). T cells are also the major immune cell type in circulating blood [33]. We performed a one-sample t-test against frequencies of immune cells reported in peripheral human blood [33], and found that the frequencies of T cells and myeloid cells in the prostate are significantly different from their frequencies in circulating blood (Figure S1).
Figure 4.
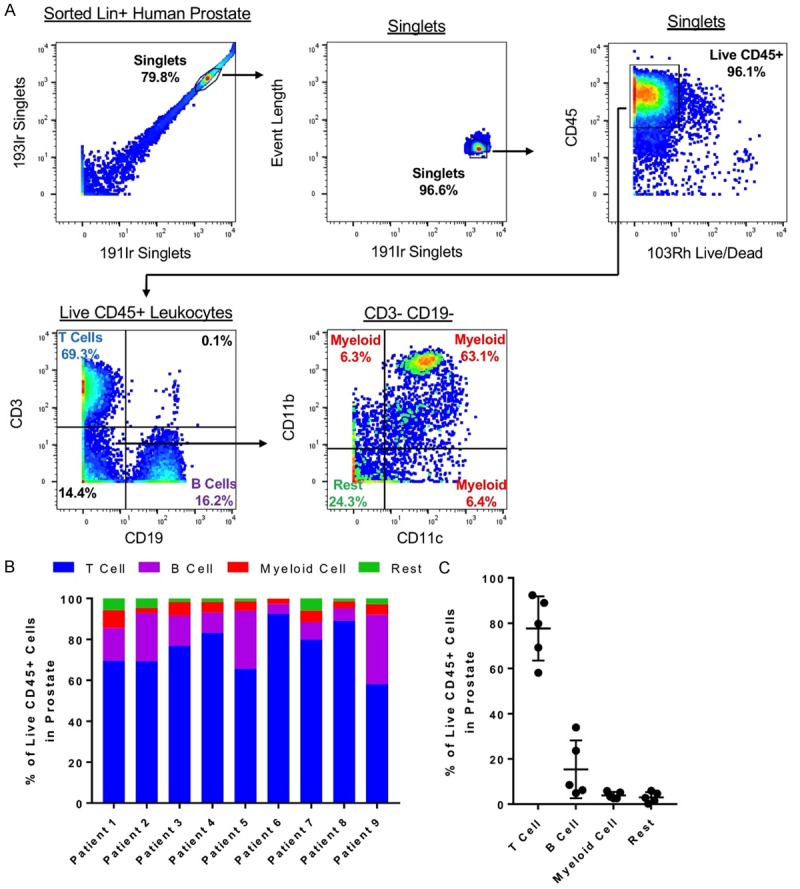
Characterization of the human prostate immune microenvironment with mass cytometry. (A) Manual gating scheme for classifying human prostate immune cells into T cells, B cells, myeloid cells, and rest. (B, C) Immune microenvironment composition of 9 de-identified benign human prostates shown individually (B) and as a group (C). P<0.01.
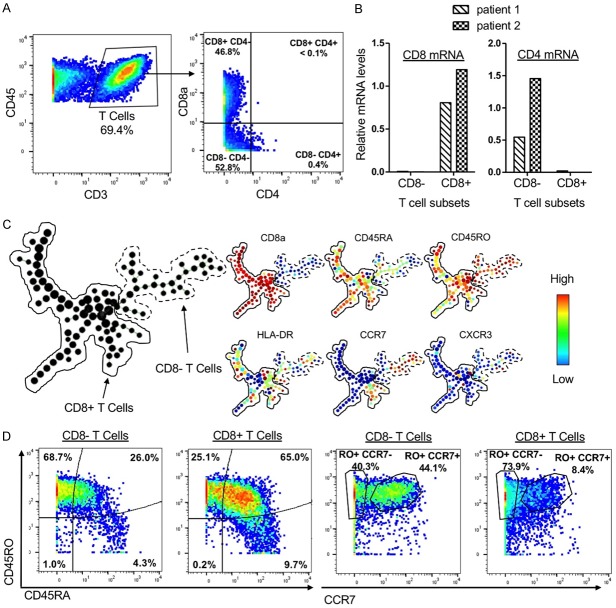
We were unable to detect some markers using this assay, notably CD4. T cells, identified as expressing CD3, can be further subdivided into CD4+ helper T cells and CD8+ cytotoxic T cells. However, we could not detect CD3+ CD4+ cells using this assay (Figure 5A). Enzymes required to digest human prostate tissue, such as collagenase and dispase, are known to cleave lymphocyte surface markers, rendering them undetectable by CyTOF [34]. We hypothesized that CD4 may be cleaved during the dissociation protocol, resulting in an inability to detect CD4+ T cells. We used FACS to sort populations of CD3+ CD8+ and CD3+ CD8- cells and evaluate expression of CD4 and CD8 mRNA. We found that CD3+ CD8- cells express CD4 mRNA while CD3+ CD8+ cells express CD8 mRNA in a mutually exclusive manner (Figure 5B). Spanning-tree progression analysis of density-normalized events (SPADE) was used to cluster CD3+ T cells in the human prostate and to observe the heterogeneity of marker expression. Many CD8- clusters expressed high levels of CCR7 (Figure 5C), which is known to be expressed on peripheral CD4+ T cells in humans [35], providing additional evidence that CD3+ CD4- CD8- cells contain CD4+ T cells that lose the CD4 antigen during tissue sample processing.
Figure 5.
The human prostate contains a heterogeneous T cell compartment. A. Gating showing T cell subsets and an absence of CD3+ CD4+ T cells. B. Expression of CD4 and CD8 mRNA in FACS-sorted CD3+ CD8- and CD3+ CD8+ T cells from 2 patients. Values are normalized to GAPDH. C. Left: SPADE plot containing 200 nodes representing T cells from a single patient. Right: SPADE plots with heatmap of expression for markers found on T cells. D. Comparison of CD8- and CD8+ T cells from a single patient in expression of CD45RA, CD45RO, and CCR7.
We found that the human prostate contains a heterogeneous T cell compartment. Using other markers found on subsets of T cells, we were able to identify many clusters within the T cell compartment that differed in expression of CD45RA, CD45RO, HLA-DR, CCR7, and CXCR3 (Figure 5C). The diversity of T cells suggests that the T cell compartment of the prostate may perform many different functions. We also identified differential expression of the naïve/memory markers CD45RA and CD45RO between CD8+ and CD8- T cells. The majority of CD8- T cells were CD45RA- CD45RO+ (Figure 5D), corresponding to the phenotype reported for memory T cells [26]. Meanwhile, the majority of CD8+ T cells were CD45RA+ CD45RO+ (Figure 5D), suggestive of a less differentiated stem cell memory phenotype [26]. Memory T cells, which express CD45RO, can be further subdivided into effector memory T cells, which are CCR7-, and central memory T cells, which are CCR7+ [35]. CD8- T cells had relatively similar proportions of CD45RO+ CCR7- (40.3%) and CD45RO+ CCR7+ (44.1%) cells, showing that the human prostate contains effector and central memory CD8- T cell phenotypes (Figure 5D). CD8+ T cells were found to be comprised of mostly effector memory T cells (73.9%), with central memory T cells making up only a small fraction of the total CD8+ T cells (8.4%) (Figure 5D).
Unsupervised clustering of human prostatic immune cells reveals differences between patients
In order to visualize the complexity of immune cells in the human prostate, we used the tSNE algorithm to cluster the immune cells from 5 patients into a two-dimensional plot based on similarity in marker expression. Each immune cell is represented by a point, and the proximity of cells in two-dimensional space reflects similarities in marker expression across multi-dimensional space. Viewing heat maps of marker expression overlayed onto the tSNE plot, we observe clear clustering of cells with similar expression, supporting successful clustering of immune cells and dimension reduction using the tSNE algorithm (Figure 6).
Figure 6.
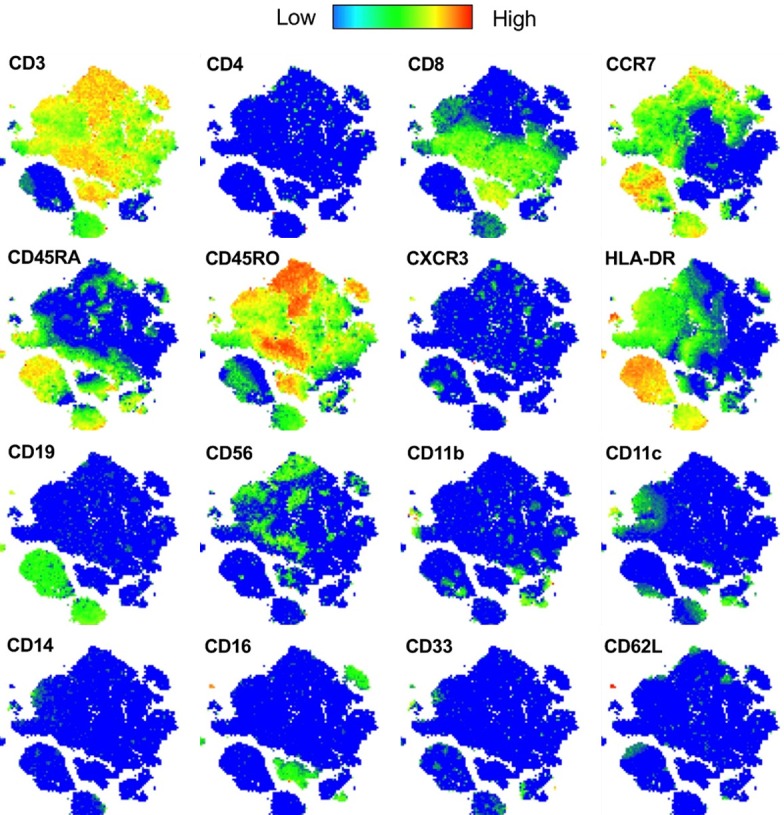
Human immune cell tSNE plot marker expression. tSNE plot generated from the prostate immune cells from 5 human patients. Heat maps of expression for each marker visualized using the Color Map Function.
To visualize contributions of each patient to the total pool of immune cells, we differentially colored cells from each patient on the tSNE plot (Figure 7A). The immune cells from each patient mostly clustered separately, with some overlap between patients (Figure 7A). These results show patient-specific differences in prostate immune cells. Manual gating to identify subpopulations, as shown in Figure 3A, becomes unfeasible as the number of markers used increases. Therefore, we compared more specific subsets of immune cells identified through unsupervised clustering using the X-shift algorithm [36]. Samples from 5 patients (Patients 2, 6, 7, 8, and 9) were used for this analysis because they shared the same panel of markers. Leukocytes from each sample were gated in FlowJo using CD45 then exported to the VorteX environment for clustering.
Figure 7.
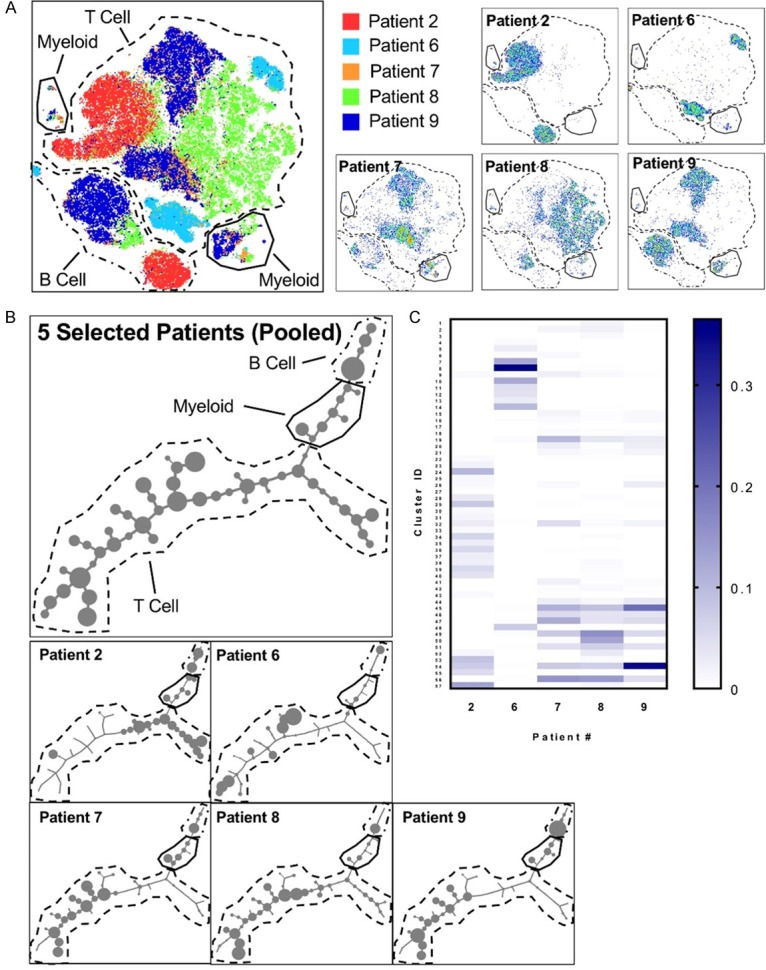
CyTOF reveals differences between patients at the immune cell subset level. A. Left: tSNE plot generated from the prostate immune cells from 5 human patients and equal numbers of immune cells from each patient. Right: tSNE-clustered immune cells from 5 individual patients shown separately. B. Minimum spanning tree (MST) of 57 human prostate immune cell clusters generated using the X-shift algorithm. Each cluster is represented by a node. Node size is proportional to number of cells in the cluster. Top: MST of 5 pooled patients. Bottom: MSTs showing each patient separately. C. Heatmap showing frequencies of 57 X-shift-generated human prostate immune cell clusters in 5 patients. Color bar represents the cluster’s abundance in a single patient as a fraction of total immune cells in the prostate.
The X-shift algorithm uses k-nearest-neighbor density estimation to automatically group cells based on similarity of expression of each marker and to choose the optimal number of clusters to prevent generalization and over-fragmentation [36]. Clustering was performed using the markers CD11b, CD56, CD45RO, CD3, CD103, CD62L, CD33, CD14, CD16, CD19, CD4, CD8a, CD11c, CCR7, CD45RA, and HLA-DR. With an elbow point at k = 65, the algorithm produced 57 clusters of immune cells representing all immune subsets found in the prostates of these 5 patients. Clusters were arranged into a minimum spanning tree (MST), allowing for visualization of relationships between groups of immune cells (Figure 7B). T cells made up the largest number of clusters, owing to the T cell majority in the human prostate and to differential expression of surface markers on the T cells (Figure 7B). Looking at the MST for each patient individually, we found that individual patients differed in the prevalence of many of the clusters, showing that patients had substantial differences in the subsets of immune cells found in their prostates (Figure 7B, 7C). Some patients showed similar patterns, but none of the 5 patients used for this analysis had the same signature (Figure 7C). While each benign human prostate had a relatively similar proportion of major immune lineages (T cells, B cells, myeloid cells), we identified patient-specific differences within subsets of these lineages using CyTOF (Figures 4B and 7C).
Discussion
In order to gain a better understanding of the prostate immune microenvironment, we used CyTOF to characterize immune cells in the prostates of mice and humans with a level of complexity not previously reported. In the mouse, we first verified that the assay is consistent between replicates (Figure 2C). Myeloid cells were found to comprise the majority of the mouse prostate. Using 4 myeloid markers, CD11b, CD11c, Ly6C, and F4/80, we identified 8 different myeloid populations in the mouse prostate (Figure 3B). Some of these myeloid populations can be further subdivided with CD25 and CD117 (Figure 3A). These results demonstrate that the immune microenvironment of the mouse prostate is highly heterogeneous. Without using such a multiplexed analysis, we would have overlooked this complexity in the myeloid compartment. Whether these immune subsets are preferentially localized to different regions or lobes within the mouse prostate has yet to be determined.
Using CyTOF to characterize immune cells in the human prostate, we found surprising consistency in the frequencies of major immune lineages (T cells, B cells, myeloid cells) between patients (Figure 4B, 4C). Using X-Shift clustering on 5 of the human samples, 57 phenotypically distinct clusters of immune cells were identified in the human prostate, with the abundance of specific immune cell clusters varying considerably between patients (Figure 7C). The heterogeneity of the prostate immune cell compartment shown through CyTOF demonstrates the need for multiplexed phenotyping when studying immune cells in the prostate.
While CyTOF is a powerful technique for characterizing the immune compartment of the prostate, it is dependent on the assumption that the single cell suspension that is stained and run on the mass cytometer is representative of the tissue from which it is dissociated. As there are blood vessels running through and surrounding the prostate, some immune cells in the peripheral blood may be taken up with the rest of the sample. When we flushed out the blood, we found no difference in the populations of immune cells in the mouse prostate detected with CyTOF (Figure 2B). These findings suggest that the contribution of immune cells passing through blood vessels to the total pool of immune cells detected in the prostate is negligible.
To produce a single cell suspension from the human prostate, a dense gland with a lot of connective tissue [37], we performed mechanical dissociation and overnight enzymatic digestion [31], which may preferentially select for cells that can withstand the protocol. The overnight enzymatic digestion, which uses collagenase and dispase, may also disrupt certain surface proteins used to phenotype immune cells, impairing their detection by CyTOF [34]. Finally, immune cells detected in the benign human prostate could be affected by the disease state of neighboring tissue [38].
We found fundamental differences in the immune cell composition of the prostates of mice and humans. While mouse prostate is myeloid dominant, T cells account for the majority of immune cells in the human prostate (Figure 3B). Since the human samples come from older patients, the biological age of the prostate tissue may be a factor in the difference between species. In addition to age, the observed difference between species may also be attributed to fundamental differences in tissue structure. While the human prostate is one dense gland containing multiple distinct regions, the mouse prostate is comprised of four lobes [39]. Each of these lobes is anatomically and biochemically distinct, and it is unknown whether a certain lobe is more relevant as a model for the human prostate [40]. Because CyTOF profiles a dissociated, single-cell suspension of the tissue, this assay does not capture information about localization within the tissue. Additionally, it only provides a snapshot of the prostate immune microenvironment at a single time point. Populations of immune cells in the prostate may change over time, and future studies will be needed to examine the dynamics of the prostate immune microenvironment.
Acknowledgements
Special thanks to Donghui Cheng, Preston Crowell, Miriam Guemes, and Jack Mottahedeh. JJF is supported by a scholarship from UCLA Minor in Biomedical Research and the UCLA Undergraduate Research Scholars Program. HIN is supported by the National Science Foundation Graduate Research Fellowship and the Eugene V. Cota-Robles Fellowship. ASG is supported by the Spitzer Family Foundation Fund and the Gill Endowment. This work was supported by the American Cancer Society (RSG-17-068-01-TBG), Department of Defense (W81XWH-13-1-0470), Margaret E. Early Medical Research Trust, NIH/NCI (P50CA092131/UCLA SPORE in Prostate Cancer), Rose Hills Foundation, and support from UCLA’s Jonsson Comprehensive Cancer Center, Broad Stem Cell Research Center, Clinical and Translational Science Institute, and Institute of Urologic Oncology. Mass cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards P30 CA016042 and 5P30 AI028697, and by the James B. Pendleton Charitable Trust, the JCCC, the UCLA AIDS Institute, the David Geffen School of Medicine at UCLA, the UCLA Chancellor’s Office, and the UCLA Vice Chancellor’s Office of Research.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 2.Castle PE, Hillier SL, Rabe LK, Hildesheim A, Herrero R, Bratti MC, Sherman ME, Burk RD, Rodriguez AC, Alfaro M, Hutchinson ML, Morales J, Schiffman M. An association of cervical inflammation with high-grade cervical neoplasia in women infected with oncogenic human papillomavirus (HPV) Cancer Epidemiol Biomarkers Prev. 2001;10:1021–1027. [PubMed] [Google Scholar]
- 3.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol. 2013;35:229–244. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dostert C, Pétrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinarello CA. The paradox of pro-inflammatory cytokines in cancer. Cancer Metastasis Rev. 2006;25:307–313. doi: 10.1007/s10555-006-9000-8. [DOI] [PubMed] [Google Scholar]
- 6.Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurel B, Lucia MS, Thompson IM Jr, Goodman PJ, Tangen CM, Kristal AR, Parnes HL, Hoque A, Lippman SM, Sutcliffe S, Peskoe SB, Drake CG, Nelson WG, De Marzo AM, Platz EA. Chronic inflammation in benign prostate tissue is associated with high-grade prostate cancer in the placebo arm of the prostate cancer prevention trial. Cancer Epidemiol Biomarkers Prev. 2014;23:847–856. doi: 10.1158/1055-9965.EPI-13-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Marzo AM, Marchi VL, Epstein JI, Nelson WG. Proliferative inflammatory atrophy of the prostate: implications for prostatic carcinogenesis. Am J Pathol. 1999;155:1985–1992. doi: 10.1016/S0002-9440(10)65517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Grönberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putzi MJ, De Marzo AM. Morphologic transitions between proliferative inflammatory atrophy and high-grade prostatic intraepithelial neoplasia. Urology. 2000;56:828–832. doi: 10.1016/s0090-4295(00)00776-7. [DOI] [PubMed] [Google Scholar]
- 12.McNeal JE, Bostwick DG. Intraductal dysplasia: a premalignant lesion of the prostate. Hum Pathol. 1986;17:64–71. doi: 10.1016/s0046-8177(86)80156-3. [DOI] [PubMed] [Google Scholar]
- 13.Dai J, Lu Y, Roca H, Keller JM, Zhang J, McCauley LK, Keller ET. Immune mediators in the tumor microenvironment of prostate cancer. Chin J Cancer. 2017;36:29. doi: 10.1186/s40880-017-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ammirante M, Luo JL, Grivennikov S, Nedospasov S, Karin M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature. 2010;464:302–305. doi: 10.1038/nature08782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia AJ, Ruscetti M, Arenzana TL, Tran LM, Bianci-Frias D, Sybert E, Priceman SJ, Wu L, Nelson PS, Smale ST, Wu H. Pten null prostate epithelium promotes localized myeloid-derived suppressor cell expansion and immune suppression during tumor initiation and progression. Mol Cell Biol. 2014;34:2017–2028. doi: 10.1128/MCB.00090-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon OJ, Zhang L, Ittmann MM, Xin L. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci U S A. 2014;111:E592–600. doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HH, Wang L, Jerde TJ, Chan BD, Savran CA, Burcham GN, Crist S, Ratliff TL. Characterization of autoimmune inflammation induced prostate stem cell expansion. Prostate. 2015;75:1620–1631. doi: 10.1002/pros.23043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mimeault M, Batra SK. Development of animal models underlining mechanistic connections between prostate inflammation and cancer. World J Clin Oncol. 2013;4:4–13. doi: 10.5306/wjco.v4.i1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boehm BJ, Colopy SA, Jerde TJ, Loftus CJ, Bushman W. Acute bacterial inflammation of the mouse prostate. Prostate. 2012;72:307–317. doi: 10.1002/pros.21433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haley PJ. Species differences in the structure and function of the immune system. Toxicology. 2003;188:49–71. doi: 10.1016/s0300-483x(03)00043-x. [DOI] [PubMed] [Google Scholar]
- 21.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 22.Crowell PD, Goldstein AS. Functional evidence that progenitor cells near sites of inflammation are precursors for aggressive prostate cancer. Mol Cell Oncol. 2017;4:e1279723. doi: 10.1080/23723556.2017.1279723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Grogan TR, Hieronymus H, Hashimoto T, Mottahedeh J, Cheng D, Zhang L, Huang K, Stoyanova T, Park JW, Shkhyan RO, Nowroozizadeh B, Rettig MB, Sawyers CL, Elashoff D, Horvath S, Huang J, Witte ON, Goldstein AS. Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome. Cell Rep. 2016;17:2596–2606. doi: 10.1016/j.celrep.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strasner A, Karin M. Immune infiltration and prostate cancer. Front Oncol. 2015;5:128. doi: 10.3389/fonc.2015.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flammiger A, Bayer F, Cirugeda-Kuhnert A, Huland H, Tennstedt P, Simon R, Minner S, Bokemeyer C, Sauter G, Schlomm T, Trepel M. Intratumoral T but not B lymphocytes are related to clinical outcome in prostate cancer. APMIS. 2012;120:901–908. doi: 10.1111/j.1600-0463.2012.02924.x. [DOI] [PubMed] [Google Scholar]
- 26.Golubovskaya V, Wu L. Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers (Basel) 2016;8:36. doi: 10.3390/cancers8030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons YA, Wu SY, Overwijk WW, Baggerly KA, Sood AK. Immune cell profiling in cancer: molecular approaches to cell-specific identification. NPJ Precis Oncol. 2017;1:26. doi: 10.1038/s41698-017-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leipold MD, Newell EW, Maecker HT. Multiparameter phenotyping of human PBMCs using mass cytometry. Methods Mol Biol. 2015;1343:81–95. doi: 10.1007/978-1-4939-2963-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 30.Yao Y, Liu R, Shin MS, Trentalange M, Allore H, Nassar A, Kang I, Pober J, Montgomery RR. CyTOF supports efficient detection of immune cell subsets from small samples. J Immunol Methods. 2014;415:1–5. doi: 10.1016/j.jim.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldstein AS, Drake JM, Burnes DL, Finley DS, Zhang H, Reiter RE, Huang J, Witte ON. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat Protoc. 2011;6:656–667. doi: 10.1038/nprot.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu YR, O’Koren EG, Hotten DF, Kan MJ, Kopin D, Nelson ER, Que L, Gunn MD. A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PLoS One. 2016;11:e0150606. doi: 10.1371/journal.pone.0150606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Autissier P, Soulas C, Burdo TH, Williams KC. Evaluation of a 12-color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytometry A. 2010;77:410–419. doi: 10.1002/cyto.a.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abuzakouk M, Feighery C, O’Farrelly C. Collagenase and Dispase enzymes disrupt lymphocyte surface molecules. J Immunol Methods. 1996;194:211–216. doi: 10.1016/0022-1759(96)00038-5. [DOI] [PubMed] [Google Scholar]
- 35.Campbell JJ, Murphy KE, Kunkel EJ, Brightling CE, Soler D, Shen Z, Boisvert J, Greenberg HB, Vierra MA, Goodman SB, Genovese MC, Wardlaw AJ, Butcher EC, Wu L. CCR7 expression and memory T cell diversity in humans. J Immunol. 2001;166:877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- 36.Samusik N, Good Z, Spitzer MH, Davis KL, Nolan GP. Automated mapping of phenotype space with single-cell data. Nat Methods. 2016;13:493–496. doi: 10.1038/nmeth.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison C, Thornhill J, Gaffney E. The connective tissue framework in the normal prostate, BPH and prostate cancer: analysis by scanning electron microscopy after cellular digestion. Urol Res. 2000;28:304–307. doi: 10.1007/s002400000123. [DOI] [PubMed] [Google Scholar]
- 38.Magi-Galluzzi C, Maddala T, Falzarano SM, Cherbavaz DB, Zhang N, Knezevic D, Febbo PG, Lee M, Lawrence HJ, Klein EA. Gene expression in normal-appearing tissue adjacent to prostate cancers are predictive of clinical outcome: evidence for a biologically meaningful field effect. Oncotarget. 2016;7:33855–33865. doi: 10.18632/oncotarget.8944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira DS, Dzinic S, Bonfil AI, Saliganan AD, Sheng S, Bonfil RD. The mouse prostate: a basic anatomical and histological guideline. Bosn J Basic Med Sci. 2016;16:8–13. doi: 10.17305/bjbms.2016.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R, Ward JM, Cardiff RD. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar harbor meeting of the mouse models of human cancer consortium prostate pathology committee. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.