Abstract
Background: De-regulation of Wnt signaling pathways has been shown to be associated with progression of castration-resistant prostate cancer and more recently, studies indicate that both canonical and non-canonical Wnt pathways may mediate resistance to anti-androgen therapies such as enzalutamide. However, the mechanisms by which Wnt signaling is altered in prostate cancer remain poorly understood. Wnt pathway function begins with Wnt biogenesis and secretion from Wnt signal sending cells. While previous studies have investigated downstream mechanisms of Wnt pathway alterations in prostate cancer, little is known on the role of Wnt secretion mediating proteins. Wntless (WLS) is thought to be essential for the secretion of all Wnts. In this study, we sought to understand the role of WLS in prostate cancer. Methods: RNA-seq and gene set enrichment analysis were used to understand expression profile changes in enzalutamide-resistant C4-2B-MDVR (MDVR) cells versus parental C4-2B cells. Quantitative-PCR and western blot were used to confirm RNA-seq data and to assess expression changes of gene targets of interest. Rv1 cells were used as a separate model of enzalutamide-resistant prostate cancer. RNAi was used to inhibit WLS expression. Cell viability, colony formation, and PSA ELISA assays were used to assess cell growth and survival. Results: Transcriptomic profiling revealed enriched Wnt pathway signatures in MDVR versus parental C4-2B cells. We further show that MDVR cells upregulate Wnt signaling and overexpress WLS. Inhibition of WLS decreases Wnt signaling, markedly attenuates prostate cancer cell viability, induces apoptosis, and re-sensitizes enzalutamide-resistant cells to enzalutamide treatment. Lastly, we show that inhibition of WLS reduces AR and AR-variants expression and downstream signaling. Conclusions: Our findings support a role for WLS in the progression of prostate cancer to a treatment-resistant state. Further efforts to understand Wnt signaling pathway alterations in this disease may lead to the development of novel treatments.
Keywords: Wnt signaling, castration-resistant prostate cancer, enzalutamide resistance, wntless, wls, porcupine, porcn
Introduction
Castration-resistant prostate cancer (CRPC) is a significant cause of cancer-related death in men worldwide. Next-generation anti-androgen therapies (NGATs) such as enzalutamide have improved outcomes [1-3]. However, resistance to therapy persists as an impediment to further progress. Understanding the mechanisms by which tumors evade NGAT resistance will enhance our ability to treat patients with CRPC.
Wnt signaling has been shown to be altered in the development and progression of cancer [4,5]. Wnt signaling exerts downstream effects through two separate signaling cascades; 1) the beta-catenin dependent canonical pathway and 2) the beta-catenin independent non-canonical pathways. Both Wnt signaling pathways have been shown to be involved in prostate cancer progression and recently, it was demonstrated that both canonical and non-canonical Wnt signaling may play a role in resistance to NGATs [6,7]. However, the mechanisms underlying Wnt-dependent treatment resistance remain poorly understood.
While studies suggest that altered Wnt signaling may be responsible for progression to an anti-androgen resistant state, these studies have largely focused on downstream pathway components such as altered expression or function of beta-catenin [6]. In contrast, little is known regarding the putative role of altered Wnt secretion in prostate cancer. Wnt secretion is largely controlled by two key players, Porcupine (PORCN) and Wntless (WLS) [8]. Porcupine is a membrane-bound-O-acyl-transferase responsible for lipidation of Wnt proteins [9-12]. Lipidated Wnts then become substrates for WLS which mediates transport of Wnts for extracellular export where they can engage various receptors to elicit downstream signaling [10,13]. Both PORCN and WLS have been shown to play a role in different cancer contexts, but the role of both PORCN and WLS in prostate cancer remains to be elucidated.
In this current study, we present evidence for increased Wnt signaling in a model of enzalutamide resistant CRPC. We also show that both PORCN and WLS are overexpressed in resistant cells. As the increase in WLS was much greater than that of PORCN, we focused on WLS and further demonstrate that WLS mediates Wnt signaling and regulates cellular viability. Inhibition of WLS re-sensitizes enzalutamide-resistant cells to treatment. Finally, we show that WLS regulates AR and AR-variant expression and downstream signaling. These findings expand our understanding of Wnt signaling in advanced prostate cancer.
Materials and methods
Cell lines and reagents
C4-2B cells were kindly provided and authenticated by Dr. Leland Chung (Cedars-Sinai Medical Center, Los Angeles, CA). CWR22Rv1 cells were obtained from the American Type Culture Collection (ATCC). ATCC uses short tandem repeat profiling for testing and authentication of cell lines. All cell lines are routinely tested for mycoplasma every 6 months using ABM mycoplasma PCR detection kit (Cat#: G238). All experiments with these cell lines and their derivatives were conducted within 6 months of receipt or resuscitation after cryopreservation. Cells were maintained in RPMI 1640 media supplemented with 10% fetal bovine serum, 100 IU penicillin and 0.1 mg/ml streptomycin. Enzalutamide-resistant C4-2B cells (C4-2B-MDVR) were described previously and maintained in complete RPMI 1640 supplemented with 20 µM enzalutamide [14]. C4-2B cells were cultured alongside C4-2B-MDVR cells during their creation as an appropriate control. All cells were maintained at 37°C in a humidified incubator with 5% carbon dioxide. Enzalutamide (Cat#: S1250) was purchased from Selleckchem. All transfections were performed using Lipofectamine RNAiMAX (Cat#: 13778150) purchased from ThermoFisher according to manufacturer’s instructions using 10-50 nM siRNA. Non-targeting control siRNA (Cat#: 12935112) and Stealth Wntless (WLS) targeting siRNA were purchased from ThermoFisher. WLS-F-Oligo (Cat#: 10620318): GGUAUUGGAGGAGGAUCACCAUGAU. WLS-R-Oligo (Cat#: 10620319): AUCAUGGUGAUCCUCCUCCAAUACC.
Cell viability assay
Cells were plated at a density of 10,000-40,000 cells/well in 24-well plates in complete RPMI 1640 media without any selection agent. After 24 hours, cells were subjected to indicated treatments. At completion of assay, cell growth was determined using CCK-8 reagent (Dojindo) according to manufacturer’s instructions. For assays testing the effect of WLS-targeting siRNA alone on cellular viability, assay was performed 96 hours after transfection. For assays combining siRNA transfection with drug treatment, cells were transfected 24 hours after plating, subsequently treated with indicated drugs the following day, and viability assays were conducted at 5 (MDVR) and 3 (Rv1) days after drug treatment. Data is displayed as percent of control cell growth. All conditions were performed in triplicate. All experiments were performed at least twice.
Colony formation assay
Cells were plated at 500 cells/well in 6-well plates in complete RPMI 1640 with no selection agent. Plated cells were subsequently treated 24 hours later as indicated. Colonies formed for 14 days. At the completion of the assay, cell colonies were fixed and stained using the following solution for 20 minutes; 0.05% w/v crystal violet, 1% of 37% formaldehyde, 1% methanol, 1X PBS. After staining, colonies were rinsed, allowed to air dry, and counted. Data is displayed as a percent of control cell colony growth. All conditions were performed in duplicate. All experiments were performed at least twice.
RNA-sequencing and GSEA analysis
Transcriptome analysis of the C4-2B and C4-2B-MDVR cell lines was performed with next-generation sequencing (NGS). For this, indexed RNA-Sequencing (RNA-Seq) libraries were prepared from 1 µg total RNA using the TruSeq RNA Library Prep Kit purchased from Illumina according to the manufacturer’s standard protocol, and as previously described [15]. The libraries were then combined for multiplex sequencing on an Illumina HiSeq 2500 System (2x100 bp, paired-end; ~30 million reads/sample). A standard TopHat-Cufflinks workflow was utilized for analysis of gene and transcript isoform expression [16]. For this, sequence reads (FASTQ format) were aligned to the reference human genomic sequence (Feb. 2009, GRCh37/hg19) and splice junction mapping performed with TopHat [17], and allowing for a maximum of two mismatches. The Cufflinks and Cuffdiff tools were used for transcript assembly, quantitation, and differential expression [18]. Transcript abundance/expression values were expressed as FPKM (fragments per kilobase per million fragments mapped), and were normalized with Cuffnorm [16]. GSEA was performed using the Java desktop software (http://software.broadinstitute.org/gsea/index.jsp) [19,20]. The PID_WNT_SIGNALING_PATHWAY, KEGG_WNT_SIGNALING_PATHWAY, and GO_WNT_SIGNALING_PATHWAY datasets were downloaded from the Molecular Signatures Database and were used in the GSEA analysis.
Preparation of whole cell lysates
Cells were harvested, washed with PBS, and lysed in RIPA buffer supplemented with 5 mM EDTA, 1 mM NaV, 10 mM NaF, and 1X Halt Protease Inhibitor Cocktail (Cat#: 78430) purchased from ThermoFisher. Protein concentration was determined with Pierce Coomassie Plus (Bradford) Assay Kit (Cat#: 23236) purchased from ThermoFisher.
Western blot
Protein extracts were resolved by SDS-PAGE and indicated primary antibodies were used. Antibodies; LRP6 (CS-3395S, rabbit monoclonal antibody, 1:1000 dilution), p-LRP6 (CS-2568S, rabbit polyclonal antibody, 1:1000 dilution), and cleaved-PARP (CS-9541S, rabbit monoclonal antibody, 1:1000 dilution) were purchased from Cell Signaling Technology. WLS (MABS87, mouse monoclonal antibody, 1:1000 dilution) was purchased from Millipore Sigma. AR-441 (SC-7305, mouse monoclonal antibody, 1:1000 dilution) was purchased from Santa Cruz Biotechnology. GAPDH (MAB374, mouse monoclonal antibody, 1:10000) and Tubulin (T5168, mouse monoclonal antibody, 1:6000 dilution) were purchased from EMD Millipore and Sigma-Aldrich respectively and were used to monitor the amounts of samples applied. Proteins were visualized with a chemiluminescence detection system (Cat#: WBLUR0500) purchased from Millipore.
Quantitative PCR (qPCR)
Total RNA was extracted using TRizol reagent (Cat#: 15596018) purchased from ThermoFisher. RNA was digested with RNase-free DNase 1 (Cat#: EN05216101) purchased from ThermoFisher. cDNAs were prepared using ImProm-II reverse transcriptase (Cat#: M314C) purchased from Promega. The cDNAs were subjected to quantitative-PCR (qPCR) using SsoFast EvaGreen Supermix (Cat#: 172-5205) purchased from Bio-Rad according to the manufacturer’s instructions. Triplicates of samples were run on default settings of Bio-Rad CFX-96 real-time cycler. Each reaction was normalized by co-amplification of Actin. Data was calculated using the efficiency corrected method. Primers used for qPCR were; ACTIN: F-CCCAGCCATGTACGTTGCTA, R-AGGGCATACCCCTCGTAGATG; PORCN: F-TACCTGAAGCATGCAAGCAC, R-CGGTGTCTACCATGTGCATC; WLS: F-TCATGGTATTTCAGGTGTTTCG, R-GCATGAGGAACTTGAACCTAAAA; LEF1: F-CAGTCATCCCGAAGAGGAAG, R-AGGGCTCCTGAGAGGTTTGT; BAMBI: F-TGCACGATGTTCTCTCTCCT, R-GAAGTCAGCTCCTGCACCTT; AR: F-CCTGGCTTCCGCAACTTACAC, R-GGACTTGTGCATGCGGTACTCA; AR-v7: F-AACAGAAGTACCTGTGCGCC, R-TCAGGGTCTGGTCATTTTGA; PSA: F-GCCCTGCCCGAAAGG, R-GATCCACTTCCGGTAATGCA; NKX3.1: F-CCGAGACGCTGGCAGAGACC, R-GCTTAGGGGTTTGGGGAAG; UBE2C: F-TGGTCTGCCCTGTATGATGT, R-AAAAGCTGTGGGGTTTTTCC.
PSA ELISA
PSA levels were measured in conditioned media from MDVR cellular viability assay testing WLS-targeting siRNA against non-targeting siRNA using a PSA ELISA Kit (KA0208, Abnova, Inc.) according to the manufacturer’s instructions. Data is displayed as percent of control well PSA either with or without normalization to well cell count. All conditions were performed in triplicate. All experiments were performed at least twice.
Statistical analysis
All quantitated data is displayed as percent of control mean ± standard deviation. Significance was assessed using a two tailed two sample equal variance students t-test. A p-value of ≤ 0.05 was accepted as significant.
Results
Wnt signaling is up-regulated in enzalutamide-resistant prostate cancer cells
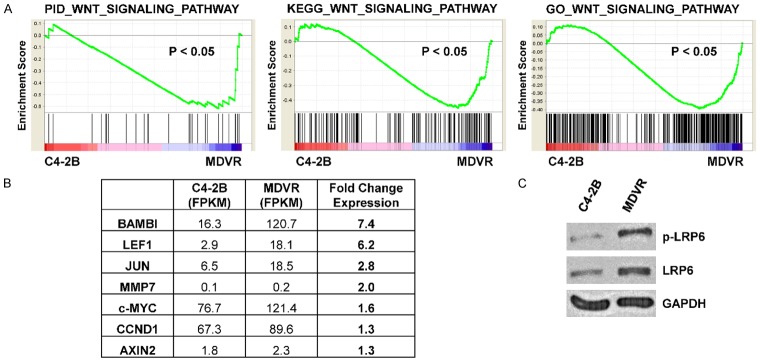
We have previously created and characterized the C4-2B-derived enzalutamide-resistant cell line C4-2B-MDVR (MDVR) as a model of CRPC with acquired resistance to enzalutamide treatment [14,21]. We performed RNA-sequencing to assess transcriptomic changes between MDVR and parental C4-2B cells and utilized gene set enrichment analysis (GSEA) to determine pathway alterations which may mediate resistance to enzalutamide [19,20]. Notably, we found Wnt signaling pathway gene sets were significantly enriched in MDVR cells from three separate data bases; PID_WNT_SIGNALING_PATHWAY, KEGG_WNT_SIGNALING_PATHWAY, and GO_WNT_SIGNALING_PATHWAY (Figure 1A). Enrichment of these gene sets suggests increased Wnt signaling activity in MDVR cells versus parental C4-2B cells. In line with this finding, we indeed found several key Wnt targets up-regulated in our RNA-sequencing data (Figure 1B). We also found increased LRP6 and phospho-LRP6, which is a key component in transmission of Wnt signaling (Figure 1C). These data indicate that increased Wnt signaling may play a role in enzalutamide resistance and suggest that upregulated autocrine Wnt signaling may mediate this change.
Figure 1.
Enzalutamide-resistant MDVR cells display increased Wnt signaling. A. GSEA analysis of RNA-sequencing data comparing MDVR to C4-2B demonstrates that MDVR cells display enrichment of Wnt pathway gene sets. B. RNA-sequencing FPKM values were compared and show increased expression of several Wnt pathway targets in MDVR versus C4-2B cells. C. Western blot shows that LRP6 and phospho-LRP6 expression is increased in MDVR cells versus C4-2B cells. GAPDH served as a loading control.
WLS regulates Wnt signaling in prostate cancer
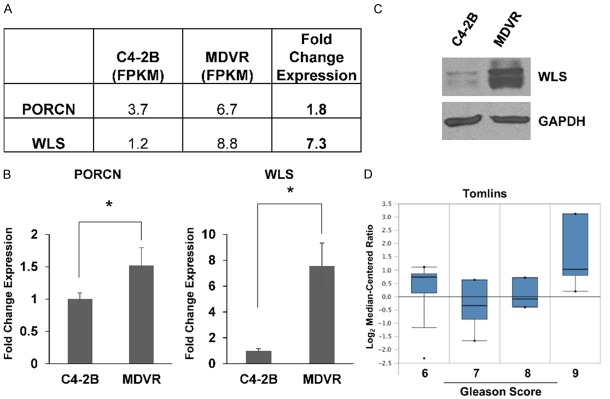
We next sought to determine how Wnt signaling may be altered in MDVR cells. For Wnt proteins to be secreted for induction of downstream signaling, they must first be modified and transported out of signaling cells. PORCN is responsible for lipidation of Wnts, which makes them substrates for WLS, which has been found to play a critical, if not indispensable, role in the secretion of all Wnts [22]. We hypothesized that an increase in PORCN and WLS expression may be involved in the Wnt signaling pathway phenotype observed in MDVR cells. Indeed, both PORCN and WLS were found represented in gene sets used for our GSEA analysis. Interestingly, we did find increased expression of both PORCN and WLS in our RNA-sequencing data comparing MDVR to C4-2B cells (Figure 2A). These results were confirmed by qPCR (Figure 2B). We chose to focus on WLS and show by western blot that WLS protein level is markedly increased in MDVR cells (Figure 2C). Analysis of publicly available data comparing enzalutamide-resistant LNCaP and C4-2 cells to parental controls is consistent with our data showing WLS expression is increased 3.8 and 1.5 fold (data not shown) [6]. We also found that increased WLS expression is associated with high Gleason score prostate cancer in an Oncomine dataset (Figure 2D). These data suggest that WLS may play a key role in prostate cancer progression.
Figure 2.
MDVR cells display increased expression of PORCN and WLS. A. RNA-sequencing analysis shows that both PORCN and WLS expression is increased in MDVR versus parental C4-2B cells. B. qPCR confirms that both PORCN and WLS are overexpressed in MDVR cells. C. Western blot demonstrates increased WLS protein expression in MDVR versus C4-2B cells. GAPDH served as a loading control. D. Analysis of the Tomlins dataset in the Oncomine database demonstrates that WLS expression is associated with Gleason score 9 prostate tumors. * = p-value ≤ 0.05.
We next asked whether WLS regulates Wnt signaling in MDVR cells. Inhibition of WLS by siRNA decreased both LRP6 and phospho-LRP6 protein levels, suggesting a decrease in autocrine Wnt signaling (Figure 3A). Using qPCR, we found that inhibition of WLS lead to decreased transcript levels of two highly expressed Wnt targets found in MDVR cells, LEF1 and BAMBI (Figure 3B). Taken together, these data suggest that increased WLS expression mediates upregulated Wnt signaling in enzalutamide-resistant prostate cancer cells.
Figure 3.
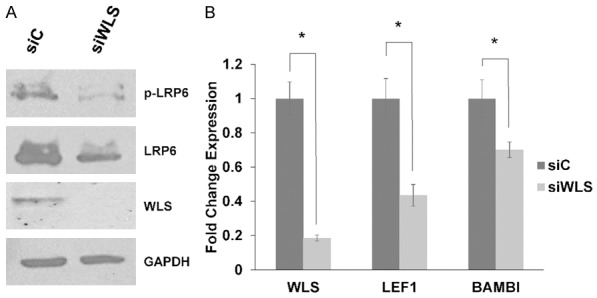
Inhibition of WLS decreases Wnt signaling in MDVR cells. A. Western blots were used to assay for protein levels of WLS, LRP6, and phospho-LRP6 in MDVR cells transfected with WLS targeting siRNA (siWLS) or a non-targeting control siRNA (siC) for 72 hours. GAPDH served as a loading control. B. MDVR cells were transfected with siC or siWLS and harvested after 48 hours. qPCR demonstrates that siRNA mediated inhibition of WLS results in decreased expression of WLS, LEF1, and BAMBI in MDVR cells. * = p-value ≤ 0.05.
Inhibition of WLS decreases proliferation and induces apoptosis
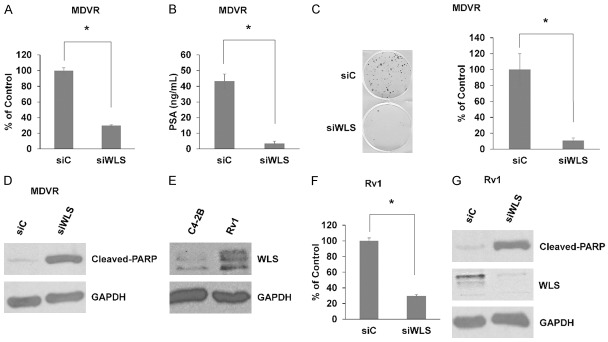
Currently, no other studies have tested the role of WLS in prostate cancer. Thus, we sought to understand how WLS may regulate prostate cancer cell viability. Using cell growth assays, we found that siRNA mediated inhibition of WLS expression significantly decreased MDVR cellular viability (Figure 4A). Decreased viability in response to WLS knockdown was also coincident with significantly lower levels of secreted PSA (Figure 4B). Colony formation assays were performed and show that inhibition of WLS markedly decreases clonogenic potential of MDVR cells (Figure 4C). We also found increased levels of cleaved-PARP in response to WLS inhibition in MDVR cells, indicating induction of apoptosis (Figure 4D).
Figure 4.
Inhibition of WLS attenuates cellular viability and induces apoptosis in enzalutamide-resistant prostate cancer cells. A. Cell viability assay demonstrates that siRNA-mediated inhibition of WLS (siWLS) versus non-targeting siRNA (siC) decreases MDVR cellular viability. B. PSA ELISA shows that inhibition of WLS reduces PSA secretion in MDVR cells. C. Colony formation assays demonstrate that inhibition of WLS reduces clonogenic potential of MDVR cells. D. MDVR cells were treated with siC or siWLS and harvested after 72 hours. Western blot for cleaved-PARP suggests induction of apoptosis in response to decreased WLS expression in MDVR cells. GAPDH served as a loading control. E. Western blot shows that Rv1 cells have increased WLS expression versus C4-2B cells. GAPDH served as a loading control. F. Cell viability assay demonstrates that siRNA-mediated inhibition of WLS decreases Rv1 cellular viability. G. Rv1 cells were treated with siC or siWLS and harvested after 72 hours. Western blot for cleaved-PARP suggests induction of apoptosis in response to decreased WLS expression in Rv1 cells. GAPDH served as a loading control. * = p-value ≤ 0.05.
Our previous work has demonstrated that 22Rv1 (Rv1) prostate cancer cells harbor intrinsic resistance to enzalutamide. Interestingly, we found that Rv1 cells also have increased WLS expression versus C4-2B cells (Figure 4E). Similar to MDVR cells, inhibition of WLS significantly decreases Rv1 cellular viability and leads to increased cleaved-PARP indicating apoptosis (Figure 4F, 4G). These data demonstrate that WLS is a critical component to maintaining viability and promoting survival in enzalutamide-resistant prostate cancer cells.
Inhibition of WLS sensitizes resistant cells to enzalutamide treatment
We’ve shown increased WLS expression in models of enzalutamide-resistant prostate cancer. Whether higher levels of WLS potentiate enzalutamide resistance is unknown. To test this, we used a cell growth assay assessing response to siRNA mediated inhibition of WLS (siWLS), enzalutamide (20 µM), or a combination of both (Figure 5A and 5B). Our results demonstrate that the combination is significantly more effective than either single treatment in both MDVR and Rv1 cells. These data indicate that increased WLS expression represents a novel mechanism of enzalutamide resistance.
Figure 5.
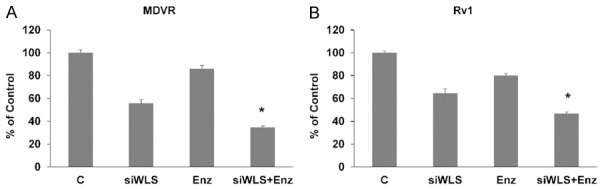
Inhibition of WLS re-sensitizes enzalutamide-resistant prostate cancer cells to enzalutamide treatment. A. Cell viability assay was used to test MDVR cell response to control treatment (C = non-targeting siRNA (siC) and drug vehicle (DMSO)), inhibition of WLS (siWLS), enzalutamide (Enz = 20 µM), or a combination of both. B. Cell viability assay was used to test Rv1 cell response to inhibition of WLS, enzalutamide (20 µM), or a combination of both. * = p-value ≤ 0.05.
WLS regulates expression of AR and downstream signaling
The AR is the primary target for treatment of prostate cancer. It has been demonstrated that there is interplay between Wnt signaling and the AR and downstream AR signaling. Furthermore, Wnt signaling has been demonstrated to positively regulate AR expression [23]. Thus, we sought to test whether WLS may regulate the AR and AR signaling. Western blot demonstrates that siRNA knockdown of WLS inhibits AR protein expression (Figure 6A). Interestingly, we also found decreased levels of AR-variants, which have been demonstrated to mediate enzalutamide resistance through constitutive AR signaling. qPCR further demonstrates that WLS inhibition results in decreased AR expression and also AR-v7 expression (Figure 6B). Inhibition of WLS results in down-regulation of key AR target genes, PSA and NKX3.1, as well as AR-v7 target gene UBE2C (Figure 6C). PSA ELISA results from Figure 4B normalized to numbers of cells remaining in the wells at the time of the assay show that despite lower cell numbers in siWLS wells, PSA secretion is still significantly decreased, in line with qPCR results (Figure 6D). We also found that inhibition of WLS decreases both AR and AR-variants expression (including that of AR-v7) in Rv1 cells (Figure 6E, 6F). These data demonstrate that WLS potentiates both AR expression and signaling and may regulate transition to enzalutamide resistance in part through regulation of AR-variants.
Figure 6.
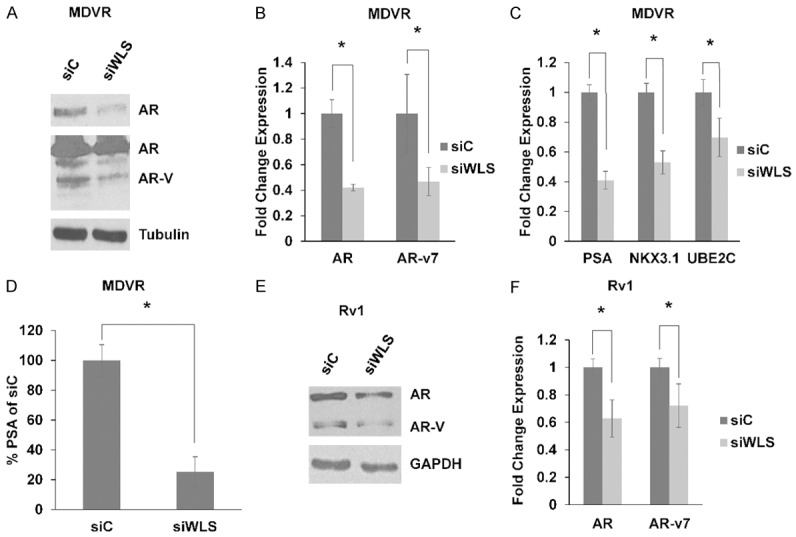
WLS regulates AR expression and AR signaling. (A) MDVR cells were treated with non-targeting siRNA (siC) or WLS-targeting siRNA (siWLS) and harvested after 72 hours. Western blot shows decreased AR (top panel, light exposure) and AR-variant (AR-V) expression (lower panel, darker exposure) in response to siWLS in MDVR cells. Tubulin served a loading control. qPCR shows decreased (B). AR and AR-v7 expression and (C). PSA, NKX3.1, and UBE2C expression in response to siWLS in MDVR cells harvested 48 hours after treatment. (D) PSA ELISA with normalizing PSA by cell number demonstrates decreased PSA secretion in MDVR cells transfected with siWLS. (E) Western blot shows decreased AR and AR-variant expression in response to siWLS treatment in Rv1 cells. GAPDH served as a loading control. (F) qPCR shows decreased AR and AR-v7 expression in response to siWLS treatment in Rv1 cells. * = p-value ≤ 0.05.
Discussion
Despite progress in the field of CRPC treatment, resistance to therapy appears inevitable. Wnt signaling has been implicated in resistance to anti-androgen therapy but the underlying mechanisms remain largely unknown. In the current study, we present evidence that Wnt signaling and resistance to enzalutamide can be potentiated by increased expression of the Wnt secretion mediator WLS. We specifically show that WLS regulates Wnt signaling and promotes tumor cell viability and resistance to treatment. We also show that WLS plays a key role in regulating AR expression and signaling. These results further our understanding of Wnt signaling in advanced prostate cancer and highlight a potential new therapeutic target for this disease.
It is thought that WLS is an essential component for the trafficking of all Wnts and thus, it represents a critical player in downstream Wnt signaling. In support of this, inhibition of WLS results in Wnt loss-of-function phenotypes in several models. Relatively little is known regarding the role of WLS in disease states, but studies have demonstrated a role for WLS in various cancer contexts. Astrocytic gliomas were shown to overexpress WLS and inhibition of WLS decreased tumor viability and in vivo tumor growth [24]. WLS was shown to play a role in bladder cancer progression presumably through regulation of the canonical Wnt signaling pathway [25]. WLS was also shown to 1) be overexpressed in a subset of ovarian, gastric, and breast cancers, 2) be associated with a poor clinical outcome in gastric cancer, and 3) be associated with HER2 expression, although the significance of this last association is poorly understood [26]. Additional studies further implicate WLS in the progression of leukemia and colorectal cancer [27,28].
In our study, we demonstrate that WLS inhibition decreases Wnt signaling, inhibits prostate cancer cell viability, and re-sensitizes resistant cells to treatment. As WLS is thought to regulate the secretion and subsequent function of all Wnts, it is reasonable to hypothesize that WLS serves as a master regulator of both canonical and non-canonical Wnt signaling in prostate cancer [22]. Both canonical and non-canonical Wnt signaling are thought to play a role in prostate cancer and progression to a castration and therapeutic resistant state [29]. Zhang et al presented evidence for increased beta-catenin expression and downstream signaling in enzalutamide resistant prostate cancer, resulting in blunted response to treatment [6]. Miyamoto et al utilized single-cell RNA-sequencing of circulating tumor cells to show that non-canonical Wnt signaling was activated in those patients progressing after treatment [7]. Additional studies to fully understand how WLS regulates Wnt signaling in advanced prostate cancer are warranted.
Our studies further demonstrated that inhibition of WLS results in decreased AR expression and downstream signaling. Interplay between the AR and Wnt signaling pathways has been documented in prostate cancer [30]. Wnt signaling has been shown to promote AR expression and a physical interaction involving the AR and beta-catenin has been demonstrated and well characterized [23,31-34]. Interestingly, the interplay between the AR and beta-catenin signaling pathways may differ by stage of the disease [29,30,35]. In androgen sensitive prostate cancer, it is thought that AR binding to beta-catenin leads to increased AR signaling with a subsequent decrease in Wnt targets expression possibly due to preferential beta-catenin binding to AR rather than TCF/LEF transcription factors. However, in CRPC, it is thought that AR and beta-catenin signaling positively work together to promote expression of a gene signature which potentiates a castration-resistant phenotype. Our work suggests that Wnt/beta-catenin signaling continues to work with AR signaling in enzalutamide-resistant prostate cancer.
Recent efforts have highlighted the important role of AR-variants in mediating a resistant phenotype in CRPC patients [36-38]. Our data indicates that inhibition of WLS results in decreased AR-variant expression and suggests that it also decreases downstream AR-variant signaling. This novel and exciting finding suggests that targeting Wnt signaling may alleviate AR-variant mediated therapeutic resistance making it a highly attractive therapeutic strategy. Understanding how Wnt signaling promotes the expression of AR-variants will require further study.
Conclusions
Our findings show that WLS is upregulated in enzalutamide-resistant prostate cancer and potentiates viability and a resistant phenotype. While further study is needed to fully understand the complex role of Wnt signaling in prostate cancer progression, our data suggests that targeting Wnt secretion and downstream signaling may represent an attractive strategy for the treatment of resistant CRPC.
Acknowledgements
This work was supported in part by grants NIH/NCI CA168601, CA179970, CA225836, DOD PC150229, DOD PC150040, and the U.S. Department of Veterans Affairs, Office of Research & Development BL&D grant number I01BX0002653 (A.C. G), and a Research Career Scientist Award (A.C.G).
Disclosure of conflict of interest
None.
References
- 1.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, Wasielewska T, Welsbie D, Chen CD, Higano CS, Beer TM, Hung DT, Scher HI, Jung ME, Sawyers CL. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, Armstrong AJ, Flaig TW, Flechon A, Mainwaring P, Fleming M, Hainsworth JD, Hirmand M, Selby B, Seely L, de Bono JS AFFIRM Investigators. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 3.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, Iversen P, Bhattacharya S, Carles J, Chowdhury S, Davis ID, de Bono JS, Evans CP, Fizazi K, Joshua AM, Kim CS, Kimura G, Mainwaring P, Mansbach H, Miller K, Noonberg SB, Perabo F, Phung D, Saad F, Scher HI, Taplin ME, Venner PM, Tombal B for the PREVAIL Investigators. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murillo-Garzon V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;14:683–696. doi: 10.1038/nrurol.2017.144. [DOI] [PubMed] [Google Scholar]
- 5.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Cheng L, Li J, Farah E, Atallah NM, Pascuzzi PE, Gupta S, Liu X. Inhibition of the wnt/beta-catenin pathway overcomes resistance to enzalutamide in castration-resistant prostate cancer. Cancer Res. 2018;78:3147–3162. doi: 10.1158/0008-5472.CAN-17-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herr P, Hausmann G, Basler K. WNT secretion and signalling in human disease. Trends Mol Med. 2012;18:483–493. doi: 10.1016/j.molmed.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- 10.Coombs GS, Yu J, Canning CA, Veltri CA, Covey TM, Cheong JK, Utomo V, Banerjee N, Zhang ZH, Jadulco RC, Concepcion GP, Bugni TS, Harper MK, Mihalek I, Jones CM, Ireland CM, Virshup DM. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J Cell Sci. 2010;123:3357–3367. doi: 10.1242/jcs.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka K, Okabayashi K, Asashima M, Perrimon N, Kadowaki T. The evolutionarily conserved porcupine gene family is involved in the processing of the Wnt family. Eur J Biochem. 2000;267:4300–4311. doi: 10.1046/j.1432-1033.2000.01478.x. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka K, Kitagawa Y, Kadowaki T. Drosophila segment polarity gene product porcupine stimulates the posttranslational N-glycosylation of wingless in the endoplasmic reticulum. J Biol Chem. 2002;277:12816–12823. doi: 10.1074/jbc.M200187200. [DOI] [PubMed] [Google Scholar]
- 13.Herr P, Basler K. Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev Biol. 2012;361:392–402. doi: 10.1016/j.ydbio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, Gao AC. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014;20:3198–3210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan CX, Zhang H, Tepper CG, Lin TY, Davis RR, Keck J, Ghosh PM, Gill P, Airhart S, Bult C, Gandara DR, Liu E, de Vere White RW. Development and characterization of bladder cancer patient-derived xenografts for molecularly guided targeted therapy. PLoS One. 2015;10:e0134346. doi: 10.1371/journal.pone.0134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Armstrong C, Zhu Y, Lou W, Gao AC. Niclosamide enhances abiraterone treatment via inhibition of androgen receptor variants in castration resistant prostate cancer. Oncotarget. 2016;7:32210–20. doi: 10.18632/oncotarget.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Najdi R, Proffitt K, Sprowl S, Kaur S, Yu J, Covey TM, Virshup DM, Waterman ML. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation. 2012;84:203–213. doi: 10.1016/j.diff.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Wang L, Zhang M, Melamed J, Liu X, Reiter R, Wei J, Peng Y, Zou X, Pellicer A, Garabedian MJ, Ferrari A, Lee P. LEF1 in androgen-independent prostate cancer: regulation of androgen receptor expression, prostate cancer growth, and invasion. Cancer Res. 2009;69:3332–3338. doi: 10.1158/0008-5472.CAN-08-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Augustin I, Goidts V, Bongers A, Kerr G, Vollert G, Radlwimmer B, Hartmann C, Herold-Mende C, Reifenberger G, von Deimling A, Boutros M. The Wnt secretion protein Evi/Gpr177 promotes glioma tumourigenesis. EMBO Mol Med. 2012;4:38–51. doi: 10.1002/emmm.201100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid SC, Sathe A, Guerth F, Seitz AK, Heck MM, Maurer T, Schwarzenbock SM, Krause BJ, Schulz WA, Stoehr R, Gschwend JE, Retz M German Bladder Cancer Network. Nawroth R. Wntless promotes bladder cancer growth and acts synergistically as a molecular target in combination with cisplatin. Urol Oncol. 2017;35:544.e1–544.e10. doi: 10.1016/j.urolonc.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Stewart J, James J, McCluggage GW, McQuaid S, Arthur K, Boyle D, Mullan P, McArt D, Yan B, Irwin G, Harkin DP, Zhengdeng L, Ong CW, Yu J, Virshup DM, Salto-Tellez M. Analysis of wntless (WLS) expression in gastric, ovarian, and breast cancers reveals a strong association with HER2 overexpression. Mod Pathol. 2015;28:428–436. doi: 10.1038/modpathol.2014.114. [DOI] [PubMed] [Google Scholar]
- 27.Chiou SS, Wang LT, Huang SB, Chai CY, Wang SN, Liao YM, Lin PC, Liu KY, Hsu SH. Wntless (GPR177) expression correlates with poor prognosis in B-cell precursor acute lymphoblastic leukemia via Wnt signaling. Carcinogenesis. 2014;35:2357–2364. doi: 10.1093/carcin/bgu166. [DOI] [PubMed] [Google Scholar]
- 28.Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, Hundsrucker C, Kerr G, Sandmann T, Anchang B, Demir K, Boehm C, Leible S, Ball CR, Glimm H, Spang R, Boutros M. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun. 2013;4:2610. doi: 10.1038/ncomms3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokoyama NN, Shao S, Hoang BH, Mercola D, Zi X. Wnt signaling in castration-resistant prostate cancer: implications for therapy. Am J Clin Exp Urol. 2014;2:27–44. [PMC free article] [PubMed] [Google Scholar]
- 30.Pakula H, Xiang D, Li Z. A tale of two signals: AR and WNT in development and tumorigenesis of prostate and mammary gland. Cancers (Basel) 2017;9 doi: 10.3390/cancers9020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Truica CI, Byers S, Gelmann EP. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 2000;60:4709–4713. [PubMed] [Google Scholar]
- 32.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, Sun Z. Linking beta-catenin to androgen-signaling pathway. J Biol Chem. 2002;277:11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
- 33.Song LN, Herrell R, Byers S, Shah S, Wilson EM, Gelmann EP. Beta-catenin binds to the activation function 2 region of the androgen receptor and modulates the effects of the N-terminal domain and TIF2 on ligand-dependent transcription. Mol Cell Biol. 2003;23:1674–1687. doi: 10.1128/MCB.23.5.1674-1687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masiello D, Chen SY, Xu Y, Verhoeven MC, Choi E, Hollenberg AN, Balk SP. Recruitment of beta-catenin by wild-type or mutant androgen receptors correlates with ligand-stimulated growth of prostate cancer cells. Mol Endocrinol. 2004;18:2388–2401. doi: 10.1210/me.2003-0436. [DOI] [PubMed] [Google Scholar]
- 35.Lee E, Ha S, Logan SK. Divergent androgen receptor and beta-catenin signaling in prostate cancer cells. PLoS One. 2015;10:e0141589. doi: 10.1371/journal.pone.0141589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wadosky KM, Koochekpour S. Androgen receptor splice variants and prostate cancer: from bench to bedside. Oncotarget. 2017;8:18550–18576. doi: 10.18632/oncotarget.14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohli M, Ho Y, Hillman DW, Van Etten JL, Henzler C, Yang R, Sperger JM, Li Y, Tseng E, Hon T, Clark T, Tan W, Carlson RE, Wang L, Sicotte H, Thai H, Jimenez R, Huang H, Vedell PT, Eckloff BW, Quevedo JF, Pitot HC, Costello BA, Jen J, Wieben ED, Silverstein KAT, Lang JM, Wang L, Dehm SM. Androgen receptor variant AR-V9 is coexpressed with AR-V7 in prostate cancer metastases and predicts abiraterone resistance. Clin Cancer Res. 2017;23:4704–4715. doi: 10.1158/1078-0432.CCR-17-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]