Abstract
Objective
Human blood metabolites are influenced by a number of lifestyle and environmental factors. Identification of these factors and the proper quantification of their relevance provides insights into human biological and metabolic disease processes, is key for standardized translation of metabolite biomarkers into clinical applications, and is a prerequisite for comparability of data between studies. However, so far only limited data exist from large and well-phenotyped human cohorts and current methods for analysis do not fully account for the characteristics of these data. The primary aim of this study was to identify, quantify and compare the impact of a comprehensive set of clinical and lifestyle related factors on metabolite levels in three large human cohorts. To achieve this goal, we improve current methodology by developing a principled analysis approach, which could be translated to other cohorts and metabolite panels.
Methods
63 Metabolites (amino acids, acylcarnitines) were quantified by liquid chromatography tandem mass spectrometry in three cohorts (total N = 16,222). Supported by a simulation study evaluating various analytical approaches, we developed an analysis pipeline including preprocessing, identification, and quantification of factors affecting metabolite levels. We comprehensively identified uni- and multivariable metabolite associations considering 29 environmental and clinical factors and performed metabolic pathway enrichment and network analyses.
Results
Inverse normal transformation of batch corrected and outlier removed metabolite levels accompanied by linear regression analysis proved to be the best suited method to deal with the metabolite data. Association analyses revealed numerous uni- and multivariable significant associations. 15 of the analyzed 29 factors explained >1% of variance for at least one of the metabolites. Strongest factors are application of steroid hormones, reticulocytes, waist-to-hip ratio, sex, haematocrit, and age. Effect sizes of factors are comparable across studies.
Conclusions
We introduced a principled approach for the analysis of MS data allowing identification, and quantification of effects of clinical and lifestyle factors with metabolite levels. We detected a number of known and novel associations broadening our understanding of the regulation of the human metabolome. The large heterogeneity observed between cohorts could almost completely be explained by differences in the distribution of influencing factors emphasizing the necessity of a proper confounder analysis when interpreting metabolite associations.
Keywords: Amino acids, Acylcarnitines, Metabolomics, Clinical factors, Lifestyle factors, Network analysis
Abbreviations: AA, amino acid; AC, acylcarnitine; BMI, body mass index; T2D, diabetes mellitus type 2; ATC, anatomical therapeutic chemical (code for medication); LDL, low-density lipoprotein; HDL, high-density lipoprotein; INT-LR, inverse normal transformation (INT) followed by linear regression (LR); WHR, waist-to-hip ratio; MS, liquid chromatography-mass spectrometry; BP, blood pressure
Graphical abstract
Highlights
-
•
Amino-acids and acylcarnitines analyzed in three studies with >16,000 individuals.
-
•
Develop a generic and adaptable bioinformatics workflow.
-
•
Analysis of the impact of 29 clinical and life-style factors on blood metabolites.
-
•
Analysis of network between factors and metabolites.
-
•
Comparison of results between studies.
1. Introduction
Targeted, high-throughput metabolomics using liquid chromatography-mass spectrometry (MS) increasingly gains momentum in epidemiology. Important fields of investigations are the understanding of the molecular basis of metabolism-related phenotypes and diseases and studying biomarkers for diagnostic and prognostic purposes [1], [2], [3], [4]. Furthermore, analysis of metabolomic features in relation to other molecular-genetic functional layers of the organism, e.g. genomics and transcriptomics, is a promising approach to extend our knowledge of regulatory pathways and associated patho-mechanisms [5], [6], [7].
Proper identification of factors affecting metabolite levels across multiple studies is highly relevant for standardized translation of metabolite biomarkers into clinical applications and to understand possible confounders of disease associations. However, only limited data exist regarding kind, number, and relevance of possible influencing factors. Furthermore, currently applied analysis methods do not fully account for the characteristics of MS data. Here, zero inflation (considerable proportion of measurements below the detection limit) is one of the issues for which limited guidelines exist. Many studies simply exclude these data, which may result in biased estimates and conclusions.
In this study, we investigated the effects of 29 clinical and lifestyle related factors on metabolite levels in dried whole blood derived from MS in three large human studies with different designs comprising a total of 16,222 subjects. We developed a generic and adaptable workflow and made it publicly available so that it can be used for other cohorts and metabolite panels. We interpreted the discovered associations biologically by applying pathway-based methods and compared their strength across studies.
2. Methods
Study design and flow of our analyses is shown in Supplementary Figure 1.
2.1. Study characteristics
Three different studies are investigated in the present work:
2.1.1. LIFE-Adult
LIFE-Adult is a population-based study of 10,000 randomly selected individuals from the city of Leipzig, Germany [8]. Individuals were phenotyped for several lifestyle diseases and corresponding lifestyle associated risk factors. Data of metabolite and clinical/lifestyle parameters were available for 9,481 participants and blood samples are collected after an over-night fast.
2.1.2. LIFE-Heart
LIFE-Heart is an observational study of 7,000 patients with suspected and confirmed coronary artery disease collected from the Heart Center, Leipzig, Germany (ClinicalTrials.gov No NCT00497887 [9]). Patients originate mainly from Leipzig and surrounding areas. Combined metabolite data and clinical and lifestyle parameters were available for 5,767 patients. Patients were not required to be at fasting state.
2.1.3. Sorbs study
The Sorb study is a convenience sample of individuals recruited in the self-contained population of the Sorbs, an ethnic minority of Slavic origin residing in the Upper Lusatia region of Eastern Saxony [10], [11]. Data of metabolite and clinical/lifestyle parameters were available for 974 participants. Blood was also collected after an overnight fast.
All studies conform to the ethical standards of the Declaration of Helsinki and were approved by the ethics committee of the University of Leipzig (LIFE-Adult: Reg. No 263-2009-14122009, LIFE-Heart: Reg. No 276e2005, Sorbs: Reg. No: 088–2005). Written informed consent was obtained from all participants.
2.2. Factors studied in relation to blood metabolite levels
We selected a number of parameters for which an impact on whole blood metabolite levels is supposed. First, blood composition can be supposed to affect measured metabolite levels derived from dried whole blood. We here considered hematocrit, hemoglobin, erythrocytes, reticulocytes, platelets, leucocytes, neutrophils, lymphocytes, and monocytes.
Second, previously applied covariates in metabolome association studies were included. The most frequently considered factors were age, sex, log-BMI, smoking status [6], [12], [13], [14], [15], [16], and, to a lesser extent, type-2-diabetes (T2D) and application of sex hormones [6], [7], [15], [17].
Third, we included waist-to-hip ratio (WHR) [18], systolic and diastolic blood pressure (BP) [12], [13] as well as the pulse pressure, defined as the difference of systolic and diastolic BP. Additionally, we considered parameters of lipid metabolism as there is a well-known relation to certain AAs [13], [14]. Regarding medication, we considered the effects of statin treatment and other lipid modifying agents (defined as Anatomical Therapeutic Chemical (ATC) classification category C10) and sex hormones or modulators of the reproductive system (ATC G03). Diabetes status was defined in our study as either self-reported consumption of type-II-diabetes-specific medication (ATC A10), self-reported diagnosis of T2D, or measured HBa1c level of >6.5%. Fasting hours were available in LIFE-Adult and LIFE-Heart. In the Sorbs study, it was required that fasting was >8 h.
Distribution of the considered clinical and lifestyle parameters of the three cohorts is presented in Table 1.
Table 1.
Subject characteristics of the three cohorts considered. For continuous variables, median and IQR are shown. For binary variables, total numbers and percentages are provided.
| LIFE-Adult | LIFE-Heart | Sorbs | |
|---|---|---|---|
| Area of collection | Leipzig, Germany | Leipzig, Germany | Upper Lusatia |
| N | 9481 | 5767 | 974 |
| Sex (female/male) | 4952 (52.2%) | 1712 (29.7%) | 574 (58.9%) |
| age (years) | 57.91 [47.7–68.2] | 63.11 [54.4–71.7] | 48.7 [35.6–60.9] |
| WHR | 0.93 [0.863–0.994] | 0.98 [0.909–1.04] | 0.87 [0.804–0.949] |
| BMI (kg/m2) | 26.58 [23.9–29.9] | 28.41 [25.7–31.8] | 26.5 [23.3–29.7] |
| fasting hours (hours) | 12 [11–14] | 3 [1.67–12.3] | >8 |
| Lipid modifying agents (yes/no) | 1272 (13.4%) | 2066 (35.8%) | 176 (18.1%) |
| sex hormones (yes/no) | 751 (7.9%) | 52 (0.9%) | 111 (11.4%) |
| diabetes status (yes/no) | 1090 (11.5%) | 1720 (29.8%) | 86 (8.8%) |
| HBa1c (%) | 5.32 [5.08–5.59] | 5.7 [5.38–6.18] | 5.4 [5.1–5.7] |
| self-reported diabetes (yes/no) | 996 (10.5%) | 1547 (26.8%) | 71 (7.3%) |
| diabetes medication (yes/no) | 840 (8.9%) | 1258 (21.8%) | 57 (5.9%) |
| smoking status (current, previous, never) | 2034 (21.5%)/2706 (28.5%)/4483 (47.3%) | 1581 (27.4%)/2108 (36.6%)/2063 (35.8%) | 150 (15.4%)/195 (20%)/616 (63.2%) |
| Blood pressure (systolic) | 127 [117–138] | 136 [125–150] | 132 [121–145] |
| Blood pressure (diastolic) | 75 [68.5–81.5] | 83.5 [76–90.5] | 80 [73–87] |
| Pulse pressure | 51 [44–60] | 53 [44–63] | 52 [44–61] |
| Cholesterol (mmol/l) | 5.52 [4.85–6.26] | 5.18 [4.4–6.01] | 5.25 [4.63–5.94] |
| LDL-Cholesterol (mmol/l) | 3.45 [2.84–4.11] | 3.15 [2.48–3.87] | 3.32 [2.71–3.98] |
| HDL-Cholesterol (mmol/l) | 1.57 [1.28–1.9] | 1.22 [1.01–1.48] | 1.57 [1.33–1.89] |
| Blood hemoglobin (mmol/l) | 14 [13.2–15] | 14.3 [13.2–15.3] | 8.8 [8.3–9.3] |
| Erythrocytes (10ˆ12/l) | 4.66 [4.38–4.94] | 4.67 [4.34–4.97] | 4.73 [4.47–4.98] |
| Reticulocytes (per 1000) | 12.1 [9.6–14.8] | 12.9 [10.5–16.1] | 10.6 [8.4–13] |
| Hematocrit (%) | 41 [39.2–43.6] | 42 [39–44] | 42 [39.2–43.8] |
| Platelets (10ˆ9/l) | 237 [204–275] | 230 [194–271] | 229 [201–263] |
| Leucocytes (10ˆ9/l) | 5.94 [5–7.1] | 7.9 [6.4–9.9] | 5.25 [4.4–6.23] |
| Neutrophils (%) | 57.6 [51.9–63.2] | 66.5 [59.9–72.8] | 54.65 [48.7–60.5] |
| Lymphocytes (%) | 30.2 [25.1–35.5] | 22.3 [16.8–28.2] | 33.3 [27.9–38.6] |
| Monocytes (%) | 8 [6.8–9.4] | 8.5 [7.1–10.1] | 8.1 [6.9–9.5] |
| Basophils (%) | 0.6 [0.4–0.8] | 0.3 [0.2–0.5] | 0.03 [0.02–0.04] |
| Eosinophils (%) | 2.5 [1.6–3.6] | 1.4 [0.7–2.5] | 0.14 [0.09–0.21] |
2.3. Metabolite measurement
In LIFE-Adult and LIFE-Heart, 40 μl of EDTA whole blood were spotted on filter paper WS 903 (Schleicher and Schüll, Germany). In the Sorbs, 40 μl of the plasma free cell suspension was spotted after centrifugation at 3500 ×g for 10 min.
Spots were dried for 3 h and stored at −80 °C until analysis. To prepare samples for tandem mass spectrometric analysis, blood spots with a 3.0 mm diameter (corresponds to 3 μl of blood) were punched out and extracted via methanol containing isotope labeled internal standards. After butylation, sample derivatives were analyzed by flow injection analysis with an API 2000 tandem mass spectrometer (Applied Biosystems, Germany) in 96-well plates. Each plate included two quality control samples, from which inter-assay coefficients of variation were estimated. A detailed description of sample preparation and the measurement method can be found elsewhere [19], [20], [21]. In consequence, 63 metabolites (27 amino acids (AAs), 34 acylcarnitines (ACs), free carnitine (C0), and the sum of total ACs, Supplementary Table 1) were quantified using the software ChemoView 1.4.2 (Applied Biosystems, Germany).
2.4. Statistical analysis of the three cohorts
Metabolites were pre-processed prior to analysis. In order to stabilize regression analysis, outliers were removed cohort-wise by applying a cutoff of mean + 5 × SD of the logarithmized data. Zero values were excluded for this purpose. In our hands, outlier analysis removed a maximum of 0.3% of measurements per metabolite and cohort. Remaining metabolite data were inverse-normal-transformed. Effects of known technical batches (e.g. analysis plate ID) are removed by a non-parametric empirical method as implemented in function ‘ComBat’ [22] of the R-package ‘sva’ [23]. We considered the plate ID of the mass-spectrometer sample plate (96 well plates including two analytical controls) as batch variable, resulting in 71, 68, and 15 batches for LIFE-Adult, LIFE-Heart, and the Sorbs, respectively. Since the ‘ComBat’ procedure requires complete data, missing values were mean-imputed, using within-batch data or all data when a certain metabolite was completely missing in a batch. After batch correction, imputed data points were set missing again. For Asparagine and Cis-11,14,17-eicosatrienoic acid methyl ester (C20:3) in LIFE- Adult and LIFE-Heart, batch affects were removed by residualization via a linear model due to small batch variance.
Following batch correction, relatedness among Sorb subjects was accounted for as described elsewhere [7], i.e. by fitting a generalized linear model as implemented in the ‘polygenic’ function of the ‘GenABEL′ package [24]. We used a kinship matrix estimated from SNP data for this purpose [25].
Prior to association analysis with metabolites, all continuous clinical or lifestyle parameters were mean-centered and scaled to one standard deviation (SD). For association analysis, inverse-normal transformed batch-adjusted metabolites were univariately associated with the clinical/lifestyle parameters by linear regression analysis. For multivariable analysis, correlated factors were pruned to avoid collinearity and to improve interpretation (default Pearson's |r| > 0.75 in any cohort [26]). Correlation structure between factors is shown in Supplementary Figures 3–5. In case of correlated factors, we preferred those which are clinically more often evaluated. In detail, we preferred diabetes status over diabetes medication and anamnestic diabetes, hematocrit over blood hemoglobin levels and erythrocytes, LDL-Cholesterol over total cholesterol, systolic BP over pulse pressure, and neutrophils over lymphocytes. To account for multiple testing of all metabolites and factors, we implemented a Bonferroni correction [27] in a hierarchical way, considering each tested factor as a family of hypotheses regarding metabolite association [28], [29].
Effect sizes of metabolite associations are assessed by the explained variance (r2) of the considered factor in a univariable model or as partially explained variance in a multivariable regression model. For every factor, we quantify the difference in the distribution of r2 between cohorts by Friedman test followed by Benjamini-Hochberg correction for multiple testing. When two distributions were compared, the Wilcoxon signed rank test was used. R-scripts of our analyses are available at https://github.com/cfbeuchel/Metabolite-Investigator.
For every factor, we performed a pathway analysis considering all metabolites for which the factor explains at least 1% of the metabolite's variance in at least one cohort. Enrichment was tested with MetaboAnalyst [30] using the intersection of all representable metabolites (M = 58) and KEGG-metabolic pathways as background. Bi-partite networks, connecting metabolite nodes, and factor nodes with edges representing the partial explained variance were created using ‘visNetwork’ [31].
2.5. Simulation study to justify the analysis approach
In our analysis approach, we applied inverse normal transformation of metabolite data in combination with linear regression analyses (INT-LR approach), i.e. no removal of measurements below the detection limit is applied. We conducted a simulation study to compare this approach with possible alternatives. In detail, we simulated data mirroring typical issues of MS data, including zero-inflation, skewness (by assuming a log-normal distribution) and batch effects and imposed different effect sizes of factors on simulated metabolite levels. In the preprocessing steps, we considered different data transformation methods [area sinus hyperbolicus, inverse-normal-transformation, dichotomization (zero vs. non-zero measurements), categorization (quantile or range-based equal spaced categories), and creation of ranks]. Accordingly, we performed univariate linear modeling, binary logistic regression, proportional odds logistic regression, and Spearmans’ Rank correlation to perform hypotheses testing in accordance to the chosen transformation method. Performance was rated according to the ability of the individual method to discover the imposed effect of a factor on a metabolite and the ability to correctly control the number of false positives at the expected 5% level. A schematic workflow of the simulations is presented in Supplementary Figure 2 and a detailed description of the simulations can be found in the Supplementary Methods.
3. Results
3.1. Justification of metabolite analysis method
To evaluate its performance, we compared our INT-LR approach in a simulation study with three other analysis strategies (rank correlation, binary, and ordinal logistic regression) and three other data transformations (categorization, dichotomisation, inverse sinus hyperbolicus transformation, see Supplementary Figure 2 for the design of the simulation study).
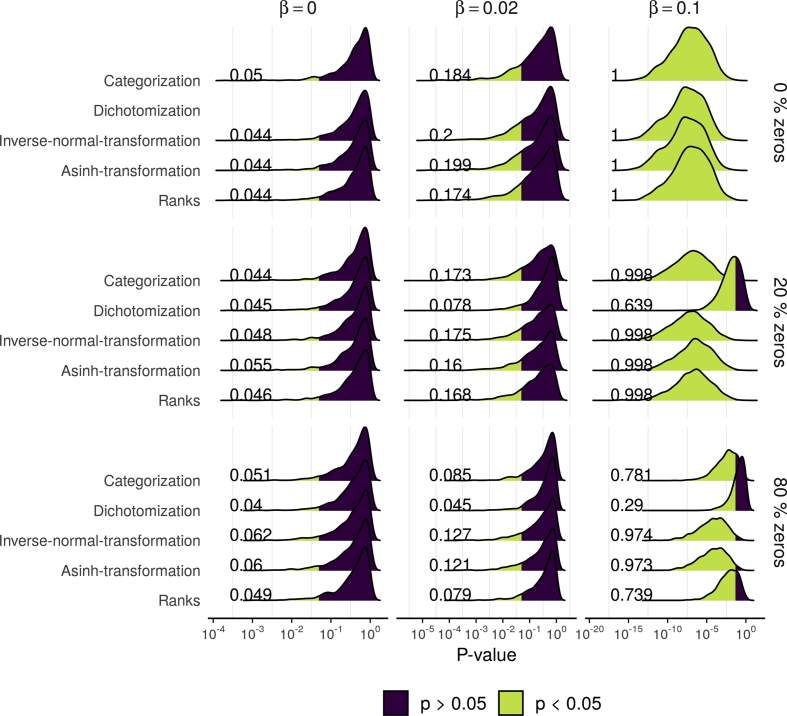
We found that INT-LR controlled false positives sufficiently well as the number of identified associations was close to 5% in all scenarios with no effect (β = 0, Figure 1 and Supplementary Table 5). Furthermore, no other approach had better power to identify true associations of factors with metabolite levels, especially in scenarios with high zero inflation (see scenarios with β > 0 in Figure 1 and Supplementary Table 5). As expected, increased zero-inflation resulted in decreased observed vs. true effect size (Supplementary Figure 7).
Figure 1.
Comparison of INT-LR method with alternatives – selected results of simulation study: Shown is the distribution of p-values from the simulation study comparing INT-LR approach (Linear regression with inverse-normal transformation) with other methodological approaches (binary/ordinal logistic regression for binary/categorical data, asinh-transformation followed by linear regression and Spearman's Correlation Coefficient for rank data). Results from nine different simulated scenarios are presented, differing in the simulated effect β (no effect: β = 0, small effect: β = 0.02, and large effect β = 0.1) and variable numbers of measurements below the detection limit (0%, 20%, and 80%). The percentage of hypotheses with nominal significance (i.e. p < 0.05) is shown (based on 1000 replications). For scenarios with β = 0, this number is required to be 0.05 (false positive control) and for scenarios with β > 0 as large as possible (good power). The binary model is only applicable in case of zeros. Overall, method INT-LR performed best. Results of additional scenarios are reported in Supplementary Figure 6 and Supplementary Table 5.
3.2. Identification and characterization of clinical and lifestyle related factors affecting metabolite levels
We applied the INT-LR approach to determine the effect of 29 individual clinical and lifestyle related factors on metabolite levels in our studies.
3.2.1. Univariate analysis
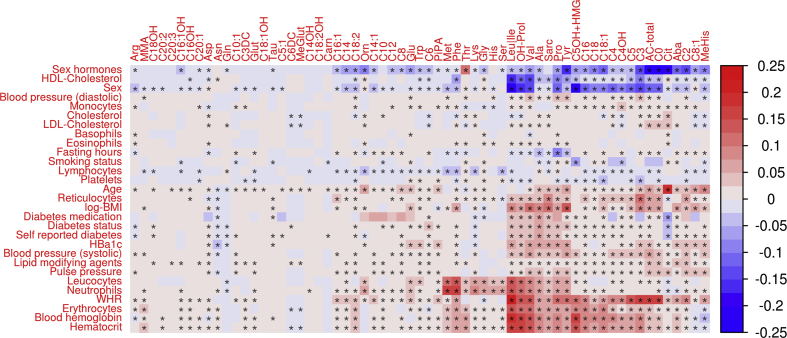
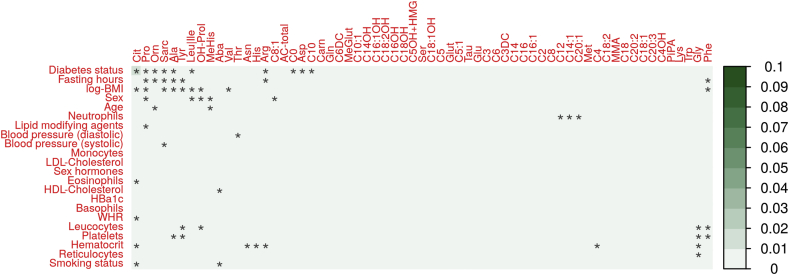
We observed statistically significant associations for all 29 analyzed parameters with at least one metabolite (multiple testing padjusted ≤ 0.05, Figure 2 and Supplementary Table 2). The overall highest explained variances were found for sex on C0 and total ACs (Sorbs, r2 = 0.25 for both), on Leucine/Isoleucine (LIFE-Adult; r2 = 0.22), and on C5OH + HMG (Sorbs; r2 = 0.21); followed by the effect of WHR on Leucine/Isoleucine (LIFE-Adult; r2 = 0.21).
Figure 2.
Heat map of univariable associations between metabolite levels and clinical or lifestyle-related factors. Explained variance by the single factor is color-coded (1 ≙ 100%) with direction of effect (red = positive correlation, blue = negative correlation). Maximum values across the three cohorts are presented. Stars indicated associations significant after adjusting for multiple testing. Rows and columns are ordered according to a hierarchical clustering. Cohort-specific plots can be found in Supplementary Figures 8–10.
The top-five factors affecting most metabolites (padjusted ≤ 0.05 and explained variance ≥1%) were WHR, sex, application of sex hormones, age, and hematocrit, influencing 44, 41, 40, 38, and 36 metabolites, respectively. Factors affecting the fewest number of metabolites at the same level were smoking status (10), eosinophils (9), cholesterol (8), fasting hours (6), and basophils (1), respectively.
To evaluate how strongly the metabolites are affected by the considered factors, we averaged corresponding explained variances over all factors and cohorts. The five most strongly affected metabolites are leucine/isoleucine (mean explained variance 3.65%), valine (3.42%), propionylcarnitine (2.70%), hydroxyproline (2.69%) and total ACs (2.46%); the metabolites with the lowest amount of explained variance (<0.14%) comprise nine ACs and the dipeptide carnosine. Of note, these are low abundant metabolites with at least 40% of values below the detection limit in at least one of the cohorts.
3.2.2. Independent effects of clinical and lifestyle related parameters on metabolite levels
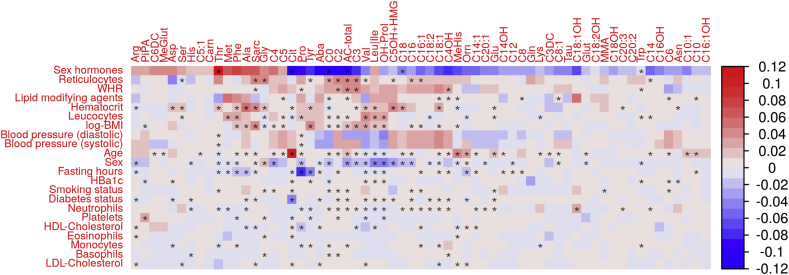
Next, we were interested in the variances independently explained by the clinical and lifestyle related factors. Therefore, we performed multivariable linear regression analysis considering all parameters simultaneously for each study. This requires elimination of correlated parameters to avoid collinearity (see methods). Thus, a total of 22 parameters were considered. Again, all parameters showed significance for at least on metabolite after adjusting for multiple testing. However, maximum partial explained variance was approximatively halved compared to univariable association analysis (Figure 3, Supplementary Table 3).
Figure 3.
Heat map of multivariable association results between clinical and lifestyle-related factors and metabolite levels. Partial explained variance (1 ≙ 100%) is color-coded according to the direction of the effect (positive = red, negative = blue). Maximum values across the three cohorts are presented. Rows and columns are ordered according to a hierarchical clustering. To avoid collinearity, strongly correlated factors were pruned (see methods). Cohort-specific plots can be found in Supplementary Figures 11–13.
The largest partial explained variance was found for sex hormones on total ACs (r2 = 0.13), threonine (r2 = 0.13), citrulline (r2 = 0.12), C0 (r2 = 0.12), and aminobutyric acid (r2 = 0.11) in the Sorbs. This is very similar to unadjusted analysis where all these association (with the exception of threonine) were among the strongest effects, too. Application of sex hormones, reticulocytes, WHR, sex, haematocrit, and age were relevant for the highest numbers of metabolites as they independently explained more than 1% of variance for 58, 18, 14, 11, and 9 metabolites, respectively. Again, this is similar to univariate analysis. Vice versa, leukocytes and platelets are the least relevant parameters in multivariate analysis explaining 1% variance for only one metabolite each (hydroxyproline (3.3%) and pipecolic acid (3.2%), respectively).
Overall, a highest percentage of variance explained by the multivariable models was observed for leucine/isoleucine in two studies (adjusted-r2 = 0.37 and 0.38 in LIFE-Adult and Sorbs, respectively). Additionally, adjusted-r2 per metabolite was for the six metabolites valine (adjusted-r2 = 0.36, Sorbs), hydroxyproline (adjusted-r2 = 0.35, Sorbs), propionylcarnitine (adjusted-r2 = 0.34, Sorbs), phenylalanine (adjusted-r2 = 0.31, Sorbs), citrulline (adjusted-r2 = 0.31, Sorbs) and total acyl-carnitines (adjusted-r2 = 0.31, Sorbs) (Supplementary Table 3). We observed that AA were more strongly affected by the investigated factors than AC by mean explained variance (p = 0.039), but not by median explained variance (p = 0.19, Wilcoxon-Test, Supplementary Figure 14).
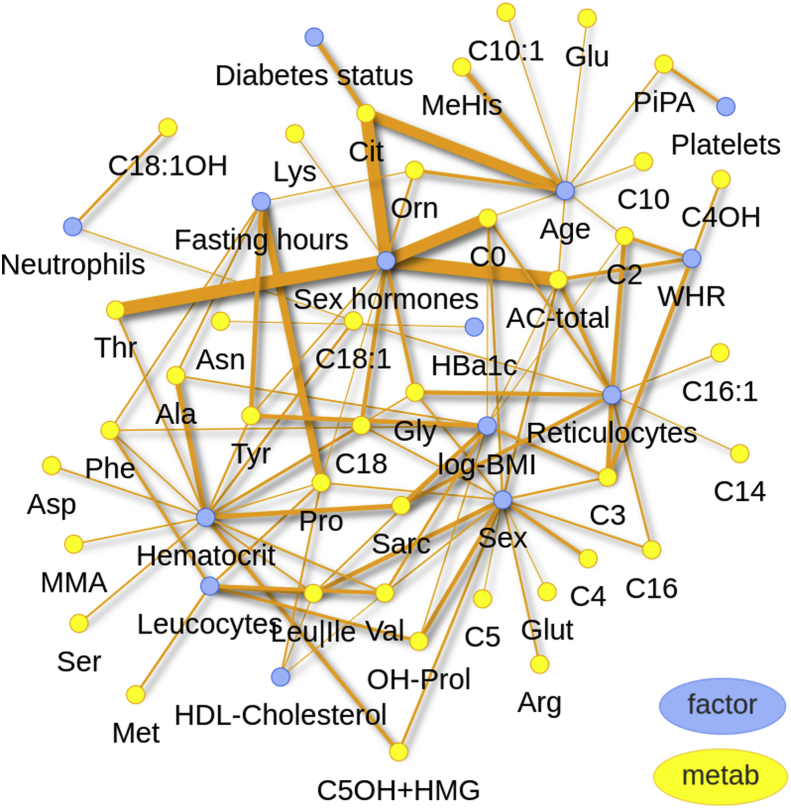
We selected factors explaining at least 1% variance in multivariable analysis of at least one metabolite in one of the cohorts. 14 such factors where identified resulting in 94 factor-metabolite relationships involving 39 metabolites. A bi-partite network of these relationships is shown in Figure 4, and interactively online, at https://cfbeuchel.shinyapps.io/interactivefig4/.
Figure 4.
Bi-partite network of metabolites (yellow) and factors (blue) based on multivariable associations explaining at least 1% of variance. Thickness of edges corresponds to the maximum partial explained variance over the three cohorts. An interactive version of this plot is available at https://cfbeuchel.shinyapps.io/interactivefig4/.
To obtain further biological insights, we analyzed which metabolite pathways are affected by the single factors analyzed. For this purpose, we selected the same associations as for the bipartite network analysis and performed formal enrichment analyses with respect to metabolite pathways implemented in ‘MetaboAnalyst’ (Supplementary Table 4). Strongest enrichment was observed for WHR. Among others, WHR is associated with the metabolites carnitine, acetylcarnitine, and propionylcarnitine resulting in an over-representation of the pathway “oxidation of branched-chain fatty acids” (p = 2.0 × 10−4).
3.2.3. Comparison of cohorts
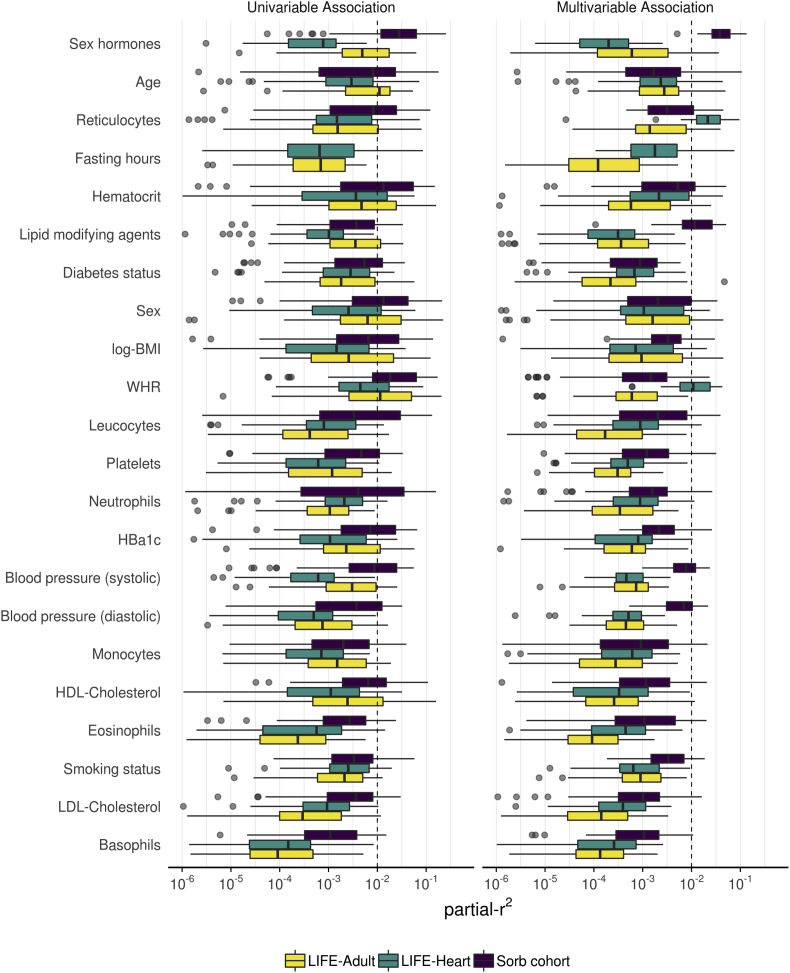
Distributions of effect sizes of the single studies are shown in Figure 5 for the 22 factors included in multivariable analysis. Agreement of distribution of effect sizes are stronger in univariable analyses compared to multivariable analysis. In univariable analyses, 13 factors had effect sizes >1% explained variance in all three cohorts. In contrast, this applies to only five factors in multivariable analysis.
Figure 5.
Distributions of uni- and multivariable explained variances of clinical and lifestyle-related factors and comparison between cohorts. Boxplots show the distribution of explained variances (respectively partial explained variances for multivariable models) for the different metabolites. The dashed line represents an exemplarily r2 cutoff (1%) to mark strong effects.
Among the 29 factors, 27 were associated significantly (padjusted ≤ 0.05) with at least one metabolite in all three studies. Exceptions were fasting hours and diabetes medication, which are not available in the Sorbs. We analyzed differences in effect sizes of our factors between cohorts by formal interaction analysis considering study as interaction partner. It revealed that only a few such interactions were significant and that only one of the interactions explains more than 1% of variability of the metabolite, namely the interaction of diabetes (and study) regarding citrulline (partial-r2 = 0.015, padjusted = 1.5 × 10−56, Figure 6). Further, interaction effects were found for fasting hours regarding proline, tyrosine and alanine, of log-BMI regarding sarcosine and tyrosine and of sex regarding hydroxyproline and leucine/isoleucine. These interactions explain 0.8% down to 0.2% of the respective metabolites variances. All interactions are presented in Supplementary Figure 15.
Figure 6.
Heatmap of partial-r2of interaction effects of study with the 22 factors and study regarding the 63 metabolites. Significance is indicated as an asterisk and was computed via likelihood-ratio test of multivariable linear regression models. The full model includes main effects for each factor and study and their interactions. It is compared with a reduced model not containing the considered interaction effect. Correction for multiple testing was applied by a hierarchical Bonferroni procedure (see methods).
4. Discussion
In this study, we comprehensively analyzed the impact of 29 clinical and lifestyle related factors on plasma levels of 37 AA, 24 AC, C0, and the sum of total ACs measured by the same tandem mass-spectrometric method in three large cohorts over 10 years. For this purpose, we propose a principled workflow of data preprocessing and analysis which can be applied to other studies and metabolite panels. A major finding is that the large heterogeneity of metabolite levels across cohorts can almost completely be explained by the different distributions of influencing factors rather than their effect size on metabolites, i.e. there were almost no interactions between study and factors. We also detected a number of known and novel associations broadening our understanding of the regulation of the human metabolome, which we discuss in the following.
Within the identified 14 strongest multivariable associating factors (defined as explaining at least 1% variance for at least one metabolite), we could confirm several previously reported AA- and AC-affecting factors. These factors included sex [32], [33], [34], [35], [36], medication with sex hormones (e.g. contraceptives, [32], [37], [38]), hematocrit [39], and medication with lipid modifying agents (e.g. statins, [40], [41]). Additionally, our work provided novel support for relationships for which contradicting results are present in the literature. Exemplarily, rising levels of proline were reported for prolonged fasting recently, contradicting an earlier study [42], [43]. Our results support the earlier studies, as we identified strong negative associations of proline levels with prolonged fasting in LIFE-Adult (, padjusted = 8.7 × 10−15) and LIFE-Heart (, padjusted = 6.4 × 10−108). This observation is also in line with research linking proline catabolism with lipid utilization during fasting [44].
We also identified a number of novel findings. Among the 33 significant (padjusted ≤ 0.05) associations with sex hormones, negative associations with glycine (, padjusted = 2.2 × 10−77, LIFE-Adult) and arginine (, padjusted = 7 × 10−12, LIFE Adult) were observed. Such an interaction of sex hormones with the creatine formation pathway is plausible given the role of estrogen in the upregulation of the l-arginine:glycine amidinotransferase [45], [46]. Additionally, the negative association of sex hormones with ornithine (, padjusted = 3.46 × 10−54, LIFE- Adult) is corroborated by research in animal studies linking sex hormones to increased ornithine decarboxylase activity [47], [48], [49], but was to the best of our knowledge not yet described for human cohorts. It needs to be acknowledged that these associations, despite being plausible, do not retain satisfactory evidence for causal relationships. Further experimental validation of interesting associations is required to unravel underlying causal mechanisms. Relevance of these mechanisms for patho-mechanisms of diseases should be investigated in specifically designed studies.
Pathway enrichment analysis revealed plausible results, all at nominal significance level (Supplementary Table 4). For instance, metabolites associated with reticulocyte counts (carnitine, acetylcaritine, propionylcarnitine) showed a significant enrichment in the metabolism of branched chain fatty acids (p = 0.017). This is in line with knowledge on the fatty acid catabolism in reticulocyte mitochondria [50], [51]. Moreover, associations of carnitine and acetylcarnitine with WHR showed an enrichment in beta oxidation of very long chain fatty acids (p = 0.06) in line with knowledge on the peroxisomes [50], [51].
Overall, we observed a stronger impact of the considered factors on AAs rather than ACs. However, since ACs show a higher rate of zero-inflation than AAs (Supplementary Table 1) and higher zero-inflation could result in underestimation of the observed explained variance (Supplementary Figure 7), we considered metabolites with less than 10% zero inflation in a sensitivity analysis. For this subset, no clear trend regarding differences in mean or median of explained variances per factor were observed (mean: p = 0.63, median p = 0.092, Wilcoxon-Test, Supplementary Figure 14).
When analyzing heterogeneity of effects across our three cohorts, we observed strong similarities. 45/63 metabolites and all but three factors associated significantly in all three studies. Limited sample size and thus power issues could be a reason for the lower number of strong associations in the Sorbs study. The low number and low effect size of interaction effects between study and factors in a pooled analysis supports reproducibility of our findings across multiple cohorts and suggest excellent between-study comparability required for mega- or meta-analyses.
The few differences in effect sizes could be explained by the different study designs. Relevance of sex hormones was highest in the Sorbs, in line with the younger age of this cohort and the higher percentage of females before menopause in this cohort (Table 1). Another example is the higher importance of blood parameters in the Sorbs, in line with the different type of blood specimen used here. Whereas dried whole blood was used in LIFE-Adult and LIFE-Heart, cell suspension after plasma removal was used in the Sorbs reducing the influence of cell-free plasma-specific metabolites, providing a clearer picture of the intracellular metabolism. Thus, associations with intracellular metabolic actors, especially ACs, are stronger than in the cohort utilizing plasma-free cell suspension as a tissue source, leading to the strongest associations found in all three studies (Supplementary Table 3). Finally, the higher effects of fasting in LIFE-Heart is also plausible due to the effect that a considerable percentage of patients were not at fasting state (Table 1). Hence, for the purpose of selecting relevant factors, we recommend study specific analyses first, which can be efficiently done with the help of our preprocessing and analysis tool provided online.
Funding source declaration
LIFE-Heart and LIFE-Adult are funded by the Leipzig Research Center for Civilization Diseases (LIFE). LIFE is an organizational unit affiliated to the Medical Faculty of the University of Leipzig. LIFE is funded by means of the European Union, by the European Regional Development Fund and by funds of the Free State of Saxony within the framework of the excellence initiative. Initial funding of LIFE-Heart was supported by the Roland-Ernst Foundation.
The Sorbs study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation – Projektnummer 209933838 – SFB 1052; B03, C01; SPP 1629 TO 718/2- 1), from the German Diabetes Association and from the DHFD (Diabetes Hilfs-und Forschungsfonds Deutschland). IFB Adiposity Diseases is supported by the Federal Ministry of Education and Research, Germany, FKZ: 01EO1501 (AD2-060E, AD2-06E95, AD2-7123).
MS received funding from the Federal Ministry of Education and Research, Germany, FKZ: 01EO1501.AD2-7117.
Availability of data and material
Data of the LIFE studies are available upon reasonable request on the LIFE Research Centre for Civilization Diseases.
Author's contributions
Study data collection: AT, MStu, ML, JT, FB, MSch.
MS analysis and assessments: UC, SB, JD.
Data analyses: CB, HK.
Drafting of manuscript: CB, HK, MS.
Critical revision of manuscript: AT, MStu, ML, JT, UC.
Acknowledgements
We thank the participants of LIFE-Adult and the Sorbs study and the patients of LIFE-Heart very much for their time and blood samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.08.010.
Conflicts of interest
Markus Scholz receives funding from Pfizer Inc. for a project not related to this research. Uta Ceglarek receives funding from Roche for a project not related to this research.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Methods: Details outlining methods used in the data simulation employed prior to the analysis.
References
- 1.Gao X., Ke C., Liu H., Liu W., Li K., Yu B. Large-scale metabolomic analysis reveals potential biomarkers for early stage coronary atherosclerosis. Scientific Reports. 2017;7(1):11817. doi: 10.1038/s41598-017-12254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirulli E.T., Guo L., Swisher C.L., Shah N., Huang L., Napier L.A. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metabolism. 2018;0(0) doi: 10.1016/j.cmet.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farshidfar F., Weljie A.M., Kopciuk K.A., Hilsden R., McGregor S.E., Buie W.D. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. British Journal of Cancer. 2016;115(7):848–857. doi: 10.1038/bjc.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kühn T., Floegel A., Sookthai D., Johnson T., Rolle-Kampczyk U., Otto W. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Medicine. 2016;14:13. doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly R.S., Croteau-Chonka D.C., Dahlin A., Mirzakhani H., Wu A.C., Wan E.S. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics. 2017;13(1):7. doi: 10.1007/s11306-016-1149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nath A.P., Ritchie S.C., Byars S.G., Fearnley L.G., Havulinna A.S., Joensuu A. An interaction map of circulating metabolites, immune gene networks, and their genetic regulation. Genome Biology. 2017;18 doi: 10.1186/s13059-017-1279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkhardt R., Kirsten H., Beutner F., Holdt L.M., Gross A., Teren A. Integration of genome-wide SNP data and gene-expression profiles reveals six novel loci and regulatory mechanisms for amino acids and acylcarnitines in whole blood. PLoS Genetics. 2015;11(9) doi: 10.1371/journal.pgen.1005510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeffler M., Engel C., Ahnert P., Alfermann D., Arelin K., Baber R. The LIFE-adult-study: objectives and design of a population-based cohort study with 10,000 deeply phenotyped adults in Germany. BMC Public Health. 2015;15:691. doi: 10.1186/s12889-015-1983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutner F., Teupser D., Gielen S., Holdt L.M., Scholz M., Boudriot E. Rationale and design of the Leipzig (LIFE) heart study: phenotyping and cardiovascular characteristics of patients with coronary artery disease. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0029070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross A., Tönjes A., Kovacs P., Veeramah K.R., Ahnert P., Roshyara N.R. Population-genetic comparison of the Sorbian isolate population in Germany with the German KORA population using genome-wide SNP arrays. BMC Genetics. 2011;12:67. doi: 10.1186/1471-2156-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veeramah K.R., Tönjes A., Kovacs P., Gross A., Wegmann D., Geary P. Genetic variation in the Sorbs of eastern Germany in the context of broader European genetic diversity. European Journal of Human Genetics. 2011;19(9):995–1001. doi: 10.1038/ejhg.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz M., Labarthe F., Fortier A., Bouchard B., Thompson Legault J., Bolduc V. Circulating acylcarnitine profile in human heart failure: a surrogate of fatty acid metabolic dysregulation in mitochondria and beyond. American Journal of Physiology Heart and Circulatory Physiology. 2017;313(4):H768–H781. doi: 10.1152/ajpheart.00820.2016. [DOI] [PubMed] [Google Scholar]
- 13.Würtz P., Havulinna A.S., Soininen P., Tynkkynen T., Prieto-Merino D., Tillin T. Metabolite profiling and cardiovascular event risk: a prospective study of three population-based cohorts. Circulation. 2015;131(9):774–785. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floegel A., Kühn T., Sookthai D., Johnson T., Prehn C., Rolle-Kampczyk U. Serum metabolites and risk of myocardial infarction and ischemic stroke: a targeted metabolomic approach in two German prospective cohorts. European Journal of Epidemiology. 2018;33(1):55–66. doi: 10.1007/s10654-017-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen A.-K., Zeilinger S., Kastenmüller G., Römisch-Margl W., Brugger M., Peters A. Epigenetics meets metabolomics: an epigenome-wide association study with blood serum metabolic traits. Human Molecular Genetics. 2014;23(2):534–545. doi: 10.1093/hmg/ddt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrachelli V.G., Rentero P., Mansego M.L., Morales J.M., Galan I., Pardo-Tendero M. Genomic and metabolomic profile Associated to clustering of cardio-metabolic risk factors. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0160656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl S., Vogt S., Stückler F., Krumsiek J., Bartel J., Kacprowski T. Multi-omic signature of body weight change: results from a population-based cohort study. BMC Medicine. 2015;13 doi: 10.1186/s12916-015-0282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachlechner U., Floegel A., Steffen A., Prehn C., Adamski J., Pischon T. Associations of anthropometric markers with serum metabolites using a targeted metabolomics approach: results of the EPIC-potsdam study. Nutrition and Diabetes. 2016;6(6):e215. doi: 10.1038/nutd.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brauer R., Leichtle A.B., Fiedler G.M., Thiery J., Ceglarek U. Preanalytical standardization of amino acid and acylcarnitine metabolite profiling in human blood using tandem mass spectrometry. Metabolomics. 2011;7(3):344–352. doi: 10.1007/s11306-010-0256-1. [DOI] [Google Scholar]
- 20.Ceglarek U., Leichtle A., Brügel M., Kortz L., Brauer R., Bresler K. Challenges and developments in tandem mass spectrometry based clinical metabolomics. Molecular and Cellular Endocrinology. 2009;301(1–2):266–271. doi: 10.1016/j.mce.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Ceglarek U., Müller P., Stach B., Bührdel P., Thiery J., Kiess W. Validation of the phenylalanine/tyrosine ratio determined by tandem mass spectrometry: sensitive newborn screening for phenylketonuria. Clinical Chemistry and Laboratory Medicine. 2002;40(7):693–697. doi: 10.1515/CCLM.2002.119. [DOI] [PubMed] [Google Scholar]
- 22.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 23.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn M.C. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 25.Wang J. An estimator for pairwise relatedness using molecular markers. Genetics. 2002;160(3):1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vatcheva K.P., Lee M., McCormick J.B., Rahbar M.H. Multicollinearity in regression analyses conducted in epidemiologic studies. Epidemiology (Sunnyvale) 2016;6(2) doi: 10.4172/2161-1165.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noble W.S. How does multiple testing correction work? Nature Biotechnology. 2009;27(12):1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson C.B., Bogomolov M., Benjamini Y., Sabatti C. Many phenotypes without many false discoveries: error controlling strategies for multitrait association studies. Genetic Epidemiology. 2016;40(1):45–56. doi: 10.1002/gepi.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Q.Q., Ritchie S.C., Brozynska M., Inouye M. Power, false discovery rate and Winner's curse in eQTL studies. Nucleic Acids Research. 2018;46(22):e133. doi: 10.1093/nar/gky780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia J., Wishart D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nature Protocols. 2011;6:743 EP. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 31.Almende B.V., Benoit Thieurmel, Robert Titouan. visNetwork: network Visualization using 'vis.js' Library. 2018. https://CRAN.R-project.org/package=visNetwork Available from:
- 32.Ruoppolo M., Campesi I., Scolamiero E., Pecce R., Caterino M., Cherchi S. Serum metabolomic profiles suggest influence of sex and oral contraceptive use. American Journal of Translational Research. 2014;6(5):614–624. [PMC free article] [PubMed] [Google Scholar]
- 33.Proenza A.M., Crespí C., Roca P., Palou A. Gender related differences in the effect of aging on blood amino acid compartmentation*. The Journal of Nutritional Biochemistry. 2001;12(7):431–440. doi: 10.1016/s0955-2863(01)00157-7. [DOI] [PubMed] [Google Scholar]
- 34.Takiyama N., Matsumoto K. Age-and sex-related differences of serum carnitine in a Japanese population. Journal of the American College of Nutrition. 1998;17(1):71–74. doi: 10.1080/07315724.1998.10720458. [DOI] [PubMed] [Google Scholar]
- 35.Li K., Sun Q.-B., Liu X.-Z., Shi Y.-H. Correlation of serum carnitine levels with age and sex among Chinese adults in Nanjing. Zhonghua Nan Ke Xue. 2009;15(4):337–340. [PubMed] [Google Scholar]
- 36.Li Z., Zhang Y., Hu T., Likhodii S., Sun G., Zhai G. Differential metabolomics analysis allows characterization of diversity of metabolite networks between males and females. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0207775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amatayakul K., Laokuldilok T., Koottathep S., Dejsarai W., Prapamontol T., Srirak N. The effect of oral contraceptives on protein metabolism. Journal of the Medical Association of Thailand. 1994;77(10):509–516. [PubMed] [Google Scholar]
- 38.Moller S.E., Moller B.M., Olesen M., Fjalland B. Effects of oral contraceptives on plasma neutral amino acids and cholesterol during a menstrual cycle. European Journal of Clinical Pharmacology. 1996;50(3):179–184. doi: 10.1007/s002280050089. [DOI] [PubMed] [Google Scholar]
- 39.Holub M., Tuschl K., Ratschmann R., Strnadová K.A., Mühl A., Heinze G. Influence of hematocrit and localisation of punch in dried blood spots on levels of amino acids and acylcarnitines measured by tandem mass spectrometry. Clinica Chimica Acta. 2006;373(1–2):27–31. doi: 10.1016/j.cca.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Iacobazzi V., Convertini P., Infantino V., Scarcia P., Todisco S., Palmieri F. Statins, fibrates and retinoic acid upregulate mitochondrial acylcarnitine carrier gene expression. Biochemical and Biophysical Research Communications. 2009;388(4):643–647. doi: 10.1016/j.bbrc.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Trupp M., Zhu H., Wikoff W.R., Baillie R.A., Zeng Z.-B., Karp P.D. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0038386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinhauser M.L., Olenchock B.A., O'Keefe J., Lun M., Pierce K.A., Lee H. The circulating metabolome of human starvation. JCI Insight. 2018;3(16) doi: 10.1172/jci.insight.121434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felig P., Owen O.E., Wahren J., Cahill G.F. Amino acid metabolism during prolonged starvation. Journal of Clinical Investigation. 1969;48(3):584–594. doi: 10.1172/JCI106017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pang S., Lynn D.A., Lo J.Y., Paek J., Curran S.P. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nature Communications. 2014;5 doi: 10.1038/ncomms6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hjelmervik H., Hausmann M., Craven A.R., Hirnstein M., Hugdahl K., Specht K. Sex- and sex hormone-related variations in energy-metabolic frontal brain asymmetries: a magnetic resonance spectroscopy study. NeuroImage. 2018;172:817–825. doi: 10.1016/j.neuroimage.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 46.Zhu Y., Evans M.I. Estrogen modulates the expression of L-arginine:glycine amidinotransferase in chick liver. Molecular and Cellular Biochemistry. 2001;221(1–2):139–145. doi: 10.1023/A:1010946414017. [DOI] [PubMed] [Google Scholar]
- 47.Bordallo J., Secades L., Bordallo C., Cantabrana B., Sánchez M. Influence of gender and sex hormones on 5alpha-dihydrotestosterone elicited effect in isolated left atria of rats: role of beta-adrenoceptors and ornithine decarboxylase activity. European Journal of Pharmacology. 2009;604(1–3):103–110. doi: 10.1016/j.ejphar.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 48.Zhu Q., Jin L., Casero R.A., Davidson N.E., Huang Y. Role of ornithine decarboxylase in regulation of estrogen receptor alpha expression and growth in human breast cancer cells. Breast Cancer Research and Treatment. 2012;136(1):57–66. doi: 10.1007/s10549-012-2235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin C., Samudio I., Ngwenya S., Safe S. Estrogen-dependent regulation of ornithine decarboxylase in breast cancer cells through activation of nongenomic cAMP-dependent pathways. Molecular Carcinogenesis. 2004;40(3):160–170. doi: 10.1002/mc.20030. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava A., Creek D.J., Evans K.J., Souza D. de, Schofield L., Müller S. Host reticulocytes provide metabolic reservoirs that can Be exploited by malaria parasites. PLoS Pathogens. 2015;11(6) doi: 10.1371/journal.ppat.1004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrader M., Fahimi H.D. The peroxisome: still a mysterious organelle. Histochemistry and Cell Biology. 2008;129(4):421–440. doi: 10.1007/s00418-008-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data of the LIFE studies are available upon reasonable request on the LIFE Research Centre for Civilization Diseases.