Abstract
Pathogenesis of Clostridium difficile has been linked to production of toxins, including the large toxins A and B as well as the binary toxin CDT. Until recently, toxin A was only found in combination in clinical strains with the toxin B, unlike toxin B or CDT, which were found alone in toxigenic variants. New toxigenic variants of C. difficile detected in our laboratory from patients with diarrhoea or severe colitis, including a variant producing only toxin A, were tested for virulence in the hamster model, which displays the clinical features of C. difficile disease. Hamsters infected with a strain producing only toxin B induced similar clinical signs, time to death from infection and histologic damage compared to the hypervirulent strain 027. No mortality or clinical signs of infection but caecal histologic damage was found with the variant producing only toxin A. The C. difficile variant strain producing only CDT was able to kill one hamster out of seven; nevertheless, the surviving animals had few alteration of the caecum.
Keywords: Clostridium difficile, tcdA, tcdB, CDT, hamster
Introduction
Clostridium difficile is a Gram-positive anaerobic and spore-forming bacterium responsible for nosocomial digestive infections in most developed countries. The symptoms of C. difficile infections (CDI) range from mild self-limited diarrhoea to life-threatening pseudomembranous colitis [1].
The major virulence factors of C. difficile are two large protein toxins which share the same structural features and the same mechanism of action: the glycosylation of the Rho and Ras proteins [2], [3]. This leads to the disruption of the epithelial cell cytoskeleton and tight junctions between cells as well as the loss of epithelial integrity and more extensive tissue damage, mainly in the colon. Despite their similarities in size and modular structure, those two toxins do not recognize the same receptor for their binding [2].
The genes tcdA and tcdB, which code for TcdA and TcdB, respectively, are part of a 19.6 kb region called the pathogenicity locus (PaLoc). The PaLoc also harbours 3 other genes, tcdR and tcdC, involved in the positive and negative transcriptional regulation of the toxin genes, respectively, and tcdE, which encodes a putative holin required for the efficient secretion of TcdA and TcdB [4], [5]. PaLoc is found mostly in the same genomic location and is replaced in nontoxigenic strains by a highly conserved 75 of 115 bp noncoding region [6]. Further, the genetic polymorphism found in the PaLoc can be explored by a PCR-restriction method called toxinotyping [7].
Most C. difficile clinical strains usually produce both toxins, but some strains only produce TcdB (strains A−B+). These strains belong to different toxinotypes, and the most frequent ones (toxinotype VIII) are characterized by the presence within the PaLoc of a modified tcdA gene leading to a truncated nonactive toxin A. They have been described worldwide and have caused large outbreaks across several Asian countries [8]. In 2015, Janezic et al. [9] described a new toxinotype strain (toxinotype XXXII) characterized by a lack of whole genes tcdA and tcdC and an atypical integration site of the PaLoc. More recently, a new variant strain, A+B−, harbouring tcdA but not tcdB [10], was isolated for the first time in humans and showed a different site of integration.
The emergence of genetic variants of the tcdA and tcdB genes raised the question of the role of TcdA and TcdB in C. difficile pathogenesis. The contribution of the two toxins to the disease has been evaluated through diverse in vitro or in vivo models. One such model, the hamster model, emerged because it mirrors many clinical aspects of human CDI, with haemorrhagic caecitis and eventually death. Using this model and different mouse infection models, Lyras et al. [11] and Carter et al. [12] analysed isogenic toxin mutants from two different C. difficile strains, including JIR8094, a derivative of strain 630, demonstrated that the A+B− mutant was less virulent than both the parental A+B+ strain and the A−B+ mutant strain, whereas in studies performed by Kuehne and colleagues [13], [14] in the hamster model, the A+B− mutant from strains 630Δerm or R20291 were almost as virulent as the parental strain and the A−B+ mutant strain. Despite this discrepancy, which has been putatively related to single nucleotide polymorphisms occurring between the two genetically close parental strains, these results strongly suggest that TcdB plays a major role in the occurrence of clinical signs.
A third toxin, the binary toxin CDT, is also produced by 23% of the toxigenic strains, including the hypervirulent clones 027 and 078 [15]. Briefly, the binary toxin of C. difficile targets the small G protein and its actin-specific ADP-ribosylating activity, resulting in the disorganization of cytoskeleton of the cells [16]. Although its precise role in pathogenesis remains unclear, it is recognized that it could act synergistically with the TcdA and TcdB toxins. In particular, it has been shown that CDT enhances the virulence of C. difficile in mice via the suppression of a protective host eosinophilic response [17]. It has also been shown that CDT induces microtubule protrusions at the cell surface, which wrap and embed bacterial cells, thereby largely increasing the adherence of C. difficile [18].
The role of the different toxins has been evaluated by testing isogenic mutants of C. difficile in the hamster model. However, to our knowledge, the virulence of clinical strains producing only toxin A or the binary toxin has never been tested in this model. Therefore, the objective of this study was to test the ability of two clinical variant strains, putatively involved in human infections, to reproduce its symptomatology in hamsters.
Materials and methods
C. difficile strains and spore preparation
Four toxigenic C. difficile clinical isolates—CD13-125 (A+B+CDT+), CD10-165 (A−B+CDT+), RA09-070 (A+B−CDT−) and CD13-073 (A−B−CDT+)—were used in this study. One nontoxigenic C. difficile clinical isolate, CD15-159 (A−B−CDT− or No Toxin (NT)), isolated from a liver abscess in 2015, was added as a negative control (Table 1). Strain CD13-125 (A+B+CDT+) belongs to the epidemic PCR-ribotype 027 and was isolated from severe CDI cases in Marseille in 2013. Strain CD13-073 (A−B−CDT+) belongs to PCR ribotype 288 and was isolated from a patient with pseudomembranous colitis in Bordeaux in 2012. Strains RA09-070 (A+B−CDT−) and CD10-165 (A−B+CDT+), isolated from patients with from antibiotic-associated diarrhoea and pseudomembranous colitis, respectively, were characterized by whole genome sequencing, as reported elsewhere [10].
Table 1.
Clinical and biological data of five study strains
| Strain | Ribotype; ST; clade | Age (years) | Clinical symptoms; treatment | Origin, year of isolation | Outcome | Phenotype | Genotype (PaLoc) |
|---|---|---|---|---|---|---|---|
| CD10-165 | New; new; clade C–I | 74 | PMC; none | Villefranche sur Sâone (France), 2010 | Death attributable to CDI day after CDI diagnosis | A−B+CDT+ | tcdR+, tcdB+, tcdE+, tcdA−, tcdC− |
| RA09-070 | New; new; clade 5 | 60 | AAD; oral MTZ 500 mg × 3 for 10 days | Cambrai (France), 2009 | Resolution of diarrhoea; no relapse | A+B−CDT− | tcdR+, tcdB−, tcdE−, tcdA+, tcdC− |
| CD13-125 | RT 027; ST 1; clade 2 | 73 | AAD; unknown | Marseille (France), 2013 | Unknown | A+B+CDT+ | tcdR+, tcdB+, tcdE+, tcdA+, tcdC+, −18 pb deletion in tcdC |
| CD15-159 | 010; ST 15; clade 1 | 54 | Liver abscess; MTZ 500 mg × 3 IV | Créteil (France), 2015 | Cure | A−B−CDT− | PaLoc replaced by 115 bp |
| CD13-073 | 288; ST 11; clade 5 | 56 | PMC; oral MTZ 250 mg × 4 | Bordeaux (France), 2012 | Resolution of diarrhoea; no relapse | A−B−CDT+ | tcdR−, tcdB−, tcdE−, tcdA−, tcdC+, −39 bp deletion in tcdC |
AAD, antibiotic-associated diarrhoea; CDI, Clostridium difficile infection; IV, intravenous; MTZ, metronidazole; PaLoc, pathogenicity locus; PMC, pseudomembranous colitis; ST, sequence type.
The identification of the strains was confirmed by MALDI-TOF MS (Brucker) and the glutamate dehydrogenase component of the C. diff Quik Chek Complete assay (Abbot).
Spores were prepared following the method described by Siani et al. [19]. Briefly, strains were plated on solid-media Wilkins-Chalgren agar for 48 hours at 37°C in an anaerobic chamber. Then plates were incubated aerobically at room temperature for 5 days. The entire bacterial lawn was scraped off using a sterile swab and then resuspended in 1 mL sterile water. To kill the vegetative cells, bacterial suspensions were mixed with ethanol (vol/vol) and incubated for 30 minutes at room temperature. Then the suspension was centrifuged (3000 r/min for 20 minutes), and the pellet was resuspended in 1 mL ethanol, sonicated for 1 minute, and washed in cold water three times. Spore stocks were stored at 4°C and were enumerated by plating on Wilkins-Chalgreen agar containing taurocholate 0.1%.
Typing
Multilocus sequence typing (MLST) was performed according to the scheme developed by Griffith et al. [20]. Briefly, seven housekeeping genes (adk, atpA, dxr, glyA, recA, sodA and tpi) were amplified by PCR and sequenced as previously described [20]. The sequence type (ST) was determined as a combination of alleles identified by comparing sequences with sequences available in the C. difficile MLST database (http://pubmlst.org/cdifficile/). Toxinotyping was performed according to the scheme developed by Rupnik et al. [21].
Determination of MIC
Susceptibility testing for erythromycin, clindamycin, moxifloxacin, metronidazole and vancomycin was carried out using the Etest (bioMérieux) on Brucella blood agar (bioMérieux), as described elsewhere [22].
Hamster infection model
Adult Mesocricetus auratus female hamsters (weight, 80–100 g) were obtained from Charles River Laboratories and were housed individually in polypropylene isolator cages fitted with filter covers holding disposable polyester air filters. All food, water, bedding, cages, wire lids and filter covers were autoclaved before use, and food and water were provided ad libitum.
Animals were randomly distributed into five groups (minimum of seven animals per group). Five days before infection, each animal provided by orogastric administration a single dose of clindamycin 50 mg/kg as described previously [23]. On day 0, each animal received 104 spores of the adequate strain, and animals were monitored daily for signs of disease such as weight loss, lack of activity, wet tail and diarrhoea until death or for 14 days for surviving hamsters, which were then humanely killed. Bacterial colonization was monitored at 2, 4, 7, 10 and 14 days after challenge for surviving hamsters. Briefly, a suspension at 10 mg/mL of stools was homogenized in phosphate-buffered saline and then diluted from 10−1 to 10−4; 100 μL of each dilution was inoculated on ChromID agar (bioMérieux) in duplicate, and plates were incubated in anaerobic atmosphere at 37°C for 48 hours before counting the colonies.
All animal experiments were conducted according to the European Union guidelines for the handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm) and were approved by the Central Animal Care Facilities and Use Committee of University Paris-Sud (agreement C-92-019-01; protocol 2012-108).
Histologic staining and scoring
Caecal sections were collected from hamsters and fixed in 10% formalin for a minimum of 2 hours, transferred to 70% ethanol for 18 hours, and embedded in paraffin. Sections of 3 μm were stained with haematoxylin and eosin, and were analysed in a blinded manner using the histopathologic scoring scheme from Buckley et al. [24]. Briefly, the extent and depth of inflammation were evaluated with four different criteria on a scale ranging from 0 to 3: neutrophil infiltration (0, no neutrophil accumulation; 1, local acute neutrophilic infiltrate; 2, extensive submucosal neutrophil accumulation; 3, transmural neutrophilic infiltrate), haemorrhagic congestion (0, normal tissue; 1, engorged mucosal capillaries; 2, submucosal congestion with unclotted blood; 3, transmural congestion with unclotted blood), hyperplasia (0, no epithelial hyperplasia; 1, twofold increase in thickness; 2, threefold increase in thickness; 3, fourfold increase in thickness) and percentage of epithelial barrier involvements (0, no damage; 1, less than 10% mucosal barrier involved; 2, between 10% and 50% of mucosal barrier involved; 3, more than 50% mucosal barrier involved).
Statistical analysis
All analysis were performed by GraphPad Prism 5 (GraphPad Software). Results of colonization kinetics were expressed as mean number of total CFU per gram of faecal pellets. Results of histology were expressed as mean (±standard deviation) histology score per strain.
Results
Typing and antimicrobial susceptibility of C. difficile strains
After MLST determination, the five C. difficile strains were classified into five different sequence STs: ST 11 (clade 5), ST 1 (clade 2) and ST 15 (clade 1) for CD13-073 (A−B−CDT+); CD13-125 (A+B+CDT+) and CD15-159 (NT), respectively; and two new ST for RA09-070 (A+B−CDT−) and CD10-165 (A−B+CDT+) (Table 1).
The allele profile of the new ST of the strain RA09-070 (A+B−CDT−) was a follows: adk = 15, atpA = 21, dxr = 31, glyA = 38, recA = 21, sodA = 36 and tpi = 30. The allele profile of the new ST of CD10-165 (A−B+CDT+) was as follows: adk = 13, atpA = 18, dxr = 22, glyA = 33, recA = 18, sodA = 31 and tpi = 26.
C. difficile CD10-165 (A−B+CDT+) and RA09-070 (A+B−CDT−) were susceptible to all antibiotics tested (erythromycin, clindamycin, moxifloxacin, metronidazole, vancomycin). C. difficile CD13-073 (A−B−CDT+) was susceptible to the same antibiotics except moxifloxacin (MIC = 12 mg/L). C. difficile CD13-125 (A+B+CDT+) and CD15-159 (NT) were susceptible to metronidazole and vancomycin but highly resistant to erythromycin (MIC > 256 mg/L). C. difficile CD15-159 (NT) was also resistant to clindamycin (MIC > 256 mg/L) and CD13-125 (A+B+CDT+) but was resistant to moxifloxacin (MIC > 32 mg/L).
The colonization rates of the hamsters is similar for the five C. difficile strains
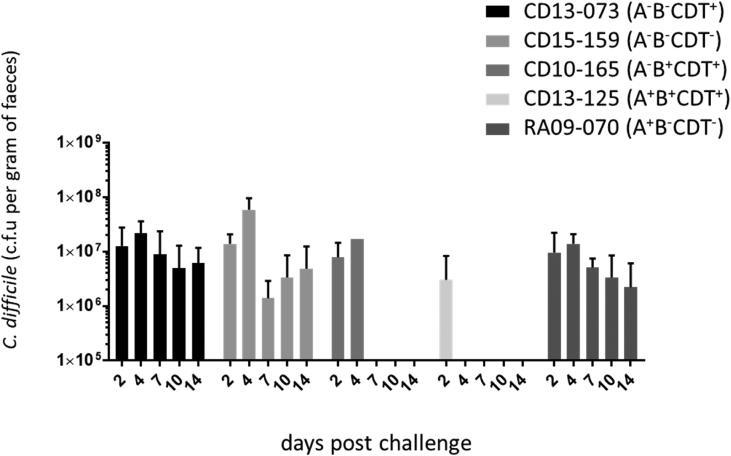
Clindamycin-pretreated hamsters were challenged by orogastric administration with spores of each C. difficile strain. We evaluated the degree of colonization by the five C. difficile strains by enumerating both the vegetative forms and spores in faecal pellets. At day 2 after infection, the level of colonization, evaluated by the faecal clearance of the bacteria (CFU/g faeces), was not significantly different for the five strains: 6.1 × 106 for RA09-070 (A+B−CDT−) to 1.5 × 107 for CD13-073 (A−B−CDT+). In hamsters infected with RA09-070 (A+B−CDT−), CD13-073 (A−B−CDT+) and CD15-159 (NT), the C. difficile colonization level remained stable, above 106 CFU/g faeces at day 14, when the hamsters were humanely killed (Fig. 1).
Fig. 1.
Colonization kinetics of Clostridium difficile strains in hamsters. Colonization of each C. difficile strain was monitored in faecal samples by enumerating both vegetative and spore forms at days 2, 4, 7, 10 and 14 for surviving hamsters.
Clinical monitoring of hamsters
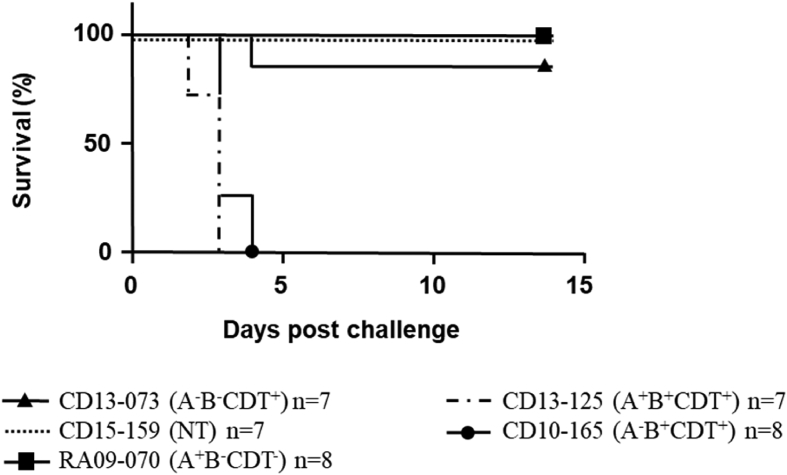
All hamsters provided CD13-125 (A+B+CDT+)or CD10-165 (A−B+CDT+) died within 96 hours, mostly between 48 and 72 hours, and were subject to wet tail, diarrhoea and loss of weight. In contrast, all hamsters provided RA09-070 (A+B−CDT−) or CD15-159 (NT) were still alive at day 14 (Fig. 2). No differences were found in terms of clinical observations between the nontoxigenic strain and the RA09-070 (A+B−CDT−) strain; in particular, an increase of about 10% of weight was observed at the end of the experiment for the hamsters infected with these two strains (Supplementary Table S1). However, hamsters infected with strain CD13-073 (A−B−CDT+) presented two phenotypes. Whereas six of seven hamsters presented no clinical signs and survived until the end of the assay, one died at day 4 without clinical signs before death.
Fig. 2.
Kaplan-Meier survival plots for hamsters infected with five different strains after single dose of clindamycin (n = 7 or 8 per group).
Histologic changes
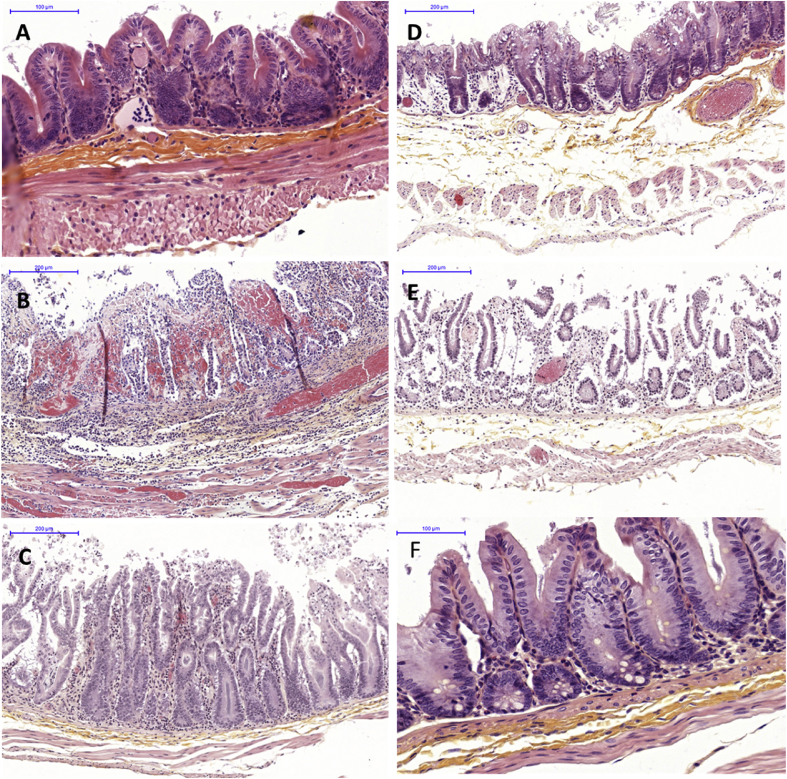
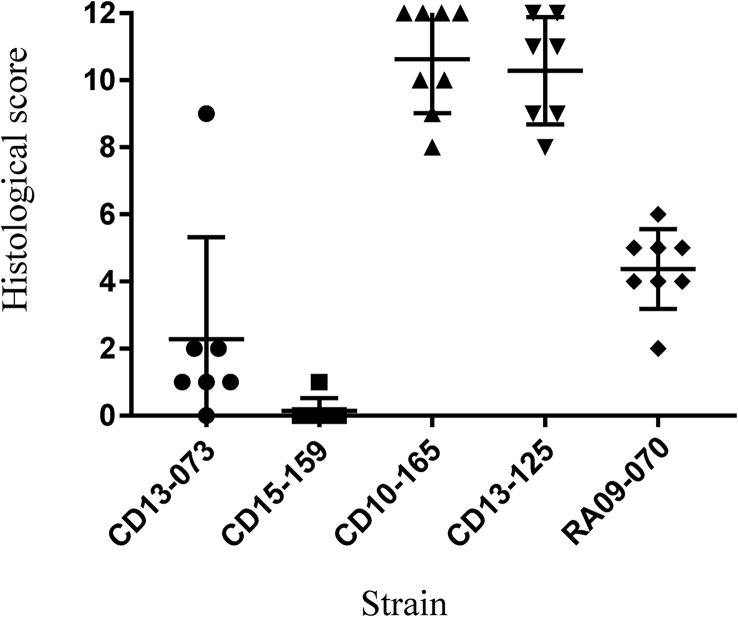
In order to assess the impact on caecal histology of the five C. difficile strains, sections of caecal tissues were sampled at the time of death for all hamsters. Strains CD10-165 (A−B+CDT+) and CD13-125 (A+B+CDT+) produced similar major histologic changes; representative histopathology in the caecum is shown in Fig. 3. There was an acute haemorrhagic congestion resulting in oedema within the submucosa and the lamina propria, a neutrophil infiltration and an acute hyperplasia; finally, we found extensive epithelial damage with disruption of the villi (mean histologic score = 10.63 and 10.29 for strains CD10-165 (A−B+CDT+) and CD13-125 (A+B+CDT+) respectively; Fig. 4).
Fig. 3.
Caecal histology of hamsters showed significant changes according to different strains used for infection. (A) Cecum of hamster infected with strain CD15-159 (NT). (B) Cecum of hamster infected with strain CD10-165 (A−B+CDT+). (C) Cecum of hamster infected with strain CD13-125 (A+B+CDT+). (D) Cecum of hamster infected with strain RA09-070 (A+B−CDT−). (E) Cecum of hamster infected with strain CD13-073 (A−B−CDT+), which died at day 4. (F) Cecum of hamster infected with strain CD13-073 (A−B−CDT+), humanely killed at day 14. All micrographs are of haematoxylin and eosin–stained tissue at original magnification × 100. Bars represent 100 or 200 μm.
Fig. 4.
Histologic differences in caeca of hamsters infected with various strains of Clostridium difficile. Criteria were neutrophil infiltration, haemorrhagic congestion, hyperplasia and percentage of epithelial barrier involvements with maximal score of 12. Bars represent mean scores for each group of animals and standard deviations.
As expected, hamsters infected with the nontoxigenic strain presented an intact caecum (Fig. 3, Fig. 4). In contrast, despite a lack of mortality and the presence of characteristic clinical signs of CDI, neutrophilia infiltration was observed with an extensive hyperplasia for hamsters infected with RA09-070 (A+B−CDT−) (Fig. 3). Epithelial cell loss was sometimes present, but unclotted red blood cells were rarely found. The mean histologic score of 4.5 was significantly different (p < 0.0001) compared to the score of the TcdB producers CD10-165 (A−B+CDT+) and CD13-125 (A+B+CDT+). Interestingly, six of the seven hamsters infected with strain CD13-073 (A−B−CDT+) that survived the challenge exhibited little alteration of the caecum, with essentially some neutrophilia infiltration; the mean histologic score was significantly higher compared to the score of the nontoxigenic strain (mean histologic score, 1.1 and 0.3, respectively; p < 0.05). The seventh hamster, which died within 48 hours after infection, exhibited marked inflammation and acute hyperplasia, and had an individual histologic score of 9 (Fig. 3, Fig. 4).
Discussion
C. difficile can produce three different toxins, and it has been established that human CDI is a multifactorial occurrence which depends of the C. difficile strain and host susceptibility. The purpose of this study was to assess the pathogenesis of new variant strains isolated from symptomatic patients using the hamster model [11], [13]. Our panel of strains included a strain producing only the binary toxin CDT; a strain producing only TcdA; and, as controls for the validation of the model, a nontoxigenic strain, a hypervirulent strain which produced the three toxins TcdA, TcdB and CDT, and a strain producing TcdB and CDT.
The validation of our model was assessed because clinical TcdB-producing strains CD13-125 (A+B+CDT+) and CD10-165 (A−B+CDT+) induced similar clinical signs in hamsters, time to death from infection and severe histologic damages, thereby highlighting the fact that TcdB clearly appears sufficient to induce CDI in the hamster model. The pathogenicity of the two variants producing only one toxin, TcdA or CDT, was different, however.
Therefore, RA09-070 (A+B−CDT−), which has been characterized as the first clinical strain producing TcdA without TcdB [10], was isolated from a patient with an antibiotic-associated diarrhoea who recovered after metronidazole treatment, thus strongly suggesting that C. difficile was implicated in the disease. The genome of the strain possesses no copy of the classical PaLoc, but a gene coding tcdA within a PaLoc encompassing tcdR and a new putative Coding DNA Sequence or CDS coding a complete domain related to the prophage BhlA/UviB located far from the usual integration site of the PaLoc in C. difficile. Also, this strain does not harbour genes coding the binary toxin CDT.
Regarding its virulence in the hamster model, no mortality was found; in addition, clinical signs were absent, notably the absence of diarrhoea from a strain that caused diarrhoea in a human patient. However, challenge of hamsters with this strain showed a kinetics of colonization identical to the four other strains used in this study, indicating that its in vivo fitness was not implicated in the lack of the virulence of the strain. These observations are consistent with the study of Lyras et al. [11] in which a tcdB toxin gene mutant of a strain JIR8094, a derivative of C. difficile strain 630, was not associated with mortality in the hamster model. However, Carter et al. [12] found lethality with a significant delay for an isogenic mutant A+B− of a Canadian epidemic strain (strain M7404). Other genetic factors could modulate the virulence of the strain, thus highlighting the notion that the criteria of lethality in the hamster model for a strain producing only TcdA should be used carefully.
RA09-070 (A+B−CDT−) infection was associated with caecal histologic damage, but to a lesser extent than TcdB-producing strains (whether or not strains produced TcdA or CDT), with significant neutrophil infiltration and hyperplasia as well as signs of inflammation. Batah et al. [25] demonstrated in a mouse model that the two toxins of C. difficile strains 630 and R20291 act in synergy with the flagellin, the major component of the filament of the flagella of the bacteria, to produce the strong proinflammatory response observed during the pathogenesis of C. difficile. Because RA09-070 (A+B−CDT−) possesses different regulons coding the proteins of the flagella (GenBank accession no. JPPA00000000), we suggest that TcdA and the flagellin of this strain are responsible of the inflammation observed in our model. The fact that a strain producing only TcdA is involved in a human CDI must be taken into account in terms of diagnostic strategy. This new variant also illustrates the genetic heterogeneity of C. difficile, and further studies must be performed to explain its pathogenicity versus what is observed in hamsters.
Strain CD13-073 (A−B−CDT+) is a C. difficile strain producing only CDT. The A−B−CDT+ strains are classified mostly as toxinotype XI and are seldom isolated in human infection [26], [27]. However their prevalence in animals, as observed in calves with diarrhoea, is more important [28]. The strain was isolated from a 56-year-old patient treated with imipenem and gentamicin for pneumonia. He developed pseudomembranous colitis, and after metronidazole treatment, the clinical signs resolved. Using the hamster model, Geric et al. [29] found no mortality or symptoms with three different strains producing CDT only. In contrast, CD13-073 (A−B−CDT+) infection resulted in death for one animal out of seven with histologic damage similar to those found with TcdB-producing strains, and was able to produce mild histologic damage in the remaining surviving animals. Death of three of eight hamsters was also observed by Kuehne et al. [14], with a mutant strain producing CDT only; animals manifested atypical symptoms of CDI with moderate clinical signs such as wet tail. Variation of virulence of strains producing only CDT in the hamster model suggests that individual susceptibilities and/or the presence of bacterial factors may modulate the effect of binary toxin and may further explain why these variants of C. difficile are rarely isolated in humans.
Overall, we found that severe inflammation of the caeca of infected hamsters is correlated with mortality. In addition, using clinical strains and not laboratory mutants, our results confirmed the major role of TcdB and a less important role of TcdA for C. difficile virulence in the hamster model. Finally, the capacity of a clinical strain to reproduce all the aspects of the human disease was not found. Several factors could explain this observation, including degree of dysbiosis of the host, presence in the microbiota of other species of bacteria promoting the effect of the pathogen and role of innate immunity. This highlights the limitations of the animal model.
Acknowledgements
We sincerely appreciate the technical assistance of V. Domergue (Region Ile de France, IFR141 IPSIT) for animal facilities. We thank A. Youssouf (AP-HP, Hôpital Saint Antoine), S. Lambert-Bordes (EA 7359, Université Paris-Sud) and S. Hoys for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2019.100590.
Conflict of Interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lo Vecchio A., Zacur G.M. Clostridium difficile infection: an update on epidemiology, risk factors, and therapeutic options. Curr Opin Gastroenterol. 2012;28:1–9. doi: 10.1097/MOG.0b013e32834bc9a9. [DOI] [PubMed] [Google Scholar]
- 2.Aktories K., Schwan C., Jank T. Clostridium difficile toxin biology. Annu Rev Microbiol. 2017;71:281–307. doi: 10.1146/annurev-micro-090816-093458. [DOI] [PubMed] [Google Scholar]
- 3.Belyi Y., Aktories K. Bacterial toxin and effector glycosyltransferases. Biochim Biophys Acta. 2010;1800:134–143. doi: 10.1016/j.bbagen.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Govind R., Dupuy B. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin-Verstraete I., Peltier J., Dupuy B. The regulatory networks that control Clostridium difficile toxin synthesis. Toxins (Basel) 2016;8:E153. doi: 10.3390/toxins8050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun V., Hundsberger T., Leukel P., Sauerborn M., Eichel-Streiber C von. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene. 1996;181:29–38. doi: 10.1016/s0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 7.Rupnik M., Janezic S. An update on Clostridium difficile toxinotyping. J Clin Microbiol. 2016;54:13–18. doi: 10.1128/JCM.02083-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J., Kim Y., Pai H. Clinical characteristics and treatment outcomes of Clostridium difficile infections by PCR ribotype 017 and 018 strains. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janezic S., Marín M., Martín A., Rupnik M. A new type of toxin A–negative, toxin B–positive Clostridium difficile strain lacking a complete tcdA gene. J Clin Microbiol. 2015;53:692–695. doi: 10.1128/JCM.02211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monot M., Eckert C., Lemire A., Hamiot A., Dubois T., Tessier C. Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci Rep. 2015;5:15023. doi: 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyras D., O’Connor J.R., Howarth P.M., Sambol S.P., Carter G.P., Phumoonna T. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458(7242):1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter G.P., Chakravorty A., Pham Nguyen T.A., Mileto S., Schreiber F., Li L. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio. 2015;6 doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuehne S.A., Cartman S.T., Heap J.T., Kelly M.L., Cockayne A., Minton N.P. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467(7316):711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 14.Kuehne S.A., Collery M.M., Kelly M.L., Cartman S.T., Cockayne A., Minton N.P. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer M.P., Notermans D.W., Benthem BHB van, Brazier J.S., Wilcox M.H., Rupnik M. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377(9759):63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 16.Gerding D.N., Johnson S., Rupnik M., Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbe. 2014;5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowardin C.A., Buonomo E.L., Saleh M.M., Wilson M.G., Burgess S.L., Kuehne S.A. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol. 2016;1:16108. doi: 10.1038/nmicrobiol.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwan C., Stecher B., Tzivelekidis T., Ham M van, Rohde M., Hardt W.-D. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siani H., Cooper C., Maillard J.Y. Efficacy of ‘sporicidal’ wipes against Clostridium difficile. Am J Infect Control. 2011;39:212–218. doi: 10.1016/j.ajic.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths D., Fawley W., Kachrimanidou M., Bowden R., Crook D.W., Fung R. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupnik M., Avesani V., Janc M., Eichel-Streiber C von, Delmée M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36:2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbut F., Decré D., Burghoffer B., Lesage D., Delisle F., Lalande V. Antimicrobial susceptibilities and serogroups of clinical strains of Clostridium difficile isolated in France in 1991 and 1997. Antimicrob Agents Chemother. 1999;43:2607–2611. doi: 10.1128/aac.43.11.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambol S.P., Tang J.K., Merrigan M.M., Johnson S., Gerding D.N. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J Infect Dis. 2001;183:1760–1766. doi: 10.1086/320736. [DOI] [PubMed] [Google Scholar]
- 24.Buckley A.M., Spencer J., Maclellan L.M., Candlish D., Irvine J.J., Douce G.R. Susceptibility of hamsters to Clostridium difficile isolates of differing toxinotype. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batah J., Kobeissy H., Bui Pham P.T., Denève-Larrazet C., Kuehne S., Collignon A. Clostridium difficile flagella induce a pro-inflammatory response in intestinal epithelium of mice in cooperation with toxins. Sci Rep. 2017;7:3256. doi: 10.1038/s41598-017-03621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckert C., Emirian A., Le Monnier A., Cathala L., De Montclos H., Goret J. Prevalence and pathogenicity of binary toxin–positive Clostridium difficile strains that do not produce toxins A and B. New Microbe. New Infect. 2015;3:12–17. doi: 10.1016/j.nmni.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Androga G.O., Hart J., Foster N.F., Charles A., Forbes D., Riley T.V. Infection with toxin A–negative, toxin B–negative, binary toxin–positive Clostridium difficile in a young patient with ulcerative colitis. J Clin Microbiol. 2015;53:3702–3704. doi: 10.1128/JCM.01810-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneeberg A., Neubauer H., Schmoock G., Grossmann E., Seyboldt C. Presence of Clostridium difficile PCR ribotype clusters related to 033, 078 and 045 in diarrhoeic calves in Germany. J Med Microbiol. 2013;62:1190–1198. doi: 10.1099/jmm.0.056473-0. [DOI] [PubMed] [Google Scholar]
- 29.Geric B., Carman R.J., Rupnik M., Genheimer C.W., Sambol S.P., Lyerly D.M. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J Infect Dis. 2006;193:1143–1150. doi: 10.1086/501368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.