Abstract
p53 is an intrinsically disordered protein with a large number of post-translational modifications and interacting partners. The hierarchical order and subcellular location of these events are still poorly understood. The activation of p53 during the DNA damage response (DDR) requires a switch in the activity of the E3 ubiquitin ligase MDM2 from a negative to a positive regulator of p53. This is mediated by the ATM kinase that regulates the binding of MDM2 to the p53 mRNA facilitating an increase in p53 synthesis. Here we show that the binding of MDM2 to the p53 mRNA brings ATM to the p53 polysome where it phosphorylates the nascent p53 at serine 15 and prevents MDM2-mediated degradation of p53. A single synonymous mutation in p53 codon 22 (L22L) prevents the phosphorylation of the nascent p53 protein and the stabilization of p53 following genotoxic stress. The ATM trafficking from the nucleus to the p53 polysome is mediated by MDM2, which requires its interaction with the ribosomal proteins RPL5 and RPL11. These results show how the ATM kinase phosphorylates the p53 protein while it is being synthesized and offer a novel mechanism whereby a single synonymous mutation controls the stability and activity of the encoded protein.
Keywords: synonymous mutations, intrinsically disordered proteins, cell signaling, MDM2, p53 messenger RNA, ATM kinase
Introduction
p53 carries out a variety of different functions in response to numerous different signaling pathways to co-ordinate the cellular response to various stresses and damages. To achieve this multi-functionality, p53 is intrinsically disordered and this allows its interaction with over 300 cellular factors that determine the physiological outcome of p53 activation (Meek and Anderson, 2009; Wright and Dyson, 2009; Joerger and Fersht, 2010; Cheok et al., 2011; MacLaine and Hupp, 2011; Kannan et al., 2016; Uversky, 2016; Lopez et al., 2017). The specificity of the p53 interactome is directed by allosteric changes that are induced by post-translational modifications (PMs) within disordered domains (Tompa, 2002, 2005). This structure-function continuum requires a strict hierarchy and hence, where and when the PMs take place on p53 have consequences for how p53 activity is differentiated in response to specific changes in the cellular environment. The best-studied pathway for p53 activation is the double-stranded DNA breaks response pathway (DDR), which leads to the activation of the ataxia telangiectasia mutated (ATM). ATM is a non-covalently linked inactive dimer under normal conditions. Upon activation, ATM dissociates into kinase-active monomers via acetylation modifications of the C-terminus and auto-phosphorylation at serine 1981 (Morgan and Kastan, 1997; Scott et al., 2002; Takagi et al., 2004; Stracker et al., 2013; Lee et al., 2015). ATM controls p53 activity via phosphorylation on p53 serine 15 p53(S15) as well as on the key p53 regulatory factors MDM2 and its non-redundant homolog MDMX (Banin et al., 1998; Canman et al., 1998; Loughery et al., 2014).
Under non-stressed conditions, the N-terminus of the E3 ubiquitin ligase MDM2 binds the BOX-1 domain of p53 and promotes p53 polyubiquitination and degradation. However, phosphorylation by ATM on MDM2(S395) switches the activity of MDM2 from a negative to a positive regulator of p53 by allowing the interaction between its C-terminal RING domain with the box-1 p53 mRNA sequence with a subsequent increase in p53 protein synthesis (Haupt et al., 1997; Kubbutat et al., 1997; Maya et al., 2001; Chene, 2003; Chen et al., 2005; Pereg et al., 2005; Naski et al., 2009; Gajjar et al., 2012; Malbert-Colas et al., 2014; Coffill et al., 2016; Karakostis et al., 2016). In addition to controlling p53 rate of synthesis, ATM also controls p53 stabilization via direct phosphorylation on p53(S15) or indirectly, via Chk2 (Thr18 and Ser20 phosphorylations) that prevent MDM2 from binding to the p53 protein (Meek, 2009; Cheng and Chen, 2010; Loughery et al., 2014). MDM2 interacts with the ribosomal factors RPL5, PRPL11, RPL23, RPL26, and the 5S rRNA and is implicated in sensing dysfunctional ribosomal biogenesis that can lead to the activation of p53 (Lindstrom et al., 2007a, b; Zhang et al., 2011; Donati et al., 2013; Bursac et al., 2014).
The interplay between p53 and MDM2 is well conserved during evolution and the p53 mRNA–MDM2 interaction is detected in pre-vertebrates while the protein–protein interaction has evolved in the vertebrates (Coffill et al., 2016; Karakostis et al., 2016). Animal models show that the phosphorylation of MDM2(Ser394) (Ser395 in human) and a p53-mediated induction of mdm2 expression are required during the DDR, suggesting a critical role of MDM2’s negative and positive activities towards p53 (Gannon et al., 2012; Pant et al., 2013).
The role of synonymous mutations in the origin of different diseases, such as cancer, is becoming increasingly clear (Sauna and Kimchi-Sarfaty, 2011; Gartner et al., 2013; Supek et al., 2014). However, apart from altering the splicing of the pre-mRNA, the underlying molecular mechanism(s) of how silent mutations can affect the encoded protein are still unclear (Gartner et al., 2013; Supek et al., 2014; Fahraeus et al., 2016). In this study, we have addressed how the p53 mRNA affects the encoded protein during the DDR. We show that MDM2 via its interaction with the p53 mRNA guides ATM to the nascent p53 protein and that this is required for p53 stabilization. MDM2’s function as a carrier depends on its binding to the p53 mRNA as well as on ribosomal proteins. The presented data shed light on the hierarchal order of the regulation of intrinsically disordered proteins and illustrate how the coding sequence of an mRNA and the encoded protein are functionally interconnected.
Results
p53 stabilization following genotoxic stress is prevented by a cancer-derived synonymous mutation in codon 22
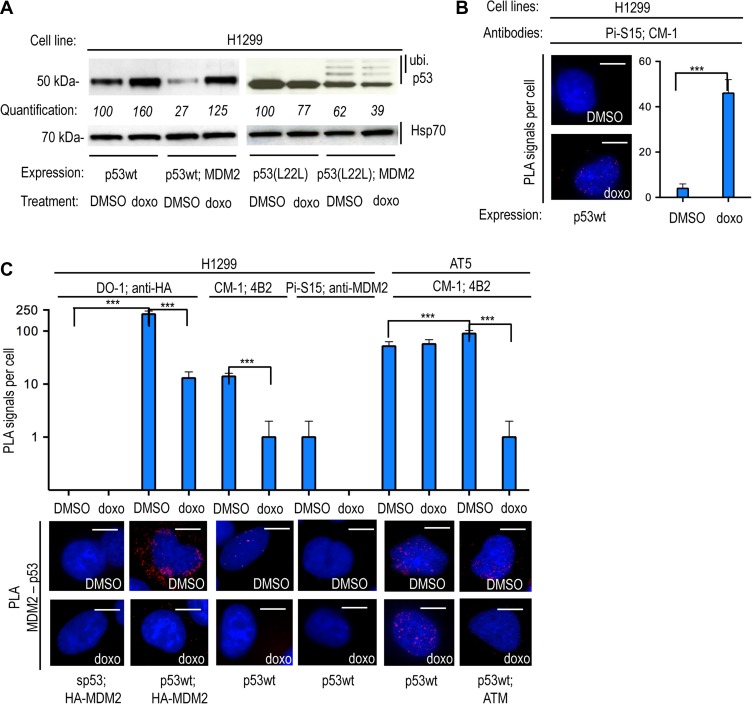
Expression of the wild-type p53 protein (p53wt) in H1299 (p53-null) cells showed the expected MDM2-dependent down-regulation of p53 under normal conditions and p53 stabilization following genotoxic stress induced by 0.1 μM doxorubicin (doxo) treatment for 12 h (Figure 1A). However, when the p53wt protein was expressed from an mRNA carrying a silent cancer-derived mutation in codon 22 (p53(L22L) CUA to CUG) which averts the binding of the p53 mRNA to MDM2 (Candeias et al., 2008), we instead observed a further MDM2-dependent degradation of p53 following DNA damage. This increase in MDM2-dependent degradation of p53 proteins expressed from the p53(L22L) mRNA was accompanied by an increase in monoubiquitinated p53 proteins that are not targeted for 26S proteasome-dependent degradation (Figure 1A). Phosphorylation of p53 on p53(S15) by ATM plays an important role in averting the interaction of p53 with MDM2. Western blot (WB) using the phosphospecific mAb (Pi-S15) showed, as expected, that the stabilization of p53 derived from the wild-type p53 mRNA in the presence of MDM2 and doxo treatment was accompanied by an increase in Ser15 phosphorylation (Supplementary Figure S1A). However, the phosphorylation was ~50% lower for the same p53wt protein derived from the p53(L22L) mRNA while the levels of p53wt expression were ~70% lower when expressed by p53(L22L) mRNA. The reduced Ser15 phosphorylation levels of p53(L22L) are linked to the total p53 levels (Figure 1A and Supplementary Figure S1A). It should be pointed out that other events, such as phosphorylation on Ser20, are also implicated in preventing MDM2 from binding p53. Due to the estimated ratio of RNA:protein (1:3000) (Teufel et al., 2009), it is unlikely that the p53(L22L) mRNA can affect the stability of the p53 protein outside the p53 polysome. In order to investigate which factors might control the activity of ATM towards p53 at its polysome, we employed the proximity ligation assay (PLA), which provides an in situ semi-quantitative estimation of endogenous molecular interactions/associations of low abundance (Soderberg et al., 2006; Gullberg et al., 2011; Schwanhausser et al., 2011). This allows us to determine the location of known protein–protein and protein–RNA interactions in the p53 pathway, as well as the location of the p53(S15) phosphorylation. Employing the PLA in a series of experiments, such as transforming cell lines deficient of p53 (H1299) or ATM (AT5 BIVA) with mutant constructs and inducing gene knockdown, allows us to simulate the induced stress conditions and explore the hierarchy of the events and their interdependence. We used the Pi-S15 mAb and the polyclonal p53 rabbit CM-1 sera to show, by PLA, the expected increase of the phosphorylation at p53 Ser15 following doxo treatment (Figure 1B). The specificity of each antibody employed in PLA was confirmed by immunofluorescence (IF), showing the expected immunostainings (Supplementary Figure S1B and C). IF confirmed the predominant nuclear localization of the DO1, the CM1 and the Pi-S15 epitopes of p53 during the DDR. Under similar conditions, the DO-1 mAb predominately cross-reacted with the nuclear fraction of p53, whereas the polyclonal CM-1 detected some cytoplasmic p53. This discrepancy might indicate a partial masking of the DO-1 epitope (residues 11–25) in the cytoplasm (Supplementary Figure S1B). Additionally, we observed the expected IF staining of MDM2, ATM, RPL5, RPS6, and the p53-binding protein 1 (p53BP1) (Supplementary Figures S1C). In control PLA experiments, we observed the expected PLA signals using a variety of p53 targets (CM-1, Pi-S15), the p53 mRNA, p53BP1, and MDM2 (Supplementary Figure S1D). Using a variety of p53 and MDM2 antibody combinations (DO-1, CM1, and 4B2, anti-MDM2 (Abcam: ab87134), anti-HA tagged MDM2), we observed an average of 70% reduction of p53–MDM2 interactions following treatment with 1 μM doxo for 90 min. Under the same conditions, there was no reduction in the number of interactions in the ATM-null AT5 cells unless an exogenous ATM was introduced (Figure 1C). In line with the notion that Ser15 phosphorylation prevents the p53–MDM2 interaction, we did not observe any PLA signal using the Pi-S15 and anti-MDM2 antibodies under any conditions (Figure 1C). These data support previous works showing that phosphorylation of p53(S15) by ATM is incompatible with an MDM2–p53 interaction. They also show that the synonymous mutation p53(L22L) prevents the stabilization of p53 following DNA damage, suggesting a connection between the p53 mRNA, the phosphorylation of p53(S15) by ATM and the p53–MDM2 interaction.
Figure 1.
The synonymous p53(L22L) mutation prevents ATM-mediated phosphorylation of the nascent p53 protein and p53 stabilization following genotoxic stress. (A) WB using the anti-p53 DO-1 antibody (N-terminal epitope), monitoring the levels of wild-type p53 protein expressed from the wild-type p53 mRNA or from the L22L mRNA in p53-null H1299 in the absence and presence of MDM2 under normal conditions (DMSO) or DNA damage induced by 0.1 mM doxorubicin (doxo). The p53(L22L) mRNA exhibits a low affinity for MDM2 (Malbert-Colas et al., 2014) and the encoded p53wt protein shows a high rate of degradation in the presence of MDM2 following doxo treatment. The quantification values show the relative density of the bands. (B) The PLA using the phosphospecific Pi-S15 mAb together with the anti-p53 rabbit CM-1 shows the subcellular localization of the phosphorylated p53(S15) in transfected H1299 cells. The graph shows that the number of PLA signals per cell increases following DNA damage. (C) PLA shows the subcellular localization of the MDM2–p53 protein–protein interaction in transfected H1299 (p53-null) and AT5 (ATM-null) cells. The silent p53 (sp53) mRNA carrying mutated AUG codons does not express the p53 protein and was used as a control. The number of MDM2–p53 interactions is reduced following DNA damage induced by 0.1 mM doxorubicin (doxo). This was shown using two different pairs of antibodies (DO-1 for p53 and anti-HA for HA-MDM2 or CM-1 for p53 and 4B2 for MDM2). The phosphospecific anti-p53(S15) mAb gave no PLA signal with anti-MDM2 antibody. In AT5 cells, the MDM2–p53 PLA signal was diminished following doxo treatment only after expression of exogenous ATM. The asterisks represent P-values of three independent experiments: n/s, P > 0.05; *P ≤ 0.05; **P ≤ 0 01; ***P ≤ 0.001. Scale bar, 10 μm. The statistical analyses are based on results obtained by at least 50 cells of each experiment. The antibodies used for the PLA are indicated.
ATM phosphorylates the nascent p53 peptide
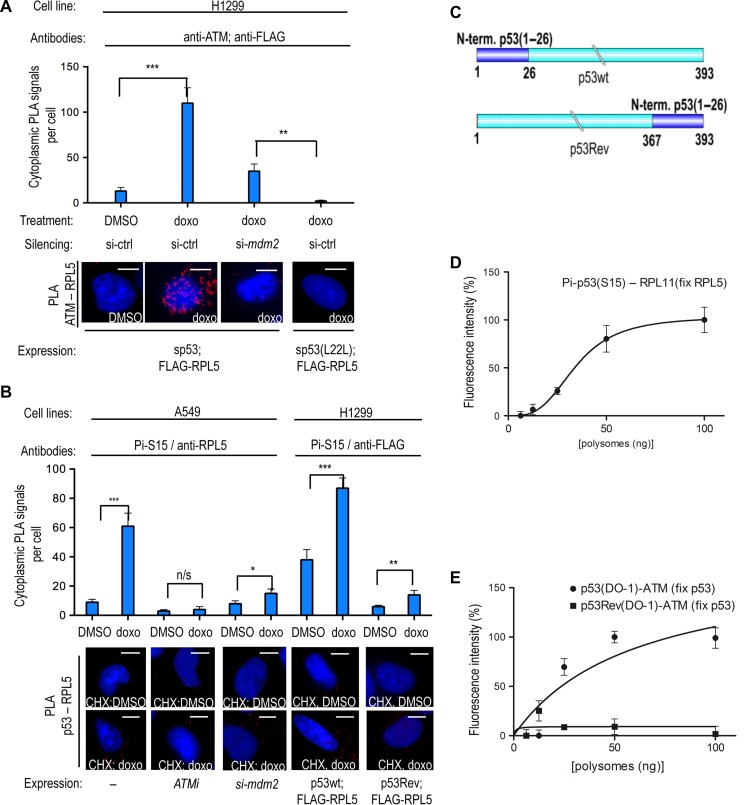
We next tested if ATM-dependent phosphorylation of p53 takes place at the polysome. We first tested if ATM is present at the p53 polysomes by PLA and antibodies against ATM and FLAG-tagged RPL5. The ATM–RPL5 signal was not observed under non-gentoxic conditions in H1299 cells (Figure 2A). Expression of a p53 mRNA that lacks the initiation codons (silent p53 mRNA or sp53) and thus does not express the p53 protein, resulted in a cytoplasmic ATM–RPL5 PLA signal following doxo treatment that was severely perturbed when MDM2 expression was suppressed using siRNAs (Figure 2A). The efficiency of the mdm2 silencing was confirmed by IF and by WB (Supplementary Figure S2A and B). Similar results were obtained by testing the ATM–RPL11 interaction (Supplementary Figure S2C). Interestingly, when we used the sp53(L22L) mRNA, we did not observe the ATM–RPL5 PLA signal at the cytoplasm following doxo treatment (Figure 2A). These results suggest that ATM’s interaction with ribosomal factors in the cytoplasm following DNA damage is p53 mRNA- and MDM2-dependent.
Figure 2.
ATM phosphorylates the nascent p53(S15) peptide. (A) PLA showing that ATM associates with the ribosomal RPL5 protein in the presence of sp53 in H1299 cells in the cytoplasm following doxo treatment. Knocking down mdm2 using siRNA prevents the ATM–RPL5 interaction. The sp53(L22L) mRNA has low affinity for MDM2 and does not support the ATM–RPL5 interaction. (B) PLA using the phosphospecific anti-p53(S15) mAb shows the cytoplasmic association of phosphorylated p53(S15) with RPL5 following DNA damage, using endogenous factors, in A549 cells treated with cycloheximide (CHX). This PLA signal is reproduced in H1299 cells expressing p53wt and the FLAG-RPL5, by using the anti-p53(Pi-S15) and anti-FLAG antibodies. Inhibition of ATM or silencing of mdm2 prevents the (Pi-S15)–RPL5 PLA signal in A549 cells. Following doxo and CHX treatment of p53Rev-expressing cells, there is an ~6.5-fold decrease in the PLA signal between RPL5 and p53(Pi-S15) epitope, as compared to p53wt. Following CHX treatment, the Ser15 epitope of the p53Rev is buried within the ribosome on stalled ribosomes. (C) The first 36 aa of p53wt were switched to the C-terminus to generate the p53Rev construct. The cartoon illustrates the p53wt and p53Rev constructs. (D) PLEA showing the association of p53(Pi-S15) epitope with RPL11 on purified polysomes from doxo-treated cell lysates fixed with anti-RPL5 antibodies. (E) PLEA showing the association of p53 with ATM on polysomes treated with doxo and captured with anti-p53 antibodies. The p53Rev shows no association, confirming that the association between the p53 Ser15 epitope and ATM occurs at the p53 polysome. Scale bar, 10 μm. n/s, P > 0.05; *P ≤ 0.05; **P ≤ 0 01; ***P ≤ 0.001. The statistical analyses are based on results obtained by at least 50 cells from three independent experiments.
We then treated A549 (p53wt) or H1299 cells with cycloheximide (CHX), which fixes emerging nascent peptides on the ribosome to test the Ser15–RPL5 interaction on endogenous targets or when expressed by exogenous p53wt and FLAG-tagged RPL5. Using the Pi-S15 and anti-RPL5/anti-FLAG antibodies, we observed an ~2.3-fold increase in cytoplasmic PLA signals following doxo treatment as compared to normal conditions (Figure 2B; Supplementary Figures S2D and S3A). Similar results were obtained using the Pi-S15 and anti-RPL5 in H1299 cells expressing p53 and treated with etoposide, or using the endogenous p53wt and RPL5 interaction in A549 cells treated with doxo (Grover et al., 2009; Hosp et al., 2015) (Supplementary Figure S2D). Inhibition of ATM using KU55933 (ATMi), or silencing of mdm2, prevented the doxo-dependent increase of endogenous p53–RPL5 PLA signals in A549 cells (Figure 2B). In line with this, ATM gave an increased PLA signal following DNA damage with both MDM2 and RPL5, indicating that these targets co-localize at the polysome (Supplementary Figure S2D).
As free ribosomal factors are instantly imported to the nucleus, these results suggest that ATM is associated with p53 polysomes. To determine if the observed cytoplasmic phosphorylation of Ser15 by ATM indeed occurs on the nascent p53 peptide and it is not derived from free cytoplasmic p53 associated with ribosomes, we deleted codons 1–26 of p53wt and instead fused this sequence to the p53 C-terminus (p53Rev) (Ren et al., 2009) (Figure 2C). This construct expresses the Ser15 epitope at the C-terminus of the protein and thus it may interact with ATM after its release from the ribosome. The abundance of these interactions on CHX-treated cells will be lower compared to the p53wt where the Ser15 epitope is at the N-terminus. Treating cells with CHX and using the same antibodies as for the p53wt–FLAG-RPL5 interaction (see Figure 2B), the p53Rev gave ~8-fold fewer PLA signals as compared to the p53wt following CHX treatment (Figure 2B and C). The conservation of the epitopes and the expression levels of p53Rev and the p53wt were confirmed and found to be similar by WB (Supplementary Figure S2E) and by IF (Supplementary Figure S2F). This indicates that the emerging nascent p53 peptide is phosphorylated by ATM at serine 15, while it is still attached to the ribosome. To further test this, we used sucrose gradients to isolate polysomes from cells expressing p53wt and treated with CHX. We initially fixed polysomes on a 96-well ELISA plate using goat anti-RPL5 antibodies. Using the mouse anti-p53 (Pi-S15) antibody and a rabbit anti-RPL11, we carried out a proximity ligation ELISA (PLEA) on the 96-well plate to demonstrate the presence of the Ser15 epitope on the polysomes (Kd of 35 nM) (Figure 2D and Supplementary Figure S3B). Furthermore, we captured p53 polysomes using a chicken anti-p53 serum and we observed PLEA signal with the p53 mAb DO-1 and a rabbit anti-ATM antibody (Figure 2E and Supplementary Figure S3C). However, this interaction was not detected when we instead expressed the p53Rev construct, in line with the notion that the C-terminus of the p53Rev protein is largely buried within the exit channel of the ribosome. This shows that ATM is associated with the nascent p53 at the polysomes (Figures 2E; Supplementary Figure S3C and D). Similar results were obtained by performing the PLA using the DO-1 and anti-ATM antibodies on polysomes captured with goat anti-RPL5 sera (Supplementary Figure S3E). Taken together, the results so far show that following DNA damage, ATM phosphorylates the nascent p53 peptide while still attached on the translating ribosome in an MDM2- and p53 mRNA-dependent manner.
The p53 mRNA and ribosomal factors govern MDM2’s trafficking to the p53 polysome
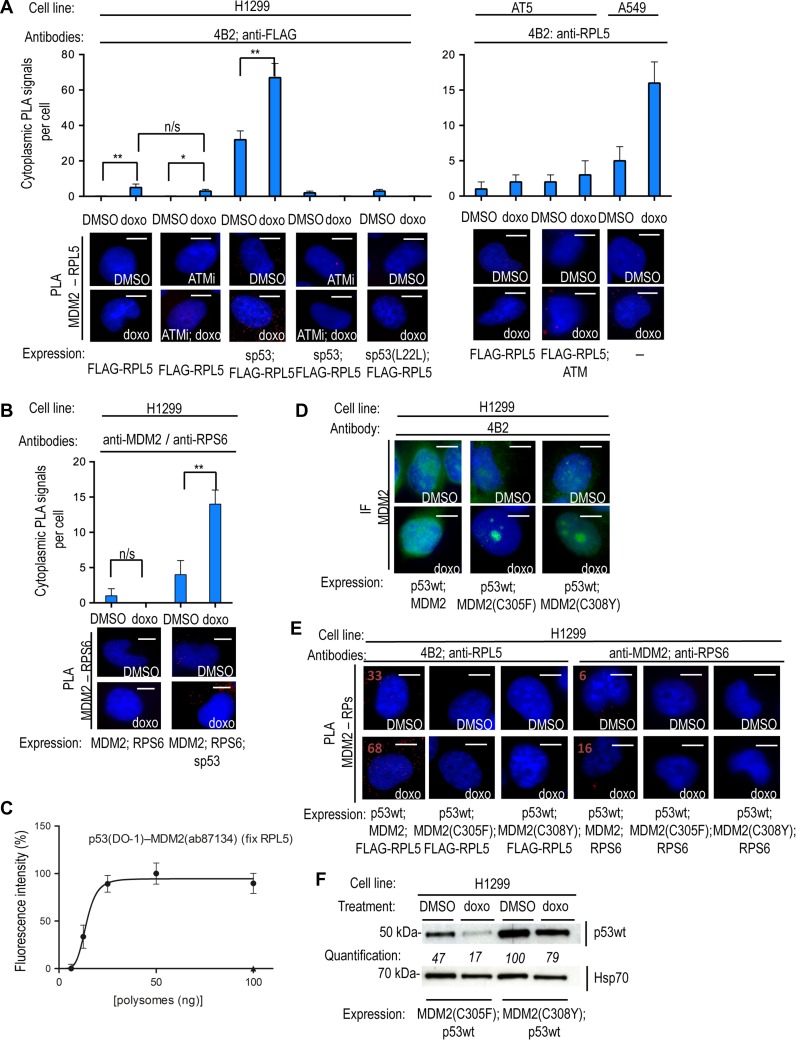
MDM2 interacts with the ribosomal precursor complex RPL5-RPL11-5S and the ribosomal factors RPL23 and RPL26 and since loss of MDM2, or a reduced p53 mRNA–MDM2 affinity, prevent the phosphorylation of Ser15 and p53 stabilization, we next set out to investigate whether MDM2 may guide ATM to the p53 polysome. (Yang et al., 2006; Gannon et al., 2012; Donati et al., 2013; Malbert-Colas et al., 2014). To test if MDM2’s interactions with ribosomal factors play a role in transporting ATM to the p53 polysome, we first tested if MDM2 is present at p53 polysomes following DNA damage. We used the anti-MDM2 4B2 mAb and a rabbit anti-FLAG antibody to carry out a PLA on H1299 cells expressing FLAG-RPL5 and treated with doxo (Figure 3A). This resulted in a few PLA cytoplasmic signals. However, when we co-transfected sp53 under these conditions, we observed a dramatic increase in cytoplasmic signals, showing that the p53 mRNA promotes the presence of MDM2 in complex with RPL5 in the cytoplasm (Figure 3A). Similar results were obtained using the p53(L22L) construct (Supplementary Figure S3F). Treating cells with ATM inhibitors (ATMi), or using the sp53(L22L) mRNA, significantly reduced MDM2’s interaction with RPL5 in the cytoplasm (Figure 3A). Using the anti-MDM2 mouse 4B2 and the anti-RPL5 rabbit antibodies, we observed no endogenous PLA signals in AT5 cells (ATM-null) following doxo treatment unless the exogenous ATM was introduced. Importantly, we observed cytoplasmic PLA signals from endogenous MDM2 and RPL5 in A549 cells following doxo treatment, underlining the interaction between ATM and RPL5 in the cytoplasm is not due to over-expression phenomena (Figure 3A).
Figure 3.
The p53 mRNA and ribosomal factors govern MDM2’s trafficking to the p53 polysome. (A, left) PLA on H1299 cells transfected with FLAG-RPL5, using the anti-MDM2 mAb 4B2 and anti-FLAG. Following DNA damage, the p53 mRNA and ATM kinase activity are required for MDM2 to associate with RPL5 in the cytoplasm. The p53 mRNA promotes the MDM2–RPL5 interaction in the cytoplasm, which is prevented by inhibition of ATM kinase activity (ATMi) or the sp53(L22L) mutant. (A, right) PLA on AT5 and A549 cells using 4B2 and anti-RPL5 antibodies on transfected and endogenous targets, respectively, confirms that the MDM2–RPL5 interaction requires ATM. (B) PLA on transfected H1299 cells using anti-RPS6 (54D2) and anti-MDM2 (ab87134) antibodies. RPS6 does not bind to MDM2 directly and the p53 mRNA induces the association of MDM2 with RPS6 at the cytoplasm, showing that the p53 mRNA is required for MDM2 to reach ribosomal factors during DDR. (C) PLEA showing the association of nascent p53 with MDM2 on polysomes from H1299 lysates treated with doxo and captured with anti-RPL5, confirming that MDM2 is present at the p53 polysome. (D) IF on H1299 cells comparing the predominately nuclear immuno-staining of MDM2wt with the accumulation of MDM2(C305F) and MDM2(C308Y) mutants in nucleoli following doxo treatment. These mutants do not interact with RPL5 or RPL11. (E) PLA showing that compared to the MDM2wt, the MDM2(C305F) or MDM2(C308Y) mutants do not interact with RPL5 or RPS6, in the presence of p53wt, on H1299 cells under normal conditions or following doxo treatment. PLA signal numbers are noted for the MDM2wt interactions (positive). Also compare with MDM2wt interactions with RPL5 and RPS6 in A and B, respectively. The interaction of MDM2 and ribosomal proteins is required for MDM2 to reach the p53 polysome (see B). (F) WB shows that p53wt is not stabilized in H1299 cells expressing MDM2(C305F) or MDM2(C308Y) following DNA damage. Scale bar, 10 μm. n/s, P > 0.05; *P ≤ 0.05; **P ≤ 0 01; ***P ≤ 0.001. The statistical analyses are based on results obtained by at least 50 cells each from three independent experiments.
In order to confirm that MDM2 interacts with RPL5 at the ribosomes, rather than with free RPL5 proteins, we employed the PLA to test the interaction of MDM2 with the ribosomal protein RPS6. MDM2 does not bind RPS6 directly (Zhou et al., 2012) and the observed PLA signals in H1299 cells transfected with the sp53, MDM2 and RPS6 and treated with doxo is therefore likely derived from MDM2 in association with the ribosomes (Figure 3B). This interaction was not observed in the absence of sp53 mRNA (Figure 3B). Furthermore, the PLEA assay using the mAb DO-1 and rabbit anti-MDM2 was used on polysomes captured with anti-RPL5 goat antibody to confirm the presence of MDM2 at p53 polysomes (Figure 3C). These results support the idea that the p53 mRNA is required for MDM2 to reach the p53 polysome following DNA damage.
We next addressed if ribosomal proteins play a role in transporting MDM2 to the p53 polysome. We introduced the MDM2(C305F) and the MDM2(C308Y) mutations that prevent the interactions with RPL5 and RPL11 (Lindstrom et al., 2007a, b). Expression of either of these constructs, in the presence of p53, led to the accumulation of MDM2 in the nucleoli following genotoxic stress (as observed by IF) and the loss of cytoplasmic MDM2–RPL5 and MDM2–RPS6 PLA signals (Figure 3D and E). These results show that the interactions of MDM2 with RPL5 and RPL11 are required for the cytoplasmic translocation of MDM2. In addition, WB showed that MDM2(C305F) and MDM2(C308Y) mutants failed to stabilize p53 following DNA damage in a similar fashion as the L22L synonymous mutation (Figure 3F, compare with Figure 1A). Together with previous results, these data suggest that the interactions of MDM2 with the p53 mRNA and RPs are required for MDM2 and ATM to reach the p53 polysome following DNA damage and for the stabilization of p53.
MDM2 traffics the active ATM to the p53 polysome
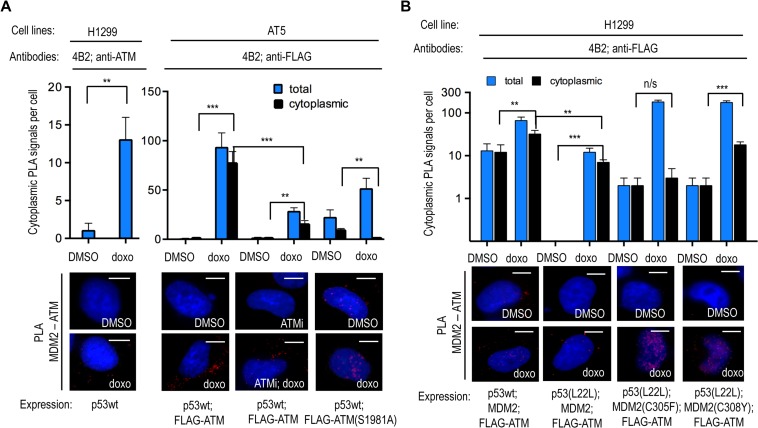
We next addressed the role of MDM2 in bringing ATM to the p53 polysome. Following doxo treatment, we observed an enhancement of endogenous cytoplasmic ATM–MDM2 complexes in H1299 cells expressing p53wt, as well as in AT5 cells co-expressing ATM (Figure 4A). This was prevented when AT5 cells were treated with ATMi or when the ATM(S1981A) mutation, which codes for an inactive ATM protein, was introduced (Hickson et al., 2004) (Figure 4A and Supplementary Figure S1C); suggesting that an active ATM is required to take the ATM–MDM2 complex to the p53 polysome. When we used the p53(L22L) mRNA and the MDM2(C305F) and MDM2(C308Y) mutants, we observed a loss of the MDM2–ATM complexes in the cytoplasm (Figure 4B). It should be noted that MDM2(C308Y) was less efficient in preventing the cytoplasmic MDM2–ATM interaction, as compared to MDM2(C305F). Importantly, when we used the p53(L22L) mRNA in combination with the MDM2(C305F) and MDM2(C308Y), we observed a sharp increase in the nuclear ATM–MDM2 interaction following doxo treatment. This shows that when MDM2 does not interact with ribosomal factors and the p53 mRNA, the stereochemistry of the ATM–MDM2 interaction is preserved, but the translocation of the complex to the cytoplasm is prevented (Figure 4B). Taken together, these results are consistent with a model whereby MDM2 serves as a carrier to bring ATM to the nascent p53 following DNA damage and that the trafficking of MDM2 to the p53 polysome requires MDM2 interacting with the p53 mRNA and ribosomal factors.
Figure 4.
The trafficking of MDM2 is required for recruiting ATM to the p53 polysome following DNA damage. (A) PLA on transfected H1299 (left) and AT5 (right) cells using 4B2 and anti-ATM or anti-FLAG antibodies show MDM2 together with ATM predominately in the cytoplasm following doxo treatment. The cytoplasmic MDM2–ATM association is prevented by ATM inhibitors or by inserting the inactive ATM(S1981A) mutation. Black bars represent cytoplasmic PLA signals and blue bars show the total number of PLA signals. (B) The MDM2–ATM association in the cytoplasm requires the interaction of MDM2 with the p53 mRNA and ribosomal factors. Preventing the MDM2–p53 mRNA interaction by using p53(L22L) prevents the induction of the cytoplasmic and total association of MDM2 with ATM following doxo treatment. The MDM2(C305F) mutant shows a strong induction of PLA signal with ATM in the nucleus, but not in the cytoplasm, following doxo treatment. Similarly, the MDM2(C308Y) mutation shows less interaction with ATM in the cytoplasm following doxo treatment. Scale bar, 10 μm. n/s, P > 0.05; *P ≤ 0.05; **P ≤ 0 01; ***P ≤ 0.001. The statistical analyses are based on results obtained by at least 50 cells each of three independent experiments.
Discussion
The activation of p53 following DNA damage is orchestrated by ATM and includes an increase in MDM2-mediated p53 synthesis and the suppression of MDM2-mediated p53 degradation. Previous studies have shown that ATM-dependent induction of p53 synthesis involves phosphorylations on MDM2, at residues 395 (394 in mice), which result in its binding to the p53 mRNA and to a consequent increase in p53 synthesis (Kubbutat et al., 1997, 1999; Morgan and Kastan, 1997; Maya et al., 2001; Marine et al., 2007; Naski et al., 2009; Marine and Lozano, 2010; Gajjar et al., 2012; Malbert-Colas et al., 2014; Tournillon et al., 2016). ATM-mediated phosphorylation events at p53 N-terminal residues prevent MDM2 binding and the degradation of p53 via the 26S proteasomal pathway. It was previously shown that the artificial silent mutant p53TriM mRNA, which carries synonymous mutations in codons 17, 18 and 19 and has an increased affinity for MDM2 under normal conditions, exhibits a higher rate of MDM2-dependent translation and interestingly, also a higher rate of MDM2-dependent p53 degradation (Candeias et al., 2008). Hence, an increase in the MDM2–p53 mRNA interaction and the consequent increase in MDM2-dependent p53 synthesis also give MDM2 access to the nascent p53 protein. However, the increase in the p53 mRNA–MDM2 affinity that occurs following genotoxic stress and ATM activation, results in a higher rate of p53 synthesis and to the inhibition of MDM2-mediated degradation of p53. Hence, during the DDR, MDM2-dependent synthesis of p53 does not result in an increase of the p53 turnover. The observation that the p53(L22L), which exhibits a poor affinity for MDM2, expresses a p53wt protein that is degraded via MDM2 following DNA damage underlines the role of the p53 mRNA in controlling MDM2-dependent degradation of p53 (Malbert-Colas et al., 2014). In fact, during genotoxic stress, MDM2 degrades p53 more efficiently when it is expressed by the p53(L22L) mRNA, compared to the p53wt mRNA. This increase in p53 degradation was initially puzzling but a recent study shows that the phosphorylation at Ser395 promotes MDM2 E3 ligase activity towards p53 in the absence of the p53 mRNA (Medina-Medina et al., 2016). This indicates that p53 proteins that are not prevented to interact with MDM2 are more efficiently degraded during the DDR.
We employed different approaches to show that ATM indeed phosphorylates the nascent p53 at Ser15. The semi-quantitative PLA data show the association between the Ser15 epitope ribosomal factors in the cytoplasm. The PLEA data confirm that these interactions are associated with the p53 polysome. By placing the Ser15 epitope at the p53 C-terminus (p53Rev) so that it is hidden inside the ribosome’s exit channel in CHX-treated cells, we could show that the phosphorylation indeed takes place on the nascent p53 peptide. The presented data on how ATM is brought to the p53 polysome where it phosphorylates p53 Ser15, which results in less MDM2-mediated p53 degradation, offer an explanation to how MDM2 can stimulate p53 synthesis while not targeting p53 for degradation during the DDR. This study focuses on Ser15 and the phosphorylation of this site by ATM but it should be pointed out that other residues surrounding Ser15 are also implicated in preventing MDM2 binding, such as Ser20, and this has not been investigated here. These data add further details to the existing model by which ATM activity controls MDM2’s switch from being a negative to becoming a positive regulator of p53 following genotoxic stress.
An important aspect of this work was to investigate how ATM reaches the p53 polysome following DNA damage. There are several data supporting the notion that MDM2 bound to the p53 mRNA brings ATM to the p53 polysome. For example, by silencing mdm2, or by averting the MDM2–p53 mRNA interaction, ATM was prevented from reaching the p53 polysome. Furthermore, ATM is activated by auto-phosphorylation Ser1981 and phosphorylates MDM2 at Ser395 which develops an affinity for the p53 mRNA hence following DNA damage, we could observe a p53 mRNA-dependent cytoplasmic association of MDM2 together with RPL5 and RPL11.
The presented data also shed light on the interaction between MDM2 and ribosomal factors RPL5 and RPL11. It has previously been shown that free RPL5 and RPL11 can prevent MDM2-mediated degradation of p53 via direct interactions. However, the fact that the PLA signals of the MDM2–RPLs interactions were observed in the cytoplasm we initially hypothesized that these interactions can also take place on the ribosome. To address this, we observed that MDM2 associates with the RPS6 in the cytoplasm, despite the fact that RPLS6 does not bind MDM2 directly. Additionally, PLEA data confirmed that MDM2 is at the p53wt polysome and that this requires ATM activity. When MDM2 does not interact with the p53 mRNA and ribosomal factors, it accumulates together with ATM in the nucleoplasm, showing that the p53 mRNA is not controlling the interaction between ATM and MDM2 but, rather, that the RNA is required for MDM2–ATM trafficking. This is in line with a previous report showing that SUMOylated MDM2 is localized to the nucleoli in a p53 mRNA-dependent fashion (Gajjar et al., 2012).
The fact that MDM2 is associated with the p53 polysome and was previously shown to bind to the RPL5–RPL11–5S pre-initiation complex suggests that ribosomal factors play a role in bringing MDM2–ATM to the p53 polysome. In support of this, we observed that mutations in MDM2 that prevent MDM2 from binding RPL5 and RPL11 (C305 and C308, respectively; Zhang et al., 2011) resulted in an accumulation of MDM2 in the nucleoli and prevented MDM2-mediated induction of p53 synthesis. Importantly, these mutations also prevented MDM2-mediated synthesis of p53 and the stabilization of p53 following DNA damage in a similar fashion as the p53(L22L) mRNA. We noticed that even though both C305F and C308Y mutations show qualitatively similar results, the MDM2(C305F) mutant shows a more dramatic effect on the stabilization of p53, as compared to the effect of C308Y mutation. This could be due to differences in the affinity of each mutant for RPL5 and RPL11. This is also suggested by data showing that the MDM2(C308Y) mutant is less effective in preventing MDM2 from bringing ATM to the p53 polysome.
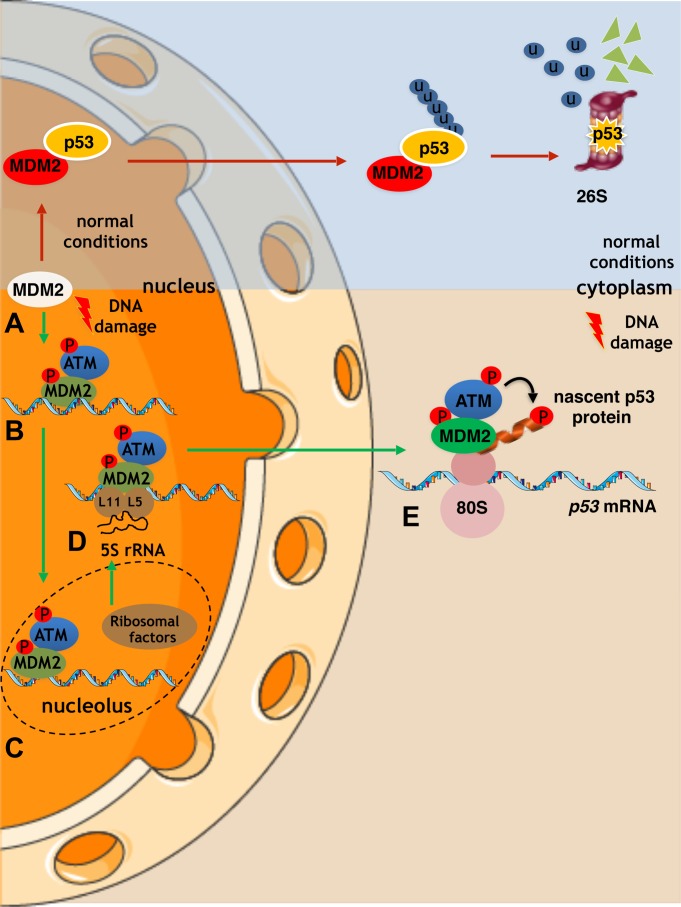
Altogether, these data suggest a model of how ATM reaches the p53 polysome in order to phosphorylate p53 at Ser15 to allow MDM2 to stimulate p53 synthesis and at the same time to prevent MDM2 from targeting the nascent p53 for degradation during the DDR (Figure 5). This model starts with ATM phosphorylating MDM2 at Ser395 thus inducing allosteric changes in MDM2 that allow its binding to the nascent p53 mRNA. This phosphorylation event also induces a conformational change to promote an MDM2–ATM interface. The p53 mRNA is required for MDM2 to reach the nucleoli where the interaction between MDM2 and ribosomal factors takes the p53 mRNA–MDM2–ATM complex to the p53 polysome. It is plausible that these ribosomal factors function as part of the pre-initiation complex (RPL5–RPL11–5S), which has previously been reported to bind MDM2. This model suggests that not only does ATM act on the nascent p53 mRNA by promoting MDM2 binding but it also acts directly on the nascent p53 protein (Figure 5).
Figure 5.
A model describes how the p53 mRNA dictates the stability of the encoded protein following DNA damage by guiding MDM2 and ATM to the p53 polysome. (A) Under normal conditions, MDM2 binds to p53 and catalyses its polyubiquitination and the subsequent degradation via the 26S proteasomal pathway. (B) Following DNA damage, ATM phosphorylates MDM2 at Ser395 promoting its binding to the p53 mRNA. (C and D) The ATM–MDM2–p53 mRNA complex is trafficked between the nucleoli and the cytoplasm and associates with the ribosome precursor complex RPN (RPL5–RPL11–5S). This association facilitates the export of the complex to the cytoplasm and the p53 polysome. (E) ATM phosphorylates the nascent p53 peptide at Ser15 and prevents MDM2 from binding to the emerging p53 protein. This model explains how MDM2 can stimulate the translation of p53 without degrading the newly synthesized p53 protein and how a single synonymous mutation can affect the stability of the encoded protein.
P53 is an intrinsically disordered protein that is post-translationally modified on over 60 different residues by different classes of enzymes in response to different signaling pathways and interacts with several hundreds of cellular factors. Post-translational modification patterns of p53 are hierarchical and synergistic and govern the interaction with cellular factors in a chain of events characteristic for functional differentiation of intrinsic disordered proteins (Lambert et al., 1998; Tompa, 2002; Uversky, 2016). Hence, the timing and location of p53’s post-translational modifications have consequences to what factors p53 interacts with and, ultimately, to the activity of p53 in response to different stress pathways. For example, the Ser15 phosphorylation plays a key in controlling phosphorylation events on p53 that control the interaction with MDM2 but also in promoting the interaction with CBP/p300 and the acetylation of C-terminal lysine residues. Ongoing studies aim to address how these events and p53 activity are affected by the MDM2- and ATM-dependent phosphorylation of the nascent p53 (Saito et al., 2003; Marechal and Zou, 2013). One interesting possibility is that the activation of MDM2 E3 ligase activity towards p53 via phosphorylation of MDM2 at Ser395 and in the absence of the p53 mRNA may stimulate the degradation of p53 proteins that are not differentiated towards the DDR.
Finally, it is worth mentioning that synonymous mutations are increasingly associated with cancer and other diseases (Fahraeus et al., 2016). However, apart from splicing and microRNA, the underlying molecular mechanisms of how synonymous mutations can affect the encoded proteins remain poorly understood. A single synonymous mutation alters the activity of the MDR gene which has been attributed to an effect of slow versus fast codon usage impacting on the protein folding, without though ruling out that changes in protein folding may be attributed to the mRNA that employs factors to the nascent protein, thus playing a role in its folding (Kimchi-Sarfaty et al., 2007). The fact that the p53 mRNA affects the stability of the encoded protein by bringing an enzyme to modify the nascent peptide offers a new mechanism whereby mRNAs can affect the encoded proteins and sheds light on the role of mRNAs as regulatory molecules in cell signaling pathways. These data suggest that analyses of disease-producing genomes using next-generation sequencing might reconsider the evaluation of how synonymous mutations impact on the phenotype.
Materials and methods
Plasmids and recombinant proteins
Plasmids were cloned using the pcDNA vector for eukaryotic expression. In occasion, FLAG tags were also fused at the 5′ of the CDS as indicated. Each cloning was confirmed by direct sequencing. The plasmids, pcDNA-p53 and pcDNA-mdm2 containing the full-length CDSs, have been described previously (Komatsu et al., 1996; Malbert-Colas et al., 2014), while the plasmids pcDNA-mdm2-C305F and pcDNA-mdm2-C308Y have been previously prepared and kindly gifted by Prof. Hua Lu (Department of Biochemistry & Molecular Biology and Tulane Cancer Center, Tulane University School of Medicine).
Transient transfection of H1299, AT5, and A549 cells
The cell lines H1299 (human non-small cells lung cancer, not expressing p53, CRL-5803, ATCC), AT5 (not expressing ATM) (Matsuura et al., 1997; Gandin et al., 2014), and A549 (human lung carcinoma, CCL-185, ATCC) (Komatsu et al., 1996) were used. H1299 cells have a basal ATM activity level that is further induced following DNA damage. Each cell line was tested authenticated by PCR and was periodically tested for mycoplasma infection. Cells were incubated at 37°C, 5% CO2 in RPMI medium (for H1299) or DMEM medium (for AT5 or A549), supplemented with antibiotics, and with 2 mM L-glutamine (Gibco/Invitrogen) and 10% fetal serum (Hyclone). Cells were transfected with small amounts of DNA (a total of 100 ng/ml plasmid DNA). In the compared samples, the cells were transfected with the same transfection mix and the total pcDNA3 concentration was equilibrated. The comparisons were exclusively made in samples of individual experiments using the same cell line and run under the same conditions. The expression of individual constructs was determined using IH. Cells were treated with DMSO or 0.5–1 μM Doxorubicin for 1–16 h causing genotoxic stress with or without 15 μM ATM inhibitor (KU55933) (Ofir-Rosenfeld et al., 2008) for 16 h, or treated with 100 μM 4-Thiouridine (4SU) for 14 h if used in UV fixation, in the presence or absence of 25 μg/ml protease inhibitor MG132 for 2 h, or treated with 5 μg/ml Cycloheximide (CHX) for 1 h for translation inhibition and ribosome fixation. Subsequently, they were either fixed in 4% PFA and used in immunohistochemistry or PLA or cross-linked by irradiation with 0.15 J/cm2 total energy of 365 nm UV light and stored at −80°C before used in western blotting or co-immunoprecipitation assays.
siRNA transfection
Cells were transfected with siRNAs targeting mdm2 or a control siRNA at a final concentration of 5 nM using the HiPerFect transfection reagent (Qiagen) and the FlexiTube GeneSolution siRNA against mdm2 (GS4193, Qiagen).
Polysome fractionation
Sucrose solutions (5% and 50% wt/vol) were prepared in polysome lysis buffer (50 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2). The 5%–50% wt/vol linear sucrose gradients were freshly casted on SW41 ultracentrifuge tubes (Beckmann) using the Gradient master (BioComp instruments) following manufacturer’s instructions (Guo et al., 2010). Twenty-four hours post-transfection, H1299 cells (of 80%-90% confluency) were treated with 100 μg/ml CHX at 37°C for 5 min and then washed twice with 1× PBS (Dulbecco modified PBS, GIBCO) containing 100 μg/ml CHX. Cells were then scrapped and lysed with polysome lysis buffer (50 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 100 μg/ml CHX) using narrow gauge syringes and spin at 5000 rpm at 4°C for 10 min. Lysates were then loaded on a sucrose gradient and centrifuged with a SW41 rotor at 36000 rpm, at 4°C for 2 h. Polysomes were then fractionated in 0.5 min intervals, using the Foxy R1 fraction collector (Teledyne ISCO). The polysomes were pooled and concentrated by Millipore tubes at 4000 rpm.
Immunochemistry and the PLA
Cells were grown on sterilized glass slides in a 24-well plate, transfected and fixed in 4% PFA for IF and PLA. After three washes with PBS for 10 min and incubation with blocking buffer (3% BSA, 0.1% saponin in PBS), samples were incubated with primary antibodies for 2 h at room temperature (RT). The antibodies used were: the DO-1 mouse antibody recognizing the N-terminal of p53; the CM-1 rabbit antibody cross-reacting with p53; a chicken anti-p53 antibody, prepared by Biosan in collaboration with Prof. Mónica Marín (Biochemistry-Molecular Biology, Facultad de Ciencias, Universidad de la República, Montevideo); the mouse Ser15 phosphospecific anti-p53(S15) antibody (16G8; Cell Signalling); the 4B2 anti-MDM2 mouse (prepared in collaboration with Dr Borivoj Vojtesek at RECAMO, Brno) and rabbit (ab87134; Abcam) antibodies; the anti-HA tag antibody (Abcam), the anti-FLAG tag antibodies (2368 S; Cell Signalling and ab18230; Abcam), the anti-ATM antibody (Thermofisher); the rabbit anti-RPL5 antibodies (PA5-27539; Invitrogen and C2114; Santa Cruz); the rabbit anti-RPL11 antibody (K1210; Santa Cruz) and the anti-RPS6 antibody (54D2; Cell Signalling). All antibodies were used in a dilution of 1:200. For IF detection, cells were incubated with Alexa-488-conjugated (Invitrogen) anti-rabbit antibody and Alexa Fluor-633-conjugated (Invitrogen) anti-mouse antibody. For the PLA, the PLA RED kit (Sigma) was used and the method described previously was followed (Weibrecht et al., 2013; Koos et al., 2014). DAPI was used for the labeling of the nucleus. Images were captured using 640 nm filters; or, when higher resolution was needed, by confocal microscopy. Each sample was tested in triplicates and the PLA signal dots were counted by the ImageJ software. For each sample, the counted cytoplasmic signals were localized in individual cells, as observed by visible-light microscopy, during the capturing of each image. DAPI staining was used to determine nuclear signals and for each sample, we counted dots in 50 cells randomly selected during the imaging. This was repeated in at least three independent experiments. As a transfection control, cells were co-transfected with GFP. In the cases, transfection was required for the assay, non-transfected cells were excluded from the statistical analyses. The values were used for the preparation of a graph and for statistical analysis to calculate the mean, the standard deviation and the one-tailed P-values using the GraphPad Prism 5 software. The asterisks in the graphs of the figures represent P-values of three independent experiments, as follows: ‘n/s’ for P > 0.05; ‘*’ for values P ≤ 0.05; ‘**’ for values P ≤ 0 01; and ‘***’ for values P ≤ 0.001. The statistical analyses are based on results obtained by at least 50 cells of each experiment.
PLA ELISA
The concept of the PLEA is to combine the PLA with the ELISA in order to obtain quantitative values, which can be used for the preparation of a graph and the calculation of the affinity of the interactions under study. The PLEA technique offers the possibility to study the interaction of three molecules, by using three primary antibodies. The capture antibody is used to capture the complex on the well plate and a set of two more antibodies is used for the amplification of the PLA signal. Here we used either a chicken anti-p53 antibody (home-made, against full-length p53wt) or the goat anti-RPL5 antibody (C2114; Santa Cruz). The 96-well ELISA plates were incubated with the capture antibody at a dilution of 1:200, o/n, at 4°C. Then the plates were blocked with 5% BSA in PBS, o/n, at 4°C, and were incubated with serial dilutions of the polysomal fractions in 3% BSA in PBST, o/n at 4°C. The plates were then washed six times with PBST (0.1 M tween in PBS) and a set of primary antibodies developed in rabbit and mouse was used at a dilution of 1:200, for 2 h, at RT. After washing six times with PBST, samples were incubated with the PLA secondary antibodies and the PLA RED kit (Sigma) was used, following the manufacturers’ instructions with modifications. Samples were incubated with the set of the detection of secondary PLA antibodies (anti-mouse and anti-rabbit) for 1 h at 37°C. The excess of antibodies was washed out six times with PBST and samples were incubated with 0.5 μl ligase for 30 min at 37°C and washed six times with PBST. Samples were incubated with the Φ29 DNA polymerase for 2 h, at 37°C and washed six times with PBST. Each sample was tested in triplicates. The fluorescence (640 nm) excitation and emission was measured by the FLUOstar plate reader. The values from three sample replicates and three independent experiments were used for the preparation of a graph and for statistical analysis to calculate the mean and the standard deviation using the GraphPad Prism 5 software.
Western blotting
Cells were harvested and identical amounts of whole lysates (prepared in lysis buffer: 20 mM HEPES-KOH pH 7.5, 50 mM beta-glycerophosphate, 1 mM EDTA pH 8, 1 mM EGTA pH 8, 0.5 mM Na3CO4, 100 mM KCl, 1% Triton X-100, 10% glycerol) and resolved in 10% SDS/PAGE gels (Invitrogen) and transferred onto nitrocellulose membranes. Blots were incubated with one of the primary antibodies: anti-p53 (DO-1; CM-1; RECAMO), anti-MDM2 (4B2; RECAMO); anti-p21 Waf1/Cip1 (2947; Cell Signalling) and anti-Hsp70 (C92F3A-5, Santa Cruz) and subsequently incubated with corresponding secondary HRP antibodies (DAKO). The films were developed and scanned or the membranes were analyzed by the My ECL Imager (Thermo Scientific). The quantifications were calculated by ImageJ and they express the relative density of the bands (relative values normalized with the Hsp70 values).
Protein–protein sandwich ELISA
For the protein–protein sandwich ELISA, the protocol by Abcam was followed. The DO-1, 4B2, and anti-His primary antibodies were used with the corresponding secondary anti-IgG HRP antibodies. Each sample was tested in triplicates in three independent experiments and the Prism software was used for the calculation of the SD and the preparation of the graphs.
Supplementary Material
Acknowledgements
We thank Dr Borivoj Vojtesek (RECAMO, Brno) for developing and providing antibodies used in this work (GACR P206/12/G151). We thank Dr Niclas Setterblad (Plateforme Technologique de l’IUH’ and the University of Paris-Diderot 7, Paris) for providing spectrophotometer and microscopy services and for his valuable advices. We thank Biosan (Uruguay) and Prof. Mónica Marín (Biochemistry-Molecular Biology, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay) for chicken anti-p53 antibodies.
Conflict of interest
none declared.
Author contributions
K.K. and R.F. conceived and designed the research; K.K. performed the experiments; S.V.G., I.L., and A.T. performed sampling; L.W., K.N., and V.O.-I. analyzed results; K.K. and R.F. analyzed the data, prepared the figures, and wrote the manuscript.
References
- Banin S., Moyal L., Shieh S., et al. (1998). Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677. [DOI] [PubMed] [Google Scholar]
- Bursac S., Brdovcak M.C., Donati G., et al. (2014). Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim. Biophys. Acta 1842, 817–830. [DOI] [PubMed] [Google Scholar]
- Candeias M.M., Malbert-Colas L., Powell D.J., et al. (2008). P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol. 10, 1098–1105. [DOI] [PubMed] [Google Scholar]
- Canman C.E., Lim D.S., Cimprich K.A., et al. (1998). Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679. [DOI] [PubMed] [Google Scholar]
- Chen L., Gilkes D.M., Pan Y., et al. (2005). ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 24, 3411–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chene P. (2003). Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer 3, 102–109. [DOI] [PubMed] [Google Scholar]
- Cheng Q., and Chen J. (2010). Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle 9, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok C.F., Verma C.S., Baselga J., et al. (2011). Translating p53 into the clinic. Nat. Rev. Clin. Oncol. 8, 25–37. [DOI] [PubMed] [Google Scholar]
- Coffill C.R., Lee A.P., Siau J.W., et al. (2016). The p53–Mdm2 interaction and the E3 ligase activity of Mdm2/Mdm4 are conserved from lampreys to humans. Genes Dev. 30, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G., Peddigari S., Mercer C.A., et al. (2013). 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 4, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus R., Marin M., and Olivares-Illana V. (2016). Whisper mutations: cryptic messages within the genetic code. Oncogene 35, 3753–3759. [DOI] [PubMed] [Google Scholar]
- Gajjar M., Candeias M.M., Malbert-Colas L., et al. (2012). The p53 mRNA–Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell 21, 25–35. [DOI] [PubMed] [Google Scholar]
- Gandin V., Sikstrom K., Alain T., et al. (2014). Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J. Vis. Exp. 87, e51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon H.S., Woda B.A., and Jones S.N. (2012). ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell 21, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner J.J., Parker S.C., Prickett T.D., et al. (2013). Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc. Natl Acad. Sci. USA 110, 13481–13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover R., Candeias M.M., Fahraeus R., et al. (2009). p53 and little brother p53/47: linking IRES activities with protein functions. Oncogene 28, 2766–2772. [DOI] [PubMed] [Google Scholar]
- Gullberg M., Goransson C., and Fredriksson S. (2011). Duolink-‘In-cell Co-IP’ for visualization of protein interactions in situ. Nat. Methods. 8, 982. [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., et al. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., et al. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C.J., et al. (2004). Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64, 9152–9159. [DOI] [PubMed] [Google Scholar]
- Hosp F., Vossfeldt H., Heinig M., et al. (2015). Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. 11, 1134–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger A.C., and Fersht A.R. (2010). The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2, a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S., Lane D.P., and Verma C.S. (2016). Long range recognition and selection in IDPs: the interactions of the C-terminus of p53. Sci. Rep. 6, 23750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakostis K., Ponnuswamy A., Fusee L.T., et al. (2016). p53 mRNA and p53 protein structures have evolved independently to interact with MDM2. Mol. Biol. Evol. 33, 1280–1292. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., Oh J.M., Kim I.W., et al. (2007). A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528. [DOI] [PubMed] [Google Scholar]
- Komatsu K., Matsuura S., Tauchi H., et al. (1996). The gene for Nijmegen breakage syndrome (V2) is not located on chromosome 11. Am. J. Hum. Genet. 58, 885–888. [PMC free article] [PubMed] [Google Scholar]
- Koos B., Andersson L., Clausson C.M., et al. (2014). Analysis of protein interactions in situ by proximity ligation assays. Curr. Top. Microbiol. Immunol. 377, 111–126. [DOI] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones S.N., and Vousden K.H. (1997). Regulation of p53 stability by Mdm2. Nature 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Kubbutat M.H., Ludwig R.L., Levine A.J., et al. (1999). Analysis of the degradation function of Mdm2. Cell Growth Differ. 10, 87–92. [PubMed] [Google Scholar]
- Lambert P.F., Kashanchi F., Radonovich M.F., et al. (1998). Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273, 33048–33053. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Lan L., Peng G., et al. (2015). Tyrosine 370 phosphorylation of ATM positively regulates DNA damage response. Cell Res. 25, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom M.S., Deisenroth C., and Zhang Y. (2007. a). Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle 6, 434–437. [DOI] [PubMed] [Google Scholar]
- Lindstrom M.S., Jin A., Deisenroth C., et al. (2007. b). Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol. Cell. Biol. 27, 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez I., Tournillon A.S., Prado Martins R., et al. (2017). p53-mediated suppression of BiP triggers BIK-induced apoptosis during prolonged endoplasmic reticulum stress. Cell Death Differ. 24, 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughery J., Cox M., Smith L.M., et al. (2014). Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 42, 7666–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaine N.J., and Hupp T.R. (2011). How phosphorylation controls p53. Cell Cycle 10, 916–921. [DOI] [PubMed] [Google Scholar]
- Malbert-Colas L., Ponnuswamy A., Olivares-Illana V., et al. (2014). HDMX folds the nascent p53 mRNA following activation by the ATM kinase. Mol. Cell 54, 500–511. [DOI] [PubMed] [Google Scholar]
- Marechal A., and Zou L. (2013). DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspect. Biol. 5, a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine J.C., Dyer M.A., and Jochemsen A.G. (2007). MDMX: from bench to bedside. J. Cell Sci. 120, 371–378. [DOI] [PubMed] [Google Scholar]
- Marine J.C., and Lozano G. (2010). Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 17, 93–102. [DOI] [PubMed] [Google Scholar]
- Matsuura S., Weemaes C., Smeets D., et al. (1997). Genetic mapping using microcell-mediated chromosome transfer suggests a locus for Nijmegen breakage syndrome at chromosome 8q21-24. Am. J. Hum. Genet. 60, 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya R., Balass M., Kim S.T., et al. (2001). ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15, 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Medina I., Garcia-Beltran P., de la Mora-de la Mora I., et al. (2016). Allosteric interactions by p53 mRNA govern HDM2 E3 ubiquitin ligase specificity under different conditions. Mol. Cell. Biol. 36, 2195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D.W. (2009). Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer 9, 714–723. [DOI] [PubMed] [Google Scholar]
- Meek D.W., and Anderson C.W. (2009). Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harbor Perspect. Biol. 1, a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan S.E., and Kastan M.B. (1997). p53 and ATM: cell cycle, cell death, and cancer. Adv. Cancer Res. 71, 1–25. [DOI] [PubMed] [Google Scholar]
- Naski N., Gajjar M., Bourougaa K., et al. (2009). The p53 mRNA–Mdm2 interaction. Cell Cycle 8, 31–34. [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y., Boggs K., Michael D., et al. (2008). Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 32, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V., Xiong S., Jackson J.G., et al. (2013). The p53-Mdm2 feedback loop protects against DNA damage by inhibiting p53 activity but is dispensable for p53 stability, development, and longevity. Genes Dev. 27, 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y., Shkedy D., de Graaf P., et al. (2005). Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc. Natl Acad. Sci. USA 102, 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., et al. (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273. [DOI] [PubMed] [Google Scholar]
- Saito S., Yamaguchi H., Higashimoto Y., et al. (2003). Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 278, 37536–37544. [DOI] [PubMed] [Google Scholar]
- Sauna Z.E., and Kimchi-Sarfaty C. (2011). Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12, 683–691. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., et al. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Scott S.P., Bendix R., Chen P., et al. (2002). Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. Proc. Natl Acad. Sci. USA 99, 925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O., Gullberg M., Jarvius M., et al. (2006). Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000. [DOI] [PubMed] [Google Scholar]
- Stracker T.H., Roig I., Knobel P.A., et al. (2013). The ATM signaling network in development and disease. Front. Genet. 4, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Minana B., Valcarcel J., et al. (2014). Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335. [DOI] [PubMed] [Google Scholar]
- Takagi M., Tsuchida R., Oguchi K., et al. (2004). Identification and characterization of polymorphic variations of the ataxia telangiectasia mutated (ATM) gene in childhood Hodgkin disease. Blood 103, 283–290. [DOI] [PubMed] [Google Scholar]
- Teufel D.P., Bycroft M., and Fersht A.R. (2009). Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene 28, 2112–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27, 527–533. [DOI] [PubMed] [Google Scholar]
- Tompa P. (2005). The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579, 3346–3354. [DOI] [PubMed] [Google Scholar]
- Tournillon A.S., Lopez I., Malbert-Colas L., et al. (2016). p53 binds the mdmx mRNA and controls its translation. Oncogene 36, 723–730. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. (2016). p53 proteoforms and intrinsic disorder: an illustration of the protein structure-function continuum concept. Int. J. Mol. Sci. 17, 1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibrecht I., Lundin E., Kiflemariam S., et al. (2013). In situ detection of individual mRNA molecules and protein complexes or post-translational modifications using padlock probes combined with the in situ proximity ligation assay. Nat. Protoc. 8, 355–372. [DOI] [PubMed] [Google Scholar]
- Wright P.E., and Dyson H.J. (2009). Linking folding and binding. Curr. Opin. Struct. Biol. 19, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.Q., Halaby M.J., and Zhang Y. (2006). The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25, 4613–4619. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Xiao H., Chai S.C., et al. (2011). Hydrophilic residues are crucial for ribosomal protein L11 (RPL11) interaction with zinc finger domain of MDM2 and p53 protein activation. J. Biol. Chem. 286, 38264–38274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liao J.M., Liao W.J., et al. (2012). Scission of the p53-MDM2 loop by ribosomal proteins. Genes Cancer 3, 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.