Abstract
Class II HDACs, such as HDAC4, are critical regulators of the immune response in various immune cells; however, its role in innate immunity remains largely unknown. Here, we report that the overexpression of HDAC4 suppresses the production of type I interferons triggered by pattern-recognition receptors (PRRs). HDAC4 repressed the translocation of transcription factor IRF3 to the nucleus, thereby decreasing IRF3-mediated IFN-β expression. In particular, we also determined that HDAC4 can be phosphorylated and simultaneously block the phosphorylation of IRF3 at Ser386 and Ser396 by TBK1 and IKKε, respectively, by interacting with the kinase domain of TBK1 and IKKε. Furthermore, IFN-β may stimulate the expression of HDAC4. Our findings suggest that HDAC4 acts as a regulator of PRR signaling and is a novel mechanism of negative feedback regulation for preventing an over-reactive innate immune response.
Keywords: HDAC4, antiviral response, IRF3, TBK1, IKKε
Introduction
Type I interferons (IFNs) family of cytokines, including IFN-β and IFN-α, have critical roles in host defense against viral and bacterial pathogens. Toll-Like receptor (TLR3) and TLR4 recognize viral double-stranded (ds) RNA or bacterial lipopolysaccharide (LPS), respectively, triggering a signaling pathway that results in the production of IFN-β. Following infection, viral RNA in the endosome is recognized by TLRs, while cytoplasmic viral ds RNA is sensed by RNA helicase RIG-I-like receptor (RLRs), melanoma differentiation associated gene 5 (MDA 5), or protein kinase R (PKR) (Yamamoto et al., 2003). In addition, sensors of viral DNA include cGAS and DAI (Paludan and Bowie, 2013). Upon pathogen recognition, these sensor molecules trigger signaling events that activate transcription factors such as NF-κB, IRF3, or IRF7, resulting in the production of IFNs (Fitzgerald et al., 2003). Excessive IFN production can induce autoimmune diseases, whereas insufficient levels of IFN can result in chronic infection. Therefore, a balanced production of IFNs likely plays a key role in the pathogenesis of autoimmune diseases, as well as in immune responses to viral infection.
IRF3 is a master transcription factor that stimulates the production of IFN-β and is essential for the innate immune response. The phosphorylation of IRF3 at Ser386 and Ser396 by the kinases TBK1 and IKKε induces its dimerization and translocation to the nucleus where it stimulates transcription of IFN-β-encoding genes (Fitzgerald et al., 2003). Pathogens have acquired various evasive mechanisms to overcome the host immune response (Liew et al., 2005; Haller et al., 2006). In particular, at the level of IRF-mediated transcription, Ebola virus, Borna disease virus (BDV), and human papilloma virus (HPV) interfere with IRF3 activation (Ronco et al., 1998; Garcia-Sastre, 2004; Unterstab et al., 2005). However, the complex regulatory mechanism regulating IRF3 activation and translocation remains elusive.
Histone-modifying enzymes are the major epigenetic regulators involved in the control of inflammatory processes during pathogen infection. Histone acetyltransferases (HATs) acetylate lysine residues on histones, while histone deacetylases (HDACs) offset HAT activity by deacetylating histones. The delicate equilibrium between the acetylated state and the deacetylated state of chromatin organizes gene transcription (Wolffe and Pruss, 1996). Furthermore, HATs and HDACs also regulate the acetylation status of non-histone proteins, thereby regulating signaling proteins and transcription factors to influence signal pathway activation and cellular function (Mowen and David, 2014). So far, HDACs, including HDAC1, 6, 8, and 9, have been implicated in the regulation of systemic innate immunity either directly or indirectly. In fact, HDAC1 and HDAC8 are recruited by Rb to attenuate the acetylation of Histone H3/H4 at the Ifnb1 promoter, resulting in the suppression of Ifnb1 transcription (Meng et al., 2016). HDAC6 transiently binds to RIG-I and deacetylates it at lysine 909 in the presence of viral RNAs, promoting RIG-I sensing of viral RNAs (Choi et al., 2016). Furthermore, HDAC9 directly maintains the deacetylated status of the key PRR signaling molecule TBK1 and enhances its kinase activity (Li et al., 2016). Therefore, identification of other HDACs that may be involved in the antiviral innate immune response is of great importance.
Here, we show that HDAC4 can decrease type I IFN production. A detailed analysis revealed a mechanism whereby HDAC4 was phosphorylated by the signaling kinase TBK1/IKKε, thus preventing IRF3 phosphorylation. Therefore, our findings suggest that HDAC4 and the IRF3-activating kinases TBK1/IKKε are connected by a negative feedback loop.
Results
The inhibitory role of HDAC4 in type I IFN production
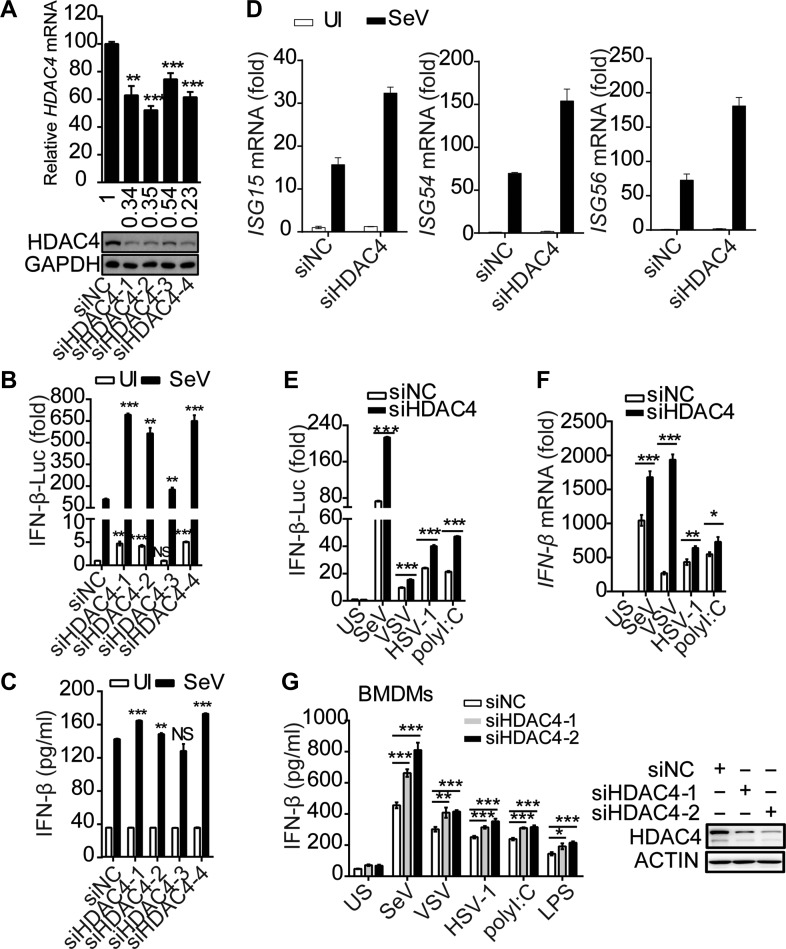
A shRNA library screen revealed that HDAC4 potentiated virus-induced activation of the IFN-β promoter (IFNB1) (data not shown). To verify this phenomenon, a siRNA-mediated knockdown of HDAC4 was performed in HEK293T cells (Figure 1A). HDAC4 knockdown increased promoter activity and induction of IFN-β after Sendai virus (SeV) infection (Figure 1B and C). Simultaneously, knockdown of HDAC4 resulted in increased transcription of IFN-stimulated genes (ISGs), including Isg15, Isg54, and Isg56, induced by SeV (Figure 1D). Similarly, knockdown of HDAC4 using shRNA validated these results (Supplementary Figure S1A–D). In addition, after stimulation with various inducers, including vesicular stomatitis virus (VSV), herpes simplex virus (HSV-1), and polyinosinic–polycytidylic acid (poly(I:C)), the IFNB1 promoter activity and IFNB1 mRNA levels in the HDAC4-knockdown samples were significantly higher compared to the control samples (Figure 1E and F). We next investigated whether HDAC4 was required for various stimuli-induced type I IFN responses in primary cells. Knockdown of HDAC4 via siRNA dramatically upregulated the secretion of IFN-β in bone marrow-derived macrophages (BMDMs) compared with IFN-β secretion from HDAC4-wild-type BMDMs under the condition of stimulation by various inducers, including RNA virus, DNA virus, poly(I:C), or bacterial LPS (Figure 1G). Altogether, these results demonstrated that the production of IFN-β induced by stimuli was increased when HDAC4 was silenced.
Figure 1.
Deficiency in HDAC4 arrestingly facilitates the production of IFN-β. (A) Quantitative RT-PCR analysis of HDAC4 mRNA (top) and immunoblot (IB) analysis of HDAC4 and GAPDH as loading control throughout (bottom). (B and C) HEK293T cells (2 × 105) were transfected for 36 h with an IFN-β firefly luciferase reporter (IFN-β-Luc), along with non-targeting control siRNA (siNC) or siRNA targeting HDAC4 (siHDAC4-1, -2, -3, -4), and then left uninfected (UI) or infected for another 8 h with SeV (50 hemagglutination units per ml). (B) Luciferase assay analysis of IFNB1 promoter activity. Luciferase reporter activity is normalized to that of renilla luciferase. (C) ELISA of IFN-β. (D) Quantitative RT-PCR analysis of ISG15, ISG54, and ISG56 mRNA in HEK293T cells transfected for 36 h with siNC or siHDAC4 (siHDAC4-4) and then infected for another 8 h with SeV. The reference gene actin serves as a loading control. (E and F) Luciferase assay of IFNB1 promoter activity (E) and quantitative RT-PCR analysis of IFNB1 mRNA (F) in HEK293T cells transfected for 36 h with siNC or siHDAC4-4 and then treated for 8 h with SeV, VSV, HSV-1, and poly(I:C). (G) ELISA of IFN-β in culture medium of siNC or siHDAC4-treated BMDMs (4 × 105) incubated for 5 days with the cytokine M-CSF and then left unstimulated (US), infected with SeV (12 h), VSV (12 h), or HSV-1 (18 h), or stimulated with LPS (10 h) or poly(I:C) (10 h). NS, not significant (P > 0.05), *P < 0.05, **P < 0.01, and ***P < 0.001 (unpaired t-test). Data are from three independent experiments (mean and SD of three independent biological replicates per group) or are representative of three independent experiments (A, bottom).
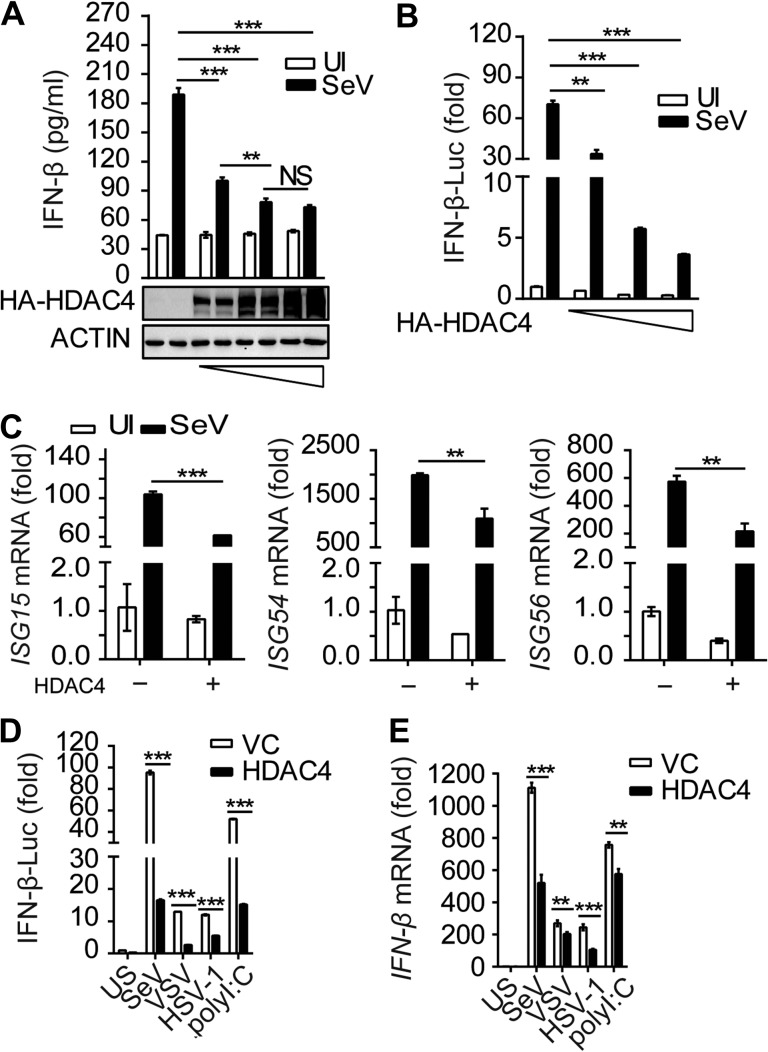
To further confirm these results, we stimulated HDAC4-overexpressing HEK293T cells with SeV. We found that IFN-β production and IFNB1 promoter activity were significantly lower when HDAC4 was overexpressed compared to samples transfected with the empty vector, and the inhibition occurred in a dose-dependent manner after infection with SeV (Figure 2A and B). In parallel experiments, the mRNA levels of the ISGs mentioned above were also suppressed in samples overexpressing HDAC4 (Figure 2C). Accordingly, the IFNB1 promoter activity and IFNB1 mRNA levels were lower in HDAC4-overexpressing HEK293T cells than the empty vector counterparts stimulated with other inducers, including VSV, HSV-1, and poly(I:C) (Figure 2D and E). Similarly, the promoter activity and mRNA expression level of IFNB1 were also markedly downregulated in HDAC4-overexpressing mouse embryonic fibroblasts (MEFs) in a dose-dependent manner following SeV infection (Supplementary Figure S2A and B). In addition, the reporter activity of IFNB1 was also significantly suppressed in HDAC4-overexpressing mouse macrophages (RAW264.7) after stimulation with SeV, VSV, HSV-1, poly(I:C), and LPS (Supplementary Figure S2C). Collectively, these data reinforced the conclusion the HDAC4 could decrease the production of type I IFN in vitro.
Figure 2.
Opulence of HDAC4 selectively impairs the production of type I interferon. (A and B) HEK293T cells were transfected with an IFN-β firefly luciferase reporter, along with control vector or gradient concentrations of HDAC4 (0.2, 0.5, and 1 μg), and then left uninfected or infected for another 8 h with SeV. (A) ELISA of IFN-β. (B) Luciferase assay analysis of IFNB1 promoter activity. (C) Quantitative RT-PCR analysis of ISG15, ISG54, and ISG56 mRNA in HEK293T cells with control vector or HDAC4 (500 ng). (D and E) Luciferase assay of IFNB1 promoter activity (D) and quantitative RT-PCR analysis of IFNB1 mRNA (E) in HEK293T cells transfected for 36 h with control vector or HDAC4 (500 ng) and then treated for 8 h with inducers. NS, not significant (P > 0.05), *P < 0.05, **P < 0.01, and ***P < 0.001 (unpaired t-test). Data are representative of three independent experiments with similar results (mean ± SD).
HDAC4 decreases IRF3 nuclear transport by inhibiting its phosphorylation
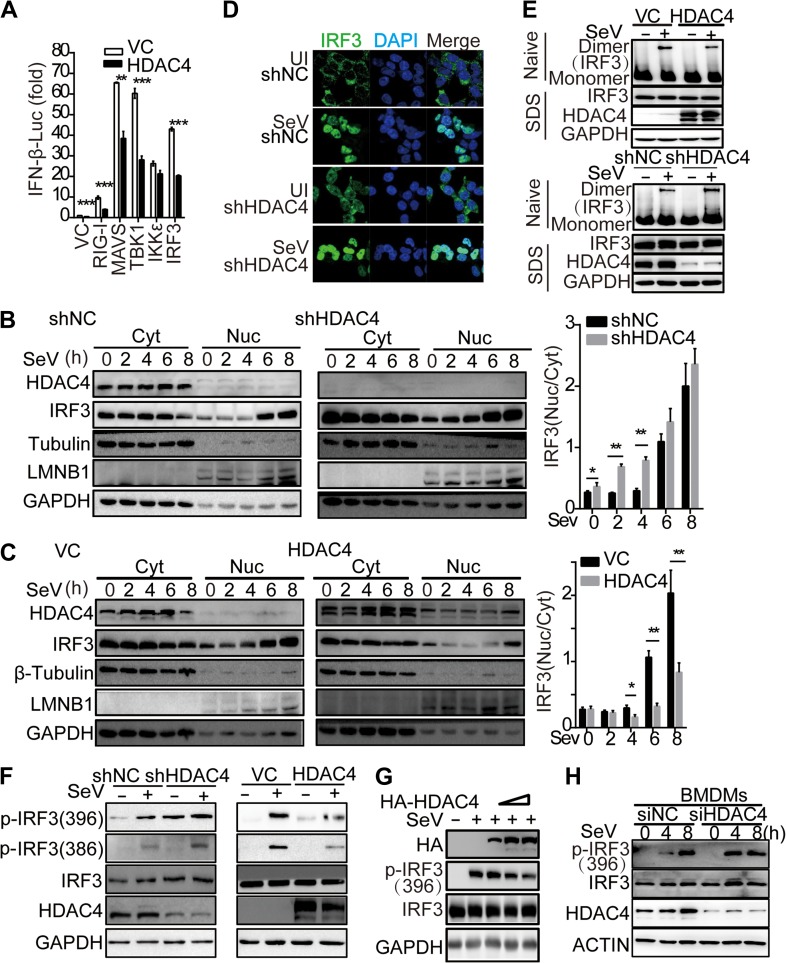
To understand the mechanism by which HDAC4 regulated innate antiviral signaling, we investigated whether the level of HDAC4 influenced the levels of the PRR-triggered signaling molecules using luciferase reporter assays. RIG-I has been reported to induce the production of type I IFN by forming a signaling complex with VISA, TBK1, and IKKε (Fitzgerald et al., 2003; Rehwinkel and Reis e Sousa, 2010). Our results demonstrated that the overexpression of HDAC4 significantly inhibited IFNB1 reporter activity induced by upstream activators (RIG-I, VISA, TBK1, and IRF3) but not IKKε (Figure 3A). These data indicated that HDAC4 might be involved in the IRF3-mediated innate immune response by targeting to IKKε and affecting the activation of IRF3.
Figure 3.
HDAC4 alleviated the import of IRF3 into the nucleus. (A) Luciferase assay analysis of IFNB1 promoter activity in HEK293T cells cotransfected with empty vector or HDAC4 plasmid and the cDNA encoding Flag-tagged RIG-I, MAVS, TBK1, IKKε, or IRF3 plasmid. (B) IB analysis of shNC or shHDAC4 HEK293T cell lines infected with SeV, followed by nucleus–cytoplasm extraction (5% of cytoplasmic extracts and 10% of nuclear extracts separated by SDS-PAGE). Histogram statistics show the nuclear–cytoplasmic ratio of IRF3 band intensity, normalized to that of Lamin B or β-tubulin. (C) IB analysis of HEK293T cells transfected for 36 h with control vector or HDAC4 plasmid and infected with SeV for the indicated times, followed by nucleus–cytoplasm extraction as in B. Histogram statistics are carried out as in B. (D) Confocal microscopy of shNC or shHDAC4 HEK293T cell lines uninfected (first and third rows) or infected for 8 h with SeV (second and fourth rows), probed with the DNA-binding dye DAPI (blue) and anti-IRF3 (green). Scale bar, 10 μm. (E) IB analysis of IRF3 in dimer or monomer form (upper) or total IRF3, HDAC4, and GAPDH (lower) in HEK293T cells transfected for 48 h with empty vector or cDNA encoding HDAC4 or in shNC and shHDAC4 HEK293T cells left uninfected or infected for 8 h with SeV, followed by native PAGE (top) or SDS-PAGE (bottom). (F and G) IB analysis of IRF3 phosphorylation at Ser386 and Ser396, respectively, total IRF3, GAPDH, and HDAC4 or HDAC4(HA) in shNC or shHDAC4 HEK293T cell lines transfected for 36 h with control vector or HDAC4 plasmid and then uninfected or infected for another 8 h with SeV or transfected gradient concentrations of HDAC4. (H) IB analysis of IRF3 phosphorylated at Ser396, total IRF3, HDAC4, and β-actin (loading control) in the wild-type or knockdown of HDAC4 BMDMs infected for various times with SeV. The numbers under the WB lines mean the gray value corresponding to the lane. NS, not significant (P > 0.05), *P < 0.05, **P < 0.01, and ***P < 0.001 (unpaired t-test). Data are representative of three independent experiments with similar results (B−H) or from three independent experiments (A and histograms in B and C; mean ± SD).
IKKε plays key roles in integrating innate receptor signaling and phosphorylating IRF3 at Ser396, inducing its nuclear translocation. Thus, we further analyzed the effect of HDAC4 on the cellular localization of IRF3. Interestingly, the nuclear localization of IRF3 was obviously higher in HDAC4-knockdown samples relative to control samples during early SeV infection (2, 4 hpi), and data further demonstrated that IRF3 could translocate to the nucleus in the HDAC4-knockdown samples in the absence of SeV challenge (Figure 3B). Consistent with these results, the import of IRF3 into the nucleus after SeV challenge was inhibited in HDAC4-overexpressing samples relative to samples transfected with the control vector, in particularly, 4, 6, and 8 h after infection with SeV (Figure 3C). We confirmed these results using confocal microscopy. Interestingly, IRF3 was found in the nucleus in the HDAC4-knockdown cells in the absence of SeV challenge (Figure 3D). Simultaneously, the degree of IRF3 dimerization was also lower in HDAC4-overexpressing HEK293T cells infected with SeV than in the control counterparts (Figure 3E). Similarly, the dimerization of IRF3 was considerably potentiated in HEK293T cells in which HDAC4 was knocked down relative to its transcription in the HDAC4-sufficient counterparts, after infection with SeV (Figure 3E). It is well known that IRF3 phosphorylation induces its dimerization and nuclear translocation. We thus deduced that the inhibition of IRF3 activation was likely due to its hindered phosphorylation. To confirm this supposition, we observed that upon SeV challenge, the IRF3 phosphorylation at Ser386 and Ser396 was upregulated in HDAC4-knockdown samples, while the phosphorylation levels at Ser386 and Ser396 were decreased in the HDAC4-overexpressing samples in a dose-dependent manner (Figure 3F and G). Interestingly, IRF3 was phosphorylated at Ser396 in HDAC4-knockdown samples even in the absence of SeV infection, which was essential for IRF3 translocation to the nucleus (Figure 3D and F). Accordingly, IRF3 phosphorylation was enhanced in BMDMs in which HDAC4 was knocked down and were infected with SeV for the indicated times (4 h), relative to that in similarly treated cells with normal levels of HDAC4 (Figure 3H). These data demonstrated that HDAC4 could inhibit IRF3 phosphorylation and thereby impairing the import of IRF3 into the nucleus to initiate transcriptional activity.
HDAC4 interacts with TBK1/IKKε and IRF3
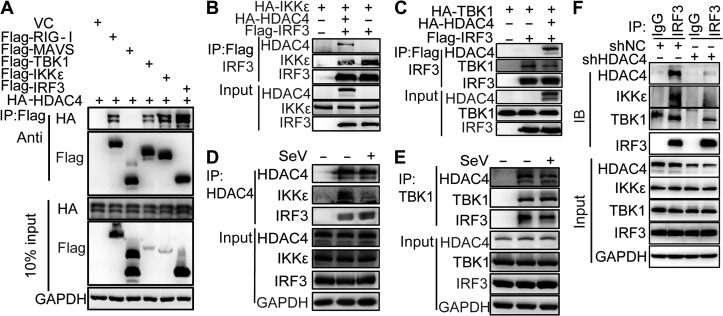
We next investigated the underlying mechanisms by which HDAC4 affected IRF3 phosphorylation. In addition to RIG-I (Liu et al., 2016), an immunoprecipitation (IP) assay indicated that HDAC4 also interacted with TBK1, IKKε, and IRF3 (Figure 4A). When we overexpressed HA-tagged IKKε, HA-tagged HDAC4, and FLAG-tagged IRF3 together in HEK293T cells, IRF3 immunoprecipitated with both HDAC4 and IKKε, indicating that HDAC4, IRF3, and IKKε associated together in a complex. The level of IKKε co-immunoprecipitated with IRF3 was much lower when HDAC4 was overexpressed compared to control samples (Figure 4B). The data suggested that HDAC4 affected the interaction between IKKε and IRF3. We obtained consistent results when HA-tagged IKKε was replaced with HA-tagged TBK1 (Figure 4C). Endogenous IP analysis suggested that HDAC4 constitutively interacted with TBK1, IKKε, or IRF3 and SeV infection led to HDAC4 dissociation from TBK1/IKKε-IRF3 (Figure 4D and E). In addition, we found that TBK1/IKKε-IRF3 associations were lost in shHDAC4 HEK293T cells (Figure 4F). Collectively, these data suggested that HDAC4 could hinder the direct interaction between IRF3 and TBK1/IKKε to prevent the phosphorylation and activation of IRF3.
Figure 4.
HDAC4 interacts with TBK1, IKKε, and IRF3. (A) IP and IB analyses of lysates of HEK293T cells overexpressing vector (VC) or plasmids encoding Flag-tagged RIG-I, MAVS, TBK1, IKKε, and IRF3, plus HA-tagged HDAC4, probed with anti-HA or anti-Flag. Bottom, IB analysis of lysates without IP (10% of input). (B and C) IP and IB analyses of lysates of HEK293T cells overexpressing plasmids encoding HA-tagged IKKε or HA-tagged TBK1, HA-tagged HDAC4, and Flag-tagged IRF3, probed with various combinations of anti-HA or anti-Flag. (D and E) Immunoassay of lysates of HEK293T cells (5 × 106) left uninfected or infected for 8 h with SeV, followed by IP analysis with immunoglobulin G (IgG), as a control (first lane), or with antibody to HDAC4 or TBK1 and IB analysis with antibodies to IKKε and IRF3 (D) or HDAC4 and IRF3 (E). Bottom, IB analysis of the samples above (input) without IP. (F) The endogenous association of TBK1, IKKε, and IRF3 in the presence or absence of HDAC4. IP analysis with anti-IRF3 and IB analysis with anti-IRF3, anti-TBK1, anti-IKKε, anti-HDAC4, or anti-β-actin of HEK293T cells infected with SeV for 6 h. Data are representative of three independent experiments.
The Ser246 and NES domains of HDAC4 are required for the inhibition of IFN-β production
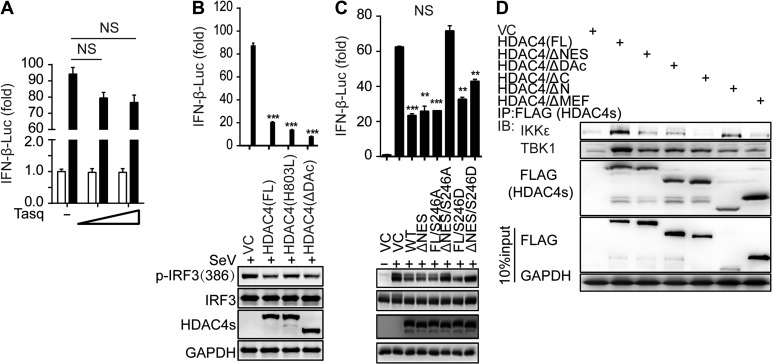
To analyze the mechanistic details, we first considered whether the deacetylation activity of HDAC4 played a critical role in inhibiting TBK1/IKKε-mediated IRF3 activation. A reporter assay indicated that Tasquinimod, a specific inhibitor of HDAC4, did not affect IFNB1 transcriptional activity (Figure 5A). The expression plasmids encoding the HDAC4-H803L mutant (abolishing HDAC activity) and the HDAC4-ΔDAc mutant (lacking the deacetylation domain) (Supplementary Figure S3A) were still capable of inhibiting IFNB1 promoter activity and the phosphorylation of IRF3 by TBK1/IKKε in HEK293T cells (Figure 5B). Moreover, the deletion of the MEF2-binding domain of HDAC4 (Supplementary Figure S3B) was also capable of suppressing IFNB1 promoter activity, similar to wild-type HDAC4. These results suggested that the deacetylation activity and the transcription regulation activity of HDAC4 were dispensable for the negative regulation of IFNs transcription.
Figure 5.
Mapping the binding domain of HDAC4 interaction with TBK1/IKKε. (A) Luciferase assay analysis of IFNB1 promoter activity in HEK293T cells transfected for 24 h with a luciferase reporter plasmid, plus the HDAC4-specific inhibitor Tasquinimod (Tasq) (wedge:50 and 200 μM), and then left uninfected or infected for 8 h with SeV. (B and C) Luciferase assay analysis of IFNB1 promoter activity and IB analysis of phosphorylation of IRF3 at Ser386, IRF3, mutant HDAC4 containing various domain combinations (as in Supplementary Figure S4A) and GAPDH in HEK293T cells transfected for 24 h with luciferase reporter plasmid of IFN-β (IFN-β-Luc), together with various mutant HDAC4, and then infected for 8 h with SeV. (D) IB analysis of HEK293T cells transiently transfected for 48 h with vector encoding Flag-tagged full-length or mutant HDAC4 (as in Supplementary Figure S4A) and HA-tagged IKKε or TBK1, followed by IP analysis with Flag antibody and IB analysis with HA antibodies to IKKε and TBK1. NS, not significant (P > 0.05), *P < 0.05, **P < 0.01, and ***P < 0.001 (unpaired t-test). Data are representative of three independent experiments with similar results (B−D) or from three independent experiments (A and histograms in B and C; mean ± SD).
HDAC4 shuttles between the nucleus and the cytoplasm, a process that is dependent on its nuclear localization signal (NLS) and nuclear export sequence (NES) domains (Wang and Yang, 2001). The export of HDAC4 to the cytoplasm is dependent on its phosphorylation at Ser246, Ser467, and Ser632 by CaMK4 and SIK1 (Wang et al., 2000). However, the deletion of the NES domain of HDAC4 was still capable of suppressing the IFNB1 promoter activity, as wild-type HDAC4 (Supplementary Figure S3B). Thus, we constructed several mutants (S246A, S467A, S632A, and S246/467/632A) based on the full-length or specific NES domain deletion in the HDAC4-encoding sequence. In a reporter assay, the plasmids harboring a single mutation or several mutations in the full-length HDAC4-encoding sequence could not rescue the suppression of HDAC4, but the plasmids harboring the S246A mutation and the NES deletion lost the inhibitory capacity on IFN expression (Supplementary Figure S3C and D). The concurrence of the S246A mutation and deletion of the NES in HDAC4 also rescued IRF3 phosphorylation at Ser386 and Ser396 induced by SeV compared to wild-type HDAC4 (Figure 5C). Thus, our data implied that the S246 residue and the NES domain of HDAC4, which are the major motifs for the cytoplasmic localization of HDAC4, were functionally important for HDAC4-mediated IRF3 regulation.
Furthermore, we mapped the domain of HDAC4 that interacts with IKKε/TBK1. A deletion of every single domain including the NLS, MEF binding domain, HDAC domain, and NES from the HDAC4-encoding sequence still permitted its interaction with IKKε, although to a lesser degree than what was observed for wild-type HDAC4 (Figure 5D). Simultaneous deletion of the HDAC and NES (HDAC4/ΔC) domains resulted in the loss of interaction between HDAC4 and IKKε, which was further supported by the observation that HDAC4/ΔN, which contains the HDAC domain and the NES sequence, could interact with IKKε, similar to wild-type HDAC4. Thus, the C-terminal domain of HDAC4 was essential for the interaction between HDAC4 and IKKε, although we could not exclude the possibility that other domains may also be involved. Interestingly, we observed that all the HDAC4 deletion mutants still co-immunoprecipitate with TBK1 (Figure 5D, second row), indicating the likelihood of multiple interaction sites.
HDAC4 interacts with the kinase domain of TBK1/IKKε
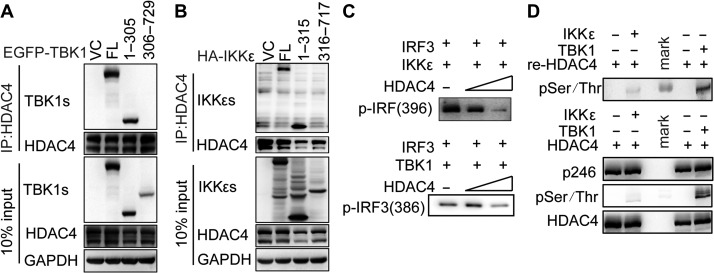
To investigate how TBK1 or IKKε interacted with HDAC4, we constructed various TBK1 or IKKε mutants for the kinase domain of TBK1 (aa1–305) and IKKε (aa1–315), or deletion of the kinase domain of TBK1 (aa306–729) and IKKε (aa316–717). We transfected vectors encoding these mutants into HEK293T cells together with a vector encoding FLAG-tagged HDAC4. Co-immunoprecipitation experiments revealed that the kinase domains of TBK1 (aa1–305) or IKKε (aa1–315) were necessary for the interaction of HDAC4 and TBK1 or IKKε (Figure 6A and B). These results indicated that HDAC4 binds to the kinase domain of TBK1 and IKKε, which may hinder their ability to phosphorylate substrates such as IRF3.
Figure 6.
HDAC4 interacts with TBK1/IKKε via the TBK1/IKKε kinase domain and HDAC4 impairs the phosphorylation of IRF3 by TBK1/IKKε. (A and B) IB analysis of HEK293T cells transiently cotransfected for 48 h with Flag-tagged HDAC4 and EGFP-tagged wild-type TBK1, HA-tagged wild-type IKKε, or their mutants, assessed with whole-cell lysates (10% input) or IP with anti-Flag antibody. (C and D) In vitro kinase assay of IRF3 phosphorylated at Ser386 or Ser396 and HDAC4 phosphorylated at Ser246 or other serine or threonine sites (pSer/Thr). The peptide of IRF3 full-length protein at its carboxyl terminus to GST and recombinant protein IKKε or TBK1 with kinase activity were introduced into a mixture containing overexpressed and immunoprecipitated Flag-tagged HDAC4.
HDAC4 affects the ability of TBK1/IKKε to phosphorylate IRF3
To further investigate the effect of HDAC4 on the regulation of IRF3 phosphorylation by TBK1 or IKKε, we performed an in vitro kinase assay using separately purified IRF3 and IKKε or TBK1, with or without HDAC4. As expected, immunoblot (IB) analysis of IRF3 phosphorylation at Ser396 revealed that the phosphorylation of IRF3 by IKKε was reduced in the presence of HDAC4 in a dose-dependent manner (Figure 6C, top). Similarly, experiments on IRF3 phosphorylation at Ser386 by TBK1 were performed and showed that phosphorylation at this residue was decreased by HDAC4 (Figure 6C, bottom). We concluded that HDAC4 hindered IRF3 phosphorylation by TBK1/IKKε.
As HDAC4 can interact with TBK1/IKKε, we suspected that HDAC4 might be phosphorylated as a substrate of TBK1/IKKε. To test this hypothesis, we introduced the fusion proteins containing HDAC4 (aa612–1084) and glutathione S-transferase (GST)-tagged IKKε or GST-tagged TBK1 with kinase activity in an in vitro kinase assay. IB analysis of phospho-serine/threonine showed that HDAC4 could be phosphorylated by IKKε and TBK1 directly. We utilized overexpressed full-length immunoprecipitated HA-tagged HDAC4, replacing the separately purified HDAC4 in kinase assay. The results showed that IKKε or TBK1 had no significant influence on phosphorylation at Ser246 but at other Ser or Thr sites of HDAC4 (Figure 6D). These experiments verified that the HDAC4 could be phosphorylated by TBK1/IKKε.
Physiological regulation of HDAC4 in the innate immune response
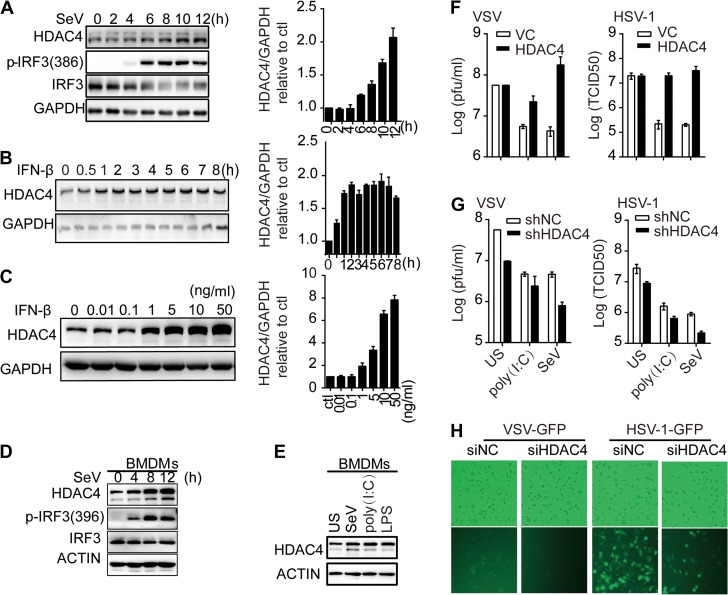
We investigated whether HDAC4 was a negative regulator of the cellular antiviral response. First, we analyzed the expression of HDAC4 at the protein level in HEK293T cells infected with SeV. The HDAC4 level increased significantly at 6 hpi and above, achieving a 2-fold increase at 12 hpi (Figure 7A). The upregulation of HDAC4 was consistent with the levels of IFN-β, implying that HDAC4 may be regulated by IFN-β in a positive feedback loop. Indeed, low doses of IFN-β (0.01 and 0.1 ng/ml) were unable to induce HDAC4 expression, while higher doses (1, 5, 10, and 50 ng/ml) significantly increased HDAC4 expression in a dose-dependent manner (Figure 7B). In addition, treatment with 1 ng/ml IFN-β immediately induced HDAC4 expression, which reached its peak at 1 h post-treatment (Figure 7C). To determine whether HDAC4 protein was also regulated in primary cells, we treated BMDMs with SeV for the indicated times. The results indicated that SeV dramatically induced HDAC4 protein levels in mouse BMDMs (Figure 7D). Furthermore, the level of HDAC4 in BMDMs was markedly upregulated after poly(I:C) or LPS stimulation (8 h) compared with unstimulated (US) BMDMs (Figure 7E). These data indicated that HDAC4 protein might be regulated by the IFN-β level. We next analyzed the effects of HDAC4 on viral replication. The overexpression of HDAC4 significantly reversed cytoplasmic poly(I:C)- or SeV-mediated inhibition of VSV and HSV-1 replication (Figure 7F). Conversely, the knockdown of HDAC4 significantly inhibited VSV and HSV-1 replication and further enhanced the inhibitory effect triggered by poly(I:C) or SeV (Figure 7G). Furthermore, we analyzed the effects of HDAC4 knockdown on viral replication in primary cells. The replication of VSV-GFP or HSV-1-GFP was severely compromised in siHDAC4 BMDMs than in siNC BMDMs as monitored by the GFP percentages and intensities (Figure 7H). Taken together, these results suggested that HDAC4 restricted viral infection-induced secretion of IFNs and thereby promoting viral replication.
Figure 7.
Roles of HDAC4 in cellular antiviral response. (A) IB analysis with antibodies to p-IRF3(386), total IRF3, HDAC4, and GAPDH in HEK293T cells infected with SeV for 2, 4, 6, 8, 10, and 12 h. (B and C) HEK293T cells were treated with gradient concentrations of IFN-β (0.01, 0.1, 1, 5, 10, 50 ng/ml) for 4 h or with IFN-β (1 ng/ml) for 0.5, 1, 2, 3, 4, 5, 6, 7, and 8 h, and then analyzed by IB analysis with antibodies to HDAC4 and GAPDH. The histograms in A−C show the gray intensity of HDAC4 relative to GAPDH normalized to the control or zero timepoint. (D) Induction of murine HDAC4 after SeV infection for the indicated times. BMDMs were stimulated with SeV (50 hemagglutination units per ml) for the indicated times and then IBs were performed with the indicated Abs. (E) Induction of murine HDAC4 after SeV, poly(I:C),or LPS stimulation. BMDMs were stimulated with SeV (50 hemagglutination units per ml), poly(I:C) (10 μg/ml), or LPS (100 ng/ml) for 8 h and then IBs were performed with the indicated Abs. (F) Plaque assay of VSV (left) or HSV-1 (right) in HEK293T cells (2 × 105) transfected for 48 h with empty vector (VC) or HDAC4 expression plasmid and transfected or infected for another 12 h with poly(I:C) or SeV, and then infected for another 24 h with VSV or HSV-1 (M.O.I. = 0.01). The supernatants were harvested and analyzed for VSV or HSV-1 production by standard plaque assays. (G) Knockdown of HDAC4 weakens VSV or HSV-1 replication. Plaque assays were performed as in F except that the shNC or shHDAC4 HEK293T cell lines were used. (H) Microscopy imaging of siNC- and siHDAC4-treated BMDMs uninfected or infected with VSV-GFP (0.1 M.O.I.) or HSV-1-GFP (0.1 M.O.I.) for 24 h. Data are representative of three independent.
Discussion
We have shown here that HDAC4 prevents the phosphorylation and activation of IRF3 by TBK1/IKKε and subsequently reduces the production of type I IFN in response to various stimuli. The overexpression of HDAC4 inhibited SeV-triggered the activation of IRF3 and IFNB1 promoter, whereas the knockdown of HDAC4 exhibited the opposite effect. Consistently, the overexpression of HDAC4 reversed poly(I:C)- or SeV-induced inhibition of VSV and HSV-1 replication. In contrast, the knockdown of HDAC4 inhibited VSV and HSV-1 replication. Our findings indicate that an epigenetic regulator specifically suppresses antiviral responses and provide new insight into how an epigenetic regulator, such as HDAC4, influences IRF3 activation and antiviral responses.
Unlike class I HDACs, class II-a HDACs, such as HDAC4, shuttle between the nucleus and the cytoplasm. Nuclear export prevents class II-a HDACs from acting as transcriptional repressors and thus promotes gene expression. HDACs can act as regulators of TLR signaling in innate immunity, as evidenced by its effects on the expression of target genes downstream of TLR signaling (Aung et al., 2006; Halili et al., 2010). In antiviral signaling, in addition to HDAC1 and HDAC8, which repress the transcription of IFNB1 mRNA, HDAC6 activates IFN-β expression and mediates the deacetylation of RIG-I and β-catenin to promote IRF3-activated transcription (Nusinzon and Horvath, 2006; Zhu et al., 2011; Choi et al., 2016; Meng et al., 2016). HDAC9, a class II-a member, can enhance the kinase activity of TBK1 via its deacetylated status during viral infection (Li et al., 2016). These studies demonstrate that HDACs regulate the transcription of IFNB1 mRNA or the activity of key regulatory factors via deacetylation. Here, we uncovered another mechanism of HDAC4 (a class II-a HDAC), by which it influences the phosphorylation on IRF3 by TBK1 and IKKε directly via interaction with TBK1 and IKKε.
Induction of viral stress-inducible genes, including IFNs, is blocked by HDAC4 by inhibiting IRF3 phosphorylation, which is required for its activation, as HDAC4 is itself phosphorylated by TBK1/IKKε. Because HDAC4 and IRF3 are substrates of the same kinases, one possibility is that HDAC4 out-competes IRF3 as substrate. In the in vitro kinase assay, quantitation of phosphorylation showed that IRF3 and HDAC4 were equally phosphorylated by IKKε. However, the phosphorylation of IRF3 was reduced when HDAC4 was present. HDAC4 may have a stronger affinity for TBK1/IKKε, thus decreasing their association with IRF3.
Our results strongly suggest that the host can fight viral infection by regulating the subcellular localization of HDAC4, an event that is important for HDAC4 function. Just as the phosphorylation of IRF3 leads to its nuclear localization, the phosphorylation of HDAC4 is accompanied by its nuclear-to-cytoplasmic trafficking. Proteins destined for export from the nucleus contain amino acid targeting sequences termed NESs, which mediate the passage of cargo proteins out of the nucleus, conferring recognition by specific members of the nuclear export proteins (Poon and Jans, 2005; Lange et al., 2007). Depending on its subcellular localization, kinases can either induce the nuclear export or block the nuclear import of HDAC4. Phosphorylation of serine 246, 467, and 632 creates the 14-3-3 binding sites necessary for nuclear export and may further block nuclear import (Backs et al., 2006). In fact, the concurrence of the S246A mutation with a deleted NES-encoding domain in the HDAC4 coding sequence was capable of restoring the production of IFN-β and IRF3 activity. The phosphorylation of Ser246, Ser467, and Ser632 mediated HDAC4 localization to the cytoplasm. HDAC4 (S246A) substantially translocated to the nucleus and lost its ability to interact with TBK1/IKKε, whereas HDAC4 (S246D), which mimics phosphorylation on Ser246, enhanced the interaction with TBK1/IKKε and translocated to the cytoplasm (Supplementary Figure S4A and B).
Moreover, our data suggest that Ser246 of HDAC4 is not the target site of TBK1/IKKε, which was analyzed with the only available antibody that is specific to phospho-Ser246 (Figure 6D). Previous reports identified several phospho-Serines of HDAC4 (such as Ser210, Ser246, Ser350, Ser467, Ser565, Ser632, Ser633) that are important for HDAC4 function (Wang et al., 2000). Besides Ser246, we also detected other Ser/Thr phosphorylation of HDAC4. The Ser/Thr phosphorylation level caused by IKKε was weaker than TBK1 (Figure 6D), which might be due to HDAC4 not affecting IKKε-mediated IFNB1 promoter activation but inhibited TBK1- or IRF3-mediated IFNB1 promoter activation (Figure 3A). The difference might also due to different phosphorylation sites of HDAC4 mediated by TBK1 and IKKε, based on the different domains of HDAC4 that interacted with IKKε or TBK1 (Figure 5D). Although IKKε and TBK1 behave similarly on HDAC4 phosphorylation, the detail mechanisms might be different, which need further investigation.
At the same time, the C-terminus of HDAC4, which contains the deacetylation domain, was important for interaction with TBK1/IKKε. Acetylation is involved in the regulation of TBK1 activation and deacetylation at Lys241 of TBK1 by HDAC9 is critical for its kinase activity (Li et al., 2016). An in vitro kinase assay using the purified HDAC4 protein, we observed that HDAC4 can repress TBK1/IKKε-mediated IRF3 activation. Although the HDAC4-specific inhibitor Tasquinimod could not rescue the inhibitory effect on the production of IFN-β, it is interesting to speculate that the acetylation of TBK1, IKKε, or even IRF3, may be regulated by HDAC4. We will investigate this possibility in a future study.
To prevent harmful effects resulting from spontaneous type I IFNs production in uninfected (UI) cells or the overproduction of type I IFNs during an acute infection, host cells have developed distinct strategies to control excessive antiviral innate immune responses. Several members of the E3 ubiquitin ligase family, including TRIM26 and FoxO1, target key components of the virus-induced type I IFNs signaling pathways for degradation (Zhang et al., 2008; Lei et al., 2013; Wang et al., 2015). Furthermore, some inhibitory proteins, such as PDE12 and NLRX1, are physically associated with key components of virus-induced type I IFNs signaling pathways to sequester them in their inactive forms (Takeuchi and Akira, 2009; Wood et al., 2015). Viral infection leads to the release of these inhibitors, resulting in the suppression of these signaling components. HDAC4 negatively regulates virus-triggered induction of type I IFNs and cellular antiviral responses by disrupting the TBK1/IKKε–IRF3 complex. Constitutively expressed HDAC4, which is at a low level, provides initial negative regulation of TBK1/IKKε-mediated signaling. Herein, we demonstrated that the interactions between HDAC4 and TBK1/IKKε were impaired and activated IRF3 at early phase of SeV infection, whereas high levels of HDAC4 induced by type I IFNs further inhibited TBK1/IKKε-mediated responses in a feedback negative-regulatory manner during the late phase of infection (Supplementary Figure S5). The detail mechanisms on how IFN-β induces HDAC4 expression and how HDAC4 inhibits TBK1/IKKε-mediated responses during the late phase of infection need further investigation.
Altogether, our findings indicate that HDAC4 is a precise regulator of the cellular antiviral response by controlling IRF3 activation and IFN-β production. These results provide a new perspective on the functions of the HDAC family of enzymes. Moreover, the results of our study suggest that HDAC4 may be a potential target for the treatment of chronic viral infection and autoimmune diseases.
Materials and methods
Reagents and antibodies
Poly(I:C) (P9582, Sigma), lipopolysaccharide (LPS, S1732), and Cell Lysis Buffer (P0013) were from Beyotime Biotechnology. Tasquinimod (S7617) were from Selleck. Protein G-agarose (#16-266) used for IP was from Millipore. Other reagents were as previously described (Chen et al., 2015). β-actin (sc-47778) was from Santa Cruz Biotechnology. GAPDH (#60004-1-lg) and IRF3 (#11312-1-AP) were from Proteintech. Mouse monoclonal antibodies against Flag (F1804) and HA (H9658) were from Sigma-Aldrich. Antibodies to IRF3 phospho-S386 (#ab76493) and phospho-serine (#ab 6639) were from Abcam. Rabbit polyclonal antibody against Flag (#2368S) and rabbit monoclonal antibodies against HA (#3724), HDAC4 (#7628), HDAC4 phospho-S246 (#3443), Lamin B1 (#13435), β-Tubulin (#2128), TBK1 (#3013S), IKKε (#2905), and IRF3 phospho-S396 (#29047) were all from Cell Signaling Technology.
Cells and virus
Human embryonic kidney 293T cells, African green monkey kidney epithelial vero cells and mouse RAW264.7 cell lines were from American Type Culture Collection. All cells were maintained in Dulbecco’s modified Eagle medium (DMEM, Invitrogen) supplemented with 2 mM L-glutamine, nonessential amino acids, 10% fetal bovine serum (Invitrogen), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in a 5% CO2 incubator. SeV, VSV-GFP (provided by Prof. Hanzhong Wang), and HSV-1 BAC with GFP (provided by Prof. Chunfu Zheng) (Li et al., 2011) were amplified and titrated as previously described (Chen et al., 2014). Cells were infected with SeV (1 M.O.I.), VSV (1 M.O.I.), or HSV-1 (10 M.O.I.) for the indicated hours. Cells were stimulated with poly(I:C) (10 μg/ml) or LPS (100 ng/ml) for the indicated time periods.
Plasmid constructs and transfection
Recombinant vectors encoding human HDAC4 (GenBank accession no. NM_006037.3) were amplified from cDNA of 293T cells, and then cloned into pXJ40-HA or pXJ40-Flag vectors. All deletion mutations and site-directed mutagenesis of HDAC4 were amplified and inserted into the pXJ40-Flag vectors. All constructs were confirmed by sequencing and a complete list of primers was provided in Supplementary Table S1. Mammalian expression plasmids for RIG-I, VISA, TBK1, IKKε, and IRF3 were provided by Prof. Hong-Bing Shu. IFN-β, NF-κB, interferon-stimulated response element, and IRF3 promoter luciferase reporter plasmids were purchased from Clontech. pRL-TK and pRL-CMV were purchased from Promega. Plasmids were transiently transfected into HEK293T cells using lipofectamine 2000 reagents (Invitrogen) following the manufacturer instructions.
Luciferase assay
HEK293T cells were seeded on 24-well plates (1 × 105 cells per well) and transiently transfected with 100 ng of luciferase reporter plasmid and pRL-TK plasmid together with a total of 500 ng of target plasmid or empty control plasmid using Lipofectamine 2000 (Invitrogen) for 24 h. Luciferase activity was measured with a Dual-Luciferase Reporter Assay System (Promega).
RNAi and lentiviral transduction
HEK293T cells were seeded into 6-well plates (4 × 105 cells per well) 1 day prior to transfection. The cells were transfected with siRNA (20 nM) using Lipofectamine RNAiMAX reagent (Invitrogen). The specific siRNAs targeting HDAC4 were designed and synthesized by Qiagen. The siRNA sequences specific for human HDAC4 were 5′-CCAAUGUAUUCCAAGCUAA-3′ and 5′-GGCGUGGGUUUCAACGUCA-3′. The siRNA sequences specific for mouse HDAC4 were 5′-UCUCUGAUUGAGGCGCAAA-3′ and 5′-GGCACAGUUGCAUGAACAU-3′. The control siRNA sequence was 5′-UUCUCCGAACGUGUCACGU-3′. After 48 h, the cells were harvested for western blot and real-time PCR analysis. The lentiviruses were produced in 293T cells by co-transfection of the shNC or shHDAC4 and packaging vectors psPAX2 and pMD2.G (purchased from Addgene). At 48 and 72 h post-transfection, the supernatant was collected and applied to infect target cells in the presence of polybrene (5 μg/ml). The infected cells were selected by puromycin for 3–4 days before the conduction of experiments.
Quantitative RT-PCR and enzyme-linked immunosorbent assay
Total RNA was isolated from HEK293T cells using Trizol reagent (Invitrogen) according to the manufacturer’s protocols. Specific mRNAs were quantified by one-step real-time RT-PCR using the QuantiFast SYBR Green RT-PCR kit (Qiagen). The data were normalized to levels of β-actin for each individual sample. The 2−ΔΔCt method was used to calculate relative expression changes. The primer sequences for quantitative RT-PCR are provided in Supplementary Table S2. Secreted IFN-β in cell culture supernatants from SeV stimulus was analyzed using human IFN-β (Cusabio Biotechnology) ELISA kit according to the manufacturer’s instructions.
Subcellular fractionation
Nuclear and cytoplasmic extracts were prepared with a nuclear–cytoplasmic extraction kit (Beyotime Biotechnology) according to the manufacturer’s instructions.
Preparation of BMDMs
Bone marrow cells were isolated from mouse tibia and femur. For the preparation of BMDMs, bone marrow cells were cultured for 5 days in medium containing 20% supernatants of L929 mouse fibroblasts containing the cytokine M-CSF.
Detection of cytokine production
The concentration of IFN-β in culture supernatants was measured with a human IFN-β ELISA kit (CSB-E09889h, Cusabio Biotechnology) or a mouse IFN-β ELISA kit (CSB-E04945m, Cusabio Biotechnology).
Western blot and co-immunoprecipitation
Cells for both western blot and IP were lysed in IP buffer containing 50 mM Tris, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 100 μM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Complete Mini, Roche) for 30 min. Cell lysates were centrifuged at 5000× g for 10 min at 4°C and quantified using the Bradford method (#500-0006, BioRad). For western blotting, the supernatants were recovered and boiled in loading buffer. For IP, the supernatants were recovered and mixed with 2 μg of primary antibody per 1 mg protein samples and then incubated overnight at 4°C. The reaction mixtures were then mixed with protein G agarose and incubated for an additional 2 h at 4°C. Protein G agarose-bound immune complexes were collected by centrifugation at 14000× g for 1 min, washed at least five times with IP buffer, and boiled in loading buffer. Then the samples were centrifuged at 14000× g for 1 min. Samples were separated by SDS-PAGE and the gels containing proteins were transferred onto a nitrocellulose filter membrane (Millipore). Membranes were blocked by 5% nonfat milk, and proteins were detected by using primary antibodies as specified and secondary antibodies conjugated with horseradish peroxidase. The proteins were visualized using suitable HRP-conjugated secondary antibodies (Jackson Immuno Research) and SuperSignal-Femto chemiluminescent substrate (Pierce).
IRF3 dimer assay
Native PAGE was performed with an 8% acrylamide gel without SDS. The gel was pre-run for 60 min at 200 V on ice with 25 mM Tris-HCl (pH 8.4) and 192 mM glycine with or without 0.5% deoxycholate in the cathode buffer and anode buffer, respectively. Samples in the 1× loading buffer (50 mM Tris-HCl, pH 6.8, 0.002% bromphenol blue, and 15% glycerol) were applied on the gel and underwent electrophoresis for 60 min at 200 V on ice, followed by IB analysis.
Immunofluorescent confocal microscopy
HEK293T cells plated on glass coverslips in 6-well plates were fixed for 10 min with 4% paraformaldehyde in PBS and then permeabilized for 15 min with 0.5% Triton X-100 in PBS. After blockade of nonspecific binding by incubation of cells for 30 min with 1% goat serum in PBS, coverlips were incubated with the appropriate primary antibodies. Alexa Fluor 561/488-conjugated secondary antibodies (Invitrogen) were added for 1 h. Then the coverslips were washed three times with PBS and stained with DAPI (Invitrogen). Images of the samples were taken using a Perkin Elmer UltraView Vox confocal microscope.
In vitro kinase assay
HEK293T whole-cell lysate (1000 μg) with HA-tagged empty vector or HA-HDAC4 overexpression plasmid were incubated for 4 h at 4°C with anti-HA-agarose (Millpore). Immunocomplexes were washed at least five times with cell lysis buffer and then twice with 1× kinase buffer (#9802, Cell Signaling Technology). Kinase reactions were performed by incubation of 1.0 μg purified GST-fused IRF3 peptide (H00003661-P02, Novus) as the substrate with 1× kinase buffer, 1 mM ATP (#9804, Cell Signaling Technology), immunoprecipitated HA-HDAC4 or 1.0 μg purified GST-fused HDAC4 peptide (amino acids 612–1084, ab104029, Abcam), and TBK1(ab85276, Abcam) or IKKε (ab201367, Abcam) at 30°C for 90 min in 50 μl reaction mixture. Reaction was stopped by addition of 2× SDS loading buffer and samples were separated by SDS-PAGE, and analyzed by immunoblotting with anti-phospho-IRF3 or anti-phospho-Thr.
Statistical analysis
Differences between groups were evaluated using the two-tailed, unpaired Student’s t-test available in the GraphPad Prism 5 software package (GraphPad Software, Inc.). Coprecipitation efficiency and fluorescence images were analyzed using ImageJ (NIH). P-values were calculated, and statistical significance was reported as highly significant with *P < 0.05. Analytic results are presented as mean ± SD.
Supplementary Material
Acknowledgements
We would like to thank Dr Hong-Bing Shu (Wuhan University), members of Chen’s lab, and the Core Facilities Center of Wuhan Institute of Virology for technical help. We thank LetPub for its linguistic assistance during the preparation of this manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFA0507201 to X.C. and 2018YFA0507202 to Y.Z.) and the Program for Youth Innovation Promotion Association in Chinese Academy of Science to J.C.
Conflict of interest
none declared.
Author contributions
J.C. and X.C. designed and supervised the study; Q.Y. and J.T. performed the experiments and analysis; Y.W. provided reagents and suggestions; J.C., X.C., and Q.Y. wrote the paper; all authors analyzed data.
References
- Aung H.T., Schroder K., Himes S.R., et al. (2006). LPS regulates proinflammatory gene expression in macrophages by altering histone deacetylase expression. FASEB J. 20, 1315–1327. [DOI] [PubMed] [Google Scholar]
- Backs J., Song K., Bezprozvannaya S., et al. (2006). CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116, 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Pei R., Zhu W., et al. (2014). An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J. Immunol. 192, 1162–1170. [DOI] [PubMed] [Google Scholar]
- Chen J., Wang N., Dong M., et al. (2015). The metabolic regulator histone deacetylase 9 contributes to glucose homeostasis abnormality induced by hepatitis C virus infection. Diabetes 64, 4088–4098. [DOI] [PubMed] [Google Scholar]
- Choi S.J., Lee H.C., Kim J.H., et al. (2016). HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J. 35, 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.A., McWhirter S.M., Faia K.L., et al. (2003). IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A. (2004). Identification and characterization of viral antagonists of type I interferon in negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283, 249–280. [DOI] [PubMed] [Google Scholar]
- Halili M.A., Andrews M.R., Labzin L.I., et al. (2010). Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J. Leukoc. Biol. 87, 1103–1114. [DOI] [PubMed] [Google Scholar]
- Haller O., Kochs G., and Weber F. (2006). The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Lange C.J., et al. (2007). Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282, 5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C.Q., Zhang Y., Xia T., et al. (2013). FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J. Biol. Chem. 288, 12596–12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang S., Zhu H., et al. (2011). Cloning of the herpes simplex virus type 1 genome as a novel luciferase-tagged infectious bacterial artificial chromosome. Arch. Virol. 156, 2267–2272. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang Q., Ding Y., et al. (2016). Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 17, 806–815. [DOI] [PubMed] [Google Scholar]
- Liew F.Y., Xu D., Brint E.K., et al. (2005). Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–458. [DOI] [PubMed] [Google Scholar]
- Liu H.M., Jiang F., Loo Y.M., et al. (2016). Regulation of retinoic acid inducible gene-I (RIG-I) activation by the histone deacetylase 6. EBioMedicine 9, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Liu X., Zhang P., et al. (2016). Rb selectively inhibits innate IFN-β production by enhancing deacetylation of IFN-β promoter through HDAC1 and HDAC8. J. Autoimmun. 73, 42–53. [DOI] [PubMed] [Google Scholar]
- Mowen K.A., and David M. (2014). Unconventional post-translational modifications in immunological signaling. Nat. Immunol. 15, 512–520. [DOI] [PubMed] [Google Scholar]
- Nusinzon I., and Horvath C.M. (2006). Positive and negative regulation of the innate antiviral response and β interferon gene expression by deacetylation. Mol. Cell. Biol. 26, 3106–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan S.R., and Bowie A.G. (2013). Immune sensing of DNA. Immunity 38, 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon I.K., and Jans D.A. (2005). Regulation of nuclear transport: central role in development and transformation? Traffic 6, 173–186. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., and Reis e Sousa C. (2010). RIGorous detection: exposing virus through RNA sensing. Science 327, 284–286. [DOI] [PubMed] [Google Scholar]
- Ronco L.V., Karpova A.Y., Vidal M., et al. (1998). Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12, 2061–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., and Akira S. (2009). Innate immunity to virus infection. Immunol. Rev. 227, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterstab G., Ludwig S., Anton A., et al. (2005). Viral targeting of the interferon-β-inducing Traf family member-associated NF-{kappa}B activator (TANK)-binding kinase-1. Proc. Natl Acad. Sci. USA 102, 13640–13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.H., Kruhlak M.J., Wu J., et al. (2000). Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20, 6904–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.H., and Yang X.J. (2001). Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21, 5992–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhao W., Zhao K., et al. (2015). TRIM26 negatively regulates interferon-β production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLoS Pathog. 11, e1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P., and Pruss D. (1996). Targeting chromatin disruption: transcription regulators that acetylate histones. Cell 84, 817–819. [DOI] [PubMed] [Google Scholar]
- Wood E.R., Bledsoe R., Chai J., et al. (2015). The role of phosphodiesterase 12 (PDE12) as a negative regulator of the innate immune response and the discovery of antiviral inhibitors. J. Biol. Chem. 290, 19681–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., et al. (2003). Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643. [DOI] [PubMed] [Google Scholar]
- Zhang M., Tian Y., Wang R.P., et al. (2008). Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 18, 1096–1104. [DOI] [PubMed] [Google Scholar]
- Zhu J., Coyne C.B., and Sarkar S.N. (2011). PKCα regulates Sendai virus-mediated interferon induction through HDAC6 and β-catenin. EMBO J. 30, 4838–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.