Abstract
Fat mass and obesity-associated (FTO) protein is a ferrous ion (Fe2+)/2-oxoglutarate (2-OG)-dependent demethylase preferentially catalyzing m6A sites in RNA. The FTO gene is highly expressed in the hypothalamus with fluctuation in response to various nutritional conditions, which is believed to be involved in the control of whole body metabolism. However, the underlying mechanism in response to different nutritional cues remains poorly understood. Here we show that ketogenic diet-derived ketone body β-hydroxybutyrate (BHB) transiently increases FTO expression in both mouse hypothalamus and cultured cells. Interestingly, the FTO protein represses Fto promoter activity, which can be offset by BHB. We then demonstrate that FTO binds to its own gene promoter, and Fe2+, but not 2-OG, impedes this binding and increases FTO expression. The BHB-induced occupancy of the promoter by FTO influences the assembly of the basal transcriptional machinery. Importantly, a loss-of-function FTO mutant (I367F), which induces a lean phenotype in FTOI367F mice, exhibits augmented binding and elevated potency to repress the promoter. Furthermore, FTO fails to bind to its own promoter that promotes FTO expression in the hypothalamus of high-fat diet-induced obese and 48-h fasting mice, suggesting a disruption of the stable expression of this gene. Taken together, this study uncovers a new function of FTO as a Fe2+-sensitive transcriptional repressor dictating its own gene switch to form an auto-regulatory loop that may link with the hypothalamic control of body weight.
Keywords: FTO, transcriptional repressor, hypothalamus, ferrous ion, obesity
Introduction
A wealth of attention has been recently paid to the fat mass and obesity-associated (FTO) gene because of its intimate connection with obesity, which constitutes a global public health concern. A major link is established by several intronic single nucleotide polymorphisms (SNPs) in the FTO gene that closely correlate with body mass and composition phenotypes (Dina et al., 2007; Frayling et al., 2007; Scuteri et al., 2007; Cecil et al., 2008; Yang et al., 2012). It has been reported that FTO transcripts with obesity-associated variants exhibit a connection with the promoter of the homeobox gene IRX3, which was already implied in regulating body mass and composition (Smemo et al., 2014). Given that the FTO gene is quite distant (mega-bases) from the IRX3 gene, it is plausible that the obesity-associated FTO intronic SNPs are important constituents of a long-rang enhancer for IRX3 (Smemo et al., 2014). Recently, another study reported that two obesity-associated SNPs in the intron of FTO gene are involved in the regulation of both FTO gene and retinitis pigmentosa GTPase regulator-interacting protein-1 like (RPGRIP1L) gene and cause obesity in mice (Stratigopoulos et al., 2016), indicating a direct linkage between the SNPs and metabolic homeostasis. A more direct role was also implicated: FTO overexpression in mice leads to increased food intake and obesity (Church et al., 2010), whereas Fto knockout mice or Fto knock-in mice expressing a loss-of-function mutant result in lean phenotypes (Church et al., 2009; Fischer et al., 2009).
FTO is a ferrous ion (Fe2+)/2-oxoglutarate (2-OG)-dependent demethylase that prefers 3-methylthymidine in single-stranded DNA and 3-methyluracil and N6-methyladenosine (m6A) in single-stranded RNA (Gerken et al., 2007; Han et al., 2010; Jia et al., 2012b), and predominantly functions as demethylase of RNA (Jia et al., 2011). The latter modification is a mark in quite a few RNA transcripts and is dynamically regulated in response to diverse cues including energy and nutrient availabilities (Cheung et al., 2013; Ronkainen et al., 2015; Mizuno et al., 2017; Nowacka-Woszuk et al., 2017), which establishes a link between FTO enzymatic function and obesity (Jia et al., 2012a, b; Meyer et al., 2012); given that this mark in mRNAs is involved in regulating genetic readouts (Meyer et al., 2012; Fu et al., 2014), FTO may play a role in regulating gene expression at a post-transcriptional level. Another study shows that FTO is a transcriptional co-activator that facilitates the transcription of other genes by interacting with CCAAT/enhancer binding proteins (C/EBPs) and core promoter elements (Wu et al., 2010). Thus, FTO may play multi-functional roles in regulating gene expression at both transcriptional and post-transcriptional levels.
The FTO gene is widely expressed in diverse tissues including the brain (Gerken et al., 2007; Lein et al., 2007), with high levels in the hypothalamus that fluctuate in response to various nutritional conditions (Gerken et al., 2007; Fredriksson et al., 2008; Tung et al., 2010; Poritsanos et al., 2011; Karra et al., 2013). Given that hypothalamic FTO is involved in regulating energy and food intake (Olszewski et al., 2009; Tung et al., 2010; Poritsanos et al., 2011; Karra et al., 2013), the nutrition-triggered fluctuation of hypothalamic FTO expression could be involved in the central control of whole body metabolism. However, a description of the underlying mechanism in response to different nutritional cues remains poorly understood.
Here, we show that upon 1-month feeding with ketogenic diet (KD), instead of high-fat diet (HFD), mouse hypothalamus tissues exhibit transiently increased levels of Fto transcript and FTO protein presumably via elevated levels of serum β-hydroxybutyrate (BHB). Cellular assays reveal a negative feedback loop in line with a transiently upregulated FTO expression pattern. We further show that FTO binds to its own promoter to decrease the promoter activity, and free Fe2+ impedes FTO binding to the promoter in a dose-dependent manner to increase Fto expression. Interestingly, a mutant (I367F) FTO, which causes a lean phenotype in FTOI367F mice (Church et al., 2009), exhibits an augmented promoter-binding in conjunction with an increased repression on Fto expression. Furthermore, the auto-regulatory loop of the Fto gene is disrupted in the hypothalamus of obese mice after 3-month feeding with HFD or KD. This newly discovered function of FTO in an auto-regulatory loop is involved in its own homeostatic hypothalamic expression, which might implicate a potential role in the control of body weight.
Results
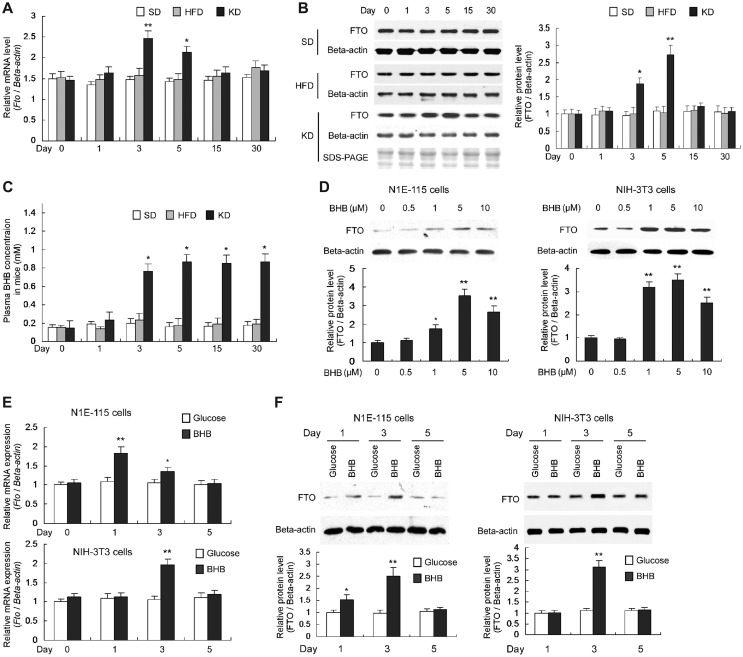
BHB induces transiently elevated levels of FTO expression
High-energy intake is associated with elevated body fat (McCrory et al., 1999; Astrup et al., 2000) and Fto expression is nutritionally regulated (Gerken et al., 2007), which prompted us to test the abilities of different HFDs on altering Fto expression. In a time course, 3-week-old mice were fed with HFD or KD, and qPCR and immunoblot were used to assess, respectively, the Fto mRNA and FTO protein levels in the hypothalamus. At Days 3 and 5, the Fto mRNA levels transiently increased in the hypothalamus of KD-fed mice but not altered in HFD-fed mice, compared to the mice fed with standard diet (SD) (Figure 1A); similarly, correspondingly altered FTO protein levels were observed during the time course (Figure 1B). Animals on KD, instead of HFD, exhibit elevated serum levels of ketone bodies, of which BHB plays a role in gene expression owing to its activity as an endogenous histone deacetylase inhibitor (Shimazu et al., 2013). The BHB levels in the sera of KD-fed mice increased at Day 3 and remained high (Figure 1C), which is not in line with transiently increased Fto mRNA and FTO protein levels. A few genes, reported by a previous study to be induced by BHB (Shimazu et al., 2013), were also upregulated in the hypothalamus of KD-fed mice (Supplementary Figure S1), in line with the increase of serum BHB levels.
Figure 1.
KD-derived BHB transiently increases Fto expression. (A) Fto mRNA levels in the hypothalamus, normalized to β-actin mRNA level. n = 5, *P < 0.01, **P < 0.001, compared with the mRNA levels in SD-fed mice at the same time point. (B) FTO protein levels in the hypothalamus, normalized to β-actin protein. n = 3, *P < 0.01, **P < 0.001, compared with the protein levels in mice at Day 0 (as ‘1’). (C) BHB levels in the serum of peripheral blood. n = 5, *P < 0.001, compared with that in SD-fed mice at the same time point. (D) Mouse N1E-115 and NIH-3T3 cells were cultured in the medium with BHB instead of glucose and the protein levels were determined using western blot after 48 h. FTO protein levels (normalized to the β-actin protein) gradually increased in response to the rise of BHB concentrations. n = 3, *P < 0.05, **P < 0.01, compared with the protein levels in cells at Day 0 (as ‘1’). (E and F) N1E-115 and NIH-3T3 cells were cultured in the medium with BHB or glucose. BHB transiently increased Fto mRNA (E) and FTO protein (F) levels in N1E-115 cells after 1- and 3-day culture while in NIH-3T3 cells only after 3-day culture. The Fto mRNA and FTO protein levels were normalized to that of β-actin gene and β-actin protein, respectively. n = 3, *P < 0.01, **P < 0.001, compared with the mRNA or protein level in cells with glucose medium at the same time point.
To explore a potentially direct role of BHB on Fto expression, we employed cultured N1E-115 cells and NIH-3T3 cells treated with BHB titrated from 0 to 10 μM, and observed a gradual increase of the FTO protein levels (Figure 1D), upon BHB titration. Next, the Fto mRNA and FTO protein levels in cells cultured with glucose or BHB as energy source were compared; transiently upregulated Fto mRNA and FTO protein levels were observed in the two cell lines treated with BHB (Figure 1E and F). These results are in accordance with those in the hypothalamus of KD-fed mice.
BHB increases the Fto promoter activity
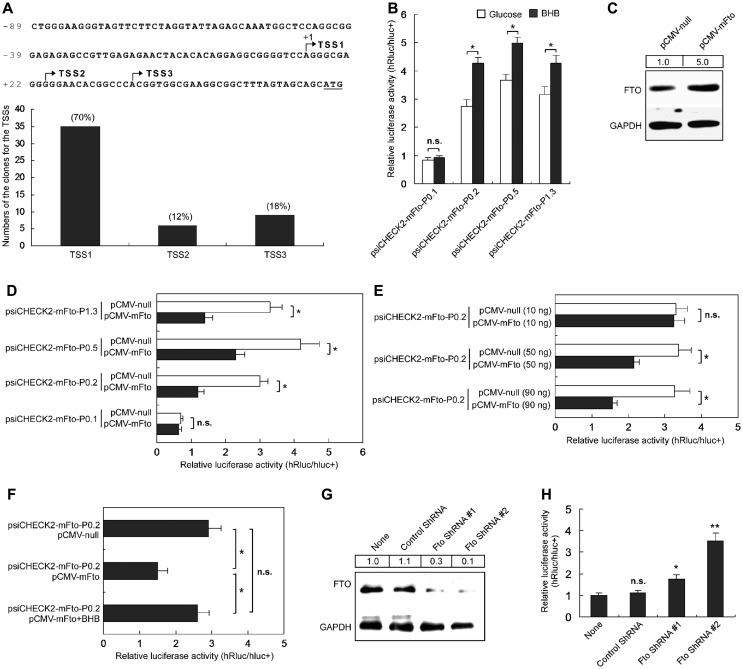
To determine whether the BHB-induced increase of Fto expression is a result of its promoter activation, we identified transcription start sites (TSSs) of the Fto gene in the mouse brain using 5′ full RACE. Three TSSs were identified from sequences of randomly selected 5′-RACE clones, and the most distal one (TSS1, relative to ATG) is 51 nucleotides (nt) upstream of ATG with the highest frequency (35 out of 50 clones, 70%), representing the most-used one (Figure 2A). Using Dual Luciferase Report System, we then analyzed relative strengths of diverse lengths of Fto promoter fragments fused to the reporter, and found that the promoter fragment (P0.2) that covers nt −161 to +50 was sufficient for promoter activation in N1E-115 cells (Supplementary Figure S2). This fragment contains three TSSs (Figure 2A) and essential upstream sequences that fully support BHB-induced transcription from the Fto promoter in N1E-115 cells (Figure 2B).
Figure 2.
BHB and FTO expression change alters the Fto promoter activity. (A) PCR-sequencing analyses show three TSSs of the Fto gene (arrows), identified by 5′ full RACE in the mouse brain. ‘+1’ indicates that the TSS is the most distal one. TSS1 is the most-used site for Fto transcription initiation in mouse brain, which was found in up to 70% of the total sequenced clones, and only 12% and 18% of clones for TSS2 and TSS3, respectively. (B) BHB significantly increased the promoter activity of the Fto gene in mouse N1E-115 cells. Luciferase assay showed that the relative luciferase activities of the serial promoter report constructs in cells with glucose medium were only ~60%−70% of that with BHB medium. n.s. represents no significance. n = 10, *P < 0.001, compared with that in glucose medium. (C) FTO expression was higher in N1E-115 cells transfected with the FTO overexpression construct pCMV-mFTO than cells transfected with the control construct pCMV-null. GAPDH expression was detected as the loading control. Values at the top show the densitometric quantification of FTO-immunoreactive bands. (D) Overexpression of FTO protein decreased the Fto promoter activity in N1E-115 cells. n.s. represents no significance. n = 10, *P < 0.001. (E) The repressive effects on the Fto promoter were dependent on the usage of pCMV-mFTO. n.s. indicates no significance. n = 20, *P < 0.001. (F) The repressive effect of FTO overexpression on the Fto promoter was offset by BHB. n.s. indicates no significance. n = 20, *P < 0.001. (G) FTO expression was lower in N1E-115 cells transfected with the Fto-specific shRNA constructs than cells transfected with control shRNA construct or untransfect cells (None). GAPDH expression was also detected as the loading control. Values at the top show the densitometric quantification of FTO-immunoreactive bands. (H) FTO knockdown increased the Fto promoter activity in N1E-115 cells. n.s. indicates no significance. n = 10, *P < 0.01, **P < 0.001.
Overexpression or knockdown of Fto alters the Fto promoter activity
The data showed that BHB transiently increased the Fto mRNA and FTO protein levels (Figure 1), and FTO is implicated in a transcription-factor function (Wu et al., 2010), we proposed that the FTO protein may play a role in repressing its own promoter. We then generated construct pCMV-mFTO to overexpress FTO in cultured cells (Figure 2C), which repressed the reporter gene activity (Figure 2D) in a dose-dependent manner (Figure 2E). The promoter activity repressed by FTO overexpression could be offset by BHB (Figure 2F). Thus, the transiently increased Fto expression induced by BHB is likely to be resulted from a negative auto-regulatory mechanism.
We also used shRNA-expressing constructs for specific knockdown of FTO expression in cultured N1E-115 cells; the FTO levels in cells expressing Fto-specific shRNA #1 and #2 were, respectively, reduced by ~70% and ~90% as compared to that in control cells or untransfected cells (Figure 2G). Following co-transfection of the reporter and shRNA-expressing constructs into N1E-115 cells, we observed that the relative reporter activity (hRluc/hluc+) in Fto-specific shRNA-expressing cells was significantly higher than that in control shRNA-expressing cells or cells transfected with just reporter (Figure 2H), suggesting that the increased promoter activity was due to a decreasing repressive function of FTO. Results in Figure 2 suggest that FTO is involved in an auto-regulatory mechanism dictating its own genetic readout.
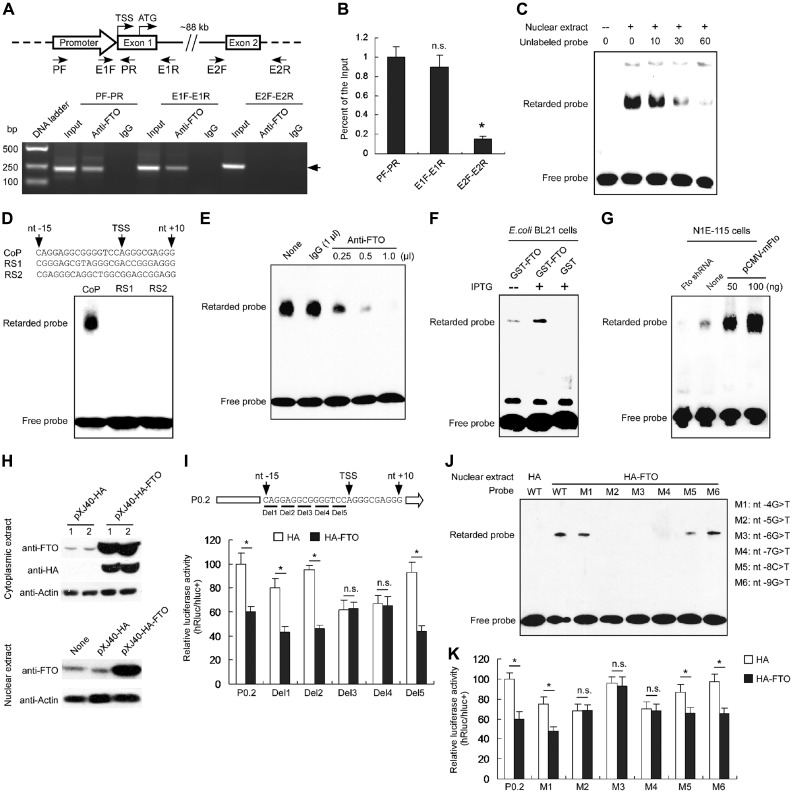
FTO binds to its own promoter
Given that FTO might function as a transcriptional cofactor for other promoters (Wu et al., 2010) and our above results, we proposed that FTO might exert a transcriptional repressor function on its own promoter. We therefore performed chromatin immunoprecipitation PCR (ChIP-PCR) assays with, respectively, anti-FTO and control IgG antibodies on mouse hypothalamic tissues. Two fragments containing, respectively, the Fto promoter and exon 1 were amplified from the DNA extricated from the anti-FTO, but not from the immuno-precipitates with the control IgG; no PCR bands corresponding to exon 2 were observed (Figure 3A). In addition, qPCR confirmed that anti-FTO ChIP precipitates were much more enriched for Fto promoter and exon 1 than for exon 2 sequences (Figure 3B), suggesting that the FTO protein might bind to its own promoter at the overlapped region between the locations of primers E1F and PR shown in Figure 3A.
Figure 3.
FTO binds to the promoter region of its own gene. (A) Identification of the binding of FTO to the mouse Fto promoter sequence using ChIP analysis with anti-FTO antibody. The ChIP precipitates were detected by PCR with primers for amplifying the Fto promoter, Fto exon 1 and exon 2, respectively. The Fto P0.2 promoter (nt −161 to nt + 50) and the exon 1 were amplified using anti-FTO instead of IgG; however, the Fto exon 2 was not amplified using either anti-FTO or IgG. The image represents one of three independent biological repeats. (B) The ChIP samples were detected by qPCR with primers for the Fto promoter, Fto exon 1 and exon 2. n.s. indicates no significance. n = 3, *P < 0.0001. (C) EMSA was performed using biotin-labeled double-strand DNA probes and the nuclear extracts of the mouse hypothalamus. A retarded band was observed in the reactions with the nuclear extracts and addition of unlabeled probes could compete with the labeled probes. (D) Hypothalamic nuclear proteins specially bound to the Fto promoter. EMSA was performed using the nuclear extracts of the mouse hypothalamus and the core promoter probes (CoP), random sequence 1 (RS1), and random sequence 2 (RS2). A retarded band was observed only in the reactions with CoP. (E) Supershift EMSA assay using anti-FTO antibody (0.25, 0.5, or 1 μl) and IgG (1 μl), respectively. The shift band was decreased following increased anti-FTO antibody, but not altered by IgG. (F) EMSA showing the specific retarded band after overexpression of FTO in E. coli BL21 (DE3) cells with IPTG induction. (G) Knockdown or overexpression of FTO decreased or increased the binding capability to the probes, respectively, in N1E-115 cells. (H) Western blot analyses of HA-FTO expression in N1E-115 cells transfected with the HA-FTO overexpression construct pXJ40-HA-FTO or the control construct pXJ40-HA. The β-actin expression was detected as the loading control. (I) In N1E-115 cells, overexpression of HA-FTO fusion protein decreased the activities of the Fto promoter, Del1 (deleting nt −13 to −15) Del2 (deleting nt −10 to −12), or Del5 (deleting nt −1 to −3), but did not alter the activities of Del3 (deleting nt −7 to −9) or Del4 (deleting nt −4 to −6). n.s. represents no significance. n = 10, *P < 0.001. (J) EMSA showing retarded bands to the promoter probe and the probes containing mutations M1, M5, and M6 with the nuclear extracts of N1E-115 cells transfected with the HA-FTO overexpression construct. (K) In N1E-115 cells, HA-FTO overexpression decreased the activities of the Fto promoter and the promoters with mutations M1, M5, and M6. n.s. represents no significance. n = 10, *P < 0.001.
We next used electrophoretic mobility shift assays (EMSA) with overlapped synthetic and biotin-labeled double stranded DNA fragments encompassing the P0.2 promoter (nt −161 to +50) region to localize where FTO binds. An element that covers the core promoter (nt −15 to +10), upon incubation with nuclear extracts from mouse hypothalamus, generated a retarded signal; in a dose-dependent manner, the intensities of the signal reduced if the labeled probe was co-incubated with increasing amounts of unlabeled oligonucleotides of the same sequence (Figure 3C). No retarded signals were observed in the reactions using the probes with the same GC contents as the core promoter (Figure 3D). When antibodies were involved, the retarded band became weak in the reaction with an increase usage of anti-FTO, but not in the reactions using the same level of IgG or no competitor (Figure 3E), suggesting that the anti-FTO antibody could compete with the binding of the complex to the probes.
We also generated bacterial extracts comprising expressed GST-FTO or GST-alone fusion protein and used them in EMSA analyses with the promoter probes. After IPTG induction, a strong retarded band was observed in reactions using the extracts from the bacteria transformed with the construct expressing GST-FTO, but not GST (Figure 3F), suggesting that the bacterially expressed FTO could also bind to the core promoter. Further EMSA showed stronger retarded bands in reactions using nuclear extracts of N1E-115 cells transfected with Fto-overexpressing constructs, as compared with that of cells containing the endogenous FTO (Figure 3G); when cells were transfected with Fto shRNA#2 (Figure 2G), the retarded signal was diminished (Figure 3G).
To further identify the exact site for FTO binding to the core promoter, we firstly generated construct pXJ40-HA-FTO to overexpress HA-FTO fusion protein in cultured N1E-115 cells (Figure 3H). We then made a series of reporter constructs containing the Fto promoter by sequentially deleting 3 nts upstream of the TSS (Figure 3I). Following co-transfections of the reporter and HA-FTO-expressing constructs into N1E-115 cells, we observed that the relative reporter activities (hRluc/hluc+) of the deleted constructs Del3 (deleting nt −7 to −9) and Del4 (nt −4 to −6) in HA-FTO-expressing cells was similar to that in control shRNA-expressing cells (Figure 3I), suggesting that the FTO binding site should locate at nt −4 to −9. Further EMSA showed no retarded bands in the reactions using the mutated probes M2 (−5G>T), M3 (−6G>T), and M4 (−7G>T) (Figure 3J). Correspondingly, following luciferase assays showed that these mutations M2, M3, and M4 in the Fto promoter led to a loss of the repressive effect of FTO on the promoter activity (Figure 3K), suggesting that the GGG triplet (covering nt −5 to −7) is critical for FTO binding and in negative regulation of gene expression. Together, these results support that FTO binds to its own promoter in vivo and in vitro.
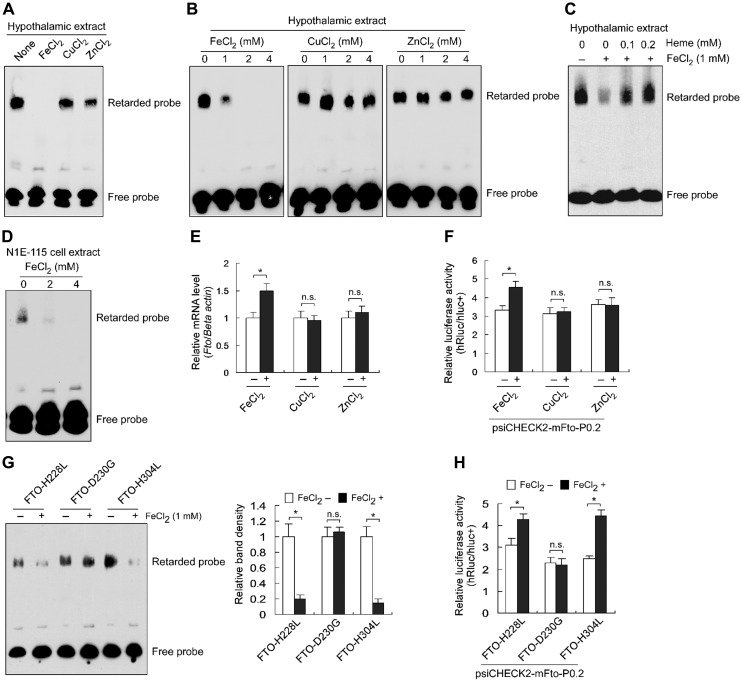
Fe2+ weakens the FTO binding to the promoter and enhances gene expression
FTO is a Fe2+-binding protein and Fe2+ is essential for its function as a demethylase (Gerken et al., 2007; Sanchez-Pulido and Andrade-Navarro, 2007; Han et al., 2010); we proposed that this divalent ion may as well affect the binding of FTO to the promoter sequence. Indeed, in EMSA with hypothalamic nuclear extracts, addition of FeCl2, but not of CuCl2 and ZnCl2, impeded FTO binding to the promoter (Figure 4A), and the efficacy of FTO binding to the probe gradually decreased with a titration of FeCl2 from 0 to 4 mM but not with that of CuCl2 and ZnCl2 in the same range (Figure 4B). This could be relieved by Fe2+-free heme that chelates Fe2+ (Figure 4C). Additionally, the inhibitory effect of FeCl2 on FTO binding to the promoter was observed using nuclear extracts of N1E-115 cells (Figure 4D). In N1E-115 cells treated with divalent ions, Fto mRNA levels were elevated by FeCl2 but not by CuCl2 or ZnCl2 (Figure 4E); in reporter assays, FeCl2 but not CuCl2 or ZnCl2 exhibited stimulatory effect (Figure 4F), suggesting that FeCl2-faciliated Fto expression is due to increased promoter activity.
Figure 4.
FTO is a Fe2+-sensitive transcriptional repressor. (A) EMSA showing a repressive effect of Fe2+, instead of Cu2+ or Zn2+, on FTO binding to the promoter sequence. (B) EMSA showing a decreasing binding ability of FTO to the core promoter in response to the increasing Fe2+, instead of Cu2+ or Zn2+. (C) The repressive effect of Fe2+ on FTO binding to the probe was deprived by Fe2+-free heme. (D) EMSA showing a decreasing binding ability of FTO to the core promoter in response to the increasing Fe2+ with the nuclear extracts of N1E-115 cells. (E) Fe2+ increased Fto mRNA levels in N1E-115 cells. The Fto mRNA levels were normalized to the β-actin level and compared to the relative mRNA levels in cells without ion treatment (as ‘1’). n = 3, *P < 0.01. n.s. represents no significant. (F) Fe2+ increased Fto promoter activity in N1E-115 cells. n = 3, *P < 0.01. n.s. represents no significant. (G) The FTO binding to the promoter in response to Fe2+ was abolished by the mutant FTO-D230G, but not FTO-H228L or FTO-H304L. The image is representative of three experiments. The average shift band density of the reaction without FeCl2 was set as ‘1.’ n = 3, *P < 0.001. n.s. indicates no significance. (H) The repressive effect of FTO on its own promoter in response to Fe2+ was abolished by the mutant FTO-D230G, but not FTO-H228L or FTO-H304L. n = 10, *P < 0.001. n.s. indicates no significance.
We used EMSA to compare response patterns of three FTO mutants H228L, D230G, and H304L, which have altered binding to Fe2+ (Han et al., 2010); Fe2+ decreased the binding of the mutants H228L and H304L to the probe whereas the mutant D230G lost response to Fe2+ (Figure 4G). Reporter assays showed that the presence of Fe2+ increased the promoter activities in cells co-transfected with mutants H228L or H304L, but not D230G (Figure 4H). These data suggest that the D230 residue is crucial for FTO function as a Fe2+-sensitive transcriptional repressor and cellular free Fe2+ homeostasis may actively participate in ensuring homeostatic Fto expression.
2-OG does not affect FTO binding to the promoter element
Along with Fe2+, 2-OG is also essential for FTO to function as a demethylase (Gerken et al., 2007). To test whether 2-OG plays a role in FTO-mediated transcriptional repression, we carried out EMSA with a 2-OG titration from 0 to 640 μM, and found that it did not affect the efficacy of FTO binding to the probes (Supplementary Figure S3A). R313 is a key residue in the 2-OG binding pocket of FTO, and an R313Q mutant abrogates its enzyme activity (Gerken et al., 2007). We made a mutant FTO-R313Q and found that it retained normal Fto promoter occupancy (Supplementary Figure S3B) and repressive effect on the reporter (Supplementary Figure S3C). Thus, 2-OG does not participate in FTO function as a transcriptional repressor.
FTO modulates the basal transcription machinery assembly that impacts on all TSSs
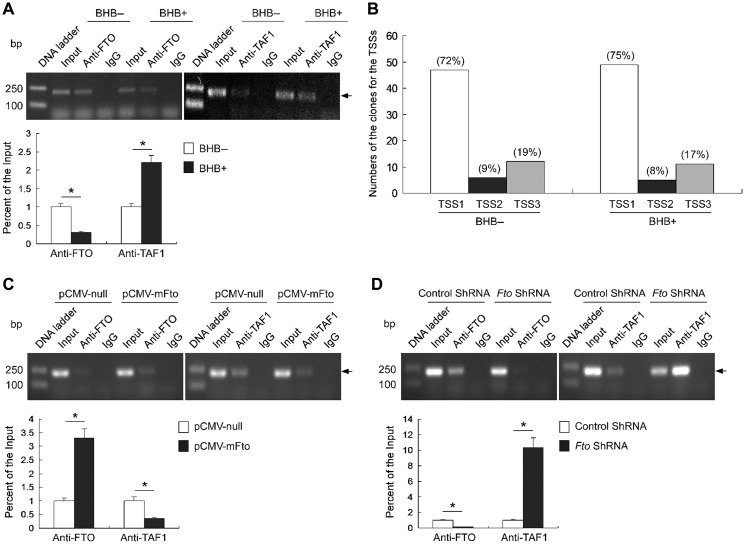
Mechanistically, transcriptional repressors modulate gene expression by affecting the assembly of the basal transcription machinery (or its function thereof) at initiation stage in particular (Roeder, 2003). We treated N1E-115 cells with BHB for slightly less than 24 h, at which point the Fto mRNA level reached its peak (see Figure 1E) but the FTO protein level was only slightly increased (see Figure 1F). ChIP assays showed that the Fto promoter occupancy by FTO reduced by ~2.5-fold and that by TAF1 increased by ~2-fold upon BHB treatment (Figure 5A), suggesting that BHB impacts FTO occupancy and promotes the assembly of the basal transcription machinery on the Fto promoter.
Figure 5.
The effect of BHB on FTO binding to the promoter affects the TAF1 binding and impacts on all TSSs. (A) The ChIP samples detected by semi-quantitative PCR (upper) and qPCR (lower) showing that BHB decreased FTO binding and increased TAF1 binding to the promoter, respectively. n = 3, *P < 0.001. (B) 5′ Full RACE showing that BHB affects all the three Fto TSSs in N1E-115 cells. The percentages of clones from the TSSs were similar in the cells treated with BHB and control cells. (C) FTO overexpression increased FTO binding and decreased TAF1 binding to the promoter. n = 3, *P < 0.001. (D) FTO knockdown decreased FTO binding and increased TAF1 binding to the promoter. n = 3, *P < 0.001.
Assembly of the basal transcription machinery is potentiated by the recruitment of TAF1, an essential general transcription factor for both TATA-directed and TATA-less promoters; the Fto promoter belongs to the latter (see Figure 2A) which typically have multiple TSSs. The nutrition-modulated Fto expression fluctuates at ~2- to 3-fold (Figure 1), which can be realized in theory by switching on/off the major site TSS1 (Figure 2A) or all sites in proportion. We used 5′-RACE to assess the relative usage of three TSSs in N1E-115 cells untreated or treated with BHB, and found that the percentiles did not significantly deviated among them (Figure 5B) and from those assessed in the brain (Figure 2A), suggesting that fluctuating Fto expression involves alteration of efficacies of all three TSSs.
To further determine the impact of FTO on the assembly of the basal transcription machinery, we performed ChIP analyses using the N1E-115 cells transfected with pCMV-mFto or Fto shRNA constructs, respectively. We found that FTO overexpression increased the binding of FTO to the promoter and decreased the binding of TAF1 to the promoter (Figure 5C). On the contrary, FTO knockdown decreased FTO binding and increased TAF1 binding to the promoter (Figure 5D). Taken together, these data suggest that FTO occupancy negatively impacts upon the assembly of the basal transcription machinery on the Fto promoter.
A loss-of-function mutant I367F augments the repressive effect of FTO
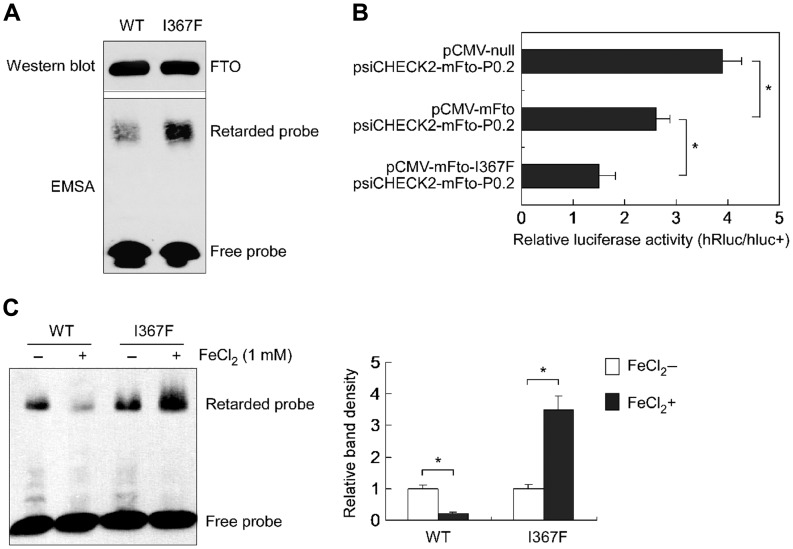
The FTO mutant I367F has altered crystal structure (Han et al., 2010), and the FTOI367F mice exhibit reduced fat mass (Church et al., 2009). We tested the abilities of FTO I367F to bind to the Fto promoter and repress Fto transcription to make a firm link between the FTO function as a transcriptional repressor and FTO-related obese/lean phenotypes. Compared to wild-type FTO, a stronger retarded band was observed in EMSA for this mutant (Figure 6A) that suggests an increased affinity; reporter assays showed that it augmented the repressive effect on Fto promoter (Figure 6B). Intriguingly, we observed that, as opposed to a repressing effect on wild-type FTO, Fe2+ increased the binding of the I367F mutant to the probe (Figure 6C). Taken together, these data bridge an auto-regulatory loop of Fto gene expression with its role in body weight control.
Figure 6.
A loss-of-function FTO mutant increases the repressive effect on its own promoter. (A) EMSA showing an increased binding of the mutant FTO-I367F to its own promoter. (B) Luciferase assays showing that the mutant FTO-I367F increased the repressive effect on its own promoter. n = 10, *P < 0.001. (C) Luciferase assays showing that Fe2+ increased the mutant binding to its own promoter. n = 3, *P < 0.001.
Upregulation of FTO expression and decrease of FTO binding to the promoter in the hypothalamus of obese mice and fasting mice
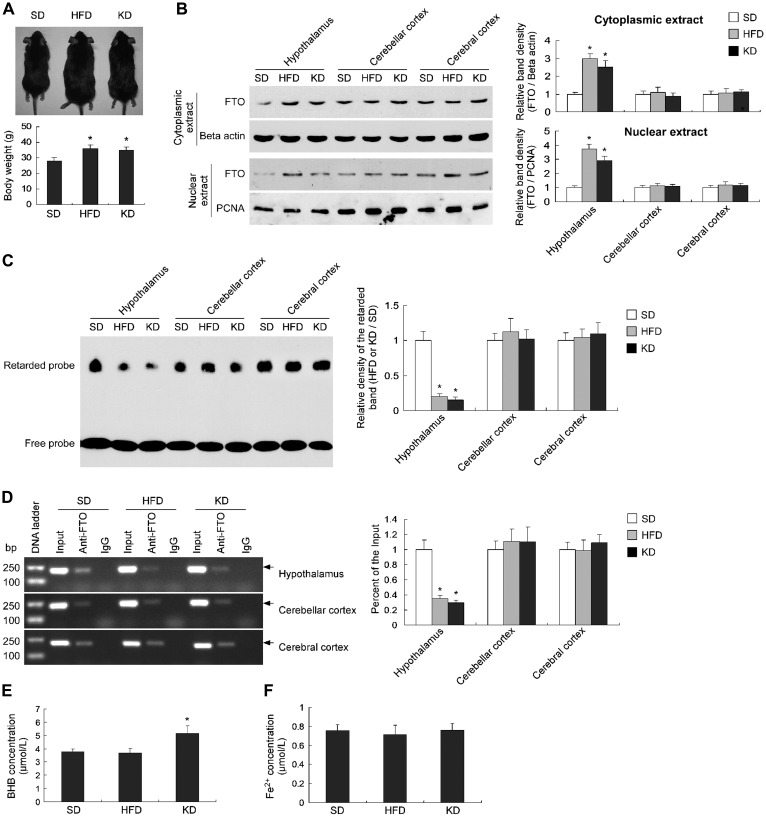
To determine the relationship between the stable expression of the Fto gene and obesity, we fed animals with HFD and KD for 3 months to generate obese mice. We found that the body weights of HFD- and KD-fed mice increased by ~30% of the SD-fed mice (Figure 7A). Compared with the control mice, both cytoplasmic and nuclear FTO protein levels significantly increased in the hypothalamus of HFD- and KD-fed mice, but not in the cerebellar cortex and cerebral cortex tissues (Figure 7B). EMSA analyses (Figure 7C) and ChIP assays (Figure 7D) showed that the capability of FTO binding to the probe or promoter significantly decreased in the hypothalamic nuclear extracts of the HFD- and KD-induced obese mice, compared with the control mice, but not altered in the cerebellar cortex and cerebral cortex tissues. These suggest the disrupted auto-regulatory loop of Fto gene in the hypothalamus of HFD- and KD-induced obese mice, which results in the upregulation of FTO expression.
Figure 7.
Upregulated FTO expression and decreased FTO binding to its own promoter in the hypothalamus of HFD- and KD-induced obese mice. (A) The body weights of the mice fed with HFD and KD for 3 months increased by ~30% compared to the SD-fed mice. n = 5, *P < 0.01. (B) Western blot showing a significant increase of the cytoplasmic and nuclear FTO protein levels in the hypothalamus of the HFD- and KD-induced obese mice, but not in cerebellar cortex or cerebral cortex. n = 3, *P < 0.001. (C) ESMA assays showing a significant decrease of the capability of FTO binding to its own promoter in the hypothalamus of HFD- and KD-induced obese mice, but not in cerebellar cortex or cerebral cortex. n = 3, *P < 0.001. (D) The ChIP samples detected by semi-quantitative PCR (left) and qPCR (right) showing a significant decrease of the capability of FTO binding to its own promoter in the hypothalamus of HFD- and KD-fed mice, but not in cerebellar cortex and cerebral cortex. n = 3, *P < 0.001. (E) BHB significantly increased in the hypothalamus of KD-fed but not HFD-fed mice. n = 5, *P < 0.01, compared to the SD-fed mice. (F) No alteration of Fe2+ levels was observed in the hypothalamus of HFD- or KD-fed mice, compared with the SD-fed mice. n = 5.
To determine the potential effects of BHB and Fe2+ on the disruption of the auto-regulatory loop of Fto gene in the hypothalamus of HFD- and KD-induced obese mice, we tested the concentrations of BHB and Fe2+ in the hypothalamus. We found that BHB levels were significantly increased in the hypothalamus of KD-fed but not HFD-fed mice (Figure 7E). No alterations in the Fe2+ levels were observed in the hypothalamus of both HFD- and KD-fed mice (Figure 7F). Therefore, BHB and Fe2+ might not be the impact factors for decreased FTO binding to its own promoter under long-term HFD conditions.
To further investigate the relationship between the Fto auto-feedback loop and fasting, we detected the FTO levels in the brain tissues of 48-h fasting mice. Compared with the control (regular feeding) mice, the FTO levels increased in the hypothalamus of the fasting mice, but not in the cerebellar cortex and cerebral cortex (Supplementary Figure S4A). Correspondingly, EMSA assays showed that the binding capability of FTO to its own promoter decreased in the extract of hypothalamus, but not in the extracts of cerebellar cortex and cerebral cortex (Supplementary Figure S4B). In addition, no alterations of the BHB and Fe2+ levels were observed in the hypothalamus upon fasting, which is similar to that upon HFD (Figure 7E and F). These data further suggest an interruption of the auto-feedback loop of the Fto gene in hypothalamus under the fasting condition.
Discussion
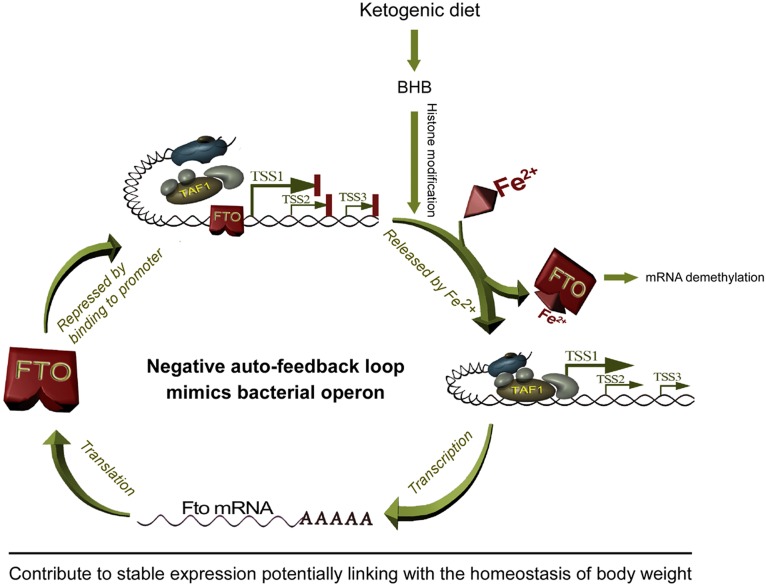
Here, we unraveled a new function of the FTO protein that is operative in a negative auto-regulatory loop impacting upon its own expression in hypothalamus (Figure 8). Given that hypothalamic FTO at least in part controls eating behavior, this novel finding provides a link between homeostatic hypothalamic FTO expression and the control of body weight.
Figure 8.
A schematic illustration of the negative auto-feedback loop of Fto gene that potentially contributes to FTO stable expression and body weight homeostasis.
A large body of studies have reported that Fto mRNA levels fluctuate in hypothalamus, a vital control center for food intake and energy homeostasis (Olszewski et al., 2009; Tung et al., 2010; Wang et al., 2011; Vujovic et al., 2013). The present study showed a BHB-induced transient upregulation of Fto expression in the hypothalamus of KD mice and culture cells (Figure 1), and this phenomenon is confirmed by following in vitro data that BHB could induce the reporter gene expression through the Fto promoter fragment that covers nt −161 to +50 (Figure 2B). Given that KD-derived BHB transiently increases Fto mRNA and FTO protein levels in mouse hypothalamus and cultured cells (Figure 1) and that starvation potentiates fatty acid oxidation to meet energy demand that also accumulates BHB, the observations that short-term starvation increases Fto mRNA levels (Gerken et al., 2007; Fredriksson et al., 2008) whereas long-term energy restriction squelches FTO expression (Wang et al., 2011) in hypothalamus and/or brainstem can be logically explained by an exquisite feedback loop that we proposed here (Figure 8). In mammals, the early response to fasting is mobilization of energy-rich molecules, such as gluconeogenic amino acids, prior to that of fats, which explains the inability of short-time (≤16 h) fasting to alter Fto expression (McTaggart et al., 2011). BHB upregulates the expression of a number of genes by increasing histone acetylation at their promoters (Shimazu et al., 2013). Such an epigenetic change could dynamically modulate the chromatin architecture, which in turn facilitates functional interplay between upstream transcription factors and the basal machinery, including attenuating occupancy of the Fto promoter by FTO, to transiently augment Fto expression. Given that in mice FTO overexpression leads to obesity and inactivation of the gene protects from obesity (Fischer et al., 2009; Church et al., 2010), the negative auto-regulation loop might be involved in controlling body weight by maintaining homeostatic expression of this gene under diverse nutritional changes, especially in long term.
FTO is a demethylase preferentially targeting the m6A residues in RNA transcripts, which play a role in post-transcriptional regulation gene expression (Jia et al., 2011; Wang et al., 2014; Alarcon et al., 2015). FTO was implicated to be a co-activator that reactivates transcription from methyl-repressed genes (Wu et al., 2010). Intriguingly, the present study demonstrates that FTO binds to its own gene promoter and functions as a repressor (Figures 2 and 3). Given that the FTO occupies a DNA element encompassing the first TSS, its repressive function mimics that of bacterial repressors via interaction with operators reminiscent of, e.g. the lac operon (Weaver, 2011). The FTO-occupied promoter may exhibit not only attenuated assembly of the basal machinery but also impeded initiation from all three TSSs by productively assembled basal machinery (Figure 5). Hence, FTO is involved in the regulation of gene expression both at transcriptional and post-transcriptional levels that expand the one-protein-multiple function paradigm.
FTO is a Fe2+-dependent demethylase preferably for RNA molecules (Gerken et al., 2007; Jia et al., 2012b). The present study shows that Fe2+ also exhibits a capacity to increase Fto expression by relieving binding of FTO to its promoter (Figure 4), hinting a role of Fe2+ in fine-tuning FTO expression and functions. Fe2+ could be a messenger that keeps functions of FTO, as a demethylase and a transcriptional repressor, in check to realize homeostatic control. Given that FTO mutants H228L, D230G, and H304L exhibit altered binding to Fe2+ and deprived demethylase activity (Han et al., 2010), of which mutant D230G shows abolished sensitivity to Fe2+, the phenotypic behaviors of these mutants in vivo deserve further investigation.
Although the activities of a number of eukaryotic transcription factors are modulated by Fe2+, the regulation is largely by association with Fe2+-coordinated heme (e.g. the ecdysone receptor) (Arbeitman and Hogness, 2000); however, the function of FTO as a transcription repressor appears to be modulated by free Fe2+ but not Fe2+-coordinated heme (Figure 4). Similar modes have been reported in prokaryotes. For instance, a corynebacterial Fe2+-sensitive protein DtoxR auto-regulates its expression by binding to its own promoter (Fourel et al., 1989), and free Fe2+ represses mntH expression via facilitating association of a Fur-binding factor to the mntH promoter in bacterial cells (Kehres et al., 2002). To our knowledge, this study provides the first evidence that free Fe2+ plays a direct role in regulating gene expression in eukaryotic cells.
It has been reported that BHB could upregulate gene expression via histone modification, i.e. at chromosome level (Shimazu et al., 2013), and our data showed that Fe2+ affects FTO binding to the promoter, i.e. at protein level. Therefore, we speculate that both BHB and Fe2+ should play roles in regulating the Fto gene expression at different levels. Furthermore, the present study shows that the Fe2+ levels did not change under different nutrient conditions (HFD or fasting), suggesting that BHB might play a responsive role and Fe2+ might play a basal role in the transiently upregulation of FTO expression upon short-term high-fat-diet or fasting treatment.
Diverse studies indicated that the FTO gene is associated with obesity, but the biological role of FTO is poorly known. It has been reported that FTOI367F mice exhibit an altered metabolic profile and reduced fat mass (Church et al., 2009). As compared to the wild-type, this mutant shows altered crystal structure (Han et al., 2010), much increased affinity for the Fto promoter with enhanced repressive efficacies, and opposing sensitivity to Fe2+ (Figure 6), which deserves further investigation. The fact that the FTOI367F mutant represses FTO expression to a greater degree is in line with a lean phenotype in the FTOI367F mice, as well as in line with the observation that FTO downregulation protects animals from obesity (Fischer et al., 2009). The present study showed that the FTO levels significantly increased only in hypothalamus of the high-fat-diet-induced obese mice and fasting mice, but not in other brain tissues such as cerebellar cortex and cerebral cortex, suggesting a hypothalamic-specific disruption of the auto-regulatory loop of Fto gene (Figure 7 and Supplementary Figure S4). It is known that there is a blood–brain barrier (BBB) in the brain, and only the hypothalamus region lacks an intact BBB, thereby providing an environment for interplay between peripheral organs and the brain (Meister, 2007; Norsted et al., 2008). The structural difference between the hypothalamus and other brain tissues may be used to explain this phenomenon that KD or HFD diet only induces hypothalamic FTO expression. Furthermore, given that our EMSA data using the same probes and the nuclear extracts from different brain tissues showed that the FTO binding to the promoter only altered in hypothalamus (Figure 7C and Supplementary Figure S4B), and no correlation between the changes of BHB and Fe2+ levels and the alteration of FTO binding to the promoter were observed, we speculate that long-term high fat diets or fasting may lead to hypothalamic-specific FTO modifications that interrupt the auto-feedback loop of this gene. Future studies are needed to explore the underlying mechanism.
In the present study, HFD and KD were used to feed 3-week old (postweaning period) mice which are in the process of development, thus the two HFDs should affect the development of whole body including body size. These phenomena have been confirmed by several previous studies both in developmental and adult animals (Parente et al., 2008; Bor et al., 2017; Chakraborty et al., 2018; Lanham et al., 2018), suggesting that multiple factors or pathways may play roles in the increase of body length. It is known that childhood obesity is associated with a higher chance of premature death and disability in adulthood (Franks et al., 2010), and the variants in FTO gene are strongly associated with child obesity (Dina et al., 2007; Frayling et al., 2007; Cecil et al., 2008), thus we think that investigation of the Fto expression regulation at the developmental stage should have a potential clinical significance.
Given that loss of Fto in mice leads to postnatal growth retardation and a significant reduction in body length (Fischer et al., 2009), and FTO overexpression leads to an increase in body and fat mass (Church et al., 2010), we speculate that HFD- and KD-induced hypothalalmic increase of FTO may play a role in the development of body length. Taken together, this work may provide a link between the negative auto-regulation of the Fto gene and the hypothalamic control of body weight.
Materials and methods
Animals and diet administration
The C57BL/6 mice were provided by the Experimental Animal center of Sun Yat-sen University (Guangzhou, China). The animals were fed and reproduced at the Experimental Animal Center of Guangzhou Medical University in a 12 h-light/12 h-dark cycle with food and water provided ad libitum. This study was approved by The Ethical Committee for the use of experimental animals of Guangzhou Medical University.
For high-fat-diet treatment, 3-week-old male mice were randomly divided into three groups: two experimental groups freely fed with HFD of 60% fat, 20% carbohydrate, and 20% protein (Cat#: D12492, Research Diets) or KD of 89.5% fat, 0.1% carbohydrate, and 10.4% protein (Cat#: D12369B, Research Diets), and the control group fed with SD of 10% fat, 70% carbohydrate, and 20% protein (Cat#: D12450B, Research Diets), respectively. The mice were allowed free access to water and food.
For fasting treatment, 3-month-old male mice were randomly divided into two groups: the fasting group with only free access to water for 48 h and the control group allowed free access to water and SD.
Serum and hypothalamic BHB analyses
The BHB concentrations in serum and hypothalamus were measured as previously described (Leino et al., 2001; Inagaki et al., 2007). Briefly, animals from the distinct diet groups described above were anesthetized with 5% halothane. For serum assay, the bloods were collected from heart and deproteinized immediately using perchloric acid at 0°C. For hypothalamic assay, the samples were isolated and homogenized in PBS buffer (pH 7.4) by grinder. The samples were centrifuged and the clear supernatant was analyzed using β-Hydroxybutyrate Assay Kit (Abcam).
Hypothalamic Fe2+ assays
Animals were anesthetized with 5% halothane. The tissues were isolated and homogenized in 0.1 mol/L Tris-HCl buffer (pH 7.4) by grinder. The samples were then centrifuged and the supernatant analyzed using an automatic analyzer (7600 series, HITACHI) with Iron Testing Kit (H309W, Medical System).
Cell culture and BHB treatment
Mouse neuroblastoma N1E-115 cells and mouse embryonic fibroblast NIH-3T3 cells were obtained from ATCC (CRL 2263 for N1E-115 and CRL6361 for NIH-3T3) and maintained in DMEM (Gibco) with 10% Fetal Bovine Serum (Gibco) and HyClone Penicilllin–Streptomycin Solution (Fermantas) at 37°C in a 5% CO2 condition. For BHB treatment, cells were cultured in glucose-free DMEM with β-hydroxybutyric acid (Sigma).
Gene transcript and protein expression analyses
Total RNA samples were extracted from hypothalamus and cultured cells using GeneJET™ RNA Purification Kit (Fermantas) and was treated with DNase I (Qiagen). The first stand of cDNA was synthesized using 2 μg of RNA using RevertAid Premium First Strand cDNA Synthesis Kit (Fermantas) with oligo (dT). The RT-PCR was performed using Dream Taq Green PCR Master Mix 2× (Fermantas). Primer sequences were 5′-GCAGAGCAGCCTACAACGTGAC-3′ and 5′-CCAACATGCCAAGTATCAGGATCTC-3′ (for Fto), 5′-TTCAAGGATAAGGGCGACAGC-3′ and 5′-GGCTCTTGGTGTACTTGTTGCTG-3′ (for Foxo3a), 5′-CTCGCCATGGACCCCAACTG-3′ and 5′-AGCTGCACTTGTCGGAAGCCTC-3′ (for Mt2), 5′-GCAGCTTTACGATGTACAGCACC-3′ and 5′-CGAACTGGTTGTAGTCCGTGG-3′ (for Lcn2), and 5′-TGGTCGTCGACAACGGCTC-3′ and 5′-CCATGTCGTCCCAGTTGGTAAC-3′ (for β-actin). The qPCR was performed using SYBR® qPCR Mix Kit (ToYoBo) and Rotor-Gene™Q instrument (Qiagen). The relative cycle threshold (CT) values were normalized by β-actin.
For protein expression analysis, the mouse tissues, N1E-115 cells, or NIH-3T3 cells were lysed in RIPA lysis buffer (100 mm NaCl, 20 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0, 0.5% Triton X-100, 0.5% Nonidet P-40) and quantified using the BCA Protein Assay Kit (Pierce). The same amount of protein was separated on a 12% SDS-PAGE under a denaturing condition and was electroblotted to nitrocellulose membrane. After incubated with FTO antibody (Cat#: 98768, Santa Cruz Biotech.), β-actin antibody (Cat#: 60008-1-Ig, ProtenTech Group), GAPDH antibody (Cat#: 10494-1-AP, ProtenTech Group), PCNA antibody (Cat#: 10205-2-AP, ProtenTech Group), or anti-HA tag antibody (Cat#: ab9110, Abcam), the bands were exposed by enhanced chemiluminescence (Pierce) and X-ray film.
Protein digestion and purification
The expression of GST-FTO fusion protein in Escherichia coli BL21 (DE3) cells was induced by 0.5 mM IPTG for 3 h at 22°C. The prokaryotic extracts were purified using BugBuster®GST.BindTM Purification Kit (Novagen) according to the manufacturer’s protocol. The GST tag was removed using GST Fusion Protein Thrombin Digestion Kit (Baomanbio).
5′ Full RACE
The TSS of mouse Fto gene was determined using a 5′ Full RACE Kit (TaKaRa) according to the manufacturer’s instructions. The PCR products were cloned into pGEM®-T easy vector (Promega). Over 50 colonies for each transfection were confirmed by restriction enzyme digestion and sequencing.
Plasmid constructions and site-directed mutagenesis
Genomic DNA was extracted from mouse brain using a DNeasy Blood & Tissue Kit (Qiagen). The different-length genomic fragments upstream of the Fto translation initiation site were amplified by PCR using TaKaRa LA Taq Kit. The primer sequences for these PCR experiments are available upon request. The PCR products were cloned into the psiCHECK2 vector (Promega) to generate a serial of luciferase constructs, respectively.
To generate an FTO overexpression construct, Fto-coding sequence was amplified from the cDNA sample (described above) using TaKaRa LA Taq Kit. The primer pairs were 5′-TGGCTAGCATGAAGCGCGTCCAG-3′ and 5′-GCGGTCGACGGATCTTGCTTCCAGC-3′ for cloning into a eukaryotic vector pCMV-null (modified from pCMV-GFP, a vector from Addgene), and 5′-TTGAATTCATGAAGCGCGTCCAGAC-3′ and 5′-TTGAATTCATGAAGCGCGTCCAGAC-3′ for cloning into a prokaryotic vector pGEX-4T1 (Amersham) to express GST-FTO fusion protein. To generate an HA-FTO overexpression construct, Fto coding sequence was amplified from pCMV-FTO described above. The primer pairs were 5′-TGAAGCTTATGAAGCGCGTCCAG-3′ and 5′-GTCTCGAGGATCTTGCTTCCAGC-3′ for cloning into a eukaryotic vector pXJ4-HA (Biovector NTCC Inc.).
Mutations were carried out using KOD-Plus-Mutagenesis Kit (TOYOBO). The mutation primers were as follows: 5′-CTGGCTTCACGATGAGAACCTGGTGGAC-3′ and 5′-CTCACCGCCATCTTCCCCATGCC-3′ (for H228L), 5′-GGTGAGAACCTGGTGGACAGGTCAGCC-3′ and 5′-GTGATGCCAGCTCACCGCCATCTTC-3′ (for D230G), 5′-CTCTGT GTTTTGGCTGGCTCACAGCC-3′ and 5′-CTCACCGCCATCTTCCCCATGCC-3′ (for H304L), 5′-CTCAGTTTAGTTCCACTCACCGTGTGGCAG-3′ and 5′-GCTGTGAGCCAGCCAAAACACAGTGC-3′ (for R313Q), and 5′-AGAGGAATTCCATAATGAGGTGGAGTTTGAG-3′ and 5′-GCTGTGAGCCAGCCAAAACACAGTGC-3′ (for I367F).
Luciferase assays
Total 200 ng construct was transfected to the N1E-115 cells in 96-well plates. For co-transfection, 100 ng promoter report construct was used in each well with the other one (100 ng) or two (50 ng each) constructs (except for specially indication). Fe2+ was added to the medium before transfection. After 48-h transfection, the luciferase activities of all transfections were detected using a Dual-Luciferase® Reporter Assay System (Promega) as previously described (Zeng et al., 2014).
ChIP analyses
ChIP experiment was performed using Millipore EZ-Magna ChIP A/G Kit (Millipore) according to the manufacturer’s protocol and previous description (Dong et al., 2014). Briefly, hypothalamic tissue and cells were incubated with 37% formaldehyde to cross-link the protein and DNA, following by quenching formaldehyde with glycine. The chromatins of lysed cells or tissues were sheared by Nuclear Lysis Buffer and Ultrasonic Cell Disruptor (Scientz-II D). The sliced chromatin was immunoprecipitated with anti-FTO (LSBio), anti-TAF1 (Abcam), and IgG (as a negative control, Millipore). The immunoprecipitated DNA was used as a template for PCR to amplify the Fto promoter. The primers were as follows (also shown in Figure 3A): 5′-ACTACGCTAGCCCTGCTAGCTG-3′ and 5′-GCTGCTACTAAAGCCGCCTTC-3′ (for Fto promoter), 5′-CGTTGAGAGAACTACACACAGGAGG-3′ and 5′-CCTTGGCTTAACGCCGAGC-3′ (for Fto exon 1), and 5′-AAGATTATTGTGTGTGTTTTGTTCGAAG-3′ and 5′-TGCAATTTGTACCCTAGCAATTCC-3′ (for Fto exon 2). The PCR products were purified and confirmed by direct sequencing.
Electrophoretic mobility shift assay
EMSA was conducted using LightShift® Chemiluinescent EMSA Kit (Thermo) and the Gel Shift Assay System (Bio-Rad) according to the manufacturer’s instructions. Nuclear proteins were isolated from mouse hypothalamus or E. coli cells using NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo). The DNA double-strand oligonucleotide was synthesized with biotin labeled or without labeled at Sangon Biotech Co. The core promoter oligonucleotide 5′-CAGGAGGCGGGGTCCAGGGCGAGGG-3′, two random oligonucleotides, and a series of deletion or mutation oligonucleotides are shown in Figure 3D, I, and J, respectively. The identical unlabeled double-strand oligonucleotides with 6- to 60-fold excess of the labeled oligonucleotides were used for competition analysis. To determine the binding of FTO to the probes, supershift assay was performed using anti-FTO and anti-IgG (as a control), respectively. Addition of FeCl2, ZnCl2, CuCl2, 2-OG, or heme (Sigma) to the protein extracts was done at room temperature 20 min before loading in the gel. The detailed procedures of these experiments were described previously (Carrion et al., 1999; Dong et al., 2014).
Statistical analyses
All experimental data were analyzed using SPSS 13.0 software and all results were expressed as mean ± SEM. Statistical significance of differences among groups was determined by ANOVA and Student’s t-test. P-values <0.05 were considered as a statistical significance.
Supplementary Material
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by grants from the Innovative Academic Teams of Guangzhou Education System (1201610025), the National Natural Science Foundation of China (81671112), the Guangzhou Science and Technology Project (201804020046), and the Science and Technology Project of Guangdong Province (2017B090904036).
Conflict of interest
none declared.
References
- Alarcon C.R., Lee H., Goodarzi H., et al. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman M.N., and Hogness D.S. (2000). Molecular chaperones activate the Drosophila ecdysone receptor, an RXR heterodimer. Cell 101, 67–77. [DOI] [PubMed] [Google Scholar]
- Astrup A., Ryan L., Grunwald G.K., et al. (2000). The role of dietary fat in body fatness: evidence from a preliminary meta-analysis of ad libitum low-fat dietary intervention studies. Br. J. Nutr. 83(Suppl 1), S25–S32. [DOI] [PubMed] [Google Scholar]
- Bor A., Nishijo M., Nishimaru H., et al. (2017). Effects of high fat diet and perinatal dioxin exposure on development of body size and expression of platelet-derived growth factor receptor β in the rat brain. J. Integr. Neurosci. 16, 453–470. [DOI] [PubMed] [Google Scholar]
- Carrion A.M., Link W.A., Ledo F., et al. (1999). DREAM is a Ca2+-regulated transcriptional repressor. Nature 398, 80–84. [DOI] [PubMed] [Google Scholar]
- Cecil J.E., Tavendale R., Watt P., et al. (2008). An obesity-associated FTO gene variant and increased energy intake in children. N. Engl. J. Med. 359, 2558–2566. [DOI] [PubMed] [Google Scholar]
- Chakraborty T.R., Gomez V., Adhikari D., et al. (2018). The synergism in hormonal and cellular changes in male mice on long term high fat exposure. J. Am. Coll. Nutr. 37, 328–335. [DOI] [PubMed] [Google Scholar]
- Cheung M.K., Gulati P., O’Rahilly S., et al. (2013). FTO expression is regulated by availability of essential amino acids. Int. J. Obes. 37, 744–747. [DOI] [PubMed] [Google Scholar]
- Church C., Lee S., Bagg E.A., et al. (2009). A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 5, e1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C., Moir L., McMurray F., et al. (2010). Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C., Meyre D., Gallina S., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726. [DOI] [PubMed] [Google Scholar]
- Dong Z.F., Tang L.J., Deng G.F., et al. (2014). Transcription of the human sodium channel SCN1A gene is repressed by a scaffolding protein RACK1. Mol. Neurobiol. 50, 438–448. [DOI] [PubMed] [Google Scholar]
- Fischer J., Koch L., Emmerling C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature 458, 894–898. [DOI] [PubMed] [Google Scholar]
- Fourel G., Phalipon A., and Kaczorek M. (1989). Evidence for direct regulation of diphtheria toxin gene transcription by an Fe2+-dependent DNA-binding repressor, DtoxR, in Corynebacterium diphtheriae. Infect. Immun. 57, 3221–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks P.W., Hanson R.L., Knowler W.C., et al. (2010). Childhood obesity, other cardiovascular risk factors, and premature death. N. Engl. J. Med. 362, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling T.M., Timpson N.J., Weedon M.N., et al. (2007). A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R., Hagglund M., Olszewski P.K., et al. (2008). The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology 149, 2062–2071. [DOI] [PubMed] [Google Scholar]
- Fu Y., Dominissini D., Rechavi G., et al. (2014). Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 15, 293–306. [DOI] [PubMed] [Google Scholar]
- Gerken T., Girard C.A., Tung Y.C., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Niu T., Chang J., et al. (2010). Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 464, 1205–1209. [DOI] [PubMed] [Google Scholar]
- Inagaki T., Dutchak P., Zhao G., et al. (2007). Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metab. 5, 415–425. [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., and He C. (2012. a). Reversible RNA adenosine methylation in biological regulation. Trends Genet. 29, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., et al. (2012. b). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karra E., O’Daly O.G., Choudhury A.I., et al. (2013). A link between FTO, ghrelin, and impaired brain food-cue responsivity. J. Clin. Invest. 123, 3539–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres D.G., Janakiraman A., Slauch J.M., et al. (2002). ). Regulation of Salmonella enterica serovar typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J. Bacteriol. 184, 3151–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanham S.A., Cagampang F.R., and Oreffo R.O.C. (2018). The influence of a high fat diet on bone and soft tissue formation in Matrix Gla Protein knockout mice. Sci. Rep. 8, 3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Leino R.L., Gerhart D.Z., Duelli R., et al. (2001). Diet-induced ketosis increases monocarboxylate transporter (MCT1) levels in rat brain. Neurochem. Int. 38, 519–527. [DOI] [PubMed] [Google Scholar]
- McCrory M.A., Fuss P.J., McCallum J.E., et al. (1999). Dietary variety within food groups: association with energy intake and body fatness in men and women. Am. J. Clin. Nutr. 69, 440–447. [DOI] [PubMed] [Google Scholar]
- McTaggart J.S., Lee S., Iberl M., et al. (2011). FTO is expressed in neurones throughout the brain and its expression is unaltered by fasting. PLoS One 6, e27968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister B. (2007). Neurotransmitters in key neurons of the hypothalamus that regulate feeding behavior and body weight. Physiol. Behav. 92, 263–271. [DOI] [PubMed] [Google Scholar]
- Meyer K.D., Saletore Y., Zumbo P., et al. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T.M., Lew P.S., Luo Y., et al. (2017). Negative regulation of hepatic fat mass and obesity associated (Fto) gene expression by insulin. Life Sci. 170, 50–55. [DOI] [PubMed] [Google Scholar]
- Norsted E., Gomuc B., and Meister B. (2008). Protein components of the blood-brain barrier (BBB) in the mediobasal hypothalamus. J. Chem. Neuroanat. 36, 107–121. [DOI] [PubMed] [Google Scholar]
- Nowacka-Woszuk J., Pruszynska-Oszmalek E., Szydlowski M., et al. (2017). Nutrition modulates Fto and Irx3 gene transcript levels, but does not alter their DNA methylation profiles in rat white adipose tissues. Gene 610, 44–48. [DOI] [PubMed] [Google Scholar]
- Olszewski P.K., Fredriksson R., Olszewska A.M., et al. (2009). Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci. 10, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente L.B., Aguila M.B., and Mandarim-de-Lacerda C.A. (2008). Deleterious effects of high-fat diet on perinatal and postweaning periods in adult rat offspring. Clin. Nutr. 27, 623–634. [DOI] [PubMed] [Google Scholar]
- Poritsanos N.J., Lew P.S., Fischer J., et al. (2011). Impaired hypothalamic Fto expression in response to fasting and glucose in obese mice. Nutr. Diabetes 1, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R.G. (2003). Lasker Basic Medical Research Award. The eukaryotic transcriptional machinery: complexities and mechanisms unforeseen. Nat. Med. 9, 1239–1244. [DOI] [PubMed] [Google Scholar]
- Ronkainen J., Huusko T.J., Soininen R., et al. (2015). Fat mass- and obesity-associated gene Fto affects the dietary response in mouse white adipose tissue. Sci. Rep. 5, 9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L., and Andrade-Navarro M.A. (2007). The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A., Sanna S., Chen W.M., et al. (2007). Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T., Hirschey M.D., Newman J., et al. (2013). Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339, 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S., Tena J.J., Kim K.H., et al. (2014). Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratigopoulos G., Burnett L.C., Rausch R., et al. (2016). Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J. Clin. Invest. 126, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung Y.C., Ayuso E., Shan X., et al. (2010). Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS One 5, e8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujovic P., Stamenkovic S., Jasnic N., et al. (2013). Fasting induced cytoplasmic Fto expression in some neurons of rat hypothalamus. PLoS One 8, e63694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Yang F.J., Du H., et al. (2011). Involvement of leptin receptor long isoform (LepRb)-STAT3 signaling pathway in brain fat mass- and obesity-associated (FTO) downregulation during energy restriction. Mol. Med. 17, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R.F. (2011). Operons: fine control of bacterial transcription. In: Molecular Biology (5th edn). New York: McGraw-Hill, 167–195. [Google Scholar]
- Wu Q., Saunders R.A., Szkudlarek-Mikho M., et al. (2010). The obesity-associated Fto gene is a transcriptional coactivator. Biochem. Biophys. Res. Commun. 401, 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Loos R.J., Powell J.E., et al. (2012). FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T., Dong Z.F., Liu S.J., et al. (2014). A novel variant in the 3′ UTR of human SCN1A gene from a patient with Dravet syndrome decreases mRNA stability mediated by GAPDH’s binding. Hum. Genet. 133, 801–811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.