Abstract
Objective
This study investigated the role of microRNAs generated from adipose tissue macrophages (ATMs) during adipose tissue remodeling induced by pharmacological and nutritional stimuli.
Methods
Macrophage-specific Dicer knockout (KO) mice were used to determine the roles of microRNA generated in macrophages in adipose tissue remodeling induced by the β3-adrenergic receptor agonist CL316,243 (CL). RNA-seq was performed to characterize microRNA and mRNA expression profiles in isolated macrophages and PDGFRα+ adipocyte stem cells (ASCs). The role of miR-10a-5p was further investigated in cell culture, and in adipose tissue remodeling induced by CL treatment and high fat feeding.
Results
Macrophage-specific deletion of Dicer elevated pro-inflammatory gene expression and prevented CL-induced de novo beige adipogenesis in gonadal white adipose tissue (gWAT). Co-culture of ASCs with ATMs of wild type mice promoted brown adipocyte gene expression upon differentiation, but co-culture with ATMs of Dicer KO mice did not. Bioinformatic analysis of RNA expression profiles identified miR-10a-5p as a potential regulator of inflammation and differentiation in ATMs and ASCs, respectively. CL treatment increased levels of miR-10a-5p in ATMs and ASCs in gWAT. Interestingly, CL treatment elevated levels of pre-mir-10a in ATMs but not in ASCs, suggesting possible transfer from ATMs to ASCs. Elevating miR-10a-5p levels inhibited proinflammatory gene expression in cultured RAW 264.7 macrophages and promoted the differentiation of C3H10T1/2 cells into brown adipocytes. Furthermore, treatment with a miR-10a-5p mimic in vivo rescued CL-induced beige adipogenesis in Dicer KO mice. High fat feeding reduced miR-10a-5p levels in ATMs of gWAT, and treatment of mice with a miR-10a-5p mimic suppressed pro-inflammatory responses, promoted the appearance of new white adipocytes in gWAT, and improved systemic glucose tolerance.
Conclusions
These results demonstrate an important role of macrophage-generated microRNAs in adipogenic niches and identify miR-10a-5p as a key regulator that reduces adipose tissue inflammation and promotes therapeutic adipogenesis.
Keywords: Dicer, microRNA, Adipose tissue macrophages, Adipocyte stem cells, Adipogenic niches
Highlights
-
•
Macrophage-specific Dicer KO prevented CL-induced beige adipogenesis in gWAT.
-
•
Bioinfomatic analysis of RNAseq identified miR-10a-5p as a key player in AT remodeling.
-
•
CL treatment upregulated miR-10a-5p in AT macrophages and in activated ASCs.
-
•
miR-10a-5p promoted beige adipogenesis in vivo and in vitro, in part by targeting Rora.
-
•
miR-10a-5p treatment reduced HFD-induced inflammation and improved glucose tolerance.
1. Introduction
Adipose tissue macrophages (ATMs) are a major immune cell type in adipose tissue that are thought to play important roles in adipose tissue remodeling, yet the mechanisms involved are poorly understood [1]. The phenotypic character of ATMs can change dramatically in response to diverse nutritional, metabolic, environmental, and pharmacological stimuli [1], [2], [3], [4]. It is well known, for instance, that pro-inflammatory macrophages infiltrate adipose tissue of mice fed high fat diets, which leads to chronic low grade systemic inflammation that contributes to the pathogenesis of obesity-related metabolic syndrome [3], [5]. On the other hand, thermogenic stimuli, such as cold exposure and β3-adrenergic agonist treatment, trigger anti-inflammatory polarization and proliferation of ATMs that participate in tissue remodeling and beige adipogenesis [2], [6], [7], [8]. Interestingly, although both high fat diet feeding and β3-adrenergic receptor stimulation produce adipocyte death and associated-macrophage efferocytosis, β3-adrenergic receptor stimulation results in quick resolution along with the generation of beige adipocytes, whereas high fat diet feeding is associated with a persistent inflammatory state [2], [3], [5].
Presently, little is known about the mechanisms that regulate macrophage phenotypes in the context of adipose tissue remodeling. However, our previous work with remodeling induced by β3-adrenergic receptor agonist CL316,243 (CL) treatment demonstrated that beige adipogenesis requires the recruitment of ATMs to adipogenic niches and their close interactions with adipocyte stem cells (ASCs) [2], [6], [8]. Nonetheless, the mechanisms that regulate ATM polarization and the impact it has on beige adipogenesis remain incompletely understood.
MicroRNAs (miRNAs) are small noncoding RNAs that generally repress gene expression by binding to the 3′ untranslated region (UTR) of targeted mRNAs, leading to their degradation [9]. Adipose tissue miRNAs have been identified as regulatory players that can contribute to adipose tissue inflammation, macrophage polarization, and metabolism [10], [11], [12], [13]. However, the role of ATM-derived miRNAs during adipose tissue remodeling has not been investigated.
Here, we investigated the roles of miRNAs generated from ATMs in CL-induced tissue remodeling of mouse gonadal WAT (gWAT). Using macrophage-specific knockout of Dicer (DicerCsf1rKO), we found that generation of miRNAs in macrophages is necessary for the anti-inflammatory polarization of macrophages and proliferation/differentiation of platelet-derived growth factor receptor alpha (PDGFRα)+ ASCs into brown adipocytes during β3-adrenergic receptor agonist treatment. Characterization of the macrophage miRNA signatures identified miR-10a-5p as a key miRNA that facilitates anti-inflammatory polarization of macrophages, suppresses RAR-Related Orphan Receptor alpha (Rora) expression and promotes beige adipogenesis. Treatment of DicerCsf1rKO mice with a miR-10a-5p mimic suppressed inflammatory signaling and restored brown adipogenesis in the CL model. Furthermore, high fat feeding reduced miR-10-5p levels in gWAT, and treatment with the miR-10-5p mimic suppressed pro-inflammatory gene expression, facilitated tissue remodeling and improved glucose tolerance, indicating the therapeutic potential of miR-10a-5p mimics.
2. Material and methods
2.1. Animals
All animal protocols were approved by the Institutional Animal Care and Use Committees at Yonsei University and Seoul National University. Mice were fed a standard chow diet and were housed at 22 °C and maintained on a 12-h light/12-h dark cycle with free access to food and water at all times. C57BL/6 mice were purchased from Central Lab. Animal Inc. Csf1r-CreER [14] (stock# 019098), Pdgfra-CreER (stock# 018280), Dicerflox/flox [15] (stock# 006366), Rosa-LSL-tdTomato (stock# 007909), and Rosa-mT/mG (stock # 007676) mice were purchased from the Jackson Laboratory. Csf1r-CreER mice and Dicerflox/flox mice were crossed to produce inducible macrophage-specific Dicer (Csf1r-CreER/Dicer flox/flox) knockout mice. Male mice were used for all experiments. For wild type (WT) control, Dicer floxed mice (WT/Dicerflox/flox) without CreER were used. Csf1r-CreER mice were crossed with Rosa-mT/mG to test efficiency and specificity of Cre recombinase expression in macrophages. Pdgfra-CreER mice were crossed with Rosa-LSL-tdTomato to trace PDGFRα+ cells during CL treatment and high fat diet feeding, as described previously [6]. For Cre recombination, double transgenic mice and WT controls were treated with tamoxifen dissolved in sunflower oil (Sigma, 75 mg/kg) by oral gavage on each of 5 consecutive days. Also, we included vehicle (oil)-treated control groups to confirm specific induction of Cre recombinase activity upon tamoxifen treatment in adipose tissue. Experiments were started 10 days after the last dose of tamoxifen. For β3 adrenergic receptor stimulation, mice were treated with CL316,243 (Sigma, St. Louis, MO) (1 mg/kg/day) by intraperitoneal injection for up to 5 days. For the high fat diet (HFD) experiments, 60% fat diet (D12492, Central Lab. Animal Inc.) was introduced at 7 weeks of age and continued for 8 weeks. For EdU labeling of proliferating cells, mice were injected with EdU (Thermo Fisher, 0.4 μmol/mouse) at indicated times described in the text. Energy expenditure was measured using an indirect calorimetry system (PhenoMaster, TSE system, Bad Homburg, Germany), as described previously [16]. For in vivo overexpression of miRNA, mice were injected with miR-10a-5p mimics (2′-Fluoro dsRNA, Genepharma) or microRNA mimics double strand negative control (Genepharma) in liposomal formulation using In Vivo Jet-PEI (Polyplus Transfection) according to the manufacturer's instructions. Briefly, 40 μg of miR-10a-5p mimics and 6 μl of in vivo-jet PEI reagent were used in a final volume of 200 μl of 5% glucose solution per injection. Mice were treated with miRNA liposomal solution (200 μl/20 g mouse, i.p.) at two week intervals during HFD feeding.
Total adipocyte numbers in eWAT of mice from the HFD study were calculated as previously described [17]. Briefly, eWAT adipocyte cell diameters (>200/mouse) were determined from 100× images of H/E stained paraffin sections, and mean triglyceride (TG) mass/adipocyte was calculated as 0.4790/106 × (3 × σ2 × mean diameter + [mean diameter]3). Tissue adipocyte cellularity was determined by dividing tissue TG content by mean fat cell TG mass.
2.2. WAT stromovascular cell fractionation and flow cytometry
Stromovascular cell (SVC) fractions from mouse gWAT were isolated, as previously described [6]. For EdU detection, fixed SVCs were processed for Click-it reaction first, followed by cell-surface marker staining. Antibodies used for flow cytometry analysis were the following: anti-PDGFRα-APC (Biolegend, cat # 135907) for ASCs, CD44-FITC (Biolegend, cat # 103021), and F4/80-APC (Biolegend, cat # 123115) for ATMs. Analytic cytometry was performed using BD LSR III (BD Biosciences) flow cytometers. Raw data were processed using FlowJo software (Tree Star). For gene expression analyses by qPCR or RNAseq, ATMs and ASCs were isolated by magnetic cell sorting (MACS) with anti-F4/80-FITC/anti-FITC-microbeads and anti-PDGFRα-PE/anti-PE-microbeads, respectively (Miltenyi Biotech).
2.3. Quantitative PCR analysis, western blot, and RNAseq
Quantitative PCR (qPCR) analyses, western blot, and RNAseq analyses were performed as previously described [6], [18] (see supplemental materials).
2.4. Cell culture
For miRNA overexpression, Raw 267.4 cells (ATCC) or C3H10T1/2 cells (ATCC) were plated at a density of 2.5 × 104 cells/well in 24 well-plates and transfected with 10 nM syn-mmu-miR-10a-5p miScript miRNA mimic or a negative control miRNA (Qiagen) using INTERFERin (Polyplus Transfection). Two days after transfection, cells were subjected to qPCR analysis or were treated with adipogenic induction medium DEME supplemented with 10% FBS, indomethacin (0.125 mM, Cayman), isobutylmethylxanthine (IBMX) (2.5 mM, Cayman), dexamethasone (1 μM, Cayman), insulin (10 μg/mL, Sigma) and triiodothyronine (T3, 1 nM, Cayman), as previously described [19]. For co-culture using trans-well plates, MACS-isolated PDGFRα+ ASCs from gWAT were placed in the lower chamber (1 × 105cells/well) and co-cultured with MACS-isolated F4/80 + ATMs from gWAT placed in the upper chamber (1 × 105 cells/well) using a trans-well plate (0.4 μm polycarbonate filter, Corning) for 24 h. For adipogenic differentiation, co-cultures were treated with adipogenic induction medium for 3 days. (See supplemental materials for plasmid construction and luciferase reporter assays.)
2.5. Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA.). Data are presented as mean ± standard errors of the means (SEMs). Statistical significance between two groups was determined by unpaired t-test. Comparisons among multiple groups were performed using a one-way or two-way analysis of variance (ANOVA), with Bonferroni post hoc tests to determine p values. Principal Components Analysis (PCA) was performed with XLStat software (Addinsoft, New York, NY) Heatmaps were generated by PermutMatrix program, as previously described [18].
3. Results
3.1. Recruitment of ATMs during β3-adrenergic receptor stimulation is specific to gWAT
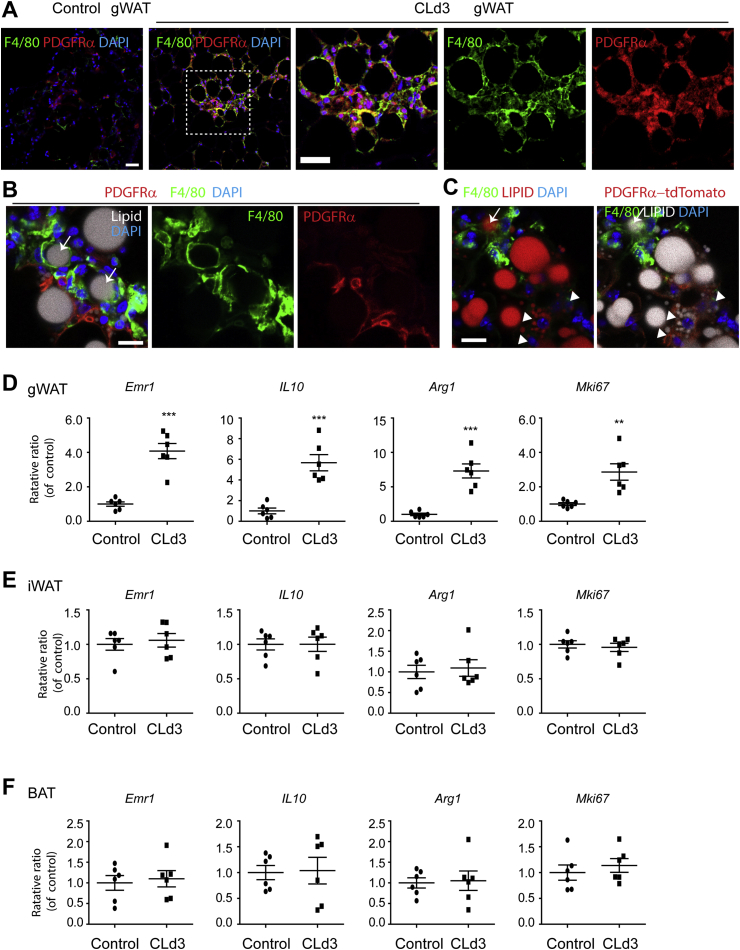
Treatment of mice with CL remodels gWAT in a process that involves white adipocyte death and removal by anti-inflammatory ATMs, and proliferation and differentiation of PDGFRα+ ASCs at the sites of adipocyte efferocytosis (i.e., “crown-like structures”) [2]. Consistent with the previous reports, three days of CL treatment recruited ATMs that were closely associated with PDGFRα+ ASCs in gWAT (Figure 1A and B). In addition, the ATMs formed tightly-associated networks that surrounded lipid+ adipocytes (indicated by arrows in Figure 1B and C). These ATM clusters create an adipogenic niche where ASCs proliferate and differentiate into adipocytes [2]. As shown in Figure 1C, lineage tracing using PDGFRα reporter mice (Pdgfra-CreER/Rosa26-LSL-tdTomato) demonstrated the generation of multilocular adipocytes from PDGFRα+ ASCs near ATM clusters (indicated by arrow-heads in Figure 1C). This close co-localization suggested a cellular interplay between the ASCs and ATMs in adipogenic niches. Next, we examined expression levels of a pan-macrophage marker (Emr1), anti-inflammatory markers (IL10, Arg1), and a proliferation marker (mKi67) in gWAT, iWAT, and BAT. qPCR analysis confirmed that recruitment of anti-inflammatory ATMs by CL treatment was specific to gWAT (Figure 1D–F) [2], [8], [20], further supporting gWAT-specific roles of ATMs in CL-induced adipose tissue remodeling.
Figure 1.
CL treatment recruited macrophages that were closely associated with PDGFRα+ ASCs in gWAT. A. Immunostaining for F4/80 and PDGFRα in paraffin sections obtained from mice treated with CL for 3 days and vehicle control mice. B–C. Immunostaining of F4/80, neutral lipid staining, and Pdgfra-tdTomato fluorescence in cryosections obtained from tamoxifen induced PdgfraCreER/Rosa-LSL-tdTomato mice treated with CL for 3 days arrows indicated lipid+ adipocytes that were surrounded by F4/80+ macrophages. Arrow heads indicate newly generated multilocular adipocytes that were positive for PDGFRα+ reporter (tdTomato+). Nuclei were counterstained with DAPI. (size of bar = 20 μm) D–E. qPCR analysis of gWAT, iWAT, and BAT of mice treated with CL for 3 days and vehicle control mice (n = 6 per condition, mean ± S.E.M, **p < 0.01, ***p < 0.001).
3.2. Macrophage-specific KO of Dicer increases pro-inflammatory ATM polarization and reduces beige adipogenesis of PDGFRα+ ASCs in gonadal white adipose tissue
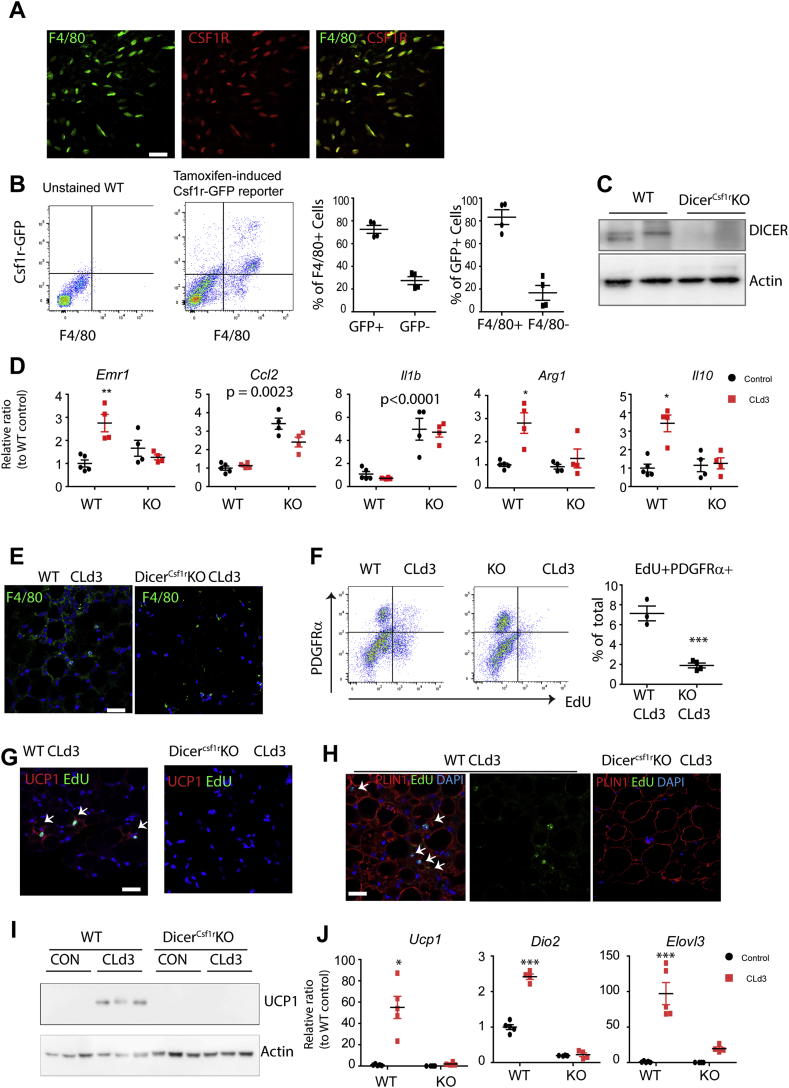
miRNAs are known to be important regulators of macrophage function that act in part by regulating macrophage polarization [21]. Our previous work demonstrated that CL treatment alters the polarization state of ATMs to an anti-inflammatory, pro-restorative phenotype [2], [8], suggesting that ATM-derived miRNAs could play a role in CL-induced adipose tissue remodeling. To investigate this possibility, we deleted Dicer, an essential gene for RNA processing, from macrophages by crossing mice harboring floxed Dicer loci with those expressing tamoxifen responsive CreER under the control of the colony stimulating factor 1 receptor (Csf1r) promoter (DicerCsf1rKO) [14]. We chose this drug-inducible conditional KO system to avoid possible chronic or developmental effects of Dicer deletion. Tamoxifen-treated floxed Dicer mice lacking Csf1r-CreER served as WT controls for deletion and tamoxifen treatment. Macrophage-specific expression of Csf1r in gWAT was confirmed by double label immunofluorescence staining of Csf1r and F4/80 (Figure 2A). In addition, we also used double transgenic mice that expressed CreER under Csf1r promoter control and a Cre-responsive GFP reporter (Csrf1-CreERT2/Rosa26_mTomato-loxP-stop-loxP-eGFP). As shown in Figure 2B, flow cytometry analysis indicated Csf1r-GFP reporter expression in 72.5 ± 0.03% of F4/80 + macrophages, and F4/80 expression in 83.3 ± 0.07% of Csf1r-GFP reporter expressing cells, confirming that the majority of F4/80 + macrophages expressed Cre recombinase under Csf1r promoter control in gWAT.
Figure 2.
Macrophage-specific deletion of Dicer led to pro-inflammatory gene expression of gWAT and prevented CL-induced beige adipogenesis from PDGFRα+ ASCs in gWAT. A. Immunostaining of F4/80 and CSF1R in cryosection of gWAT. (size of bar = 20 μm) B. Flow cytometry analysis of F4/80 and Csf1r-GFP reporter double positive cells and quantification (mean ± S.E.M, n = 4). Tamoxifen-induced Csf1r-GFP reporter mice were used to confirm specificity of Csf1r expression in F4/80+ cells. C. Immunoblot analysis of Dicer1 expression in isolated F4/80+ cells from gWAT of tamoxifen-induced Csf1r-CreER/Dicer1-floxed mice (DicerCsf1rKO: KO) and wild type control mice (WT). D. qPCR analysis of genes related to macrophage polarization (mean ± S.E.M, Bonferroni posttests, n = 4–5 per group, *p < 0.05, **p < 0.01; two-way ANOVA test: p values indicate significance of Dicer KO effects). E. F4/80 staining in paraffin sections of gWAT of WT and KO mice treated with CL for 3 days. F. Flow cytometry analysis of EdU+ PDGFRα+ cells in gWAT of tamoxifen-induced Csf1r-CreER/Dicer-floxed mice (DicerCsf1rKO) and WT control mice after 3 days of CL treatment. (mean ± S.E.M, n = 3, t-test, ***p < 0.001) G–H. EdU and UCP1 (G) or Perilipin (Plin1, H) double staining in paraffin sections of gWAT of WT and KO mice treated with CL for 3 days. I. Immunoblot analysis of UCP1 expression in gWAT of WT and KO mice treated with CL for 3 days. J. qPCR analysis of Ucp1, Dio2 and Elovl3 expression in gWAT of WT and KO mice treated with CL for 3 days (mean ± S.E.M, Bonferroni posttests, n = 4–5 per group, *p < 0.05, **p < 0.01, ***p < 0.001).
Next, we examined the effect of Dicer KO in adipose tissue remodeling during CL treatment. Macrophage-specific KO of Dicer was confirmed by immunoblot analysis of Dicer expression in magnetic cell sorting (MACS)-isolated F4/80 + macrophages from gWAT of DicerCsf1rKO and WT mice after tamoxifen treatment (Figure 2C). Interestingly, Dicer KO increased pro-inflammatory gene expression in gWAT of both control and CL-treated mice (Figure 2D). CL treatment did not induce anti-inflammatory marker expression in Dicer KO mice or formation of crown-like structures (Figure 2D and E). In contrast, macrophage-specific Dicer KO did not affect pro-inflammatory marker expression in inguinal white adipose tissue (iWAT) or brown adipose tissue (BAT) (Figs. S1A and S1B).
CL-induced proliferation of ASCs in gWAT of DicerCsf1rKO mice was greatly reduced compared to controls, confirmed by FACS analysis of EdU incorporation (Figure 2F). In addition, the number of EdU+UCP1+ adipocytes (Figure 2G) and EdU+Plin1+ adipocytes (Figure 2H) was reduced in DicerCsf1rKO mice, indicating impairment of de novo beige adipogenesis. Immunoblot analysis of UCP1 (Figure 2I) and qPCR analysis of brown adipocyte markers (Ucp1, iodothyronine deiodinase 2 (Dio2), elongation of very long chain fatty acids (Elovl3)) confirmed that macrophage-specific deletion of Dicer greatly impaired beige adipogenesis in gWAT (Figure 2J). The effects of macrophage-specific Dicer KO on CL-induced browning were specific to gWAT, as UCP1 induction by CL treatment was similar in iWAT and BAT of WT and Dicer KO mice (Figs. S1C and S1D). In addition, indirect calorimetry indicated that Dicer KO did not affect whole body energy expenditure levels under basal conditions or after CL injection (Figs. S2A and S2B). However, we noted that Dicer KO slightly elevated respiratory exchange ratio (RER) after CL treatment (Figs. S2C and S2D), suggesting a relative reduction in fatty acid oxidation. Collectively, these data indicate that biosynthesis of miRNAs in macrophages is required for anti-inflammatory macrophage recruitment and in vivo brown/beige adipogenesis in gWAT.
3.3. Global miRNA profiling identifies miR-10a-5p as a key microRNA differentially regulated in ATMs from gWAT of mice treated with CL
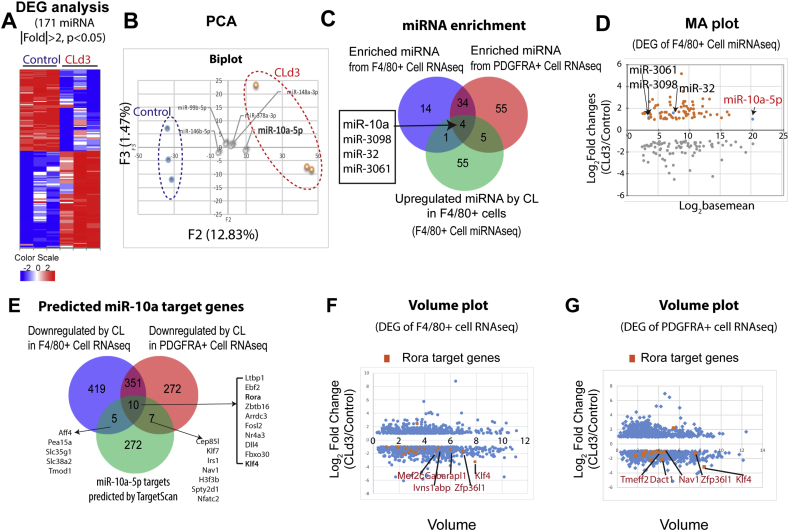
To determine whether CL treatment regulates ATM miRNAs, we performed RNA-seq analysis of gWAT ATMs from control mice and mice treated with CL for 3 days and identified 171 differentially regulated miRNAs (Figure 3A). Principal component analysis (PCA) indicated that controls and CL-treated groups were mainly segregated by Factor 2 (Figure 3B, Tables S1 and S2). Top five miRNAs that had the highest contributions to Factor 2 were miR-10a-5p, miR-146b-5p, miR-378a-3p, 99b-5p and 148a-3p (Figure 3B). miR-10a-5p, whose expression was increased by 2-fold by CL treatment, was the greatest contributor to Factor 2 (68%). Additionally, the MA plot indicated that the expression level of miR-10a-5p was the highest among miRNAs that were differentially regulated in macrophages by CL treatment (Figure 3D).
Figure 3.
Transcriptomic profiling predicted mir-10a-5p to be a major upstream regulator of down-regulated genes in macrophages and PDGFRα+ cells isolated from gWAT of mice treated with CL316,243 for 3 days. A. Heat map of 171 miRNAs differentially regulated in F4/80+ cells isolated from gWAT of mice treated with CL for 3 days compared to vehicle-treated control conditions from micro RNA sequencing data (|fold change| ≥ 2, p < 0.05). B. PCA analysis of miRNA of control F4/80 + and CLd3 F4/80+ cells. F2 and F3 of PCA components are shown. Distinct two clusters of Control and CLd3 were separated by F2. The biplot indicates factor scores of miRNAs with top 5 contribution (miR-10a-5p, miR-146b-5p, miR-378a-3p, miR-99b-5p, and miR-148a-3p). C. Venn diagram of enriched miRNAs in F4/80+ cells and PDGFRα+ cells, and downregulated miRNAs in F4/80 + cells. D. MA-plot of miRNAs that were differentially regulated in F4/80 + cells by 3 days of CL treatment (4 overlapping miRNAs are indicated in the plot as blue dots) (|fold change| ≥ 2, p < 0.05). X axis: log2 (Basemean), Y axis: log2 (Fold Change). E. Venn diagram of downregulated genes by CL in F4/80+ cells and PDGFRα+ cells, and predicted miR-10a-5p targets. F-G. Volume plots of differentially expressed genes from RNAseq of F4/80+ cells (F) and PDGFRα+ cells (G) (|fold change|>2, p < 0.05). Predicted Rora target genes are indicated.
Analysis of potential targets of these miRNAs by miRbase [22], indicated that one putative target gene of miR-10a-5p was RAR-related orphan receptor alpha (Rora), which has been shown to promote pro-inflammatory responses and suppress adipogenesis and brown adipogenic pathways [23], [24] (Table S4). miR-146-5p displayed the second highest contribution to factor 2 (22%), which was decreased 4-fold in the CL-treated groups. miR-378a-3p was upregulated 3-fold and was predicted to suppress the expression of Pde1b [25], which could elevate cAMP levels. miR-143-3p, which has also been reported to be anti-adipogenic [26], was downregulated 2.5-fold, and miR-155-5p, which targets adipogenic transcription factor CCAAT/enhancer-binding protein beta (Cebpb) [27], [28], was downregulated 18-fold.
3.4. Transcriptomic profiles predicts that miR-10a-5p represses remodeling-related mRNA levels in macrophages and PDGFRα+ ASCs isolated from gWAT
We next performed mRNA profiling of MACS-isolated F4/80+ macrophages and PDGFRα+ progenitors from gWAT to identify genes regulated by CL treatment and determine whether those downregulated might be targets of the miRNAs identified above. (Fig. S3). miRNA target enrichment analysis of downregulated genes (∣fold change∣ > 2, p < 0.05) in both F4/80 + cells and PDGFRα+ cells using TargetScan algorithm [29], [30], [31], [32] (Figs. S4A and 4B) identified 4 upregulated miRNAs (i.e. miR-10a, miR-3098, miR-32, miR-3061) in F4/80 + cells (∣fold change∣ >2, p < 0.05) that overlapped with the TargetScan-predicted miRNAs in both cell types (Figure 3C). Of these candidate miRNAs, miR-10a-5p expression levels were highest (Figure 3D).
Next, we examined the expression levels of predicted targets of miR-10a-5p in mRNA sequencing data of ATMs and ASCs isolated from gWAT [22] (Figure 3E and Table S4). Importantly, 10 predicted targets of the miR-10a-5p were significantly downregulated in ATMs and ASCs after CL treatment (Figure 3E). CL-downregulated miR-10a target genes included anti-adipogenic molecules, such as Rora [23], [24], Delta-like 4 (Dll4) [33], and nuclear receptor subfamily 4, group A, member 3 (Nr4a3) [34], as confirmed by qPCR (Fig. S4C). Rora is a candidate miR-10a-5p target transcription factor [35] that regulates multiple genes in a coordinated manner and has been reported to suppress adipogenesis [36], [37]. Several Rora target genes (predicted by TRANSFAC Curated Transcription Factor Targets [32]) were down-regulated by CL treatment in both F4/80+ cells and PDGFRα+ cells (Figure 3F and G and Fig. S4D-S4G) including Klf4, Arntl, Zfp36l1, Fbxo30, Tmeff2, and Spock2.
3.5. miR-10a-5p is upregulated in PDGFRα+CD44 + adipocyte progenitors in gWAT of mice treated with CL
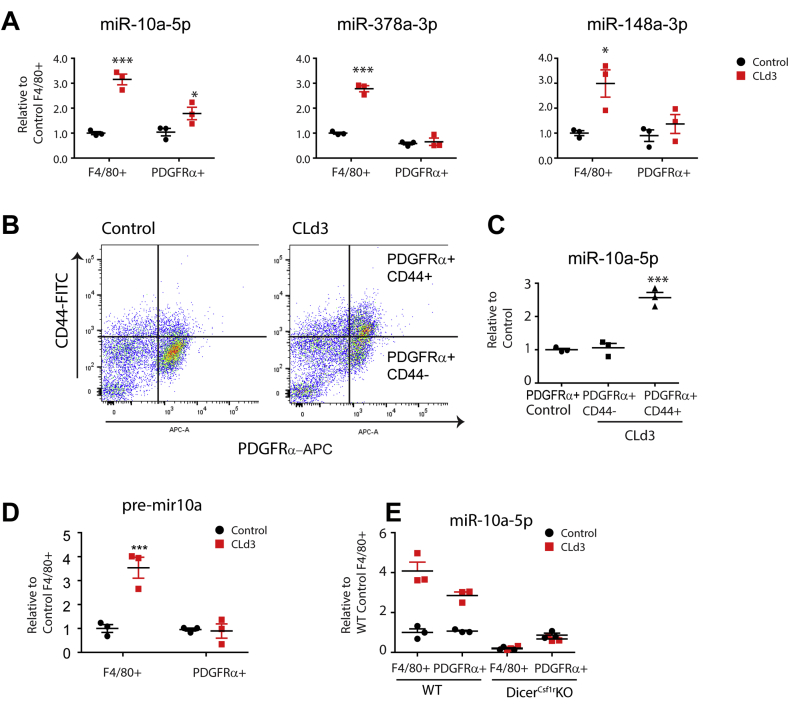
To confirm the miRNA sequencing data, we performed qPCR analysis of magnetic cell sorting (MACS)-isolated F4/80+ ATMs and PDGFRα+ ASCs (Figure 4A). CL treatment upregulated expression of miR-10a-5p, miR-378a-3p and miR-148a-3p in isolated macrophages. Interestingly, miR-10a-5p levels were also elevated in PDGFRα+ ASCs after CL treatment (Figure 4A). We previously identified CD44+ PDGFRα+ cells as a subpopulation of ASCs that closely associates with ATMs in crown-like structures, where they proliferate and differentiate into brown adipocytes during CL treatment [2]. Significantly, we found that miR-10a-5p levels were selectively increased in the activated CD44+PDGFRα+ ASC subpopulation (Figure 4B and C).
Figure 4.
CL316,243 treatment increased miR-10a-5p expression in macrophages and adipocytes progenitors in gWAT. A. qPCR analysis of miRNAs in F4/80+ ATMs and PDGFRα+ ASCs isolated from gWAT of mice treated with CL for 3 days and controls (mean ± S.E.M, stromovascular cells (SVCs) were pooled from 4 mice per sample and biological triplicates (n = 3) per condition were analyzed for qPCR. *p < 0.05, ***p < 0.001). B. Flow cytometry analysis CD44 and PDGFRα expression in SVCs obtained from gWAT of mice treated with CL for 3 days and control mice. C. qPCR analysis of miR-10a-5p expression in PDGFRα+CD44 + cells compared to PDGFRα+CD44-cells. D. qPCR analysis of pre-mir10a expression in F4/80+ macrophages and PDGFRα+ ASCs isolated from gWAT of mice treated with CL for 3 days and controls. E. qPCR analysis of miR-10a-5p expression in F4/80+ macrophages and PDGFRα+ ASCs obtained from gWAT of DicerCsf1rKO and WT mice treated with CL (mean ± S.E.M, Bonferroni posttests, n = 3 per group, *p < 0.05, **p < 0.01, ***p < 0.001).
To determine the cellular source of miR-10a-5p biosynthesis in the adipogenic niches, we analyzed expression levels of the precursor form of mir-10a (pre-mir10a) in MACS isolated macrophages and PDGFRα+ ASCs. Interestingly, only F4/80 + cells increased pre-mir10a expression after CL treatment, suggesting that miRNAs might have been transferred to ASCs from macrophages (Figure 4D). In addition, macrophage-specific Dicer KO suppressed CL-induction of miR-10a-5p expression in PDGFRα+ cells (Figure 4E), indicating that miRNA generation in macrophages was required for upregulation of miR-10a-5p in PDGFRα+ cells.
3.6. miR-10a-5p regulates anti-inflammatory polarization of ATMs and beige adipogenic differentiation
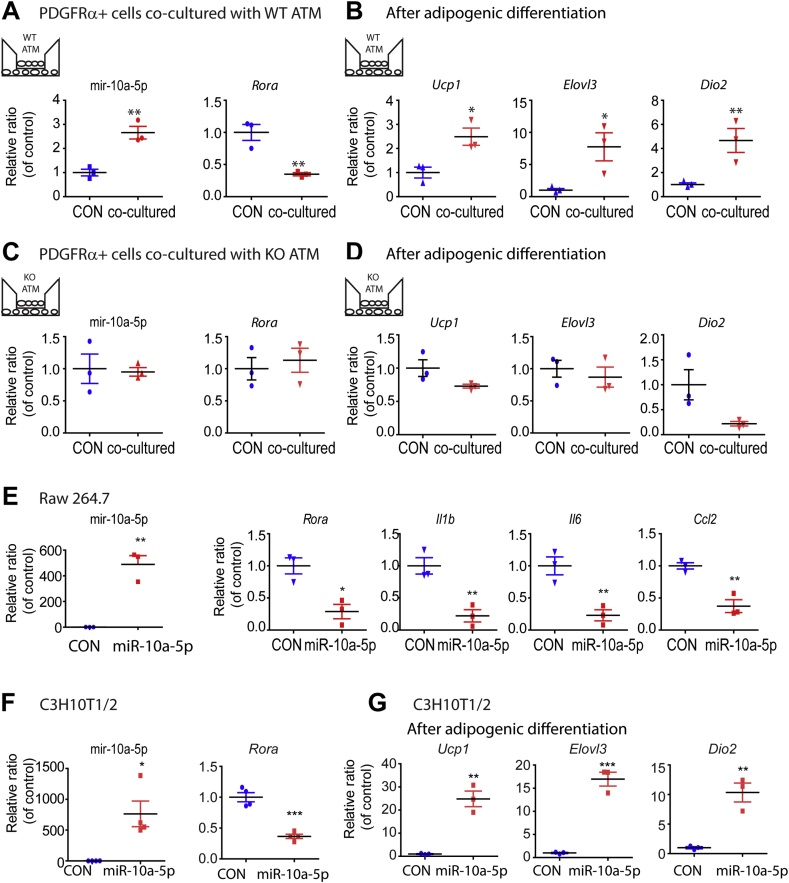
The above data suggest that macrophage-derived miR-10a-5p plays a key role in macrophage polarization and beige adipogenesis. To explore this possibility in a defined system, we examined the effects of co-culturing WT or Dicer KO ATMs isolated from gWAT of CL-treated mice on adipogenic gene expression of ASCs. As expected, co-culture with WT ATMs increased levels of miR-10a-5p in PDGFRα+ ASCs and this was associated with suppression of Rora and induction of beige adipocyte marker expression (Figure 5A and B). Each of these effects was absent in co-cultures of Dicer KO ATMs and ASCs (Figure 5C and D).
Figure 5.
In vitro overexpression of miR-10a-5p reduced Rora mRNA and inhibited inflammatory responses in ATMs and facilitated beige adipogenesis of ASCs. A,B. Co-culture of PDGFRα+ ASCs with F4/80+ ATMs obtained from gWAT of CL-treated WT mice and qPCR analysis of miR-10a-5p, Rora and beige adipocyte marker gene expression in co-cultured PDGFRα+ ASCs. C,D. Co-culture of PDGFRα+ ASCs with F4/80+ ATMs obtained from gWAT of CL-treated macrophage specific Dicer KO mice and qPCR analysis of miR-10a-5p, Rora and beige adipocyte marker gene expression in co-cultured PDGFRα+ ASCs. E. qPCR analysis of miR-10a-5p, Rora, pro-inflammatory gene expression in Raw264.7 cells transfected with miR-10a-5p and negative controls. F. qPCR analysis of miR-10a-5p and Rora expression in C3H10T1/2 cells transfected with miR-10a-5p and negative controls. G. qPCR analysis of beige adipocyte marker expression in C3H10T1/2 cells transfected with miR-10a-5p and negative controls, and then treated with adipogenic induction media for 3 days (mean ± S.E.M, t-test, n = 3 per group, **p < 0.01, ***p < 0.001).
To dissect the specific role of miR-10a-5p in macrophage polarization and adipogenesis, we turned to cell culture models employing Raw 264.7 and C3H10T1/2 cells, respectively. As shown in Figure 5E, overexpression of miR-10a-5p suppressed inflammatory gene expression in Raw 264.7 cells. Furthermore, miR-10a-5p suppressed expression of Rora in C3H10T1/2 cells and strongly promoted expression of the brown adipocyte markers Ucp1, Elovl3, and Dio2 in differentiated adipocytes (Figure 5F and G).
3.7. Rora mRNA is target of miR10-a-5p and overexpression of Rora blocks adipogenesis in CH310T1/3 cells
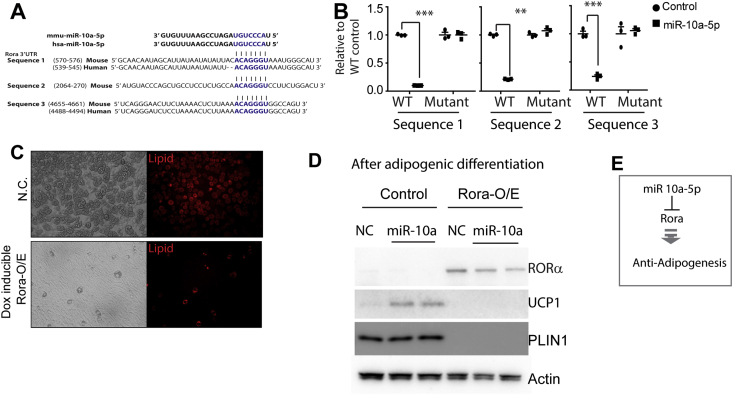
Bioinformatic analysis above predicted that Rora, a transcription factor that might oppose adipogenesis [36], [37], is a target of miR10-a-5p (Figure 3F and G). To determine whether miR-10a-5p targets Rora mRNA, we identified 3 highly conserved putative target sequences in the 3′ UTR of Rora were identified (Figure 6A). To test the whether miR10a-5p targets Rora mRNA, we cloned the 3 putative sequences and corresponding mutants into a luciferase reporter and co-transfected cells with the reporter and a miR-10a-5p mimic. As shown in Figure 6B, expression of miR-10a-5p repressed expression of the reporter gene containing WT, but not mutant, Rora 3′ UTR.
Figure 6.
Overexpression of Rora in C3H10T1/2 cells inhibits adipogenesis. A. Putative miR-10a-5p binding site in Rora 3′UTR. B. Luciferase reporter assay of Rora mRNA 3′UTR. (mean ± S.E.M, Bonferroni posttests, n = 3 per group, **p < 0.01, ***p < 0.001) C. Lipid staining (BODIPY). C3H10T1/2 cells were infected and cultured with or without doxycycline (Dox, 2 μg/ml) during the course of differentiation. D. Western blot analysis. C3H10T1/2 were infected with doxycycline (dox) inducible pINDUCER21-RORA using lentivirus infection, and then cells were cultured with/without dox. miR-10a-5p mimics were treated before adipogenic differentiation. E. Anti-adipogenic role of Rora.
Next we used an inducible lentiviral system to investigate the effect of Rora overexpression on adipogenic differentiation of C3H10T1/2 cells [38]. Consistent with previous reports [36], [37], Rora overexpression suppressed differentiation as indicated by reduced lipid accumulation (Figure 6C) and adipocyte marker expression (i.e., Plin1) (Figure 6D). In addition, treatment with a miR-10a mimic before induction of differentiation upregulated UCP1 protein expression, and this effect was abrogated by Rora overexpression (Figure 6D) owing to suppression of adipogenesis.
3.8. In vivo treatment with miR-10a-5p mimic reduces inflammatory response and restores CL-induced beige adipogenesis in gWAT of macrophage-specific Dicer KO mice
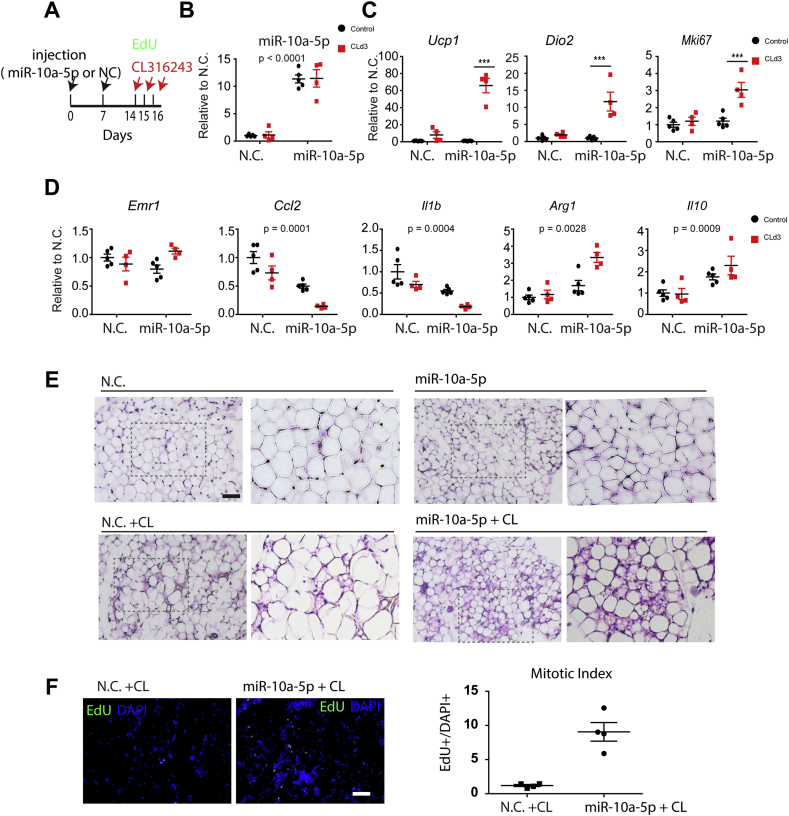
To test in vivo function of miR-10a-5p, macrophage-specific Dicer KO mice were injected with liposomes containing miR-10a-5p mimics (Figure 7A). qPCR analysis confirmed increased levels of miR-10a-5p in gWAT (Figure 7B). miR-10a-5p treatment restored CL-induced beige adipogenesis, as indicated by upregulation of Ucp1, Dio2, and Mki67 expression (Figure 7C) and increased mitotic index (EdU+ cells/DAPI+ cells) (Figure 7F) after 3 days of CL treatment. In addition, miR-10a-5p treatment reduced inflammatory gene expression (i.e., Ccl2, Il1b) in controls and CL-treated mice (Figure 7D) and increased the appearance of multilocular adipocytes (Figure 7E). The results indicate that miR-10a reduces pro-inflammatory signaling in adipogenic niches and promotes beige adipogenesis.
Figure 7.
miR-10a-5p reduced inflammation and restored CL-induced beige adipogenesis in gWAT of macrophage specific Dicer KO mice. A. miR-10a-5p mimic and CL injection B-D. qPCR analysis of miR-10a-5p, Ucp1, Dio2, Mki67 and pro-/anti-inflammatory gene expression in gWAT of DicerCsf1rKO mice injected with miR-10a-5p mimics liposome and CL for 3 days (negative controls: N.C., mean ± S.E.M, Bonferroni posttests, n = 4–5 per group,***p < 0.001; two-way ANOVA: p values indicate significance of miR-10a-5p treatment effects). E. Representative images of H/E staining of paraffin sections of gWAT. F. EdU staining of paraffin sections of gWAT and quantification (size of bar = 40 μm).
3.9. Treatment with miR-10a-5p facilitates adipose tissue remodeling and improves HFD-induced glucose intolerance
As mentioned, high fat feeding, like CL treatment, results in adipocyte death and replacement, yet unlike CL treatment, HFD produces persistent inflammation that can disrupt systemic glucose homeostasis. The results with CL-induced remodeling prompted us to investigate whether miR-10a-5p plays a role in adipose tissue inflammation and remodeling during high fat feeding.
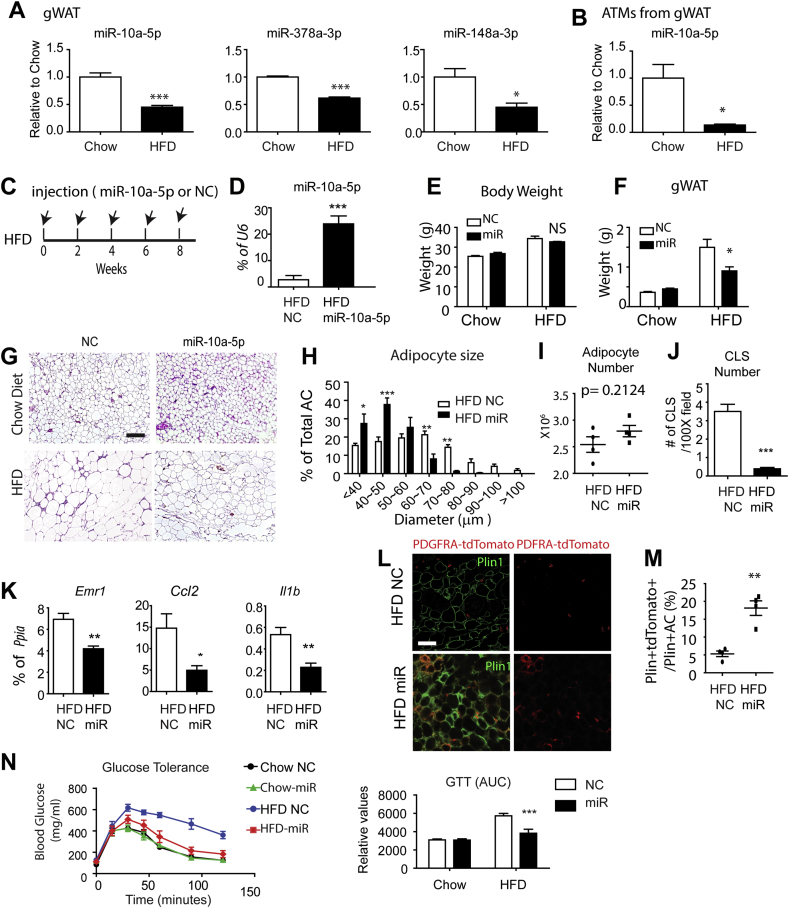
Eight weeks of high fat feeding significantly suppressed levels of miR-10a-5p expression in gWAT and ATMs isolated from gWAT compared to chow-fed control mice (Figure 8A and B). Other major miRNAs (miR378a-3p, mir-148a-3p) that were upregulated by CL treatment were also down-regulated in gWAT by high fat feeding (Figure 8A).
Figure 8.
miR-10a-5p treatment increased de novo adipogenesis in gWAT and prevented HFD-induced glucose intolerance. A. qPCR analysis of miRNAs in gWAT of mice fed chow or high fat diet for 8 weeks (mean ± S.E.M, t-tests, n = 4 per group,*p < 0.05, ***p < 0.001). B. qPCR analysis of miR-10a-5p in ATM isolated from gWAT of mice fed chow or high fat diet for 8 weeks. Stromovascular cells were pooled from 4 mice per sample and biological triplicates (n = 3) per condition were analyzed for qPCR. (mean ± S.E.M, t-tests, *p < 0.05). C. miR-10a-5p mimic injection and HFD feeding. D. qPCR analysis of miRNA in gWAT of mice fed with HFD for 8 weeks and treated with miR-10a-5p mimic or negative controls (NC). E. Body weight. F. Fat mass analysis. G. H/E staining of gWAT. H. Adipocyte size analysis of H/E stained paraffin sections of gWAT. I. Adipose cellularity analysis. J. Crown-like structure (CLS) analysis of H/E stained paraffin sections of gWAT. CLS were defined as circular regions with continuous rim of myeloid cells. K. qPCR analysis of gWAT of mice fed with HFD for 8 weeks and treated with miR-10a-5p mimics or NC. L. Plin1 and tdTomato staining in paraffin sections of gWAT of tamoxifen-induced Pdgfra-CreER/tdTomato reporter mice fed with HFD and treated with miR-10a-5p mimics or NC. M. Quantification of Plin1+tdTomato + adipocytes (n = 4, t-test, **p < 0.01) N. Glucose tolerance test and quantification. (negative controls: N.C., mean ± S.E.M, Bonferroni posttests, n = 4–5 per group,***p < 0.001; two-way ANOVA: p values indicate significance of miR-10a-5p treatment effects).
Biweekly treatment with miR-10a-5p mimic (Figure 8C) increased levels of miR-10a-5p (Figure 8D). miR-10a-5p treatment did not significantly alter body weight (Figure 8E), but reduced gWAT mass in high fat-fed mice (Figure 8F). The reduction in fat mass was accompanied by reduced adipocyte size (Figure 8G and H) and greatly reduced number of crown-like structures (Figure 8J), which are indicative of cell death and inefficient efferocytosis [3]. Consistent with the histological observations, we found that miR-10a-5p treatment reduced proinflammatory gene expression of in gWAT of high fat fed mice (Figure 8K).
To determine the impact of miR-10a-5p treatment on diet-induced adipogenesis, we traced the appearance of new fat cells from progenitors using tamoxifen-induced Pdgfra-CreER/tdTomato mice during 8 weeks of HFD feeding. Surprisingly, we observed that miR-10a-5p treatment significantly upregulated de novo adipogenesis, as indicated by the 3–4 fold increase in the appearance of PLIN1+tdTomato+ adipocytes (Figure 8L and M).
We also assessed the impact of miR-10a-5p treatment on glucose tolerance. As expected, high fat feeding greatly impaired glucose tolerance compared to chow-fed controls (Figure 8N). Biweekly treatment with miR-10a-5p did not affect glucose tolerance of chow-fed mice, but strongly improved tolerance in mice fed high fat diet (Figure 8N). Together, these observations indicate that miR-10a-5p treatment promotes adipose tissue remodeling by reducing persistent inflammation and promoting new fat cell formation, and these effects are correlated with improvements in systemic glucose metabolism.
4. Discussion
ATMs have been recognized as key regulators that support healthy adipose tissue remodeling [1], [2], [8], [39], [40]. By using macrophage-specific deletion of Dicer in a robust model of de novo adipogenesis, our results demonstrate that generation of miRNAs in ATMs is required for both anti-inflammatory activation of ATMs and induction of beige adipogenesis in vivo.
miRNA profiling identified miR-10a-5p as a key miRNA that is upregulated in ATMs of CL-treated mice. Indeed, in vitro overexpression of miR-10a-5p effectively suppressed pro-inflammatory polarization of ATMs and promoted brown/beige differentiation of adipocyte precursor cells. Interestingly, co-culture with ATMs of WT, not Dicer-KO mice, increased miR-10a-5-p levels in ASCs, indicating mediation by a Dicer-dependent factor from ATMs. Notably, co-culture with WT ATMs raised miR-10a-5p levels in ASCs without increasing levels of pre-miR-10a-5p. While speculative, these observations suggest that miRNAs derived from ATMs might be transferred to ACSs to facilitate adipogenic differentiation, perhaps via exosomal transfer [35], [41]. In this regard, we have previously demonstrated the close contact of ATMs and ASCs in adipogenic niches, and functional interactions involving release of chemotactic ligands and generation PPARγ ligands that facilitate ASC recruitment and differentiation [42].
The effects of miR-10a-5p on adipogenic remodeling likely involve suppressing expression of gene network, and while the current study did not conclusively identify the all relevant targets of miR-10a, we demonstrate that Rora, a transcriptional inhibitor of adipogenesis [36], [37], is a direct target of miR-10a-5p whose suppression is required for brown adipogenesis in vitro. Physiologic roles of Rora expression in adipose tissue are not well understood. Mice with global inactivation of Rora (i.e, staggerer mice) are lean and exhibit enhanced browning of WAT [23], [43]; however, these mice exhibit strong neurological defects, and it is presently unclear whether this enhanced thermogenesis reflects the direct actions of RORA in adipocytes. In this regard, the beneficial roles of miR-10a-5p expression in adipose tissue remodeling should be interpreted in the context where multiple targets of miR-10a-5p are coordinately regulated. Other putative targets of miR-10a-5p that have been implicated in suppression of adipogenesis include Nr4a3 [34] and Dll4 [33]. In addition to miR-10a-5p, CL treatment upregulated other miRNAs that have been previously linked to beige/brown adipogenesis, such as miR-378a [25].
miRNA biosynthesis is known to be involved in monocyte differentiation and efficient phagocytosis [44]. The current study indicates that miR-10a-5p treatment might have therapeutic potential by promoting ATM phenotypes that resolve inflammation and restore beneficial adipocyte phenotypes. As mentioned, both high fat feeding and CL treatment result in adipocyte cell death, yet in the case of CL treatment, dead cells are rapidly removed (within 2–3 days) and replaced with beneficial beige adipocytes. By contrast, cell death induced by high fat feeding results in persistent inflammation, including hallmarks reminiscent of ‘frustrated phagocytosis’ [3], [45]. Treatment with miR-10a-5p reduced the levels of inflammation and numbers of crown-like structures in visceral WAT. The ability to reduce levels of crown-like structures while promoting adipogenesis strongly indicates that miR-10a-5p enhances tissue restoration and repair. During preparation of this manuscript, miR-10a-5p was reported to prevent atherosclerosis by altering macrophage polarization and activating mitochondrial oxidative lipid metabolism in macrophage subtypes that give rise to foam cells [46]. Similarities between the ATMs that comprise the crown-like structures and foam cells have been noted previously [47]. Moreover, recent work from our labs [2], [8] and others [48] have demonstrated the importance of the subpopulation of lipid-handling macrophages in healthy adipose tissue remodeling. The current study extends these concepts by demonstrating the importance of miRNAs in ATMs, and specifically the role of miR-10a in ATMs and ASCs in suppressing inflammation and promoting healthy tissue remodeling. Future studies will be required to determine the clinical relevance of miR-10a function in visceral adipose tissue of humans.
In summary, our results clearly demonstrate the importance of ATM miRNAs in adipose tissue cellular plasticity and suggest that miR-10a-5p mimics might serve as a new approach to support healthy adipose tissue remodeling and improving glucose homeostasis.
Author contributions
YHL and JGG conceived and designed the study. SNK, YHS, YKC, SYA, HDS, AS, JHP, YSJ, and YHL conducted the experiments. YHL, YKC, YHS, and SNK performed FACS analysis, and cell isolation for RNA sequencing, qPCR and western blot. YKC, YHS, HDS, and AS performed Dicer KO mouse experiment including liposome injection, and lineage tracing mouse experiment. YKC and YHS performed in vitro overexpression experiments and co-culture experiments. YHS performed luciferase assay. YHL, YKC, and JGG wrote the manuscript. All authors reviewed the manuscript.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) (2014R1A6A3A04056472 (Y.H.L.), 2019R1C1C1002014 (Y.H.L), 2018R1A5A2024425), the Bio & Medical Technology Development Program of the NRF (2016M3A9D5A01953818 (Y.H.L.), 2013M3A9D5072550), and the United States Public Health Service (NIH R01 DK-062292 to J.G.G.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.08.015.
Contributor Information
James G. Granneman, Email: jgranne@med.wayne.edu.
Yun-Hee Lee, Email: yunhee.lee@snu.ac.kr.
Conflict of interest
The authors declare no conflicts of interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hill A.A., Reid Bolus W., Hasty A.H. A decade of progress in adipose tissue macrophage biology. Immunological Reviews. 2014;262(1):134–152. doi: 10.1111/imr.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y.H., Petkova A.P., Granneman J.G. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metabolism. 2013;18(3):355–367. doi: 10.1016/j.cmet.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. Journal of Clinical Investigation. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen K.D., Qiu Y., Cui X., Goh Y.P., Mwangi J., David T. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480(7375):104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lumeng C.N., Saltiel A.R. Inflammatory links between obesity and metabolic disease. Journal of Clinical Investigation. 2011;121(6):2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y.H., Petkova A.P., Mottillo E.P., Granneman J.G. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metabolism. 2012;15(4):480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui X., Gu P., Zhang J., Nie T., Pan Y., Wu D. Adiponectin enhances cold-induced browning of subcutaneous adipose tissue via promoting M2 macrophage proliferation. Cell Metabolism. 2015;22(2):279–290. doi: 10.1016/j.cmet.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Burl R.B., Ramseyer V.D., Rondini E.A., Pique-Regi R., Lee Y.H., Granneman J.G. Deconstructing adipogenesis induced by beta3-adrenergic receptor activation with single-cell expression profiling. Cell Metabolism. 2018;28(2):300–309. doi: 10.1016/j.cmet.2018.05.025. e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausser J., Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nature Reviews Genetics. 2014;15(9):599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 10.Arner P., Kulyte A. MicroRNA regulatory networks in human adipose tissue and obesity. Nature Reviews Endocrinology. 2015;11(5):276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 11.Shamsi F., Zhang H., Tseng Y.H. MicroRNA regulation of Brown adipogenesis and thermogenic energy expenditure. Frontiers in Endocrinology. 2017;8:205. doi: 10.3389/fendo.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rottiers V., Naar A.M. MicroRNAs in metabolism and metabolic disorders. Nature Reviews Molecular Cell Biology. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arner E., Mejhert N., Kulyté A., Balwierz P.J., Pachkov M., Cormont M. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61(8):1986–1993. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S.N., Jung Y.S., Kwon H.J., Seong J.K., Granneman J.G., Lee Y.H. Sex differences in sympathetic innervation and browning of white adipose tissue of mice. Biology of Sex Differences. 2016;7:67. doi: 10.1186/s13293-016-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch J., Gallian E. Methods for the determination of adipose cell size in man and animals. The Journal of Lipid Research. 1968;9(1):110–119. [PubMed] [Google Scholar]
- 18.Lee Y.H., Kim S.N., Kwon H.J., Granneman J.G. Metabolic heterogeneity of activated beige/brite adipocytes in inguinal adipose tissue. Scientific Reports. 2017;7:39794. doi: 10.1038/srep39794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S.N., Kwon H.J., Akindehin S., Jeong H.W., Lee Y.H. Effects of epigallocatechin-3-gallate on autophagic lipolysis in adipocytes. Nutrients. 2017;9(7) doi: 10.3390/nu9070680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y.H., Petkova A.P., Konkar A.A., Granneman J.G. Cellular origins of cold-induced brown adipocytes in adult mice. The FASEB Journal. 2015;29(1):286–299. doi: 10.1096/fj.14-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Jiang T., Li M.Q., Zheng X.L., Zhao G.J. Transcriptional regulation of macrophages polarization by MicroRNAs. Frontiers in Immunology. 2018;9:1175. doi: 10.3389/fimmu.2018.01175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research. 2011;39(Suppl. 1):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau P., Tuong Z.K., Wang S.C., Fitzsimmons R.L., Goode J.M., Thomas G.P. Roralpha deficiency and decreased adiposity are associated with induction of thermogenic gene expression in subcutaneous white adipose and brown adipose tissue. American Journal of Physiology. Endocrinology and Metabolism. 2015;308(2):E159–E171. doi: 10.1152/ajpendo.00056.2014. [DOI] [PubMed] [Google Scholar]
- 24.Tuong Z.K., Fitzsimmons R., Wang S.M., Oh T.G., Lau P., Steyn F. Transgenic adipose-specific expression of the nuclear receptor RORα drives a striking shift in fat distribution and impairs glycemic control. EBioMedicine. 2016;11:101–117. doi: 10.1016/j.ebiom.2016.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan D., Mao C., Quattrochi B., Friedline R.H., Zhu L.J., Jung D.Y. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nature Communications. 2014;5:4725. doi: 10.1038/ncomms5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esau C., Kang X., Peralta E., Hanson E., Marcusson E.G., Ravichandran L.V. MicroRNA-143 regulates adipocyte differentiation. Journal of Biological Chemistry. 2004;279(50):52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y., Siegel F., Kipschull S., Haas B., Frohlich H., Meister G. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nature Communications. 2013;4:1769. doi: 10.1038/ncomms2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaudet A.D., Fonken L.K., Gushchina L.V., Aubrecht T.G., Maurya S.K., Periasamy M. miR-155 deletion in female mice prevents diet-induced obesity. Scientific Reports. 2016;6:22862. doi: 10.1038/srep22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinfeld I., Navon R., Ach R., Yakhini Z. miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic Acids Research. 2013;41(3) doi: 10.1093/nar/gks1142. e45-e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bi P., Shan T., Liu W., Yue F., Yang X., Liang X.R. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nature Medicine. 2014;20(8):911–918. doi: 10.1038/nm.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao L.C., Bensinger S.J., Villanueva C.J., Wroblewski K., Tontonoz P. Inhibition of adipocyte differentiation by Nur77, Nurr1, and Nor1. Molecular Endocrinology. 2008;22(12):2596–2608. doi: 10.1210/me.2008-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X., Qiu W., Liu Q., Qian M., Wang S., Zhang Z. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene. 2018;37(31):4239–4259. doi: 10.1038/s41388-018-0261-9. [DOI] [PubMed] [Google Scholar]
- 36.Duez H., Duhem C., Laitinen S., Patole P.S., Abdelkarim M., Bois-Joyeux B. Inhibition of adipocyte differentiation by RORalpha. FEBS Letters. 2009;583(12):2031–2036. doi: 10.1016/j.febslet.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 37.Ohoka N., Kato S., Takahashi Y., Hayashi H., Sato R. The orphan nuclear receptor RORalpha restrains adipocyte differentiation through a reduction of C/EBPbeta activity and perilipin gene expression. Molecular Endocrinology. 2009;23(6):759–771. doi: 10.1210/me.2008-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doulatov S., Vo L.T., Chou S.S., Kim P.G., Arora N., Li H. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13(4):459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coats B.R., Schoenfelt K.Q., Barbosa-Lorenzi V.C., Peris E., Cui C., Hoffman A. Metabolically activated adipose tissue macrophages perform detrimental and beneficial functions during diet-induced obesity. Cell Reports. 2017;20(13):3149–3161. doi: 10.1016/j.celrep.2017.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolf Y., Boura-Halfon S., Cortese N., Haimon Z., Sar Shalom H., Kuperman Y. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nature Immunology. 2017;18(6):665–674. doi: 10.1038/ni.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171(2):372–384. doi: 10.1016/j.cell.2017.08.035. e312. [DOI] [PubMed] [Google Scholar]
- 42.Lee Y.H., Kim S.N., Kwon H.J., Maddipati K.R., Granneman J.G. Adipogenic role of alternatively activated macrophages in beta-adrenergic remodeling of white adipose tissue. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2016;310(1):R55–R65. doi: 10.1152/ajpregu.00355.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monnier C., Auclair M., Le Cam G., Garcia M.P., Antoine B. The nuclear retinoid-related orphan receptor RORα controls circadian thermogenic programming in white fat depots. Physics Reports. 2018;6(8) doi: 10.14814/phy2.13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Self-Fordham J.B., Naqvi A.R., Uttamani J.R., Kulkarni V., Nares S. MicroRNA: dynamic regulators of macrophage polarization and plasticity. Frontiers in Immunology. 2017;8:1062. doi: 10.3389/fimmu.2017.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haka A.S., Barbosa-Lorenzi V.C., Lee H.J., Falcone D.J., Hudis C.A., Dannenberg A.J. Exocytosis of macrophage lysosomes leads to digestion of apoptotic adipocytes and foam cell formation. The Journal of Lipid Research. 2016;57(6):980–992. doi: 10.1194/jlr.M064089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y., Corbalan-Campos J., Gurung R., Natarelli L., Zhu M., Exner N. Dicer in macrophages prevents atherosclerosis by promoting mitochondrial oxidative metabolism. Circulation. 2018;138(18):2007–2020. doi: 10.1161/CIRCULATIONAHA.117.031589. [DOI] [PubMed] [Google Scholar]
- 47.Russo L., Lumeng C.N. Properties and functions of adipose tissue macrophages in obesity. Immunology. 2018;155(4):407–417. doi: 10.1111/imm.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaitin D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178(3):686–698. doi: 10.1016/j.cell.2019.05.054. e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.