Abstract
Endothelial NO synthase (eNOS) expression is regulated by a number of transcriptional and post-transcriptional mechanisms, but the effects of competing endogenous RNAs (ceRNAs) on eNOS mRNA and the underlying mechanisms are still unknown. Our bioinformatic analysis revealed three highly expressed eNOS-targeting miRNAs (miR-15b, miR-16, and miR-30b) in human endothelial cells (ECs). Among the 1103 mRNA targets of these three miRNAs, 15 mRNAs share a common disease association with eNOS. Gene expression and correlation analysis in patients with cardiovascular diseases identified insulin receptor substrate 2 (IRS2) as the most correlated eNOS-ceRNA. The expression levels of eNOS and IRS2 were coincidentally increased by application of laminar shear but reduced with eNOS or IRS2 siRNA transfection in human ECs, which was impeded by Dicer siRNA treatment. Moreover, luciferase reporter assay showed that these three miRNAs directly target the 3′UTR of eNOS and IRS2. Overexpression of these three miRNAs decreased, whereas inhibition of them increased, both mRNA and protein levels of eNOS and IRS2. Functionally, silencing eNOS suppressed the Akt signal pathway, while IRS2 knockdown reduced NO production in ECs. Thus, we identified eNOS and IRS2 as ceRNAs and revealed a novel mechanism explaining the coincidence of metabolic and cardiovascular diseases.
Keywords: eNOS, ceRNA, miRNAs, IRS2, endothelial dysfunction
Introduction
The endothelium is a selective permeable barrier between the bloodstream and the outer vascular wall (Antonetti et al., 1998; Mehta and Malik, 2006). Endothelial dysfunction is considered the initial phase of the development of cardiovascular diseases (CVDs) (Cines et al., 1998; Suwaidi et al., 2000). In disease conditions, including the presence of cardiovascular risk factors, the endothelium undergoes functional and structural alterations, thus losing its protective roles in maintaining vessel homeostasis (Cines et al., 1998; Versari et al., 2009a).
Endothelial NO synthase (eNOS) is involved in the synthesis of NO, one of the most important protective mediators in vessels, by relaxing media smooth muscle, inhibiting leukocyte adhesion and migration into the arterial wall, smooth muscle cell proliferation, platelet adhesion and aggregation, and adhesion molecule expression (Sneddon and Vane, 1988; Kubes et al., 1991). In the early stage of CVDs, endothelial dysfunction is mainly caused by impaired eNOS activity and subsequent diminished NO release (Sena et al., 2013). Therapeutic strategies reversing endothelial dysfunction by enhancing the release of NO from the endothelium could be beneficial for multiple conditions. Although numerous studies have concentrated on the transcriptional and post-translational regulation of eNOS, including the activity of the eNOS promoter, changes in eNOS mRNA expression, amino acid phosphorylation, and interactions with other proteins, several recent studies have reported that eNOS is also regulated by other mechanisms such as mRNA stability (Brownlee, 2001; Balligand et al., 2009). Therefore, its regulation mechanisms are extremely complex and still not fully understood, which requires more attention to explore these other novel regulators.
MicroRNAs (miRNAs), identified as a class of conserved 19- to 25-nt non-coding RNAs, regulate gene expression at the post-transcriptional level (Kim, 2005). miRNAs are largely known as negative regulators of gene expression by binding with the miRNA response elements (MREs) in target transcripts (Salmena et al., 2011). Many studies of endothelial cells (ECs) have demonstrated miRNAs as key regulators of endothelial function. For instance, miR-10a expression was found decreased in athero-susceptible regions of the aorta and suppressed a pro-inflammatory EC phenotype by regulating the expression of inflammatory factors (Fang et al., 2010). Similarly, downregulation of Let-7g led to endothelial activation and subsequent vessel injury (Liao et al., 2014). miR-320a plays an important role in mediating the effects of metastasis-associated lung adenocarcinoma transcript 1 on human EC proliferation by suppressing FOXM1 expression (Sun et al., 2017). In addition, miR-155 regulates endothelial function by increasing autophagic activity via inhibiting Rho b-mediated mTOR-P70S6 kinase-4EBP signaling (Lv et al., 2017).
miRNAs are able to regulate the abundance of target genes, which can crosstalk with each other by competing for their shared miRNAs (Tay et al., 2014). The crosstalk transcripts are termed as ‘competitive endogenous RNAs’ (ceRNAs) (Salmena et al., 2011). CeRNAs reveal a new mechanism of the interaction of RNAs and represent a new gene expression regulation mode. Many validated ceRNA pairs participate in the initiation and progression of cancers; the diagnosis, therapy, and prognosis of cancers have been investigated by systemic ceRNA network analysis (Hayes et al., 2014; Xu et al., 2015). For example, in gastric cancer, the long non-coding RNA (lncRNA) HOTAIR functions as a ceRNA to regulate HER2 expression by sponging miR-331-3p, which may contribute to a better understanding of gastric cancer pathogenesis and facilitate the development of lncRNA-directed diagnostics and therapeutics against this disease (Liu et al., 2014). However, the ceRNAs in CVDs have been rarely investigated. Recently, Let-7e was found to promote the inflammatory responses of ECs by activating the NF-κB pathway via the crosstalk between Lnc-MKI67IP3, Let-7e, and its target IκBβ (Lin et al., 2017). As a critical regulator of EC function, ceRNA-mediated eNOS regulation in CVDs has not been reported.
In the present study, we predicted the ceRNAs for eNOS and identified insulin receptor substrate 2 (IRS2) as a major miRNA-mediated ceRNA in ECs and human arteries. In ECs, the expression of eNOS and IRS2 regulating each other depended on miRNAs via ceRNA crosstalk. Thus, our study provides a new regulation mode of eNOS and uncovers a possible explanation of the initiation of CVDs.
Results
Bioinformatic analysis of eNOS ceRNAs
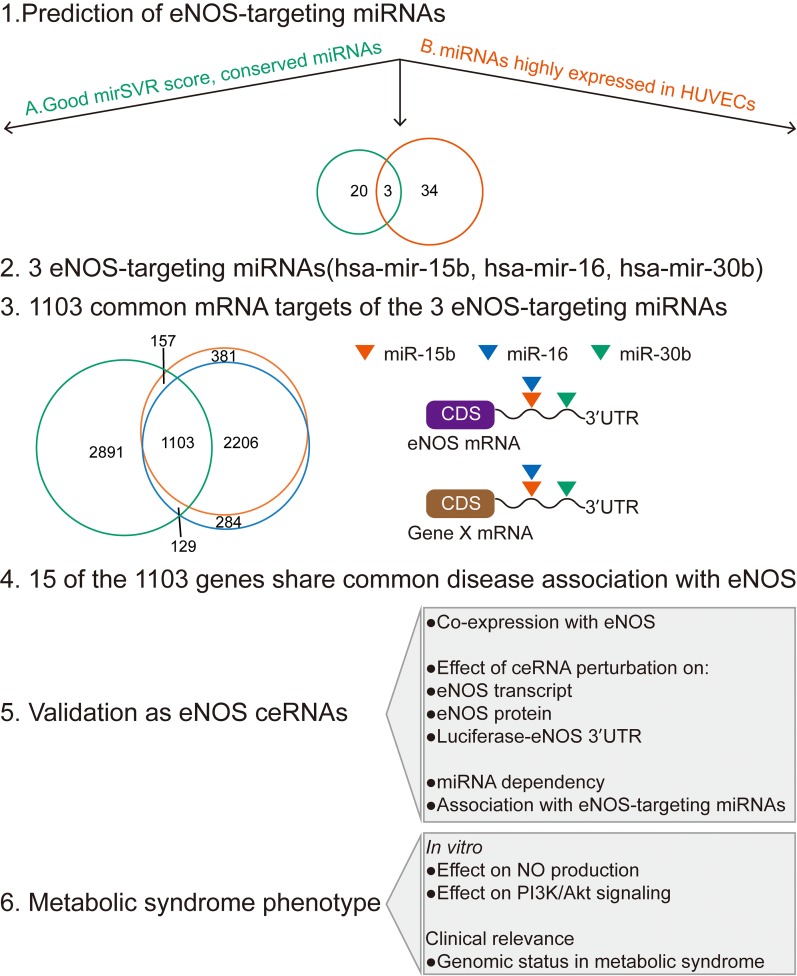
To identify and characterize the eNOS-ceRNA network in the human genome, we integrated computational analysis and experimental validation approaches (Figure 1). We identified 23 eNOS-targeting conserved miRNAs with good mirSVR score from the human mRNA-miRNA target database at microRNA.org (Supplementary Table S1). Because human umbilical vein ECs (HUVECs) are the most commonly used primary cultured human vascular ECs, we adopted data from Kuehbacher et al. (2007). We first ranked miRNAs according to their expression levels and then focused on the highly expressed 37 miRNAs in ECs (Supplementary Table S2). We compared the 23 eNOS-targeting miRNAs with these 37 miRNAs in ECs and identified three candidates (miR-15b, miR-16, and miR-30b). All mRNA targets of miR-15b, miR-16, and miR-30b were downloaded from microRNA.org; the three miRNAs shared 1103 common targets (Supplementary Table S3). We further explored the disease association of these 1103 genes by searching the Disgenet.org database and compared with eNOS-associated diseases: 15 genes shared a common diseases (e.g. CVDs and diabetes) association with eNOS (Supplementary Figure S1).
Figure 1.
The bioinformatics pipeline for analyzing eNOS ceRNAs. (1) Human microRNA target predictions with good mirSVR score and conserved miRNA targeting were downloaded from the microRNA.org database to obtain eNOS-targeting miRNAs (A). miRNA expression profiles in HUVECs were from Kuehbacher et al. (2007)(B). (2) eNOS-targeting miRNAs were confirmed by filtering miRNA–target interactions with good mirSVR score, miRNAs conserved among species, and miRNAs highly expressed in HUVECs. (3) All mRNA targets of miR-15b, miR-16, and miR-30b were downloaded from microRNA.org; the three miRNAs share 1103 common targets. (4) Disease association of these 1103 genes was summarized by using the Disgenet.org database and compared with eNOS-associated diseases; 15 genes with disease score >0.2 were confirmed. (5) In vitro approaches to validate ceRNAs. (6) Biological function investigation of ceRNAs and patient sample expression of ceRNAs.
Seven predicted genes are identified as inferential eNOS ceRNAs in human arteries
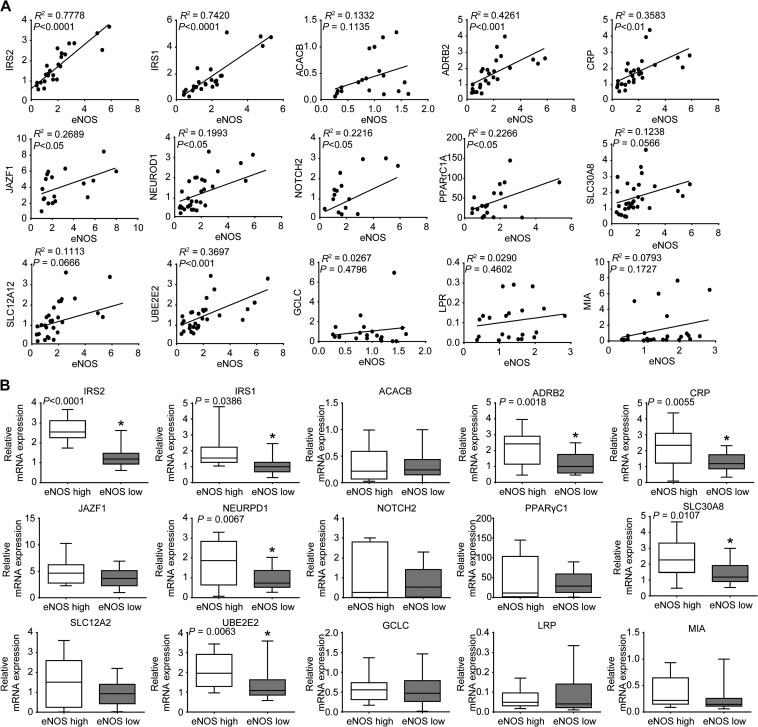
To validate the predicted eNOS-ceRNA coregulation in the human genome, we examined whether the predicted 15 eNOS ceRNAs were co-expressed with eNOS. Human coronary arteries and femoral arteries from 35 patients with CVDs were collected for gene expression analysis. The mRNA levels of eNOS and the 15 predicted ceRNAs were detected by qPCR. The Pearson correlation coefficients between eNOS and its predicted ceRNAs were calculated. Significantly, we found a significant correlation between eNOS and seven of the predicted ceRNAs: IRS2, IRS1, β-2 adrenergic receptor (ADRB2), C-reactive protein (CRP), neurogenic differentiation 1 (NEUROD1), solute carrier family 30 Member 8 (SLC30A8), and ubiquitin-conjugating enzyme E2 (UBE2E2) (Figure 2A). Moreover, we found an increase in correlation between eNOS and its seven ceRNAs when we divided patients into groups of eNOS low and eNOS high by average eNOS mRNA level in all individuals. The expression of these seven genes was significantly decreased by 52.3% (IRS2), 46.5% (IRS1), 40.2% (ADRB2), 44.0% (CRP), 47.3% (NEUROD1), 40.2% (SLC30A8), and 39.0% (UBE2E2) in eNOS low group, compared with eNOS high group, which suggested that IRS2 was the most relevant ceRNA for eNOS (Figure 2B).
Figure 2.
Correlation analysis of coexpression of eNOS and eNOS ceRNAs in human arteries. (A) The mRNA levels of eNOS and its ceRNAs were detected by real-time PCR in human CVD arteries (n = 35). Spearman correlation analysis was used to analyze the coexpression of eNOS and eNOS ceRNAs. (B) Patients were divided into eNOS low and eNOS high groups by mean value of eNOS mRNA level in all individuals. The expression of eNOS ceRNAs in human arteries was measured by real-time PCR. Horizontal lines are mean, box edges are SD, and whiskers are range.
The crosstalk between eNOS and IRS2 depends on miRNAs
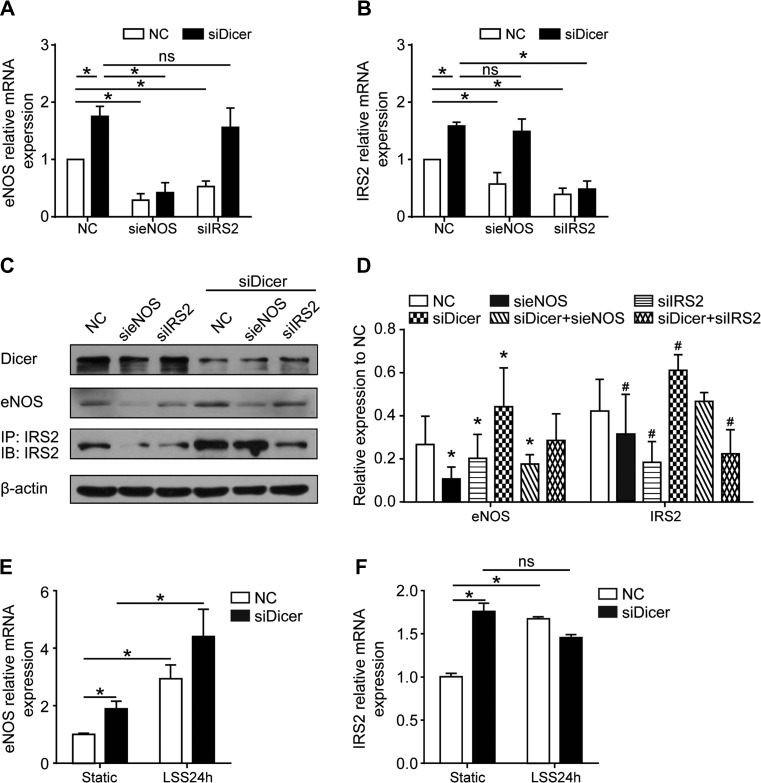
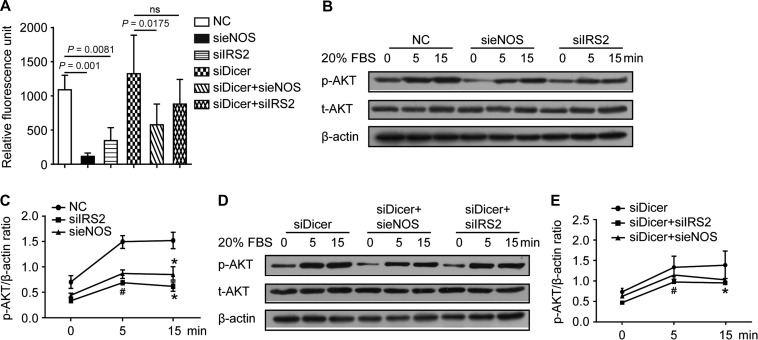
To further characterize the eNOS-ceRNA network, we choose IRS2, the most significant eNOS ceRNAs from human sample data, to study the regulation between eNOS-ceRNA. Knockdown eNOS or IRS2 by siRNAs significantly decreased both IRS2 and eNOS mRNA and protein levels (Figure 3A–C). It was reported that fluid shear physiologically upregulated eNOS expression (Jin et al., 2003; Zhou et al., 2014). We utilized laminar shear stress (LSS) to transcriptionally upregulate the expression of eNOS in ECs for 24 h as we previously reported (Fu et al., 2011). As shown in Figure 3E and F, LSS also increased the expression of IRS2. To investigate whether this observed ceRNA coexpression depended on miRNAs, we transfected Dicer siRNA into HUVECs to block the miRNA mature process (Supplementary Figure S2A). Depletion of Dicer abolished the mutual regulation of eNOS and IRS2 and the response to the stimulation of LSS in human ECs (Figure 3A–F). Therefore, the crosstalk between eNOS and IRS2 depends on miRNAs.
Figure 3.
Predicted NOS ceRNAs regulate eNOS expression dependent on miRNAs. (A–D) HUVECs were transfected with siRNAs including control, eNOS, IRS2, and Dicer (100 nmol/L) for 48 h. (A and B) Real-time PCR was performed to analyze the mRNA levels of Dicer, eNOS, and IRS2. ANOVA followed by the Bonferroni post hoc test, n = 6, *P < 0.0001. (C) Western blot analysis and quantification of the expression levels of Dicer, eNOS, and IRS2. Unpaired Student t-test, n = 4, *P < 0.05, #P < 0.05. (E and F) HUVECs were transfected with 20 nM Dicer siRNA or control RNA for 24 h and then exposed to LSS (12 dyne/cm2) for 24 h. Quantitative real-time PCR was performed to detect the mRNA level of eNOS (E) and IRS2 (F). n = 4, *P < 0.05. Data are mean ± SEM normalized to control. ns, no significance.
eNOS and IRS2 are post-transcriptionally regulated by miR-15b, miR-16, and miR-30b
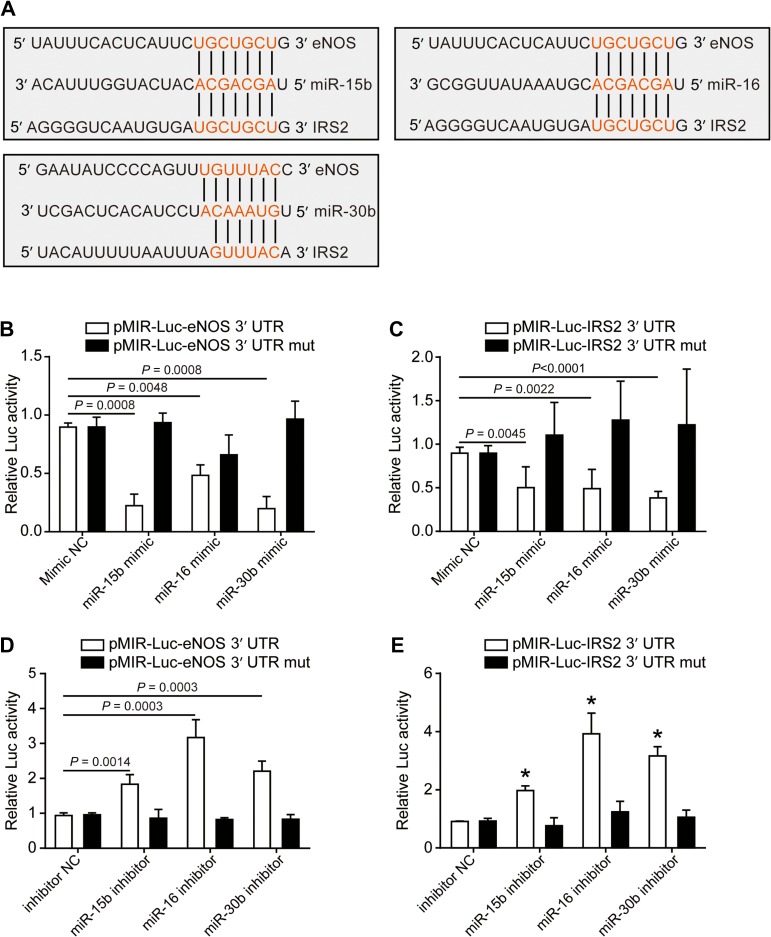
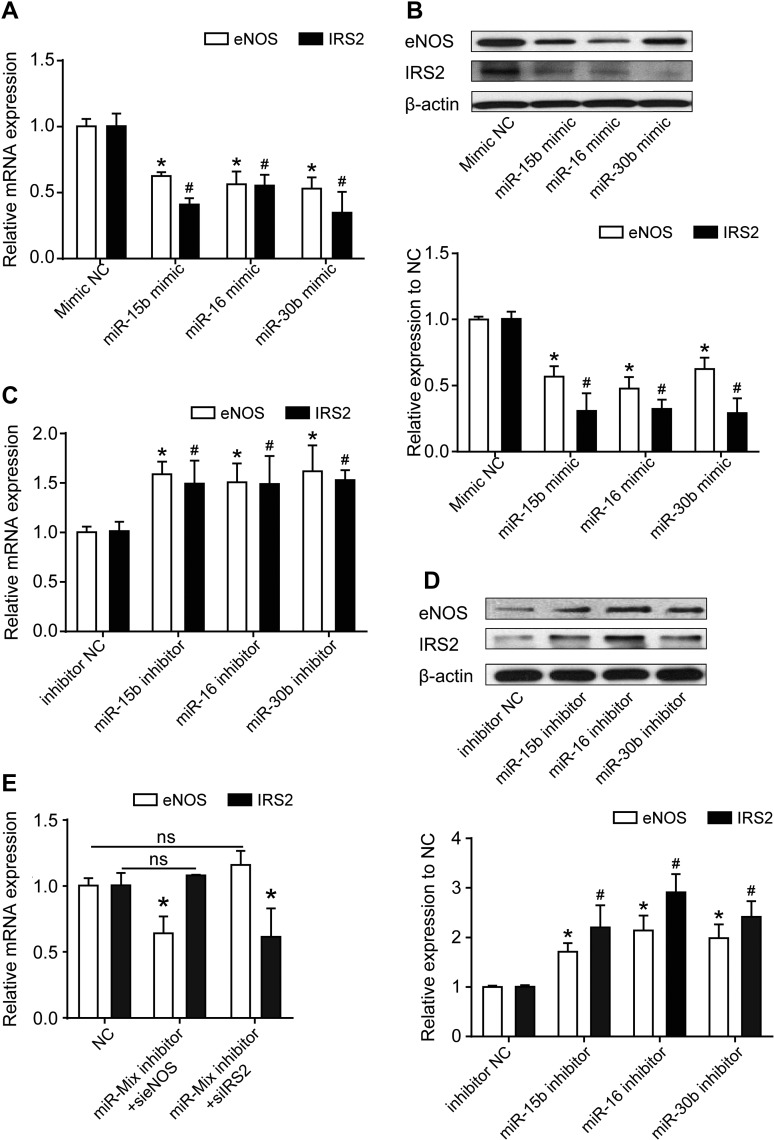
According to the bioinformatics analysis, we predicted that miR-16, miR-15b, and miR-30b target both eNOS and IRS2. Here, we analyzed the sequence of eNOS-3′UTR and IRS2-3′UTR and identified the binding sites of these three miRNAs by using Targetscan 7.0 (www.targetscan.org) (Figure 4A). Expression of these three miRNAs was significantly increased by microRNA mimics and reduced by microRNA inhibitors (Supplementary Figure S2B and C). To determine the direct targeting of these miRNAs on eNOS and IRS2 mRNAs, we generated luciferase reporter constructs containing the 3′UTR fragments of eNOS (pMIR-Luc-eNOS 3′UTR) and IRS2 (pMIR-Luc-IRS2 3′UTR). pMIR-Luc-eNOS 3′UTR or pMIR-Luc-IRS2 3′UTR was cotransfected with miRNA mimics in HUVECs. All three mimics were able to reduce the luciferase activities of both pMIR-Luc-eNOS 3′UTR and pMIR-Luc-IRS2 3′UTR (Figure 4B and C). To further validate the binding specificity, we deleted the miRNA seeds on the constructs (pMIR-Luc-eNOS 3′UTR mut and pMIR-Luc-IRS2 3′UTR mut). Suppression of eNOS and IRS2 3′UTR activities by miRNA mimics was aborted when miRNA seeds were deleted (Figure 4B and C). In contrast, inhibition of miR-15b, miR-16, and miR-30b significantly upregulated the activity of eNOS and IRS2 Luc-3′UTR (Figure 4D and E), which was diminished by miRNA seed deletion (Figure 4D and E). Similarly, the individual or mixed mimics repressed the pMIR-Luc-eNOS 3′UTR and pMIR-Luc-IRS2 3′UTR luciferase activities in HEK293 cells (Supplementary Figure S3A–D), whereas inhibitors of miR-30b rather than miR-15b and miR-16 induced the activities (Supplementary Figure S4A–D), which might be due to the low expression of the latter two miRNAs in HEK293 cells. In contrast, the constructs with miRNA seed deletion barely responded to mimics or inhibitors (Supplementary Figures S3E, F and S4E, F).
Figure 4.
eNOS and IRS2 are post-transcriptionally regulated by miR-15b, miR-16, and miR-30b. (A) Schematic indicates that the three miRNAs (miR-15b, miR-16, miR-30b) share common MREs on both eNOS and IRS2 3′UTR. (B–E) HUVECs were cotransfected with scramble control (NC), miRNA mimics (15bmi, 16mi, or 30bmi) (B and C), or miRNA inhibitors (15bi, 16i, or 30bi) (D and E) with pMIR-Luc-eNOS 3′UTR, pMIR-Luc-IRS2 3′UTR, pMIR-Luc-eNOS 3′UTR mut, or pMIR-Luc-IRS2 3′UTR mut. CMV–β-gal was cotransfected in all groups as a transfection control. Luciferase activity was normalized to that of β-gal. Data are mean ± SEM. n = 3, *P < 0.0001 compared with control.
To further test whether these three miRNAs were able to affect the expression of eNOS and IRS2, we analyzed the mRNA and protein levels of eNOS and IRS2 in ECs with miRNA mimic or inhibitor transfection. miR-15b, miR-16, and miR-30b coincidentally downregulated the mRNA and protein levels of eNOS and IRS2 (Figure 5A and B). In contrast, the expression of eNOS and IRS2 was significantly upregulated by inhibitors of these three miRNAs (Figure 5C and D). By using three miRNA inhibitor pools, IRS2 and eNOS almost completely lose their ceRNA function, i.e. mutual regulation of eNOS and IRS2 mRNA levels (Figure 5E). Therefore, miR-15b, miR-16, and miR-30b could post-transcriptionally regulate both eNOS and IRS2.
Figure 5.
eNOS and IRS2 are post-transcriptionally regulated by miR-15b, miR-16, and miR-30b. (A and B) The mRNA (A) and protein (B) levels of eNOS and IRS2 in HUVECs transfected with scramble control (NC) or miRNA mimics (15bmi, 16mi, or 30bmi). (C and D) The mRNA (C) and protein (D) levels of eNOS and IRS2 in HUVECs transfected with scramble control (NC) or miRNA inhibitors (15bi, 16i, or 30bi). (E) The mRNA levels of eNOS and IRS2 were detected with RT-PCR in HUVECs cotransfected with siRNA of eNOS or IRS2 with miRNA inhibitor mix for 48 h. Data are mean ± SEM. Two-way ANOVA and t-test, n = 3, *P < 0.05 compared with control; #P < 0.05 compared with control.
eNOS or IRS2 deficiency inhibits both NO production and Akt signaling pathway
We explored the biological function of the crosstalk between eNOS and IRS2 in terms of ceRNAs. Consistent with the ceRNA effect on eNOS protein levels, abrogation of IRS2 reduced NO production (Figure 6A). Consistently, knockdown of eNOS significantly decreased Akt (protein kinase B) phosphorylation (Figure 6B and C). Notably, the inhibitory effects of IRS2 and eNOS on NO production and Akt activation were both abrogated when Dicer was knockdown (Figure 6A, D, and E). Our results demonstrate that downregulation of IRS2 and eNOS functionally inhibits NO production and the Akt pathway, which is miRNA-dependent.
Figure 6.
Depletion of IRS2 attenuates NO production and eNOS knockdown reduces AKT activity. (A) NO production was measured by the conversion of L-arginine to NO in HUVECs transfected with the indicated siRNA for 48 h. (B–E) The protein levels of p-Akt and t-Akt detected by western blot analysis in HUVECs transfected with the indicated siRNA for 24 h, starved overnight, then treated with or without 20% FBS for 5 or 15 min. Data are mean ± SEM normalized to control. ANOVA followed by the Bonferroni post hoc test was used for analysis. n = 5, *P < 0.05 compared with control; #P < 0.05 compared with control; ns, no significance.
Discussion
Over the years, the vascular endothelium lining the entire inner surface of the blood vessels has been found to have very distinct functions indispensable for the maintenance of vasomotor balance and vascular homeostasis (Cines et al., 1998; Carmeliet, 2000). Endothelial dysfunction is characterized by reduced bioavailability of eNOS-derived NO, which results in impaired endothelium-dependent vasodilation or endothelial activation-mediated inflammatory response (Versari et al., 2009b). Considering the importance of endothelial dysfunction in CVDs, we focused on eNOS, the central molecule in endothelial biology, as an example for a new mode- the whole ceRNA hypothesis- of regulation of eNOS. The prediction analysis and qPCR results from patient arteries suggested that levels of eNOS and its ceRNAs, especially IRS2, were significantly correlated with each other in disease conditions. In vitro, eNOS and IRS2 regulated each other’s expression; three common miRNAs were involved in regulation of these two genes, as evidenced by their ability to reduce the luciferase activity of Luc-NOS3-3′UTR and Luc-IRS2-3′UTR, whereas their inhibition increased the luciferase activity; silencing of eNOS significantly reduced the Akt signal pathway and silencing of IRS2 decreased eNOS activity. This crosstalk between these two genes depended on miRNAs and the 3′UTR, because silencing Dicer abolished the interdependence. Therefore, our study provides evidence of aberrant regulation of eNOS via miRNA competition with its ceRNA IRS2, which might contribute to complications of CVDs such as atherosclerosis.
The ceRNA regulation theory was based on the fact that mRNAs can harbor specific MREs (i.e. miRNA seeds) on the 3′UTR region (Guil and Esteller, 2015). Therefore, theoretically, any nucleotide molecules that act as miRNA decoys such as pseudogenes may function as ceRNAs. As a model for the functional relation, PTEN tumor suppressor gene possesses a regulatory role on its pseudogene PTENP1 independent of protein coding (Poliseno et al., 2010). Although other examples of ceRNAs functioning in cancer biology have been described (Karreth et al., 2011; Xu et al., 2015), little is known about whether and how ceRNAs work together with miRNAs contributing to the pathogenesis of CVDs. Recently, another group, investigating the actions of Let-7e in HUVECs, found that Let-7e increased the inflammatory responses of ECs by activating NF-κB, which depended on a ceRNA (Lin et al., 2017). In this study, we combined bioinformatics and in vitro experimental validation in human arteries to characterize the ceRNAs of eNOS. We predicted eNOS-targeting miRNAs that are conserved in human (Supplementary Table S1). In turn, we screened ceRNAs highly expressed in human ECs sharing the same set of conserved miRNAs with eNOS (Supplementary Table S2). To screen the ceRNAs with more biological relevance with eNOS, we further narrowed down ceRNAs with the same high disease score as eNOS in the same disease conditions (Supplementary Figure S1). By this approach, we generated a list of putative eNOS ceRNAs sharing three common miRNAs (miR-15b, miR-16, and miR-30b).
By detecting the mRNA levels of the 15 putative eNOS ceRNAs in human arteries from patients with CVDs, we identified seven genes with levels significantly correlated with eNOS level, including IRS2, IRS1, ADRB2, CRP, NEUROD1, SLC30A8, and UBE2E2 (Figure 2A). Four of these, IRS1, IRS2, SLC30A8, and UBE2E2, were associated with diabetes (Taniguchi et al., 2005; Thaker et al., 2006), and ADRB2 and NEUROD1, were involved in the central nervous system (Thaker et al., 2006; Kazantsev and Thompson, 2008). IRS1 and IRS2 are mainly involved in insulin signaling; their deficiency caused a diabetes phenotype (Taniguchi et al., 2006). SLC30A8 and UBE2E2 are related to β-cell function and insulin secretion in humans (Flannick and Florez, 2016; Thomsen et al., 2016). ADRB is essential for physiological responses to the hormones/neurotransmitters epinephrine and norepinephrine in the nervous system (Armaiz-Pena et al., 2013), and NeuroD1 encodes a basic helix-loop-helix transcription factor involved in the development of neural and endocrine structures, including the retina and pineal gland (Ochocinska et al., 2012). CRP is a pentameric protein found in blood plasma, whose levels increase in response to inflammation (Pepys and Hirschfield, 2003). Identifying these genes highly indicated the possible interaction between eNOS and diabetes-associated genes. Because of the modest change in levels of the other genes between eNOS high and low groups, IRS2 was considered the most relevant ceRNAs for eNOS in our following study. Further functional analysis showed that IRS2 has a regulatory property on NO production as a ceRNA of eNOS (Figure 6A). This observation functionally links a number of unexpected biological pathways to NO production via the ceRNA language.
The predicted three common miRNAs for eNOS and IRS2 functioning as ceRNAs are miR-16, miR-15b, and miR-30b. The ‘canonical’ miR-16 family shares the same ‘seed’ binding sequence AGCAGC (Hausser et al., 2013), so the members likely have functional redundancy. As defined by sequence similarity, members of the miR-16 family include miR-15 (a and b) and miR-16 (1 and 2) (Liu et al., 2008). Among them, miR-15 and miR-16 are co-transcribed from one of two different intragenic loci, miR-15a/16-1 and miR-15b/16-2, which are highly conserved among mammalian species (Cimmino et al., 2005). Although previous studies demonstrated that miR-15/16 clusters act as tumor suppressors (Bonci et al., 2008) and play a critical role in the control of cardiomyocyte proliferation and heart regeneration (Mendell and Olson, 2012), the role of miR-15/16 in ECs is hardly reported. Meanwhile, miR-30b regulates angiogenesis by targeting DLL4 in vitro and in vivo (Bridge et al., 2012). Therefore, our study revealed a unique mechanism of the post-transcriptional regulation of eNOS and new functions of the three miRNAs in ECs.
Clinically, individuals with insulin resistance have compromised EC function and increased frequency and severity of CVDs (Cersosimo and DeFronzo, 2006). Although the global consequences of insulin resistance such as dyslipidemia and hyperglycemia are sufficient to induce cardiovascular dysfunction (American Diabetes, 2014), the local actions of insulin resistance on ECs in blood vessels are also thought to be significant. Insulin directly stimulates NO release from the vascular endothelium via a cascade that involves activating phosphatidylinositol 3-kinase (PI3K)–Akt signaling and downstream phosphorylation of eNOS (Zeng et al., 2000; Kawasaki et al., 2003). eNOS-knockout mice showed significantly reduced whole-body glucose disposal rates and insulin resistance in liver and peripheral tissues (Shankar et al., 2000). Type 2-like diabetes develops in mice deficient for IRS2. In apoE−/−IRS2−/− mice, atherosclerosis was significantly enhanced as compared with apoE−/− mice (Gonzalez-Navarro et al., 2007). Further, atherosclerotic lesion sizes were significantly higher in apoE−/− mice lacking endothelial insulin receptor (Rask-Madsen et al., 2010). However, 3′UTR comparison analysis revealed that the binding sites of the three miRNAs in IRS2 3′UTR are highly conserved among different species, but those binding sites in eNOS are only conserved among humans, nonhuman primates and other large animals (pig/cow/horse), not in mouse and rat (Supplementary Figure S5). It limited us to verify the results in common laboratory animal models in vivo. Thus, our study provides evidence to support the connection between CVDs and diabetes in human.
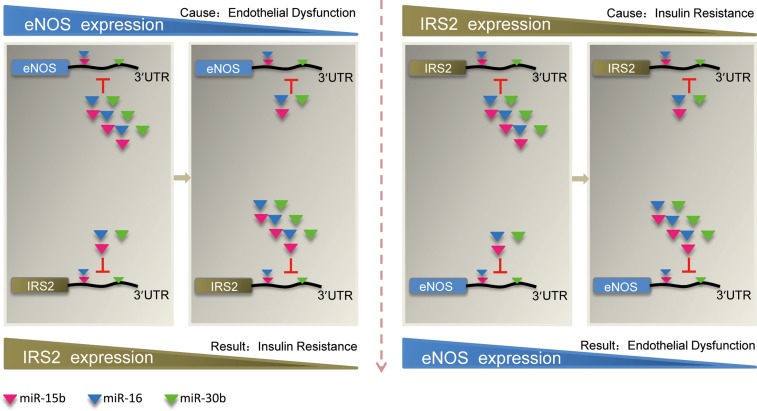
We propose that the ceRNA language may play an important role in physiological processes and pathophysiological conditions such as metabolic syndrome. Here we identified that eNOS and IRS2 regulate each other dependent on miRNAs and function as ceRNAs in human (see the proposed model in Figure 7), which might explain the clinical observations of the association of CVDs and diabetes.
Figure 7.
A model of eNOS and IRS2 coregulation as ceRNAs. Both eNOS and IRS2 contain MREs that can bind to miR-15b, miRmiR-16, and miR-30b. When the abundance of eNOS is downregulated (endothelial dysfunction), more miRNAs are bound by IRS2, and the repression conferred by its targeting microRNAs would result in decreased expression of IRS2 (insulin resistance), and vice versa.
Materials and methods
Human samples
Our studies were approved by the Research Ethics Committee of Tianjin Medical University. Human arteries including coronary arteries and femoral arteries from patients with CVDs (n = 22) and diabetes (n = 13) were collected during coronary artery bypass graft surgery and amputation from the Tianjin Medical University General Hospital. Participants provided informed written consent. No organs were obtained from executed prisoners or other institutionalized persons.
Antibodies and reagents
MirVana mimics or inhibitors for miR-15b, miR-16, and miR-30b are synthesized double-stranded or single-stranded RNAs that mimic or inhibit endogenous miRNAs (Ambion). All siRNA molecules (siDicer, sieNOS, and siIRS2) were from Invitrogen. The antibodies for western blotting including anti-eNOS, anti-IRS2, anti-p-AKT, and anti-t-AKT were from Cell Signaling Technology. Lipofectamine RNAimax, lipofectamine 2000, and Trizol reagent were from Invitrogen.
Cell cultures
HUVECs and HEK293 cells were isolated and cultured as described (Zhu et al., 1998). All HUVECs were used before passage 5 and were cultured in EC medium till confluence for transfection.
Shear stress experiment
The flow experiments were performed as previously described (Fu et al., 2011). The applied laminar flow was the steady shear stress of 12 dyne/cm2.
RNA extraction and real-time PCR of miRNA
RNA from cultured cells or tissues was extracted by use of TRIzol (Invitrogen). Total RNA was reverse-transcribed by use of the TaqMan microRNA Reverse Transcription Kit. GAPDH was an internal control for cultured cells and human arteries. Real-time PCR of miRNA assay involved the TaqMan microRNA Assay (Life Technologies). U6 was detected as an internal control in miRNA assays. Primers for qPCR are in Supplementary Table S4.
Western blot analysis
HUVECs were washed with PBS and harvested in lysis buffer containing protease inhibitors (Roche) on ice. Proteins were separated by SDS-PAGE and electrophoretically transferred to PVDF membranes (Millipore), which were incubated with primary antibodies overnight at 4°C with gentle shaking, then the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature (Proteintech); bands were visualized with enhanced chemiluminescence reagents (Thermo Scientific).
RNA interference
All siRNA molecules were from Invitrogen. The following primer sequences were used: eNOS sense: 5′-GCAGGUCUGCACAGGAAAUTT-3′, eNOS anti-sense: 5′-AUUUCCUGUGCAGACCUGCTT-3′; IRS2 sense: 5′-CCGGCGAGUACAUCAACAUTT-3′, IRS2 anti-sense: 5′-AUGUUGAUGUACUCGCCGGTT-3′; Dicer sense: 5′-GCAAUUCAUGAUAACAUUU-3′, Dicer anti-sense: 5′-AAAUGUUAUCAUGAAUUGC-3′; Control siRNA sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, Control anti-sense: 5′-ACGUGACACGUUCGGAGAATT-3′.
Control RNA, siRNAs, pre-miR-15b, miR-16, and miR-30b or anti-miR-15b, miR-16, and miR-30b were transfected by using Lipofectamine RNAiMAX (Invitrogen) into HUVECs and HEK293 cells.
Plasmid constructs and luciferase activity assay
To generate p-MIR-Luc-eNOS-3′UTR and p-MIR-Luc-IRS2-3′UTR, full-length human eNOS 3′UTR and IRS2 3′UTR were subcloned into the pMIR-REPORT vector (Ambion). Then p-MIR-Luc-eNOS-3′UTR (Mut) and p-MIR-Luc-IRS2-3′UTR (Mut) were created by trimming regions complementary to miR-15b, miR-16, and miR-30b seed sequences. Luciferase activity was measured by using the Dual-Glo Luciferase Reporter Assay Kit (Promega).
NO production measurement
Cellular eNOS activity was measured by the conversion of L-arginine to NO by use of a nitric-oxide synthase assay kit (Beyotime Institute of Biotechnology), combined with a specific inhibitor of eNOS (NG-Nitro-L-arginine methyl ester, hydrochloride) (Beyotime Institute of Biotechnology). Briefly, HUVECs were transfected with siIRS2 for 48 h and seeded into 96-well plates. After culture for 12 h, cell confluence was about 80%–90%. The medium was discarded, and 0.1 ml working solution containing NADPH (0.1 mM), L-arginine, DAF-FMDA, and L-NAME (100 μM) was added to each well, then 0.1 ml reaction buffer was added, and plates were incubated at 37°C in the dark for 1 h. The relative fluorescence intensity was detected by using a fluorescence microplate reader, with excitation and emission wavelengths 488 and 515 nm, respectively. Values were corrected for the blank control. The fluorescence intensity of the control was defined as 100%, and the relative eNOS activity was expressed as relative fluorescence unit compared to the control value.
Bioinformatics analysis
eNOS-targeting miRNAs and ceRNAs were predicted by using the microRNA.org database (Human Good mirSVR score, conserved miRNA). A mirSVR cutoff of ≤−1.2 was defined as good mirSVR score. This value represents the top 5% of miRSVR scores. Also, miRBase and TargetScan Human were used to confirm the miRNA–eNOS relation. The mRNA–disease association with disease score >0.2 was searched on the DisGeNET database. The workflow of the disease relative ceRNA enrichment analysis is shown in Supplementary Figure S6.
Statistical analysis
Data are expressed as mean ± SEM from three or more independent experiments. Two groups were compared by unpaired Student t-test. Differences among multiple groups were evaluated by ANOVA followed by the Bonferroni post hoc test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
The authors gratefully thank the Tianjin Medical University General Hospital for providing human samples for this work.
Funding
This work was supported in part by grants from the Ministry of Science and Technology of China (2016YFC0903000), the National Natural Science Foundation of China (81420108003 and 81730014), and the Tianjin Municipal Science and Technology Project (14JCYBJC41800).
Conflict of interest
none declared.
Author contributions
X.S., Q.C., P.Z., and Y.Z designed the research; S.F. and M.W. contributed the human samples; X.S., H.L., P.Z., and Q.C. performed the research; X.S., H.L., and P.Z. analyzed data; X.S., H.L., P.Z., and J.H. formatted the figures; X.S., H.L., P.Z., and Y.Z. wrote the paper.
References
- American Diabetes Association. (2014). Diagnosis and classification of diabetes mellitus. Diabetes Care 37(Suppl 1), S81–S90. [DOI] [PubMed] [Google Scholar]
- Antonetti D.A., Barber A.J., Khin S., et al. (1998). Vascular permeability in experimental diabetes is associated with reduced endothelial occludin content: vascular endothelial growth factor decreases occludin in retinal endothelial cells. Penn State Retina Research Group. Diabetes 47, 1953–1959. [DOI] [PubMed] [Google Scholar]
- Armaiz-Pena G.N., Cole S.W., Lutgendorf S.K., et al. (2013). Neuroendocrine influences on cancer progression. Brain Behav. Immun. 30(Suppl 1), S19–S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balligand J.L., Feron O., and Dessy C. (2009). eNOS activation by physical forces: from short-term regulation of contraction to chronic remodeling of cardiovascular tissues. Physiol. Rev. 89, 481–534. [DOI] [PubMed] [Google Scholar]
- Bonci D., Coppola V., Musumeci M., et al. (2008). The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 14, 1271–1277. [DOI] [PubMed] [Google Scholar]
- Bridge G., Monteiro R., Henderson S., et al. (2012). The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood 120, 5063–5072. [DOI] [PubMed] [Google Scholar]
- Brownlee M. (2001). Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. (2000). Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 6, 389–395. [DOI] [PubMed] [Google Scholar]
- Cersosimo E., and DeFronzo R.A. (2006). Insulin resistance and endothelial dysfunction: the road map to cardiovascular diseases. Diabetes Metab. Res. Rev. 22, 423–436. [DOI] [PubMed] [Google Scholar]
- Cimmino A., Calin G.A., Fabbri M., et al. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA 102, 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cines D.B., Pollak E.S., Buck C.A., et al. (1998). Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood 91, 3527–3561. [PubMed] [Google Scholar]
- Fang Y., Shi C., Manduchi E., et al. (2010). MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc. Natl Acad. Sci. USA 107, 13450–13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J., and Florez J.C. (2016). Type 2 diabetes: genetic data sharing to advance complex disease research. Nat. Rev. Genet. 17, 535–549. [DOI] [PubMed] [Google Scholar]
- Fu Y., Hou Y., Fu C., et al. (2011). A novel mechanism of γ/δ T-lymphocyte and endothelial activation by shear stress: the role of ecto-ATP synthase β chain. Circ. Res. 108, 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Navarro H., Vila-Caballer M., Pastor M.F., et al. (2007). Plasma insulin levels predict the development of atherosclerosis when IRS2 deficiency is combined with severe hypercholesterolemia in apolipoprotein E-null mice. Front. Biosci. 12, 2291–2298. [DOI] [PubMed] [Google Scholar]
- Guil S., and Esteller M. (2015). RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem. Sci. 40, 248–256. [DOI] [PubMed] [Google Scholar]
- Hausser J., Syed A.P., Bilen B., et al. (2013). Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 23, 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J., Peruzzi P.P., and Lawler S. (2014). MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 20, 460–469. [DOI] [PubMed] [Google Scholar]
- Jin Z.G., Ueba H., Tanimoto T., et al. (2003). Ligand-independent activation of vascular endothelial growth factor receptor 2 by fluid shear stress regulates activation of endothelial nitric oxide synthase. Circ. Res. 93, 354–363. [DOI] [PubMed] [Google Scholar]
- Karreth F.A., Tay Y., Perna D., et al. (2011). In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K., Smith R.S. Jr., Hsieh C.M., et al. (2003). Activation of the phosphatidylinositol 3-kinase/protein kinase Akt pathway mediates nitric oxide-induced endothelial cell migration and angiogenesis. Mol. Cell. Biol. 23, 5726–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A.G., and Thompson L.M. (2008). Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 7, 854–868. [DOI] [PubMed] [Google Scholar]
- Kim V.N. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385. [DOI] [PubMed] [Google Scholar]
- Kubes P., Suzuki M., and Granger D.N. (1991). Nitric oxide: an endogenous modulator of leukocyte adhesion. Proc. Natl Acad. Sci. USA 88, 4651–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher A., Urbich C., Zeiher A.M., et al. (2007). Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 101, 59–68. [DOI] [PubMed] [Google Scholar]
- Liao Y.C., Wang Y.S., Guo Y.C., et al. (2014). Let-7g improves multiple endothelial functions through targeting transforming growth factor-β and SIRT-1 signaling. J. Am. Coll. Cardiol. 63, 1685–1694. [DOI] [PubMed] [Google Scholar]
- Lin Z., Ge J., Wang Z., et al. (2017). Let-7e modulates the inflammatory response in vascular endothelial cells through ceRNA crosstalk. Sci. Rep. 7, 42498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fu H., Sun F., et al. (2008). miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 36, 5391–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.H., Sun M., Nie F.Q., et al. (2014). Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 13, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Yang L., Guo R., et al. (2017). Ox-LDL-induced microRNA-155 promotes autophagy in human endothelial cells via repressing the Rheb/mTOR pathway. Cell. Physiol. Biochem. 43, 1436–1448. [DOI] [PubMed] [Google Scholar]
- Mehta D., and Malik A.B. (2006). Signaling mechanisms regulating endothelial permeability. Physiol. Rev. 86, 279–367. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., and Olson E.N. (2012). MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochocinska M.J., Munoz E.M., Veleri S., et al. (2012). NeuroD1 is required for survival of photoreceptors but not pinealocytes: results from targeted gene deletion studies. J. Neurochem. 123, 44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepys M.B., and Hirschfield G.M. (2003). C-reactive protein: a critical update. J. Clin. Invest. 111, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., et al. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Madsen C., Li Q., Freund B., et al. (2010). Loss of insulin signaling in vascular endothelial cells accelerates atherosclerosis in apolipoprotein E null mice. Cell Metab. 11, 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., et al. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena C.M., Pereira A.M., and Seica R. (2013). Endothelial dysfunction—a major mediator of diabetic vascular disease. Biochim. Biophys. Acta 1832, 2216–2231. [DOI] [PubMed] [Google Scholar]
- Shankar R.R., Wu Y., Shen H.Q., et al. (2000). Mice with gene disruption of both endothelial and neuronal nitric oxide synthase exhibit insulin resistance. Diabetes 49, 684–687. [DOI] [PubMed] [Google Scholar]
- Sneddon J.M., and Vane J.R. (1988). Endothelium-derived relaxing factor reduces platelet adhesion to bovine endothelial cells. Proc. Natl Acad. Sci. USA 85, 2800–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.-Y., Zhao Z.-W., Li W.-M., et al. (2017). Knockdown of MALAT1 expression inhibits HUVEC proliferation by upregulation of miR-320a and downregulation of FOXM1 expression. Oncotarget 8, 61499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwaidi J.A., Hamasaki S., Higano S.T., et al. (2000). Long-term follow-up of patients with mild coronary artery disease and endothelial dysfunction. Circulation 101, 948–954. [DOI] [PubMed] [Google Scholar]
- Taniguchi C.M., Emanuelli B., and Kahn C.R. (2006). Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96. [DOI] [PubMed] [Google Scholar]
- Taniguchi C.M., Ueki K., and Kahn R. (2005). Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J. Clin. Invest. 115, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tay Y., Rinn J., and Pandolfi P.P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker P.H., Han L.Y., Kamat A.A., et al. (2006). Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat. Med. 12, 939–944. [DOI] [PubMed] [Google Scholar]
- Thomsen S.K., Ceroni A., van de Bunt M., et al. (2016). Systematic functional characterization of candidate causal genes for type 2 diabetes risk variants. Diabetes 65, 3805–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versari D., Daghini E., Virdis A., et al. (2009. a). Endothelial dysfunction as a target for prevention of cardiovascular disease. Diabetes Care 32(Suppl 2), S314–S321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versari D., Daghini E., Virdis A., et al. (2009. b). Endothelium-dependent contractions and endothelial dysfunction in human hypertension. Br. J. Pharmacol. 157, 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li Y., Lu J., et al. (2015). The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 43, 8169–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Nystrom F.H., Ravichandran L.V., et al. (2000). Roles for insulin receptor, PI3-kinase, and Akt in insulin-signaling pathways related to production of nitric oxide in human vascular endothelial cells. Circulation 101, 1539–1545. [DOI] [PubMed] [Google Scholar]
- Zhou J., Li Y.S., and Chien S. (2014). Shear stress-initiated signaling and its regulation of endothelial function. Arterioscler. Thromb. Vasc. Biol. 34, 2191–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Lin J.H., Liao H.L., et al. (1998). LDL induces transcription factor activator protein-1 in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 18, 473–480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.