Abstract
Our aim was to introduce and validate qPSMA, a semiautomatic software package for whole-body tumor burden assessment in prostate cancer patients using 68Ga-prostate-specific membrane antigen (PSMA) 11 PET/CT. Methods: qPSMA reads hybrid PET/CT images in DICOM format. Its pipeline was written using Python and C++ languages. A bone mask based on CT and a normal-uptake mask including organs with physiologic 68Ga-PSMA11 uptake are automatically computed. An SUV threshold of 3 and a liver-based threshold are used to segment bone and soft-tissue lesions, respectively. Manual corrections can be applied using different tools. Multiple output parameters are computed, that is, PSMA ligand–positive tumor volume (PSMA-TV), PSMA ligand–positive total lesion (PSMA-TL), PSMA SUVmean, and PSMA SUVmax. Twenty 68Ga-PSMA11 PET/CT data sets were used to validate and evaluate the performance characteristics of qPSMA. Four analyses were performed: validation of the semiautomatic algorithm for liver background activity determination, assessment of intra- and interobserver variability, validation of data from qPSMA by comparison with Syngo.via, and assessment of computational time and comparison of PSMA PET–derived parameters with serum prostate-specific antigen. Results: Automatic liver background calculation resulted in a mean relative difference of 0.74% (intraclass correlation coefficient [ICC], 0.996; 95%CI, 0.989;0.998) compared with METAVOL. Intra- and interobserver variability analyses showed high agreement (all ICCs > 0.990). Quantitative output parameters were compared for 68 lesions. Paired t testing showed no significant differences between the values obtained with the 2 software packages. The ICC estimates obtained for PSMA-TV, PSMA-TL, SUVmean, and SUVmax were 1.000 (95%CI, 1.000;1.000), 1.000 (95%CI, 1.000;1.000), 0.995 (95%CI, 0.992;0.997), and 0.999 (95%CI, 0.999;1.000), respectively. The first and second reads for intraobserver variability resulted in mean computational times of 13.63 min (range, 8.22–25.45 min) and 9.27 min (range, 8.10–12.15 min), respectively (P = 0.001). Highly significant correlations were found between serum prostate-specific antigen value and both PSMA-TV (r = 0.72, P < 0.001) and PSMA-TL (r = 0.66, P = 0.002). Conclusion: Semiautomatic analyses of whole-body tumor burden in 68Ga-PSMA11 PET/CT is feasible. qPSMA is a robust software package that can help physicians quantify tumor load in heavily metastasized prostate cancer patients.
Keywords: PSMA, tumor segmentation, qPSMA, PET/CT
Prostate cancer is a significant health problem, being the leading malignancy diagnosed in men and the second-to-third leading cause of cancer death in men (1,2). In the locoregional stage, the relative 5-y survival rate is 99%, whereas in patients with metastatic prostate cancer the survival rate dramatically decreases to 29% (3). Therefore, metastatic disease has become an important entity in the diagnosis and treatment of prostate cancer. PET imaging plays an important role in evaluating disease extent and treatment response. The Prostate Cancer Working group 3 (4) recommends determining radiographic response to treatment using RECIST 1.1 (5) for extraskeletal disease evaluation and qualitative interpretation of bone scans as the standard for bone lesion assessment. The development of a quantitative image-derived biomarker exploiting recent advances in PET imaging to assess tumor burden based on tumor activity is an unmet clinical need in prostate cancer. Such a biomarker is expected to be crucial for accurate evaluation of therapy response.
Prostate-specific membrane antigen (PSMA), also known as glutamate carboxypeptidase II, is a nonsecreted transmembrane glycoprotein (6,7) that is weakly expressed in healthy prostate tissue but strongly upregulated in prostate cancer cells (7). In the last few years, molecular imaging targeting PSMA has become of great interest in prostate cancer diagnostics (8). 68Ga-PSMA11 PET/CT is a novel imaging technique that showed enhanced accuracy compared with conventional imaging modalities in detecting prostate cancer lesions (9–11). Consequently, it is a promising technique to allow tumor load quantification as a candidate image-based biomarker in prostate cancer. However, in patients with a high tumor load, manual quantification is time-consuming and a semiautomatic tool would considerably reduce the segmentation time. A first step toward a semiautomatic tumor burden assessment in prostate cancer was described by Bieth et al. (12), who proposed a tool that quantifies involvement of the skeleton in prostate cancer inspired by the bone scan index using 68Ga-PSMA11 PET/CT.
We introduce qPSMA as a further development of this first approach into a software that allows whole-body semiautomatic tumor burden assessment, that is, skeletal, lymph node, and visceral metastases. We aimed to describe and validate its use in metastatic prostate cancer.
MATERIALS AND METHODS
General Description
qPSMA reads images in DICOM format. PET and CT are coregistered automatically using the information contained in DICOM headers. The pipeline was written using Python and C++ languages. The software runs on Ubuntu and can be installed via a virtual machine on Windows (Microsoft) or Macintosh (Apple) operating systems. After the computation is finished, the entire work, including the PET, the CT, and the labels, can be saved into a single MATLAB file (.mat). Considering the intra- and intervariability in reconstructing different types of PET/CT data sets, an image interpolation algorithm using cubic B-spline curves was implemented (13). Because of a lack of standardization in protocol scanning, anatomic segments (i.e., head, arms, and legs) are not always entirely contained in the field of view of PET/CT images. To allow for intra- and interpatient comparison, the reader can define a specific volume between certain slices to be included in the final statistics. At the end of computation, the maximum-intensity projection PET image, including the segmentation labels, can be displayed. The software includes the possibility of using different SUV thresholds for skeleton and soft-tissue lesions. This thresholding is based on the observation that bone metastases reveal lower PSMA expression than lymph nodes metastases (14). Accordingly, 68Ga-PSMA11 PET imaging showed higher PSMA uptake for lymph node metastases than skeleton lesions (15).
Software Workflow
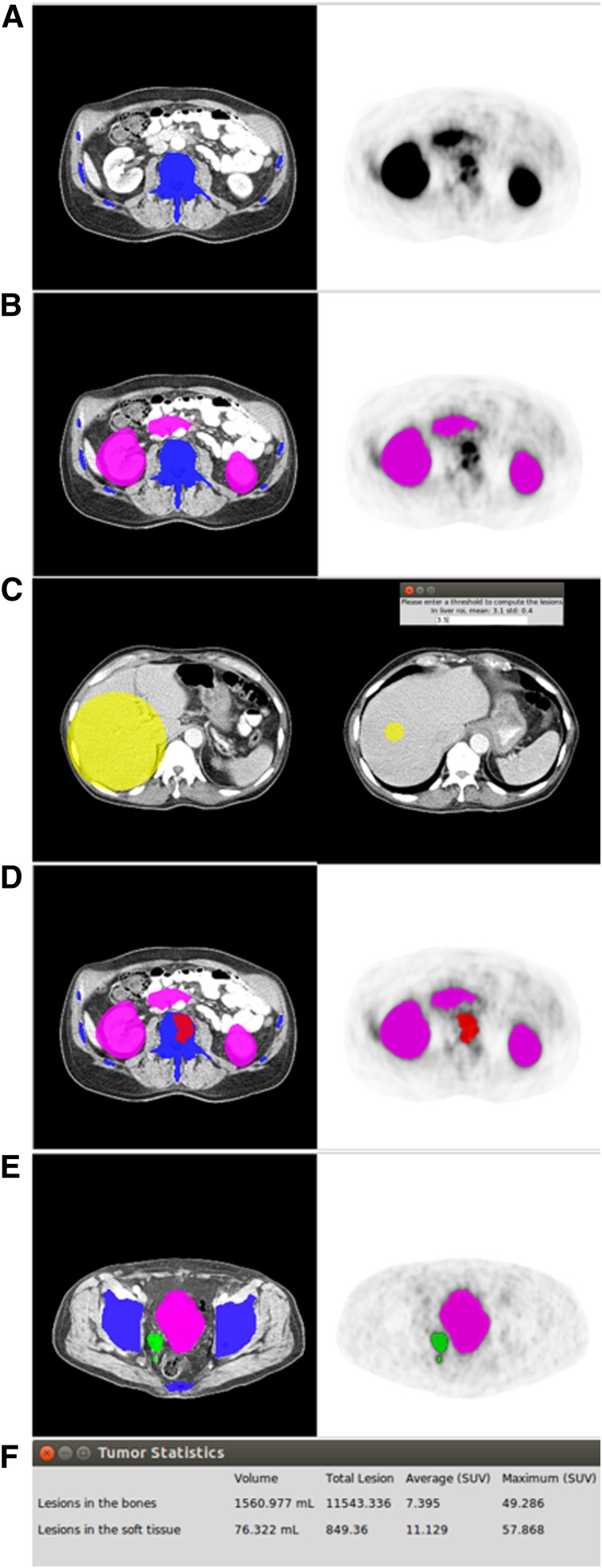
Figure 1 displays the proposed 6-step workflow of qPSMA for whole-body tumor segmentation.
FIGURE 1.
The 6-step workflow of qPSMA. First, bone mask (A) and normal-uptake mask (B) are automatically computed. Then, SUVthr_st is semiautomatically computed from liver background activity (C). Bone lesions are segmented using SUVthr_bone (D), whereas soft-tissue lesions are segmented using SUVthr_st, previously calculated at third step (E). Finally, output parameters are obtained by performing general statistics (F).
Bone Mask
The bone mask incorporates the skeleton, which is first segmented on the CT scan. The segmentation method relies on pixel intensities and different morphologic operations, as described previously (12,16). When necessary, manual corrections can subsequently be applied. Because the CT and PET are coregistered, the bone mask is automatically transposed to PET images to determine the location of the skeleton.
Normal-Uptake Mask
The normal-uptake mask includes the organs that typically exhibit high physiologic PSMA uptake, namely the salivary glands, liver, spleen, kidneys, and urinary bladder. A Vantage Point Forest algorithm was trained to automatically compute the normal uptake mask, as described previously (16).
Liver Background Activity
In accordance with PERCIST 1.0 (17), the SUVmean within a 3-cm spheric volume of interest (VOI) within the right liver lobe is used to obtain the liver background activity. To minimize intra- and interuser variability, this 3-cm VOI is semiautomatically placed using an algorithm that showed high reliability and reproducibility in evaluating liver background activity (18).
Bone Lesion Segmentation
68Ga-PSMA11 does not usually lead to relevant unspecific uptake within the skeleton. Bieth et al. (12), described a separate fixed SUV threshold for soft-tissue lesions (SUVthr_st) of 3 as useful for bone lesion segmentation to omit low background uptake. Application of this threshold is restricted to voxels within the transposed bone mask from CT. Because of the spillover effect and frequent misalignment between the CT and PET images (e.g., because of breathing), some of the bone lesions may be located outside the bone mask and consequently will be improperly segmented as soft-tissue lesions. To overcome this issue, an automated algorithm adds uptake outside the bone mask, but in conjunction with a bone lesion, to this bone lesion. Supplemental Figure 1 displays an example of a bone lesion located partially outside the bone mask, before and after application of the “extend bone lesion” algorithm (supplemental materials are available at http://jnm.snmjournals.org).
Soft-Tissue Lesion Segmentation
An SUVthr_st can be applied. The software automatically displays the measured liver background activity (SUVmean + SD) to assist the user in choosing the SUVthr_st. The rationale for choosing a liver-based threshold parallels the recommendation of liver uptake as background as established in the PROMISE criteria (19) and its physiologic 68Ga-PSMA11 uptake, with no detectable PSMA expression by immunohistochemistry (7).
This approach allows use of a patient- and scan-individualized threshold. To obtain an accurate value taking into account the so-called tumor sink effect, the following formula is recommended. The value 4.30 represent the average liver SUVmean obtained in 80 consecutive patients.
After determination of SUVthr_st, all voxels that show an SUV greater than SUVthr_st and located outside bone and normal-uptake masks are automatically segmented as soft-tissue lesions.
Manual Corrections
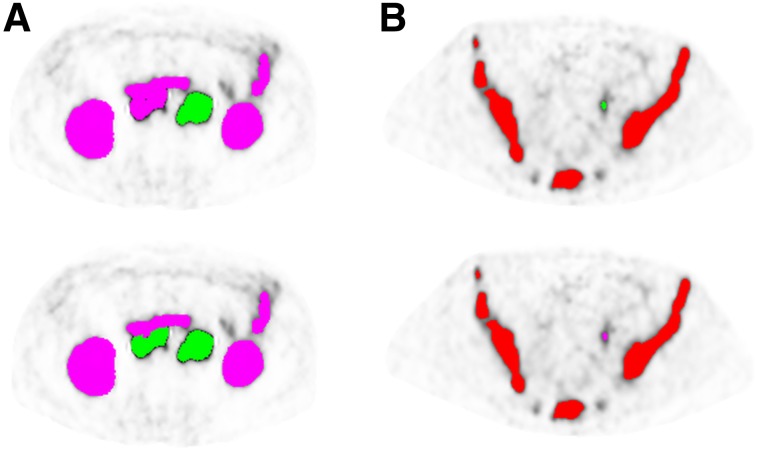
Usually, manual corrections are necessary to delineate the intestine from abdominal PSMA ligand–positive lymph nodes and to remove false-positive uptake within structures with unspecific uptake, such as the aorta, esophagus, ureter, and rectum. Typical pitfalls in PSMA ligand PET imaging (e.g., celiac and other ganglia, adrenal glands) should be considered (20). Different tools, such as “brush,” “erase,” “remove in contour,” and “remove whole structure,” assist the user in manual corrections. Figure 2 displays 2 examples of manual corrections that are often required.
FIGURE 2.
Examples of manual corrections in 2 metastatic castration-resistant prostate cancer patients. (A) Because of their large connections with intestine, retroperitoneal lymph nodes were wrongly classified as having normal uptake and not considered when SUVthr_st was applied. After correction of normal-uptake label, lymph nodes were segmented as soft-tissue lesions. (B) Ureter segmented as soft-tissue lesions and manually changed to normal-uptake label.
Output Parameters
Multiple output parameters are possible and specified in the algorithm. PSMA tumor volume (PSMA-TV), similar to metabolic tumor volume from 18F-FDG PET, represents the volume of all PSMA ligand–positive tumor voxels. PSMA total lesion (PSMA-TL), similar to total-lesion glycolysis from 18F-FDG PET, represents the total PSMA activity from all tumor voxels. PSMA SUVmean is the average SUV in all PSMA ligand–positive tumor voxels, and PSMA SUVmax is the voxel with the highest PSMA expression in the tumor. They are calculated as follows, where N is the number of v tumor voxels.
All 4 PET-derived parameters can be calculated separately for soft-tissue lesions (stPSMA-TV, stPSMA-TL, stPSMA SUVmean, and stPSMA SUVmax) and skeleton lesions (bonePSMA-TV, bonePSMA-TL, bonePSMA SUVmean, and bonePSMA SUVmax). They are added up to the parameters describing whole-body tumor load (wbPSMA-TV, wbPSMA-TL, wbPSMA SUVmean, and wbPSMA SUVmax).
Technical Validation
Four analyses were performed to validate and evaluate the performance characteristics of qPSMA using 20 68Ga-PSMA11 PET/CT data sets.
Liver Threshold Validation
The purpose of liver threshold validation was to validate proper implementation of the semiautomatic algorithm. The liver-based SUVthr_st was calculated using both qPSMA and the open-source software METAVOL, in which it was originally implemented (18).
Intra- and Interobserver Variability
The objective of the intra- and interobserver variability analyses was to evaluate reliability using qPSMA. For intervariability evaluation, 2 trained readers used qPSMA and applied manual corrections independently. To evaluate intraobserver reliability, 1 trained reader analyzed the data sets twice at an interval of 4 wk. For both analyses, all computational steps and recommendations were followed as described above. An SUVthr_bone of 3 and a liver-based SUVthr_st were used. To allow for intrapatient comparison, only slices including the trunk between the first thoracic vertebra and the lower end of the ischium (both easily recognized on CT) were included.
Value Validation
The objective of the value validation analysis was to validate the values of the outcome parameters. Lesions selected from the 20 68Ga-PSMA11 PET/CT scans were individually segmented using qPSMA and commercially available software (Syngo.via; Siemens Medical Solutions).
Feasibility
To evaluate the practicability and learning curve of using qPSMA, the time spent analyzing the datasets included in the intrauser variability for both reads was counted. Computational time was counted from loading of the bone mask until the output parameters were obtained. To assess the feasibility of introducing PSMA ligand PET–derived tumor burden parameters into a clinical setting, correlations between serum prostate-specific antigen (PSA) value and wbPSMA-TV were evaluated. PSA values were obtained at ±2 wk from the 68Ga-PSMA11 PET/CT acquisition.
68Ga-PSMA11 PET/CT Data Sets
All 68Ga-PSMA11 PET/CT scans were performed at our institution before 177Lu-PSMA RLT for metastatic castration-resistant prostate cancer. Patient characteristics, including age and metastasis sites, are presented in Table 1. All patients gave written consent for evaluation of their data. The institutional review board of the Technical University Munich approved this retrospective analysis (permit 5665/13). 68Ga-PSMA11 was administered in compliance with the German Medicinal Products Act, AMG §13(2b), and in accordance with the responsible regulatory body (Government of Oberbayern). 68Ga-PSMA11 was synthesized and PET/CT images obtained as described previously (9). The transaxial pixel size was 4.07 mm for PET and 1.52 mm for CT, with a 5-mm slice thickness for both.
TABLE 1.
Patient Characteristics
| Characteristic | Data |
| Patients (n) | 20 |
| Age (y) | |
| Mean | 73 |
| Range | 65–84 |
| PSA (ng/mL) | |
| Mean | 369 |
| Range | 1–2,222 |
| Site of metastasis (n) | |
| Lymph node, overall | 12 |
| Lymph node only | 1 |
| Bone, overall | 19 |
| Bone only | 1 |
| Bone and lymph node | 12 |
| Local recurrence | 4 |
| Visceral, overall | 3 |
Statistical Analysis
Values are reported as mean and range. Percentage relative differences were calculated by dividing the absolute value of the differences within the measurements by the average of the 2 measurements, all multiplied by 100. Means and 95% confidence intervals (95%CIs) of the relative differences are reported. Paired t testing was used when the values were considered paired. Intraclass correlation coefficient (ICC) estimates and 95%CI were calculated on the basis of a single rater, absolute agreement, and 2-way mixed-effect model (21). Spearman rank correlations were performed to assess the correlations between tumor burden parameters. In each analysis, a P value of less than 0.05 was considered statistically significant. All statistical analyses were performed using IBM SPSS Statistics, version 22.0 (IBM Corp.).
RESULTS
Liver Threshold Validation
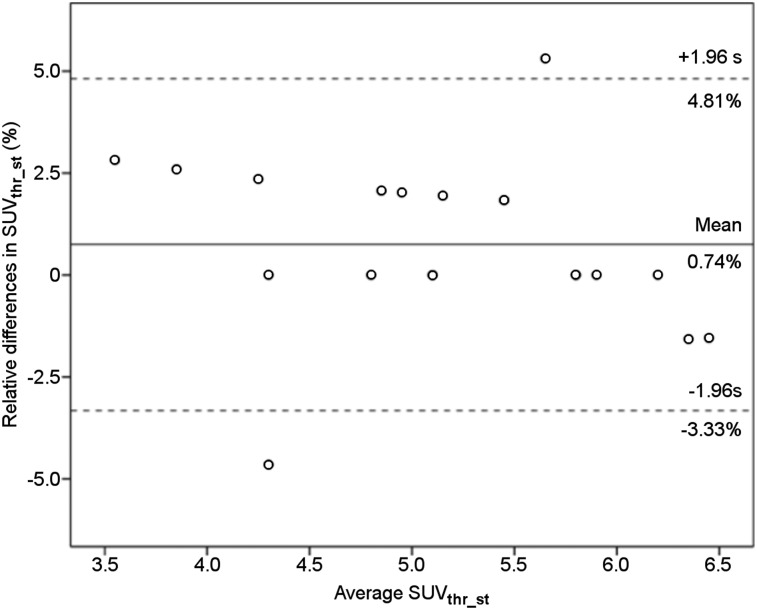
The mean of SUVthr_st computed with METAVOL and qPSMA was 5.18 and 5.14, respectively. Paired t testing did not show a significant difference (P = 0.14). The mean relative difference was 0.74% (95%CI, −0.22%;1.72%). The ICC estimate was 0.996 (95%CI, 0.989;0.998). Figure 3 illustrates the Bland–Altman plot of the differences between SUVthr_st obtained with both software packages.
FIGURE 3.
Bland–Altman plot of qPSMA and METAVOL agreement on semiautomatic computation of SUVthr_st. Solid line indicates average mean difference, and dotted lines delineate 95% limits of agreement (mean ± 1.96 × SD). No systematic difference between the 2 software programs was found.
Intra- and Interobserver Variability
The absolute mean values, the mean and 95%CI of the differences, and the ICC and 95%CI in PSMA-TV, PSMA-TL, PSMA SUVmean, and PSMA SUVmax for bone and soft tissue are presented in Table 2.
TABLE 2.
Intra- and Interobserver Analyses
| Intraobserver analysis |
Interobserver analysis |
|||||
| Output parameter | Read 1 vs. 2 | Difference (%) | ICC | User 1 vs. 2 | Difference (%) | ICC |
| bPSMA-TV (mL) | 801.9 vs. 800.4 | −2.22 (−5.72;1.25) | 1.000 (0.999;1.000) | 801.9 vs. 800.1 | 2.53 (−2.60;7.68) | 1.000 (1.000;1.000) |
| bPSMA-TL | 6397 vs. 6393 | −2.94 (−7.75;1.86) | 1.000 (0.999;1.000) | 61397 vs. 6392 | 2.37 (−1.93;6.68) | 1.000 (1.000;1.000) |
| bPSMA SUVmean | 7.34 vs. 7.39 | −0.73 (−2.24;0.77) | 0.998 (0.998;1.000) | 7.34 vs. 7.33 | −0.16 (−1.99;1.67) | 0.998 (0.996;0.999) |
| bPSMA SUVmax | 38.43 vs. 38.45 | −0.07 (−0.38;0.22) | 1.000 (0.999;1.000) | 38.43 vs. 38.43 | 0.05 (−0.20;0.31) | 1.000 (1.000;1.000) |
| stPSMA-TV (mL) | 67.8 vs. 68.6 | 3.10 (−8.24;14.45) | 1.000 (0.999;1.000) | 67.8 vs. 67.1 | 9.05 (−1.49;19.61) | 0.999 (0.998;1.000) |
| stPSMA-TL | 1026 vs. 1033 | 5.22 (−4.87;15.31) | 1.000 (0.999;1.000) | 1026 vs. 1016 | 8.70 (−1.37;18.77) | 1.000 (0.999;1.000) |
| stPSMA SUVmean | 9.95 vs. 9.96 | −0.43 (−1.86;0.98) | 1.000 (0.999;1.000) | 9.95 vs.9.92 | −0.49 (−1.88;0.90) | 0.999 (0.996;0.999) |
| stPSMA SUVmax | 31.23 vs. 31.20 | 0.18 (−0.15;0.53) | 1.000 (1.000;1.000) | 31.23 vs. 33.21 | 0.32 (−0.25;0.89) | 1.000 (1.000;1.000) |
Data are mean; 95%CIs are in parentheses. P values on paired t testing are all >0.05
Value Validation
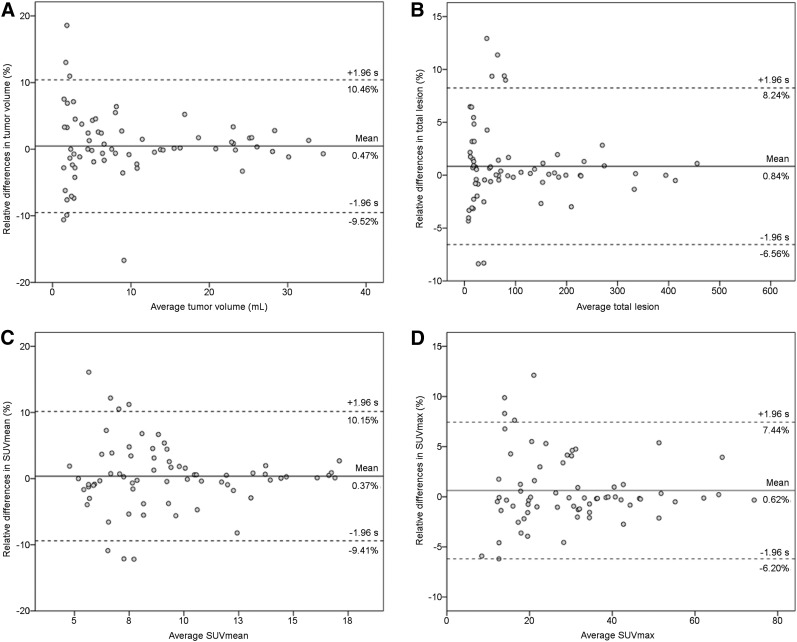
Sixty-eight representative lesions were segmented using both qPSMA and Syngo.via software. The mean values obtained for PSMA-TV, PSMA-TL, SUVmean, and SUVmax were 10.27 versus 10.23 mL, 108.4 versus 107.8, 9.33 versus 9.31, and 30.03 versus 29.87, respectively. Paired t testing did not show significant differences (P > 0.05). Mean relative differences were 0.47% (95%CI, −0.78%;1.72%), 0.84% (95%CI, −0.07%;1.75%), 0.37% (95%CI, −0.83%;1.57%), and 0.62% (95%CI, −0.21%;1.47%) for PSMA-TV, PSMA-TL, SUVmean, and SUVmax, respectively. ICC estimates were 1.000 (95%CI, 0.999;1.000), 1.000 (95%CI, 1.000;1.000), 0.997 (95%CI, 0.994;0.998), and 0.999 (95%CI, 0.999;1.000), respectively. Figure 4 illustrates the Bland–Altman plots of the differences between values obtained with both software packages.
FIGURE 4.
Bland–Altman plots for tumor volume (A), total lesion (B), SUVmean (C), and SUVmax (D) from 68 lesions segmented with qPSMA and Syngo.via software. Solid lines indicate average mean difference, and dotted lines delineate 95% limits of agreement (mean ± 1.96 × SD).
Feasibility
The mean computational time was 13.63 min (range, 8.22–25.45 min) and 9.27 min (range, 8.10–12.15 min) for the first and second reads, respectively. The differences in computational time were significantly different (P = 0.001). For the hardware used (Intel Core i7, 4.2-GHz, 32-GB random-access memory), the time loading masks and liver threshold was approximatively 6 min. At baseline, the mean serum PSA, wbPSMA-TV, and wbPSMA-TL were 369 ng/mL (range, 1–2,222 ng/mL), 827 mL (range, 7–3,076 mL), and 7,005 (range, 62–26,304), respectively. Highly significant correlations were found between PSA and both wbPSMA-TV (r = 0.72, P < 0.001) and wbPSMA-TL (r = 0.66, P = 0.002).
DISCUSSION
To the best of our knowledge, the introduction of qPSMA as a software tool for whole-body tumor segmentation is a novel approach toward semiautomatic analysis of PET data in prostate cancer. Basically, qPSMA integrates various segmentation procedures and PET quantification into a single package to facilitate PSMA ligand assessment of PET tumor burden. In detail, the software uses a fixed SUV threshold having liver background activity as a reference for physiologic PSMA ligand uptake, as recommended in PROMISE (19), and computed using a 3-cm VOI, as recommended in PERCIST (17). To minimize interuser variability, a semiautomatic algorithm was incorporated to place the VOI within the right liver lobe (18). As a novelty, we introduced 2 different thresholds for bone and soft-tissue lesion segmentation that consider their different PSMA ligand uptake levels (14,15).
The results of our investigation indicate that semiautomatic evaluation of bone, soft-tissue, and whole-body tumor load in heavily metastasized prostate cancer patients is feasible. qPSMA is a robust software package with a considerable training effect for the user. Values obtained with our in-house–developed tool agree strongly with commercial software. Its application is of high interest in the setting of PSMA theranostics, in which PSMA ligand PET imaging is typically used for pretreatment evaluation as well as treatment response assessment.
There is increasing use of 18F-FDG PET imaging quantification applying metabolic tumor volume and total-lesion glycolysis as predictor parameters for treatment outcome (22,23).
A first approach in whole-body tumor burden assessment using 68Ga-PSMA11 PET/CT was introduced by Schmuck et al. (24) using an isocontour SUV threshold method. Because the process was time-consuming, only patients with a low tumor load (<10 lesions) were manually analyzed. This resulted in a relatively low mean PSMA-TV and PSMA-TL of 3.4 mL and 33.2 per patient, respectively. Schmidkonz et al. (25) extended the work by analyzing patients with a higher tumor burden (PSMA-TV and PSMA-TL of 7.4 mL and 73.8, respectively). No segmentation time was reported in either work. Nevertheless, such manual segmentation methods are time-consuming, making whole-body tumor burden assessment in heavily metastasized patients not feasible. Therefore, a semiautomatic method would pave the way toward a whole-body tumor load quantification in prostate cancer patients. In the present study, the mean analyzed PSMA-TV and PSMA-TL were 823 mL and 7,273, respectively, indicating that our patient cohort was much more advanced than previous reports in the literature using manual segmentation (24,25). Furthermore, this factor could also explain why we obtained stronger correlations between serum PSA and PSMA ligand PET–derived parameters than those previously described in 2 reports (24,25). However, those 2 reports analyzed patients with biochemical recurrence, and our study investigated patients with metastatic castration-resistant prostate cancer.
Our data indicate that a certain amount of training can reduce the time needed for manual correction; for the second analysis of the same data set, the time was significantly reduced (13.63 vs. 9.27 min). Though further improvements are warranted, the current time needed to analyze each patient is likely to represent a strong advantage over manual approaches. Notably, as qPSMA uses fully automatic steps as well as manual correction, the time needed to process a data set also depends on the specific hardware.
The technical validation of qPSMA resulted in reliable and reproducible data for different items. The implementation of automatic liver background calculation for our data exhibited excellent agreement (ICC of 0.996) with METAVOL as a first tool for this task and only a minimal deviation of 0.74% using a Bland–Altman plot. Quantitative data produced by qPSMA are in strong concordance (all ICC > 0.997) with output from a commercial system (Syngo.via). In data shown using Bland–Altman plots, a bias below 1% was found in all 4 parameters. Interestingly, by increasing lesion volume and uptake, the bias decreases, which shows that lesions with high volumes and high SUVs had a stronger concordance. This finding confirms that small lesions exhibit higher SUV variability (26). For SUVmax, we found a mean difference of 0.6%, which is in concordance with a mean difference of up to 0.5% obtained in a comparison among 4 commercial software packages (27). In terms of reliability, qPSMA offers high intra- and interobserver agreement, with superb ICC estimates and no significant differences between users.
No segmentation method has been yet established for PET as the gold standard. Fixed threshold and isocontour relative threshold have been largely used in PET quantification (22–24). However, it has been shown that despite different results using various segmentation methods or partial-volume effect correction, no significant impact on the predictive or prognostic power of PET-derived parameters could be found (28,29). As a consequence, we focused more on developing a semiautomatic algorithm that allows for whole-body tumor burden assessment in heavily metastasized patients than on deeply analyzing differences between segmentation methods.
We are aware that the presence of liver metastases can interfere with the liver-based threshold because, on the basis of our experience, the semiautomatic algorithm has always placed the 3-cm VOI in healthy tissue. In patients with diffuse liver involvement, this placement can lead to incorrect results. Supplemental Figure 2 displays 2 examples of computation of SUVthr_st in patients with liver metastases. The average liver SUVmean obtained in the present study (4.30) and used to compute SUVthr_st is in concordance with data reported by Gaertner et al. (4.19) (30).
Currently, EBONI (31) is the only software tool available and published for automatic PSMA ligand PET image assessment. However, EBONI was introduced as a tool focusing on skeletal tumor load based only on its approach to fully automatically extracting PET data based on its location within the skeleton on the corresponding CT scan. A fixed SUV threshold of 2.5 was found to be the optimal cutoff for bone lesion segmentation. This is in concordance with the SUVthr_bone of 3 that has been proposed in qPSMA. A drawback of EBONI is the fact that it does not correct for misalignments between the PET and CT scans and thus can miss parts of bone lesions that project outside the bone mask because of breathing or movement. In qPSMA, a specific algorithm is implemented that allows automatic recognition of those parts of bone lesion that lie outside the bone mask because of misalignment. Second, use of EBONI is limited to PET/CT with no intravenous or positive oral contrast agents because they increase the density (Hounsfield units) of certain structures (e.g., intestine and vessels) and will be falsely annotated as bone lesions. This is related to its implementation using automatic masking from the CT data set with no further corrections. In qPSMA, a machine-learning algorithm for bone masks has been trained using multiple features (e.g., Hounsfield units and shape) (16) in addition to manual corrections.
Compared with Syngo.via, qPSMA introduces bone and normal-uptake masks as a novelty. The software package can be installed via a virtual machine on any computer. Because of the interpolation algorithm, any PET/CT data can be open and analyzed. Therefore, the tool can be used at any institution, which could enable a more accurate assessment of radiographic response in the framework of PSMA-targeted radioligand therapies. However, because of the use of different PET/CT scanners and reconstruction methods, it is recommended that each institution first assess the average liver SUVmean in a sufficiently large cohort and subsequently adapt the SUVthr_st formula.
Several limitations of the current version of qPSMA and of our analyses have to be noted. The first drawback is the use of a liver-based threshold, which limits its use in diffuse liver involvement. Additionally, adaptions might be necessary for PSMA ligands that are mainly liver-excreted (e.g., PSMA-1007). Second, lesions with a lower PSMA uptake than liver background activity are missed by the algorithm. Even though the SUV-based threshold is currently the state of the art in PET segmentation, SUV is susceptible to the use of different scanners and reconstruction methods (32). To overcome these issues, for the next version of the software we will focus on shifting from thresholding to convolutional neural networks, which have already shown enhanced accuracy in PET segmentation, as compared with conventional methods (33).
CONCLUSION
qPSMA is a user-friendly and reliable semiautomatic software tool that allows assessment of bone, soft-tissue, and whole-body tumor burden in prostate cancer. Its application to 68Ga-PSMA11 PET/CT is feasible and robust and can help physicians quantify whole-body tumor load in heavily metastasized patients within an acceptable time frame. It is the first step toward further analyses of PSMA ligand PET imaging as a potential image-derived biomarker in prostate cancer, especially within the PSMA theranostics framework.
DISCLOSURE
Matthias Eiber received funding from the SFB 824 (DFG Sonderforschungsbereich 824, project B11) from the Deutsche Forschungsgemeinschaft, Bonn, Germany. Siemens Medical Solutions (Erlangen, Germany) provided the continuous bed motion option for the Biograph mCT as part of an academic collaboration. No other potential conflict of interest relevant to this article was reported.
Supplementary Material
KEY POINTS
QUESTION: Is it possible to develop a semiautomatic tool that allows for whole-body tumor burden assessment in prostate cancer using PSMA PET imaging?
PERTINENT FINDINGS: Using a retrospective cohort of 20 patients with prostate cancer, we showed that semiautomatic quantification of PSMA PET is feasible. The output values obtained using qPSMA were in high correlation with that obtained with a commercial software, while whole-body PSMA-avid tumor volume was significantly correlated with PSA levels.
IMPLICATIONS FOR PATIENT CARE: Whole-body tumor burden assessment using PSMA PET is feasible and might allow for patient outcome prediction or therapeutic response evaluation.
REFERENCES
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. [DOI] [PubMed] [Google Scholar]
- 2.Key Statistics for Prostate Cancer. American Cancer Society website. https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html. Accessed April 26, 2019.
- 3.Survival rates for prostate cancer. American Cancer Society website. https://www.cancer.org/cancer/prostate-cancer/detection-diagnosis-staging/survival-rates.html. Revised February 7, 2019. Accessed April 26, 2019.
- 4.Scher HI, Morris MJ, Stadler WM, et al. Trial design and objectives for castration-resistant prostate cancer: updated recommendations from the prostate cancer clinical trials working group 3. J Clin Oncol. 2016;34:1402–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz LH, Litiere S, de Vries E, et al. RECIST 1.1: update and clarification—from the RECIST committee. Eur J Cancer. 2016;62:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Israeli RS, Powell CT, Corr JG, Fair WR, Heston WD. Expression of the prostate-specific membrane antigen. Cancer Res. 1994;54:1807–1811. [PubMed] [Google Scholar]
- 7.Silver DA, Pellicer I, Fair WR, Heston WD, Cordon-Cardo C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin Cancer Res. 1997;3:81–85. [PubMed] [Google Scholar]
- 8.Eiber M, Fendler WP, Rowe SP, et al. Prostate-specific membrane antigen ligands for imaging and therapy. J Nucl Med. 2017;58(suppl):67S–76S. [DOI] [PubMed] [Google Scholar]
- 9.Eiber M, Maurer T, Souvatzoglou M, et al. Evaluation of hybrid 68Ga-PSMA ligand PET/CT in 248 patients with biochemical recurrence after radical prostatectomy. J Nucl Med. 2015;56:668–674. [DOI] [PubMed] [Google Scholar]
- 10.Afshar-Oromieh A, Avtzi E, Giesel FL, et al. The diagnostic value of PET/CT imaging with the 68Ga-labelled PSMA ligand HBED-CC in the diagnosis of recurrent prostate cancer. Eur J Nucl Med Mol Imaging. 2015;42:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahbar K, Weckesser M, Huss S, et al. Correlation of intraprostatic tumor extent with 68Ga-PSMA distribution in patients with prostate cancer. J Nucl Med. 2016;57:563–567. [DOI] [PubMed] [Google Scholar]
- 12.Bieth M, Kronke M, Tauber R, et al. Exploring new multimodal quantitative imaging indices for the assessment of osseous tumor burden in prostate cancer using 68Ga-PSMA PET/CT. J Nucl Med. 2017;58:1632–1637. [DOI] [PubMed] [Google Scholar]
- 13.Pan J. Image interpolation using spline curves. CiteSeerX website. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.517.3701. Published 2003. Accessed April 26, 2019.
- 14.Schmittgen TD, Teske S, Vessella RL, True LD, Zakrajsek BA. Expression of prostate specific membrane antigen and three alternatively spliced variants of PSMA in prostate cancer patients. Int J Cancer. 2003;107:323–329. [DOI] [PubMed] [Google Scholar]
- 15.Freitag MT, Radtke JP, Hadaschik BA, et al. Comparison of hybrid 68Ga-PSMA PET/MRI and 68Ga-PSMA PET/CT in the evaluation of lymph node and bone metastases of prostate cancer. Eur J Nucl Med Mol Imaging. 2016;43:70–83. [DOI] [PubMed] [Google Scholar]
- 16.Bieth M, Peter L, Nekolla SG, et al. Segmentation of skeleton and organs in whole-body CT images via iterative trilateration. IEEE Trans Med Imaging. 2017;36:2276–2286. [DOI] [PubMed] [Google Scholar]
- 17.Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med. 2009;50(suppl 1):122S–150S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirata K, Kobayashi K, Wong KP, et al. A semi-automated technique determining the liver standardized uptake value reference for tumor delineation in FDG PET-CT. PLoS One. 2014;9:e105682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eiber M, Herrmann K, Calais J, et al. Prostate cancer molecular imaging standardized evaluation (PROMISE): proposed miTNM classification for the interpretation of PSMA-ligand PET/CT. J Nucl Med. 2018;59:469–478. [DOI] [PubMed] [Google Scholar]
- 20.Hofman MS, Hicks RJ, Maurer T, Eiber M. Prostate-specific membrane antigen PET: clinical utility in prostate cancer, normal patterns, pearls, and pitfalls. Radiographics. 2018;38:200–217. [DOI] [PubMed] [Google Scholar]
- 21.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogasch JMM, Hundsdoerfer P, Hofheinz F, et al. Pretherapeutic FDG-PET total metabolic tumor volume predicts response to induction therapy in pediatric Hodgkin’s lymphoma. BMC Cancer. 2018;18:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikhaeel NG, Smith D, Dunn JT, et al. Combination of baseline metabolic tumour volume and early response on PET/CT improves progression-free survival prediction in DLBCL. Eur J Nucl Med Mol Imaging. 2016;43:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmuck S, von Klot CA, Henkenberens C, et al. Initial experience with volumetric 68Ga-PSMA I&T PET/CT for assessment of whole-body tumor burden as a quantitative imaging biomarker in patients with prostate cancer. J Nucl Med. 2017;58:1962–1968. [DOI] [PubMed] [Google Scholar]
- 25.Schmidkonz C, Cordes M, Schmidt D, et al. 68Ga-PSMA-11 PET/CT-derived metabolic parameters for determination of whole-body tumor burden and treatment response in prostate cancer. Eur J Nucl Med Mol Imaging. 2018;45:1862–1872. [DOI] [PubMed] [Google Scholar]
- 26.Brendle C, Kupferschlager J, Nikolaou K, la Fougere C, Gatidis S, Pfannenberg C. Is the standard uptake value (SUV) appropriate for quantification in clinical PET imaging? Variability induced by different SUV measurements and varying reconstruction methods. Eur J Radiol. 2015;84:158–162. [DOI] [PubMed] [Google Scholar]
- 27.Arain Z, Lodge M, Wahl R. A comparison of SUV parameters across four commercial software platforms [abstract]. J Nucl Med. 2015;56(suppl):580.25698781 [Google Scholar]
- 28.Kanoun S, Tal I, Berriolo-Riedinger A, et al. Influence of software tool and methodological aspects of total metabolic tumor volume calculation on baseline [18F]FDG PET to predict survival in Hodgkin lymphoma. PLoS One. 2015;10:e0140830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatt M, Le Pogam A, Visvikis D, Pradier O, Cheze Le Rest C. Impact of partial-volume effect correction on the predictive and prognostic value of baseline 18F-FDG PET images in esophageal cancer. J Nucl Med. 2012;53:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaertner FC, Halabi K, Ahmadzadehfar H, et al. Uptake of PSMA-ligands in normal tissues is dependent on tumor load in patients with prostate cancer. Oncotarget. 2017;8:55094–55103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammes J, Tager P, Drzezga A. EBONI: a tool for automated quantification of bone metastasis load in PSMA PET/CT. J Nucl Med. 2018;59:1070–1075. [DOI] [PubMed] [Google Scholar]
- 32.Adams MC, Turkington TG, Wilson JM, Wong TZ. A systematic review of the factors affecting accuracy of SUV measurements. AJR. 2010;195:310–320. [DOI] [PubMed] [Google Scholar]
- 33.Hatt M, Laurent B, Ouahabi A, et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal. 2018;44:177–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.