ABSTRACT
Most essential physiological functions in mammals show a 24-h rhythmic pattern, which includes sleep-wake, feeding-non-feeding cycles and energy metabolism. Recent studies indicate that macroautophagy/autophagy also displays a robust circadian rhythmicity following the daily feeding pattern in adult mammals. We discovered that MiT-TFE transcription factors TFEB and TFE3, master regulators of autophagy and lysosomal biogenesis, are activated in a circadian manner and drive the expression of NR1D1/REV-ERBα, a key component of the core clockwork, thus revealing a molecular link between the nutrient-driven circadian cycle and the light-induced molecular clock. The dynamic balance between TFEB and TFE3 activation and NR1D1 expression is responsible for the modulation and oscillation of autophagy and metabolism genes.
KEYWORDS: Autophagy, circadian rhythm, gene oscillation, NR1D1/REV-ERBα, TFEB/TFE3
Biological clocks are responsible for the 24-h cycles in behavioral and biochemical processes operating in all cells and allowing physiological anticipation of daily environmental changes. Organisms evolved to restrict their activity to night or day by developing an endogenous circadian clock to ensure that physiological processes are performed at optimal times and to minimize futile reactions.
In mammals, circadian rhythms are generated by the central circadian clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus, which constantly synchronizes with environmental cues via its own circadian pathways, thus controlling the peripheral clock through circadian output pathways. Secondary oscillators are present in many brain regions and peripheral organs (e.g. liver, muscle, white adipose tissue). SCN clock genes are synchronized by light, while clock oscillations in peripheral tissues largely depend on feeding time.
Under homeostatic conditions, the clock acts as a driver of organismal metabolism. Perhaps not surprisingly, perturbations of either the circadian or metabolic system (e.g., shiftwork or jet-lag) can result in the disruption of metabolic pathways and circadian clock function leading to the development of metabolic disorders such as obesity and diabetes. Increasing evidence suggests that this timekeeping mechanism controls a much greater proportion of rhythmic physiology that have no obvious links to the 24-h day, such as cell proliferation, inflammation and DNA damage response.
Autophagy, a highly conserved mechanism of recycling in the cell, displays a robust circadian rhythmicity in mouse tissues. A tight and synchronized regulation of metabolism and autophagy is necessary to optimize nutrient storage or consumption. However, the mechanisms that regulate autophagy oscillation are still under investigation.
TFEB and TFE3 are master controllers of autophagy and lysosomal biogenesis, and energy metabolism. MTOR (mechanistic target of rapamycin kinase) negatively regulates the activity of TFEB and TFE3 in a nutrient-dependent manner, contributing to the switch between anabolic and catabolic pathways. In our recent study [1], we hypothesized that TFEB and TFE3 contribute to the circadian modulation of autophagy activation and regulate, directly or indirectly, the molecular clock. We showed that in mice TFEB and TFE3 are rhythmically activated in peripheral tissues in a nutrient-dependent/clock-independent manner during the light/rest phase, consistent with the activation of the autophagy pathway and with the inactivation of MTOR. Depletion of TFEB and TFE3 in liver and muscle results in blunted autophagy activation during the light phase as demonstrated by impaired induction of autophagy genes and autophagy flux measured by LC3-II accumulation after autophagy inhibition (Figure 1).
Figure 1.
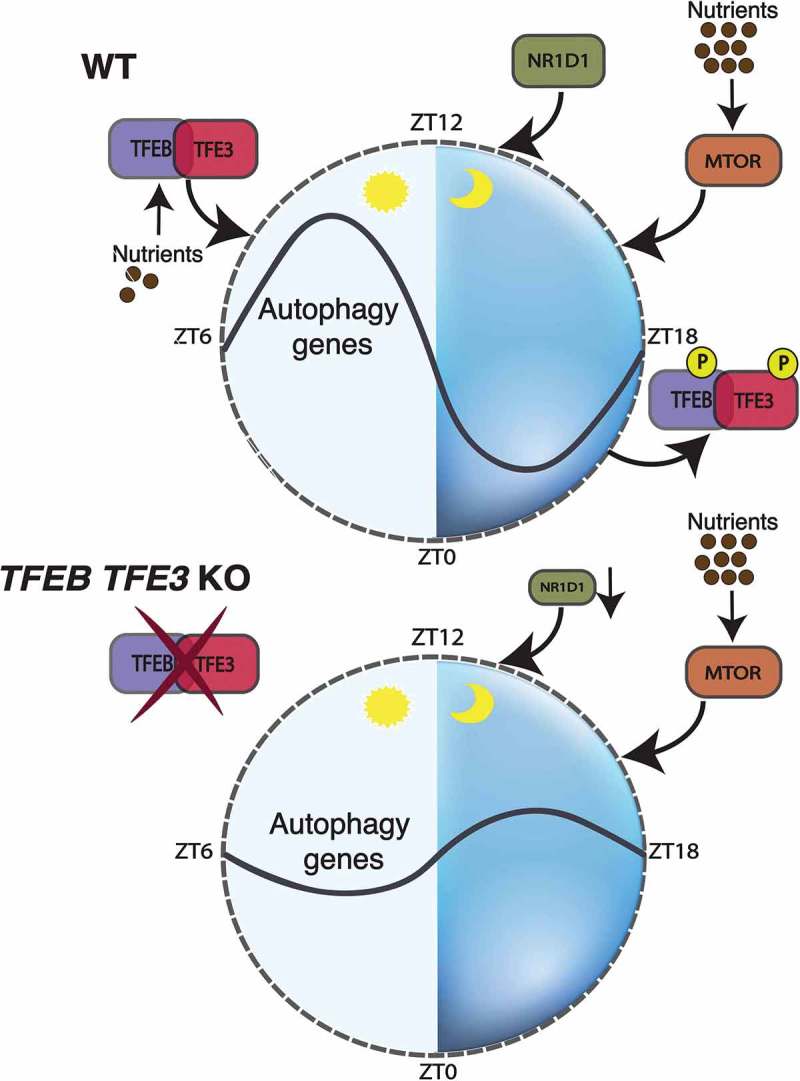
The nutrient-sensitive factors TFEB and TFE3 link autophagy to the circadian molecular clock. In normal conditions, TFEB and TFE3 translocate to the nucleus in the absence of nutrients (light phase) and induce the expression of genes involved in autophagy and metabolism. NR1D1, activated by both circadian signals and TFEB or TFE3 at Zeitgeber time (ZT) 8, inhibits the expression of its targets including autophagy genes. Nutrients activate MTOR in the dark phase, contributing to the inactivation of TFEB and TFE3 and the complete inhibition of autophagy. In the absence of TFEB and TFE3, autophagy is blunted during the light phase and loses the oscillatory pattern due to the reduced expression of NR1D1. Thus, the crosstalk between TFEB and TFE3, NR1D1 and MTOR contributes to circadian oscillation of autophagy.
We then sought to determine whether TFEB- and TFE3-driven regulation of gene expression has an influence on the circadian clockwork. By comparing previously published ChIP-seq data for TFEB and TFE3, we found that TFEB and TFE3 directly bind to the promoter of Nr1d1, a negative regulator of several clock (e.g., Arntl/Bmal1, Npas2), metabolism (e.g., Srebf1/Srebp1, Fasn) and autophagy (e.g., Atg5, Ulk1) genes. Interestingly, TFEB and TFE3 do not require formation of a complex with ARNTL-CLOCK to regulate Nr1d1 expression, suggesting that they act in parallel to the core components of the clock machinery. Comparative analysis of the cistromes of TFEB and TFE3, and NR1D1 shows an extensive overlap of their binding sites, particularly in genes involved in autophagy and metabolic functions. Because NR1D1 is a transcriptional inhibitor, we postulated that the presence of both E-boxes/CLEAR sites and NR1D1/REV-ERBα-responsive elements (ROREs) in the same promoters resulted in a coordinated regulation of gene expression leading to a tight control of cell homeostasis over the diurnal cycle. Indeed, overexpression of TFEB and TFE3 in cells with the concomitant knockdown of the endogenous Nr1d1 results in enhanced expression of the autophagy genes and the autophagy flux. Thus, opposite but interconnected forces driven by TFEB and TFE3 and NR1D1 dictate the timing of autophagy activation. Their dynamic interaction over the diurnal cycle allows the cell to anticipate environmental changes. In line with our findings, mice deficient for TFEB and TFE3 show an altered free-running period after 2 weeks of constant darkness, decreased activity during the light phase and deregulated expression of the core components of the circadian clock compared with control animals. These results strongly support the tight connection between TFEB and TFE3 and circadian physiology.
In conclusion, our study identifies a novel mechanism linking nutrient availability and autophagy with the cell-autonomous circadian clock through the regulation of gene expression.
Funding Statement
This work was supported by a grant from the US National Institutes of Health (R01-NS078072), AIRC (Italian Association for Cancer Research) (IG 2015-17639), Foundation Louis-Jeantet and Telethon Foundation to AB.
Disclosure statement
A. Ballabio is a co-founder of CASMA Therapeutics, Boston, MA, USA.
Reference
- [1].Pastore N, Vainshtein A, Herz NJ, et al. Nutrient-sensitive transcription factors TFEB and TFE3 couple autophagy and metabolism to the peripheral clock. Embo J. 2019;38:e101347. [DOI] [PMC free article] [PubMed] [Google Scholar]


