Abstract
Background
The role of genetic polymorphisms in latent tuberculosis (TB) infection and progression to active TB is not fully understood.
Methods
We tested the single-nucleotide polymorphisms (SNPs) rs5743708 (TLR2), rs4986791 (TLR4), rs361525 (TNFA), rs2430561 (IFNG) rs1143627 (IL1B) as risk factors for tuberculin skin test (TST) conversion or development of active TB in contacts of active TB cases. Contacts of microbiologically confirmed pulmonary TB cases were initially screened for longitudinal evaluation up to 24 months, with clinical examination and serial TST, between 1998 and 2004 at a referral center in Brazil. Data and biospecimens were collected from 526 individuals who were contacts of 177 active TB index cases. TST conversion was defined as induration ≥5 mm after a negative TST result (0 mm) at baseline or month 4 visit. Independent associations were tested using logistic regression models.
Results
Among the 526 contacts, 60 had TST conversion and 44 developed active TB during follow-up. Multivariable regression analysis demonstrated that male sex (odds ratio [OR]: 2.3, 95% confidence interval [CI]: 1.1–4.6), as well as SNPs in TLR4 genes (OR: 62.8, 95% CI: 7.5–525.3) and TNFA (OR: 4.2, 95% CI: 1.9–9.5) were independently associated with TST conversion. Moreover, a positive TST at baseline (OR: 4.7, 95% CI: 2.3–9.7) and SNPs in TLR4 (OR: 6.5, 95% CI: 1.1–36.7) and TNFA (OR: 12.4, 95% CI:5.1–30.1) were independently associated with incident TB.
Conclusions
SNPs in TLR4 and TNFA predicted both TST conversion and active TB among contacts of TB cases in Brazil.
Keywords: single-nucleotide polymorphism, tuberculin skin test, Mycobacterium tuberculosis, tumor necrosis factor, Toll-like receptor
Immune-related genetic polymorphisms were screened in a large cohort of contacts of active tuberculosis patients from Brazil. Single-nucleotide polymorphisms of TLR4 and TNFA were independently associated with increased risk for tuberculin skin test conversion and development of active tuberculosis.
Approximately 1.7 billion individuals are infected with Mycobacterium tuberculosis (Mtb), representing one-quarter of the global population [1]. Because bacille Calmette-Guerin (BCG) vaccine does not protect either against infection or tuberculosis (TB) disease in adults, the only currently effective strategy to prevent active TB in adults is treatment of latent TB infection (LTBI). Treatment is efficacious in decreasing TB risk; however, compliance is low, and effectiveness therefore decreased, particularly with longer-course regimens [2]. Although the World Health Organization has recently emphasized the need to treat LTBI, high burden countries are unable to implement full-scale contact investigations and LTBI treatment. Of note, if left untreated, only a small proportion (5–10%) of infected persons will develop active disease [3]. Although some risk factors for developing TB disease have been recognized, such as human immunodeficiency virus (HIV) coinfection, diabetes, young age, and recently acquired Mtb infection [4], many TB patients do not have any known risk factors. To identify those who would most benefit from LTBI treatment, biomarkers for susceptibility have been investigated. Interferon-gamma release assays have been widely tested as a marker of LTBI and, to a lesser extent, susceptibility to TB disease [5]. However, these tests do not discriminate between active disease and LTBI and, more importantly, have a low predictive value for progression to TB [6].
In addition, not all contacts of pulmonary TB patients acquire Mtb infection. A meta-analysis reported great variability in the proportion of infected household contacts with a positive tuberculin skin test (TST) [7]. Transmission of Mtb depends on index case-related factors, such as bacillary burden and duration of cough [7] and on contact-related factors, such as degree of exposure and individual genetic susceptibility [8]. Mtb infection and progression to TB disease may have distinct genetic influences that underlie the biological mechanisms involved in individual susceptibility [9]. Robust activation of the innate immune response is considered an essential prerequisite for protective immunity and vaccine efficacy. However, data published to date provide an incomplete view of the functional importance of innate immunity in TB [10].
Some key genetic components of protective immunity in human TB include Toll-like receptor (TLR)2, TLR4, tumor necrosis factor (TNF)A, interferon (IFN)G and interleukin (IL)1B [11–14]. Indeed, immune-related single-nucleotide polymorphisms (SNPs) such as TLR2 rs5743708 [15], TLR4 rs4986791 [13],TNFA rs361525 [16], IFNG rs2430561 [17], IL1B rs1143627 [18], and many others, have all been suggested to influence susceptibility to TB, but the functional immunologic correlates are still unclear. The objective of this study was to evaluate potential genetic biomarkers of susceptibility to Mtb infection and TB disease. We studied close contacts of microbiologically confirmed pulmonary TB patients to estimate the risk of Mtb infection (TST conversion) and development of active TB according to the presence of 5 immune-related SNPs, while also accounting for clinical and epidemiological factors.
MATERIALS AND METHODS
Ethics Statement
Written informed consent was obtained from all participants or their legally responsible guardians, and all clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Clementino Fraga Filho University Hospital (HUCFF), Federal University of Rio de Janeiro Ethics Review Board. The anonymity of study subjects was preserved, and all study specimens were de-identified.
Study Design
We performed a longitudinal study of contacts of pulmonary TB patients at the time of diagnosis from November 1998 through March 2004. TB was diagnosed by acid-fast bacilli (AFB) smear and/or culture, according to Brazilian Ministry of Health Guidelines [19]. All TB index cases diagnosed at HUCFF >18 years old. Investigation of TB cases included data on cough, AFB sputum grade, and chest radiographs. After identifying TB cases, we searched for their close contacts. TB contacts were defined as living in the same household or reporting contact with the TB index case for ≥20 hours weekly for 2 months. All individuals identified who were ≥9 years old were invited to participate in the study and were evaluated and screened for active TB following Brazilian guidelines [19]. Prevalent TB cases among close contacts were excluded from analyses.
Procedures
Close contacts were evaluated at baseline and also 4 and 12 months after identification of the TB index case. At first visit, a standardized questionnaire was administered to obtain demographic and clinical data, including type and duration of contact with the index case, and history of risk factors for TB (eg, HIV, diabetes, hematologic malignancies, and use of immunosuppressant drugs). If a contact was a grandparent, parent or sibling of the index case, they were considered to have consanguinity (this definition extended to children with the index case), whereas spouses or other relationships did not. At study baseline, a medical visit and chest radiograph were scheduled. BCG scar was assessed, and TST performed by a trained nurse using the Mantoux technique [19], with 2 tuberculin units of the purified protein derivative RT 23 (Statens Serum Institute, Copenhagen, Denmark). TST reading was performed 48–72 hours after administration. Additional TST screening was performed at months 4 and 12 to evaluate for possible TST conversion.
TST Interpretation and TB Diagnosis
A positive TST was defined as ≥5 mm induration, according to the Brazilian Ministry of Health [19]. A positive TST at the first visit was considered to represent LTBI. Contacts with any TST ≥5 mm were not retested with TST. During the study period, the Brazilian National TB Guidelines indicated that treatment of TST-positive individuals was not mandatory, and assessment of cost-benefit of therapy with isoniazid for 6 months was performed by healthcare workers prior to a decision to treat [19]. If treatment was not initiated, individuals were followed up with periodic examinations to identify development of active TB disease. Twenty-nine participants received isoniazid.
Contacts with signs or symptoms suggestive of active TB underwent medical visits and investigation for TB disease by acid-fast bacilli (AFB) smear and culture in Lowenstein Jensen (LJ) medium. Active TB was diagnosed when ≥1 specimen yielded a positive microbiologic (AFB smear or culture) result. An incident active TB case was defined as TB diagnosed after baseline study assessment. All patients (n = 526) were contacted after 12 additional months (24 months after study enrollment) to assess for incident TB disease. Data on TB incidence from all individuals who could not be contacted at month 24 (n = 168) were collected by searching the Brazil’s Information System for Notifiable Diseases (SINAN). Of 44 incident TB cases, 8 (18%) had TB diagnosis extracted from SINAN rather than at month 24 interview.
Genotyping
Genomic DNA was extracted from peripheral blood collected from TB contacts at study enrollment. DNA extraction and genotyping were performed using the FlexiGene kit (Qiagen, Germany). Genotypes of 5 gene polymorphisms TLR2 (rs5743708), TLR4 (rs4986791), TNFA (rs361525), IFNG (rs2430561), and IL1B (rs1143627) were detected using polymerase chain reaction (PCR) restriction fragment length polymorphism (RFLP) method [20, 21]. The primer sequences are in Supplementary Table 1. The PCR products were digested by the enzymes Msp I for TLR2, Hinf I for TLR4, BamHI for TNFA, AvaII for IFNG, and AluI for IL1B.
Data Analysis
Categorical data were presented as proportions and continuous data as medians and interquartile ranges (IQR). The frequency distributions of alleles (wild type vs variant) for each polymorphism were compared. The Fisher and χ2 tests were used to compare categorical variables between study groups. Continuous variables were compared using the Mann-Whitney U test. A multivariable regression model using variables with univariate P-value ≤ .2 was performed to assess the odds ratios (OR) and 95% confidence intervals (CIs) of the associations with TST conversion and incident active TB. For analysis of TLR4 in the multivariable model, there was no event among participants who remained TST negative; thus for OR calculation, we added “1” to the group without detected events. In addition, we employed Bayesian Network modeling [22] to infer causal relationships between TST conversion and active TB disease and sociodemographic, clinical, laboratory, and genetic parameters, with 100× bootstrapping. Only associations which remained statistically significant in >20 of 100× bootstraps were considered significant. A P-value < .05 was considered statistically significant.
RESULTS
Characteristics of the Study Participants
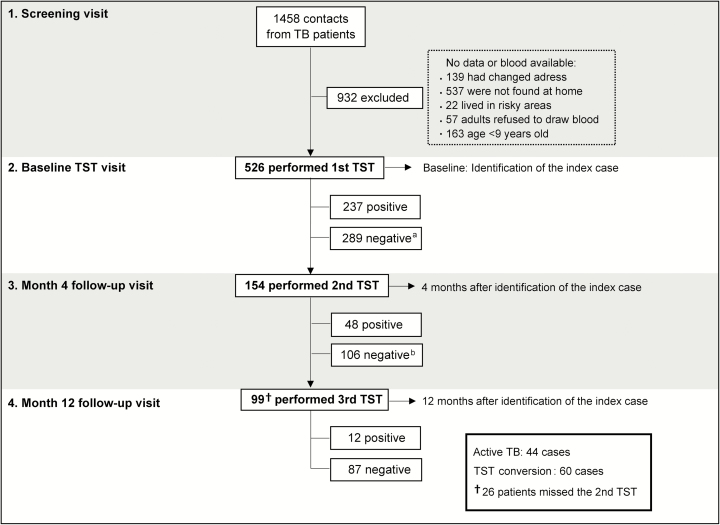
We approached 1458 contacts of 1191 microbiologically confirmed TB index cases who attended HUCFF between 1998 and 2004. Of those, 932 persons were excluded for the reasons listed in Figure 1. The final study population, from which we collected data and samples, included 526 contacts of 177 TB index cases. The description of the study population is in Table 1. The study population was mostly female, household contacts, and consanguineous with the index case. Indeed, 474 persons (90.5%) were household contacts, with a high rate of consanguinity with the index case (62.5%). There were low frequencies of HIV infection, alcohol use, illicit drug use, and use of immunosuppressant drugs. Only 8 persons (1.8%) had a history of TB. At baseline, few reported cough for more than 4 weeks, and of these, only 3 had a positive AFB smear and were then treated for TB. During the evaluation of the index cases associated with the contacts, almost all had TB diagnosis confirmed by culture and cough for more than 4 weeks. TB index cases frequently exhibited high bacterial loads in sputum (41.1% had AFB grade ≥ +2). In addition, 84 index TB patients had cavitary lesions on chest radiograph.
Figure 1.
Study flow chart. Index case: first tuberculosis case identified in the household. aMissing 2nd TST: 135 cases; bMissing 3rd TST: 33 cases and 26 people who missed the 2nd TST showed up. Abbreviation: TST, tuberculin skin test.
Table 1.
Clinical and Demographic Characteristics of the Study Population
| Characteristics of Contact | n/N | n | (%) |
|---|---|---|---|
| Age, median (IQR) | 526/526 | 35 | (33–38) |
| Male | 526/526 | 181 | (34.4) |
| Consanguinity with index case | 526/526 | 329 | (62.5) |
| BCG vaccination | 521/526 | 177 | (33.9) |
| HIV infection | 31/526 | 4 | (12.9) |
| IDU | 439/526 | 7 | (1.6) |
| Smoking | 524/526 | 131 | (25.0) |
| Alcohol use | 444/526 | 1 | (0.2) |
| Prior tuberculosis | 440/526 | 8 | (1.8) |
| Household contacta | 523/526 | 474 | (90.5) |
| Frequency of contact (>20 hours) | 526/526 | 489 | (93.0) |
| Comorbid conditionsb | 500/526 | 127 | (25.4) |
| Immunosuppressant drugs | 444/526 | 3 | (0.7) |
| Cough (> 4 weeks) | 518/526 | 19 | (3.6) |
| Positive AFB screening | 429/526 | 3 | (0.7) |
| Characteristics of TB index case | |||
| Cavities on chest x-ray | 517/526 | 84 | (16.2) |
| Cough (> 4 weeks) | 518/526 | 470 | (90.7) |
| ≥2+ AFB | 444/526 | 200 | (41.1) |
| Positive culture | 367/526 | 352 | (95.9) |
Abbreviations: AFB, acid fast bacilli; BCG, bacille Calmette-Guerin; HIV, human immunodeficiency virus; IDU, illicit drug use; IQR, interquartile range; n, number of persons for whom such data were available; N, number total that participants from the study available; TB, tuberculosis.
aHousehold contact is defined as living in the same household or reporting contact with the TB index case for >20 hours weekly for 2 months.
bComorbidities: renal failure, diabetes, heart failure, and/or hypertension, chronic obstructive pulmonary disease, neoplasia, systemic lupus erythematous, and hepatitis.
Variant alleles of IFNG were the most common polymorphism in the study population, present in 82.9% of the participants (Table 2). Variations in the IL1B gene were also common (47%), whereas polymorphisms in TLR2, TLR4, and TNFA genes were less common (Table 2).
Table 2.
Gene Polymorphisms of the Study Participants
| SNP | n | (%) |
|---|---|---|
| rs5743708 (TLR2) | ||
| GG | 365 | (83.9) |
| GA+AAa | 70 | (16.1) |
| rs4986791 (TLR4) | ||
| CC | 410 | (96.7) |
| CT+TTa | 14 | (3.3) |
| rs361525 (TNFA) | ||
| GG | 447 | (85.8) |
| GA+AAa | 74 | (14.2) |
| rs2430561 (IFNG) | ||
| TT | 69 | (17.1) |
| TA+AAa | 335 | (82.9) |
| rs1143627 (IL1B) | ||
| TT | 254 | (53.0) |
| TC+CCa | 225 | (47.0) |
Data on 526 individuals are shown.
Abbreviations: IFNG, interferon gamma; IL1B, interleukin-1 beta; SNP, single-nucleotide polymorphism, TLR, Toll-like receptor, TNFA, tumor necrosis factor α.
aVariant alleles of SNP.
Association Between Polymorphisms and TST Conversion
Exposure to Mtb at the time of study enrollment was examined by TST screening of the 526 individuals; 237 (45.1%) had a positive TST (Figure 1). There were 135 individuals who missed the month 4 visit, and 154 (53.3% of those TST negative at baseline) were retested. A positive TST was detected in 48 individuals, representing 16.6% of the participants with an initially negative TST. At month 12, a third TST was performed in TB contacts who remained TST negative at month 4. In addition, 26 participants who missed TST testing at month 4 were tested at month 12. A total of 99 individuals were tested. Twelve individuals had a positive TST at this time point. Thus, during the study period, 60 persons converted to a positive TST, suggesting recent Mtb infection.
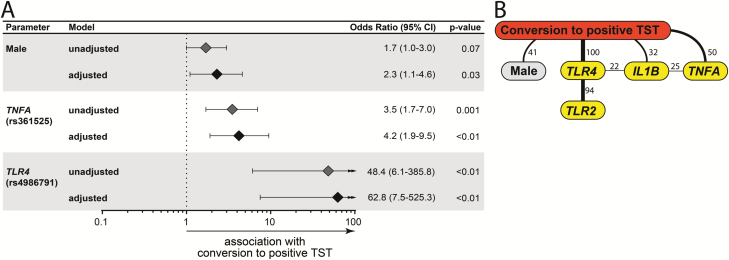
TST converters were more commonly male and more frequently household contacts than nonconverters (Table 3). Other characteristics were similar between converters and nonconverters. Univariate analyses indicated that variant alleles in TLR2 (P = .03), TLR4 (P < .01), and TNFA (P = .001) were associated with TST conversion, whereas mutant IL1B (P = .006) alleles were more common in those who did not convert (Table 4). Multivariable regression analysis confirmed that male sex and genetic variants in TLR4 and TNFA were all independently associated with increased odds of TST conversion (Figure 2A), whereas IL1B SNP was not significant (adjusted OR: 0.6, 95% CI: 0.28–1.29, P = .191).
Table 3.
Characteristics of the Study Participants Evaluated for Conversion From TST Negative to TST Positive
| Characteristic | Conversion | TST Negative | OR (95% CI) | P-Value | |
|---|---|---|---|---|---|
| n/N | n = 60 | n = 224 | |||
| Age, median (IQR) | 284/284 | 37 (15.59) | 34 (21–53) | .85 | |
| Male | 284/284 | 28 (46.7) | 76 (33.9) | 1.7 (1.0–3.0) | .072 |
| Consanguinity with index case | 284/284 | 36 (60.0) | 142 (63.4) | 0.9 (0.5–1.6) | .65 |
| BCG vaccination | 281/284 | 17 (28.8) | 74 (33.3) | 0.8 (0.4–1.5) | .54 |
| HIV infection | 20/284 | 1 (5.9) | 2 (66.7) | 0.03 (0.0–0.7) | .05 |
| Nonwhite race | 275/284 | 28 (48.3) | 117 (53.9) | 0.8 (0.4–1.4) | .46 |
| IDU | 230/284 | 0 (0) | 3 (1.5) | … | 1 |
| Smoking | 283/284 | 15 (25.0) | 56 (25.1) | 1 (0.5–1.9) | 1 |
| Alcohol consumption | 231/284 | 0 (0) | 0 (0) | … | … |
| Prior tuberculosis | 229/284 | 1 (2.9) | 0 (0) | … | … |
| Household contact | 282/284 | 50 (83.3) | 195 (87.8) | 0.7 (0.3–1.5) | .39 |
| Frequency of contact (>20 hours) | 284/284 | 56 (93.3) | 206 (92.0) | 1.2 (0.4–3.8) | 1 |
| Comorbid conditions | 266/284 | 9 (16.1) | 54 (25.7) | 0.5 (0.3–1.2) | .16 |
| Immunosuppressant drugs | 231/284 | 0 (0) | 0 (0) | … | |
| Cough (> 4 weeks) | 283/284 | 2 (3.3) | 5 (2.2) | 1.5 (0.3–7.9) | .64 |
| Positive AFB | 231/284 | 0 | 1 (0.2) | … | .92 |
| Characteristics of TB index case | |||||
| Cavities on chest x-ray | 276/284 | 4 (7.1) | 24 (10.9) | 0.6 (0.2–1.9) | .62 |
| Cough (> 4 weeks) | 283/284 | 30 (50.0) | 88 (39.5) | 1.5 (0.9–2.7) | .18 |
| ≥2 AFB | 256/284 | 16 (30.8) | 79 (38.7) | 0.7 (0.4–1.4) | .37 |
| Positive culture | 201/284 | 39 (95.1) | 151 (94.4) | 1.2 (0.2–5.6) | 1 |
Data represent no. (%). Comorbidities: diabetes, heart failure, and/or hypertension, chronic obstructive pulmonary disease, neoplasia, systemic lupus erythematous, and hepatitis.
Abbreviations: AFB, acid fast bacilli; CI, confidence interval; IDU, illicit drug use; n, number of persons for whom such data were available; N, number total that participants from the study available; OR, odds ratio; TB, tuberculosis; TST, tuberculin skin test.
Table 4.
Gene Polymorphisms of the Study Participants Evaluated for Conversion From TST Negative to TST Positive
| SNP | Conversion | TST Negative | OR | 95% CI | P-Value |
|---|---|---|---|---|---|
| n = 60 | n = 224 | ||||
| rs5743708–TLR2 | 15 (29.4) | 27 (15.1) | 2.3 | (1.1–4.9) | .03 |
| rs4986791–TLR4 | 11 (21.6) | 0 (0) | … | … | <.01 |
| rs361525–TNFA | 18 (30.0) | 24 (10.9) | 3.5 | (1.7–7.0) | .001 |
| rs2430561–IFNG | 36 (78.3) | 140 (83.8) | 0.6 | (0.3–1.6) | .38 |
| rs1143627–IL1B | 14 (25.5) | 94 (46.3) | 0.4 | (0.2–0.8) | .006 |
Data represent no. (%).
Abbreviations: CI, confidence interval; IFNG, interferon gamma; IL1B, interleukin-1beta; OR, odds ratio; SNP: single-nucleotide polymorphism; TLR, Toll-like receptor; TNFA, tumor necrosis factor α; TST, tuberculin skin test.
Figure 2.
Factors associated with TST conversion. A, Multivariable regression model of variables shown in Tables 3 and 4, which displayed univariate P-value ≤ .2. B, Bayesian network with bootstrap (100×) was used to illustrate the statistically significant associations between the parameters and the presence of TST conversion in the study population. Lines represent direct associations. Associations that remained statistically significant on ≥20 of 100 bootstraps are plotted. Numbers of times each association persisted during bootstrap are shown. Bold lines highlight the strongest associations. All parameters from Table 3 were included. Only those displaying significant associations are shown. Abbreviation: TST, tuberculin skin test.
Furthermore, we applied Bayesian network modeling to infer causal relationships between the presence of polymorphisms and TST conversion, and all recorded statistically relevant demographic, epidemiologic, and behavioral information from univariate analyses were cited above. This approach confirmed the strong direct associations between male sex, polymorphisms in TLR4 and TNFA, in addition to IL1B, with TST conversion (Figure 2B). The TLR2 polymorphism was not directly connected to TST conversion but was associated with TLR4 SNP using the Bayesian network approach. In fact, 10 out of 11 individuals with TST conversion and the TLR4 SNP also had the TLR2 polymorphism.
Individuals who were TST positive at study baseline (n = 203) were similar to those who were TST negative and did not convert nor develop active TB during study follow-up (n = 224) with regard to most of the characteristics evaluated, including the SNPs (Supplementary Table 2). Cavitary lesions as well as cough in the index TB cases were more frequent in participants who were TST positive at the first visit compared to those who remained TST negative (P = .005 and P = .009, respectively).
Association Between Polymorphisms and Incident TB
Incident TB was higher in those who were TST positive at baseline (Table 5). Only 2 of the 29 individuals who received isoniazid therapy developed incident TB. In addition, index cases from participants who developed active TB more frequently had cavitary lung lesions identified on chest x-ray compared to index cases of contacts who did not develop TB (Table 5). Lastly, incident TB was more frequent in participants who had allelic variants in both TLR4 and TNFA genes (Table 6).
Table 5.
Characteristics of Contacts of Pulmonary TB Cases Evaluated for Development of Active TB Disease
| Characteristic | n/N | Active TB n = 44 |
No Active TB n = 482 |
OR (95% CI) | P-Value |
|---|---|---|---|---|---|
| Age – median (IQR) | 526/526 | 32 (29–39) | 39 (34–40) | … | .04 |
| Male | 526/526 | 18 (40.9) | 163 (33.8) | 1.4 (0.7–2.5) | .4 |
| Consanguinity with index case | 526/526 | 31 (70.5) | 299 (62.0) | 1.5 (0.8–2.6) | .3 |
| BCG vaccination | 521/526 | 14 (31.8) | 163 (34.1) | 0.9 (0.5–1.7) | .8 |
| HIV infection | 31/526 | 2 (22.2) | 2 (9.1) | 2.9 (0.3–24.3) | .6 |
| Nonwhite race | 505/526 | 21 (46.9) | 258 (55.8) | 0.8 (0.4–1.4) | .4 |
| IDU | 439/526 | 1 (3.4) | 6 (1.5) | 2.4 (0.3–20.7) | .4 |
| Smoking | 524/526 | 12 (27.3) | 119 (24.8) | 1.1 (0.6–2.3) | .7 |
| Alcohol use | 444/526 | 1 (3.1) | 0 | … | .07 |
| Prior TB | 440/526 | 6 (19.4) | 2 (0.5) | 48.8 (9.4–254.4) | <.01 |
| Household contact | 523/526 | 41 (93.2) | 433 (92.9) | 1.5 (0.4–5.0) | .8 |
| Frequency of contact (>20 hours) | 526/526 | 42 (95.5) | 448 (92.9) | 1.6 (0.4–6.9) | .4 |
| Comorbid conditions | 500/526 | 10 (25.0) | 117 (25.4) | 1.0 (0.4–2.1) | 1.0 |
| Immunosuppressant drugs | 444/526 | 1 (3.1) | 2 (0.5) | 6.6 (0.6–75.0) | .2 |
| Cough (> 4 weeks) | 518/526 | 8 (18.2) | 7 (1.5) | 15.0 (5.2–43.8) | <.01 |
| Conversion | 526/526 | 5 (11.4) | 55 (11.4) | 1.0 (0.4–1.1) | 1.0 |
| Positive TST at baseline | 526/526 | 34 (77.3) | 203 (42.1) | 4.7 (2.3–9.7) | <.01 |
| Characteristics of TB index case | |||||
| Cavities on chest x-ray | 517/526 | 13 (29.5) | 71 (15.0) | 2.4 (1.2–4.8) | .04 |
| Cough (> 4 weeks) | 518/526 | 43 (97.7) | 427 (90.1) | 4.7 (0.6–35.2) | .1 |
| ≥2+ AFB | 444/526 | 21 (47.7) | 179 (40.4) | 1.3 (0.7–2.5) | .4 |
| Positive culture | 367/526 | 29 (96.7) | 323 (95.8) | 1.3 (0.2–9.9) | 1.0 |
Data represent no. (%). Comorbidities: diabetes, heart failure, and/or hypertension, chronic obstructive pulmonary disease, neoplasia, systemic lupus erythematous, and hepatitis.
Abbreviations: AFB, acid fast bacilli, CI, confidence interval; HIV, human immunodeficiency virus; IDU, illicit drug use; n, number of persons for whom such data were available; N, number total that participants from the study available; OR, odds ratio; TB, tuberculosis.
Table 6.
Gene Polymorphisms of Contacts of Pulmonary TB Cases Evaluated for Development of Active TB Infection
| SNP | Active TB n = 44 |
No Active TB n = 482 |
OR | 95% CI | P-Value | ||
|---|---|---|---|---|---|---|---|
| rs5743708–TLR2 | 8 | (23.5) | 62 | (15.5) | 1.7 | (0.7–3.9) | .2 |
| rs4986791–TLR4 | 5 | (14.7) | 9 | (2.3) | 7.3 | (2.3–23.2) | <.01 |
| rs361525–TNFA | 23 | (52.3) | 51 | (10.7) | 9.0 | (4.7–17.7) | <.01 |
| rs2430561–IFNG | 25 | (78.1) | 310 | (83.3) | 0.7 | (0.3–1.7) | .5 |
| rs1143627–IL1B | 17 | (44.7) | 208 | (47.2) | 0.9 | (0.5–1.8) | .8 |
Data represent no. (%).
Abbreviations: CI, confidence interval; IFNG, interferon gamma; IL1B, interleukin-1β; OR, odds ratio; SNP, single-nucleotide polymorphism; TLR, Toll-like receptor; TNFA, tumor necrosis factor α.
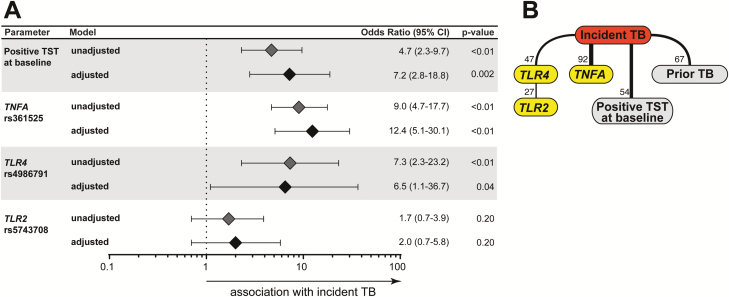
Multivariable regression analysis revealed that contacts who were TST positive at baseline had 7 times greater odds of developing active TB than those who remained TST negative (Figure 3A). Occurrence of allelic variants in either TLR4 or TNFA genes was independently associated with odds of incident TB. Bayesian networks confirmed the associations between TNFA and TLR4 polymorphisms and incident TB (Figure 3B). Three participants had both SNPs: all 3 were TST converters, of whom 2 also developed active TB. A total of 5 TST converters developed TB disease. Of these, 2 had 2 SNPs, TLR4 and TNFA, 1 had only the TLR4 variants and 1 had only the TNFA polymorphism. In addition, prior TB and being TST positive at baseline were robustly associated with development of active TB (Figure 3B). Interestingly, this model indicated that TLR2 SNPs were again indirectly associated with incident TB through TLR4 polymorphisms, suggesting that the combination of allelic variants in these genes may be associated with increased risk of Mtb infection and development of active TB.
Figure 3.
Variables associated with development of active TB among contacts of pulmonary TB. A, Multivariable regression model of variables shown in Tables 5 and 6 which displayed univariate P-value ≤ .2. B, Bayesian network with bootstrap (100×) was used to illustrate the statistically significant associations between the parameters and the occurrence of incident TB in the study population. Lines represent direct associations. Associations that remained statistically significant on ≥20 of 100 bootstraps are plotted. Numbers of times each association persisted during bootstrap are shown. Bold lines highlight the strongest associations. All parameters from Table 5 were included. Only those displaying significant associations are shown. Abbreviation: TB, tuberculosis.
DISCUSSION
In this study we tested associations between SNPs from immune related genes in a large cohort of TB contacts from a highly endemic region in Brazil. The most important finding was that TLR4 Thr399Ile (rs4986791) and TNFA-238 (rs361525) were independently associated with both TST conversion and subsequently developing TB disease. These findings highlight the importance of innate immunity, particularly of these molecules, in the pathogenesis of human Mtb infection and TB disease.
Our results are consistent with our current understanding of TB pathogenesis, in which TLRs are considered critical for host immunity against Mtb in both experimental and clinical settings. Indeed, several groups have shown that polymorphisms in TLR genes are associated with increased susceptibility to TB disease [13]. The TLR4 ectodomain plays a key role in recognition of pathogen-associated molecular patterns. Interestingly, TLR4 Thr399Ile has been associated with hypo-responsiveness to ligand interaction due its location near the central ectodomain region [23]. This polymorphism has been associated with more severe forms of pulmonary TB as quantified by sputum bacillary loads and chest radiographs [24]. Our findings on TB contacts provide additional evidence for the critical role of TLR4 in susceptibility to TB. Upon activation through interaction between Mtb ligands and TLR4, myeloid cells produce IL-12 among other proinflammatory mediators [25], which are important to drive T helper 1 (Th1) responses. Exposure to mycobacteria also triggers production of TNF-α and IL-1β [26]. Thus, TLR4 may be critical to drive the protective Th1 responses in the context of Mtb infection and hypo-responsiveness may drive increased susceptibility to TB.
TNF-α has a central role both in the host immune response to Mtb infection and in the immunopathology of TB. TNF-α is produced by many cell types and has cytotoxic synergy with human interferon [27]. Experimental studies have shown that TNF-α is required for the formation and maintenance of granulomas [28]. In humans, anti-TNF drugs are associated with heightened risk of a number of severe respiratory infections including TB [29] In a Chinese population, the TNFA-308 allele was associated with elevated odds of pulmonary TB [21]. To our knowledge, no previous study has tested the TNFA SNP in the context of TB in Brazil. While examining a Brazilian population, Rocha et al. reported that TNFA-238 (rs361525) was associated with spondylarthritis [30]. Our results argue that screening for TNFA SNPs could serve as a tool to guide implementation of preventive therapy in TB contacts.
In the present study, the LTBI cases identified at baseline may reflect a cumulative risk for infection before the programmatic contact tracing. Initial LTBI was associated with nonwhite ethnicity and with the presence of cavity on chest radiograph of the index case. Nonwhite ethnicity has been found as a risk factor for extrapulmonary TB [31], but in our study, this characteristic may be a proxy variable for socioeconomic conditions in Brazil, reflecting crowding and higher community exposure.
Both logistic regression and Bayesian network analyses demonstrated that male sex was associated with TST conversion. This relationship has been reported previously [25, 32]. Other direct associations with TST conversion found here included TLR4 and TNFA SNPs. The Bayesian network analyses refined these relationships while suggesting that TLR2 and TLR4 SNPs may sometimes act combined to increase odds of TST conversion. Both TLR2 and TLR4 are expressed on cell surface and share common intracellular signaling adaptors [33]. Our findings are intriguing and deserve additional investigations to validate the results and narrow down potential interdependency between TLR2 and TLR4 in the immune response against Mtb.
We examined the characteristics associated with development of active TB in our study population and found that polymorphisms in TLR4 and TNFA were independent risk factors. Importantly, such SNPs were also associated with TST conversion, reinforcing the idea that TLR4 signaling and TNF-α production are critically involved in TB pathogenesis. As TNF-α is important for maintenance of granulomas [34], it is possible that the SNP reported here could affect this process and favor development of active TB. The TLR4 polymorphism was also directly associated with development of active TB as well as with the TLR2 polymorphism, which although not significantly linked to this clinical outcome in logistic regression, was identified by the Bayesian network and indirectly linked through TLR4, reinforcing the idea of interdependency between these TLRs. The same analyses revealed that a prior history of TB was also a risk factor, which has already been demonstrated previously [35].
Our study has several strengths such as serial TST testing (currently recommended as the diagnostic test for LTBI in most resource-restrained countries), microbiologically confirmed TB, and SNPs closely related to immune responses against TB. This study had some limitations. Approximately 20% (n = 109) of the study population were lost to follow-up, but this proportion was lower than the average reported by studies of TB contacts [36]. In addition, most contacts were consanguineous with the index TB case, but there was no impact on the outcomes evaluated. Furthermore, we assumed that within a household all were infected by a common Mtb strain, which may not have always been true and might influence the host immune response.
In conclusion, our study provides strong evidence for associations between polymorphisms in innate immune genes and the risk of Mtb infection and development of active TB in Brazil. Further translational studies are warranted to delineate the molecular events behind these associations.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors acknowledge study participants and also the staff of the Clementino Fraga Filho University Hospital of the Federal University of Rio de Janeiro.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding. This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) / Instituto Nacional de Ciência e Tecnologia (INCT, grant number: 573548/2008-0) and Fundação de Amparo à Pesquisa do Rio de Janeiro (FAPERJ, grant E-26/110.974/2011). A. K. is the recipient of a career award from CNPq (produtividade em pesquisa) and FAPERJ (Cientistas do Nosso Estado). The work from B. B. A. and K. F. F. was supported by an intramural research program from FIOCRUZ and from the National Institutes of Health (U01AI115940). J. M. C.-A. was supported by the Organization of American States - Partnerships Program for Education and Training (OAS-PAEC) and the Coordenação de Aperfeiçoamento de pessoal de Nível Superior Brasil (CAPES, Finance Code 001). M. B. A. receives a fellowship from the Fundação de Amparo à Pesquisa da Bahia.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Houben RM , Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med 2016; 13:e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zenner D , Beer N , Harris RJ , Lipman MC , Stagg HR , van der Werf MJ. Treatment of latent tuberculosis infection: an updated network meta-analysis. Ann Intern Med 2017; 167:248–55. [DOI] [PubMed] [Google Scholar]
- 3. Sutherland I. Recent studies in the epidemiology of tuberculosis, based on the risk of being infected with tubercle bacilli. Adv Tuberc Res 1976; 19:1–63. [PubMed] [Google Scholar]
- 4. Ai JW , Ruan QL , Liu QH , Zhang WH. Updates on the risk factors for latent tuberculosis reactivation and their managements. Emerg Microbes Infect 2016; 5:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trajman A , Steffen RE , Menzies D. Interferon-gamma release assays versus tuberculin skin testing for the diagnosis of latent tuberculosis infection: an overview of the evidence. Pulm Med 2013; 2013:601737.23476763 [Google Scholar]
- 6. Rangaka MX , Wilkinson KA , Glynn JR , et al. . Predictive value of interferon-γ release assays for incident active tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis 2012; 12:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morrison J , Pai M , Hopewell PC. Tuberculosis and latent tuberculosis infection in close contacts of people with pulmonary tuberculosis in low-income and middle-income countries: a systematic review and meta-analysis. Lancet Infect Dis 2008; 8:359–68. [DOI] [PubMed] [Google Scholar]
- 8. Lienhardt C , Fielding K , Sillah J , et al. . Risk factors for tuberculosis infection in sub-Saharan Africa: a contact study in The Gambia. Am J Respir Crit Care Med 2003; 168:448–55. [DOI] [PubMed] [Google Scholar]
- 9. Commandeur S , van Meijgaarden KE , Prins C , et al. . An unbiased genome-wide Mycobacterium tuberculosis gene expression approach to discover antigens targeted by human T cells expressed during pulmonary infection. J Immunol 2013; 190:1659–71. [DOI] [PubMed] [Google Scholar]
- 10. Azad AK , Sadee W , Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun 2012; 80:3343–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milano M , Moraes MO , Rodenbusch R , et al. . Single nucleotide polymorphisms in IL17A and IL6 are associated with decreased risk for pulmonary tuberculosis in Southern Brazilian population. PLoS One 2016; 11:e0147814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou Y , Tan CY , Mo ZJ , et al. . Polymorphisms in the SP110 and TNF-α gene and susceptibility to pulmonary and spinal tuberculosis among Southern Chinese population. Dis Markers 2017; 2017:4590235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schurz H , Daya M , Möller M , Hoal EG , Salie M. TLR1, 2, 4, 6 and 9 variants associated with tuberculosis susceptibility: a systematic review and meta-analysis. PLoS One 2015; 10:e0139711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cobat A , Hoal EG , Gallant CJ , et al. . Identification of a major locus, TNF1, that controls BCG-triggered tumor necrosis factor production by leukocytes in an area hyperendemic for tuberculosis. Clin Infect Dis 2013; 57:963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo XG , Xia Y. The rs5743708 gene polymorphism in the TLR2 gene contributes to the risk of tuberculosis disease. Int J Clin Exp Pathol 2015; 8:11921–8. [PMC free article] [PubMed] [Google Scholar]
- 16. Pacheco AG , Cardoso CC , Moraes MO. IFNG +874T/A, IL10 -1082G/A and TNF -308G/A polymorphisms in association with tuberculosis susceptibility: a meta-analysis study. Hum Genet 2008; 123:477–84. [DOI] [PubMed] [Google Scholar]
- 17. Wei Z , Wenhao S , Yuanyuan M , et al. . A single nucleotide polymorphism in the interferon-γ gene (IFNG +874 T/A) is associated with susceptibility to tuberculosis. Oncotarget 2017; 8:50415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amaral EP , Riteau N , Moayeri M , et al. . Lysosomal cathepsin release is required for NLRP3-inflammasome activation by Mycobacterium tuberculosis in infected macrophages. Front Immunol 2018; 9:1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(Brasil) MdS. Manual de Recomendações para o Controle da Tuberculose no Brasil Available at: http://www.crf-rj.org.br/crf/arquivos/manual_recomendacoes_controle_tb.pdf. Accessed 10 October 2018.
- 20. Saleh MA , Ramadan MM , Arram EO. Toll-like receptor-2 Arg753Gln and Arg677Trp polymorphisms and susceptibility to pulmonary and peritoneal tuberculosis. APMIS 2017; 125:558–64. [DOI] [PubMed] [Google Scholar]
- 21. Fan HM , Wang Z , Feng FM , et al. . Association of TNF-alpha-238G/A and 308 G/A gene polymorphisms with pulmonary tuberculosis among patients with coal worker’s pneumoconiosis. Biomed Environ Sci 2010; 23:137–45. [DOI] [PubMed] [Google Scholar]
- 22. Tien I , Der Kiureghian A. Algorithms for Bayesian network modeling and reliability assessment of infrastructure systems. Reliab Eng Syst Saf 2016; 156:134–47. [Google Scholar]
- 23. Mucha R , Bhide MR , Chakurkar EB , Novak M , Mikula I Sr. Toll-like receptors TLR1, TLR2 and TLR4 gene mutations and natural resistance to Mycobacterium avium subsp. paratuberculosis infection in cattle. Vet Immunol Immunopathol 2009; 128:381–8. [DOI] [PubMed] [Google Scholar]
- 24. Najmi N , Kaur G , Sharma SK , Mehra NK. Human Toll-like receptor 4 polymorphisms TLR4 Asp299Gly and Thr399Ile influence susceptibility and severity of pulmonary tuberculosis in the Asian Indian population. Tissue Antigens 2010; 76:102–9. [DOI] [PubMed] [Google Scholar]
- 25. Barletta-Naveca RH , Naveca FG , de Almeida VA , et al. . Toll-like receptor-1 single-nucleotide polymorphism 1805T/G is associated with predisposition to multibacillary tuberculosis. Front Immunol 2018; 9:1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang ZM , Zhang AR , Xu M , Lou J , Qiu WQ. TLR-4/miRNA-32-5p/FSTL1 signaling regulates mycobacterial survival and inflammatory responses in Mycobacterium tuberculosis-infected macrophages. Exp Cell Res 2017; 352:313–21. [DOI] [PubMed] [Google Scholar]
- 27. Gardam MA , Keystone EC , Menzies R , et al. . Anti-tumour necrosis factor agents and tuberculosis risk: mechanisms of action and clinical management. Lancet Infect Dis 2003; 3:148–55. [DOI] [PubMed] [Google Scholar]
- 28. Bean AG , Roach DR , Briscoe H , et al. . Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol 1999; 162:3504–11. [PubMed] [Google Scholar]
- 29. Keane J , Gershon S , Wise RP , et al. . Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med 2001; 345:1098–104. [DOI] [PubMed] [Google Scholar]
- 30. Rocha Loures MA , Macedo LC , Reis DM , et al. . Influence of TNF and IL17 gene polymorphisms on the spondyloarthritis immunopathogenesis, regardless of HLA-B27, in a Brazilian population. Mediators Inflamm 2018; 2018:1395823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noppert GA , Wilson ML , Clarke P , Ye W , Davidson P , Yang Z. Race and nativity are major determinants of tuberculosis in the U.S.: evidence of health disparities in tuberculosis incidence in Michigan, 2004–2012. BMC Public Health 2017; 17:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diwan VK , Thorson A. Sex, gender, and tuberculosis. Lancet 1999; 353:1000–1. [DOI] [PubMed] [Google Scholar]
- 33. Cervantes JL. MyD88 in Mycobacterium tuberculosis infection. Med Microbiol Immunol 2017; 206:187–93. [DOI] [PubMed] [Google Scholar]
- 34. Algood HM , Lin PL , Yankura D , Jones A , Chan J , Flynn JL. TNF influences chemokine expression of macrophages in vitro and that of CD11b+ cells in vivo during Mycobacterium tuberculosis infection. J Immunol 2004; 172:6846–57. [DOI] [PubMed] [Google Scholar]
- 35. Chiang CY , Riley LW. Exogenous reinfection in tuberculosis. Lancet Infect Dis 2005; 5:629–36. [DOI] [PubMed] [Google Scholar]
- 36. Alsdurf H , Hill PC , Matteelli A , Getahun H , Menzies D. The cascade of care in diagnosis and treatment of latent tuberculosis infection: a systematic review and meta-analysis. Lancet Infect Dis 2016; 16:1269–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.