Abstract
Sickle cell disease (SCD) is a monogenic disorder that affects millions worldwide. Allogeneic hematopoietic stem cell transplantation is the only available cure. Here, we demonstrate the use of CRISPR/Cas9 and a short single-stranded oligonucleotide template to correct the sickle mutation in the β-globin gene in hematopoietic stem and progenitor cells (HSPCs) from peripheral blood or bone marrow of patients with SCD, with 24.5 ± 7.6% efficiency without selection. Erythrocytes derived from gene-edited cells showed a marked reduction of sickle cells, with the level of normal hemoglobin (HbA) increased to 25.3 ± 13.9%. Gene-corrected SCD HSPCs retained the ability to engraft when transplanted into non-obese diabetic (NOD)-SCID-gamma (NSG) mice with detectable levels of gene correction 16–19 weeks post-transplantation. We show that, by using a high-fidelity SpyCas9 that maintained the same level of on-target gene modification, the off-target effects including chromosomal rearrangements were significantly reduced. Taken together, our results demonstrate efficient gene correction of the sickle mutation in both peripheral blood and bone marrow-derived SCD HSPCs, a significant reduction in sickling of red blood cells, engraftment of gene-edited SCD HSPCs in vivo and the importance of reducing off-target effects; all are essential for moving genome editing based SCD treatment into clinical practice.
INTRODUCTION
Sickle cell disease (SCD) is a devastating chronic illness marked by severe pain, end organ damage and early mortality (1,2). SCD is caused by a point mutation in the β-globin gene (HBB). A single nucleotide substitution converts glutamic acid to a valine that leads to the production of sickle hemoglobin (HbS). HbS polymerizes under hypoxic conditions that impair the function of red blood cells (RBC) and markedly shortens their lifespan due to hemolysis (3). SCD affects ∼100 000 Americans and millions more worldwide (4,5); however, the average lifespan of individuals with SCD has not improved over the last few decades since treatment options for SCD remain very limited (6). Pharmacological therapy with hydroxyurea may modulate disease severity through induction of fetal hemoglobin (HbF) and reduction of HbS polymerization, but does not cure patients with SCD (7). Chronic transfusion therapy may be difficult to sustain due to high rates of alloimmunization and iron overload (8,9). The only curative therapy for SCD is a hematopoietic stem cell transplant (HSCT), typically from a matched related donor, which is available to only ∼15% of SCD patients (10,11). Morbidity and mortality from HSCT increases significantly when using matched unrelated donors (12) or haploidentical donors (13). A prospective study of unrelated donor HSCT in SCD concluded that, without modifications to existing regimens, this therapy is not safe for widespread adoption (14). Results from a recent lentiviral-vector based clinical trial of one patient with SCD using autologous hematopoietic stem/progenitor cells (HSPCs) showed the promise of gene therapy for SCD (15).
There are several possible genome editing based strategies to ameliorate SCD: (i) correction of the causative A-T point mutation in HBB via the homology-directed repair (HDR) pathway (16–18), (ii) induction of fetal hemoglobin (HbF) via gene disruption by non-homologous end-joining (NHEJ) (19,20) and (iii) gene addition of an β-globin, γ-globin or anti-sickling β-globin cassette (21). Correction of the sickle cell disease mutation in human HSPCs has been demonstrated with zinc finger nuclease (16). Using Streptococcus pyogenes (Spy) Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) (22,23), DeWitt et al. demonstrated HBB gene editing in CD34+ HSPCs from patients with SCD (SCD HSPCs) by delivery of ribonucleoprotein (RNP) complex of CRISPR guide RNA (gRNA) and Cas9 protein together with a single-stranded oligonucleotide (ssODN) template (24), achieving up to 25% of alleles corrected with a high RNP dose (200 pmol) (17). Injection of gene-edited HSPCs from healthy persons into immunodeficient NOD-SCID-gamma (NSG) mice showed engraftment at a level much higher than that using mRNA of zinc finger nuclease (ZFN) and ssODN templates for HBB gene editing (16), with a significant decrease in the percent of HDR modified cells following transplantation. Dever et al. showed an average of 50% HBB gene correction rate in HSPCs from patients with SCD when delivering gRNA/Cas9 RNPs together with rAAV6 vector packaging a donor template consisting of a GFP expression cassette flanked by homology arms for HBB. With an HBB cDNA template packaged in rAAV6, an average of 11% HDR-mediated gene correction rate was achieved in SCD HSPCs (18). Engraftment of gene-edited HSPCs from healthy donors was demonstrated using immunodeficient NSG mice (18). The studies by DeWitt et al. (17) and Dever et al. (18) employed the gRNA R-02 (or the truncated version of R-02), we previously described, which has a high on-target activity (25). In both studies, the R-02 gRNA was found to induce high levels of off-target cutting in human HSPCs (17,18); however, in these studies genome-wide unbiased off-target analysis was not performed.
In this study we systematically optimized the gRNA and ssODN template designs, quantified the gene editing rates in human CD34+ HSPCs from normal individuals and from the peripheral blood (PB) and bone marrow (BM) of patients with SCD, and performed a genome-wide unbiased analysis of off-target effects. In contrast to engraftment studies using gene-edited CD34+ HSPCs from ‘healthy persons’ (17,18), we performed two engraftment studies using gene-edited CD34+ HSPCs derived, respectively, from unmobilized peripheral blood and bone marrow of ‘patients with SCD’, aiming to provide more clinically relevant evidence on the feasibility of using CRISPR/Cas9 based gene-editing to treat SCD. We found that gene-edited SCD HSPCs were able to engraft in the bone marrow of NSG mice and the corrected alleles were stable for up to 16–19 weeks post-transplantation. Compared with previous studies, our results provide important new insights into the opportunities and challenges of using gene-editing based approaches to treat SCD, including the upregulation of fetal hemoglobin in gene-edited cells (especially those with HBB cutting only), gene conversion by the δ-globin gene (HBD), differences in engraftment of gene-edited CD34+ HSPCs from different sources (PB and BM) and different SCD patients, the risk of chromosomal rearrangements revealed by extensive off-target analysis, and the ability to significantly reduce off-target effects by using an HiFiCas9 protein (26); these findings may facilitate the translation of genome editing based SCD treatment into clinical practice.
MATERIALS AND METHODS
Cells and cell culture media
K562 (ATCC), U2OS (ATCC) and CD34+ cells were cultured at 37°C and 5% CO2. K562 cells were cultured in RPMI1640 (ATCC, Cat No. 30–2001) supplemented with 10% FBS and L-glutamine. Frozen CD34+ cells from mobilized peripheral blood of normal individuals were purchased from AllCells. Peripheral blood CD34+ cells were acquired from SCD patients undergoing therapeutic red cell exchange at the Cancer & Hematology Centers, Texas Children’s Hospital (Houston, TX), under the approved IRB protocol H-33997. CD34+ HSPCs were purified from the peripheral blood as previously described (27). Briefly, the mononuclear fraction was separated from the peripheral blood of patients with SCD by Ficoll-Hypaque (GE Healthcare) density centrifugation. CD34+ cells were extracted from the mononuclear fraction by immunomagnetic separation using the CD34 Microbeads Kit (Miltenyi Biotech, CD34 MicroBead Kit UltraPure, human), according to the manufacturers’ instructions. Then, the human erythroid progenitor cells were cultured and differentiated into erythroblasts using an ex vivo primary erythroid culture system with two phases. In expansion phase, cells were cultured in GMP SCGM (CellGenix) supplemented with 300 ng/ml SCF (Peprotech), 100 ng/ml TPO (Peprotech), 300 ng/ml Flt3 ligand (Peprotech) and 60 ng/ml IL3 (Peprotech). In differentiation phase, cells were cultured in SFEM II (StemSpan) supplemented with 20 ng/ml SCF, 10 ng/ml IL3, 3 U/ml EPO (Peprotech), 10−5 M 2-mercaptoethanol, 10−6 M dexamethasone, and 0.3 mg/ml human holo-transferrin (Sigma Aldrich). Harvested CD34+ cells were cultured in expansion phase for 2–3 days before electroporation. Forty-eight hours after the electroporation, 104 cells were transferred to 1 ml differentiation media in 24-well plates. Fresh differentiation medium was added every 2 days and cells were cultured at a density under 106 live cells/ml for 21–27 days before analysis. The cell count and viability were measured using Trypan Blue dye, 0.4% solution (Bio-Rad) and T20 Automated Cell Counter (Bio-Rad).
Plasmid construction
The SpyCas9 expressing plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (22) (Addgene plasmid #42230) was a gift from Feng Zhang. The pX330 vector was digested using BbsI, and a pair of annealed oligos was cloned into the vector before the gRNA scaffold.
RNP and ssODN delivery using electroporation
A total of 2 × 105 K562 cells (program FF-120, solution SF), U2OS cells (program CM-137, solution SE) and CD34+ cells (program CA-137, solution P3) were electroporated on a Lonza Nucleofector 4-D according to manufacturer’s instructions. In K562 cells, 1 μg of pX330 plasmid and 0–5 μM (0–100 pmol) ssODN template (Ultramer® DNA Oligonucleotides from Integrated DNA Technologies) were transfected. In U2OS, 1 μg of pX330 plasmid and 100 pmol of dsODN were transfected for GUIDE-seq analysis. In CD34+ cells, 5 μg (30.5 pmol) of Cas9 protein (Feldan Therapeutics or Integrated DNA Technologies), 2.5 μg (73 pmol) of chemically synthesized gRNAs (TriLink BioTechnologies) and 0–5 μMof ssODN were transfected. For mock-treated CD34+ cells, electroporation of the same amount of cells was performed using the same P3 buffer and CA-137 program as the treated cells, but without RNP or ssODN.
CD34+ cell colony formation assay
Forty-eight hours after electroporation, cells were diluted to 5 × 103 cells/0.3 ml in IMDM + 2% FBS. Once diluted, the cell sample was added to 2.7 ml tubes of Methylcellulose Medium for Human Cells (with Cytokines) (STEMCELL Technologies) and cultured according to the manufacturer’s protocol. The culture dishes were incubated for 14–16 days before being assayed. The proliferation and differentiation potential of these cells were measured by the observation of the numbers and types of colonies grown in culture using a microscope. Zygosity of burst-forming unit erythroid (BFU-E) colonies was assessed by droplet digital PCR assay (Supplementary Figure S1).
T7 Endonuclease I (T7E1) assay
The HBB locus was amplified from genomic DNA (Accuprime Taq HiFi) using PCR primers to amplify a 669 bp region containing the target site (Supplementary Table S1). About 200 ng of purified PCR products were annealed and digested with T7 Endonuclease I (New England Biolabs) according to the manufacture’s instruction. The digestion products were run on a 2% polyacrylamide gel with 1× GelGreen Nucleic Acid Gel Stain (Biotium). The fragmented PCR products were analyzed by ImageJ and the percent of nuclease-specific cleavage products was calculated using the formula: % Indel frequency = 100 × (1 – (1 – fraction cleaved)1/2) (28).
Droplet digital PCR (ddPCR)
As the identification of optimal genome editing conditions requires a rapid and cost-effective method to accurately measure gene-editing frequencies mediated by CRISPR/Cas9 systems, we developed a probe-based ddPCR assay that can simultaneously quantify the levels of HDR, NHEJ and unmodified alleles at the endogenous locus (Supplementary Figure S1). For the allele quantification assay, probe-based reaction mixes were prepared with 15 ng genomic DNA template, 1× ddPCR Supermix for Probes (Bio-Rad), 900 nM target primers, 250 nM target probes (Eurofins Genomics) and 10 U HindIII-HF restriction enzyme in each 20 μl reaction mix. For colony genotyping, BFU-E colonies were resuspended in 10 μl of QuickExtract DNA extraction solution (Epicentre) and 1 μl of colony lysate was used. For the chromosomal rearrangement assay, reactions were prepared with 50 ng genomic DNA template, 1× QX200 ddPCR Evagreen Supermix (Bio-Rad), 100 nM target primers, 10 U HindIII-HF restriction enzyme (New England Biolabs) in each 20 μl reaction mix. To quantify all types of editing outcomes, three PCR reactions were run simultaneously. The inversion and deletion PCR assays were carried out using a pair of primers spanning the un-rearranged on-target site, the expected junction point on inverted allele or the deleted allele. The reference PCR from non-target chromosome estimates the expected copy number of chromosome 11 in the reaction. PCR was performed according to the manufacturer’s cycling protocol with optimized annealing temperature. The primer and probe sequences for ddPCR assays are provided in Supplementary Table S1.
Hemoglobin native PAGE
Hemoglobin variants were separated by native PAGE as follows: CD34+ HSPCs were differentiated for 14–21 days prior to protein extraction. Cells were resuspended in 1× Halt Protease and Phosphatase Inhibitor Cocktails (Thermo Scientific) in water and lysed using freeze/thaw method. The hemolysates were mixed in 1:2 ratio with Native PAGE Sample Buffer (Bio-Rad). The samples were run on a 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad) in 1× Tris/Glycine running buffer (Bio-Rad) until the AFSC Hemo Control (Helena Laboratories) reaches the bottom of the gel. Proteins were transferred to a PVDF membrane using the Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was blocked at RT for 30 min in 5% non-fat dry milk (NFDM) in 1× TBS-T, incubated with primary antibodies for hemoglobin alpha subunit (Abcam, EPR3608) at 4°C overnight, then incubated with Goat Anti-Rabbit IgG H&L secondary antibody (Abcam, ab6721) at RT for 1 h. Blots were visualized using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) and imaged on ChemiDoc MP imaging system (Bio-Rad).
HPLC (high-pressure liquid chromatography)
Hemoglobin variants were identified by HPLC (29,30). Primary erythroid cells were lysed in HPLC grade water, and then centrifuged at 20 000 g for 20 min at 4°C to remove cell debris. The absorbance of total hemoglobin variants was detected at 415 nm using a 100 × 4.6mm PolyCAT A™ column (PolyLC Inc, # 104CT0315) fitted to a Waters HPLC system. The areas under the peaks were used to quantify the hemoglobin fractions. The total area under the HbS and HbF peaks was used for ratio comparisons (29,31).
RP-HPLC (reverse-phase high performance liquid chromatography)
RP-HPLC assays were performed on an Agilent 1260 Infinity II HPLC system with Diode Array Detector (32,33). The chromatographic column is Aeris™ 3.6 μm WIDEPORE XB-C18 200 Å, LC Column 250 × 4.6 mm behind a securityGuard™ ULTRA cartridge (Phenomenex). Solvent A: 0.1% trifluoroacetic acid in HPLC grade water at pH 2.9. Solvent B: acetonitrile. The linear gradient developed from 41% of solvent B to 47% of solvent B to separate and analyze the various globin chains.
In vitro sickling assay
On day 21 of culture, SCD CD34+ HSPCs were incubated in a 2% oxygen environment for 4 h. Cellular morphology imaging was performed using an Olympus ZX71 microscope at 40× magnification within 10 min of exposure to atmospheric oxygen (34–36). Sickled cells were counted and scored by manual examination by a board certified hematologist blinded to sample identity. Eight individual fields were scored per sample, and percentage sickled cells recorded.
Off-target identification
Bioinformatic prediction of potential off-target sites for R-66 and R-66 SCD gRNAs was carried out using the web-based tool CRISPR Search with Mismatches, Insertions and/or Deletions (COSMID) (37) with up to three mismatches or with up to two mismatches and an insertion or deletion allowed in the 19 PAM proximal bases. The Homo sapiens genome assembly GRCh38/hg38 genome build was used as a reference. Genome-wide, unbiased identification of DSBs enabled by sequencing (GUIDE-Seq) (38) was used for experimental identification of potential off-target sites. Briefly, a plasmid-expressing Cas9 and R-66 or R-66 SCD gRNA along with a dsDNA tag was nucleofected into U2OS cells as previously described (38). Genomic DNA was extracted 3 days after electroporation, sheared using a Covaris M220 focused ultrasonicator to an average size of 500 bp. The DNA was prepared and sequenced on an Illumina MiSeq and the resulting sequencing data analyzed using the standard pipeline (38) with a reduced gap penalty.
Quantification of on-target and off-target activity by deep sequencing
DNA flanking the gRNA on- and off-target sites was amplified using locus-specific primers (Supplementary Table S2) followed by a second PCR to introduce Illumina sequencing adaptors and sample barcodes. Each sample was normalized to 50 nM and the pooled library was quantified using KAPA QC: Library Quantification Kit (KAPABIOSYSTEMS) and sequenced on the Illumina MiSeq. Sequencing libraries were also prepared using RNase-H dependent primers to facilitate PCR multiplexing (Integrated DNA Technologies). About 50 ng of genomic DNA was used for multiplex PCR using rhAmpSeq HotStart Master Mix (Integrated DNA Technologies) and a primer pool for all R66SCD off-target sites. The blocked-cleavage primers were designed and provided by Integrated DNA Technologies, and the genome-specific sequences are presented in Supplementary Table S3. An initial enrichment amplification of 10 cycles was carried out. The PCR products were purified using SPRI beads and barcoded using unique P5 and P7 indexing primers for each amplified pool in a second PCR reaction using rhAmpSeq HotStart Master Mix for 18 cycles. The library preparation and sample loading were performed according to standard Illumina MiSeq library prep. CRISPR-Cas9 genome editing outcomes from deep sequencing data were analyzed using a custom pipeline (38), CRISPResso (39) and a MATLAB script.
Transplantation of peripheral blood SCD CD34+ HSPCs in NSG mice
Six- to eight-week-old immunodeficient non-obese diabetic (NOD)-severe combined immunodeficiency (SCID) Il2rg−/− (gamma) mice (NSG mice) (Jackson laboratory, Bar Harbor, ME 442, USA) were sub-lethally irradiated with 200 cGy 12–24 h before transplantation to clear the mouse bone marrow niche. Unmobilized peripheral CD34+ cells were isolated from patients undergoing red cell exchange therapy and frozen in cryo-preservation medium consisting of 5% DMSO in combination with 10% ES Cell FBS (ES009B, Life Technologies) in IMDM. Cells were thawed and cultured in expansion media for 2 days before electroporation. Two days post-delivery of WT Cas9/gRNA RNP and ssODN for HBB gene editing (less than a 96 h total culture period), gene edited or mock electroporated SCD CD34+ HSPCs were counted and 5 × 105 viable cells were spun down and then resuspended in 30 μl PBS for intrafemoral transplantation. For intrafemoral transplantation, a 27-gauge needle was used to create an injection site through the patella and then a 29-gauge needle was used to transplant the resuspended cells through the patella and into the right mouse femur. Mice were randomly assigned to each experimental group and analyzed in a blinded fashion. The experimental protocol was approved by Stanford University’s Administrative Panel on Laboratory Animal Care.
Transplantation of BM SCD CD34+ HSPCs in NSG mice
The frozen bone marrow cells from SCD patients were thawed shortly before CD34+ HSPC isolation. Cells were cultured for 3 days prior to electroporation to deliver HiFi Cas9/gRNA RNP and ssODN template for HBB gene correction. Two days post-electroporation, cells were cryopreserved. These cells were then thawed and injected intravenously into NSG mice within 24 h of thawing. A total of 5 × 105 viable cells were injected per mouse. Due to low post-thaw recovery of Donor1 RNP and ssODN sample, 3 × 105 viable cells were injected per mouse. The mice were euthanized at week 16 after transplantation, and the BM was harvested to determine the engraftment potential.
Human cell engraftment analysis
About 16–19 weeks after transplantation, mice were euthanized and mouse bone marrow was harvested (2× femur, 2× tibia, sternum, 2× pelvis and spine) and then crushed using a mortar and pestle in 5 ml RPMI supplemented with 10% fetal bovine serum, 100 mg/ml streptomycin, 100 unit/ml penicillin, 2 mM L-glutamine, 4 U/ml heparin and 20 U/ml DNase. Cells were filtered through a 45 μm mesh, incubated at 37°C for 10 min, and then subjected to a Ficoll gradient centrifugation for 25 min at 2000rpm (with full acceleration and no break). After centrifugation, mononuclear cells were collected from the Ficoll layer (max 5 ml), were washed with 45 ml FACS buffer (1× PBS supplemented with 2% FBS and 2 mM EDTA), spun down at 300 × g and then resuspended in the human engraftment staining cocktail that consisted of the following antibodies: monoclonal anti-human HLA-ABC APC-Cy7 (W6/32, BioLegend), anti-human CD45 V450 (HI30 BD Biosciences), anti-mouse CD45.1 PE-Cy7 (A20, eBioScience, San Diego, CA, USA), anti-human CD19 APC (HIB19, BD511 Biosciences), CD33 PE (WM53, BD Biosciences), as well as anti-mouse mTer119 PE-Cy5 (TER-119, BD Biosciences) to facilitate the exclusion of mouse RBCs from the analysis. Cells were stained for 30 min at 4°C on ice and protected from light. Stained cells were then washed and resuspended in FACS buffer supplemented with propidium iodide to detect dead cells. Cells were analyzed on a BD FACS Sort II Aria where human engraftment was defined as HLA-ABC+/CD45+ cells. Human cells were then sorted into PBS, spun down, resuspended in QuickExtract DNA extraction solution (Epicentre) and gDNA was harvested following the manufacturer’s recommendations. HBB alleles were then quantified for NHEJ, HDR or WT by deep sequencing as described above.
Statistics
SPSS Statistics (SPSS) was used for all the calculations. Data were analyzed using Student’s t-tests or one-way ANOVA and post hoc multiple comparison tests. The difference with P < 0.05 was considered statistically significant (* denotes P < 0.05; # denotes P < 0.01). Figures were prepared using GraphPad Prism (GraphPad Software).
RESULTS
Optimization of gRNA designs targeting HBB
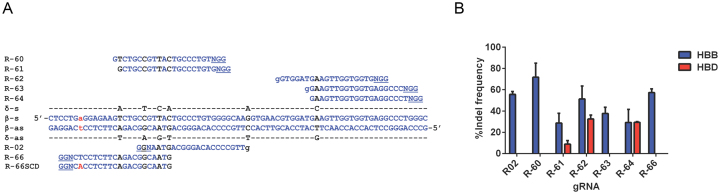
We designed Spy CRISPR gRNAs targeting the first exon of HBB with varying proximity to the SCD mutation site and used the web-based search tool COSMID (37) to select 7 gRNAs with low numbers of potential off-target sites (Figure 1A). The off-target cleavage at the region within the HBD locus (with high homology to the gRNA target sequences) was analyzed by the T7E1 assay (Figure 1B). K562 cells, an erythroid leukemia cell line, were used for gRNA and gene-correction DNA template screening and optimization since it is a widely used model for studying gene transfer into human hematopoietic cells (40). Plasmids expressing SpyCas9 and individual gRNAs were delivered into K562 cells, which gave small insertion/deletion (indel) rates of 29–72% at HBB, with 3 gRNAs having high levels of off-target cutting (9–33%) at HBD (Figure 1B). In particular, the R-64 gRNA, which has a 1 bp mismatch to HBD, had comparable activities at both HBB and HBD. The R-66 gRNA had 57% on-target indel rate without any observable off-target cutting at HBD. Compared with the R-02 gRNA that induces HBB cutting 18 bp away from the SCD mutation site, R-66 generates a DSB 1 bp away from the sickle mutation (Figure 1A). It has been reported that the HDR rate decreases as the distance between targeted insertion site and the cut-site increases (41–43). R-66 gRNA thus may facilitate an increased level of gene correction using a short ssODN template (41,44). Therefore, although R-02 gRNA (and its truncated version) was used in previous studies of HBB genome editing in human HSPCs (17,18), and the R-60 gRNA had a higher (72%) level of on-target activity, R-66 was chosen as the gRNA for use in subsequent studies. While R-66 gRNA targets the wild-type (WT) allele of HBB, R-66 SCD gRNA was designed to specifically recognize the sickle mutant allele of HBB (Figure 1A).
Figure 1.
Design of gRNAs targeting the HBB locus close to the SCD mutation site. (A) The target sites of Spy CRISPR gRNA designs near the first exon of HBB with varying proximity to the SCD mutation site (lowercase, red). The blue capital letters represent homologous sequences, blue and black lowercase letter ‘g’ indicates the addition of nucleotide G to the 5′ end of the 19 base pair guiding sequence, and the black letters indicate mismatches between HBB and the highly homologous HBD gene. For each target site, the PAM sequence is underlined. R-66 gRNA targets the WT HBB sequence and R-66 SCD gRNA targets the HBB sequence with SCD mutation. (B) Activity of gRNAs at HBB and HBD was measured by the T7E1 assay. Note that R-02, R-60 and R-66 gRNAs had high on-target indel rates and no observable HBD off-target cutting; n = 2 biological replicates.
Optimization of ssODNs to enhance HDR rate
To optimize the ssODN template design for enhanced HDR rates, we introduced the sickle mutation into WT HBB in K562 cells using short (120–121 nucleotide) ssODNs containing the A-T mutation (Supplementary Table S4). The targeted integration of the ssODNs was tested in K562 cells together with plasmid encoding SpyCas9 and R-66 gRNA. We designed ssODN templates corresponding to the gRNA target strand (t) or to the non-target strand (nt) (Figure 2A) to investigate the effect of strand preference on HDR rates. We first tested the SCDunt0 ssODN, a non-target strand template with the A-T mutation (Figure 2A) in K562 cells. The SCDunt0 ssODN is asymmetrically positioned, with the homology arms 33 nucleotide proximal and 87 nucleotide distal to the Cas9 cutting site (Figure 2A). Deep-sequencing results of the HBB locus were analyzed by CRISPResso to determine the outcomes of genome editing (39). The SCDunt0 ssODN induced an HDR rate of 9.3%, with a re-cutting rate of 8.3% of the integrated ssODN template (Figure 2B). Re-cutting of the target sequence was measured as mixed HDR-NHEJ alleles that show additional indels at the cut site, consistent with sequential cleavage and NHEJ repair of HDR-edited alleles (39). We hypothesized that introducing mismatches in the ssODN donor template within the sequence corresponding to the gRNA-specific PAM-proximal region can prevent re-cutting of the HDR-edited alleles, owing to the reduced affinity between gRNA and target sequence. To reduce the target sequence homology after ssODN insertion, we designed eight 121-nucleotide non-target strand ssODNs (SCDcnt1 to SCDcnt8) that are centered with respect to the cutting site, have different numbers and locations of silent mismatches (MM) within the gRNA target region thus are not complementary to the gRNA (Figure 2A). As expected, ssODN templates with multiple mismatches prevented the re-cutting of HDR-modified alleles (Figure 2B and Supplementary Figure S2). However, with the number of mismatches >2 in the ssODN template or putting the mismatches further away from the cutting site resulted in reduced HDR rates (Supplementary Figure S3a,b). We found that, of the 8 ssODN templates tested (SCDcnt1 to SCDcnt8), SCDcnt5, a non-target strand ssODN centered with respect to the cutting site and containing two consecutive mismatches (sickle mutation and silent mutation) next to the cutting site, gave the highest HDR rate (23.6%) (Figure 2B).
Figure 2.
Optimization of ssODN DNA template designs for gene correction. To test the gene correction approach using K562 cells, we introduced the sickle mutation into wild-type (WT) HBB gene using short ssODNs containing the A-T mutation. (A) Schematic of ssODN template designs near the SCD locus (with the black or red box showing the location of sickle mutation in SCD). Initially 11 ssODN templates (shown in blue) complementary to the non-target strand and 3 ssODN templates (shown in orange) complementary to the target strand were designed, which contain the sickle mutation (red box) and different numbers and locations of silent mismatches (yellow box). The vertical black line (position zero) indicates the Cas9 cut site and the numbers on the x-axis indicate the number of nucleotide from the cut site. These ssODNs were tested in K562 cells together with plasmid encoding SpyCas9 and R-66 gRNA. The R-66 gRNA sequence is shown in blue (with the WT base T in black indicating the SCD mutation site), and the PAM sequence is shown in orange and underlined. (B) The eight ssODN templates (SCDcnt1-SCDcnt8) with varying number of mismatches prevented the re-cutting of HDR-modified alleles. Re-cutting of the target sequence was measured as mixed HDR-NHEJ alleles that show additional indels at the cut site, consistent with sequential cleavages initially repaired by HDR and subsequently by NHEJ. Note that SCDcnt5 ssODN centered with respect to the cutting site and containing two consecutive mismatches next to the cut site was the best in terms of having a high HDR rate and low retargeting rate. Editing outcomes were measured by NGS. Data shown is representative of n = 2 biological replicates. (C) Six different ssODN template designs based on SCDcnt6 were tested, and SCDct6, a target strand ssODN symmetric with respect to the R-66 cut site, gave the best performance. The frequency of HDR was measured by NGS. Data shown are representative of n = 2 biological replicates. Combining with the results in panels (B) and (C), the best sickling ssODN template is SCDct5. (D and E) To identify the optimal ssODN concentration, K562 cells were electroporated with 0 to 5 μM (1–100 pmol) of SCDct5 ssODN along with (D) the plasmid encoding Cas9 and R-66 gRNA, or (E) the corresponding gRNA/Cas9 RNP. The allele frequencies of gene correction (HDR), Cas9 cutting but no ssODN integration (NHEJ) and no cutting (WT) events were quantified by ddPCR (Supplementary Figure S1). Note that 0.5 μM (10 pmol) and 5 μM (100 pmol) ssODN gave the highest level of HDR with plasmid and RNP delivery, respectively.
In a parallel experiment to optimize the ssODN templates, we tested three pairs of templates, all with three mismatches based on the SCDcnt6 ssODN. Each pair had one target strand template and one non-target strand template, and was either symmetrically or asymmetrically positioned with respect to the cut site (24) (Figure 2A). We found that SCDct6, a target strand ssODN symmetric with respect to the R-66 target site, gave the highest HDR rate (28.3%) (Figure 2C). Therefore, combining the results of two studies on template design, we concluded that, SCDct5, the target strand ssODN symmetric with respect to the R-66 gRNA target site with two mismatches is optimal. The optimal design of ssODN template is likely dependent on the sequence to be replaced and the direction of transcription relative to the PAM orientation (45). In all subsequent studies, we used the SCDct5 and SCDct5-wt respectively for inducing or correcting the sickle mutation in HBB. Since K562 is triploid for the HBB locus and has a silent C/C/T SNP (rs713040) located 11 bases upstream of the SCD mutation site (Supplementary Figure S3c), we tested ssODN templates containing either a C or T at this location but did not observe any difference in the HDR rate in K562 and normal CD34+ cells (data not shown). Therefore, we decided to incorporate a C at this position in the ssODN sequence for subsequent experiments, since in the African population rs713040 is 78.2% C/C, 20.3% C/T and 1.5% T/T (34) (Supplementary Table S4). As predicted, all patients with SCD in this study had a C/C genotype at this location.
In order to determine the optimal amount of ssODN templates for gene correction, K562 cells were electroporated with 0–5 μM (0–100 pmol) of SCDct5 ssODN templates, along with the optimized amount of plasmid encoding Cas9 and R-66 gRNA, or the corresponding RNP. Cells were harvested 72 h post-electroporation. The allele frequencies of HDR, NHEJ and no-cutting (WT) events were quantified by the probe-based ddPCR assay (Supplementary Figure S1). With the plasmid encoding Cas9/R-66, 0.5 μM (10 pmol) of ssODN donor template induced the highest HDR rate (Figure 2D). With the Cas9/R-66 RNP, 5 μM (100 pmol) of ssODN template induced the highest level of HDR (Figure 2E), which was used in all subsequent experiments in CD34+ HSPCs. Dose–response studies in normal CD34+ HSPCs revealed that, although high doses of ssODN template may increase HDR rates, it could also cause short-term toxicity (Supplementary Figure S4a,b).
Targeted HBB gene modification in normal CD34+ HSPCs
We next quantified the genome editing efficiency in CD34+ cells derived from peripheral blood of normal individuals using commercially available SpyCas9 proteins complexed with chemically synthesized R-66 gRNAs. Normal CD34+ HSPCs were thawed and cultured in expansion media for 2 days before electroporation. CD34+ HSPCs were electroporated with 100 pmol of SCDct5 ssODN templates, along with 5 μg (30.5 pmol) of SpyCas9 protein and 2.5 μg (73.4 pmol) of R-66 gRNA. At 72 h post-electroporation, the cells were harvested and analyzed for editing frequency via ddPCR. Single gRNA that contains 2′-O-methyl 3′-phosphorothioate at the three terminal positions at both the 5′ and 3′ ends (TriLink BioTechnologies) enhanced HDR efficiency in CD34+ cells compared to the unmodified crRNA:tracrRNA duplex gRNA (46) (Supplementary Figure S4c,d). The HDR rate in CD34+ HSPCs was 22%, with high viability (∼90%) post-electroporation with optimized conditions (Supplementary Figure S4c,d). Only a low level of toxicity was observed when RNP and ssODN were co-delivered using electroporation, and delivering RNP did not affect cell viability compared to controls; toxicity was evident only with the delivery of ssODN alone (Supplementary Figure S4e).
Normal CD34+ HSPCs were assayed as both single cell clones and in bulk culture with and without (as control) delivery of the optimized genome editing reagents. The editing frequencies at the genomic level persisted over a 4-week period in differentiation culture (Figure 3A), and colony forming unit (CFU) assay confirmed that the CD34+ HSPCs treated with gRNA/Cas9 RNP and ssODN template retained differentiation potential (data not shown). To analyze how editing frequencies in the bulk HSPC population translate to the zygosity in individual HSPCs, genotype of the single-cell derived burst-forming unit-erythroid (BFU-E) colonies was assessed by colony ddPCR assay. The allele frequency in the starting culture was 28% HDR, 31% WT and 41% NHEJ. Colony genotyping of gene-edited cells revealed that 35% of the colonies showed ssODN template mediated HDR, with 12% of colonies homozygous for HDR alleles and 23% heterozygous with a single HDR allele (Figure 3B and Supplementary Figure S5a,b) that were different from the expected frequencies of 8% homozygous HDR and 40% heterozygous HDR predicted by multi-allele Hardy–Weinberg equation (p2 + 2pq + q2 + 2pr + 2qr + r2 = 1) (Supplementary Table S5) (47). This deviation from the Hardy–Weinberg equilibrium is most likely due to the unequal distribution of HDR and NHEJ alleles in the cell population. Interestingly, we did not observe any colonies with a WT/HDR heterozygous genotype indicating that correction of one allele in the presence of a second unmodified allele is rare. The HDR allele frequencies in BFU-E colonies (23%) were comparable to that predicted based on the editing frequencies in the bulk starting culture (28%), demonstrating erythroid differentiation potential of gene corrected HSPCs (Supplementary Figure S5b). CD34+ HSPCs were differentiated for 14–21 days prior to protein extraction. HPLC and native PAGE analysis of erythroid cells confirmed the production of HbS as a result of HBB gene editing in normal CD34+ cells after delivery of gRNA/Cas9 RNP and ssODN template containing the sickle mutation (Figure 3C and D; Supplementary Figure S5c). The band observed at the HbS migration position in cells treated with RNP only is hemoglobin Leiden (α2β6/7-Glu) resulting from an in-frame 3 bp deletion, which is the predominant type of indel induced by the R-66 gRNA in CD34+ HSPCs (36) (Supplementary Figure S5c–e). These results demonstrate that site-specific modification of the HBB gene in CD34+ HSPCs can be realized with a high HDR rate.
Figure 3.
HBB gene correction in normal CD34+ HSPCs. To quantify HBB editing, both untreated CD34+ cells as control and CD34+ HSPCs treated with R-66 RNP and SCDct5 ssODN were assayed as single cell clones and in bulk culture. Normal CD34+ HSPCs were thawed and cultured in expansion media for 2 days before electroporation. Three days post-electroporation cells were transferred to differentiation phase. (A) In two-phase liquid culture, HBB editing frequencies were quantified by ddPCR to confirm persistence of editing in CD34+ cells in the heterozygous population over a 4-week period (4 days in expansion phase followed by differentiation phase). Only small fluctuation in gene editing rates was observed. The frequencies of HDR, WT and mutagenic NHEJ alleles were measured by ddPCR. Mean with SE error bars from n = 2 biological replicates. (B) The gRNA/Cas9 RNP and ssODN-treated CD34+ HSPCs that gave gene editing rates of 28% HDR, 31% WT and 41% NHEJ in bulk were used for the colony formation assays (CFU) followed by genotype characterization. The distribution of five genotypes in edited cells was determined by colony ddPCR performed on 43 BFU-E colonies, revealing that 35% of the colonies had HDR-mediated gene correction, with 12% of colonies homozygous and 23% heterozygous for gene-corrected alleles. (C) HPLC traces showing hemoglobin production after 21 days differentiation of normal CD34+ HSPCs edited with R-66 RNP and SCDct5 sickle ssODN as compared with mock-treated cells and cells treated with R-66 RNP only. (D) Quantitation of HbS, HbA, HbF and HbA2 protein levels based on HPLC, indicating induced HbS in cells differentiated from gene-edited normal HSPCs. Note the increase in HbF and HbA2 levels in cells from HSPCs treated with RNP only, and HSPCs treated with both RNP and ssODN.
HBB gene correction in SCD patients’ unmobilized peripheral blood-derived CD34+ HSPCs
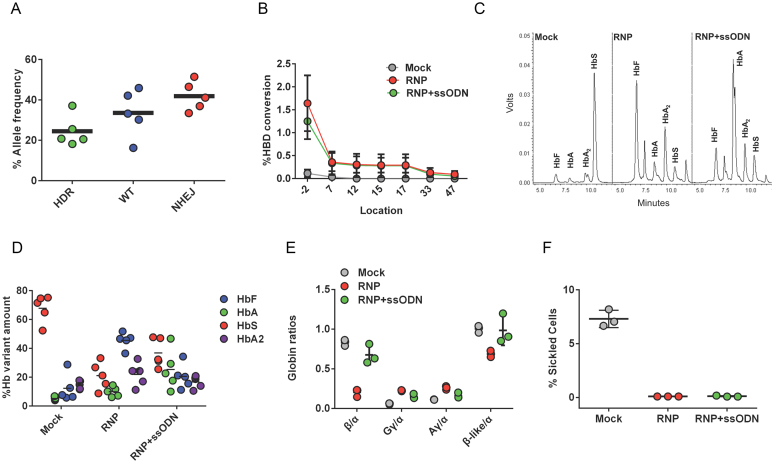
To determine the efficacy of HBB gene correction in CD34+ HSPCs derived from patients with SCD (with HBB genotype SS), CD34+ HSPCs were purified from patients’ unmobilized peripheral blood samples using a microbead kit. The R-66 SCD gRNA that targets the HBB sequence with SCD mutation was used instead of R-66 gRNA (which targets the WT HBB sequence) (Figure 1A). Co-delivery of gRNA/Cas9 RNP and SCDct5-wt ssODN into SCD CD34+ HSPCs from five different patients resulted in 24.5 ± 7.6% allele-specific gene correction (Figure 4A) without changing cell growth rate or viability compared with mock-treated and untreated cells (Supplementary Figure S6). The HBB gene had 41.9 ± 6.4% NHEJ-induced indels in SCD HSPCs treated with gRNA/Cas9 RNP and ssODN, and SCD HSPCs treated with RNP without ssODN that had 81 ± 12% NHEJ-induced indels (Supplementary Figure S7a). To determine the genotype of gene-edited cells, colony ddPCR was performed on 83 colonies from the starting culture that had 37.18% HDR (Figure 4A), indicating that 43% of colonies (24% HDR/HDR, 19% HDR/SCD) have at least one allele gene corrected due to ssODN template integration (Supplementary Figure S7b,c). Further, CRISPR/Cas9 cutting of HBB in SCD HSPCs induced DNA repair using the homologous sequences from HBD as an endogenous template (>90% sequence homology with HBB). The HBD footprints left in the repaired HBB locus was used to map the conversion track showing the decreasing conversion rate with increasing distance to the Cas9 cut site (Figure 4B and Supplementary Figure S7d,e). Because HBD does not carry the sickle mutation, the gene conversion between the HBB sequences in the vicinity of the target locus and the homologous region in HBD resulted in 1.6 ± 0.6% additional T to A gene correction mediated by HBD conversion in cells with RNP-treated cells and 1.3 ± 0.4% in RNP- and ssODN-treated cells (Figure 4B and Supplementary Figure S7d,e). The gene correction mediated by HBD conversion (GtG to GaG) can be distinguished from ssODN-mediated HDR that introduces additional silent MM (GtG to Gaa). The majority of HBD conversion events (76 ± 13%) result in repair of the sickle mutation only and these are expected to be therapeutic events. However, we also observed low frequency alleles (0.3 ± 0.2%) with partial gene-conversion that generates a chimeric HBB–HBD gene with up to three missense mutations (codon 9, 12 and 22) with minimal differences in amino acid chemistry (Figure 4B and Supplementary Figure S7e).
Figure 4.
HBB gene correction in SCD patient-derived CD34+ HSPCs. To determine the efficacy of HBB gene correction in CD34+ HSPCs derived from SCD patients, CD34+ HSPCs were purified from patients’ unmobilized peripheral blood samples. The R-66 SCD gRNA that targets the HBB sequence containing the SCD mutation was used, along with the anti-sickling SCDct5-wt ssODN template for gene correction. (A) The results of HBB gene correction in CD34+ HSPCs from five SCD patients using gRNA/Cas9 RNP and SCDct5-wt ssODN. The frequencies of HDR, WT and NHEJ alleles were measured by ddPCR. Each data point shown corresponds to editing in cells from a particular SCD patient. (B) CRISPR/Cas9 cutting of HBB induced DNA repair using the homologous sequence from the HBD gene as an endogenous template, resulting in T-A SCD mutation correction (Supplementary Figure S7). Each data point shown corresponds to the average editing in cells from six SCD patients. (C) HPLC trace showing hemoglobin production after 21 days of differentiation of SCD HSPCs edited with R-66 SCD RNP and SCDct5-wt anti-sickling ssODN, as compared with mock-treated cells and cells treated with R-66 SCD RNP only. (D) Quantification of HbS, HbA, HbF and HbA2 protein levels measured by HPLC, indicating a significant reduction in HbS and an increased level of HbA in cells differentiated from gene edited SCD HSPCs with RNP and ssODN. Note the low level of HbA (5.3 ± 1%) in mock-treated cells because CD34+ cells were acquired from the peripheral blood of SCD patients undergoing therapeutic red cell exchange. SCD HSPCs treated with RNP only showed an increased level of HbA (9.9 ± 3%) compared to mock-treated cells as a result of the sickle mutation correction via HBD conversion. Data obtained using CD34+ HSPCs from five different SCD patients. (E) Quantitation of globin chain ratios using Reverse phase (RP)-HPLC. Globin chain ratios are shown for SCD HSPCs treated with mock conditions, RNP-only, and with RNP and ssODN. Note that SCD HSPCs treated with RNP and ssODN did not alter the overall balance of β-globin-like chains to α-globin chains compared to mock-treated cells (n = 3, no statistical significance, unpaired t-test). The sickle β-globin (βS) and WT β-globin (βA) are not distinguishable by RP-HPLC. The representative RP-HPLC plot and relevant peak quantification are shown in Supplementary Figure S8d,e. Data shown are representative of n = 3 biological replicates from one SCD donor. (F) After 21 days of differentiation of gene-edited SCD HSPCs, the amount of sickle cells was quantified and their percentage determined, indicating a dramatic decrease in the amount of sickle cells as a result of gene editing.
To test if SCD phenotype can be reversed by gene editing at HBB, we induced erythroid differentiation of the gene-edited SCD CD34+ HSPCs, examined the red blood cell (RBC) progeny and quantified the normal HbA protein levels by HPLC. Compared with mock-treated cells, SCD CD34+ HSPCs treated with RNP and ssODN induced a high level (25.3 ± 13.9%) of HbA protein expression (Figure 4C–E). We found that SCD CD34+ HSPCs from transfusion-dependent patients’ peripheral blood expressed low HbA protein level (5.3 ± 1% in mock), and SCD HSPCs cells treated with RNP only showed an increased level of HbA (9.9 ± 3%) compared to mock-treated cells as a result of the sickle mutation correction via HBD conversion and removal of HbS (Figure 4C–E). We also observed a very strong correlation (R2 = 0.9374) between HBB indel rates and the change in HbF percentage compared to mock-treated cells (Supplementary Figure S8a). The percentage of HbF positive cells (F-cells) and F-cell mean fluorescent intensity (MFI) increased in cells from RNP-treated SCD HSPCs (Supplementary Figure S8b–c). In particular, the ratio of γ-globin to α-globin level in cells from SCD HSPCs treated with both gRNA/Cas9 RNP and ssODN increased by 1.8-fold, and that from SCD HSPCs treated with RNP only increased by 2.8-fold compared to mock-treated cells (Figure 4E), suggesting that HBB disruption could lead to increased production of γ-globin chains (Gγ and Aγ). Increase in γ-globin chains restored the overall balance of β-like globin to α-globin chains (Figure 4E), which may help maintain the total hemoglobin level in cells from SCD HSPCs treated with RNP and ssODN. The representative RP-HPLC plot and relevant peak quantification are shown in Supplementary Figure S8d,e. Further, we found that targeting the HBB intron region did not change HbF level, suggesting that the increase in HbF percentage is due to the loss of HBB expression (Supplementary Figure S9). However, the consequences of inducing indels near the HBB sickle mutation in SCD CD34+ HSPCs merit further investigation.
Under deoxygenated conditions, the hemoglobin of patients with SCD (HbS, α2βS2) forms rigid polymers (48). Addition of HbF to the growing HbS polymer terminates the chain; therefore, either replacing HbS with HbA through gene correction or increasing HbF concentrations in the cell can prevent sickling (49). HbF concentrations of 20% or higher have been shown to significantly reduce clinical complications of SCD and prevent sickling in vitro (50). To determine the anti-sickling effect of HBB gene correction and HbF induction on RBCs, SCD HSPCs were differentiated in a serum-free two-phase culture system for 4 weeks. At day 28, sickled RBCs were counted and their percentage determined. These cultured cells represent orthochromoblasts, not mature RBCs (51). Orthochromoblasts are larger, reducing the intracellular hemoglobin concentration compared to mature RBCs, and are not enucleated; they therefore do not sickle to the same extent as a circulating RBC. We found that, compared with mock-treated cells, the average amount of sickled RBCs resulting from gene-edited SCD HSPCs decreased from 7.3% to 0.1% in SCD HSPCs cells treated with both RNP and ssODN, and those treated with RNP only (Figure 4F). This demonstrates that removal of sickle alleles by Cas9 cutting (81 ± 12% indel rate) is sufficient to prevent RBCs from sickling as a result of delivering RNP only to SCD HSPCs cells. However, the presence of the ssODN donor is essential for correcting the sickle alleles to a functional sequence and restore normal adult hemoglobin expression (Figure 4D–E).
Evaluation of off-target effects in gene-edited CD34+ HSPCs from patients with SCD
Although CRISPR/Cas9 systems can have highly efficient gene editing at the intended target site, off-target cutting may occur due to the tolerance for base mismatches, small insertions and deletions between DNA sequences in the genome and the intended 20-nt gRNA target sequence (23,25,52,53). The off-target cutting activity of Cas9 nuclease can cause disruption of normal gene function via NHEJ-mediated indels, as well as genome instability via large chromosomal rearrangements (54), which is of serious concern in therapeutic applications of genome editing. We used a web-based in silico off-target search tool COSMID (37) to identify potential off-target sites for the R-66 SCD gRNA and the top-ranked off-target sites were labeled as OT1–OT57 (Supplementary Table S2). For the R-66 SCD gRNA, we further used a genome-wide, unbiased off-target search tool GUIDE-Seq (Genome-wide Unbiased Identification of DSBs Enabled by Sequencing) (38) to identify its off-target sites in U2OS cells (Figure 5A and Supplementary Table S6). All the GUIDE-Seq identified off-target sites, including GS_02 (OT18), GS_03 (OT1), GS_04 (OT12), GS_05 (OT10) and GS_06 (OT9), were among those predicted by COSMID (Supplementary Tables S2 and S6). After delivering the RNP of R-66 SCD gRNA with WT Cas9 into peripheral blood SCD CD34+ HSPCs, the off-target activity at the 57 COSMID predicted sites (including the 5 GUIDE-Seq identified sites) was quantified at Day 2 and 12 post-electroporation using rhAmpSeq™ (Integrated DNA Technologies), a multiplex RNase H-dependent PCR (rhPCR) approach consisting of pooled blocked-cleavage primers (45,55) (Supplementary Table S3), which allowed for multiple sites to be assessed simultaneously with high sensitivity. The use of blocked-cleavage primers requires that the primers correctly hybridize to the target sequence prior to amplification to reduce primer dimerization and increase specificity in multipexed PCR reactions (26). Of the 57 COSMID-identified potential off-target sites, we found that seven sites (OT18, OT1, OT12, OT2, OT34, OT3, OT9) were true off-target sites with detectable activity (indel rates ≥ 0.1%) (Figure 5B and Supplementary Table S7). In particular, the off-target site OT18, which has three single-base mismatches compared to the target sequence of R-66 SCD gRNA, had substantial off-target activity (36.7% indel rate at Day 2). The genome-wide unbiased search using GUIDE-Seq in U2OS cells identified the top 3 active off-target sites, OT18, OT1 and OT12, as well as OT9 that had very low activity (Figure 5B). However, GUIDE-Seq was not able to identify OT2, OT3 and OT34, which had low but measurable cutting activity (Figure 5B and Supplementary Table S6). We found that when both gRNA/Cas9 RNP and ssODN were delivered into peripheral blood CD34+ HSPCs from patients with SCD, we could achieve ∼30% HDR rate at the on-target site; however, the off-target activities at the COSMID-predicted sites are different from that with RNP only (Figure 5C and D; Supplementary Table S7), consistent with what previously reported (56). For example, the predicted off-target site OT27 had detectable activity only in the presence of the ssODN donor (Figure 5B and C). We also found that when the off-target activities of gRNA/WT Cas9 RNP at the 57 COSMID predicted sites were quantified using single-plex PCR amplification followed by NGS at Day 5 post-electroporation, both the active sites and their indel rates changed slightly (Supplementary Figure S10). This is most likely due to experimental variations (different stimulation time before extracting DNA and different patient cells).
Figure 5.
Off-target analysis for R-66 SCD gRNA and reduction of off-target effect by high-fidelity Cas9. We used a web-based in silico off-target site search tool (COSMID) and unbiased experimental off-target search method (GUIDE-Seq) to identify off-target sites for the R-66 SCD gRNA. (A) The 5 off-target sites of R-66 SCD gRNA identified using GUIDE-Seq in U2OS cells. (B) The WT SpyCas9 and a high fidelity SpyCas9 (HiFi Cas9), respectively, complexed with R-66 SCD gRNA as RNP were delivered into peripheral blood SCD CD34+ HSPCs. The cleavage activities at the on-target site and 57 COSMID predicted sites were analyzed by targeted deep sequencing. The dot-plot shows indel frequencies at the on-target site (HBB) and the 7 off-target sites with measurable cleavage activity. WT and HiFi Cas9 showed comparable on-target editing rates. However, the use of HiFi Cas9 significantly reduced off-target activity, with the average activity at four sites (OT1, OT3, OT12 and OT34) below 0.1%. Near 0.1% background indel frequencies were found in mock-treated samples at OT26, OT31 and OT45, which have Poly-C (4 Cs) stretches at the expected cut site. Insertions and deletions occur at elevated rates in homopolymeric regions and this type of sequencing errors prevents accurate indel quantitation as they are indistinguishable from CRISPR-induced deletion or insertion. (C and D) The WT SpyCas9 and a HiFi Cas9, respectively, complexed with R-66 SCD gRNA as RNP were delivered into peripheral blood SCD CD34+ HSPCs together with ssODN template. (C) The indel frequencies at 5 off-target sites with measurable activity, quantified by targeted deep sequencing. The use of HiFi Cas9 significantly reduced off-target activity in the presence of ssODN. (D) WT and HiFi Cas9 showed comparable on-target HDR and NHEJ editing rates when both RNP and ssODN are delivered into SCD CD34+ HSPCs. (E and F) The frequencies of chromosomal deletion (E) and inversion (F) events at Day 2, 7 and 12 after RNP delivery were measured by ddPCR. Note that the use of HiFi Cas9 significantly reduced both chromosomal deletion and inversion. During the 12 days post-electroporation, chromosomal inversion maintained at 3.3% (E) while large chromosomal deletion decreased from 1.8% to 0.2% (F). In (B, E and F), data shown are representative of n = 3 biological replicates from one SCD patient. In (C and D), data shown are representative of 1 biological replicate from two SCD patients.
It has been shown that two concurrent DNA double strand breaks (DSBs) produced by CRISPR/Cas9 can introduce large chromosomal rearrangements (54). Given that HBB is located on the p (short) arm of ch.11 and the highly active off-target site OT18 is located on the q (long) arm of ch.11, we assessed the 54 MB pericentric deletion or inversion between the R-66 SCD gRNA target site in HBB and OT18 in gRNA/Cas9 RNP-treated SCD HSPCs. Using primer pairs spanning the expected junction point on deleted or inverted chromosomes (Supplementary Table S1), we detected chromosomal rearrangement by end-point PCR, which was confirmed by sequencing of PCR clones (Supplementary Figure S11a–c). We next used a ddPCR assay to quantify deletion and inversion events at Day 2, 7 and 12 after WT Cas9 RNP delivery, and found that chromosomal inversion was maintained at 3.3%, while large chromosomal deletion decreased from 1.8% to 0.2% in treated samples (Figure 5E–F; Supplementary Figure S11d,e). This was most likely due to the loss of the entire chromosome during cell division, resulting in a reduction in the copy number of chromosome 11 in RNP-treated cells over time.
High fidelity Cas9 increased editing specificity in CD34+ HSPCs from patients with SCD
Off-target effects and large chromosomal rearrangements such as the 54 MB pericentric deletion and inversion we identified in this study raise safety concerns for clinical translation. In order to improve safety and specificity, we tested a recently described high-fidelity Cas9 variant (HiFi Cas9) (26), which was complexed with R-66 SCD gRNA as RNP and delivered into peripheral blood CD34+ HSPCs from patients with SCD. The off-target activities at the 57 COSMID predicted sites Day 2 and 12 post-electroporation were quantified using the rhAmpSeq™ assay (Supplementary Table S3). The chromosomal rearrangement between the on-target site and OT18 was quantified at Day 2, 7 and 12 post-electroporation using a ddPCR assay. We found that the use of HiFi Cas9 gave a similar level of on-target editing in terms of HDR and NHEJ rates compared with WT Cas9 (Figure 5D), but significantly reduced the indel rates at most of the off-target sites, with a maximum off-target indel rate of 0.23% (Figure 5B and C; Supplementary Table S7). We further found that the use of HiFi Cas9 significantly reduced the intra-chromosomal rearrangement events, while maintaining the same level of on-target gene editing (Figure 5D and F), demonstrating our ability to achieve high-efficiency gene correction with increased specificity in SCD CD34+ HSPCs.
Engraftment of WT Cas9-edited SCD patient peripheral blood CD34+ HSPCs in NSG mice
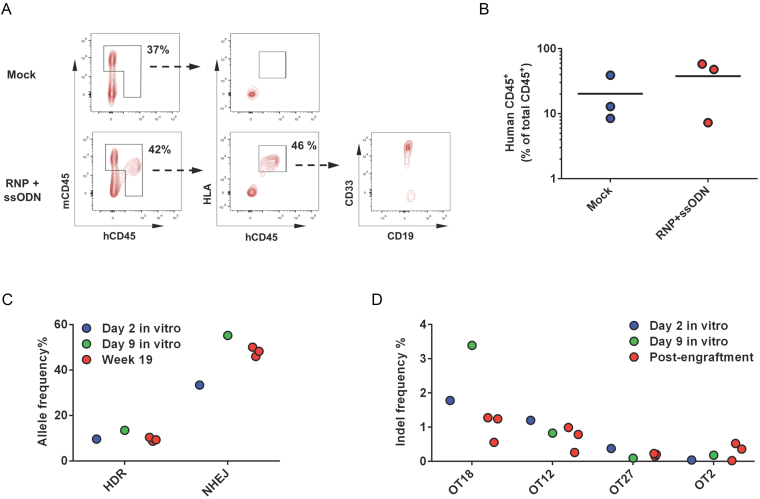
To determine if gene-corrected SCD HSPCs retain the ability to engraft, we performed the first engraftment study using the CD34+ HSPCs derived from the unmobilized peripheral blood of patients with SCD by injecting the cells intrafemorally into NSG mice. Peripheral CD34+ cells were isolated from patients undergoing red cell exchange therapy and frozen for storage until sufficient cells were collected for transplant studies. After thawing, cells were cultured for 2 days and electroporated with R-66 SCD gRNA/WTCas9 RNP and SCDct5-wt ssODN. After 2 days post-delivery of genome-editing reagents, 500 000 viable cells were intrafemorally transplanted into NSG mice. These mice were euthanized at Week 19 after transplantation and the bone marrow was harvested to determine the engraftment potential. Post-engraftment data were derived from n = 3 transplanted mice for mock electroporated cells and RNP- and ssODN-treated cells. The level of engraftment was measured as the fraction of human HLA+/CD45+ cells in the total CD45+ compartment (Figure 6A and Supplementary Figure S12). We found that the engraftment levels of gene-edited SCD HSPCs were similar to those of control cells (mock electroporated SCD CD34+ HSPCs), with an average of 30.2 ± 24.1% of cells double positive for HLA and hCD45 (Figure 6B). Almost all human cells were positive for CD33 demonstrating near-exclusive myeloid lineage (Supplementary Figures S13 and S14). This myeloid bias may reflect the biologic properties of un-mobilized CD34+ cells in the peripheral blood of sickle cell patients and does not necessarily reflect the biologic properties of CD34+ cells after mobilization, or of CD34+ harvested from bone marrow.
Figure 6.
Engraftment of patient-derived peripheral blood CD34+ HSPCs into NSG mice. To determine if gene-edited SCD peripheral blood HSPCs retain the ability to engraft, CD34+ cells isolated from the peripheral blood of a SCD patient were treated with R-66 SCD gRNA/WT Cas9 RNP and ssODN template, and injected intrafemorally into NSG mice. Bone marrow was harvested from euthanized mice at Week 19 after transplantation to analyze the engraftment rates. (A) Schematic of flow cytometry analysis of bone marrow harvested from transplanted mice at week 19. Total CD45+ cell population was selected and the presence of human cells confirmed by staining for both HLA and human CD45. Cell lineage was tested by staining for CD33 and CD19. (B) Engraftment rates of both control (mock electroporated) cells and cells treated with RNP and ssODN in the circulating blood/bone marrow at week 19. There was no significant difference in the level of engraftment in gene-edited cells when compared to control cells. (C) On-target editing rates in bulk transplanted cells at the time of engraftment (Day 2 in vitro), Day 9 in vitro and at Week 19 post transplantation. The rates of HDR and NHEJ were assessed using high-throughput sequencing. Week 19 data were derived from three mice transplanted with cells treated with RNP and ssODN. (D) Off-target cutting rates in bulk transplanted cells at the time of engraftment, at Day 9 in vitro, and at week 19 post-transplantation. Week 19 data were derived from three mice. Four genomic loci had detectable levels of off-target cutting at Week 19.
Human cells were isolated from mouse bone marrow and DNA was extracted for gene-editing analysis. Editing rates pre-transplantation (at Day 2 post-electroporation) were 10% HDR and 33.3% NHEJ (Figure 6C) as determined by high-throughput sequencing. The rate of HDR at Week 19 was unchanged compared to the pre-transplantation level; however, the rate of NHEJ increased from 33.3% to 48% at Week 19 (Figure 6C). A similar increase in the NHEJ rate was observed in the gene-edited SCD peripheral blood CD34+ HSPCs expanded in vitro at Day 9 post-electroporation (55.17% NHEJ, 13.46% HDR, 31.37% WT) (Supplementary Table S8), indicating that CRISPR/Cas9- and ssODN-mediated editing was not complete at the time of transplantation (Day 2 post-electroporation). Off-target analysis of the engrafted cells revealed off-target activity at 4 loci (OT18, OT12, OT27 and OT2) (Figure 6D) out of six active OT sites identified in the in vitro studies in RNP- and ssODN-treated cells (OT18, OT12, OT1, OT2, OT48 and OT27) (Supplementary Figure S10). The indel diversity in gene-edited cells at Week 19 in all three transplanted mice showed a similar pattern as compared to pre-engraftment levels at Day 2 post-electroporation and in vitro expanded cells at Day 9 post-electroporation (Supplementary Figure S15), as demonstrated by the relative fractions of different indels indicated by the color code. Although an increase in indel rates due to NHEJ was observed at week 19 compared to Day 2 post-electroporation, this increased level of NHEJ was also observed at Day 9 post-electroporation, suggesting that editing events had not completed at Day 2 post-delivery, and/or unrepaired DSBs were still present at Day 2 (57). The lack of increase in HDR may result from reduced stability of ssODN template since it is well known that unmodified single‐stranded DNA can be degraded intracellularly (58).
Engraftment of HiFi-Cas9 edited SCD patient bone marrow CD34+ HSPCs in NSG mice
To evaluate the impact of the source of CD34+ cells on myeloid bias after engraftment, we performed the second engraftment study using the CD34+ HSPCs derived from the bone marrow of patients with SCD by injecting the cells intravenously into NSG mice. Prior to engraftment experiments, we first validated that CRISPR/Cas9 can correct the sickle mutation in bone marrow derived SCD CD34+ cells. These cells were cultured for 3 days prior to electroporation since we had observed an increase in editing efficiency at this time point versus 2 days after freeze/thawing of SCD HSPCs (data not shown). Using R-66 SCD gRNA complexed with HiFi Cas9 (RNP), we achieved 71% on-target NHEJ rate with a low level of off-target cutting at Day 7 post electroporation (0.27% activity at OT2, 0.12% activity at OT9) (Supplementary Figure S16a and Supplementary Table S7). Further, no intra-chromosomal rearrangements were detected (Supplementary Figure S16b). Gene edited CD34+ cells were single-cell sorted from two patients with SCD (donors) into methocellulose plates to monitor colony formation. While there was a reduction in the total number of colonies formed, we did not observe a difference in the distribution of colony types between mock and treated cells (Supplementary Figure S17). To determine if gene-corrected SCD bone marrow HSPCs retain the ability to engraft, CD34+ cells isolated from the bone marrow of two SCD donors were treated with R-66 SCD gRNA/HiFiCas9 RNP and ssODN template and frozen for 48 h after treatment (Supplementary Figure S18a). For bone marrow-derived SCD CD34+ cells, cell viability was reduced by 10–25% due to freezing/thawing and by 10–35% due to electroporation (Supplementary Figure S18b,c). Due to low post-thaw recovery of SCD Donor 1 sample treated with RNP and ssODN (Supplementary Figure S18d), 3 × 105 viable cells were injected per mouse. In all other cases, after thawing, 5 × 105 viable cells were injected intravenously into each NSG mouse. Mice were euthanized at Week 16 after transplantation and the bone marrow harvested to determine the engraftment potential. The level of engraftment was measured as the fraction of human HLA+/CD45+ cells in the total CD45+ compartment (Supplementary Figures S19–S22). Post-engraftment data were derived from n = 4 transplanted mice for SCD Donor 2 cells treated with RNP only and SCD Donor 2 cells treated with RNP and ssODN, n = 3 for SCD Donor 1 cells treated with RNP and ssODN and n = 2 for untreated cells. Only a small percentage of infused cells from the SCD Donor 1 engrafted (0.1 ± 0.1% for CD34+ cells treated with RNP, 1.6 ± 1.1% for CD34+ cells treated with RNP and ssODN) (Figure 7A). In mice injected with SCD Donor 2 CD34+ cells treated with RNP and ssODN, an average of 7.5 ± 6.2% of cells were double positive for HLA and hCD45, compared to 16.8 ± 13.2% with unedited CD34+ cells, indicating a good level of engraftment of gene-edited SCD HSPCs (Figure 7A). With the same injection condition (SCD Donor 2 cells with both RNP and ssODN), a higher fraction of human cells were positive for CD19 (84 ± 15%) (Figure 7B), demonstrating a lymphoid lineage bias that has been reported for NSG mouse models of human engraftment (59). This is in contrast to the myeloid lineage bias observed in our first engraftment study using SCD peripheral blood CD34+ HSPCs.
Figure 7.
Engraftment of HiFi Cas9 gene-edited patient bone marrow CD34+ HSPCs in NSG mice. To determine if gene-edited SCD bone marrow HSPCs retain the ability to engraft, CD34+ cells isolated from the bone marrow of two SCD patients were treated with R-66 SCD gRNA/Hifi Cas9 RNP and ssODN template, and injected intravenously into NSG mice. Bone marrow was harvested from euthanized mice at Week 16 after transplantation to analyze the engraftment rates. (A) The engraftment levels for samples from mice injected with six different edited CD34+ cells showing varying levels of engraftment. The level of engraftment was measured as the fraction of human HLA+/ CD45+ cells in the total CD45+ compartment. (B) Cell lineage was tested by staining for CD33 and CD19. A higher fraction of human cells were positive for CD19, demonstrating a lymphoid lineage bias. (C) The rates of HDR and NHEJ in transplanted SCD CD34+ cells right before transplantation. (D) The rates of HDR and NHEJ in bulk transplanted cells at Week 16 post-transplantation. Note that the result from SCD Donor 1 cells treated with RNP only is not included due to very low engraftment rate and low human DNA copy number in the sample. For (C) and (D), on-target editing rates were assessed using high-throughput sequencing. (E) Off-target cutting rates at the time of transplantation, 2 days after electroporation, or before freezing edited cells. The graph shows indel frequencies at the three off-target sites with detectable cleavage activity. (F) Off-target cutting rates 16 weeks post-transplantation. The measured indel rates had a large variation among the samples. However, the overall average off-target cutting rates are quite low (most < 0.1%), demonstrating the high specificity of HiFi Cas9 editing. For (C) and (E), pre-engraftment data were derived from n = 2 biological replicates for Donor1 RNP, n = 3 for Donor2 RNP and Donor1 RNP+ssODN, and n = 4 for Donor2 RNP+ssODN and Donor2 ssODN. For (D) and (F), post-engraftment data were derived from n = 4 transplanted mice for SCD Donor 2 cells treated with RNP only and SCD Donor 2 cells treated with RNP and ssODN, n = 3 for SCD Donor 1 cells treated with RNP and ssODN and n = 2 for untreated cells.
The on-target editing rates (HDR and NHEJ) of edited CD34+ cells from each batch of electroporation were quantified before freezing (pre-engraftment) using high-throughput sequencing. Similar quantification was performed post-transplantation using DNA samples extracted from mouse bone marrow (including mouse cells and engrafted human cells) with human HBB specific primers for PCR and NGS. We found that the average HDR rate was reduced from 29.8 ± 3.9% for pre-engraftment samples to 8.8 ± 14% at Week 16 post-transplantation in SCD Donor 2 CD34+ cells treated with RNP and ssODN (Figure 7C and D). In addition to HDR-induced gene-correction, we also observed correction of the sickle mutation by gene conversion with HBD. The average rate of gene conversion was 2.4 ± 0.2% in pre-engraftment samples, and became 1.8 ± 2% at Week 16 post-transplantation (Supplementary Figure S18e,f,h), but the gene conversion rate increased respectively to 4.65% in mouse 14 that was injected with SCD Donor 2 cells treated with RNP and ssODN, and to 6.97% in mouse 15 that was injected with SCD Donor2 cells treated with RNP only (Supplementary Figures S18g and S23). The average rate of NHEJ also dropped from 42.8 ± 1.9% in SCD CD34+ cells pre-engraftment to 24.5 ± 13.8% in human cells 16 weeks post-transplantation, suggesting that in general gene-edited cells display reduced engraftment potential compared to unedited cells (Figure 7C and D). The highly variable editing rate (Figure 7D) and indel diversity (Supplementary Figure S23) in gene-edited cells from the two SCD donors at Week 16 post-transplantation may reflect the condition of the two patients with SCD whose bone marrow CD34+ cells were used in the second engraftment study, as well as the total number of CD34+ cells engrafted. Further, we found that the indel rates (NHEJ) at the predicted off-target sites have a large variation (Figure 7E) in different DNA samples extracted from mouse bone marrow 16 weeks post-engraftment (Supplementary Table S9). Compared with the indel rates pre-engraftment, at certain sites (such as OT7, OT22, OT30, OT43), the average indel rates are higher at 16 weeks post-engraftment, while at some other sites (such as OT2, OT9 and OT53), the trend is the opposite. However, the overall average off-target cutting rates at Week 16 post-engraftment are quite low (<0.1%), demonstrating the high specificity of HiFi Cas9 based genome editing (Figure 7F).
DISCUSSION
Compared with conventional gene therapy via lentiviral vector-mediated addition of anti-sickling β-globin gene into HSPCs of patients with SCD (14), the CRISPR/Cas9-based genome editing strategy adopted here has the potential to be safer and more efficient in treating SCD. This is likely due to the precision of CRISPR/Cas9 in correcting the sickling mutation, removal of the sickle allele via NHEJ and the avoidance of using a viral vector for delivery. The recent report of gene therapy in a patient with SCD using lentiviral transduction reported 2066 unique integration sites, of which the long-term effects are unknown (14). In demonstrating the potential of CRISPR/Cas9 based genome editing, DeWitt et al. (17) and Dever et al. (18) showed that a high level of HBB gene-correction in SCD CD34+ HSPCs can be achieved in vitro. However, the engraftment studies by DeWitt et al. (17) and Dever et al. (18) were carried out using CD34+ HSPCs from normal individuals, not from patients with SCD, and a genome-wide unbiased off-target analysis was not performed for the CRISPR/Cas9 systems used.
In this study, we demonstrated that our non-viral, selection-free gene-correction approach using optimized R-66 SCD gRNA and SCD5ct-wt ssODN template could achieve a high level (up to 37% in peripheral blood CD34+ cells and 33% in bone marrow CD34+ cells) of HBB gene correction in SCD-patient derived CD34+ HSPCs, and gene-edited SCD HSPCs could differentiate into erythroid cells that produced a high level of normal hemoglobin HbA in vitro (up to 47% HbA), resulting in a significant reduction of the amount of sickled red blood cells from differentiated SCD HSPCs even under extreme hypoxic conditions. We found that the increase in γ-globin chains restored the overall balance of β-like globin to α-globin chains. We further demonstrated that gene-edited CD34+ HSPCs derived from the bone marrow of patients with SCD were able to engraft in NSG mice and the corrected alleles were stable for up to 16 weeks post-transplantation. These results clearly show the promise of CRISPR/Cas9 based genome editing for curing sickle cell disease. When we used CD34+ cells from the unmobilized peripheral blood of patients with SCD, we observed a high level of myeloid bias. It is possible that the CD34+ cells, we isolated from peripheral blood of patients with SCD, were mostly myeloid progenitors, with only a small amount of true hematopoietic stem cells (HSCs).
When delivering only gRNA/Cas9 RNP (i.e. without ssODN) into SCD CD34+ HSPCs, we found that cutting at the HBB locus in the exonic region near the sickle mutation induced a large increase of HbF percentage among all hemoglobin proteins expressed. An increase in HbF percentage was also observed in SCD HSPCs when both gRNA/Cas9 RNP and ssODN template were delivered (Figure 4D). It is possible that the increase in HbF percentage is due to Cas9 cutting induced biallelic β-thalassemia mutation, or due to the increase in hemoglobin formation between α-globin and γ-globin since β-globin chains are unavailable. However, while intriguing, it is unclear if Cas9 HBB-cutting induced increase in HbF percentage would have a long-lasting effect at a therapeutically relevant level (assumed to be 20% HbF or higher), and if the resulting benefits to SCD patients would outpace the potential harm of HBB disruption, including the possibility of inducing β-thalassemia major or minor. The molecular mechanism underlying this type of HbF induction warrants further investigation.
For therapeutic genome editing, potential off-target effects need to be carefully analyzed, and significant challenges exist in both accurately predicting potential off-target sites and in performing genome-wide unbiased searches (60). Further, currently deep sequencing has a detection limit of 0.1% thus cannot capture low frequency indels (61), and gross chromosomal rearrangements due to concurrent on- and off-target DSBs or two off-target DSBs cannot be comprehensively identified or quantified by most existing methods. For R-66 SCD gRNA tested in this study, the use of in silico prediction tool COSMID has identified 57 potential off-target sites in the human genome, of which 7 were confirmed experimentally as true off-target sites in peripheral blood SCD HSPCs, including one (OT18) that caused chromosomal rearrangements. To date this is the deepest analysis of CRISPR/Cas9 induced off-target effect in SCD HSPCs. We found that the use of a recently described high-fidelity Cas9 variant (26) significantly reduced the off-target editing to background noise levels and prevented large intra-chromosomal rearrangements in CD34+ cells from patients with SCD without compromising the on-target editing rates. Further, with this HiFi Cas9, we observed only minimal levels of off-target effects at 16 weeks post-transplantation in NSG mice. This further demonstrates the potential of this high-fidelity Cas9 variant for clinical translation of CRISPR/Cas9 based genome editing. However, it is reasonable to assume that even with HiFi-Cas9, CRISPR/Cas9 based gene-editing will have some degree of off-target effects. As the CRISPR/Cas9 technology moves toward clinical applications, there is a critical need for more sensitive off-target analysis methods as well as approaches to reduce off-target effects for safer genome editing. These efforts should take into account large deletions induced by DSBs at the on-target site (62), and large chromosomal rearrangements such as inversions and translocations.
Our results demonstrated that gene-corrected CD34+ HSPCs from bone marrow of patients with SCD were able to engraft in NSG mice. However, there was a reduction of the percentage of gene-corrected alleles at 16 weeks post-transplantation, which may reflect a reduced engraftment capacity of edited cells. It is unclear if this is due to gene editing alone or a combination of the stress of gene editing and freeze/thaw of the transplanted cells. The difference between the engraftment behaviors of CD34+ cells harvested from the bone marrow of two patients with SCD may reflect their individual SCD pathology, including differences in age, gender, disease severity and treatment exposure. However, overall, CRISPR gRNA/Cas9 RNP and ssODN template treatment of bone marrow CD34+ cells from patients with SCD was well tolerated. When we used peripheral blood derived CD34+ cells from patients with SCD for engraftment, the level of gene-corrected alleles in the engrafted population was similar to the pre-transplantation levels, indicating the survival of gene-corrected CD34+ cells over the period of 19 weeks post-transplantation. We believe that the non-mobilized peripheral blood SCD CD34+ cells used in our first engraftment study have a lower number of true HSCs compared to plerixafor mobilized or bone marrow harvested CD34+ cells, resulting in a myeloid bias (63,64). In contrast, a clear lymphoid bias was evident with engraftment of bone marrow SCD CD34+ cells treated with RNP and ssODN.
In general, delivery of gRNA/Cas9 and ssODN into SCD CD34+ cells may result in six cell subpopulations in terms of HBB allele modification: (i) HDR/HDR, (ii) HDR/NHEJ, (iii) HDR/WT, (iv) WT/WT, (v) WT/NHEJ, (vi) NHEJ/NHEJ (here HDR = correction of sickle mutation, WT = sickle mutation, NHEJ = induced mutagenesis). Although CD34+ cells that had Cas9 cutting of HBB but no HDR-mediated correction of sickle mutation may offer some potential benefit, the possibility of inducing β-thalassemia major or minor phenotype in a sub-population of cells needs to be carefully studied, and the potential consequences of reduction in the level of functional β-globin in a patient with SCD need to be determined. Since the lifespan of a sickle RBC is ∼20 days but that of a normal RBC or even sickle cell trait RBC (which contains 40% HbS) is ∼120 days in circulation (65), prolonged survival of the RBCs containing a lower percentage of HbS may compensate for the reduction of functional β-globin due to HBB disruption, resulting in no change in total hemoglobin level in the body. Further, the extent of on-target mutagenesis need to be systematically studied, in light of the recent finding that on-target DNA double-strand breaks could induce large deletions and complex genomic rearrangements, which may have pathogenic consequences (62). To minimize the risk of inducing β-thalassemia (major or minor) in patients with SCD by ontarget Cas9 cutting, the optimal dose and half-life of the gRNA/Cas9 RNP need to be further investigated, aiming to reduce on-target mutations (NHEJ indels) without compromising the rate of HDR. It is possible that, with further optimization, selection using chemical/biological methods such as cell-cycle synchronization (66), NHEJ inhibition (67,68) or cell surface receptor (18) will not be necessary in order to achieve a positive clinical outcome.
DATA AVAILABILITY
The data that support the findings of this study and the custom MatLab scripts used in this study are available on request from the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Harshavardhan Deshmukh for help in optimizing the protocols for cell culture, Anirban Ray for help in sequencing experiments and Katherine King for help in bone marrow extraction. We are grateful to the sickle cell disease patients for permitting the use of discarded red cell exchange samples for HSPC isolation. M.H.P. gratefully acknowledges the support of the Amon Carter Foundation, the Laurie Kraus Lacob Faculty Scholar Award in Pediatric Translational Research and the Sutardja Foundation for their support.
Author contributions: G.B. and C.M.L. conceived the idea and designed the study. S.H.P. and C.M.L, performed gene editing experiments, HSPC differentiation and data analysis. V.A.S., A.K.C. and Y.Z. isolated patient CD34+ HSPCs and performed HPLC experiments. A.P. performed in vitro sickling assays. T.H.D. performed GUIDE-seq experiments and data analysis. S.H.P., C.M.L, D.P.D., W.S. and J.C. performed the engraftment study. G.B., S.H.P., C.M.L, V.A.S. and M.H.P wrote the manuscript.
Notes
Present address: Ciaran M. Lee, APC Microbiome Ireland, University College Cork, Ireland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Prevention and Research Institute of Texas [RR140081 to G.B.]; National Heart, Lung and Blood Institute of NIH [1K08DK110448 to V.S.]; Chao Physician Scientist Award (to V.S.); National Institute of Diabetes and Digestive and Kidney Diseases of NIH [T32 DK06044515 to A.C.]; Stanford Child Health Research Institute (CHRI) Grant and Postdoctoral Award (to D.P.D). Funding for open access charge: Cancer Prevention and Research Institute of Texas.
Conflict of interest statement. M.H.P. serves on the Scientific Advisory Board of CRISPR Therapeutics.
REFERENCES
- 1. Platt O.S., Thorington B.D., Brambilla D.J., Milner P.F., Rosse W.F., Vichinsky E., Kinney T.R.. Pain in sickle cell disease. Rates and risk factors. N. Engl. J. Med. 1991; 325:11–16. [DOI] [PubMed] [Google Scholar]
- 2. Platt O.S., Brambilla D.J., Rosse W.F., Milner P.F., Castro O., Steinberg M.H., Klug P.P.. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N. Engl. J. Med. 1994; 330:1639–1644. [DOI] [PubMed] [Google Scholar]
- 3. Ingram V.M. A specific chemical difference between the globins of normal human and sickle-cell anaemia haemoglobin. Nature. 1956; 178:792–794. [DOI] [PubMed] [Google Scholar]
- 4. Hassell K.L. Population estimates of sickle cell disease in the U.S. Am. J. Prev. Med. 2010; 38:S512–S521. [DOI] [PubMed] [Google Scholar]
- 5. Odame I. Perspective: we need a global solution. Nature. 2014; 515:S10. [DOI] [PubMed] [Google Scholar]
- 6. Lanzkron S., Carroll C.P., Haywood C.. Mortality rates and age at death from sickle cell disease: U.S., 1979–2005. Public Health Rep. 2013; 128:110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Steinberg M.H., McCarthy W.F., Castro O., Ballas S.K., Armstrong F.D., Smith W., Ataga K., Swerdlow P., Kutlar A., DeCastro L. et al.. The risks and benefits of long-term use of hydroxyurea in sickle cell anemia: A 17.5 year follow-up. Am. J. Hematol. 2010; 85:403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vichinsky E.P., Ohene-Frempong K., Thein S.L., Lobo C.L., Inati A., Thompson A.A., Smith-Whitley K., Kwiatkowski J.L., Swerdlow P.S., Porter J.B. et al.. Transfusion and chelation practices in sickle cell disease: a regional perspective. Pediatr. Hematol. Oncol. 2011; 28:124–133. [DOI] [PubMed] [Google Scholar]
- 9. Rosse W.F., Gallagher D., Kinney T.R., Castro O., Dosik H., Moohr J., Wang W., Levy P.S.. Transfusion and alloimmunization in sickle cell disease. The Cooperative Study of Sickle Cell Disease. Blood. 1990; 76:1431–1437. [PubMed] [Google Scholar]
- 10. Walters M.C., Patience M., Leisenring W., Rogers Z.R., Aquino V.M., Buchanan G.R., Roberts I.A., Yeager A.M., Hsu L., Adamkiewicz T. et al.. Stable mixed hematopoietic chimerism after bone marrow transplantation for sickle cell anemia. Biol. Blood Marrow Transplant. 2001; 7:665–673. [DOI] [PubMed] [Google Scholar]
- 11. Mentzer W.C., Heller S., Pearle P.R., Hackney E., Vichinsky E.. Availability of related donors for bone marrow transplantation in sickle cell anemia. Am. J. Pediatr. Hematol./Oncol. 1994; 16:27–29. [PubMed] [Google Scholar]
- 12. King A., Shenoy S.. Evidence-based focused review of the status of hematopoietic stem cell transplantation as treatment of sickle cell disease and thalassemia. Blood. 2014; 123:3089–3094. [DOI] [PubMed] [Google Scholar]
- 13. Dallas M.H., Triplett B., Shook D.R., Hartford C., Srinivasan A., Laver J., Ware R., Leung W.. Long-term outcome and evaluation of organ function in pediatric patients undergoing haploidentical and matched related hematopoietic cell transplantation for sickle cell disease. Biol. Blood Marrow Transplant. 2013; 19:820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shenoy S., Eapen M., Panepinto J.A., Logan B.R., Wu J., Abraham A., Brochstein J., Chaudhury S., Godder K., Haight A.E. et al.. A BMT CTN phase II trial of unrelated donor marrow transplantation for children with severe sickle cell disease. Blood. 2016; 128:2361–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ribeil J.A., Hacein-Bey-Abina S., Payen E., Magnani A., Semeraro M., Magrin E., Caccavelli L., Neven B., Bourget P., El Nemer W. et al.. Gene therapy in a patient with sickle cell disease. N. Engl. J. Med. 2017; 376:848–855. [DOI] [PubMed] [Google Scholar]
- 16. Hoban M.D., Cost G.J., Mendel M.C., Romero Z., Kaufman M.L., Joglekar A.V., Ho M., Lumaquin D., Gray D., Lill G.R. et al.. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015; 125:2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Munoz D.P., Boffelli D. et al.. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016; 8:360ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. et al.. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016; 539:384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Charache S., Dover G.J., Moore R.D., Eckert S., Ballas S.K., Koshy M., Milner P.F., Orringer E.P., Phillips G. Jr, Platt O.S. et al.. Hydroxyurea: effects on hemoglobin F production in patients with sickle cell anemia. Blood. 1992; 79:2555–2565. [PubMed] [Google Scholar]
- 20. Uda M., Galanello R., Sanna S., Lettre G., Sankaran V.G., Chen W., Usala G., Busonero F., Maschio A., Albai G. et al.. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Urbinati F., Hargrove P.W., Geiger S., Romero Z., Wherley J., Kaufman M.L., Hollis R.P., Chambers C.B., Persons D.A., Kohn D.B. et al.. Potentially therapeutic levels of anti-sickling globin gene expression following lentivirus-mediated gene transfer in sickle cell disease bone marrow CD34+ cells. Exp. Hematol. 2015; 43:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 25. Cradick T.J., Fine E.J., Antico C.J., Bao G.. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013; 41:9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vakulskas C.A., Dever D.P., Rettig G.R., Turk R., Jacobi A.M., Collingwood M.A., Bode N.M., McNeill M.S., Yan S., Camarena J. et al.. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018; 24:1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ronzoni L., Sonzogni L., Fossati G., Modena D., Trombetta E., Porretti L., Cappellini M.D.. Modulation of gamma globin genes expression by histone deacetylase inhibitors: an in vitro study. Br. J. Haematol. 2014; 165:714–721. [DOI] [PubMed] [Google Scholar]
- 28. Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J.. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010; 649:247–256. [DOI] [PubMed] [Google Scholar]
- 29. Hahn C.K., Lowrey C.H.. Eukaryotic initiation factor 2a phosphorylation mediates fetal hemoglobin induction through a posttranscriptional mechanism. Blood. 2013; 122:477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zurbriggen K., Schmugge M., Schmid M., Durka S., Kleinert P., Kuster T., Heizmann C.W., Troxler H.. Analysis of minor hemoglobins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Chem. 2005; 51:989–996. [DOI] [PubMed] [Google Scholar]
- 31. Deng W., Rupon J.W., Krivega I., Breda L., Motta I., Jahn K.S., Reik A., Gregory P.D., Rivella S., Dean A. et al.. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014; 158:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nemati H., Bahrami G., Rahimi Z.. Rapid separation of human globin chains in normal and thalassemia patients by RP-HPLC. Mol. Biol. Rep. 2011; 38:3213–3218. [DOI] [PubMed] [Google Scholar]
- 33. Loucari C.C., Patsali P., van Dijk T.B., Stephanou C., Papasavva P., Zanti M., Kurita R., Nakamura Y., Christou S., Sitarou M. et al.. Rapid and sensitive assessment of globin chains for gene and cell therapy of hemoglobinopathies. Hum. Gene Ther. Methods. 2018; 29:60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Traxler E.A., Yao Y., Wang Y.D., Woodard K.J., Kurita R., Nakamura Y., Hughes J.R., Hardison R.C., Blobel G.A., Li C. et al.. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 2016; 22:987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Du E., Diez-Silva M., Kato G.J., Dao M., Suresh S.. Kinetics of sickle cell biorheology and implications for painful vasoocclusive crisis. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Vasconcellos J.F., Fasano R.M., Lee Y.T., Kaushal M., Byrnes C., Meier E.R., Anderson M., Rabel A., Braylan R., Stroncek D.F. et al.. LIN28A expression reduces sickling of cultured human erythrocytes. PLoS One. 2014; 9:e106924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G.. COSMID: A Web-based tool for identifying and validating CRISPR/Cas Off-target sites. Mol. Ther. Nucleic Acids. 2014; 3:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. et al.. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015; 33:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C.. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016; 34:695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shayakhmetov D.M., Papayannopoulou T., Stamatoyannopoulos G., Lieber A.. Efficient gene transfer into human CD341 cells by a retargeted adenovirus vector. J. Virol. 2000; 74:2567–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elliott B., Richardson C., Winderbaum J., Nickoloff J.A., Jasin M.. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell Biol. 1998; 18:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kan Y., Ruis B., Takasugi T., Hendrickson E.A.. Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 2017; 27:1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harmsen T., Klaasen S., van de Vrugt H., Te Riele H.. DNA mismatch repair and oligonucleotide end-protection promote base-pair substitution distal from a CRISPR/Cas9-induced DNA break. Nucleic Acids Res. 2018; 46:2945–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M.. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016; 533:125–129. [DOI] [PubMed] [Google Scholar]
- 45. Clarke R., Heler R., MacDougall M.S., Yeo N.C., Chavez A., Regan M., Hanakahi L., Church G.M., Marraffini L.A., Merrill B.J.. Enhanced bacterial immunity and mammalian genome editing via RNA-Polymerase-Mediated dislodging of Cas9 from Double-Strand DNA breaks. Mol. Cell. 2018; 71:42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. et al.. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015; 33:985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez S., Gaunt T.R., Day I.N.. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am. J. Epidemiol. 2009; 169:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ingram V.M. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957; 180:326–328. [DOI] [PubMed] [Google Scholar]
- 49. Hoban M.D., Orkin S.H., Bauer D.E.. Genetic treatment of a molecular disorder: gene therapy approaches to sickle cell disease. Blood. 2016; 127:839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Powars D.R., Weiss J.N., Chan L.S., Schroeder W.A.. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia. Blood. 1984; 63:921–926. [PubMed] [Google Scholar]
- 51. Hu J., Liu J., Xue F., Halverson G., Reid M., Guo A., Chen L., Raza A., Galili N., Jaffray J. et al.. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013; 121:3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G.. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014; 42:7473–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Torres R., Martin M.C., Garcia A., Cigudosa J.C., Ramirez J.C., Rodriguez-Perales S.. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat. Commun. 2014; 5:3964. [DOI] [PubMed] [Google Scholar]
- 55. Dobosy J.R., Rose S.D., Beltz K.R., Rupp S.M., Powers K.M., Behlke M.A., Walder J.A.. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol. 2011; 11:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Richardson C.D., Ray G.J., Bray N.L., Corn J.E.. Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat. Commun. 2016; 7:12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brinkman E.K., Chen T., de Haas M., Holland H.A., Akhtar W., van Steensel B.. Kinetics and fidelity of the repair of Cas9-Induced Double-Strand DNA breaks. Mol. Cell. 2018; 70:801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fisher T.L., Terhorst T., Cao X., Wagner R.W.. Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res. 1993; 21:3857–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lamming C.E., Augustin L., Blackstad M., Lund T.C., Hebbel R.P., Verfaillie C.M.. Spontaneous circulation of myeloid-lymphoid–initiating cells and SCID-repopulating cells in sickle cell crisis. J. Clin. Invest. 2003; 111:811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee C.M., Cradick T.J., Fine E.J., Bao G.. Nuclease target site selection for maximizing On-target activity and minimizing Off-target effects in genome editing. Mol. Ther. 2016; 24:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S.. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015; 12:237–243. [DOI] [PubMed] [Google Scholar]
- 62. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wiekmeijer A.S., Pike-Overzet K., Brugman M.H., Salvatori D.C., Egeler R.M., Bredius R.G., Fibbe W.E., Staal F.J.. Sustained engraftment of cryopreserved human bone marrow CD34(+) cells in young adult NSG mice. Biores Open Access. 2014; 3:110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang K.H., Smith S.E., Sullivan T., Chen K., Zhou Q., West J.A., Liu M., Liu Y., Vieira B.F., Sun C. et al.. Long-Term engraftment and fetal globin induction upon BCL11A gene editing in Bone-Marrow-Derived CD34+ hematopoietic stem and progenitor cells. Mol. Ther. Methods Clin. Dev. 2017; 4:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barbedo M.M., McCurdy P.R.. Red cell life span in sickle cell trait. Acta Haematol. 1974; 51:339–343. [DOI] [PubMed] [Google Scholar]
- 66. Lin S., Staahl B.T., Alla R.K., Doudna J.A.. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014; 3:e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study and the custom MatLab scripts used in this study are available on request from the corresponding author.