Abstract
Objectives
To assess the ability of oxyclozanide to enhance tobramycin killing of Pseudomonas aeruginosa biofilms and elucidate its mechanism of action.
Methods
Twenty-four hour biofilms formed by the P. aeruginosa strain PAO1 and cystic fibrosis (CF) isolates were tested for susceptibility to oxyclozanide and tobramycin killing using BacTiter-Glo™ and cfu. Biofilm dispersal was measured using crystal violet staining. Membrane potential and permeabilization were quantified using DiOC2(3) and TO-PRO-3, respectively.
Results
Here we show that the ionophore anthelmintic oxyclozanide, combined with tobramycin, significantly increased killing of P. aeruginosa biofilms over each treatment alone. This combination also significantly accelerated the killing of cells within biofilms and stationary phase cultures and it was effective against 4/6 CF clinical isolates tested, including a tobramycin-resistant strain. Oxyclozanide enhanced the ability of additional aminoglycosides and tetracycline to kill P. aeruginosa biofilms. Finally, oxyclozanide permeabilized cells within the biofilm, reduced the membrane potential and increased tobramycin accumulation within cells of mature P. aeruginosa biofilms.
Conclusions
Oxyclozanide enhances aminoglycoside and tetracycline activity against P. aeruginosa biofilms by reducing membrane potential, permeabilizing cells and enhancing tobramycin accumulation within biofilms. We propose that oxyclozanide counteracts the adaptive resistance response of P. aeruginosa to aminoglycosides, increasing both their maximum activity and rate of killing. As oxyclozanide is widely used in veterinary medicine for the treatment of parasitic worm infections, this combination could offer a new approach for the treatment of biofilm-based P. aeruginosa infections, repurposing oxyclozanide as an anti-biofilm agent.
Introduction
Cystic fibrosis (CF) is the most common life-shortening genetic disease in Caucasians. It affects 70000 people worldwide and 30000 people in the USA.1 A mutation in the CF transmembrane conductance regulator gene and subsequent loss of a chloride channel and bicarbonate transport throughout the body causes CF. In the lungs, the loss of coordinated chloride and bicarbonate transport results in the airway mucus becoming thick and dry, hindering the clearance of bacteria and debris.2,3 This immunological defect makes CF patients prone to recurrent lung infections, including those caused by several members of the MDR ‘ESKAPE’ pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species).4 By their mid-to-late teens, the dominant bacterial pathogen colonizing the lungs of CF patients is P. aeruginosa.1 Central to this pathogen’s success is its ability to form a biofilm, which is a community of cells embedded in a thick matrix that provides resistance to antibacterial therapies, macrophages and neutrophils.5,6 Treatment is further hindered by numerous intrinsic antimicrobial resistance mechanisms found in P. aeruginosa, including decreased outer membrane permeability, resistance nodulation division (RND) efflux family proteins and chromosomally encoded β-lactamases.7,8
Nebulized tobramycin is currently the cornerstone therapy for Pseudomonas eradication protocols in CF patients; however, it rarely eradicates this pathogen.9 Three hundred milligrams of tobramycin is inhaled twice a day for 28 days in on–off cycles, reaching mean sputum concentrations of ∼737 μg/g (∼1576 μmol/g) per dose.10,11 It has also been found in paediatric CF patients that the mean concentration of bioactive tobramycin within the epithelial lining fluid is 80 μg/mL per dose ranging from 11 to 265 μg/mL following inhalation.12 Despite the repeated use of high concentrations of tobramycin, it is estimated that more than half of all CF patients are chronically colonized with P. aeruginosa by adulthood.9 Retrospective studies have shown that the prevention and possible eradication of transient infections by P. aeruginosa before a chronic infectious state is underway can extend the lives of CF patients.13,14 Thus, new therapies that more effectively target this pathogen, especially as it transitions from an acute to a chronic infection, would be of significant clinical benefit for CF patients.
To this end, we previously performed a high throughput screen (HTS) of 6080 compounds from four drug-repurposing libraries (Prestwick, MS2400, LOPAC and focus collection libraries) at the University of Michigan Center for Chemical Genomics to identify novel compounds that enhanced tobramycin killing of mature P. aeruginosa biofilms and demonstrated that triclosan exhibits this activity.15 Here, we report the characterization of another compound identified in the HTS, oxyclozanide. This molecule is a proton ionophore that disrupts proton motive force (PMF) and is approved for the treatment of parasitic worm infections in cattle.16 We found that oxyclozanide had weak antibacterial activity on its own, while oxyclozanide combined with tobramycin significantly enhanced the rate and degree of aminoglycoside killing of P. aeruginosa biofilms. The combination was effective against CF clinical isolates, including a tobramycin-resistant isolate and multiple S. aureus isolates growing as biofilms. We further show that oxyclozanide both permeabilized P. aeruginosa and functioned as a proton ionophore, reducing the membrane potential (Δψ) of P. aeruginosa.
Finally, we show that oxyclozanide resulted in increased tobramycin accumulation within cells in biofilms. Our findings suggest that tobramycin combined with oxyclozanide represents a potential new antimicrobial therapy for the treatment of P. aeruginosa and S. aureus biofilms in CF patients, as well as other biofilm-based infections such as diabetic foot ulcers and burn wounds.17–19
Materials and methods
Bacterial strains, culture conditions and compounds
All strains used in this study are listed in Table 1. Bacterial strains were grown in glass test tubes (18 × 150 mm) at 35°C in CAMHB (Mueller–Hinton Broth II, Sigma–Aldrich) with agitation at 210 rpm. Antibiotics and oxyclozanide were obtained from Sigma–Aldrich. Tobramycin sulphate, gentamicin sulphate and streptomycin sulphate were dissolved in autoclaved deionized water and filter sterilized using 0.22 μm filter membranes (Thomas Scientific). Oxyclozanide and tetracycline were dissolved in 95% and 75% ethanol, respectively.
Table 1.
Bacterial strains used in this study
| Strain | Characteristics | Reference |
|---|---|---|
| PAO1 | P. aeruginosa standard reference strain, isolated in 195445 | 15 |
| AMT0023_30 | P. aeruginosa early isolate, patient 6 months old, tobramycin susceptible | 46 |
| AMT0023_34 | P. aeruginosa late isolate, patient 8 years old, tobramycin resistant | 46 |
| CF_110_N | P. aeruginosa clinical CF isolate, MI, USA | 15 |
| CF_110_O | P. aeruginosa clinical CF isolate, MI, USA | 15 |
| CF_115_J | P. aeruginosa clinical CF isolate, MI, USA | 15 |
| CF_131_M | P. aeruginosa clinical CF isolate, MI, USA | 15 |
| USA_300_JE2 | MRSA, wound, CA, USA | 47 |
| COL | MRSA, Colindale, England | 48 |
| Newman (25904) | MSSA, endocarditis, ATCC | 49 |
| Wichita (29213) | MSSA, better biofilm former, ATCC | 50 |
MICs
MICs were determined as described previously.20 Briefly, microdilutions were made in a 96-well plate in 10% (v/v) CAMHB diluted in Dulbecco’s PBS with magnesium and calcium (DPBS, Sigma–Aldrich). Approximately 1 × 106 cfu/mL were added and incubated for 24 h at 35°C in a humidified chamber with agitation at 150 rpm. MICs were defined as the minimum concentration in which no turbidity greater than background was measured (absorbance at 595 nm), using a SpectraMax M5 microplate spectrophotometer system (Molecular Devices).
Biofilm susceptibility testing using BacTiter-Glo™
We used the Minimum Biofilm Eliminating Concentration (MBEC™, Innovotech) assay to measure antimicrobial susceptibility of biofilms.21 This assay consists of a lid with 96 polystyrene pegs used to grow biofilms. An overnight culture was washed and diluted to an OD600 of 0.001 and seeded into an MBEC™ plate. The plate was then incubated for 24 h at 35°C in a humidified chamber with agitation at 150 rpm. After 24 h, the lid was then washed for 5 min to remove non-adherent cells, transferred to the 96-well treatment plate and incubated for the indicated time at 35°C without agitation. After the incubation, the MBEC™ lid was washed and transferred to a black 96-well ViewPlate (PerkinElmer) filled with 40% (v/v) BacTiter-GloTM (Promega) diluted in DPBS. The BacTiter-GloTM microbial cell viability assay is a luminescence assay that determines the number of viable cells present based on quantification of ATP concentration. Cell viability was enumerated using luminescence measured by an EnVison Multilabel Plate Reader (PerkinElmer). Percent killing was calculated as:
A calibration curve was previously performed and it was found, using linear regression, that the coefficient of determination was r2 = 0.9884 for luminescence versus cfu/mL.15 Dose–response curves, chequerboard assays and time–kill curves were performed similarly. S. aureus biofilms were measured in standard 96-well plates because they do not form biofilms at the air–liquid interface.
Crystal violet staining
To study biofilm dispersal under static conditions, crystal violet staining was performed as previously described.22 Biofilms formed, as described above, and were stained with crystal violet after a 6 h incubation with treatments.
BacLight™ membrane potential assay and live/dead TO-PRO-3 staining
Twenty-four hour biofilms were formed in glass test tubes (18 × 150 mm) in 1 mL of 10% (v/v) CAMHB at 35°C and agitated at 150 rpm. Cells were then washed in DPBS to remove non-adherent cells and treated with oxyclozanide and tobramycin for 2 h or 6 h. Following treatment, cells were washed in PBS without magnesium and calcium and the biofilm was disrupted from the air–liquid interface using an autoclaved wooden stick. The cells were stained in 1 mL of PBS for 20 min using the BacLight™ bacterial membrane potential kit in combination with flow cytometry (Thermo Fisher Scientific). This assay uses the dye DiOC2(3), which fluoresces in the FITC channel within all cells. However, greater Δψ drives accumulation and self-association of the dye in the cell cytoplasm, shifting its fluorescence to the phycoerythrin (PE) channel. To this assay we added the TO-PRO-3 iodide live/dead stain that cannot penetrate live cells but can accumulate within cells that have the permeabilized membranes characteristic of dead cells (Thermo Fisher Scientific). Once inside cells with compromised membranes, this dye fluoresces in the allophycocyanin (APC) channel upon intercalating with DNA. Single cell flow cytometry was performed on an LSR II (BD Biosciences), with excitation from 488 mm and 640 mm lasers, and analysed in FITC/PE and APC channels, respectively.
Stationary phase killing assays
Cultures were grown for 20 h and 100 μL/well of stationary phase cells were added to individual wells of a 96-well plate.23 Treatments were added and the plate was incubated at 35°C without agitation. At hours 2, 4, 6, 8 and 24, aliquots were serially diluted, plated on Dey–Engley neutralizing agar plates, which neutralize the activity of disinfectants and antiseptics (Sigma–Aldrich), and cfu were enumerated. The dilutions that contained 3–30 colonies per 10 μL were used to quantify cfu/mL.
Tobramycin accumulation assay
To measure the accumulation of tobramycin within cells in biofilms, tobramycin was conjugated to Texas Red® (Sigma–Aldrich) using an amine conjugation reaction, as previously described.24,25 Briefly, conjugated tobramycin was used at a concentration of 250 μg/mL (∼500 μM) alone and in combination with 100 μM oxyclosanide against 24 h biofilms formed in glass test tubes at the air–liquid interface, as described above. Following treatments, biofilms were washed in DPBS for 3 min and then disrupted with autoclaved wooden sticks into 1 mL of 0.2% Triton X-100 to lyse cells (Sigma–Aldrich). Lysed cells were then transferred to spectrophotometer cuvettes (Thermo Fisher Scientific) and read using a SpectraMax M5 microplate spectrophotometer system (λexcite, 595 nm and λemit, 615 nm).
Results
Oxyclozanide potentiates tobramycin activity against P. aeruginosa and S. aureus biofilms
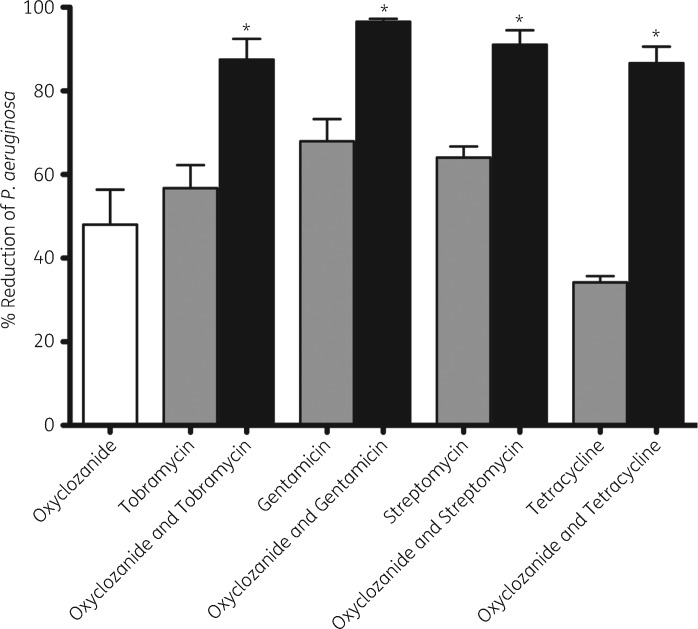
We previously performed an HTS to identify compounds that enhance tobramycin killing of P. aeruginosa biofilms15 and we report here that the anthelmintic oxyclozanide was identified as a significant hit from that screen. To confirm the results of the screen, P. aeruginosa PAO1 biofilms were exposed to 100 μM oxyclozanide or 500 μM tobramycin (∼500-fold the planktonic MIC), alone and in combination, for 6 h and the efficacy was determined using BacTiter-GloTM.15 Oxyclozanide and tobramycin alone resulted in ∼2-fold fewer viable cells within biofilms compared with untreated controls. However, the combination of oxyclozanide and tobramycin was more effective, eradicating 87% (7.7-fold reduction) of the cells within biofilms (Figure 1). Oxyclozanide combined with the aminoglycosides gentamicin or streptomycin killed 96% (25-fold reduction) and 91% (11.1-fold reduction) of the cells within biofilms, respectively.
Figure 1.
Oxyclozanide enhances aminoglycosides and tetracycline. Biofilms (24 h old) were treated for 6 h with oxyclozanide (100 μM), tobramycin (500 μM), gentamicin (100 μM), streptomycin (100 μM) or tetracycline (100 μM), alone and in combination, and the number of viable cells within the biofilms was quantified by BacTiter-GloTM. The assay was performed at least three times in triplicate. The results represent means plus the SEM. A one-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to determine statistical significance between the combination and each antibiotic alone (*P < 0.05).
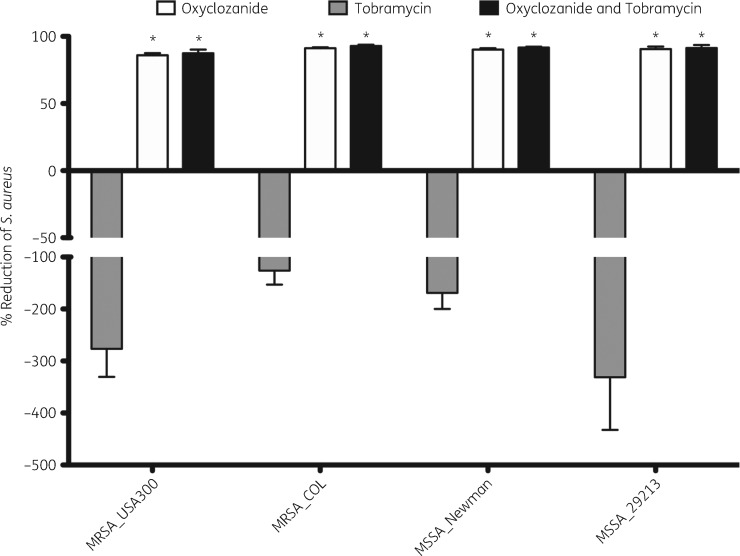
We also tested whether oxyclozanide enhanced third- and fourth-generation cephalosporins, β-lactams, fluoroquinolones or tetracycline and determined that in addition to aminoglycosides, oxyclozanide only enhanced tetracycline, increasing killing of the biofilm to 86% (7.1-fold reduction, Figure 1). We also tested the combination against four strains of S. aureus and found oxyclozanide was effective alone, killing ∼90% of each strain grown as a biofilm (Figure 2). Interestingly, S. aureus biofilms were resistant to tobramycin alone and the biofilm increased in cellular number and/or ATP after 6 h of treatment. Supporting our observation, it has been observed clinically that high doses of inhaled tobramycin have no effect on MRSA.26 This resistance was lost when tobramycin was used in combination with oxyclozanide, resulting in 90% killing, on average, for each strain of S. aureus tested and a maximum ∼36-fold reduction (MSSA_29213) compared with tobramycin treatment alone.
Figure 2.
Oxyclozanide kills S. aureus biofilms. S. aureus biofilms (24 h old) were treated with oxyclozanide (100 μM) or tobramycin (100 μM), alone and in combination, for 6 h. The number of viable cells within the biofilms was quantified by BacTiter-GloTM. The assay was performed twice in triplicate. The results represent means plus the SEM. A two-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to determine statistical significance between either oxyclozanide alone and tobramycin alone or the combination and tobramycin alone (*P < 0.05).
We also performed MIC assays to determine whether oxyclozanide and these antibiotics were more effective against planktonic cells. The MIC of tobramycin, gentamicin, streptomycin or tetracycline did not change when used in combination with oxyclozanide against PAO1 planktonic cells, suggesting oxyclozanide specifically enhanced the activity of these antibiotics against cells growing as a biofilm (Table S1 available as Supplementary data at JAC Online).
Dose–response curves of oxyclozanide and aminoglycosides
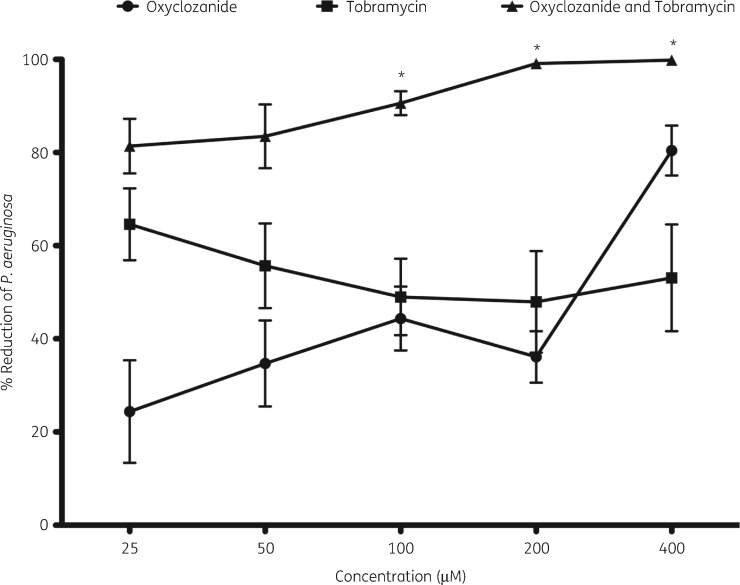
To determine the effective ranges of the combinations, we performed dose–response curves. Tobramycin treatment showed modest activity at concentrations between 25 and 400 μM, reducing the number of the cells within biofilms by between 50% and 60% after 6 h of treatment (Figure 3). Similarly, oxyclozanide treatment exhibited modest activity between 25 and 200 μM, killing ∼20%–40% of the cells within biofilms but, at 400 μM, oxyclozanide alone killed ∼80% of the cells within biofilms. The combination of oxyclozanide and tobramycin significantly increased killing of biofilms compared with tobramycin treatment alone between 100 and 400 μM for each compound, with maximum efficacy of 99% killing (100-fold reduction) seen at 200 μM and 400 μM oxyclozanide and tobramycin. Dose–response curves were also performed with gentamicin, streptomycin or tetracycline, alone or in combination with oxyclozanide ranging from 25 to 400 μM (Figure S1). The results were similar with significant killing observed when biofilms were treated with combinations at 100, 200 or 400 μM versus tobramycin alone.
Figure 3.
Dose–response curve of oxyclozanide or tobramycin alone and in combination. P. aeruginosa biofilms (24 h old) were treated for 6 h with oxyclozanide or tobramycin alone and in combination, in equal molar two-fold dilutions, and the number of viable cells within the biofilms was quantified by BacTiter-GloTM. The assay was performed at least three times in triplicate. The results represent means ± SEM. A one-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to determine statistical significance between the combination and tobramycin alone (*P < 0.05).
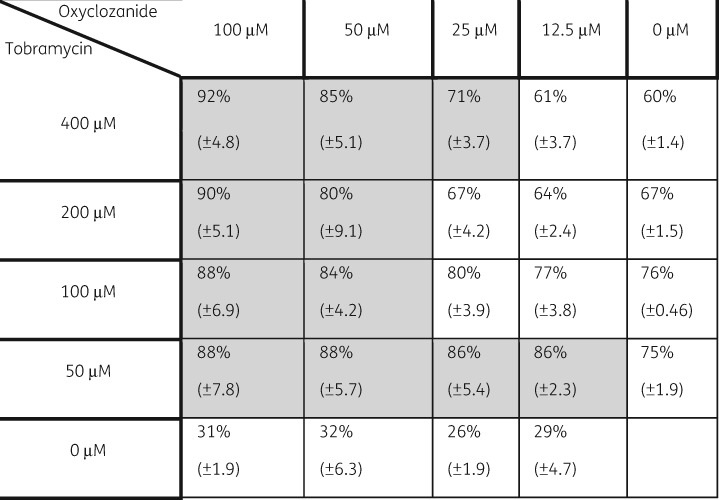
To determine the combinations of lowest possible concentrations of oxyclozanide and tobramycin that resulted in significant killing, we performed chequerboard assays (Figure 4). Biofilms were treated with dilutions of oxyclozanide, ranging from 12.5 to 100 μM, and tobramycin, ranging from 50 to 400 μM. An 86% reduction in cells within the biofilms resulted from 12.5 μM oxyclozanide and 50 μM tobramycin. The greatest reduction was seen at 100 μM oxyclozanide and 400 μM tobramycin, reducing the cells within the biofilms by 92%.
Figure 4.
A chequerboard dilution series of oxyclozanide and tobramycin. Twenty-four hour biofilms were treated with varying combinations of oxyclozanide and tobramycin for 6 h. The number of viable cells within the biofilms was quantified by BacTiter-GloTM. The assay was performed twice in triplicate. The results represent means ± SD. A two-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to determine statistical significance between the combination and tobramycin alone. Shaded cells indicate significance compared with tobramycin alone (P < 0.05).
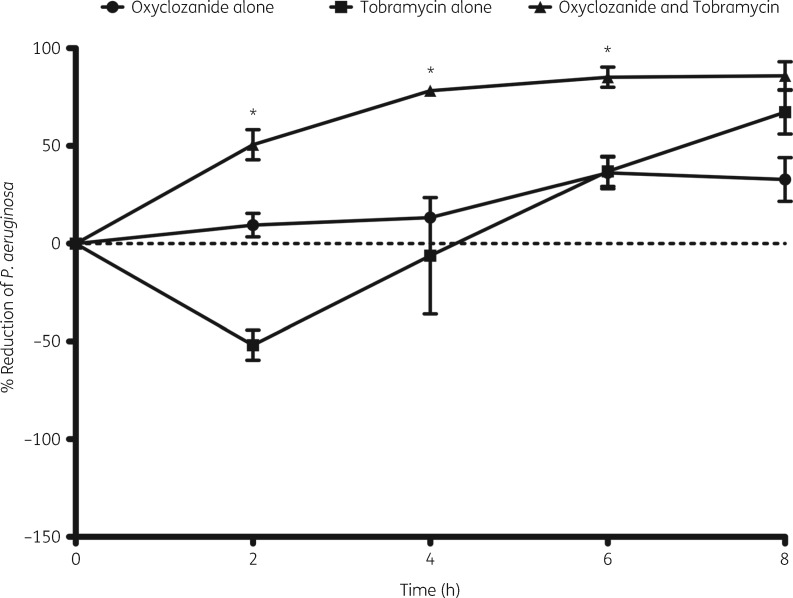
Oxyclozanide accelerates tobramycin killing of cells within biofilms
Another important property of these antimicrobials, which is highly clinically relevant, is the rate at which biofilms are killed. To determine the rate of killing of cells within biofilms by tobramycin, oxyclozanide or the combination, we measured the percentage reduction over the course of 8 h. Oxyclozanide alone did not exhibit significant killing until 6 h (Figure 5). Rather than cell death, tobramycin treatment alone led to an increase in cell number and/or ATP at 2 h, which was lost by ∼4 h. We hypothesize that this increase is due to the mechanism of adaptive resistance that occurs in P. aeruginosa in response to aminoglycosides or protein synthesis inhibitors/corruptors. Adaptive resistance, which has been demonstrated in P. aeruginosa growing planktonically, in biofilms and in human lungs, is a transient phenotype that occurs within the first 1–2 h of exposure of P. aeruginosa to translation inhibitors.27–29 This phenomenon results in reduced intracellular accumulation of aminoglycosides due to the activation of RND-type efflux pumps such as the MexXY-OprM tripartite efflux pump, among other mechanisms.28,29 Importantly, this temporary resistance was abolished in the combination treatment as oxyclozanide significantly shortened the onset of action of tobramycin from 6 h to 2 h, resulting in 50%, 78% and 85% killing of the cells within the biofilm at 2, 4 and 6 h, respectively (3-, 4- and 6-fold reductions). Thus, at 2 h the combination of oxyclozanide and tobramycin was 100-fold more effective at killing biofilms than tobramycin treatment alone, indicating this combination exhibits both enhanced activity and accelerated action.
Figure 5.
Oxyclozanide accelerates tobramycin killing of P. aeruginosa biofilms. Twenty-four hour biofilms were treated with oxyclozanide (100 μM) or tobramycin (500 μM), alone and in combination. At 0, 2, 4, 6, and 8 h, the number of viable cells within the biofilms was determined by BacTiter-GloTM. The assay was performed at least three times in triplicate. The results represent means ± SEM. A one-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to determine statistical significance between the combination and tobramycin alone (*P < 0.05).
Oxyclozanide significantly accelerated the onset of action of gentamicin or tetracycline. However, unlike the other aminoglycosides examined, streptomycin did not exhibit a delay in biofilm killing and minimal enhancement when combined with oxyclozanide was only observed at 6 h (Figure S2B).
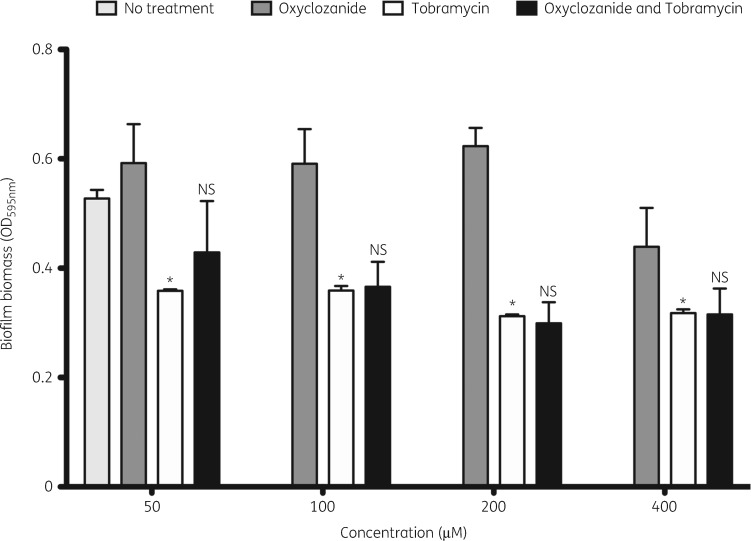
Oxyclozanide combined with tobramycin does not increase biofilm dispersal
One explanation for our results thus far is that oxyclozanide induces dispersal of P. aeruginosa biofilms. We tested the ability of oxyclozanide to disperse P. aeruginosa biofilms using a crystal violet staining assay. Biofilms were treated with oxyclozanide or tobramycin alone and in combination from 50 to 400 μM (Figure 6). Tobramycin significantly reduced biofilm biomass at all concentrations examined. Conversely, oxyclozanide did not cause biofilm dispersal and the combination of tobramycin and oxyclozanide did not cause an increase in biofilm dispersal compared with tobramycin alone. This experiment suggests that oxyclozanide is not acting by inducing biofilm dispersal.
Figure 6.
Oxyclozanide does not induce biofilm dispersal. Twenty-four hour biofilms were treated with oxyclozanide, tobramycin or the combination, in equal molar two-fold dilutions, for 6 h. The effect on biofilm biomass was quantified by staining with crystal violet. The experiment was performed twice in triplicate. The results represent the means plus SEM. A one-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to determine statistical significance between tobramycin alone and the untreated control (*P < 0.05) and between the combination and tobramycin alone. NS, not significant.
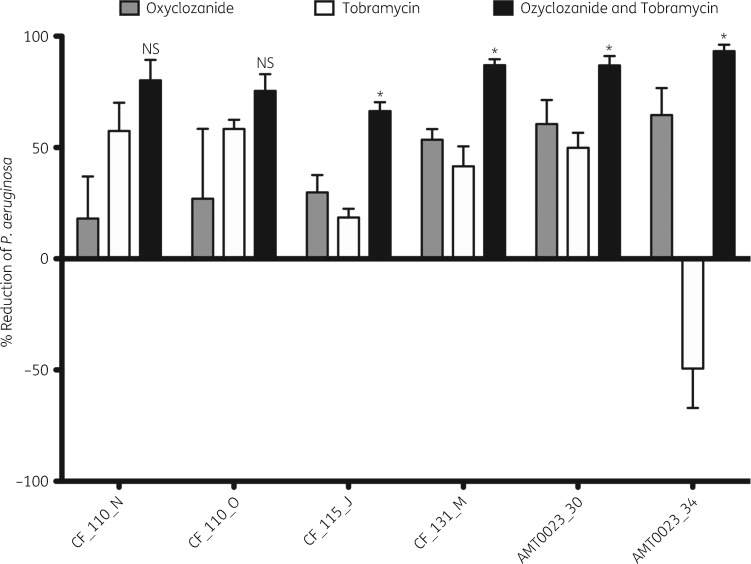
Oxyclozanide and tobramycin are effective against CF isolates of P. aeruginosa
To determine whether the combination of oxyclozanide and tobramycin exhibits widespread activity, we tested its activity against six P. aeruginosa CF clinical isolates. Two of the clinical isolates were isolated longitudinally from the same patient at 6 months of age and 8 years of age: AMT0023_30 and AMT0023_34, respectively. In addition, clinical isolates CF_110_N and CF_110_O were isolated longitudinally from the same patient 3 months apart (the strains are described in Table 1). Importantly, these clinical isolates exhibited distinct colony morphologies on blood agar plates. These six clinical isolates were treated with 100 μM oxyclozanide or 500 μM tobramycin, alone or in combination. All isolates displayed no to modest susceptibility to oxyclozanide or tobramycin treatment alone and exhibited the greatest susceptibility to the combination (Figure 7). This increase was statistically significant in 4/6 isolates. Importantly, the combination significantly enhanced killing of strain AMT0023_34, which overexpresses the RND-type MexXY-OpRM tripartite multidrug efflux pump, rendering it resistant to tobramycin,30 resulting in an ∼16-fold reduction in viable AMT0023_34 cells compared with tobramycin treatment alone (Figure 7).
Figure 7.
Oxyclozanide is broadly active against P. aeruginosa. Twenty-four hour biofilms were treated with oxyclozanide (100 μM) or tobramycin (500 μM), alone and in combination for 6 h. The number of viable cells within the biofilms was quantified by BacTiter-GloTM. The assay was performed three times in triplicate. The results represent means plus the SEM. A one-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to determine statistical significance between the combination and tobramycin alone (*P < 0.05; NS, not significant).
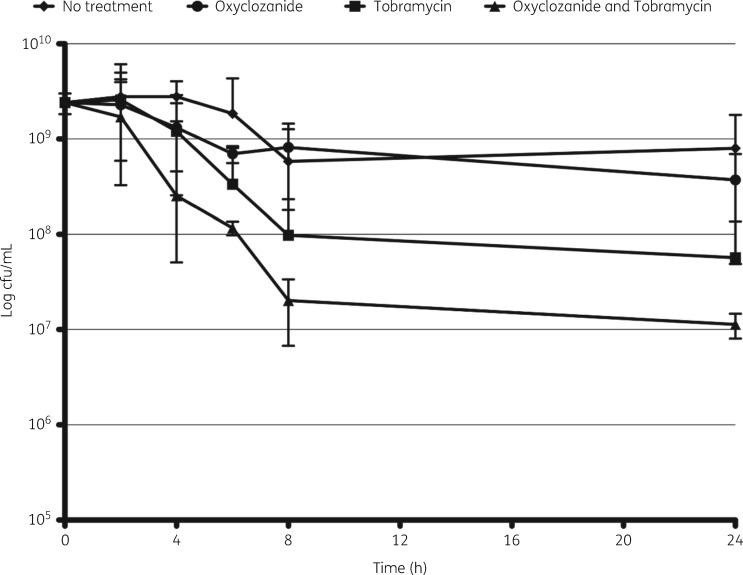
Oxyclozanide and tobramycin are more effective against stationary phase cells
We hypothesized that the combination therapy may also be effective against stationary phase cells that mimic cells growing in biofilms as they are in a lower metabolic state and enriched for persister cells. Persister cells are metabolically dormant and thus highly tolerant to antimicrobials that target actively growing cells.31,32 To test this prediction, we analysed the activity of oxyclozanide (100 μM), tobramycin (50 μM) or the combination on cultures of P. aeruginosa grown for 20 h. Similar to our results with biofilms, we found that the combination of oxyclozanide and tobramycin significantly reduced viable cells as measured by quantifying cfu, exhibiting a 10-fold reduction in viable cells compared with tobramycin alone at 8 and 24 h (Figure 8).
Figure 8.
Tobramycin combined with oxyclozanide is more effective against stationary phase cells. Twenty-hour-old stationary phase cells were treated with oxyclozanide (100 μM) or tobramycin (50 μM), alone and in combination. At 0, 2, 4, 6, 8 and 24 h post-treatment, aliquots were enumerated for cfu/mL. The experiment was performed two times in triplicate. The results represent means ± SD.
Oxyclozanide causes both cellular permeabilization and the loss of a Δψ in P. aeruginosa biofilms
The mode of action of oxyclozanide in parasitic flatworms is the uncoupling of oxidative phosphorylation by the translocation of protons through the inner mitochondrial membrane, disrupting the PMF and eliminating the Δψ.16 Additionally, oxyclozanide has been shown to induce cellular permeabilization in the Gram-positive pathogen S. aureus.33 However, neither ionophore activity nor membrane disruption by oxyclozanide has been demonstrated in Gram-negative bacteria or bacteria growing in a biofilm.
To test whether oxyclozanide exhibits ionophore activity against P. aeruginosa, biofilms exposed to tobramycin, oxyclozanide or the combination were assayed with BacLight™, which consists of the dye DiOC2(3). This dye is a marker for cellular Δψ and cells were analysed by flow cytometry (representative plots are shown in Figure S3). Because oxyclozanide has been shown to cause cell permeabilization, we also added to this assay TO-PRO-3 iodide, which stains the DNA of bacteria with disrupted outer membranes, characteristic of dead cells. The combination of these two dyes allowed us to determine both cellular permeabilization and Δψ in the same assay. For those cells that have an intact outer membrane and do not stain with TO-PRO-3, import of DiOC2(3) into the cell cytoplasm, where self-association of this dye shifts its fluorescence spectrum to the PE channel, indicates an intact Δψ (Figure S3).
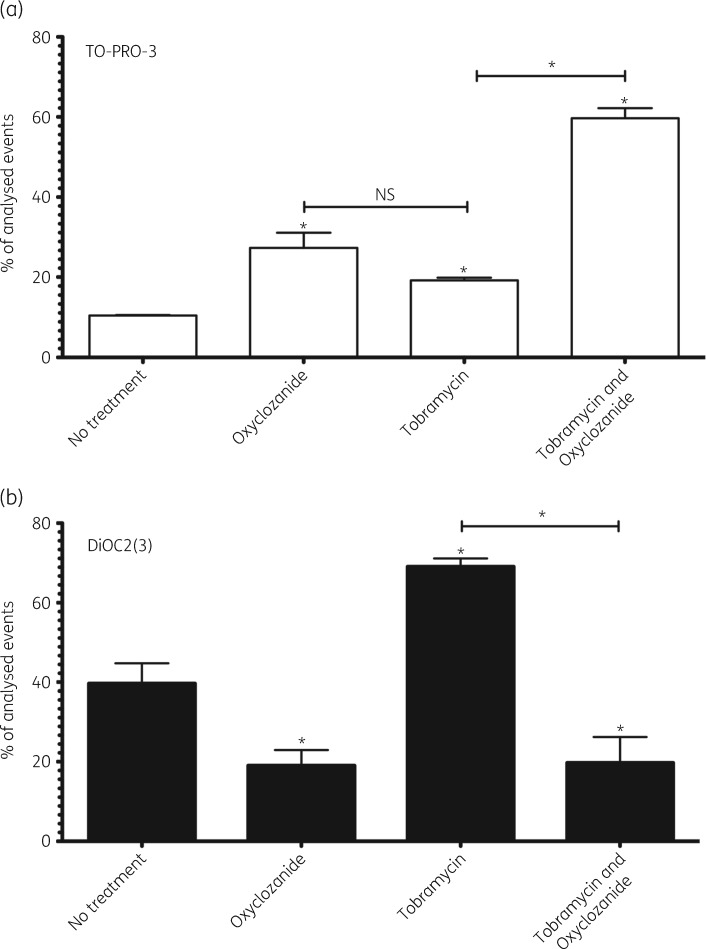
TO-PRO-3 staining indicated that oxyclozanide caused significant cellular permeabilization and death, increasing the population of permeabilized cells from 10% in untreated biofilms to 27% when treated for 2 h (Figure 9a). Tobramycin treatment alone similarly increased permeabilized cells compared with no treatment, but this increase was not statistically significant compared with oxyclozanide alone (Figure 9a). This is not unexpected, given both oxyclozanide and tobramycin treatment alone caused an ∼50% reduction of viable cells within biofilms (Figure 1). However, oxyclozanide combined with tobramycin significantly increased the population of permeabilized and dead cells compared with tobramycin treatment alone: 59% versus 19%, respectively. Similar results were obtained after 6 h of treatment. TO-PRO-3 staining indicated that oxyclozanide increased the population of permeabilized and dead cells from 2.6% in untreated biofilms to 13% when treated for 6 h (Figure S4A). Tobramycin did not significantly cause permeabilization and death. However, the combination of oxyclozanide and tobramycin significantly increased the population of permeabilized and dead cells to 38% compared with 8% of permeabilized cells with only tobramycin treatment.
Figure 9.
Oxyclozanide induces permeabilization and depolarizes the Δψ of P. aeruginosa. Twenty-four hour biofilms were treated with oxyclozanide (100 μM) or tobramycin (500 μM), alone and in combination, for 2 h. (a) Cells were stained with TO-PRO-3 and analysed by flow cytometry to determine the number of cells that were permeabilized. (b) Cells were also stained with DiOC2(3) and analysed by flow cytometry to determine the number of cells maintaining a Δψ. Permeabilized cells were excluded from Δψ analysis. The experiment was performed two separate times in duplicate. The results are percentage averages plus the SEM. Percentage values indicate the average relative abundance of events within each gate normalized to the total number of events analysed, excluding artefacts, aggregates and debris. A one-way ANOVA followed by Dunnett’s multiple comparison post hoc test was used to determine statistical significance between each treatment and the untreated control (*P < 0.05) and oxyclozanide treatment alone was compared with tobramycin treatment alone in panel (a) (NS, not significant). A one-way ANOVA followed by Bonferroni’s multiple comparison post hoc test was used to compare tobramycin to the combination in panel (a) and (b) (*P < 0.05).
Subsequent analysis of the cells that were living (i.e. TO-PRO-3 negative) by DiOC2(3) staining demonstrated that oxyclozanide showed significant ionophore activity against cells within biofilms after 2 h of treatment (Figure 9b). The population of cells maintaining a Δψ was reduced from 39% in untreated biofilms to 19% in oxyclozanide-treated biofilms. Alternatively, tobramycin treatment alone significantly increased the population of cells maintaining a Δψ to 69%. This dramatic increase in the population of cells maintaining a Δψ is, again, indicative of adaptive resistance.28,29 Oxyclozanide combined with tobramycin abolished this effect, reducing the population of cells maintaining a Δψ to 19%. Similar results were obtained after 6 h of treatment with the exception that tobramycin did not result in a significant increase in the population of cells maintaining a Δψ (Figure S4B). This outcome is expected if the increased Δψ is due to adaptive resistance as this is known to be temporal in nature and a short-lived response.34 Strikingly, by 6 h virtually no cells exhibited a Δψ when treated with the combination of oxyclozanide and tobramycin.
Dose–response curves were performed with oxyclozanide to further characterize its effects on the Δψ and its ability to permeabilize and kill cells within biofilms. Untreated controls were re-plotted for comparison. After 2 h of treatment, oxyclozanide caused permeabilization and cell death at 200 μM (Figure S5), resulting in very few cells with a Δψ. However, this effect was lost from 100 to 25 μM while a significant reduction of the Δψ, compared with no treatment, could be observed. In this experiment, significant permeabilization compared with the untreated control was not observed at 100 μM, which contrasts with our previous results observing permeabilization at this concentration (Figure 9 and Figure S4). However, permeabilization was clear at 200 μM. We interpret these results to indicate that oxyclozanide can permeabilize P. aeruginosa biofilms between 100 and 200 μM and at the lower end of these concentrations there can be variability within different experiments.
Oxyclozanide increases cell-associated tobramycin in mature P. aeruginosa biofilms
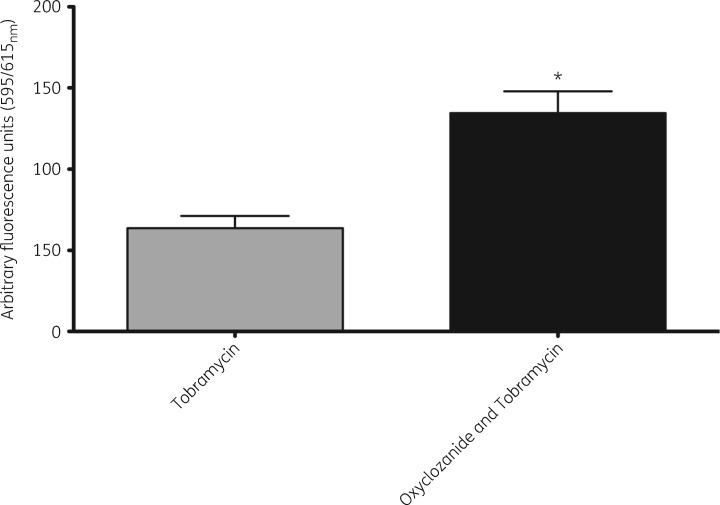
Since oxyclozanide causes cellular permeabilization, reduces Δψ and accelerates the activity of tobramycin, we sought to determine whether oxyclozanide increased the accumulation of cell-associated tobramycin. To do this, we fluorescently labelled tobramycin with Texas Red® and measured its accumulation within cells of a mature biofilm following 30 min of treatment. We found oxyclosanide in combination with tobramycin resulted in significantly more tobramycin accumulation within cells (Figure 10).
Figure 10.
Oxyclozanide results in increased cellular accumulation of tobramycin. Twenty-four hour biofilms grown in glass test tubes were treated for 30 min with oxyclozanide (100 μM) and Texas Red®-labelled tobramycin (250 μg/mL). After washing off the planktonic cells, the biofilms were lysed with 0.2% Triton-X 100® and relative fluorescence units with excitation at 595 nm and emission at 615 nm were measured to quantify labelled tobramycin. The assay was performed twice in duplicate. The results represent means plus the SEM. An unpaired t-test was performed comparing tobramycin versus oxyclosanide and tobramycin (*P < 0.05).
Discussion
Drug repurposing affords many benefits for developing novel antimicrobials, including reduced costs and accelerated deployment.35 We recently demonstrated that triclosan combined with tobramycin can be repurposed to eradicate biofilms formed by both Gram-negative and Gram-positive bacteria.14 Here, we show that the anthelmintic drug oxyclozanide enhances the activity of aminoglycosides and tetracycline to eradicate antibiotic-tolerant biofilms. The combination therapy exhibited more rapid killing and was effective against a tobramycin-resistant P. aeruginosa clinical isolate. Importantly, we found that the combination had activity against cells within biofilms formed by both Gram-negative and Gram-positive pathogens. Young CF patients are often colonized with Staphylococcus organisms, making tobramycin combined with oxyclozanide an applicable therapy for both early and late CF lung pathogens.1
Oxyclozanide is given orally at a dose of 10–15 mg/kg (∼25–37 μmol/kg).16 We envisage the use of oxyclozanide in conjunction with tobramycin at a concentration between 50 and 100 μM as an inhaled aerosolized solution for the treatment of CF. This route of administration provides many benefits including reduced side effects and enhanced activity, which could deliver higher concentrations locally while limiting systemic exposure.36,37 We also imagine the use of tobramycin combined with oxyclozanide as a topical therapy for the treatment of diabetic and burn wounds at concentrations similar to those currently used clinically in animals.
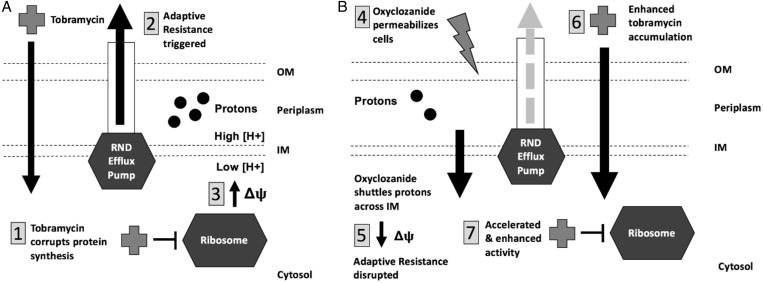
Oxyclozanide has been reported to have two modes of action. It has been shown to function as an ionophore in parasitic worms16 and it was found to permeabilize the Gram-positive bacterium S. aureus growing planktonically.29 Our results indicate that oxyclozanide exhibits both activities against P. aeruginosa biofilms, which could contribute to the observed increase of cellular-associated tobramycin in the presence of oxyclozanide, and we suggest that each activity is important for oxyclozanide and tobramycin enhanced activity (Figure 11).
Figure 11.
Oxyclozanide sensitizes P. aeruginosa to tobramycin by depolarizing the Δψ and further permeabilizing cells. (a) (1) Tobramycin corrupts protein synthesis triggering adaptive resistance, which includes the (2) induction of RND-type efflux pumps such as MexXY-OprM, resulting in reduced accumulation of tobramycin within the cytosol.42,43 (3) During this time, an increase in Δψ is observed driving RND-type efflux pump activity and providing temporary resistance to tobramycin. (b) (4) Oxyclozanide permeabilizes cells and (5) shuttles protons across the IM, depolarizing the Δψ. We hypothesize that both of these activities interfere with adaptive resistance (6), allowing for greater accumulation of tobramycin within cells resulting in (7) accelerated activity and increased effectiveness. OM, outer membrane; IM, inner membrane.
The PMF is one component of the Δψ that has been shown to contribute to efflux of antibiotics and adaptive resistance in bacteria. A gradient of protons across the inner membrane drives the production of ATP as they are imported via the phosphorylation of ADP to ATP by ATP synthase.38 Importantly, the PMF also functions as the energy source for efflux pumps to export antimicrobials, including those in the RND superfamily. The RND efflux superfamily is responsible for MDR and adaptive resistance in P. aeruginosa.28,34,39 Our results are consistent with previous results as we observed that P. aeruginosa growing in a biofilm increases the Δψ in response to tobramycin (Figure 9b), which may be responsible for the increased ATP that we observed during the first few hours of tobramycin treatment (Figure 5). Whether uncoupling of the Δψ by oxyclozanide inhibits efflux pump activity remains to be examined. However, one of the most intriguing results presented here is that the combination of oxyclozanide and tobramycin overcomes tobramycin resistance in the well-characterized AMT0023_34 clinical isolate (Figure 7). Resistance of this isolate is known to be driven by overproduction of efflux pumps.30 This result is suggestive that the combination negatively influences efflux pump activity in P. aeruginosa and, further, it can have clinical efficacy against certain classes of tobramycin-resistant isolates of P. aeruginosa.
Based on our data, we propose a model where oxyclozanide enhances tobramycin accumulation in P. aeruginosa by abolishing adaptive resistance through permeabilizing the cells and disrupting the Δψ (Figure 11). This causes increased tobramycin accumulation, yielding accelerated activity and increased effectiveness. In support of this model, oxyclozanide inhibited the large increase in the population of cells maintaining a Δψ that was induced in response to tobramycin (Figure 9b) and significantly increased the rate of killing by tobramycin (Figure 5). Uncoupling the contributions of permeabilization and inhibition of Δψ in the enhancement of tobramycin by oxyclozanide requires further investigation.
Although an intact PMF is considered one of the main mechanisms of aminoglycoside influx into cells, it is well known that, independently of respiration, aminoglycosides can accumulate within cells through a process termed self-promoted uptake.40 In this process, aminoglycosides interact with the membrane surface by displacing cations, creating ‘cracks’ or ‘fissures’ in the outer membrane of cells. This leads to the diffusion of aminoglycosides into the cytosol of cells, which in turn contributes to additional membrane damage by the insertion of misfolded proteins in the outer membrane.40,41 Thus, at the concentrations we are examining, it is likely that the increased killing in the absence of a Δψ can largely be attributed to the effects of self-promoted uptake combined with the permeabilizing effects of oxyclozanide.
We found that oxyclozanide only enhanced antimicrobials that targeted the ribosome, namely aminoglycosides and tetracycline. We hypothesize that this is due to the fact that only ribosomal inhibitors (and not antimicrobials that target other cellular pathways) trigger the induction of MexXY efflux pumps and adaptive resistance.42,43 Oxyclozanide overcomes this mechanism by both permeabilizing and disrupting the Δψ of cells within biofilms.
Many bacterial species, including S. aureus and P. aeruginosa, can be found growing as biofilms in non-healing chronic wounds such as diabetic foot ulcers and burns.17–19 Previously, it has been shown that oxyclozanide has activity against planktonic S. aureus and cancerous cells, indicating broad applicability.29 The repurposing of veterinary drugs has a proven history of success, notably the ionophore anthelmintic ivermectin, which has been repurposed for the treatment of several diseases in humans.44 Oxyclozanide combined with tobramycin could be a potential new treatment for P. aeruginosa and S. aureus infections in CF patients as well as diabetic foot ulcers and burn wounds.
Supplementary Material
Acknowledgements
We thank Martha Mulks and Neal Hammer for generously providing P. aeruginosa and S. aureus strains, respectively.
Funding
This work was supported by grants from the Hunt for a Cure Foundation, the NSF BEACON center for evolution in action (DBI-0939454), NIH grants GM109259 and GM110444, and an MSU Strategic Partnership Grant to C. M. W. and the Cystic Fibrosis Foundation Traineeship to M. M. M. In addition, M. M. M. was supported by a Wentworth Fellowship and Rudolf Hugh award from the MSU Microbial and Molecular Genetics department and by a Dissertation Completion Fellowship from the College of National Science at MSU.
Transparency declarations
None to declare.
References
- 1. O’Sullivan BP, Freedman SD.. Cystic fibrosis. Lancet 2009; 373: 1891–904. [DOI] [PubMed] [Google Scholar]
- 2. Borowitz D. CFTR, bicarbonate, and the pathophysiology of cystic fibrosis. Pediatr Pulmonol 2015; 50 Suppl 40: S24–30. [DOI] [PubMed] [Google Scholar]
- 3. Stoltz DA, Meyerholz DK, Welsh MJ.. Origins of cystic fibrosis lung disease. N Engl J Med 2015; 372: 1574–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boucher HW, Talbot GH, Bradley JS. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48: 1–12. [DOI] [PubMed] [Google Scholar]
- 5. Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother 2001; 45: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Costerton JW, Lewandowski Z, Caldwell DE. et al. Microbial biofilms. Annu Rev Microbiol 1995; 49: 711–45. [DOI] [PubMed] [Google Scholar]
- 7. Blair JMA, Webber MA, Baylay AJ. et al. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 2014; 13: 42–51. [DOI] [PubMed] [Google Scholar]
- 8. Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2011; 2: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tingpej P, Smith L, Rose B. et al. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol 2007; 45: 1697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chmiel JF, Aksamit TR, Chotirmall SH. et al. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Annals Am Thoracic Soc 2014; 11: 1120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mogayzel PJ, Naureckas ET, Robinson KA. et al. Cystic fibrosis foundation pulmonary guideline. pharmacologic approaches to prevention and eradication of initial Pseudomonas aeruginosa infection. Annals Am Thoracic Soc 2014; 11: 1640–50. [DOI] [PubMed] [Google Scholar]
- 12. Rosenfeld M, Gibson R, McNamara S. et al. Serum and lower respiratory tract drug concentrations after tobramycin inhalation in young children with cystic fibrosis. J Pediatrics 2001; 139: 572–7. [DOI] [PubMed] [Google Scholar]
- 13. Stuart B, Lin JH, Mogayzel PJ Jr.. Early eradication of Pseudomonas aeruginosa in patients with cystic fibrosis. Paediatr Respir Rev 2010; 11: 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Folkesson A, Jelsbak L, Yang L. et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 2012; 10: 841–51. [DOI] [PubMed] [Google Scholar]
- 15. Maiden MM, Hunt AMA, Zachos MP. et al. Triclosan is an aminoglycoside adjuvant for eradication of Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2018; 62: e00146–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swan GE. The pharmacology of halogenated salicylanilides and their anthelmintic use in animals: review article. J S Afr Vet Assoc 1999; 70: 61–70. [DOI] [PubMed] [Google Scholar]
- 17. Sivanmaliappan TS, Sevanan M.. Antimicrobial susceptibility patterns of Pseudomonas aeruginosa from diabetes patients with foot ulcers. Int J Microbiol 2011; 2011: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Omar A, Wright J, Schultz G. et al. Microbial biofilms and chronic wounds. Microorganisms 2017; 5: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altoparlak U, Erol S, Akcay MN. et al. The time-related changes of antimicrobial resistance patterns and predominant bacterial profiles of burn wounds and body flora of burned patients. Burns 2004; 30: 660–4. [DOI] [PubMed] [Google Scholar]
- 20. Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 2001; 48 Suppl 1: 5–16. [DOI] [PubMed] [Google Scholar]
- 21. Ceri H, Olson ME, Stremick C. et al. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 1999; 37: 1771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sambanthamoorthy K, Gokhale AA, Lao W. et al. Identification of a novel benzimidazole that inhibits bacterial biofilm formation in a broad-spectrum manner. Antimicrob Agents Chemother 2011; 55: 4369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spoering AL, Lewis K.. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 2001; 183: 6746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meylan S, Porter CBM, Yang JH. et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol 2017; 24: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sandoval R, Leiser J, Molitoris B.. Aminoglycoside antibiotics traffic to the Golgi complex in LLC-PK1 cells. Am Soc Nephrol 1997; 9: 167–74. [DOI] [PubMed] [Google Scholar]
- 26. Goss CH, Muhlebach MS.. Review: Staphylococcus aureus and MRSA in cystic fibrosis. J Cyst Fibros 2011; 10: 298–306. [DOI] [PubMed] [Google Scholar]
- 27. Taylor PK, Yeung ATY, Hancock REW.. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol 2014; 191: 121–30. [DOI] [PubMed] [Google Scholar]
- 28. Hocquet D, Vogne C, El Garch F. et al. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 2003; 47: 1371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barclay ML, Begg EJ, Chambers ST. et al. The effect of aminoglycoside-induced adaptive resistance on the antibacterial activity of other antibiotics against Pseudomonas aeruginosa in vitro. J Antimicrob Chemother 1996; 38: 853–8. [DOI] [PubMed] [Google Scholar]
- 30. Mulcahy LR, Burns JL, Lory S. et al. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 2010; 192: 6191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wood TK, Knabel SJ, Kwan BW.. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 2013; 79: 7116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 2006; 5: 48–56. [DOI] [PubMed] [Google Scholar]
- 33. Rajamuthiah R, Fuchs BB, Conery AL. et al. Repurposing salicylanilide anthelmintic drugs to combat drug resistant Staphylococcus aureus. PLoS One 2015; 10: e0124595, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gilleland LB, Gilleland HE, Gibson JA. et al. Adaptive resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. J Med Microbiol 1989; 29: 41–50. [DOI] [PubMed] [Google Scholar]
- 35. Sharlow ER. Revisiting repurposing. Assay Drug Dev Technol 2016; 14: 554–6. [DOI] [PubMed] [Google Scholar]
- 36. Lam J, Vaughan S, Parkins MD.. Tobramycin Inhalation Powder (TIP): an efficient treatment strategy for the management of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Clin Med Insights Circ Respir Pulm Med 2013; 7: 61–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Waters V, Smyth A.. Cystic fibrosis microbiology: advances in antimicrobial therapy. J Cystic Fibrosis 2015; 14: 551–60. [DOI] [PubMed] [Google Scholar]
- 38. Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961; 191: 144–8. [DOI] [PubMed] [Google Scholar]
- 39. Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005; 49: 479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hancock RE, Raffle VJ, Nicas TI.. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1981; 19: 777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Montie T, Patamasucon P.. Aminoglycosides: the complex problem of antibiotic mechanisms and clinical applications. Eur J Clin Microbiol Infect Dis 1995; 14: 85–7. [DOI] [PubMed] [Google Scholar]
- 42. Jeannot K, Sobel ML, Garch El F. et al. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J Bacteriol 2005; 187: 5341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karlowsky JA, Saunders MH.. In vitro characterization of aminoglycoside adaptive resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1996; 40: 1387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Omura S. Ivermectin: 25 years and still going strong. Int J Antimicrob Agents 2008; 31: 91–8. [DOI] [PubMed] [Google Scholar]
- 45. Holloway BW. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 1955; 13: 572–81. [DOI] [PubMed] [Google Scholar]
- 46. De Soyza A, Hall AJ, Mahenthiralingam E. et al. Developing an international Pseudomonas aeruginosa reference panel. Microbiol Open 2013; 2: 1010–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fey PD, Endres JL, Yajjala VK. et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 2012; 4: e00537–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Lencastre H, Tomasz A.. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 1994; 38: 2590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baba T, Bae T, Schneewind O. et al. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. Journal of Bacteriology 2007; 190: 300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soni I, Chakrapani H, Chopra S.. Draft genome sequence of methicillin-sensitive Staphylococcus aureus ATCC 29213. Genome Announc 2015; 3: e01095–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.