See Vidal and Atkin (doi:10.1093/brain/awz256) for a scientific commentary on this article.
ALS remains incurable due to an incomplete understanding of its molecular pathogenesis. Tyzack, Luisier et al. report that FUS protein, previously thought to mislocalise only in patients with FUS mutations, is also mislocalised in many cases of sporadic ALS. They further propose an underlying molecular mechanism for the mislocalisation.
Keywords: amyotrophic lateral sclerosis (ALS), fused in sarcoma FUS, RNA binding protein, intron retention
Abstract
Mutations causing amyotrophic lateral sclerosis (ALS) clearly implicate ubiquitously expressed and predominantly nuclear RNA binding proteins, which form pathological cytoplasmic inclusions in this context. However, the possibility that wild-type RNA binding proteins mislocalize without necessarily becoming constituents of cytoplasmic inclusions themselves remains relatively unexplored. We hypothesized that nuclear-to-cytoplasmic mislocalization of the RNA binding protein fused in sarcoma (FUS), in an unaggregated state, may occur more widely in ALS than previously recognized. To address this hypothesis, we analysed motor neurons from a human ALS induced-pluripotent stem cell model caused by the VCP mutation. Additionally, we examined mouse transgenic models and post-mortem tissue from human sporadic ALS cases. We report nuclear-to-cytoplasmic mislocalization of FUS in both VCP-mutation related ALS and, crucially, in sporadic ALS spinal cord tissue from multiple cases. Furthermore, we provide evidence that FUS protein binds to an aberrantly retained intron within the SFPQ transcript, which is exported from the nucleus into the cytoplasm. Collectively, these data support a model for ALS pathogenesis whereby aberrant intron retention in SFPQ transcripts contributes to FUS mislocalization through their direct interaction and nuclear export. In summary, we report widespread mislocalization of the FUS protein in ALS and propose a putative underlying mechanism for this process.
See Vidal and Atkin (doi:10.1093/brain/awz256) for a scientific commentary on this article.
Introduction
Amyotrophic lateral sclerosis (ALS) is a relentlessly progressive neurodegenerative condition, which remains incurable due to our incomplete understanding of the molecular pathogenesis. Genetic discoveries in ALS strongly implicate ubiquitously expressed regulators of RNA-processing (Taylor et al., 2016). Pathologically, in 97% of cases, TDP-43 protein is mislocalized from the nucleus to the cytoplasm, where it aggregates (Neumann et al., 2006). However, ALS-causing mutations in fused in sarcoma (FUS) and superoxide dismutase 1 (SOD1) conspicuously lack TDP-43 proteinopathy in the majority of cases (Mackenzie et al., 2007). FUS aggregation is a recognized feature of FUS mutation-related ALS (Vance et al., 2009). However, the concept that wild-type FUS nuclear-to-cytoplasmic mislocalization (rather than aggregation) might be a more widespread feature of other forms of ALS has not been systematically assessed to our knowledge. Indeed such mislocalization may have evaded detection thus far due to bias towards studying aggregates/inclusions rather than perturbed subcellular localization of unaggregated proteins alone. This possibility is strengthened by the facts that (i) impaired nuclear-cytoplasmic compartmentalization is increasingly recognized as a key feature of ALS (Boeynaems et al., 2016); and (ii) FUS is known to shuttle between the nucleus and cytoplasm.
Here we systematically investigated FUS protein localization across a human induced pluripotent stem cell (iPSC) model, mouse transgenic models and human post-mortem tissue from multiple cases of sporadic ALS. We find that the nuclear-to-cytoplasmic mislocalization of FUS is a more widespread feature of ALS than previously recognized. Furthermore, we present evidence that supports a putative molecular mechanism for this mislocalization through interaction between FUS protein and the aberrant intron-retaining SFPQ transcript.
Materials and methods
Induced pluripotent stem cell culture and motor neuron differentiation
IPSCs were maintained using standard protocols. Motor neuron differentiation was carried out as described previously (Hall et al., 2017; Luisier et al., 2018). See Supplementary material for more detailed information. Details of iPSC lines are provided in Supplementary Table 1.
Animals, transgenic models and tissue processing
All experiments were carried out following the guidelines of the UCL Institute of Neurology Genetic Manipulation and Ethic Committees and in accordance with the European Community Council Directive of November 24, 1986 (86/609/EEC). Animal experiments were undertaken under licence from the UK Home Office in accordance with the Animals (Scientific Procedures) Act 1986 (Amended Regulations 2012) and were approved by the Ethical Review Panel of the Institute of Neurology. The following transgenic mouse lines were used, and were analysed as different experimental groups: (i) female SOD1G93Amice [B6SJL-Tg(SOD1*G93A)1Gur/J, Jackson Laboratories], postnatal Day 93–95 (P93–95) (n = 4 mice); (ii) male over-expressing mutant human VCPA232E were generated by J. Paul Taylor et al., St Jude Children’s Research Hospital, Memphis, TN, USA and are described in Custer et al. (2010) and bred to wild-type C57 Black 6 (C57/B6) background, symptomatic, 9 months old (n = 3 mice); and (iii) wild-type C56BL/6-SJL mixed background (Jackson Laboratories) were used as control (n = 4 mice). Mice were bred and maintained at the UCL Institute of Neurology in standard individually ventilated cages with up to three mice per cage, in a temperature and humidity controlled environment with a 12-h light/dark cycle and had access to drinking water and food ad libitum. Cages were checked daily to ensure animal welfare. Body weight was assessed regularly to ensure no weight loss. For tissue collection, animals were injected with terminal anaesthesia (pentobarbital sodium, Euthatal) and were transcardially perfused with 4% paraformaldehyde. The lumbar region of the spinal cord was removed and post-fixed with 4% paraformaldehyde and cryoprotected overnight with 30% sucrose; 10 or 20 μm serial transverse cryosections were cut for immunofluorescence staining.
Human post-mortem tissue
Snap frozen tissue sections were obtained from lumbar spinal cords of eight healthy donors and 12 age and sex matched sporadic ALS patients (Supplementary Table 2). Death to snap-freezing delay times were also comparable between the groups [mean delay ± standard deviation (SD): 30.13 ± 12.87 and 27.75 ± 10.63 h, for control and sporadic ALS patients, respectively]. The spinal cord samples were obtained from the tissue bank NeuroResource, UCL Institute of Neurology, London, UK. Samples were donated to the tissue bank with written tissue donor informed consent following ethical review by the NHS NRES Committee London–Central and stored under a Research Sector Licence from the UK Human Tissue Authority (HTA). In this study, ALS cases were considered sporadic by the absence of any family history of motor neuron disease. Furthermore, all cases showed TDP-43 pathology when screened by NeuroResource, suggesting that they are not SOD1 mutants [as TDP-43 pathology is absent in SOD1-related ALS cases (Mackenzie et al., 2007)].
Immunolabelling, imaging and image analysis
For immunocytochemistry and immunohistochemistry, samples were blocked in 10% normal goat serum (NGS) or 10% normal donkey serum (NDS) as appropriate and permeabilized in 0.3% Triton™ X-100 [Sigma-Aldrich; in phosphate-buffered saline (PBS)] at room temperature for 1 h. Immunolabelling was performed with primary antibodies in NGS (5%) and Triton™ X-100 (0.1% in PBS) at 4°C overnight followed by species-specific secondary antibodies for 1 h at room temperature and DAPI nuclear counterstain (100 ng/ml) for 10 min at room temperature. For human post-mortem samples fixation and permeabilization in cold methanol (−20°C, 20 min) was performed before the immunostaining. Primary antibodies were diluted as follows: goat anti ChAT (Millipore, AB144P) 1:100; rabbit and mouse anti FUS (Abcam, ab84078 1:500; and Santa Cruz, sc-47711 1:100 respectively). Images were acquired using either a 710 Laser Scanning Confocal Microscope (Zeiss) or the Opera Phenix High-Content Screening System (Perkin Elmer). Images were acquired as confocal z-stacks with a z-step of 1 μm and processed to obtain a maximum intensity projection. For the analysis of nuclear/cytoplasmic ratio of FUS in iPSC-derived cells, images were analysed using the Columbus Image Analysis System (Perkin Elmer). The animal and post-mortem tissue sections were analysed using Fiji. Motor neurons were identified by choline acetyltransferase (ChAT) immunoreactivity, and the nuclear and cytoplasmic area were manually drawn based on DAPI and ChAT staining, respectively. For each cell, the average FUS immunoreactivity intensity in each region of interest was measured, background was subtracted, and the ratio between nuclear and cytoplasmic average intensity was calculated and used as the main experimental outcome.
Cell fractionation, RNA extraction, reverse transcription and quantitative PCR
For the biochemical fractionation of iPSC-derived neural precursors the Ambion PARIS kit (Thermo Fisher Scientific) was used following manufacturer’s instructions. A cytosolic fraction was obtained by lysing the cultures for 10 min in ice-cold cell fractionation buffer. Nuclei were lysed in 8 M Urea Nuclear Lysis Buffer, containing 50 mM Tris-HCl (pH 8), 100 mM NaCl, 0.1% SDS, 1 mM DTT. Both lysis buffer contained 0.1 U/μl RiboLock RNase Inhibitor (Thermo Fisher Scientific). RNA was extracted from both fractions using the Promega Maxwell® RSC simplyRNA cells kit including DNase treatment, alongside the Maxwell® RSC instrument. Reverse transcription was performed using the RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) using 1 μg of RNA and random hexamers. Quantitative PCR was performed using the PowerUP™ SYBR® Green Master Mix (Thermo Fisher Scientific) and the the QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems). Specific amplification was determined by melt curve analysis and agarose gel electrophoresis of the PCR products. Analysis of intron retaining transcripts was performed as previously described (Luisier et al., 2018). Levels of intron retention (primer pair F2R2) were normalized over the expression level of each individual gene (primer pair F1R1). Primers used were as follows: SFPQ F1: GCCGAATGGGCTACATGGAT; SFPQ R1: TCAGTACG CATGTCACTTCCC; SFPQ F2: GTGGATCGACTCATTGGT GA; SFPQ R2: TTCCTCTAGGACCCTGTCCA.
Single molecule fluorescence in situ hybridization
Based on Raj et al. (2008), iPSC-derived neural precursors (Day 7 after beginning of neural conversion) that were grown on Geltrex®-coated sterile 2-well µ-Slides (80286, Ibidi) were washed twice for 5 min with PBS, and fixed using 4% methanol-free formaldehyde (10321714, Thermo Fisher Scientific) for 10 min at room temperature. Following additional two wash steps, cells were permeabilized using 70% ethanol for 12 h at 4°C, and were washed twice again. Then preparations were incubated for 15 min with hybridization buffer prepared using 2× saline-sodium citrate (SSC) solution/10% deionized formamide (4610-OP, Calbiochem). Hybridization with Stellaris FISH probes was done in a total volume of 50 µl hybridization buffer containing 50 µg competitor tRNA from Escherichia coli (10109541001, Roche Diagnostics), 10% dextran sulphate (9011–18–1, VWR), 2 mg/ml UltraPure BSA (AM2616, Thermo Fisher Scientific), and 10 mM vanadyl-ribonucleoside complex (S1402S, New England Biolabs) with probes at final concentration of 1 ng/µl. Preparations were covered with parafilm and incubated at 37°C for 5 h, and afterwards washed twice with pre-warmed 2 × SCC/10% formamide for 30 min at 37°C. Finally the preparations were washed twice with PBS at room temperature, and then mounted using 10 µl ProLong® Gold Antifade Reagent containing DAPI (9071S, New England Biolabs). The slides were imaged when the mounting medium was fully cured >12 h.
Probes were designed using the Probe Designer software from Biosearch Technologies and were provided by same vendor. Probes included were designed against SFPQ intron conjugated to Quasar®570 (SMF-2037–1) and mature SFPQ conjugated to Quasar®670 (sequences of probes available upon request). Quantification of hybridization signal was performed using custom spot-intensity detection algorithm in DAPI segmented cells to separate nuclear and cytoplasmic signal.
Data analysis of FUS protein subcellular localization
We used R and lme4 (Bates et al., 2015) to perform linear mixed effects analysis of the relationship between FUS localization and VCP or SOD1 mutation, as well as with sporadic ALS, that accounts for idiosyncratic variation due to either animal or individual differences. As fixed effects, we either entered the mutation or the ALS disease variable into the model. As random effects, we had intercepts for either animals (SOD1, VCP) or patients and batches. Visual inspection of residual plots did not reveal any obvious deviations from homoscedasticity or normality. P-values were obtained by likelihood ratio tests of the full model with the effect in question against the model without the effect in question.
Mapping of iCLIP data
Raw FUS individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) data can be accessed at https://imaps.genialis.com/ (Attig et al., 2018). Before alignment, two-stage adapter removal was performed using Cutadapt according to the ENCODE iCLIP standard operating procedure. A two-stage approach was also used for alignment. First, Bowtie2 was used to remove reads aligning to rRNA or tRNA. Then, STAR was used to align the remaining reads to GRCh38, with only uniquely mapping reads retained. PCR duplicates were collapsed based on the unique molecular identifiers and mapping locations. The nucleotide-resolution crosslink position was calculated as the coordinate immediately preceding the reverse transcription truncation event.
Compliance with ethical standards
For human iPSC work, informed consent was obtained from all patients and healthy controls in this study. Experimental protocols were all carried out according to approved regulations and guidelines by UCLH’s National Hospital for Neurology and Neurosurgery and UCL Queen Square Institute of Neurology joint research ethics committee (09/0272). The human post-mortem spinal cord samples were obtained from the tissue bank NeuroResource, UCL Queen Square Institute of Neurology, London, UK. Samples were donated to the tissue bank with written tissue donor informed consent following ethical review by the NHS NRES Committee London–Central and stored under a Research Sector Licence from the UK Human Tissue Authority (HTA). All animal experiments described in this study were carried out under licence from the UK Home Office, and were approved by the Ethical Review Panel of the Institute of Neurology.
Data availability
Data supporting the findings of this study are available from the corresponding author, upon reasonable request.
Results
Nuclear-to-cytoplasmic mislocalization of FUS in human and mouse VCP-mutant ALS models
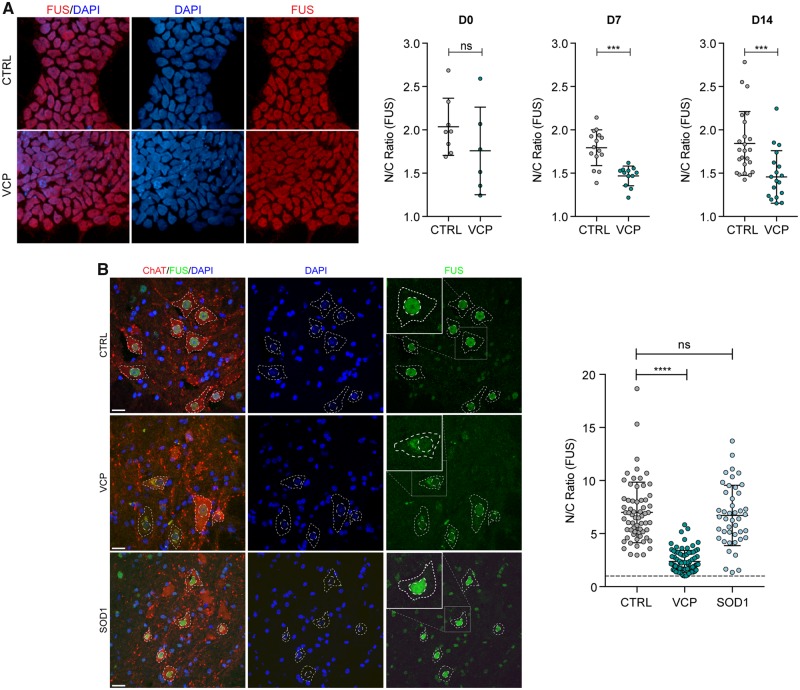
Previously, we reported robust differentiation of human iPSCs into highly enriched (>85%) cultures of comprehensively validated and functionally characterized spinal cord motor neurons (Hall et al., 2017; Maffioletti et al., 2018). Using this approach, we have found clear molecular pathogenic phenotypes in a human iPSC model of VCP-related ALS (four clones from two patients with the mutations VCPR155C and VCPR191Q) (Hall et al., 2017; Luisier et al., 2018). We harnessed this well-characterized ALS model to investigate the subcellular localization of FUS protein, which revealed a modest but statistically significant (P < 0.001) decrease in nuclear-to-cytoplasmic localization during motor neuron differentiation (Fig. 1A). We next sought to validate this finding in vivo by examining tissue sections from symptomatic transgenic VCPA232E mice. Additionally, we examined tissue from the SOD1G93A mouse model as a comparator. Using linear mixed model analysis that accounts for inter-animal variation we showed that although FUS remained within the nucleus in the SOD1 mouse model [2(1) = 0.031, P = 0.955], nuclear-to-cytoplasmic mislocalization abounded in the VCP mouse model, with a reduction in FUS nuclear to cytoplasmic ratio of −4.0176 ± 1.1775 [2(1) = 8.164, P = 0.0042] (Fig. 1B).
Figure 1.
Nuclear-to-cytoplasmic mislocalization of FUS in human and mouse VCP-mutant ALS models. (A) Subcellular localization of FUS determined by immunocytochemistry in iPSCs (D0), neural precursors (NPCs; D7) and patterned motor neuron precursors (pMNs; D14). The ratio of the average intensity of the FUS immunolabelling in the nucleus (N) versus cytoplasm (C) was automatically determined in both CTRL and VCP mutant lines cells. Data shown are average N/C ratios (±SD) per field of view from four control and four VCP mutant lines. P-value from unpaired t-test with Welch’s correction. (B) Analysis of the subcellular localization of FUS in motor neurons in the ventral spinal cord of wild-type, VCPA232E and SOD1G93A mice. Motor neuron cytoplasm was identified by ChAT staining, nuclei were counterstained with DAPI. Images were acquired as confocal z-stacks using a Zeiss 710 confocal microscope with a z-step of 1 μm, processed to obtain a maximum intensity projection and analysed using Fiji. Motor neurons were identified by ChAT staining, and the nuclear and cytoplasmic area were manually drawn based on DAPI and ChAT staining, respectively. For each region of interest, the average FUS immunoreactivity intensity was measured, background was subtracted, and the ratio between nuclear and cytoplasmic average intensity was calculated. Data shown are nuclear/cytoplasmic (N/C) ratio (mean ± SD) per cell from four wild-type, four SOD1G93A and three VCPA232E mice. Scale bar = 20 μm. Linear mixed effects analysis of the relationship between FUS localization and either VCP or SOD1 mutation to account for idiosyncratic variation due to animal differences: SOD1 mutation does not affect FUS localization [2(1) = 0.6387, P = 0.4242], lowering it by about −0.8152 ± 1.1152 (standard errors), while VCP mutation does affect FUS localization [2(1) = 8.3145, P = 0.003933], lowering it by −4.2175 ± 0.9731 (standard errors).
Nuclear-to-cytoplasmic mislocalization of FUS in human sporadic ALS
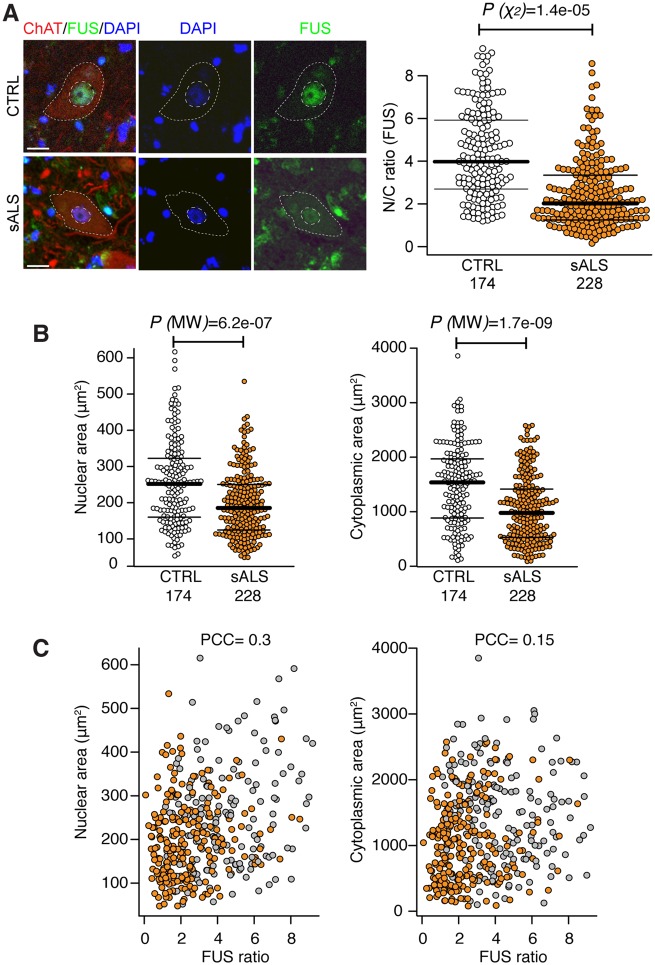
Having established that FUS is mislocalized in VCP-related ALS models, but not in SOD1, we next sought to address the generalizability of this finding across sporadic forms of ALS (which represent 90% of all cases). To this end, we examined post-mortem spinal cord tissue from 12 sporadic ALS cases and eight healthy controls (Fig. 2A). We found clear evidence of nuclear-to-cytoplasmic FUS mislocalization in these sporadic ALS cases, but in the absence of cytoplasmic FUS inclusions.
Figure 2.
Nuclear-to-cytoplasmic mislocalization of FUS in human sporadic ALS. (A) Analysis of the subcellular localization of FUS in motor neurons in the ventral spinal cord of healthy controls (n = 8) and patients with sporadic ALS (sALS) (n = 12). Motor neuron cytoplasm was identified by ChAT immunolabelling, nuclei were counterstained with DAPI. N/C ratio of FUS immunoreactivity was measured as described in Fig. 1B. Only motor neurons with a visible nucleus were considered for the analysis. We used R and lme4 (Bates et al., 2015) to perform a linear mixed effects analysis of the relationship between FUS localization and sALS disease. Scale bar = 20 μm. Data shown are N/C ratio (mean ± SD) per cell from at least eight cases per group. (B) Nuclear and cytoplasmic areas of motor neuron nuclei and cytoplasm in sALS (n = 12) versus control (n = 8). Data shown are mean ± SD. P-values from non-parametric Mann-Whitney’s test. (C) Single-cell measurement of nuclear (right) and cytoplasmic area (left) shows no correlation with FUS nuclear to cytoplasmic ratio as shown with scatter plots. PCC = Pearson correlation coefficient.
A reduction in motor neuronal soma size has been previously shown in an in vitro model of ALS which associates with increased apoptosis (Kiskinis et al., 2014). We thus investigated whether changes in FUS subcellular localization in sporadic ALS samples accompany alterations in cell morphology. Using single-cell analysis of nuclear and cell size, we demonstrated significant reductions in both nuclear and cytoplasmic areas in sporadic ALS versus control motor neurons (Fig. 2B), which did not correlate with reduction in FUS nuclear-to-cytoplasmic localization in sALS cases (Fig. 2C).
FUS binds to an aberrantly retained intron within the SFPQ transcript, which is exported from the nucleus
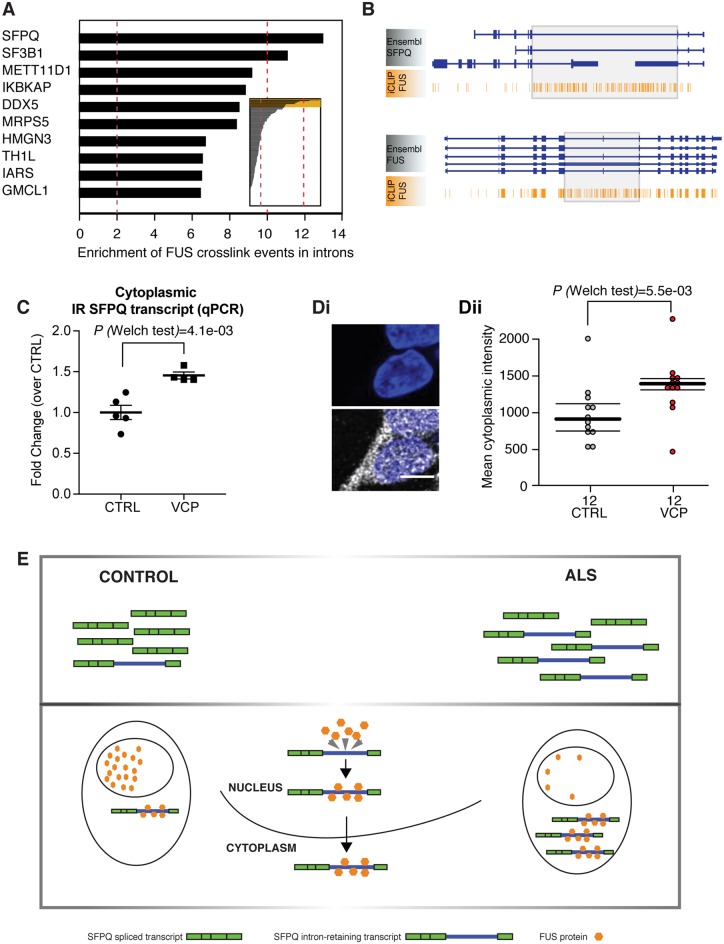
Having found clear evidence that FUS nuclear-to-cytoplasmic mislocalization is more widespread in ALS than previously recognized, we sought to understand its molecular interplay with 167 aberrant intron retaining transcripts that we recently described in ALS (Luisier et al., 2018). To this end we analysed iCLIP data, which allowed identification of RNA binding targets of the FUS protein. Using this approach, we found that FUS protein binds extensively to the aberrantly retained intron 9 within the SFPQ transcript (Fig. 3A and B), which we identified as the most significantly retained intron across diverse ALS mutations (Luisier et al., 2018). We next confirmed that the SFPQ intron-retaining transcript is exported to the cytoplasm using nuclear-cytoplasmic cellular fractionation and qPCR (Fig. 3C), showing an increased proportion of SFPQ intron retaining transcript in the cytosol of VCP mutant cultures. We used single molecule RNA fluorescence in situ hybridization (smFISH) as orthogonal validation that this transcript is exported from the nucleus (Fig. 3D).
Figure 3.
FUS binds to an aberrantly retained intron within the SFPQ transcript, which is exported from the nucleus. (A) Bar plots depicting the level of enrichment in FUS-binding to the retained intron compared with non-retained introns within the same gene. Analysis of the 167 previously reported as aberrantly retained introns in ALS. (B) Genome browser view of FUS iCLIP crosslinking events along the SFPQ and FUS transcript. Grey boxes highlight the location of retained introns. (C) Dot plot showing the levels of cytoplasmic intron retention (IR) SFPQ transcripts measured by qPCR. IR transcript levels were measured using an IR-specific primer pair and normalized over the expression levels of a constitutive SFPQ exon, as described previously (Luisier et al., 2018). n = 5 control lines and n = 4 VCP lines, mean ± standard error of the mean (SEM) from two independent replicates per line, each analysed in technical duplicate. P = 0.0041 from Welch’s test. [D(i)] Single molecule fluorescence in situ hybridization (smFISH) showing SFPQ intron-retaining transcript in the cytoplasm. The top panel shows the nuclear staining only for context; the probe clearly detects intron-retaining transcripts in the cytoplasm. [D(ii)] Quantification and statistical analysis of smFISH performed on three control lines and three VCP mutant lines using Welch’s test. (E) Schematic diagram of proposed model. Cartoon summarizing the functional consequence of aberrant IR in SFPQ transcript. The 9 kb intron 9 in SFPQ is retained and exported to the cytoplasm. The FUS protein binds extensively to this retained intron. The FUS protein itself is abundantly relocalized from the nucleus to the cytoplasm in diverse ALS models (human iPSC and mouse transgenic), as well as post-mortem samples from patients with sporadic ALS.
Discussion
The core finding of our study is that the FUS protein is mislocalized from the nucleus to the cytoplasm in cases beyond FUS mutation-related ALS. Specifically, we find FUS mislocalization in VCP mutation-related ALS and, crucially, in sporadic ALS. Our findings have been carefully cross-validated in iPSC lines (four VCP mutant and four control iPSC lines), a mouse transgenic model (three VCP mutant and three control mice), and post-mortem tissue (12 sporadic ALS cases and eight control cases). The pervasive mislocalization of FUS has likely evaded detection thus far as FUS largely remains unaggregated in the cytoplasm, rather than forming part of the TDP-43 aggregates in sporadic ALS cases.
Our findings support a model whereby FUS mislocalization from the nucleus to the cytoplasm occurs in the majority of ALS cases, but it generally does not appear to aggregate in the cytoplasm. Nuclear loss of FUS protein may impair pre-mRNA splicing whilst the possibility of a cytosolic toxic gain of function is also noteworthy in light of recent studies (López-Erauskin et al., 2018). Interestingly, TDP-43 aggregation is also observed in most familial and sporadic ALS cases, except from those caused by SOD1 mutations. Thus, FUS mislocalization appears to occur in a similar majority of cases to TDP-43 aggregation. Importantly, the mislocalized FUS is observed in mouse and iPSC models at early stages, before the onset of TDP-43 aggregates, indicating that the mislocalized unaggregated FUS might play a causative role early in the ALS disease process, perhaps by creating a more aggregation-prone cytoplasmic environment.
Our analysis of iCLIP data suggest that FUS binds avidly to the aberrantly retained intron of the SFPQ transcript in ALS. Cumulatively, our data are consistent with a working hypothesis that wild-type FUS might travel out of the nucleus when bound to the aberrantly retained intron 9 of the SFPQ transcript in ALS (Fig. 3E), though confirmation of such a mechanism will require detailed molecular follow-up work.
In our previous study, we showed SFPQ intron retention occurs in VCP, SOD1 and FUS mutant iPSC-motor neuron cultures and that this correlates with loss of nuclear SFPQ across different model systems (Luisier et al., 2018). The current study, however, adds interesting complexity to this paradigm by suggesting that RNA-binding proteins sequestered by the SFPQ intron-retaining transcript might vary in a mutation-dependent manner. Indeed although SFPQ protein is sequestered by the SFPQ intron-retaining transcript across different mutations we examined, FUS is not statistically significantly sequestered and/or exported from the nucleus by this transcript in the context of SOD1 mutations. In addition to the evidence we provided in our previous work (Luisier et al., 2018), mutant SOD1 has also been shown to bind RNA transcripts in other contexts, playing a role in their stabilization (Chen et al., 2014; Lu et al., 2007, 2009). Furthermore, SOD1 is known to form complexes with other ribonucleoproteins such as TIAR and HuR (Lu et al., 2007, 2009). To further support the concept that mutant SOD1 can influence the localization (and therefore function) of other RNA binding proteins, a recent proteomic study found a nuclear shift in proteins involved in RNA transport and processing in SOD1 mutants compared to controls (Kim et al., 2017). In aggregate, these studies raise the hypothesis that by altering RNA metabolism, mutations in SOD1 could lead to mutation-dependent changes in the molecular composition of ribonucleoprotein complexes. Specifically, mutant SOD1 might affect the interaction of specific RNA binding proteins, such as FUS, with their target RNAs. The absence of FUS mislocalization in this context, together with the lack of TDP-43 proteinopathy, seems to further substantiate a recognized pathological divergence between SOD1-ALS and other forms of familial and sporadic ALS.
In summary, we report a previously unrecognized widespread mislocalization (but not aggregation) of FUS in ALS, and propose a putative context-specific mechanism for this through its interaction with the ALS-related aberrantly retained intron 9 in SFPQ transcripts. These findings raise the prospect of targeting the nuclear-to-cytoplasmic mislocalization of unaggregated FUS as a putative therapeutic strategy in ALS.
Supplementary Material
Acknowledgements
The authors wish to thank Martina Hallegger and Roberto Simone for their input regarding primer design, Anob M. Chakrabarti for sharing BED files of aligned iCLIP data. We thank Benjamin Clarke, Mhoriam Ahmed and members of the Schiavo and Greensmith Labs for valuable technical support.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- iCLIP
individual-nucleotide resolution cross-linking and immunoprecipitation
- iPSC
induced pluripotent stem cell
Funding
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK 65 (FC010110), the UK Medical Research Council (FC010110), and the Wellcome Trust (FC010110). R.P. holds an MRC Senior Clinical Fellowship [MR/S006591/1]. N.M.L. and J.U. are supported by a Wellcome Trust Senior Investigator Award [103760/Z/14/Z], an MRC eMedLab Medical Bioinformatics Infrastructure Award to N.M.L. (MR/L016311/1), a Marie Curie Post-doctoral Research Fellowship (657749-NeuroUTR) and an Advanced Postdoc Mobility Fellowship from the Swiss National Science Foundation (P300PA_174461) to R.L. N.M.L. is a Winton Group Leader in recognition of the Winton Charitable Foundation's support towards the establishment of the Francis Crick Institute.
Competing interests
The authors report no competing interests.
References
- Attig J, Agostini F, Gooding C, Chakrabarti AM, Singh A, Haberman N, et al. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell 2018; 174: 1067–81.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4 [Internet]. J Stat Softw 2015; 67: 1–48. [Google Scholar]
- Boeynaems S, Bogaert E, Van Damme P, Van Den Bosch L. Inside out: the role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol 2016; 132: 159–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Qian K, Du Z, Cao J, Petersen A, Liu H, et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 2014; 14: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custer SK, Neumann M, Lu H, Wright AC, Taylor JP. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum Mol Genet 2010; 19: 1741–55. [DOI] [PubMed] [Google Scholar]
- Hall CE, Yao Z, Choi M, Tyzack GE, Serio A, Luisier R, et al. Progressive motor neuron pathology and the role of astrocytes in a human stem cell model of VCP-related ALS. Cell Rep 2017; 19: 1739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-E, Hong YH, Kim JY, Jeon GS, Jung JH, Yoon B-N, et al. Altered nucleocytoplasmic proteome and transcriptome distributions in an in vitro model of amyotrophic lateral sclerosis. Plos One 2017; 12: e0176462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 2014; 14: 781–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Erauskin J, Tadokoro T, Baughn MW, Myers B, McAlonis-Downes M, Chillon-Marinas C, et al. ALS/FTD-Linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron 2018; 100: 816–30.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisier R, Tyzack GE, Hall CE, Mitchell JS, Devine H, Taha DM, et al. Intron retention and nuclear loss of SFPQ are molecular hallmarks of ALS. Nat Commun 2018; 9: 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Wang S, Zheng L, Li X, Suswam EA, Zhang X, et al. Amyotrophic lateral sclerosis-linked mutant SOD1 sequesters Hu antigen R (HuR) and TIA-1-related protein (TIAR). J Biol Chem 2009; 284: 33989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zheng L, Viera L, Suswam E, Li Y, Li X, et al. Mutant Cu/Zn-superoxide dismutase associated with amyotrophic lateral sclerosis destabilizes vascular endothelial growth factor mRNA and downregulates its expression. J Neurosci 2007; 27: 7929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IRA, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol 2007; 61: 427–34. [DOI] [PubMed] [Google Scholar]
- Maffioletti SM, Sarcar S, Henderson ABH, Mannhardt I, Pinton L, Moyle LA, et al. Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep 2018; 23: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–3. [DOI] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 2008; 5: 877–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature 2016; 539: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009; 323: 1208–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author, upon reasonable request.