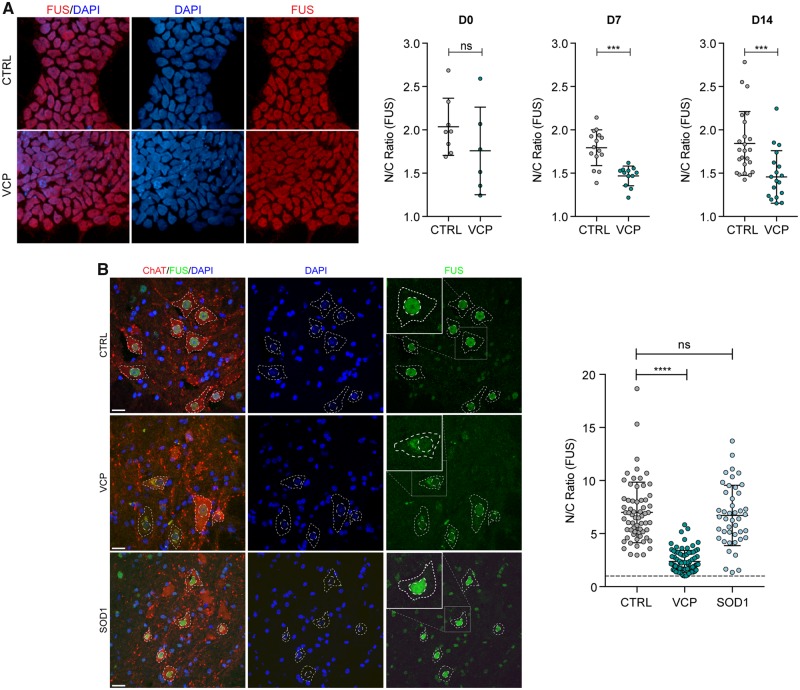
Figure 1.
Nuclear-to-cytoplasmic mislocalization of FUS in human and mouse VCP-mutant ALS models. (A) Subcellular localization of FUS determined by immunocytochemistry in iPSCs (D0), neural precursors (NPCs; D7) and patterned motor neuron precursors (pMNs; D14). The ratio of the average intensity of the FUS immunolabelling in the nucleus (N) versus cytoplasm (C) was automatically determined in both CTRL and VCP mutant lines cells. Data shown are average N/C ratios (±SD) per field of view from four control and four VCP mutant lines. P-value from unpaired t-test with Welch’s correction. (B) Analysis of the subcellular localization of FUS in motor neurons in the ventral spinal cord of wild-type, VCPA232E and SOD1G93A mice. Motor neuron cytoplasm was identified by ChAT staining, nuclei were counterstained with DAPI. Images were acquired as confocal z-stacks using a Zeiss 710 confocal microscope with a z-step of 1 μm, processed to obtain a maximum intensity projection and analysed using Fiji. Motor neurons were identified by ChAT staining, and the nuclear and cytoplasmic area were manually drawn based on DAPI and ChAT staining, respectively. For each region of interest, the average FUS immunoreactivity intensity was measured, background was subtracted, and the ratio between nuclear and cytoplasmic average intensity was calculated. Data shown are nuclear/cytoplasmic (N/C) ratio (mean ± SD) per cell from four wild-type, four SOD1G93A and three VCPA232E mice. Scale bar = 20 μm. Linear mixed effects analysis of the relationship between FUS localization and either VCP or SOD1 mutation to account for idiosyncratic variation due to animal differences: SOD1 mutation does not affect FUS localization [2(1) = 0.6387, P = 0.4242], lowering it by about −0.8152 ± 1.1152 (standard errors), while VCP mutation does affect FUS localization [2(1) = 8.3145, P = 0.003933], lowering it by −4.2175 ± 0.9731 (standard errors).