Abstract
The nucleolus is a nuclear sub-domain containing the most highly transcribed genes in the genome. Hundreds of human ribosomal RNA (rRNA) genes, located in the nucleolus, rely on constant maintenance. DNA double-strand breaks (DSBs) in rRNA genes activate the ATM kinase, repress rRNA transcription and induce nucleolar cap formation. Yet how ribosomal-DNA (rDNA) lesions are detected and processed remains elusive. Here, we use CRISPR/Cas9-mediated induction of DSBs and report a chromatin response unique to rDNA depending on ATM-phosphorylation of the nucleolar protein TCOF1 and recruitment of the MRE11–RAD50–NBS1 (MRN) complex via the NBS1-subunit. NBS1- and MRE11-depleted cells fail to suppress rRNA transcription and to translocate rDNA into nucleolar caps. Furthermore, the DNA damage response (DDR) kinase ATR operates downstream of the ATM-TCOF1-MRN interplay and is required to fully suppress rRNA transcription and complete DSB-induced nucleolar restructuring. Unexpectedly, we find that DSBs in rDNA neither activate checkpoint kinases CHK1/CHK2 nor halt cell-cycle progression, yet the nucleolar-DDR protects against genomic aberrations and cell death. Our data highlight the concept of a specialized nucleolar DNA damage response (n-DDR) with a distinct protein composition, spatial organization and checkpoint communication. The n-DDR maintains integrity of ribosomal RNA genes, with implications for cell physiology and disease.
INTRODUCTION
Genome surveillance mechanisms are constantly alert to process aberrant DNA structures to prevent changes in the genetic material transferred from mother to daughter cells. A broad spectrum of lesions challenges genome integrity with double strand breaks (DSBs) being a particularly severe type as lack or faulty repair of DSBs can lead to grave diseases including cancer (1,2). Over the last decade a growing body of evidence has described the cellular DNA damage response (DDR) and how it functions to minimize the negative impact of DSBs by regulation of processes such as DNA repair, cell-cycle arrest, transcription, replication, cell division and cell death.
In nuclear chromatin, a DSB is initially detected by the MRN complex, which facilitates the ensuing activation of the major DDR kinase Ataxia-telangiectasia mutated (ATM) (3,4). ATM kick-starts phosphorylation-dependent signaling cascades and initiates modification of the local chromatin environment (5). Chromatin modifications include phosphorylation of the histone H2AX, that binds the mediator protein MDC1, and promotes additional recruitment of the MRN complex and broader modification of DSB-flanking chromatin (6–9). Chromatin modifications at and around the damage site lead to recruitment of a large number of proteins resulting in the formation of so-called Ionizing-radiation-induced-foci (IRIF), a structure that can be recognized microscopically and used as a read-out for the damage load experienced by cells (7).
In mammalian cells, DSBs are primarily repaired by one of two pathways: non-homologous end-joining (NHEJ) or homology-directed repair (HDR). The choice of repair pathway is affected by the cell-cycle phase, complexity of the lesion and the chromatin environment, but generally DNA end-joining with minimal processing by NHEJ is the initial pathway activated followed by resection-dependent HDR when successful repair is not accomplished (10).
One challenge faced by the DDR lies in the compartmentalization of the nucleus into a variety of different chromatin structures and nuclear bodies, each with specific needs of genome maintenance depending on their functions (11–15). The nucleolus is the largest sub-structure in the nucleus functioning in ribosome biogenesis and acting as a stress sensor. The nucleolus is formed around transcribed ribosomal RNA genes (rDNA), with each cell containing hundreds of ribosomal RNA genes, distributed across the short arm of the acrocentric chromosomes in human cells (16). Multiple chromosomes can contribute with rDNA to the same nucleolus (17). At the exit of mitosis RNA Polymerase I initiates the transcription of the rDNA that leads to self-assembly of the nucleolus (18). The rDNA is intrinsically unstable and its instability is increased upon loss of genome maintenance factors, emphasizing the need for surveillance of rDNA (19). In particular, faulty recombination between rDNA sequences from different chromosomes can have detrimental consequences for the cell and must be avoided if possible.
Upon DSB-induction in the nucleolus, the ATM kinase becomes activated and leads to repression of nucleolar transcription, to nucleolar segregation and to the translocation of rDNA to nucleolar caps at the periphery (20–22). It has been suggested that restructuring of the nucleolus and localisation of rDNA to nucleolar caps serve as a mechanism to separate rDNA originating from different chromosomes to prevent inter-chromosomal recombination in response to DNA damage (14). In agreement with this HDR factors were shown to be recruited to nucleolar caps formed at the nucleolar periphery after DNA damage induction (21–23).
The rapid development in programmable gene editing tools, especially the CRISPR/Cas9 system, now allows us to introduce DSBs to almost any locus of the genome in a precise and controllable manner (24,25). These advances have also provided researchers with new possibilities to study specialized DDR pathways associated with certain chromatin conformations or specific nuclear compartments (22,26).
In this study we investigate the early events of the nucleolar DDR, directly following ATM activation, that facilitate the segregation of rDNA into nucleolar caps. We use a CRISPR/Cas9-based system to induce site-specific DSBs in rDNA combined with a fluorescently tagged version of the DNA damage protein NBS1 to visualize the behaviour of rDNA breaks. By these means, we demonstrate how rDNA breaks are initially detected in the nucleolar interior, cluster and translocate to the nucleolar periphery. We describe protein recruitment coordinated by the nucleolar protein TCOF1, a mechanism distinct from that operating in nuclear chromatin. ATM-dependent phosphorylation of TCOF1 is required for MRN recruitment that induces further inhibition of ribosomal RNA (rRNA) transcription and translocation of breaks to the nucleolar periphery. We find that the ATR kinase operates downstream of ATM and the MRN-complex, and provide evidence that supports a two-step kinase mechanism required for full segregation of the rDNA after DSBs. Unexpectedly, this response is contained within the nucleolus and, unlike the canonical checkpoint responses, this process does not lead to CHK1/CHK2 activation or delay in cell-cycle progression. Finally, we show that MRE11 and TCOF1 protect cells upon rDNA damage, as depletion of these factors sensitizes cells to rDNA DSBs ensuing pronounced increase in abnormal nuclear morphology and cell death.
MATERIALS AND METHODS
Cell culture
U2OS-Cas9-NBS1-GFP, HEK 293T-Cas9, DIvA AID and HT1080 (I-PpoI) cells were grown in Dulbecco’s-modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (Invitrogen), 100 U penicillin and 100 μg ml−1streptomycin. The medium for the U2OS-Cas9-NBS1-GFP cells contain 1 μg ml−1 puromycin and 400 μg ml−1 neomycin (G418). The medium for DIvA AID cells was containing 1 mM sodium pyruvate and 400 μg ml−1 neomycin (G418). Where indicated, the culture medium for HEK 293T-Cas9 cells was supplied with doxycycline to a final concentration of 0.2 μg ml−1 to induce Cas9 expression. The culture medium for DIvA AID was supplied with 300 nM 4-hydrotamoxifen (4-OHT) and the media for HT1080 (I-PpoI) cells with 1 mM 4-OHT to induce endonuclease activity.
Generation of cell lines
The U2OS derivative cell lines, U2OS-CAG-Cas9-NBS1-GFP, expressing constitutive SpCas9 and GFP-tagged NBS1 were generated by co-transfecting wild type U2OS cells with an NBS1-GFP2 construct, a PiggyBac CAG driven Cas9 (SpCas9) expression transposon plasmid and a PiggyBac transposase plasmid. The ratio of transposon and transposase plasmids was 10:1 to minimize the copy number of integrated CAG-Cas9 cassette per cell. Stably transfected cells were selected with puromycin (1 μg ml−1) and neomycin (G418, 400 μg ml−1) for 1 week. GFP positive (puromycin and neomycin resistance) U2OS cells, named U2OS-Cas9-NBS1-GFP, were enriched by FACS sorting (Aarhus University FACS CORE facility). Single-cell derived U2OS-CAG-Cas9-NBS1-GFP2 clones were generated by sub-cloning from single cells. The Cas9 expression was subsequently validated by quantitative polymerase chain reaction (qPCR) and western blot.
To generate the HEK293T cells stably expressing an inducible SpCas9, we firstly generated a PiggyBac transposon vector that contains TRE promoter-driven SpCas9 expression cassette and an EF1a-promoter driven Tet-on-2A-hygromycin expression cassette (PB-TRE-SpCas9-Hyg, synthesis). One day before transfection, HEK293T cells (5 × 104 cell per well) were seeded to a 24-well plate. Transfection was conducted using a commercially available transfection reagent XtremeGene 9 following the manufacturer’s instructions. To avoid insertion of multiple transgene copies, we used a ratio of 1:10 between the PiggyBac transposase vector and the PiggyBac transposon vector (PB-TRE-SpCas9-Hyg). Two days after transfection, cells were cultured in selection medium containing hygromycin (10 μg ml−1). Approximately 1-week post-transfection, single cells were manually picked with a 10 μl pipette under a stereomicroscope and expanded in a 96-well plate with medium changes every 3–4 days. Cell colonies were further expanded upon 80–90% confluence and confirmed by PCR and dual cutting assay. In this study, clone #25 was selected for subsequent CRISPR gene editing experiments.
Small molecule inhibitors
Where indicated, the culture medium was supplied with 1 μM ATM inhibitor (KU55933), 0.5 μM ATR inhibitor (AZ20), 1 μM ATR inhibitor (VE821) or 0.5 μM ATR inhibitor (BAY1895344) (Selleckchem) one hour prior to gRNA transfection.
Plasmids and transfections
Transient transfections of U2OS-Cas9-NBS1-GFP cells were carried out using Lipofectamine LTX with plus reagent (Invitrogen) according to the manufacturer's specifications and collected 0–6 h post-transfection unless otherwise stated. The four gRNA plasmids used for transfection were ampicillin resistant pMA plasmids (generated by the Luo laboratory) expressing either rDNA targeting sequences (Supplementary Table S1) (22) or a control pMA vector expressing the gRNA scaffold but no target sequence. The cloning of CRISPR gRNAs was carried out as described previously (27). Unless otherwise stated the three rDNA gRNA plasmids were pooled in a ratio 1:1:1 for each transfection. The TCOF1 wild-type (wt) construct was previously described (28).
RNA interference and site-directed mutagenesis
The siRNA transfections were performed with 50 nM siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen). Samples were collected 60 h after transfection unless otherwise stated. The siRNA oligonucleotides were obtained from Eurofins Genomics (MWG). For annotations and sequences see Supplementary Table S1. The TCOF1 STTT, S1216A+S1199A and S1216A expression plasmids were previously described (28,29) and primer sequences used for the generation of the ATM-null construct can be found in Supplementary Table S1. The shRNA against TCOF1 was generated by insertion of an oligonucleotide into the pSUPERIOR.puro vector (Oligoengine) (28). Cells were transfected in a 4:1 ratio of shTCOF1 and the rescue plasmid (either TCOF1 WT, TCOF1 STTT, TCOF1 ATM-null, TCOF1 S1216A+S1199A or TCOF1 S1216A) using Lipofectamine LTX with plus reagent (Invitrogen) according to the manufacturer's specifications. After 72 h cells were transfected with gRNAs and collected at indicated time points.
Irradiation
X-ray irradiation was done with a YXLON.SMART 160E—1.5 device (150 kV, 6 mA; YXLON International A/S) delivering 11.8 mGy s−1. Soft X-rays were largely filtered out with a 3 mm aluminium filter. Specific doses used in individual experiments: Figure 1C = 10 Gy followed by 1 h incubation, Figure 6A = 5 Gy followed by 3–6 h incubation, Figure 6B = 5 Gy followed by 3 or 15 h incubation, Supplementary Figure S6a = 5 Gy followed by 3 or 15 h incubation, Supplementary Figure S6b = 5 Gy followed by 3 or 6 h incubation.
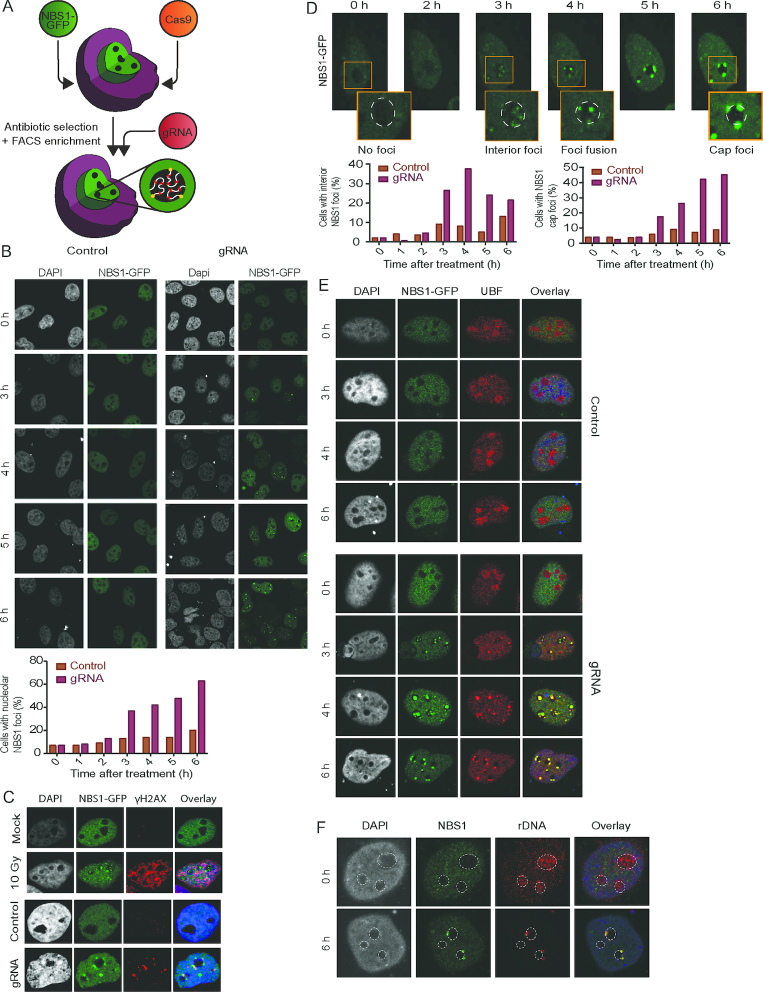
Figure 1.
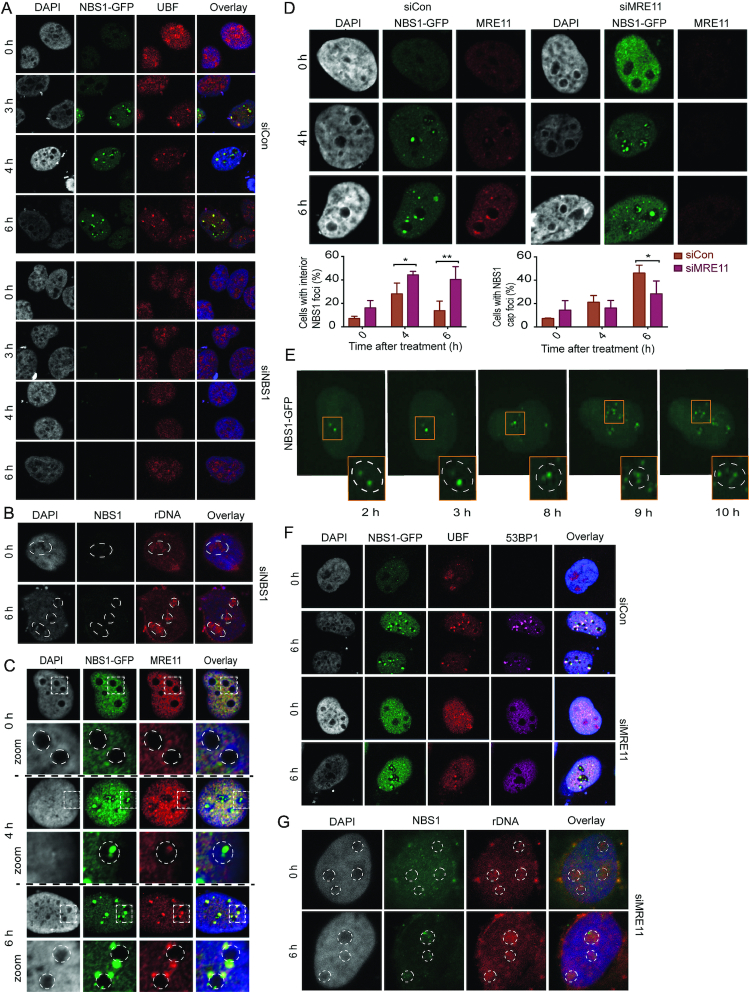
NBS1 is an early marker of rDNA-DSB. (A) Schematic drawing of the U2OS-based cell model with stable expression of the Cas9 endonuclease and GFP-tagged NBS1 (U2OS-Cas9-NBS1-GFP). Upon transient transfection of specific gRNAs, DSB induction can be targeted to rDNA. (B) Timecourse from 0 to 6 h after transfection of U2OS-Cas9 -NBS1-GFP cells with gRNAs. The percentage of cells with NBS1-positive nucleoli after transfection with gRNAs targeting rDNA or a control vector were counted and numbers are presented in the graph below (>100 cells/condition). (C) Co-localisation of NBS1 and γH2AX were examined in U2OS-Cas9-NBS1-GFP cells and immunostained with antibodies against γH2AX after IR (10 Gy, 1 h) or gRNA transfection (6 h post-transfection). (D) Timelapse images of NBS1-GFP foci up to 6 h post-transfection. Insets show enlarged individual nucleoli. Cells with NBS1-GFP interior foci and NBS1-GFP nucleolar caps were counted and the graphs below show the temporal pattern of interior and cap foci from one representative experiment (>200 cells/condition). Those cells that contain nucleoli with interior foci as well as nucleoli with caps were scored in both categories. (E) Co-localisation of NBS1 and UBF were investigated in U2OS-Cas9-NBS1-GFP cells immunostained with antibodies against UBF after 0, 3, 4 and 6 h after control vector or gRNA transfection. (F) Co-localisation of NBS1 and rDNA were investigated in U2OS-Cas9-NBS1-GFP cells by immuno-FISH after 0 and 6 h after gRNA transfection. Note, rDNA intensities cannot be compared directly between pictures as signal intensity has been adjusted to give a representative image of rDNA localisation. Dotted-lines mark the edge of nucleoli.
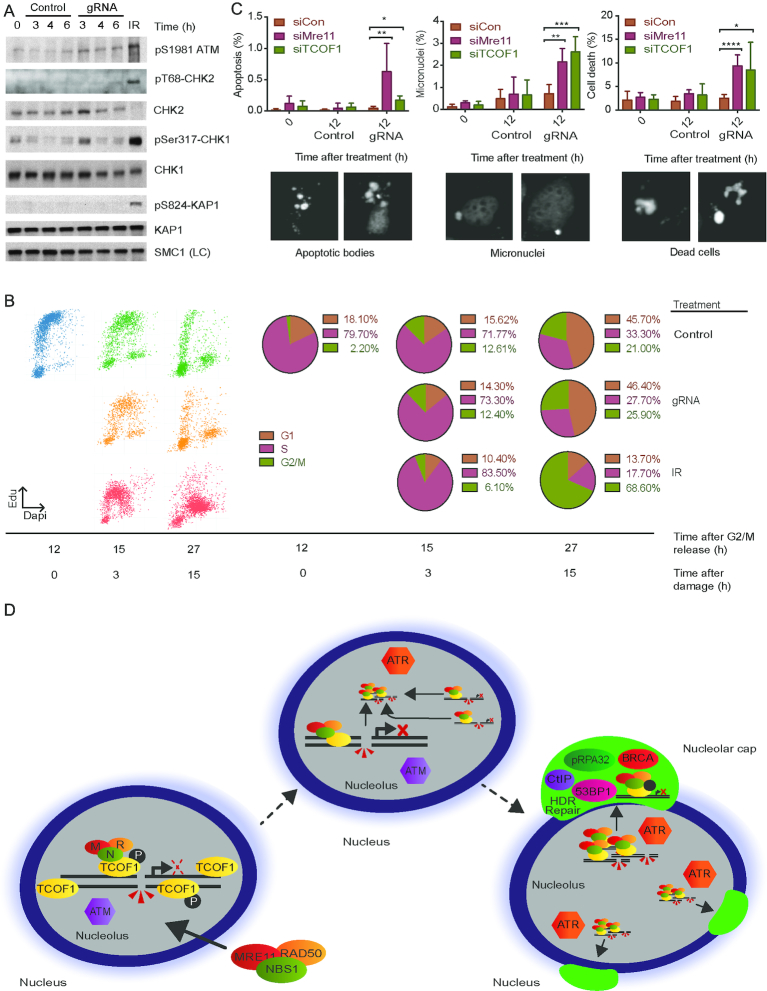
Figure 6.
Deregulation of rDNA repair undermines genomic integrity and cell viability. (A) Analysis of acute checkpoint activation in U2OS-Cas9-NBS1-GFP cells treated either with control vector or specific gRNAs. Samples were harvested at indicated time points for western blot analysis and activation of ATM was detected with an antibody against its modified form (pS1981 ATM). Activation of downstream targets of ATM and ATR were analysed using phospho-specific antibodies against CHK1, CHK2 and KAP1. SMC1 was included as a LC. IR-treated cells were included as a positive control. (B) G2/M checkpoint analysis. Cells were arrested in G2 with nocodazole, shaked off and released into the cell cycle. Twelve hours after release cells were treated with gRNAs or IR (positive control), labeled with EdU and harvested 15 and 27 h post-release. Samples were stained with DAPI and EdU incorporation was detected using Click-iT chemistry and analysed by quantitative image-based cytometry. The pie charts show the distribution of cells between G1, S and G2/M. (C) Phenotypes associated with a dysfunctional n-DDR. U2OS-Cas9-NBS1-GFP cells were treated with siRNAs against MRE11 and TCOF1, then treated with gRNAs and harvested at indicated time points. Three abnormal phenotypes were observed; micronuclei, apoptotic and dead cells represented in the image panel. These phenotypes were scored, and the data are presented in the graphs. Error bars represent SD, n = 3, multiple comparisons of the mean for all time points were carried out with unpaired t-tests and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns. (D) Proposed model of pathways coordinating the nucleolar DDR. rDNA DSBs activate the ATM kinase and induce ATM-dependent phosphorylation of TCOF1 to promote recruitment of the MRN complex and partial repression of rDNA transcription. In addition to ATM, consecutive ATR kinase activity is then required to fully repress transcription of rDNA and facilitate nucleolar cap formation.
EU incorporation
In situ detection of nascent RNA was done with the Click-iT RNA Alexa Fluor 594 Imaging Kit (Invitrogen, Molecular Probes #C10330). Briefly, cells were incubated for 25–30 min in the presence of 1 mM EU prior to harvest. Samples were fixed in 4% paraformaldehyde at room temperature for 12 min and permeabilized in 0.5% Triton X-100 for 5 min at room temperature. Samples were then processed according to the manufacturer's recommendation.
Cell synchronisation and EdU incorporation
U2OS-Cas9-NBS1-GFP cells were plated at 75% confluency and 0.04 μg ml−1 nocodazole was added to synchronise them at the entry into mitosis. Cells were incubated at 37°C for 12 h before being shaken off and spun down. Cells were washed 3 times in fresh medium and re-plated to release them from G2/M arrest. Twelve hours after release, cells were irradiated with 5 Gy or transfected with either gRNA or control vector and incubated with fresh medium for additional 0, 3 or 15 h before harvest. Prior to harvest, cells were incubated 1 h with 10 μM EdU. Samples were fixed in 4% paraformaldehyde at room temperature for 12 min and permeabilized in 0.5% Triton X-100 for 5 min at room temperature. Click-iT chemistry was used for the detection of EdU (Thermo Fischer Scientific, #C10340) and subsequently samples were incubated with 5 mg ml-1 DAPI (Invitrogen # D1206) in phosphate-buffered saline (PBS) for 2 min and analysed with Quantitative Image-based Cytometry (30). HEK 293T-Cas9 cells were plated at 75% confluency and doxycycline was added to a final concentration of 0.2 μg ml−1 to induce Cas9 expression as well as 0.04 μg ml−1 nocodazole for 12 h. Treatment and EdU incorporation was performed as stated above for the U2OS-Cas9-NBS1-GFP cells. The samples were harvested 0, 3 and 6 h after gRNA treatment by trypsinization and fixed in 75% cold ethanol. Click-iT chemistry was used for the detection of EdU (Thermo Fischer Scientific, #C10340) and subsequently samples were incubated with 1 mg ml-1 Hoechst 33342 (Invitrogen # H3570) in PBS for 30 min and analysed with BD FACSVerse (BD Biosciences). The data was processed using FlowJo.
Sample preparation, microscopy and image analysis
Cells were grown on glass coverslips and fixed in 4% paraformaldehyde at room temperature for 12 min and permeabilized in 0.5% Triton X-100 for 5 min at room temperature. Samples used for phospho-RPA staining were pre-extracted with 0.1% Triton X-100 in PBS on ice for 2 min, washed once with PBS and fixed as described above. Samples were incubated with primary antibody for 60 min at room temperature, 30 min with secondary and 5 mg ml-1 DAPI (Invitrogen # D1206) in PBS for 2 min. Cells were washed three times in PBS in between stainings, rinsed with water and mounted with Mounting media for fluorescence (VECTASHIELD, H-1000).
For analysis by spinning disc microscopy cells were plated in 1 μ-Slide 4-well ibiTreat well (IBIDI) and incubated in DMEMgfp-2 medium (EVROGEN) minimizing photobleaching. Transient transfection was carried out using Lipofectamine LTX with plus reagent (Invitrogen) and the cells were placed in UltraVIEW VoX Spinning Disk Confocal Microscope (PerkinElmer) at 37°C and 5% CO2. Cells with moderate NBS1-GFP signal were selected for imaging and movies were processed using iMovie (Apple).
Qualitative image analysis for fluorescence and co-colocalisation studies were carried out using the point scanning confocal microscope LSM700 (Zeiss), utilizing an ×63 oil objective and ZEN Software (Zeiss). The files were afterward processed using Adobe Photoshop CS6. The scoring of NBS1 nucleolar foci was performed manually including 75–200 cells per sample. For quantification in experiment 1b, 1d and 2b cells were scored depending on whether or not nucleolar NBS1 foci could be detected. For quantification in experiment 2c, S2a S2b, 3b, 3d, 3e, S3f, 4d, 5c and S5c the NBS1 foci were categorized either as interior foci or cap foci depending on their localisation. Based on the type of foci identified in each cell, they were scored as cells without NBS1 foci, cells with interior NBS1 foci and cells with NBS1 cap foci. If a cell had both interior NBS1 foci and cap foci it was included in both categories. To ensure the quality of the manual scoring-procedure it was evaluated by four independent scientists that repeatedly obtained comparable results.
Quantitative image analysis for measurement of fluorescence intensities was done as described previously (31). The images were obtained with a × 20 0.75 NA (UPLSAPO20x) dry objective, a quadruple-band filter set for DAPI and Cy3 fluorescent dyes, a MT20 Illumination system and a digital monochrome Hamamatsu C9100 EM-CCD (electron-multiplying charge-coupled device) camera. Camera resolution is 200 × 200 nM per pixel (binning 1, × 40). Image analysis was performed with Olympus propriety ScanR automated image and data analysis software using standard algorithms for detection of nuclei and sub-objects within nuclei. Typically, 49 images (corresponding to 2000–4000 sub-objects) were acquired under non-saturated conditions for each data point, allowing high-throughput measurements of experimental parameters such as intensities.
Experimental repetitions and statistics
All experiments were performed three times unless specified in the figure legends. Error bars represent the standard deviation in all figures. Where statistical tests were applied 75–200 cells were included to calculate the mean. Multiple comparisons of the mean for all time points were carried out with unpaired t-tests, and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns.
Real-time PCR
U2OS-Cas9-NBS1-GFP cells were treated with gRNA3 for 0–6 h. Cells were then collected, and DNA was isolated by DNeasy kit (Qiagen) according to the manufacturers protocol using pH adjusted (pH>9) nuclease-free water (Ambion) for elution. Real-time PCR was performed with Fast SYBR Green Master Mix (Thermo Fischer 4385612) with an amplification speed of 1 kb/min, using 2.5 ng DNA and 500 nM final primer concentration per sample. Samples were analysed on the LightCycler480 and quantified by the LightCycler480 software. The primer pair was designed to span the gRNA3 cut site, and the number of breaks was quantified by changes in the amplification of the PCR product relative to T0. Values were normalized to the GAPDH housekeeping gene. Quantifications were carried out at amplification cycle 21–23 for the gRNA3 cut site and cycle 28–30 for GAPDH. Statistical analysis was performed as described above with three repetitions in quadruplicates. For primer sequences see Suppl. Table S1.
Cell extracts and western blotting
Cell lysates, prepared in Laemmli sample buffer (with buffer volume adjusted to cell confluency) (1×LSB: 50 mM Tris–HCL pH6,8; 100 mM DTT; 2% sodium dodecyl sulphate (SDS); 0,1% bromophenol blue; 10% glycerol), were used in western blots included in Figure 2A, 2B, Suppl. Figures 1a and 3d. Alternatively, cell extracts were prepared in a clear denaturing lysis buffer (200 mM Tris–HCL (pH 6,8), 40% glycerol (99.5%), 8% SDS pure (95%)) and protein concentration was measured before adding DTT to a final concentration of 100 mM and bromophenol blue (1 μl per sample). These lystates were used for western blots included in Figure 6A, Suppl. Figures 3e, 4c and 5d. The samples were denatured by heating the samples to 95°C for 5 min. Proteins were separated using NuPAGE 3–8% Tris-Acetate gels or 4–12% Bis-Tris gels (Invitrogen) and transferred to membranes using iBlot Dry Blotting System (Invitrogen) following the manufacture's protocol.
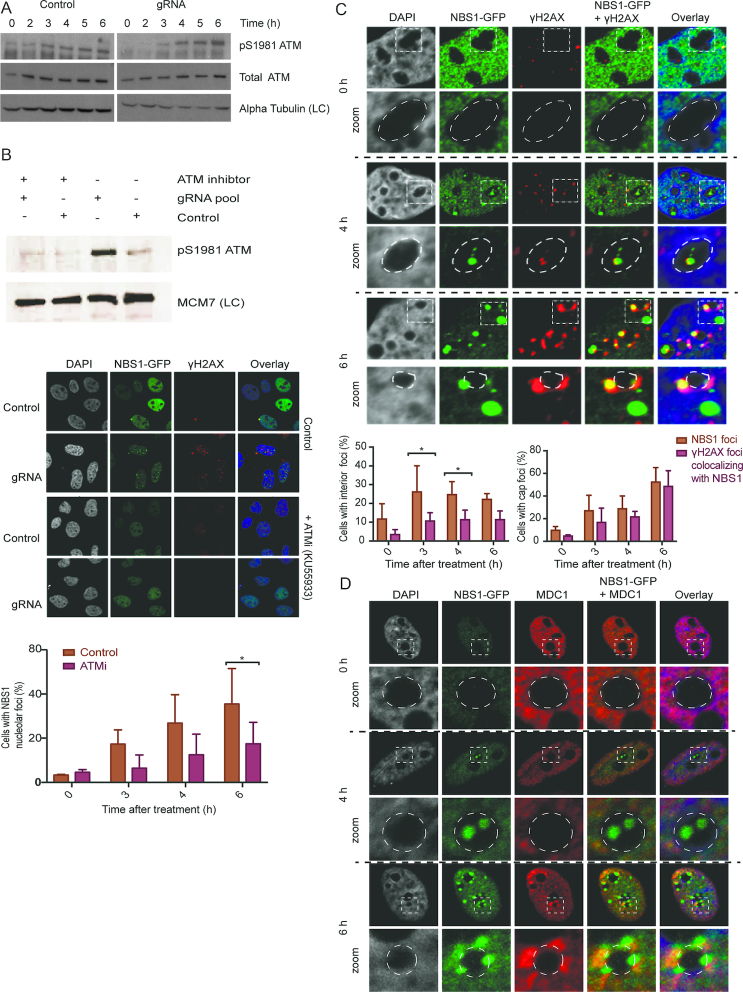
Figure 2.
The nucleolus harbours a distinct chromatin response and NBS1 recruitment mechanism after DSBs. (A) Western blot analysis to assay the activation of the ATM kinase (detected by phosphorylation of serine 1981; ATM pS1981) from 0–6 h after gRNA transfection. Total ATM shows ATM protein level. Alpha Tubulin was used as a loading control (LC). (B) Western blot analysis to evaluate the inhibition of ATM activity (ATM pS1981) by the ATM inhibitor KU55933. Cells were treated with control vector or gRNAs combined with DMSO or ATM inhibitor KU55933. MCM7 protein was used as a LC. Middle panel: IF analysis of the inhibition of NBS1-GFP foci formation in U2OS-Cas9-NBS1-GFP cells after control vector and gRNA transfection combined with DMSO or the ATM inhibitor KU55933 treatment. Lower panel: Count of cells with NBS1-GFP nucleolar foci upon ATM-inhibitor or DMSO treatment. Error bars represent SD, n = 3, multiple comparisons of the mean for all time points were carried out with unpaired t-tests. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns. (C) IF analysis was conducted to examine co-localisation of NBS1-GFP and γH2AX at indicated time points after gRNA transfection. Zoomed images show details of individual nucleoli with dotted lines marking the edge of the nucleoli. The graph below shows a count of cells with NBS1-GFP foci and cells where NBS1 and γH2AX co-localisation could be detected. Error bars represent SD, n = 4, multiple comparisons of the mean for all time points were carried out with unpaired t-tests, and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns. (D) Co-localisation of NBS1 and MDC1 as in (C) at indicated time points after gRNA transfection. Zoomed images show details of individual nucleoli with dotted lines marking the edge of the nucleoli.
ImmunoFISH
Cells grown on glass coverslips were fixed with 4% formaldehyde for 10 min at room temperature, then washed three times with PBS and permeabilized in PBS containing 0.5% saponin and 0.5% Triton X-100 for 10 min at room temperature. Next, samples were washed three times with PBS, incubated in PBS/20% glycerol for 2 h at room temperature, and snap-frozen in liquid nitrogen for 10 s. After thawing at room temperature, cells were denatured in 0.1 N HCl for 5 min at room temperature, washed briefly in 2× saline-sodium citrate (SSC) and incubated in 50% formamide/2× SSC for 15 min at 37°C. Before hybridisation, 3 μl of the Acro P-arm probe (Cytocell) were spotted on a microscope glass slide, then covered with the coverslips with the cells facing down and denatured by incubation at 73°C for 12 min after sealing the edges of the coverslip. The hybridisation was carried out at 37°C for 40 h. After the hybridisation, samples were washed twice in 0.4× SSC at 72°C and further two times in 2× SSC/0.05% Tween-20. Next, cells were washed three times with PBS and immunostained by incubating with primary antibody solution overnight at 4°C, followed by three washes with PBS and incubation with secondary antibody solution for 1 h at room temperature. At last, cells were washed three times with PBS and mounted on Vectashield containing DAPI for fluorescence.
Antibodies
Antibodies used for immunofluorescence (IF) in this study were: mouse γH2AX (Millipore, #16–193, 1:250), rabbit MDC1 (Abcam, #ab11171–50, 1:1000), rabbit TCOF1 (Sigma Life Science, #HPA038237, 1:200), mouse Mre11 (Abcam, #ab214, 1:500), rabbit phospho-RPA32 (S4/S8) (Bethyl, A300–245A, 1:200), rabbit 53BP1 (Santa Cruz #sc-22760, 1:500), rabbit NBS1 (Novus Biologicals, NB100–143, 1:200), mouse NBS1 (Genetex, Clone 1D7, GTX70224, 1:500), rabbit H2AX (Novus Biologicals, NB100–668, 1:800), mouse BRCA1 (Santa Cruz, #sc-6954, 1:100), mouse UBF (Santa Cruz, sc-13125), mouse IgG CtIP (Active motif, #61141, 1:10).
Antibodies used for western blots were: mouse NBS1 (Genetex, Clone 1D7, GTX70224, 1:500), rabbit NBS1 (Novus Biologicals, NB100–143, 1:500), mouse Cas9 (Abcam, #ab191468, 1:300), rabbit TCOF1 (Sigma Life Science, HPA038237, 1:500), mouse Mre11 (Abcam, 12D7, #ab214, 1:500), mouse MDC1 (Sigma, #M2444, 1:2000), rabbit H2AX (Novus Biologicals, NB100–668, 1:800), mouse γH2AX (Millipore, clone JBW302, #05–636, 1:500), rabbit phospho-1981-ATM (Abcam, #ab81292, 1:2000), mouse CHK1 (Santa Cruz, #sc-8408, 1:500), rabbit CHK1 pSer317 (Cell Signaling, #2344, 1:250), mouse CHK2 (Danish Cancer Society, 1:500), rabbit pCHK2 (Cell Signaling, #2661, 1:100), rabbit KAP1 (Bethyl, #A300–274A, 1:1000), rabbit pKAP1 (Abcam, #ab70369, 1:500), mouse alpha-tubulin (Santa Cruz, #sc-8035, 1:2000, IgM), mouse beta-importin (Abcam, #ab2811, 1:5000), mouse Mcm7 (Santa Cruz, DCS141, sc65469, 1:500), rabbit SMC1 (Abcam, #ab9262, 1:1000).
RESULTS
NBS1 is an early marker of rDNA-DSB
Previous studies have described how introduction of DSBs in the rDNA leads to transcriptional inhibition in the nucleolus and nucleolar cap formation (20–22). However, the events taking place during nucleolar re-organization and that are required for transcriptional inhibition remain poorly understood. To gain further insights we established a U2OS-based cell model stably expressing the endonuclease Cas9 and a GFP-tagged version of the DDR protein NBS1 (Figure 1A). Upon transfection with guide RNAs (gRNAs) targeting the rDNA (Supplementary Figure S1a and (22)) we can induce DSBs specifically in the nucleolus. We tested NBS1 as a marker of rDNA DSBs as it was previously shown to localise to the nucleolus in response to DNA damage (28,29). The expression level of GFP-tagged NBS1 in our cell model was found to be below that of the endogenous protein (Supplementary Figure S1b). In cells transfected with rDNA-specific gRNAs we observed gradual nucleolar accumulation of NBS1 (Figure 1B); 3 h after transfection the number of nucleolar NBS1 foci increased and this trend continued throughout the 6 h timecourse. This recruitment-pattern of NBS1 to nucleoli was confirmed in several sub-clones (Supplementary Figure S1c). To ensure that the nucleolar localisation reflected the behaviour of endogenous NBS1 and was not cell line specific we examined an independent cell line, the HT1080 I-PpoI cell model (32), in which the homing endonuclease I-PpoI induces DSBs in rDNA (33). In this cell line we confirmed the formation of nucleolar NBS1 foci upon induction of rDNA DSBs (Supplementary Figure S1d). Furthermore, to verify that the gRNAs specifically target rDNA, we examined the localisation of the DNA damage marker γH2AX 6 h after gRNA transfection and after IR. γH2AX could only be detected around the nucleolus where it co-localises with NBS1 in response to rDNA DSBs whereas IR generated a pan-nuclear response (Figure 1C). We then examined the spatial behaviour of NBS1 in the U2OS model in more detail using timelapse microscopy. We observed a reproducible pattern with initial formation of many small and mobile foci in the interior of the nucleolus (Figure 1D, 3 h post-transfection) that gradually fused into fewer and larger foci (Figure 1D, 4 h post-transfection) that ultimately moved to the nucleolar periphery and co-localise with nucleolar caps (Figure 1D, 6 h post-transfection). When interior and cap foci were quantified we observed an initial wave of interior foci peaking between 3 and 5 h followed by continuous accumulation of cap-foci (Figure 1D, lower panel). Similar spatial behaviour of NBS1 was confirmed using two independent cell models; the HT1080 I-PpoI cell line described above and the AID-DIvA (DIvA) cell line (15,34) expressing the AsiSI endonuclease that targets an 8-bp recognition site scattered throughout the genome and also present in rDNA (Supplementary Figure S1d and e). The kinetics of nucleolar NBS1 segregation and intensity of NBS1 foci varied between the three systems. In particular, a less pronounced nucleolar NBS1 response was observed in the DIvA cells, most likely reflecting fever rDNA DSBs as the transcriptional inhibition was also delayed compared to the U2OS-Cas9 cell model (data not shown). To confirm that NBS1 recruitment and translocation reflect the behaviour of rDNA we then stained for UBF, a protein bound to actively transcribed rDNA and performed rDNA FISH-analysis (Figure 1E and F). We found that NBS1 overlapped with UBF and rDNA throughout the timecourse and that NBS1 is present in all nucleolar caps. We generally used a pool of three gRNAs (with reduced concentration of each gRNA) but also tested that each of them individually induces rDNA damage (Supplementary Figure S1f). To estimate the number of rDNA repeats, cleaved by the Cas9 endonuclease in our cell model, we used qPCR to measure the fraction of intact sequences after transfection with gRNA3 and found that 6 h after DSB-induction 30 ± 8% (SD) of the rDNA is cleaved (Supplementary Figure S1g). The number of NBS1 foci detected, even at the earliest stages, is significantly lower suggesting that rapidly repaired breaks may not be visualized and/or that several breaks are clustered in each focus. These results show that our CRISPR/Cas9 cell model specifically induces DSBs in rDNA and that NBS1 can be used as a marker to follow the lesions at early stages within the interior of the nucleolus, through their translocation to nucleolar caps.
The nucleolus harbours a distinct chromatin response and NBS1 recruitment mechanism after DSBs
ATM is the major coordinator of the cellular response to DSBs and also regulates transcriptional inhibition and cap formation in the nucleolus (20–22). We therefore examined if induction of DSBs by the Cas9 endonuclease leads to ATM activation in our U2OS model. As expected from previous reports, we could detect activated ATM (ATM pS1981) in cells treated with gRNAs (Figure 2A). We then assayed if ATM activity is required for the formation of NBS1 nucleolar foci and found that pre-treatment of cells with the ATM inhibitor (KU55933) strongly inhibits NBS1 foci formation (Figure 2B). A previous study using MEFs from histone H2AX knockout mice indicated that nucleolar transcriptional inhibition is independent on H2AX (20). Furthermore, siRNA-mediated depletion of the mediator protein MDC1 in human cells showed only a very limited effect on rDNA transcriptional inhibition after DNA damage (28). We therefore investigated if these factors co-localise with NBS1 in the nucleolus after DSBs. In U2OS cells, we found a subset of cells showing ‘micro’ γH2AX foci in the nucleolar interior co-localising with NBS1 3 and 4 h after gRNA transfection (Figure 2C, middle panel and quantification below). However, after cap formation (Figure 2C, lower panel) an extensive γH2AX signal co-localising with NBS1 could be observed in all cells. Interestingly, at the nucleolar periphery the γH2AX signal even extends into chromatin regions not associated with NBS1. These data suggest limited phosphorylation of H2AX immediately after DSBs induction but a more prominent induction of γH2AX once the breaks have translocated to the periphery and nucleolar caps have formed. We also examined the recruitment of MDC1 (Figure 2D), which we found completely excluded from the nucleolar interior and not co-localising with NBS1 prior to nucleolar segregation. Upon cap formation MDC1 was recruited to nucleolar caps where its localisation juxtaposes NBS1 foci (Figure 2D, lower panel). Depletion of MDC1 and H2AX also confirmed that interior NBS1 foci form independently of MDC1 and γH2AX (Supplementary Figure S2a and b). These observations show that the chromatin modifications induced immediately after DSB induction, and that facilitate interior foci formation in rDNA, are distinct from those induced in nuclear chromatin.
NBS1 is recruited to nucleolar foci by ATM-phosphorylated TCOF1 via SDT-like motifs
Next, we set out to investigate the mechanism underlying NBS1 recruitment to nucleolar DSBs. We and others have previously described an interaction between NBS1 and the nucleolar protein TCOF1 and their localisation to the nucleolus after IR (28,29). Upon induction of rDNA DSBs in cells, TCOF1 and NBS1 initially co-localise in the nucleolar interior, then during translocation and also later in nucleolar caps (Figure 3A). Co-localisation of endogenous NBS1 and TCOF1 was also confirmed in the DIvA cell line (Supplementary Figure S3a). We then depleted TCOF1 using siRNA, both in U2OS-Cas9-NBS1-GFP (Figure 3B) and DIvA cells (Supplementary Figure S3b), and found DSB-induced NBS1 foci in the interior of the nucleolus abrogated in the absence of TCOF1. As anticipated cap formation was also impaired, although not completely diminished, at later time points under these conditions (Supplementary Figure S3c). These results were confirmed with two independent siRNAs against TCOF1 (Supplementary Figure S3d). To test whether loss of NBS1 recruitment in TCOF1-depleted cells reflects impaired cap formation we assayed the localisation of rDNA in TCOF1-depleted cells after rDNA DSBs. We found that TCOF1-depleted cells retain their rDNA in the interior of nucleoli and fail to segregate rDNA and form nucleolar caps (Figure 3C). However, some restructuring of rDNA still occurs after TCOF1 depletion in the interior of nucleoli, possibly reflecting ATM-mediated repression and clustering activities upstream of TCOF1. Alternatively, residual TCOF1 may result in a partial response to rDNA DSBs. To rule out that TCOF1 depletion could impair nucleolar ATM activation we examined ATM pS1981 but did not detect any impairment in the absence of TCOF1 (Supplementary Figure S3e). In nuclear chromatin, recruitment of NBS1 to DSBs occurs via binding of the FHA-domain in NBS1 to a phosphorylated SDT-motif in MDC1 (8,9). We previously identified three motifs in TCOF1 that resemble the SDT-motif of MDC1 capable of binding NBS1, and showed that alteration of the serine and threonine in these motifs, the so called STTT-mutant (Supplementary Table S1), abrogated TCOF1-NBS1 binding in vitro (28). With functional in cellulo reconstitution we could now show that this motif is required for the recruitment of NBS1 to rDNA after DSBs, as the amount of NBS1 interior and cap foci in STTT-complemented cells were similar to that in non-complemented TCOF1-depleted cells (Figure 3D and Supplementary Figure S3f). Phosphoproteomics have identified TCOF1 as a putative ATM/ATR substrate (35), and a previous study identified the SQ-site at position 1199 in TCOF1 as required for recruitment of NBS1 into the nucleolus after IR. In our hands however, mutation of serine-1199 to alanine only partially abrogated NBS1 recruitment into nucleoli after rDNA DSBs (Supplementary Figure S3g). In total TCOF1 contains 17 SQ/TQ sites and we therefore mutated the 17 SQ/TQ sites to AQ-sites, creating an ATM-null mutant (Supplementary Figure S3f and Supplementary Table S1). We then performed functional reconstitution experiments with this ATM-null version of TCOF1 and showed that NBS1 nucleolar foci formation depends on ATM phosphorylation of TCOF1 (Figure 3E). In summary, we describe how NBS1 is recruited to rDNA by binding the SDT-like motifs of ATM-phosphorylated TCOF1.
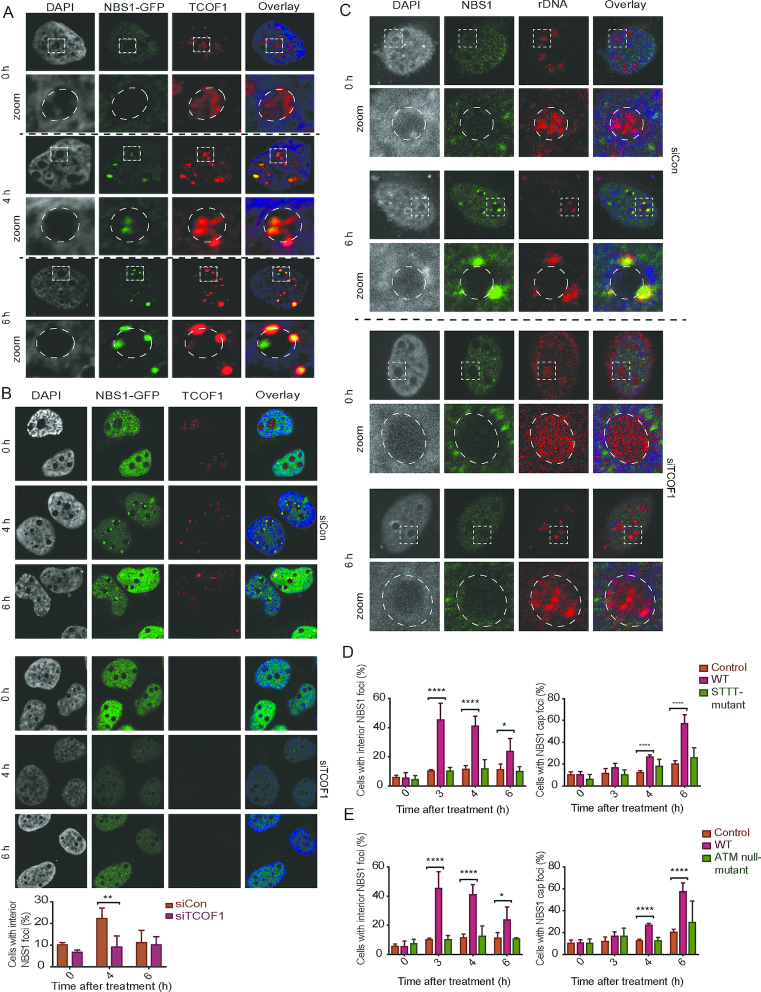
Figure 3.
NBS1 is recruited to nucleolar foci by ATM-phosphorylated TCOF1 via SDT-like motifs. (A) Co-localisation of NBS1 and TCOF1 in U2OS-Cas9-NBS1-GFP cells immunostained with antibodies against TCOF1 at indicated time points after gRNA transfection. Zoomed images show details of individual nucleoli with dotted lines marking the edge of the nucleoli. (B) Impact of TCOF1 depletion on NBS1 foci formation. Cells were treated with control siRNA or siRNA against TCOF1 for 60 h and treated with gRNAs. Samples were immunostained with antibodies against TCOF1 to confirm the knock down and NBS1-GFP nucleolar foci were examined. The graph below shows a quantification of the percentage of cells with interior NBS1 foci in TCOF1-depleted and control cells. Error bars represent SD, n = 3, multiple comparisons of the mean for all time points were carried out with unpaired t-tests, and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns. (C) Co-localisation of NBS1 and rDNA were investigated in U2OS-Cas9-NBS1-GFP cells, after depletion of TCOF1, by immuno-FISH at indicated time points after control vector or gRNA transfection. Zoomed images show details of individual nucleoli with dotted lines marking the edge of the nucleoli. Note, rDNA intensities cannot be compared directly between pictures as signal intensity has been adjusted to give a representative image of rDNA localisation. (D) Functional complementation assays. Cells were transfected with shRNA against TCOF1 for 72 h and concomitantly complemented with an shRNA resistant control vector, wt or STTT-mutant version of TCOF1. Cells were then transfected with gRNAs, fixed and analysed for nucleolar NBS1 foci. The graph displays the percentage of cells with NBS1 interior or cap foci (>100 cells/condition). Error bars represent SD, n = 3, multiple comparisons of the mean for all time points were carried out with unpaired t-tests, and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns. (E) Functional complementation as in D using the TCOF1 ATM-null mutant for complementation (>100 cells/condition). Error bars represent SD, n = 3, multiple comparisons of the mean for all time points were carried out with unpaired t-tests, and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns.
NBS1 functions as part of the MRN-complex that facilitates translocation of nucleolar DSBs to nucleolar caps
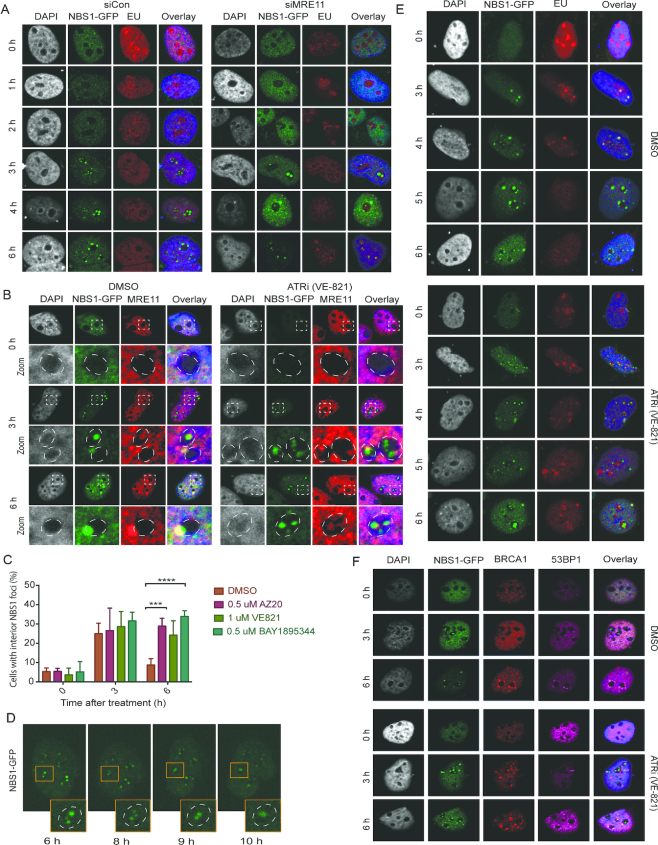
The NBS1–TCOF1 complex is required for transcriptional inhibition in the nucleolus after DSBs (20,28) and we therefore carefully correlated NBS1 focus formation and inhibition of nucleolar transcription. We find that nucleolar transcriptional inhibition is partial at the time of NBS1 foci formation and that complete inhibition of rDNA transcription occurs with delayed kinetics compared to NBS1 accumulation (Supplementary Figure S4a), suggesting that transcriptional inhibition may rely on events downstream of foci formation. We therefore tested if NBS1 is required for nucleolar cap formation by NBS1 depletion after rDNA DSBs. We assayed nucleolar caps by UBF staining (Figure 4A) and immuno-FISH detecting rDNA (Figure 4B) and found translocation of both UBF and rDNA to be impaired after NBS1 depletion. These findings support a role of NBS1 in transcriptional repression of nucleolar rDNA upon rDNA DSBs. Previous studies, including our own, suggested that the nucleolar role of NBS1, as a transcriptional regulator together with TCOF1, was independent of the MRE11–Rad50–NBS1 (MRN) complex, in which NBS1 functions as a repair protein (28–29,36). To gain further insights into the function of NBS1 after rDNA DSBs we therefore investigated if NBS1 functions as part of the MRN-complex in the nucleolus after induction of rDNA DSBs. We initially investigated MRE11 localisation in our U2OS-Cas9-NBS1-GFP cell system and despite that MRE11 foci are less pronounced we could clearly detect co-localisation of MRE11 and NBS1 in the nucleolus, both at early time points after DSB-induction, when interior foci are dominant, and also at later time points where cap formation has occurred (Figure 4C). The co-localisation of endogenous NBS1 and MRE11 could also be detected in the DIvA and HT1080 I-PpoI cell models (Supplementary Figure S4b). We then investigated the effect of MRE11 depletion on NBS1 foci formation. Unexpectedly, we found that MRE11-depleted cells show increased accumulation of interior NBS1 foci (Figure 4D) and these foci remain trapped in the interior resulting in a reduced number of NBS1 positive nucleolar caps (Figure 4D, lower panels). These findings were confirmed using an independent siRNA against MRE11 (Supplementary Figure S4c). When we follow NBS1 foci after MRE11 depletion using timelapse microscopy, we find that the initial foci-stage in the interior is sustained while the segregation of rDNA into nucleolar caps is strongly impaired (Figure 4E). To ensure that NBS1 is still bound to rDNA and represents cap formation under these conditions, we examined the accumulation of rDNA in the interior by UBF staining and rDNA FISH analysis in MRE11-depleted cells (Figure 4F and 4G). We find nucleolar cap formation strongly impaired in MRE11-depleted cells. We also investigated the recruitment of the repair protein 53BP1 that associates with rDNA after cap formation (Figure 4F) (21,22). We did not detect recruitment of 53BP1 to NBS1-positive nucleoli either in MRE11-depleted cells further supporting an impairment of nucleolar cap formation in response to rDNA DSBs. In summary, these findings show that NBS1 functions as part of the MRN-complex in nucleoli after rDNA DSBs and that, in addition to ATM activation, the MRN-complex is required for translocation of rDNA DSBs to nucleolar caps.
Figure 4.
NBS1 functions as part of the MRN-complex that facilitates translocation of nucleolar DSBs to nucleolar caps. (A) Localisation of UBF in NBS1-depleted cells. IF analysis was conducted in U2OS-Cas9-NBS1-GFP cells immunostained with antibodies against UBF at indicated time points after gRNA transfection. (B) Localisation of rDNA was analysed in NBS1-depleted U2OS-Cas9-NBS1-GFP cells by immuno-FISH at indicated time points after gRNA transfection. Note, rDNA intensities cannot be compared directly between pictures as signal intensity has been adjusted to give a representative image of rDNA localisation. Dotted-lines mark the edge of the nucleoli. (C) Co-localisation of MRE11 and NBS1 after gRNA transfection in U2OS-Cas9-NBS1-GFP cells immunostained with antibodies against MRE11 at indicated time points. Zoomed images show details of individual nucleoli with dotted lines marking the edge of the nucleoli. (D) Effect of MRE11 depletion on NBS1-GFP foci translocation after gRNA treatment. U2OS-Cas9-NBS1-GFP cells were depleted for MRE11 for 60 h and subsequently transfected with gRNAs and harvested at indicated time points. Samples were immunostained with antibodies against MRE11. The graphs below show count of cells with NBS1-GFP nucleolar foci. Error bars represent SD, n = 3, multiple comparisons of the mean for all time points were carried out with unpaired t-tests, and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns. (E) Temporal behaviour of NBS1-GFP foci up to 10 h post-gRNA transfection in MRE11-depleted U2OS-Cas9-NBS1-GFP analysed using timelapse microscopy. Insets show enlarged individual nucleoli. (F) Co-localisation of NBS1-GFP and UBF after MRE11 depletion were investigated in U2OS-Cas9-NBS1-GFP cells immunostained with antibodies against UBF after 0 and 6 h after control vector or gRNA transfection. Recruitment of repair proteins was examined by concomitant immunostaining against 53BP1. (G) Localisation of rDNA was analysed in MRE11-depleted U2OS-Cas9-NBS1-GFP cells by immuno-FISH at indicated time points after gRNA transfection. Dotted-lines mark the edge of nucleoli. Note, rDNA intensities cannot be compared directly between pictures as signal intensity has been adjusted to give a representative image of rDNA localisation.
The ATR kinase facilitates repression of nucleolar transcription and nucleolar cap formation
Restructuring of nucleoli and localisation of rDNA to nucleolar caps is driven by the inhibition of rRNA transcription (21). To understand the role of the MRN-complex in translocation of rDNA we measured repression of rRNA transcription after DNA damage. We find that in MRE11-depleted cells rRNA transcription is maintained, even though slightly reduced, even 4–6 h after induction of DSBs, in contrast to MRE11-proficient cells that repress transcription after 2 h (Figure 5A). These results show that the transcriptional repression mechanism, initiated by ATM and leading to the restructuring of nucleoli, likely operates through the MRN-complex and that this step is needed to fully repress nucleolar transcription and to translocate breaks to the periphery. MRE11 is known to promote the initial steps of the DSB resection pathway and we therefore examined if resection takes place in the interior of the nucleolus. As a read-out for resection we examined levels of RPA32 pS4/8, specifically induced by DSBs, in nucleolar NBS1 foci (Supplementary Figure S5a). We did not observe consistent co-localisation of RPA32 pS4/8 and NBS1 at the 4h time point, whereas this was much more pronounced after 6 h in nucleolar caps.
Figure 5.
The ATR kinase facilitates repression of nucleolar transcription and nucleolar cap formation. (A) Measurements of transcription by 5-EU incorporation in MRE11-depleted U2OS-Cas9-NBS1-GFP cells treated with gRNAs at indicated time points. Prior to fixation cells were labeled with 5-EU and samples were subsequently processed with Click-iT chemistry. (B) Analysis of NBS1-GFP nucleolar foci after ATR inhibition. U2OS-Cas9-NBS1-GFP cells were treated with the indicated ATR inhibitor and transfected with gRNAs. Samples were harvested at indicated time points and immunostained with antibodies against MRE11. Zoomed images show details of individual nucleoli with dotted lines marking the edge of the nucleoli. (C) Count of cells with NBS1-GFP interior nucleolar foci from experiments carried out as in b using three ATR inhibitors. Error bars represent SD, n = 3, multiple comparisons of the mean for all time points were carried out with unpaired t-tests, and the two-sided P-values were adjusted using the Holm–Sidak method. Statistical significance is depicted with stars (* = 0.05–0.01, ** = 0.01–0.001, *** = 0.001–0.0001, **** ≤ 0.0001) and no legend = ns. (D) Temporal behaviour of NBS1-GFP foci up to 10 h after gRNA transfection combined with ATR inhibition (VE821) analysed using timelapse microscopy. Insets show enlarged individual nucleoli. (E) Measurements of transcription by 5-EU incorporation in ATR inhibitor and gRNA treated U2OS-Cas9-NBS1-GFP cells. Prior to fixation cells were labeled with 5-EU and samples were subsequently processed with Click-iT chemistry. (F) Recruitment of repair proteins after ATR inhibition. Staining patterns of NBS1-GFP, BRCA1 and 53BP1 were analysed by immunostaining with antibodies against BRCA1 and 53BP1 in cells treated with ATR inhibitor and gRNA.
CtIP interacts with MRE11 and is also needed for resection (37) and we therefore examined the recruitment of CtIP to rDNA DSBs but could only detect NBS1-CtIP co-localisation in nucleolar caps (Supplementary Figure S5b). We then depleted CtIP using siRNA (Supplementary Figure S5c) and found no significant change in interior foci but a pronounced abrogation of NBS1 accumulation in nucleolar caps (Supplementary Figure S5c), suggesting that CtIP processing of rDNA DSBs promotes NBS1-GFP association with nucleolar caps after translocation of interior breaks to the periphery. As previous studies have shown a recruitment of repair factors, such as 53BP1 and BRCA1, after DNA damage-induced cap formation (21,22), we also examined the impact of CtIP depletion on accumulation of DNA repair factors. CtIP depletion abrogated both 53BP1 and BRCA1 recruitment to nucleolar caps suggesting that breaks after translocation to the nucleolar periphery are preferentially repaired by HDR pathways (Supplementary Figure S5c and d). In summary, these findings suggest that resection primarily occurs after formation of nucleolar caps allowing recognition of breaks by HDR factors but does not impact on the initial steps preceding cap formation.
These findings prompted us to consider other mechanisms by which MRE11 could promote transcriptional repression of the rDNA. Interestingly, a previous study showed that overexpression of TopBP1 can induce ATR-dependent silencing of nucleolar transcription (38). As the MRN complex can function as a recruiter and activator of the TopBP1-ATR signaling (39–42), we wondered if ATR may be the kinase activated downstream of the MRN complex leading to nucleolar transcriptional silencing. We therefore examined the translocation of NBS1 foci after treatment with an ATR inhibitor and observed a similar phenotype to that of MRE11-depleted cells with increased accumulation of rDNA in the nucleolar interior (Figure 5B). We extended these investigations using three different ATR inhibitors (VE821, AZ20 and BAY 1895344) and found that all three inhibitors led to the accumulation of interior nucleolar foci (Figure 5C). We also followed cells treated with an ATR inhibitor using timelapse microscopy and observed an extended period of small mobile interior NBS1-GFP foci resembling the behaviour of MRE11-depleted cells (Figure 5D). We then tested if ATR inhibitors impair MRE11 foci formation, but found that MRE11 is recruited to nucleolar foci as in control cells (Figure 5B), supporting the notion that ATR signaling is functioning downstream of the MRN complex. Consistently, treatment with ATR inhibitors also impaired transcriptional silencing similarly to MRE11 depletion (Figure 5A and E). Notably, when we examined BRCA1 and 53BP1 accumulation in cells treated with ATR inhibitors, we noticed an increased accumulation of BRCA1 in the interior nucleolar space but did not detect any changes in 53BP1 recruitment (Figure 5F). These findings indicate that impaired segregation may influence the compartmental association between rDNA and repair factors such as BRCA1. In summary, we identify a downstream role of ATR required for inhibition of rRNA transcription and for the formation of nucleolar caps to facilitate HDR.
Deregulation of rDNA repair undermines genomic integrity and cell viability
To understand the consequences of dysfunctional regulation of rDNA repair we examined the global cellular response to rDNA damage. Unexpectedly, immunoblot analysis of checkpoint protein activation showed that while ATM becomes activated by rDNA DSBs, it activates neither the checkpoint kinase CHK1 nor CHK2, detected by phosphorylation of serine-317 and threonine-68, respectively (Figure 6A). Likewise, we found no activating phosphorylation of KAP1 (phosphorylation of serine-824) (Figure 6A). These findings show that the n-DDR signaling in response to rDNA DSBs does not induce a global checkpoint response, contrary to DSBs that occur in nuclear chromatin. If rDNA DSBs do not initiate signaling cascades they potentially also fail to activate cellular checkpoints. To test this hypothesis, we synchronised cells using nocodazole and released them for 12 h. We then transfected them with gRNA or exposed them to 5 Gy of IR to induce DSBs and followed their progression through G2, M and into the following G1 phase (Figure 6B and Supplementary Figure S6a). Cells treated with 5 Gy of IR progress through S phase (3 h post-damage) until they reach the G2/M border where they arrest (6 and 15 h post-damage). In contrast, cells with rDNA DSBs behave similarly to control cells, progressing through mitosis and continuing into the subsequent G1 with no indications of cell-cycle arrest (6 and 15 h post-damage). To exclude that this phenotype is specific to the U2OS cell line we also examined cell-cycle progression in human embryonic kidney 293T (HEK 293T) cells following rDNA damage. In the HEK 293T cells induction of rDNA DSBs also did not activate a G2/M checkpoint response (Supplementary Figure S6b). These findings reveal that rDNA DSBs only activate a localised ATM response that is not transmitted to the surrounding nucleus and does not activate an acute checkpoint response.
The lack of checkpoint activation suggests that cells will progress through mitosis with damaged rDNA. We therefore speculate that abrogation of the n-DDR-mechanisms that ensure accurate processing of DSBs may become toxic to cells. We depleted TCOF1 or MRE11 prior to induction of rDNA DSBs and followed cells for 12 h after damage induction. Both in cells depleted of TCOF1 and MRE11, we found a clear increase in the number of cells with abnormal nuclear morphology (Figure 6C). We divided the abnormal nuclear morphology into the following categories: apoptotic cells, micronuclei and dead cells. For all three phenotypes we found that depletion of MRE11 or TCOF1 in combination with gRNAs lead to a significant increase 12 h after gRNA treatment compared to control samples. These findings suggest that both the initial coordination by TCOF1 and the downstream role of MRE11 are essential for maintenance of genome integrity and cellular survival upon rDNA DSBs.
DISCUSSION
In this study we describe the molecular pathway coordinating nucleolar DDR signaling and repair of DSBs in the highly transcribed rDNA sequences. We conclude that ATM phosphorylates the nucleolar protein TCOF1, which functions as a damage-induced docking platform for the NBS1-subunit of the MRN complex through binding of the STTT-motifs and ATM-mediated phosphorylation sites in TCOF1. Recruitment of the MRN complex is required to translocate rDNA DSB from the interior to the periphery of the nucleolus, documented by insufficient repression of rDNA transcription in MRE11-depleted cells. In the n-DDR pathway described here, in addition to ATM, the ATR kinase is required to fully suppress nucleolar transcription and facilitates nucleolar cap formation (Figure 6D). Furthermore, unlike the canonical DSB response to IR (40), we show that the nucleolar DSB-induced ATM–ATR pathway does not activate CHK1/CHK2 kinases and, consequently, fails to evoke the cellular checkpoint response. Nevertheless, the nucleolar DDR plays an important role in guarding genome integrity.
In the canonical DDR the MRN complex is recruited to sites of DSBs either by direct binding to DNA or by interaction with the mediator protein MDC1 (7). In this study we find MDC1 exclusion from the nucleolus and show that in rDNA the nucleolar protein TCOF1, recruits the MRN complex to DSBs. Depletion of TCOF1 completely abrogates NBS1 localisation to the nucleolus and direct binding of DSBs in rDNA could not be detected (Figure 3B) as it has been suggested in nuclear chromatin (43). Interestingly TCOF1 contains structural motifs that resemble the NBS1-binding domain in MDC1 (8,9) and we show that deletion of four serines/threonines prevent NBS1 recruitment to the nucleolus upon rDNA DSBs. These findings suggest that TCOF1 functions as a nucleolar paralog of MDC1 to facilitate MRN association with sites of DNA damage.
TCOF1 is abundant throughout the entire rDNA region and one question of interest is how NBS1 recruitment is limited to specific regions after rDNA DSBs. We find that abrogation of the 17 SQ/TQ motifs, representing potential ATM phosphorylation sites in TCOF1, prevents recruitment of NBS1 after DSB induction (Figure 3D). We therefore propose that activation of ATM leads to local phosphorylation of TCOF1 that enables MRN recruitment specifically to DSB-associated chromatin regions. This mechanism resembles that of γH2AX–MDC1–MRN in nuclear chromatin, but in nucleolar chromatin TCOF1 is both the ATM target as well as the direct interaction partner of NBS1, while γH2AX is less critical for the interior nucleolar response.
Previous studies have described ATM activation as the driving force leading to transcriptional inhibition and rDNA relocalisation into nucleolar caps (20–22,28,44). It was therefore unexpected that depletion of MRE11 prevented the translocation of breaks from the nucleolar interior to the periphery (Figure 4D). An obvious explanation could be impaired ATM activation, yet we see equivalent and even sustained accumulation of NBS1, a response dependent on ATM activity (Figures 2B and 4D). MRE11 could facilitate translocation by several other means and our data suggest that this likely involves the ATR kinase. However, the interplay between MRE11 and ATR is currently not clear and will be a topic for further investigation. MRE11 can activate ATR in nuclear chromatin by generation of a single-stranded substrate and recruitment of TopBP1 (39–40,42). Interestingly, high levels of TopBP1 were shown to silence nucleolar transcription in an ATR dependent manner (38) suggesting that a similar mechanism operates in the nucleolus. Several ATM/ATR targets have been identified in the nucleolus and ATR may therefore play a role downstream of ATM to silence rRNA transcription. Notably, a broadly analogous two-step activation mechanism for ATM and ATR has been described in response to IR in nuclear chromatin (40). Further investigations will clarify to what extent these observations are transferrable to the nucleolar DDR. The role of ATR in repression of nucleolar transcription does not exclude a continued requirement for the ATM kinase in formation of nucleolar caps, and the precise interplay between the kinases and their respective targets remains a topic of investigation. The experiments included in this study, examining the role of ATR in repair of nucleolar DSBs has been conducted using the Cas9/gRNA for induction of DSBs. Further investigations of this mechanism should include DSBs induced by other means including other endonucleases and/or IR to assay its applicability to a broader range of DSBs.
Interestingly, NBS1 and MRE11 have previously been linked to clustering of DNA breaks in actively transcribed sequences (45) that coincide with delayed repair. Together with our present findings, this suggests that the repair pathway described here in rDNA might generally be used by actively transcribed sequences.
At last, we show that nucleolar activation of ATM and ATR does not activate the canonical signaling cascades as judged from the lack of CHK1, CHK2 and KAP1 activation (Figure 6A) and does not induce cell-cycle arrest at the G2/M border. This may be due to local activation of ATM in nucleoli, also reported after localised micro laser-irradiation (20), leading to physical separation of ATM/ATR and their substrates, as we cannot detect neither of the CHK-kinases in purified nucleoli (data not shown). As the rDNA is intrinsically error-prone this mechanism may have developed to make ribosome biogenesis compatible with proliferation.
In previous studies, including work in which we have been involved, MRE11 could not be detected in ionizing radiation-induced nucleolar foci (28,29), whereas MRE11 co-localises with NBS1 in nucleolar foci induced by CRISPR/Cas9. An explanation for this is possibly that IR induces a wave of damage that does not immediately lead to nucleolar restructuring. Damage induced by CRISPR/Cas9 is sustained and induces break clustering, translocation and cap formation. It is therefore possible that clustering of breaks is necessary to bring MRE11 above the detection level. In addition, the repair pathway choice may influence the recruitment and detection of MRE11 in nucleolar foci.
The major advantage that can be envisaged from nucleolar restructuring is the separation of ribosomal RNA genes from different chromosomes prior to HDR repair as has already been proposed (14). Physical separation of chromosomes can prevent inter-chromosomal recombination that would have fatal consequences in mitosis and lead to the loss of large amounts of genetic material. A two-step mechanism of the repair process may reflect an initial attempt to repair the breaks by NHEJ in the interior and only when such attempts fail breaks are translocated to nucleolar caps. In agreement with this hypothesis NHEJ factors such as DNA-PKcs, XRCC4 and Ku80 can be found in the interior of nucleoli after DSB induction (46) (and data not shown). This could also explain the initial mobile and dynamic focal pattern observed prior to translocation of breaks. The consequences of a compromised nucleolar-DDR still await further investigations both with regards to cell physiology and its suggested involvement in disease (47). Our data show that the n-DDR is of outmost importance for overall cellular survival and that depletion of nucleolar repair factors lead to pronounced chromosomal abnormalities and increase in cell death.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Claus Storgaard Sørensen, Christoffel Dinant and Daniel Gómez Cabello for discussion and constructive comments for the manuscript, Louise Heidemann Jensen for technical assistance, Riccardo Vanzo for statistical advice and Signe Lilja for help with graphical design. We thank Gaelle Legube for sharing the AID-DIvA U2OS cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Cancer Society-Knæk Cancer [R209-A12925]; Danish Cancer Society [R146-A9403]; Lundbeck Foundation [R192-2015-335, R173-2014-1105, R219-2016-1375]; Danish Research Council for Independent Research [6110-00506A, DFF-1337-00128, DFF-1335-00763A and project CARD]; Innovation Fund Denmark (BrainStem); Aarhus University Strategic Grant (AU-iCRISPR); BGI. Funding for open access charge: Danish Cancer Society Research Center, Lundbeck Foundation [R192-2015-335]; Danish Cancer Society [R146-A9403]; The Independent Research Council CARD.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001; 411:366–374. [DOI] [PubMed] [Google Scholar]
- 3. Lee J.-H., Paull T.T.. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007; 26:7741–7748. [DOI] [PubMed] [Google Scholar]
- 4. Williams R.S., Williams J.S., Tainer J.A.. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007; 85:509–520. [DOI] [PubMed] [Google Scholar]
- 5. Bartek J., Lukas J.. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007; 19:238–245. [DOI] [PubMed] [Google Scholar]
- 6. Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P.. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005; 123:1213–1226. [DOI] [PubMed] [Google Scholar]
- 7. Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J.. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006; 173:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J.. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 2008; 181:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spycher C., Miller E.S., Townsend K., Pavic L., Morrice N.A., Janscak P., Stewart G.S., Stucki M.. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 2008; 181:227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shibata A., Jeggo P.A.. DNA double-strand break repair in a cellular context. Clin. Oncol. (R. Coll. Radiol.). 2014; 26:243–249. [DOI] [PubMed] [Google Scholar]
- 11. Murray J.M., Stiff T., Jeggo P.A.. DNA double-strand break repair within heterochromatic regions. Biochem. Soc. Trans. 2012; 40:173–178. [DOI] [PubMed] [Google Scholar]
- 12. Chiolo I., Minoda A., Colmenares S.U., Polyzos A., Costes S.V., Karpen G.H.. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011; 144:732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemaître C., Soutoglou E.. Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair. DNA Repair (Amst.). 2014; 19:163–168. [DOI] [PubMed] [Google Scholar]
- 14. van Sluis M., McStay B.. Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 2017; 46:81–86. [DOI] [PubMed] [Google Scholar]
- 15. Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P. et al.. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014; 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stults D.M., Killen M.W., Pierce H.H., Pierce A.J.. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008; 18:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mangan H., Gailín M.Ó, McStay B.. Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 2017; 284:3977–3985. [DOI] [PubMed] [Google Scholar]
- 18. Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D.. Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 2007; 129:13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Killen M.W., Stults D.M., Adachi N., Hanakahi L., Pierce A.J.. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 2009; 18:3417–3428. [DOI] [PubMed] [Google Scholar]
- 20. Kruhlak M., Crouch E.E., Orlov M., Montaño C., Gorski S.A., Nussenzweig A., Misteli T., Phair R.D., Casellas R.. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007; 447:730–734. [DOI] [PubMed] [Google Scholar]
- 21. Harding S.M., Boiarsky J.A., Greenberg R.A.. ATM dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. Cell Rep. 2015; 13:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Sluis M., McStay B.. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015; 29:1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warmerdam D.O., van den Berg J., Medema R.H.. Breaks in the 45S rDNA lead to Recombination-Mediated loss of repeats. Cell Rep. 2016; 14:2519–2527. [DOI] [PubMed] [Google Scholar]
- 24. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering Using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsouroula K., Furst A., Rogier M., Heyer V., Maglott-Roth A., Ferrand A., Reina-San-Martin B., Soutoglou E.. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell. 2016; 63:293–305. [DOI] [PubMed] [Google Scholar]
- 27. Vad-Nielsen J., Lin L., Bolund L., Nielsen A.L., Luo Y.. Golden Gate Assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes. Cell. Mol. Life Sci. 2016; 73:4315–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larsen D.H., Hari F., Clapperton J.A., Gwerder M., Gutsche K., Altmeyer M., Jungmichel S., Toledo L.I., Fink D., Rask M.-B. et al.. The NBS1–Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell Biol. 2014; 16:792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ciccia A., Huang J.-W., Izhar L., Sowa M.E., Harper J.W., Elledge S.J.. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18631–18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toledo L.I., Altmeyer M., Rask M.-B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J.. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013; 155:1088–1103. [DOI] [PubMed] [Google Scholar]
- 31. Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grøfte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S. et al.. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012; 150:697–709. [DOI] [PubMed] [Google Scholar]
- 32. Oka Y., Suzuki K., Yamauchi M., Mitsutake N., Yamashita S.. Recruitment of the cohesin loading factor NIPBL to DNA double-strand breaks depends on MDC1, RNF168 and HP1γ in human cells. Biochem. Biophys. Res. Commun. 2011; 411:762–767. [DOI] [PubMed] [Google Scholar]
- 33. Berkovich E., Monnat R.J., Kastan M.B.. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007; 9:683–690. [DOI] [PubMed] [Google Scholar]
- 34. Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G.. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010; 29:1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al.. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 36. Lindström M.S., Jurada D., Bursac S., Orsolic I., Bartek J., Volarevic S.. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene. 2018; 37:2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Makharashvili N., Paull T.T.. CtIP: A DNA damage response protein at the intersection of DNA metabolism. DNA Repair (Amst.). 2015; 32:75–81. [DOI] [PubMed] [Google Scholar]
- 38. Sokka M., Rilla K., Miinalainen I., Pospiech H., Syväoja J.E.. High levels of TopBP1 induce ATR-dependent shut-down of rRNA transcription and nucleolar segregation. Nucleic Acids Res. 2015; 43:4975–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams K.E., Medhurst A.L., Dart D.A., Lakin N.D.. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006; 25:3894–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C.M., Lukas J., Jackson S.P.. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006; 8:37–45. [DOI] [PubMed] [Google Scholar]
- 41. Myers J.S., Cortez D.. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2006; 281:9346–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duursma A.M., Driscoll R., Elias J.E., Cimprich K.A.. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol. Cell. 2013; 50:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lukas C., Melander F., Stucki M., Falck J., Bekker-Jensen S., Goldberg M., Lerenthal Y., Jackson S.P., Bartek J., Lukas J.. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004; 23:2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pefani D.E., Tognoli M.L., Pirincci Ercan D., Gorgoulis V., O’Neill E.. MST2 kinase suppresses rDNA transcription in response to DNA damage by phosphorylating nucleolar histone H2B. EMBO J. 2018; 37:e98760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aymard F., Aguirrebengoa M., Guillou E., Javierre B.M., Bugler B., Arnould C., Rocher V., Iacovoni J.S., Biernacka A., Skrzypczak M. et al.. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 2017; 24:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Britton S., Coates J., Jackson S.P.. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 2013; 202:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Velimezi G., Liontos M., Vougas K., Roumeliotis T., Bartkova J., Sideridou M., Dereli-Oz A., Kocylowski M., Pateras I.S., Evangelou K. et al.. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat. Cell Biol. 2013; 15:967–977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.