Abstract
Typical avian eyes are phenotypically engineered for photopic vision (daylight). In contrast, the highly derived eyes of the barn owl (Tyto alba) are adapted for scotopic vision (dim light). The dramatic modifications distinguishing barn owl eyes from other birds include: 1) shifts in frontal orientation to improve binocularity, 2) rod-dominated retina, and 3) enlarged corneas and lenses. Some of these features parallel mammalian eye patterns, which are hypothesized to have initially evolved in nocturnal environments. Here, we used an integrative approach combining phylogenomics and functional phenotypes of 211 eye-development genes across 48 avian genomes representing most avian orders, including the stem lineage of the scotopic-adapted barn owl. Overall, we identified 25 eye-development genes that coevolved under intensified or relaxed selection in the retina, lens, cornea, and optic nerves of the barn owl. The agtpbp1 gene, which is associated with the survival of photoreceptor populations, was pseudogenized in the barn owl genome. Our results further revealed that barn owl retinal genes responsible for the maintenance, proliferation, and differentiation of photoreceptors experienced an evolutionary relaxation. Signatures of relaxed selection were also observed in the lens and cornea morphology-associated genes, suggesting that adaptive evolution in these structures was essentially structural. Four eye-development genes (ephb1, phactr4, prph2, and rs1) evolved in positive association with the orbit convergence in birds and under relaxed selection in the barn owl lineage, likely contributing to an increased reliance on binocular vision in the barn owl. Moreover, we found evidence of coevolutionary interactions among genes that are expressed in the retina, lens, and optic nerve, suggesting synergetic adaptive events. Our study disentangles the genomic changes governing the binocularity and low-light perception adaptations of barn owls to nocturnal environments while revealing the molecular mechanisms contributing to the shift from the typical avian photopic vision to the more-novel scotopic-adapted eye.
Keywords: relaxed and intensified evolution, pseudogenization, eye-development, coevolution, barn owl, ocular adaptations
Introduction
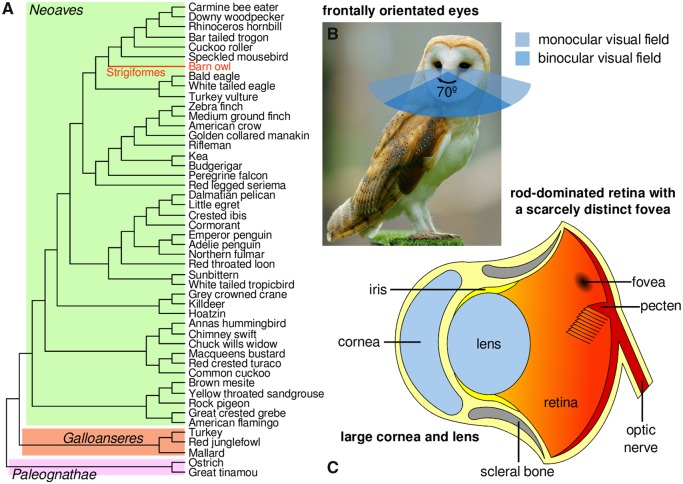
Typically, birds have photopic vision (daylight). In contrast, the barn owl Tyto alba (Strigiform order) is a nocturnal predator successfully adapted for scotopic vision (dim light). In addition, the barn owl is well-adapted to a wide range of environments, ranging from temperate to tropical climates, and has one of the most extensive geographical distributions among birds (IUCN 2014). Barn’s owl remarkable adaptations are linked with its specialized predatory behavior and nocturnal lifestyle. They prey on rodents, small birds, lizards, amphibians, and invertebrates (IUCN 2014), relying on their acute hearing to define their prey’s position in total darkness (Coles and Guppy 1988) and on their unique wing-feather design to identify obstacles through a form of ecolocation (Bachmann 2007). Barn owls are typically nocturnal, but they also can be crepuscular (active during twilight) (Lisney et al. 2012). Concordantly, the anatomy of their eye differs greatly from the standard pattern of other birds (fig. 1) (Hall 2008). The barn owl has large and elongated eyes with outsized corneas and lens to achieve maximum visual sensitivity in nocturnal settings (Lisney et al. 2012; Orlowski et al. 2012), and that are frontally located to increase the binocular visual field (Orlowski et al. 2012). Furthermore, the barn owl has a scarcely evident fovea (region of the retina that is rich in cone photoreceptors and responsible for color discrimination in bright environments), but instead has an abundance of rod cells (Harmening and Wagner 2011), which distinct from cones, gather light more efficiently in low-light (scotopic) environments (Hart 2001). Together, these features of the barn owl eye mirror patterns observed in the mammalian eye, which has also been hypothesized to have evolved specialized adaptations to nocturnal environments (Silva and Antunes 2017; Borges, Johnson, et al. 2018; Borges, Machado, et al. 2018).
Fig. 1.
—Phylogenetic context and the scotopic-adapted eye of the barn owl. (A) The avian species tree highlighting the barn owl lineage. The tree topology is from (Jarvis et al. 2014) and was employed in this study to perform the phylogenetic analyses. (B and C) The unique ocular features of the barn owl highlighting its scotopic adaptations. (C) Anatomy of the Barn owl’s eye structures. Photo of the barn owl (credits: Peter Trimming) taken from Wikipedia under the GNU Free Documentation License.
Publication of the barn owl genome (Zhang, Li, Li, Li 2014; Borges et al. 2015) afforded the opportunity to detail the evolution of its visual genes relative to other birds, revealing clues of barn owl adaptations to a nocturnal lifestyle. This included the assessment of the barn owl opsin gene family (opsins are photosensitive proteins and the major regulators of the visual and nonvisual responses in vertebrates; Hart 2001; Philip et al. 2012), which have been shown to possess adaptive signatures that significantly differ from the general patterns observed in other birds (Borges et al. 2015; Wu et al. 2016). Furthermore, early stage gene pseudogenization was documented in the barn owl green-sensitive rh2 opsin (Borges et al. 2015), suggesting that the barn owl has lower visual acuity (i.e., the ability to discriminate objects on the basis of wavelength; Hart 2001) than most birds, which typically have four visual opsins.
Here, we conducted extensive genomic analyses to infer the genetic basis and the adaptive processes underlying the unique visual system of the barn owl (fig. 1). We studied 211 vertebrate eye-development genes using comparative genomic approaches from 48 avian genomes representing most of the avian orders, including the stem lineage of barn owl (Jarvis et al. 2014; Zhang, Li, Li, Li 2014 ). We assessed selective signatures and possible associations between orbit convergence and the developmental processes of ocular structures. Using this approach, we identified 25 eye-development candidate genes with roles in the retina, lens, cornea, and optic nerve that likely interact synergistically to increase the visual sensitivity and binocular vision of the barn owl.
Materials and Methods
Eye-Development Genes Sequences
A Gene Ontology database was used to select a group of genes involved in eye-development processes (GO: 0001654) (The Gene Ontology Consortium 2015) based on human and rat gene models and products. The respective protein sequences were used to perform TBlastN searches in the barn owl genome (Avian Phylogenomics Project, Zhang, Li, Li, Gilbert 2014), from which 211 genes were identified. The same procedure was implemented for an additional 47 avian genomes, overall encompassing 48 different species of most extant orders of birds (Jarvis et al. 2014) (supplementary table S1, Supplementary Material online).
Phylogenetic and Branch-Specific Selection Analyses
Nucleotide sequences were aligned using the MUSCLE algorithm (Edgar 2004) with the amino acid sequences and with subsequent improvements by removing gap-rich sites. Because we aimed to trace gene evolution within a framework of species evolution, the total evidence genome-scale avian species tree (Jarvis et al. 2014) was used to perform the selection analysis. The branch-specific selection models were employed in PAML (Yang and Nielsen 2002; Maldonado 2016), using the ω-ratio statistic (i.e., the ratio between the nonsynonymous by the synonymous rates of substitution) as an indicator of selective pressures acting on protein-coding genes (Anisimova et al. 2001; da Fonseca et al. 2007; Machado et al. 2011; Sunagar et al. 2013; Khan et al. 2014). Branch-specific selection tests were implemented comparing the one-ratio model, which estimates a single ω-ratio for all lineages in the tree, with the two-ratio model, which assigns an additional ω-ratio parameter to branches of interest (in our case, the tip lineage of the barn owl).
Orbit Convergence and Eye-Development Gene Association Analysis
Overhead orbit convergence angles were calculated using GeoGebra from superior views of the skulls of each avian species (Hohenwarter 2015) that were retrieved from the Bird Skull Collection, DigiMorph, and ADW databases (Rowe 2002; Jansen and van Gestel 2015; Myers et al. 2015). The phylogenetic correlation between the eye-development genes and the orbit convergence were tested in COEVOL (Lartillot and Poujol 2011) using the multiple sequence alignment of each gene and the Jarvis et al. (2014) total evidence genome-scale time tree (Jarvis et al. 2014). The simultaneous reconstruction of the phylogenetic history of the orbit convergence and the ω-ratio was performed by Markov chain Monte Carlo simulations for two chains and 1,000 iterates. Converged, mixed, and independent random draws were posteriorly used to estimate the distribution of the phylogenetic correlation coefficient (ρOC) between the orbit convergence and the ω-ratio in birds. Significant correlations were assessed using Bayes factors and a threshold of 15 for the two hypotheses: ρOC >0 and ρOC <0.
Eye-Development Genes Coevolution
We employed the free-ratios model in PAML (Yang and Nielsen 2002) to compute the maximum likelihood ω-ratio trees (hereafter ω-tree) for each of the 211 eye-development genes. The degree of coevolution among eye-development genes was assessed by computing the correlation between the branch lengths of each pairwise combination of ω-trees (22,115 pairwise combinations in total). We employed the Pearson’s correlation test to identify significant coevolving pairs of genes. The statistical analyses were performed using the R statistical programing language (R Development Core Team 2008). A false discovery rate (FDR, Benjamini and Hochberg 1995) correction was employed whenever correction for multiple testing was necessary. Protein–protein interactions between gene products were assessed using STRING (Franceschini et al. 2012) with a confidence threshold of 0.400. Gene–disease associations were identified and assessed in DisGeNET (Pinero et al. 2015).
Results
Eye-Development Gene Mining in the Barn Owl Genome
A group of 348 genes involved in eye-development (i.e., the progression of the eye from conception to maturity) was defined considering all of the human and rat gene products in the Gene Ontology (GO) database (The Gene Ontology Consortium 2015). We observed that the number of genes associated with eye-development in chicken GO represented only a subset of the genes identified in human and rat. Since the eye-development gene families of birds and mammals do not differ significantly, as corroborated by previous studies (Hunt et al. 2009; and references therein), we surmised we could use an orthologous gene set. Thus, the respective protein sequences were used to perform TBlastN searches in the barn owl genome (Avian Phylogenomics Project, Zhang, Li, Li, Gilbert 2014), through which 211 genes were identified (supplementary table S1, Supplementary Material online).
Barn Owl Branch Selection
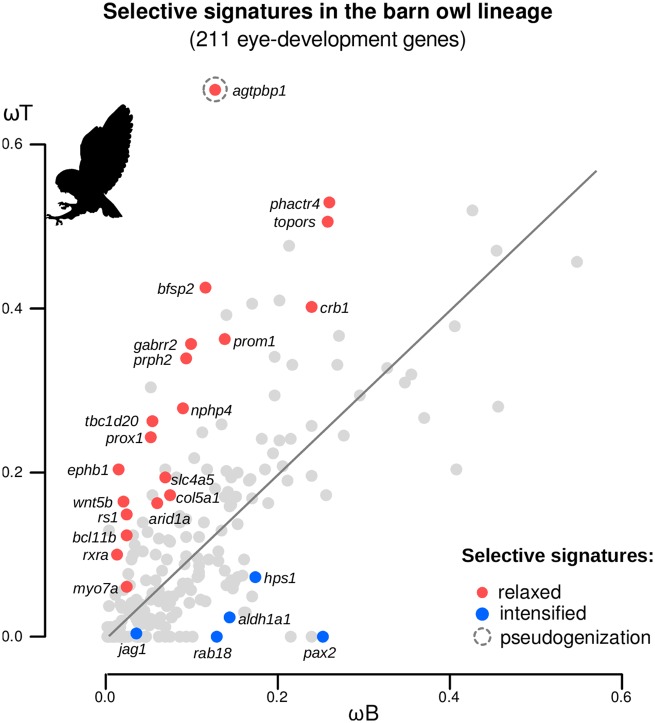
To identify the eye-development genes involved in the nocturnal adaptations of the barn owl, we studied the selective signatures in the tip branch of the barn owl compared with the evolutionary trends of the other birds: branch-specific selection models were used to estimate the ω-foreground (here, in the barn owl terminal lineage: ωT) and the ω-background (in all the other avian lineages: ωB). Out of the 211 genes tested, 25 showed signatures of either intensified or relaxed selection in the barn owl (P value <0.05, FDR corrected for 211 tests, supplementary table S2, Supplementary Material online). These two regimes of selection (more broadly defined in Wertheim et al. 2015) refer to whether the relative rate of evolution in the barn owl lineage is significantly higher (relaxed selection; ωT is significantly higher than ωB) or lower (intensified selection; ωT is significantly lower than ωB) than the general trend among birds. Specifically, agtpbp1, arid1a, bcl11b, bfsp2, col5a1, crb1, ephb1, gabrr2, myo7a, nphp4, phactr4, prom1, prox1, prph2, rs1, rxra, slc4a5, tbc1d20, topors, and wnt5b were under relaxed evolution, while aldh1a1, hps1, jag1, pax2, and rab18 showed evidence of intensified evolution (fig. 2). Furthermore, we found that these genes have previously been reported to be involved in several eye-development malfunctions, such as retinitis pigmentosa, cataracts, retinoschisis, and ocoleus albinism (table 1).
Fig. 2.
—Signatures of selection of 211 eye-development genes in the barn owl lineage. Scatterplot of the ω-ratio estimated in the barn owl terminal lineage (the ω-foreground, ωT) and the ω-ratio estimated in all the other avian lineages (the ω-background, ωB) for 211 avian eye-development genes. Colored circles indicate relaxed (ωT significantly higher than ωB, red) and intensified (ωT significantly lower than ωB, blue) evolving genes (P < 0.05, adjusted for 211 comparisons using the FDR). Branch-specific selection models were used to assess the typology of the selective signatures acting on the terminal lineage of the barn owl (Yang and Nielsen 2002).
Table 1.
Functional and Phenotypic Characterization of the Eye-Development Genes Exhibiting Evidence of Adaptive Evolution in the Barn Owl Lineage
| Gene (Protein) | Function | Eye-Related Phenotype | Adaptive Signatures | References |
|---|---|---|---|---|
| agtpbp1 (ATP/GTP binding protein 1) | agtpbp1 is a functional zinc-binding domain in the agtpbp1 is required for survival of neuron populations. | agtpbp1 is required to prevent photoreceptor degeneration in the retina. |
 ( ( ) See text for details ) See text for details |
Chakrabarti et al. (2008) |
| arid1a (AT-rich interactive domain-containing protein 1A) | arid1a is part of a large ATP-dependent chromatin remodeling complex, which is involved in transcriptional activation and repression of genes by chromatin remodeling. | arid1a mutants possess small optic cups compared with the wild type. |

|
Chandler et al. (2013) |
| bcl11b (B-cell CLL/lymphoma 11B) | bcl11b is zinc finger transcription protein involved in cell proliferation, differentiation, and apoptosis. | bcl11b knockout mice are born with eyes open. |

|
Kominami (2012) |
| bfsp2 (beaded filament structural protein 2) | bfsp2 is a structural gene involved in stabilization of lens fiber cell cytoskeleton. | Mutations in the bfsp2 gene are associated with cataracts and myopia susceptibility. |

|
Song et al. (2009) |
| col5a1 (alpha 1 type V collagen) | col5a1 is a type V collagen, which forms heterotypic fibrils with type I collagen and accounts for 10–20% of corneal collagen. | Mutations in the col5a1 genes are associated with abnormally thin and steep corneas. |

|
Segev et al. (2006) |
| crb1 (crumbs family member 1) | crb1 may be involved in the development of the cell polarization and adhesion in the retina. | Mutations in the crb1 gene are associated with severe retinal dystrophies, including the rod-cone dystrophy, also called retinitis pigmentosa. |

|
Bujakowska et al. (2012) |
| ephb1 (ephrin receptor B1) | ephb1 is a receptor tyrosine kinase which directs the axonal path through interactions with ephrin-B-type proteins following axon-cell contact. | ephb1 is responsible for the retinal axon guidance, redirecting the retinal ganglion cells axons at the optic chiasm midline. |


|
Chenaux and Henkemeyer (2011) |
| gabrr2 (gamma-aminobutyric acid receptor subunit rho-2) | gabrr2 encodes the rho2 subunits of the ligand-gated ion channels, which mediate fast synaptic inhibitory effects of the gamma-aminobutyric acid. | gabrr2 is expressed in the horizontal and bipolar cells of the retina and plays a role in retinal neurotransmission. |

|
Marcos et al. (2000) |
| myo7a (myosin VIIA) | myo7a is a member of the myosin gene family, with actin-based motor activity. It is present in the retinal pigment epithelium where it plays an important role in regulating opsin transport in retinal photoreceptors. | Mutations in the myo7a result in Usher syndrome type 1B, which is characterized by progressive retinal degeneration. |

|
Williams and Lopes (2011) |
| nphp4 (nephronophthisis 4) | rpgrip1 and nephrocystin-4 colocalize in the retina. | Mutations in nphp4 are associated with a combination of nephronophthisis and retinitis pigmentosa called Senior–Løken syndrome. |

|
Won et al. (2011) |
| phactr4 (phosphatase and actin regulator 4) | phactr4 interacts with the regulator of protein phosphatase 1 that is required for neural cell migration during development. | phactr4 regulates neural tube and optic fissure closure. |


|
Kim et al. (2007) |
| prom1 (prominin 1) | prom1 plays a role in early retinal development, acting as a key regulator of disk morphogenesis in photoreceptors. | Mutations in prom1 result in retinitis pigmentosa and cone-rod dystrophy. |

|
Michaelides et al. (2010) |
| prox1 (prospero homeobox 1) | prox1 is a member of the homeobox transcription factor family that functions as a key regulatory protein in neurogenesis. | prox1 knockout mice have defects in the elongation of lens fiber cells. prox1 is also detected in differentiating horizontal, bipolar, and amacrine cells |

|
Duncan et al. (2002), Dyer et al. (2003) |
| prph2 (peripherin-2) | prph2 encodes a photoreceptor-specific tetraspanin protein called peripherin-2, which is critical for the formation and maintenance of rod and cone outer segments. | Mutations in prph2 are associated with a variety of forms of retinitis pigmentosa and macular degeneration phenotypes. |


|
Conley and Naash (2014) |
| rs1 (retinoschisin 1) | rs1 is an extracellular protein that plays a crucial role in the cellular organization of the retina. | Mutations in rs1 are associated with progressive retinal and macular degeneration, common phenotypes of retinoschisis disease. |

|
Takada et al. (2008) |
| rxra (alpha retinoid X receptor) | rxra mediates the biological effects of retinoids by their involvement in retinoic acid-mediated gene activation. | rxra mutants show abnormal opening of the retina at the optic nerve exit point (optic disk coloboma) and also conformational alterations in the cornea and lens. |


|
Mascrez et al. (2009) |
| slc4a5 (solute carrier family 4, member 5) | slc4a5 mediate sodium- and bicarbonate-dependent cotransport, regulating the intracellular pH. | slc4a5 knockout mice develop severe retinopathy, with loss of photoreceptors and ganglion cells, and retinal detachment. |

|
Kao et al. (2011) |
| tbc1d20 (TBC1 domain family, member 20) | tbc1d20 encodes a GTPase-activating protein specific for Rab1 and Rab2 small GTPase families. | tbc1d20 mutations are associated with the Warburg Micro syndrome 4 that is characterized by eye cataracts (vacuoles present throughout the entire lens). |

|
Park et al. (2014) |
| topors (topoisomerase I-binding arginine/serine-rich) | topors functions in proteasomal degradation pathway by acting as an E3 ubiquitin ligase for p53, and is involved in the photoreceptor development and function. | Genetic variants of topors were shown to cause a form of retinal degeneration (retinitis pigmentosa). |

|
Chakarova et al. (2011) |
| wnt5b (wingless-type MMTV integration site family, member 5B) | wnt5b is a ligand for members of the frizzled family of seven transmembrane receptors and has a probable signaling role in the anterior eye-development. | wnt5b is expressed in the differentiating lens fiber cells. |

|
Fokina and Frolova (2006) |
| aldh1a1 (aldehyde dehydrogenase 1 family, A1 member) | aldh1a1 act as an enzyme that catalyzes the oxidation of the retinol (vitamin A) metabolite, retinal, to retinoic acid, and also as a crystallin in the eye. | aldh1a1 knockout mice were shown to develop cataracts and being sensitive to UV-induced damage. |

|
Chen et al. (2012) |
| hps1 (Hermansky–Pudlak syndrome 1) | hps1 encodes a protein that may play a role in melanosome biogenesis. | hps1 is associated with the Hermansky–Pudlak syndrome that is characterized by oculocutaneous albinism (iris transillumination). |

|
Jardón et al. (2015) |
| jag1 (jagged 1) | jag1 encodes a ligand that participates in the Notch pathway of the lens, transducing cell contact-mediated communication and contributing to the lens progenitor cell proliferation and differentiation. | jag1 mutants have both lens progenitor cell proliferation and differentiation deficits. |

|
Le et al. (2009) |
| pax2 (paired box 6) | pax2 is a transcription factor with a conserved DNA-binding paired box domain. | Mutations in pax2 can result in retinal coloboma syndrome manifested by the failure of optic fissure histogenesis and a damaged retina. |

|
Stanke et al. (2010) |
| rab18 (member RAS oncogene family) | rab18 may play a role in the maintenance of the cytoskeleton in lens fiber cells. | Mutations in the rab18 cause Warburg Micro Syndrome characterized by defective ophthalmological phenotypes in lens development, such as congenital nuclear cataracts and atonic pupils. |

|
Carpanini et al. (2014) |
Note.—The function and the eye-related phenotypes of the listed eye-development genes were inferred from the GeneCards database (http://www.genecards.org/; Safran et al. 2010) and specific citations referenced below. Patterns of the evolution of these genes in the barn owl lineage are summarized in the table using colored circles: relaxed selection (red circle) and intensified selection (blue circle), pseudogenization (gray circle), and association with orbit convergence (black circle).
A closer inspection of the barn owl agtpbp1 sequence revealed signs of pseudogenization, with two stop codons in positions 85 and 246 (using the chicken agtpbp1 protein as reference) and nonsynonymous amino acid substitutions in conserved regions (supplementary fig. S1, Supplementary Material online). The ω-ratio of this gene is clearly outlier in the branch-selection analyses relative to other genes (ωT/ωB = 0.668/0.138, LRT = 91.622, P value <0.001; fig. 2).
Orbit Convergence and Eye-Development Gene Associations
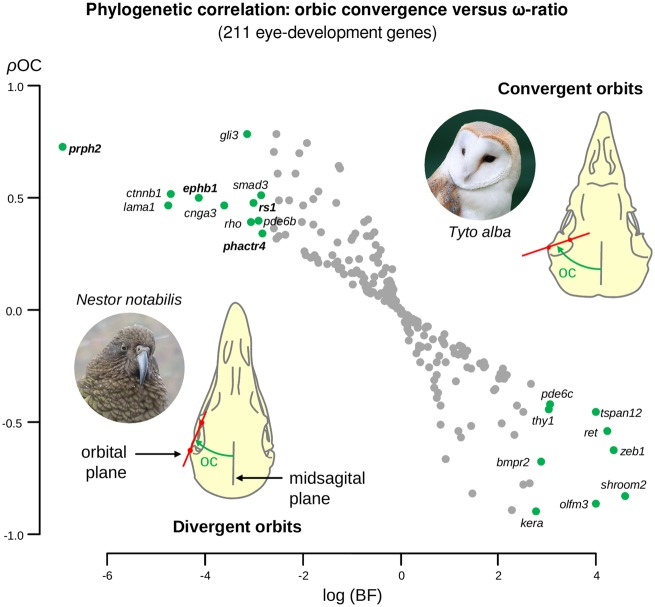
Thirty-seven avian skulls were used to measure the orbit convergence angle (in degrees; supplementary table S3 and fig. S2, Supplementary Material online). These measurements were validated using previously published measurements of the orbit convergence for 11 avian species (Menegaz and Kirk 2009): both measures are strongly correlated (r = 0.83, P value = 0.003, Pearson test). Using orbit convergence angles and the COEVOL package (COEVOL implements a probabilistic framework for testing the coupling between continuous characters and parameters of the molecular substitution process; Lartillot and Poujol 2011), we tested the phylogenetic correlation between the orbit convergence and the ω-ratio (ρOC, Pearson phylogenetic correlation coefficient) by simultaneously reconstructing the orbit convergence and the ω-ratio evolution in the avian tree for all eye-development genes. About 20 eye-development genes showed significant associations with orbit convergence in birds (Bayes factor >15 considering both the hypotheses ρOC >0 and ρOC <0, fig. 3, supplementary table S4, Supplementary Material online: bmpr2, cnga3, ctnnb1, ephb1, gli3, kera, lama1, olfm3, pde6b, pde6c, phactr4, prph2, ret, rho, rs1, shroom2, smad3, thy1, tspan12, and zeb1); the higher the ω-ratio the higher the orbit convergence angle. Among these genes, ephb1, phactr4, prph2, and rs1, (showing a positive correlation between the ω-ratio and the orbits convergence) evolved through relaxed selection in the barn owl lineage.
Fig. 3.
—Phylogenetic correlation between the orbit convergence and the ω-ratio of the 211 avian eye-development genes. Pearson’s correlation coefficient (ρOC) between the orbit convergence and ω-ratio of the 211 eye-development genes is plotted in the vertical axis. The horizontal axis represents the logarithm of the Bayes factors (BF) calculated under both of the hypotheses: ρOC >0 (positive association, upper left quadrant) and ρOC <0 (negative association, lower right quadrant). Green circles indicate genes with evidence of having a phylogenetic correlation with the orbit convergence at a BF threshold of 15. Genes in bold (ephb1, phactr4, prph2, and rs1) evolved under relaxed selection (see fig. 2) in the barn owl lineage. Photos of the barn owl (credits: Peter Trimming) and kea (Nestor notabilis; credits: Markus Koljonen) taken from Wikipedia under the GNU Free Documentation License.
Coevolution of Eye-Development Genes
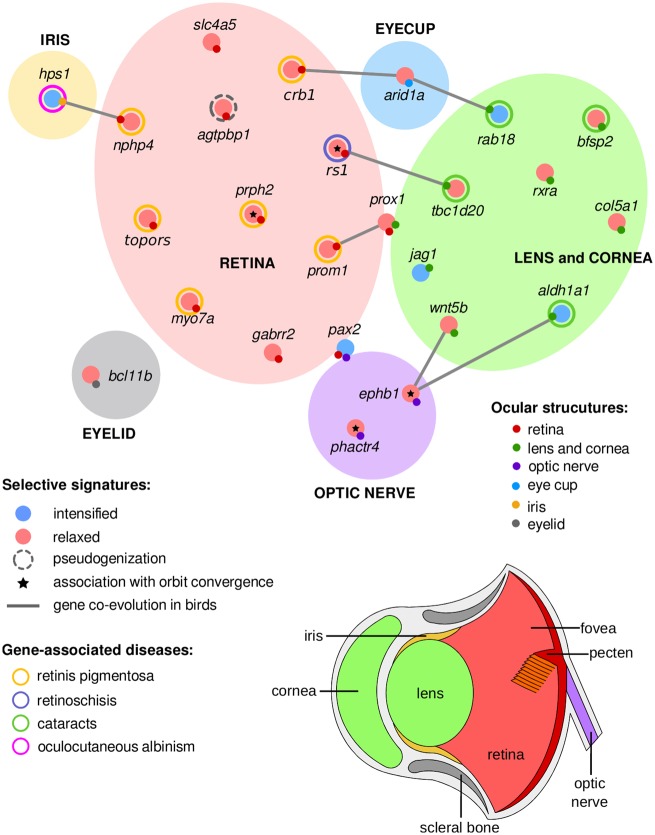
To test for evidence of gene coevolution in birds, we used the ω-tree to perform branch-to-branch ω-ratio correlations for each pair of eye-development genes (P value <0.05, Pearson correlation test, FDR corrected for 22,155 tests; supplementary table S5, Supplementary Material online). We obtained eight significant correlations (gray edges in fig. 4) between 12 of the 25 genes that evolved under regimes of relaxed or intensified selection in the barn owl genome: hps1: nphp4, crb1: arid1a: rab18, rs1: tbc1s20, prox1: prom1, and ephb1: wnt5b: aldh1a1. Most of these interactions were between genes that have different, but linked roles, in specific substructures of the avian eye, including the retina, lens, cornea, and optic nerve. None of these interactions has been linked with a previously described protein–protein interaction in birds and mammals (searched in STRING webserver; supplementary fig. S3, Supplementary Material online).
Fig. 4.
—The adaptive gene network of the barn owl eye-development genes. The eye-development genes showing adaptive evolution in the barn owl lineage, along with those correlated with the orbit convergence in birds (marked with an asterisk), were inspected for functional roles in ocular structures: lens and cornea (green), eyelid (gray), eyecup (blue), iris (yellow), retina (red), and optic nerve (purple). We determined possible roles of these 25 genes in ocular structures from previously described phenotypes, syndromes, and malfunctions with which they have been associated(table 1). Genes associated with several eye-structures are represented in the circle’s boundaries: pax2 and prox1. Signatures of coevolution among avian eye-development genes are represented in gray lines.
Discussion
Here, we applied comparative genomic approaches with 48 avian genomes representing most of the avian orders (including the stem lineage of the scotopic adapted barn owl), along with phylogenetic and phenotypic-association analyses, to identify candidate eye-development genes involved in the binocularity and low-light perception adaptations of the barn owl. We describe 25 eye-development genes that likely coevolved adaptively in the stem lineage of the barn owl. These genes have important functional roles in retina, lens, cornea, and optic nerve development. The genetic changes we found in the barn owl are candidates that give the species its unique ocular features among birds, including high visual sensitivity and frontally orientated eyes, as well as these convergent innovations in nocturnal mammals (Khan et al. 2015; Borges, Johnson, et al. 2018; Borges, Machado, et al. 2018).
Our phylogenetic analyses were all based on the genome-scale phylogeny of birds proposed in (Jarvis et al. 2014) (fig. 1); in particular, the branch-selection tests cannot be done without an explicit phylogenetic placement of the barn owl lineage relative to other birds. However, alternative phylogenies that have some incongruence with the one used by us have been proposed. For example, a polytomy zone was proposed among the core landbirds, which does not resolve the relative position of owls, eagles and vultures, and mousebirds (Suh 2016). Importantly, a recent study employed more extensive taxon sampling but much less genomic DNA (Prum et al. 2015). The incongruences between the Jarvis et al. (2014) and Prum et al. (2015) were found to the result convergent protein coding sequencing in the Prum et al. (2015), overriding the noncoding signature in the rest of the genome. But the Prum et al. (2015) study grouped owls together with Coraciimorphae (which includes mousebirds, woodpeckers, kingfishers, and trogons), which is congruent with the topology we used from Jarvis et al. (2014). Thus, we do not believe that the results would change with this alternative tree topology.
The pseudogenization of agtpbp1 gene in barn owls is particularly evocative, as this gene is crucial for the survival of neuron populations and has been associated with retinitis pigmentosa, a progressive form of retinal degeneration that in mice culminates in the marked loss of photoreceptors and thinning of the outer segment region (Chakrabarti et al. 2008). Similarly, we previously reported the loss of eye-development genes and described early signatures of pseudogenization in the green-sensitive rh2 cone photoreceptor in the barn owl (Borges et al. 2015). Given that agtpbp1 is involved in photoreceptor maintenance in the retina, we hypothesize some photoreceptor morphogenesis pathways could have degenerated as the barn owl evolved a scotopic vision. However, this hypothesis requires further validation as mouse agtpbp1 mutants have better-preserved cones than rods, which does not explain the barn’s owl rod-rich retina (Harmening and Wagner 2011).
It has long been known that the retina is very sensitive to different light environments, being one of the structures that change most when animals evolve adaptions from nocturnal/crepuscular to diurnal lifestyles (Hall 2008). We found that all the ten retina-expressing genes evolved under relaxed selection in the barn owl. These retina-associated genes are vital for the development of retinal cell-types: crb1, myo7a, nphp4, prom1, prph2, and topors were shown to be directly involved in rods and cones morphogenesis of the outer segment formation and in opsin transport (Michaelides et al. 2010; Chakarova et al. 2011; Williams and Lopes 2011; Won et al. 2011; Bujakowska et al. 2012; Conley and Naash 2014); rs1 has a key role in the development and maintenance of photoreceptor cells (Takada et al. 2008); gabrr2 and prox1 are involved in the development of the horizontal, bipolar, and amacrine cells (Marcos et al. 2000; Dyer et al. 2003); and slc4a5 is associated with the loss of photoreceptors and ganglion cells (Kao et al. 2011).
In nocturnal environments, retinas are generally more sensitive and the barn owl retina, in particular, has several features that can be associated with improved sensitivity, including the preponderance of rods over cones, scarcely distinct fovea, and a lower density of ganglion cells density (Harmening and Wagner 2011; Borges et al. 2015). Based on the function of the retina-associated genes, we have pinpointed in our analyses, we hypothesize that the scotopic-adapted of the barn owl retina evolved by: 1) regulation of the proliferation of cone and rod precursor cells (likely through evolution of rs1 and slc4a5 genes) at early stages of the retina development; 2) maintenance of a low number of ganglion cells (likely through slc4a5); and 3) differentiation of neural retina with increased ratios of rods over cones (likely through the remaining genes) at later phases of the retina development.
Eye-development genes with signatures of relaxed selection in the cornea and lens included mostly morphogenes (genes affecting morphological traits when mutated; Liao et al. 2010). For example, col5a1 is associated with corneal thickness (Segev et al. 2006); wnt5b has been shown to be expressed right before the elongation of the lens fiber cells (Fokina and Frolova 2006); tbc1d20 mice mutants have shortened and disorganized lens fiber cells (Park et al. 2014); and bfsp2 is involved in the stabilization of lens fiber cell cytoskeleton (Song et al. 2009). In contrast, genes evolving under intensified selection were mostly physiogenes (genes affecting physiological traits; Liao et al. 2010), including rab18, which is associated with lens development and closure of the lens vesicle and denucleation of fiber cells (Carpanini et al. 2014); aldh1a1, which has a metabolic role in protecting the eye from UV-induced damage (Chen et al. 2012); and jag1, which is responsible for lens progenitor cell proliferation and differentiation (Le et al. 2009). Our results suggest that the evolutionary diversification of the optic system associated-genes in the barn owl could be linked with the redesign of the lens and cornea. Consistent with these results, corneas and lens of nocturnal species are generally larger than those in diurnal species, but share the same function, that is, focusing light rays onto the back of the eye (Lisney et al. 2012).
The distinct frontal orientation of barn owl eyes is among its most dramatic phenotype. Orbit convergence is an indicator of binocularity (Heesy et al. 2011) in birds and is more pronounced in nocturnal species (Menegaz and Kirk 2009). The four eye-development genes (ephb1, phactr4, prph2, and rs1) showing significant correlations with the orbit convergence in all birds are also evolving under relaxed selection in the barn owl lineage. Thus, it is very likely that these genes have had a major role in increasing the binocular vision in the barn owl. prph2 and rs1 are responsible for the maintenance, proliferation, and differentiation of photoreceptors (Takada et al. 2008; Conley and Naash 2014) and ephb1 and phactr4 regulate the retinal axon guidance and optic fissure closing (Kim et al. 2007; Chenaux and Henkemeyer 2011). In particular, ephb1 is involved in directing the ipsilateral projection, that is, the uncrossed fibers in the optic chiasm (Chenaux and Henkemeyer 2011). A high proportion of ipsilateral retinal projections have been associated with frontal eyes in vertebrates. Owls, in particular, have a higher degree of ipsilateral retinal projections than most other birds (Larsson 2015). We suggest that the relaxed selection in the ephb1 gene contributed to permit the retinal ganglion cells axons to form ipsilateral rather than contralateral projections, which the barn owl would have needed to evolve binocular vision. Therefore, we suggest that these four genes, evolving in parallel with the orbit convergence in birds, were most likely acting at the neuronal level for binocular vision. However, as they cannot be directly linked to the morphogenesis of the orbit cavity, future experimental validation is needed to corroborate the causality of the phylogenetic correlations obtained here.
The bcl11b, a gene associated with eyelid development (the respective knockout mice is born with open eyes; Kominami 2012; Kyrylkova et al. 2015), is a candidate for controlling the development of the unusual owl eyelids. The third owl eyelid, also known as the nictitating membrane, is particularly opaque and robust, suggesting a role in regulating the light that enters the eye (Jochems and Phillips 2015). Another gene with evidence of adaptive evolution, hps1, is involved in iris development and has been associated with the oculocutaneous albinism malfunction (iris transillumination) (Jardón et al. 2015). Iris color is an important aspect of owls’ vision and there is some evidence that eye color correlates with activity patterns in owls (Passarotto et al. 2018). In the barn owl (as other nocturnal owls), the iris is typically dark brown or black with large amounts of melanin within the iris stroma, which is consistent with the signature of intensified selection found in the hps1.
Our evidence suggests that some genes expressed in different eye structures may have coevolved. The molecular roles of the coevolving genes include extracellular (rs1, Takada et al. 2008) and intracellular (crb1, rab18, prom1, and tbc1d20, Michaelides et al. 2010; Bujakowska et al. 2012; Carpanini et al. 2014; Park et al. 2014) regulators, transcription factors (arid1a and prox1, Duncan et al. 2002; Dyer et al. 2003; Chandler et al. 2013), and signaling proteins (wnt5b, aldh1a1, and ephb1, Fokina and Frolova 2006; Chenaux and Henkemeyer 2011; Chen et al. 2012). They represent potential cointeractions in the eye-development network of birds, which need further confirmation since the current state of knowledge on the avian visual pathways is only in its infancy.
The signatures of coevolution reported here suggest both adaptive synergy and compensation among various structures of the eye. For example, arid1a (arid1a mutants have smaller optic cups than the wild-type; Chandler et al. 2013) coevolves with crb1 in the retina and rab18 in the lens and cornea, suggesting that the combination of genes likely are involved in the enlargement of the barn owl’s eyes, which are more than twice as large as the average for birds of the same weight (Brooke et al. 1999). Coevolution between genes that are associated with orbit convergence and those having roles in the optic system (e.g., tbc1d20, wnt5b, and aldh1a1) support the hypothesis of parallel evolution between the orbit bone and the lens morphology in the evolution of binocularity. Finally, interactions among genes evolving with contrasting selective signatures (e.g., hps1: nphp4, arid1a: rab18, and wnt5b: aldh1a1: ephb1) would be consistent with adaptive compensation.
In conclusion, our results provide evidence that 1) pseudogenization, 2) differentiated relaxed and intensified selective signatures affecting eye-structural genes, and 3) gene coevolution were the prominent molecular mechanisms associated with adaptations of the barn owl eye to nocturnal environments.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
R.B. and C.G. were funded with a PhD grant from “Fundação para a Ciência e a Tecnologia” (FCT) (R.B.: SFRH/BD/79850/2011 and C.G.: SFRH/BD/71041/2010). W.E.J. prepared portions of this manuscript while holding a National Research Council Research Associateship Award at the Walter Reed Army Institute of Research and the published material reflects the views of the authors and should not be construed to represent those of the Department of the Army or the Department of Defense. S.J.O. was supported, in part, by the Russian Science Foundation grant (project no. 17-14-01138) and by St. Petersburg State University (Genome Russia Grant no. 1.52.1647.2016). E.D.J. was supported by the Howard Hughes Medical Institute. A.A. was partially supported by the Strategic Funding UID/Multi/04423/2019 through national funds provided by FCT and the European Regional Development Fund (ERDF) in the framework of the program PT2020, by the European Structural and Investment Funds (ESIF) through the Competitiveness and Internationalization Operational Program—COMPETE 2020 and by National Funds through the FCT under the project PTDC/AAG-GLO/6887/2014 (POCI-01-0124-FEDER-016845). We are thankful for the comments provided by the Associate Editor and two anonymous reviewers, which helped improving a previous version of this manuscript.
Literature Cited
- Anisimova M, Bielawski JP, Yang Z.. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 18(8):1585–1592. [DOI] [PubMed] [Google Scholar]
- Bachmann T. 2007. Morphometric characterisation of wing feathers of the barn owl Tyto alba pratincola and the pigeon Columba livia. Front Zool. 4(1):23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Borges R, et al. 2015. Gene loss, adaptive evolution and the co-evolution of plumage coloration genes with opsins in birds. BMC Genomics 16:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges R, Johnson WE, et al. 2018. Adaptive genomic evolution of opsins reveals that early mammals flourished in nocturnal environments. BMC Genomics 19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges R, Machado JP, Gomes C, Rocha AP, Antunes A.. 2018. Measuring phylogenetic signal between categorical traits and phylogenies. Bioinformatics. 35:1862–1869. [DOI] [PubMed] [Google Scholar]
- Brooke MDL, Hanley S, Laughlin SB.. 1999. The scaling of eye size with body mass in birds. Proc R Soc Lond B Biol Sci. 266(1417):405–412. [Google Scholar]
- Bujakowska K, et al. 2012. CRB1 mutations in inherited retinal dystrophies. Hum Mutat. 33(2):306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpanini SM, et al. 2014. A novel mouse model of Warburg Micro syndrome reveals roles for RAB18 in eye development and organisation of the neuronal cytoskeleton. Dis Model Mech. 7(6):711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarova CF, et al. 2011. TOPORS, implicated in retinal degeneration, is a cilia-centrosomal protein. Hum Mol Genet. 20(5):975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L, et al. 2008. The zinc-binding domain of Nna1 is required to prevent retinal photoreceptor loss and cerebellar ataxia in Purkinje cell degeneration (pcd) mice. Vision Res. 48(19):1999–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler RL, et al. 2013. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 33(2):265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Koppaka V, Thompson DC, Vasiliou V.. 2012. Focus on Molecules: aLDH1A1: from lens and corneal crystallin to stem cell marker. Exp Eye Res. 102:105–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenaux G, Henkemeyer M.. 2011. Forward signaling by EphB1/EphB2 interacting with ephrin-B ligands at the optic chiasm is required to form the ipsilateral projection. Eur J Neurosci. 34(10):1620–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles RB, Guppy A.. 1988. Directional hearing in the barn owl (Tyto alba). J Comp Physiol. 163(1):117–133. [DOI] [PubMed] [Google Scholar]
- Conley SM, Naash MI.. 2014. Gene therapy for PRPH2-associated ocular disease: challenges and prospects. Cold Spring Harb Perspect Med. 4(11):a017376–a017376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca RR, Antunes A, Melo A, Ramos MJ.. 2007. Structural divergence and adaptive evolution in mammalian cytochromes P450 2C. Gene. 387:58–66. [DOI] [PubMed] [Google Scholar]
- Duncan MK, Cui W, Oh D-J, Tomarev SI.. 2002. Prox1 is differentially localized during lens development. Mech Dev. 112(1–2):195–198. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G.. 2003. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 34(1):53–58. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokina VM, Frolova EI.. 2006. Expression patterns of Wnt genes during development of an anterior part of the chicken eye. Dev Dyn. 235(2):496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, et al. 2012. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41(D1):D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MI. 2008. The anatomical relationships between the avian eye, orbit and sclerotic ring: implications for inferring activity patterns in extinct birds. J Anat. 212(6):781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmening WM, Wagner H.. 2011. From optics to attention: visual perception in barn owls. J Comp Physiol A. 197(11):1031–1042. [DOI] [PubMed] [Google Scholar]
- Hart NS. 2001. The visual ecology of avian photoreceptors. Prog Retin Eye Res. 20(5):675–703. [DOI] [PubMed] [Google Scholar]
- Heesy CP, Kamilar JM, Willms J.. 2011. Retinogeniculostriate pathway components scale with orbit convergence only in primates and not in other mammals. Brain Behav Evol. 77(2):105–115. [DOI] [PubMed] [Google Scholar]
- Hohenwarter M. 2015. GeoGebra. Available from: http://www.geogebra.org/. Last accessed January, 2018.
- Hunt DM, Carvalho LS, Cowing JA, Davies WL.. 2009. Evolution and spectral tuning of visual pigments in birds and mammals. Philos Trans R Soc B. 364(1531):2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN. 2014. The IUCN red list of threatened species. 2012, BirdLife Int. Tyto alba Available from: http://www.iucnredlist.org. Last accessed May, 2018.
- Jansen J, van Gestel W.. 2015. Bird skull collection. Available from: http://www.skullsite.com/. Last accessed December, 2017.
- Jardón J, Izquierdo NJ, Renta JY, García-Rodríguez O, Cadilla CL.. 2015. Ocular findings in patients with the Hermansky-Pudlak syndrome (types 1 and 3). Ophthalmic Genet. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochems B, Phillips TE.. 2015. Histological and ultrastructural studies on the conjunctiva of the barred owl (Strix varia). PLoS One 10(11):e0142783.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao L, et al. 2011. Severe neurologic impairment in mice with targeted disruption of the electrogenic sodium bicarbonate cotransporter NBCe2 (Slc4a5 gene). J Biol Chem. 286(37):32563–32574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, et al. 2014. Mammalian keratin associated proteins (KRTAPs) subgenomes: disentangling hair diversity and adaptation to terrestrial and aquatic environments. BMC Genomics 15(1):779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, et al. 2015. Olfactory receptor subgenomes linked with broad ecological adaptations in Sauropsida. Mol Biol Evol. 32(11):2832–2843. [DOI] [PubMed] [Google Scholar]
- Kim T-H, Goodman J, Anderson KV, Niswander L.. 2007. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Dev Cell. 13(1):87–102. [DOI] [PubMed] [Google Scholar]
- Kominami R. 2012. Role of the transcription factor Bcl11b in development and lymphomagenesis. Proc Jpn Acad B. 88(3):72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrylkova K, Iwaniec UT, Philbrick KA, Leid M.. 2015. BCL11B regulates sutural patency in the mouse craniofacial skeleton. Dev Biol. 415:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson ML. 2015. Binocular vision, the optic chiasm, and their associations with vertebrate motor behavior. Front Ecol Evol. 3. :1–12. [Google Scholar]
- Lartillot N, Poujol R.. 2011. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol Biol Evol. 28(1):729–744. [DOI] [PubMed] [Google Scholar]
- Le TT, Conley KW, Brown NL.. 2009. Jagged 1 is necessary for normal mouse lens formation. Dev Biol. 328(1):118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B-Y, Weng M-P, Zhang J.. 2010. Contrasting genetic paths to morphological and physiological evolution. Proc Natl Acad Sci U S A. 107(16):7353–7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisney TJ, Iwaniuk AN, Bandet MV, Wylie DR.. 2012. Eye shape and retinal topography in owls (Aves: Strigiformes). Brain Behav Evol. 79(4):218–236. [DOI] [PubMed] [Google Scholar]
- Machado J, Johnson WE, O’Brien SJ, Vasconcelos V, Antunes A.. 2011. Adaptive evolution of the matrix extracellular phosphoglycoprotein in mammals. BMC Evol Biol. 11(1):342.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E. 2016. LMAP: lightweight multigene analyses in PAML. BMC Bioinformatics. 17:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos I, et al. 2000. Mutation analysis of GABRR1 and GABRR2 in autosomal recessive retinitis pigmentosa (RP25). J Med Genet. 37(6):5e–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascrez B, Ghyselinck NB, Chambon P, Mark M.. 2009. A transcriptionally silent RXRα supports early embryonic morphogenesis and heart development. Proc Natl Acad Sci U S A. 106(11):4272–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegaz RA, Kirk EC.. 2009. Septa and processes: convergent evolution of the orbit in haplorhine primates and strigiform birds. J Hum Evol. 57(6):672–687. [DOI] [PubMed] [Google Scholar]
- Michaelides M, et al. 2010. The PROM1 mutation p.R373C causes an autosomal dominant Bull’s eye maculopathy associated with rod, rod–cone, and macular dystrophy. Invest Ophthalmol Vis Sci. 51(9):4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers P, Jones T, Espinosa R, Dewey T, Hammond G.. 2015. The animal diversity web. Available from: http://animaldiversity.org. Last accessed December, 2017.
- Orlowski J, Harmening W, Wagner H.. 2012. Night vision in barn owls: visual acuity and contrast sensitivity under dark adaptation. J Vis. 12(13):4.. [DOI] [PubMed] [Google Scholar]
- Park AK, et al. 2014. Targeted disruption of Tbc1d20 with zinc-finger nucleases causes cataracts and testicular abnormalities in mice. BMC Genet. 15:135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarotto A, Parejo D, Cruz-Miralles A, Avilés JM.. 2018. The evolution of iris colour in relation to nocturnality in owls. J Avian Biol. 48:12. [Google Scholar]
- Philip S, et al. 2012. Fish lateral line innovation: insights into the evolutionary genomic dynamics of a unique mechanosensory organ. Mol Biol Evol. 29(12):3887–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero J, et al. 2015. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database 2015(0):bav028–bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prum RO, et al. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526(7574):569–573. [DOI] [PubMed] [Google Scholar]
- R Development Core Team R. 2008. Computational many-particle physics. Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- Rowe T. 2002. Digital morphology. Available from: http://digimorph.org. Last accessed December, 2017.
- Safran M, et al. 2010. GeneCards Version 3: the human gene integrator. Database 2010:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev F, et al. 2006. Structural abnormalities of the cornea and lid resulting from collagen V mutations. Invest Ophthalmol Vis Sci. 47(2):565. [DOI] [PubMed] [Google Scholar]
- Silva L, Antunes A.. 2017. Vomeronasal receptors in vertebrates and the evolution of pheromone detection. Annu Rev Anim Biosci. 5(1):353–370. [DOI] [PubMed] [Google Scholar]
- Song S, et al. 2009. Functions of the intermediate filament cytoskeleton in the eye lens. J Clin Invest. 119(7):1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke J, Moose HE, El-Hodiri HM, Fischer AJ.. 2010. Comparative study of Pax2 expression in glial cells in the retina and optic nerve of birds and mammals. J Comp Neurol. 518(12):2316–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A. 2016. The phylogenomic forest of bird trees contains a hard polytomy at the root of Neoaves. Zool Scr. 45:50–62. [Google Scholar]
- Sunagar K, et al. 2013. Molecular evolution of vertebrate neurotrophins: co-option of the highly conserved nerve growth factor gene into the advanced snake venom. PLoS One 8(11):e81827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, et al. 2008. Synaptic pathology in retinoschisis knockout (Rs1 −/y) mouse retina and modification by rAAV-Rs1 gene delivery. Invest Ophthalmol Vis Sci. 49(8):3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2015. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43:D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Pond SLK, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DS, Lopes VS.. 2011. The many different cellular functions of MYO7A in the retina. Biochem Soc Trans. 39(5):1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J, et al. 2011. NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum Mol Genet. 20(3):482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, et al. 2016. Retinal transcriptome sequencing sheds light on the adaptation to nocturnal and diurnal lifestyles in raptors. Sci Rep. 6:33578.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 19(6):908–917. [DOI] [PubMed] [Google Scholar]
- Zhang G, Li B, Li C, Gilbert MT, et al. 2014. Comparative genomic data of the Avian Phylogenomics Project. Gigascience 3(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.