Abstract
Background
Hepatitis E virus (HEV) infection can occur through consumption of undercooked pork meat or exposure to animal feces. Because there are scarce data only in developing countries, we assessed whether pigs might be a potential source of human HEV infections in Vietnam. In addition, we determined anti-HEV seroprevalences in the general population and in individuals professionally exposed to pigs and pork meat.
Methods
The study took place in Hanoi, Vietnam. Liver tissues from domestic pigs (n = 210) and serum samples obtained from individuals occupationally exposed to pigs and pork meat (n = 283) and from unexposed healthy controls (n = 168) were screened for HEV-ribonucleic acid (RNA) by reverse-transcription polymerase chain reaction. The exposed group was divided into pork meat vendors (n = 81), pig farmers (n = 96), and slaughterers (n = 106). Serum samples were subjected to HEV immunoglobulin (Ig)G and IgM enzyme-linked immunosorbent assays. The HEV genotypes were assessed by direct sequencing, followed by phylogenetic analyses.
Results
Hepatitis E virus seroprevalence was higher among persons occupationally exposed to pigs/pork meat compared with unexposed individuals (anti-HEV IgM 11% vs 6%, P = .07; anti-HEV IgG 53% vs 31%, P < .0001). Positivity of anti-HEV IgG among slaughterhouse staff was 66%, followed by 51% in pig-farmers and 38% in pork meat vendors (P = .00073). A similar trend was observed for IgM positivity. Of the pig liver tissues, 26 of 210 (12.4%) were positive for HEV-RNA and assessed to be HEV genotype 3.
Conclusions
Hepatitis E virus circulates in domestic pigs in Hanoi and constitutes a permanent zoonotic disease risk. The high HEV seroprevalence among occupationally exposed individuals indicates an associated risk of HEV infection.
Keywords: hepatitis E virus, occupationally exposed, pigs, pork meat, zoonoses
Hepatitis E virus (HEV) is the major cause of an enterically acquired acute hepatitis. Annually, an estimated 20 million novel human infections occur worldwide, leading to 3 million symptomatic cases and 56 000 hepatitis E-related deaths [1]. Hepatitis E virus infection mainly affects people in East and South Asia, Africa, and Latin America, in particular under conditions of poor sanitation and hygiene and restricted access to clean water and health services [2]. It is noteworthy that 60% of cases and 65% of related deaths occur in Asia, where the seroprevalence of anti-HEV antibodies may exceed 25% in certain populations [1]. In developed countries, an increasing number of locally acquired human hepatitis E cases has been recognized [3].
Most acute HEV cases are asymptomatic; however, the overall mortality rate may be 1%–3% [4]. Hepatitis E virus infection and acute fulminant hepatitis in pregnant women is a serious condition with a fatality rate of up to 30% especially in the third trimester, spontaneous abortions, and stillbirths [5]. Chronic HEV infection is observed particularly among immunocompromised patients and in patients subjected to organ transplantation or cancer chemotherapy [6].
In addition to the water-associated transmission pattern, there is clear evidence that HEV is a zoonotic pathogen, which can infect humans through consumption of undercooked meat of infected domestic pigs and wild animals [7, 8]. Phylogenetic analyses have shown that swine-derived HEV nucleotide sequences are genetically closely related to human HEV isolates, suggesting that pigs serve as reservoirs of human infections [9, 10]. Moreover, serological studies have shown a higher prevalence of HEV infections among workers exposed to pork meat, indicating that these individuals have a higher risk of HEV infection [11, 12].
Hepatitis E virus belongs to the family of Hepeviridae, which consists of the 2 genera Piscihepevirus and Orthohepevirus [13]. Orthohepeviruses include the 4 species Orthohepevirus A–D, which can infect several mammalian and avian species. Orthohepevirus A is the most important species; it has been isolated from humans, pigs, wild boars, deer, rabbits, and camels [13]. It includes 8 HEV genotypes (HEV-1 to HEV-8) that are characterized based on the phylogeny of entire viral genomes [14, 15]. The HEV genotypes have distinct geographical distributions and clinical features. Although severe liver disease in pregnancy caused by HEV-3 and -4 has so far not been reported, HEV-1 and -2 can cause critical disease in pregnant women [5]. Hepatitis E virus-1 is widely distributed in Asia, and HEV-2 predominates in Africa and Mexico. Both genotypes exclusively infect humans, and they are responsible for substantial waterborne outbreaks in developing countries. Hepatitis E virus-3 and -4 are meanwhile distributed globally, infecting animals and, through consumption of undercooked meat, also humans [14, 16]. Genotypes HEV-5 and -6 have been isolated from wild boars in Japan [13, 17], and, recently, the 2 novel HEV strains HEV-7 and HEV-8 were isolated from camels [15, 18, 19]. Hepatitis E virus-1 and HEV-2 generally cause severe acute hepatitis, but not chronic infection [20], whereas genotypes HEV-3, -4, and -7 may be the cause of acute hepatitis and of chronic hepatitis in immunocompromised patients [19, 21, 22].
In Vietnam, hepatitis E is a significant public health concern. We have previously shown that HEV circulates among healthy Vietnamese individuals and in hepatitis B virus-infected patients with anti-HEV immunoglobulin (Ig)G seroprevalences of 31% and 45%, respectively [23]. A previous study found 19.1% and 8.2% positivity of HEV-3 viral ribonucleic acid (RNA) in fecal samples and in rectal swabs from pigs, respectively, as well as a HEV seroprevalence of 16% in pig farmers in southern Vietnam [24]. However, the molecular epidemiology of HEV infection both in animals and humans is not yet completely understood. The present cross-sectional study aims to assess molecular epidemiological characteristics and seroprevalences of HEV infection in domestic pigs and, in particular, in workers occupationally exposed to pigs and pork meat and in healthy controls to determine the burden and zoonotic transmission dynamics of HEV infections in northern Vietnam.
METHODS
Study Design and Sample Collection
This study was implemented between January 2016 and June 2017. The sample size was determined assuming an expected prevalence of 10% of HEV-RNA positivity in pig liver tissues, 10% of HEV anti-IgM positivity among occupationally exposed individuals, and 8% among a sample of the healthy general population at a 95% confidence level and a 5% margin of error. We estimated that a sample size of at least 139 liver tissues from domestic pigs and 139 and 114 serum samples from occupationally exposed workers and healthy individuals, respectively, was required.
Liver tissues of domestic pigs (n = 210) were obtained from 6 markets, namely, Ngoc My, Ngoc Than, Quoc Oai, Cau Kiem, Huu Bang, and Thach That markets in the Hanoi metropolitan area. Liver tissues (~3 mm × 3 mm × 3 mm in size) from each pig were collected, preserved in 0.5 mL TRIzol reagent, and frozen until further use. In addition, blood samples from individuals (n = 283) occupationally exposed to pigs/pork meat were collected in the Quoc Oai and Thach That districts, Hanoi. Exposed individuals were further classified as pork meat vendors (n = 81), pig farmers (n = 96), and personnel employed in slaughterhouses (n = 106). Blood samples from unexposed healthy individuals (n = 168) were obtained from blood banks.
Five milliliters of venous blood were collected from all participants. Sera were separated and stored until further use. Informed written consent was obtained at the time of sampling from all study participants. The study was approved by the Institutional Review Board of Vietnam Military Medical University, Hanoi, Vietnam.
Serological Testing for Antihepatitis E Virus Antibodies
Anti-HEV IgG and IgM were determined in sera from workers exposed to domestic swine and pork meat by enzyme-linked immunosorbent assays (ELISAs) (MP Biomedicals, Santa Ana, CA) according to the manufacturer’s instructions. The MP HEV IgM ELISA 3.0 is an indirect immunoassay that utilizes a highly conserved conformational epitope derived from the open reading frame 2 (ORF2) of the virus. Immunoglobulin M antibodies were detected by monoclonal mouse antihuman IgM antibodies labeled with horseradish peroxidase. Sensitivity and specificity of the assay were 99.3% and 97.6%, respectively. The MP Diagnostics HEV-IgG ELISA utilizes recombinant HEV antigens derived from the structural region of the viral genome. The test was considered positive when the optical density was ≥0.4 + nonreactive control mean (NRCx) or ≥0.5 + NRCx for IgM and IgG, respectively.
Hepatitis E Virus-Ribonucleic Acid Detection in Pig Liver Tissues and Human Sera
Total RNA was extracted from 210 pig liver tissues with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). Efforts were made to isolate viral RNA from sera obtained from 283 workers exposed to pigs/pork meat and 168 healthy unexposed individuals (QIAamp Viral RNA Mini Kit; QIAGEN GmbH, Hilden, Germany). Hepatitis E virus-RNA was reversely transcribed into complementary deoxyribonucleic acid (QuantiTect Reverse Transcription Kit; QIAGEN GmbH).
Nested Polymerase Chain Reaction
The presence of HEV-RNA was examined using a nested polymerase chain reaction (PCR) assay. In brief, primers were designed based on the RNA-dependent RNA polymerase (RdRp) region, which belongs to the viral ORF1. Outer primer pairs were HEV-38 (sense) 5’-GAG GCY ATG GTS GAG AAR G-3’ and HEV-39 (antisense) 5’-GCC ATG TTC CAG ACR GTR TTC C-3’; the inner primers were HEV-37 (sense) 5’-GGT TCC GYG CTA TTG ARA ARG-3’ and HEV-27 (antisense) 5’-TCR CCA GAG TGY TTC TTC C-3’. All positive and suspected positive samples were either confirmed or excluded applying an additional nested-PCR, using primers that amplified a 497-base pair (bp) fragment of ORF2. Outer primers were HEV-34 (sense) 5’-CCG ACG TCY GTY GAY ATG AA-3’ and HEV-36 (antisense) 5’-TTR TCC TGC TGA GCR TTC TC-3´; inner primers were HEV-35 (sense) 5’-AAG TGA GCG CCT ACA YTA YCG-3’ and HEV-29 (antisense) 5’-CTC GCC ATT GGC TGA GAC-3’.
Hepatitis E Virus Genotyping and Phylogenetic Analysis
Amplified PCR products were purified (Exo-SAP-IT kit; USB, Affymetrix, Santa Clara, CA) and applied as templates for sequencing (Bigdye Terminatior v3.1 cycle sequencing kit; Applied Biosystems, Foster City, CA) and the ABI 3130XL sequencer system. Hepatitis E virus genotyping was performed through phylogenetic analyses based on sequences of the ORF1 RdRp region using the MEGA7 software (www.megasoftware.net). All sequences were edited and aligned using BioEdit software version 7.0 (http://bioedit.software.informer.com/7.0) and the CLUSTAL Muscle algorithm. Phylogenetic trees were constructed using the neighbor-joining method and the Kimura-2 model. Statistical robustness and reliability of the branching order was confirmed by bootstrap analysis using 1000 reiterations. All HEV reference sequences belonging to the 8 HEV genotypes were obtained from the NCBI GenBank database (HEV-1: D11093, L08816, JF443721, X98292, AF051830, M73218, AY204877, AY230202; HEV-2: M74506; HEV-3: FJ426403, AF060669, AF082843, AB089824, AB291963, B189071, AY115488, AB591734, AF455784, FJ705359, FJ705359, FJ653660, AB481226, AB248521, FJ906895; HEV-4: GU1199661, JQ655735, AJ344171, B197673, AB220974, DQ279091, Y723745, DQ450072, AB108537; HEV-5: AB573435; HEV-6: AB856243, AB602441; HEV-7: KJ496143, KJ496144; HEV-8: KX387865, KX387867).
Statistical Analysis
All analyses were performed useing the R software (https://www.r-project.org/). Fisher’s and χ 2 exact tests were used to compare the prevalence of HEV infection between groups. Mann-Whitney Wilcoxon test was used to compare the non-parametric data of quantitative variables between 2 groups. The level of significance was set at a P value of <.05.
RESULTS
Demographic Characteristics of the Study Subjects
The median age did not differ between occupationally exposed and control individuals (median age 42 [range, 18–78] vs 40 [range, 18–70], respectively; P > .05). There was a significant difference in gender distribution between the 2 groups in which one half of unexposed healthy individuals was male and 70% of occupationally exposed employees were female (P < .05).
Seroprevalence of Hepatitis E Virus Infection in Healthy Individuals and Personnel Professionally Exposed to Pigs and Pork Meat
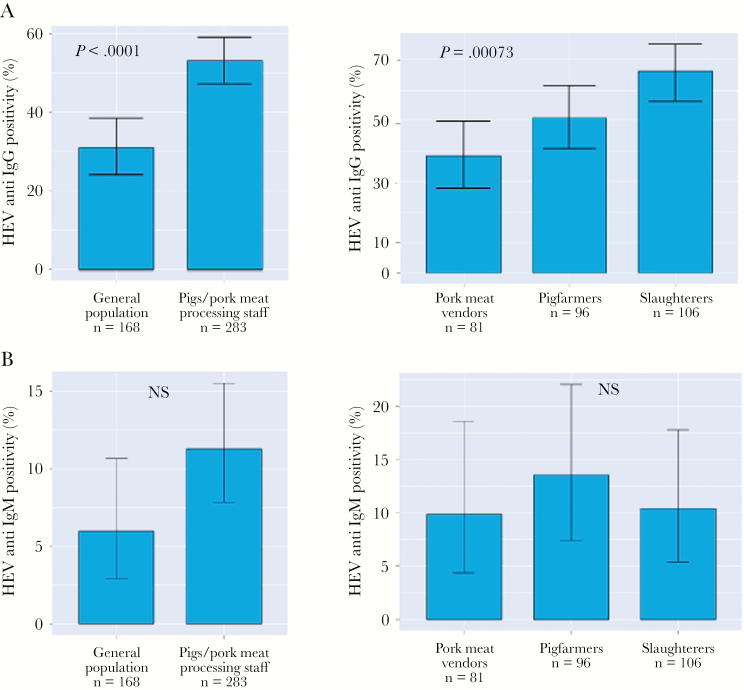
Hepatitis E virus seroprevalence rates were significantly higher in individuals routinely exposed to pigs and pork meat compared with the controls. This applied for both anti-HEV IgM (11.3%, 95% confidence interval [CI] = 7.8%–15.5% vs 6%, 95% CI = 3%–11%; P = .07) and anti-HEV IgG (53%, 95% CI = 47%–59% vs 31%, 95% CI = 24%–38%; P < .0001) (Figure 1A and B). When stratifying the 283 individuals permanently exposed to pig contacts and pork meat for the subgroups of meat vendors, slaughterers, and pig farmers, we observed positivity rates of anti-HEV IgG among slaughterers of 66% (95% CI = 56%–75%), followed by 51% of pig farmers (95% CI = 41%– 61%) and 38% of pork meat vendors (95% CI = 28%–50%; P = .00073) (Figure 1A). The prevalences of anti-HEV IgM were 13.5% in pig farmers (95% CI = 7.4%–22%), 11% in slaughterers (95% CI = 5.3%–19%), and 10% among pork meat vendors (95% CI = 5%–18%) (Figure 1B).
Figure 1.
Hepatitis E virus (HEV) seroprevalence in case and control group. Anti-HEV immunoglobulin (Ig)G positivity rates (A) and anti-HEV IgM (B) in the healthy control group, in individuals constantly exposed to pigs and pork meat, and in subgroups classified according to their specific occupation. Data are given as percentages and 95% confidence intervals. NS, not significant.
Detection of Hepatitis E Virus-Ribonucleic Acid
Liver tissue samples were randomly collected from 210 domestic pigs in markets of the Hanoi metropolitan area. All pigs were between 5 and 10 months of age. Hepatitis E virus-RNA was detected by an in-house nested PCR assay [23]. Overall, 26 of 210 pig liver samples (12.4%) were positive for HEV-RNA. Nineteen of the 26 samples were successfully sequenced, whereas the remainder of 7 samples could not be sequenced, indicating a low level of HEV replication in these liver tissues. We did not detect HEV-RNA in any of the human serum samples.
Phylogenetic Analysis
Phylogenetic analyses involving the 306-bp fragment of the RdRp of the ORF1 region revealed that the 19 isolates identified in domestic pigs belong to the HEV genotype 3 (Figure 2A). All sequences were submitted to the GenBank database (accession numbers MH777770 to MH777788). We further analyzed 19 HEV-3 isolates to characterize sub-genotypes. Most of the HEV-3 isolates (17 of 19) were HEV genotype 3b; the remaining 2 isolates belonged to genotype 3a (Figure 2B).
Figure 2.
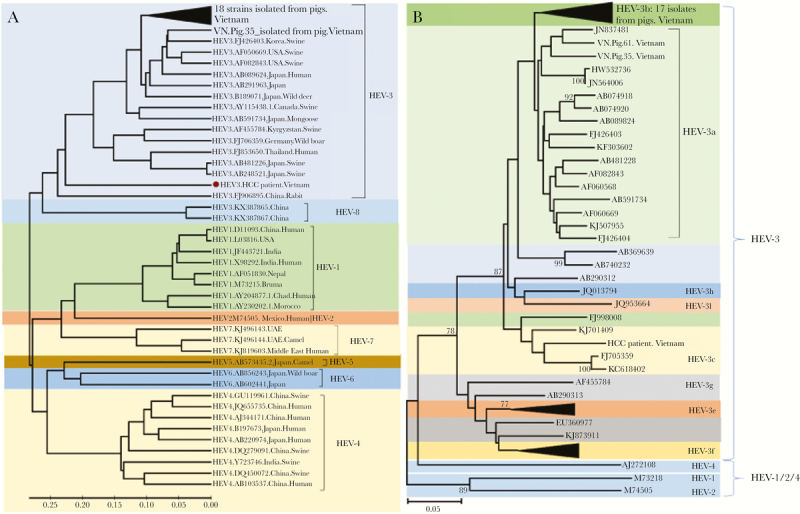
Phylogenetic analysis of 19 hepatitis E virus (HEV) strains isolated from domestic pigs. (A) Phylogenetic tree was constructed based on the alignment of 306 base pairs of the HEV RNA-dependent RNA polymerase (RdRp) region (ORF1) of 19 nucleotide sequences isolated from domestic pigs, 1 HEV strain isolated from a liver cancer patient coinfected with hepatitis B virus in Vietnam. Forty full-length HEV genomes isolated from animals and humans (HEV-1 to HEV-8) retrieved from the NCBI database along with GenBank accession numbers were included in the analysis. A neighbor-joining tree was constructed with a bootstrap of 1000 replicates. Genetic distances that are in the units of the number of base substitutions per site were computed using the Kimura-parameter method are given. The bar at the base of the tree indicates the scale for nucleotide substitutions per position. (B) Phylogenetic tree constructed for all identified HEV genotype 3 sequences. The analysis involved 34 nucleotide sequences, including 19 HEV strains isolated from domestic pigs in Vietnam. Bootstrap analysis values (percentages) are shown.
DISCUSSION
The zoonotic transmission pattern of HEV infection has been recognized in several European countries, and, meanwhile, HEV infection has become a significant public health concern globally, particularly in transplantation patients and in patients immunocompromised due to other reasons [3]. Currently, most cases of chronic hepatitis E in Europe are caused by HEV-3 [25]. In developing countries including Vietnam, HEV morbidity is mostly caused by the HEV-1 genotype, which is transmitted mainly through the fecal-oral route. Genotypes HEV-3 and -4 have been reported to exist in Vietnam; however, only scarce information on their prevalences, transmission patterns, and disease characteristics is available. In the present study, we aimed to investigate the prevalence of HEV infection and to molecularly characterize HEV strains occurring in Northern Vietnam. We could confirm that HEV is circulating among domestic pigs and constitutes a zoonotic disease risk.
Reported prevalence rates of HEV infection in Vietnam differ considerably between studies due to varying methodological approaches and study groups included [23, 26–28]. Our previous study has provided evidence of a far higher rate of anti-HEV IgG seroprevalence in the general population (31%) compared with an earlier study indicating a seroprevalence rate of 9% only [23, 26]. In addition, the seroprevalence of anti-HEV IgG and IgM in this study is slightly higher compared with a previous study observed in general and occupational populations in different parts of China [29]. This difference is attributable to the sensitivity of the ELISA test systems applied, which differ substantially with regard to their sensitivity [30, 31]. In the current study, we used the MP diagnostics HEV-IgM/IgG ELISA kit, which both have a high sensitivity and specificity, and found higher rates of HEV infection in swine-exposed workers (53%) compared with healthy unexposed individuals (31%). There is no doubt that individuals involved in handling pig and pork meat are at an increased risk of zoonotic HEV transmission [32], and an epidemiological and genetic link has been established between hepatitis E cases and consumption of undercooked pork meat, clearly indicating this zoonotic pattern of transmission [9, 10, 33]. Although this cross-sectional study provides indirect evidence of HEV transmission between human and swine, the high HEV seroprevalence among the occupationally exposed implies a potential risk of transmission from the exposed to the unexposed community in Vietnam.
Several studies on HEV-RNA prevalence from swine have utilized a nested reverse-transcription PCR (RT-PCR) methodology for HEV-RNA detection [33–37]. Although there is no consensus on a standard quantitative PCR methodology for HEV-RNA quantification, in-house methods have frequently been applied for HEV-RNA detection and subsequent quantification of viral loads [38–41]. It has been shown that detecting HEV-RNA using nested RT-PCR methodology is as highly sensitive as quantitative RT-PCR [34]. In Asia, the presence of HEV-RNA in pig livers collected in slaughterhouses or markets ranges from 0.3% to 11% [33–37]. We observed a still higher prevalence of HEV-RNA (12.4%) in retail pig liver products in Hanoi markets compared with previous studies performed in Asia. For instance, HEV positivity in pigs was 2%–5% in Japan [33, 34] and approximately 5% in China [35, 36]. However, a significantly lower prevalence of HEV-RNA in pigs was reported from Thailand (0.23%) [37] and Hong Kong (1.5%) [32]. The prevalence of HEV-RNA in pig liver-derived food products is much higher in several European countries, where HEV-3 and -4 are endemic and constitute the main cause of zoonotic infection. For instance, HEV-RNA was detected in 10 of 90 (11.1%) meat products, 7 of 37 (18.9%) liver sausages, and 3 of 53 (5.7%) raw meat sausages in Switzerland [40]. Hepatitis E virus-RNA positivity in pig livers can be as high as 20% in Germany [38], 30% in France [39], and even in 31% in Hungary [41].
Our findings are consistent with a recent study showing that all sequences retrieved from positive samples of pig livers or feces belong to the HEV-3 genotype [24]. In Vietnam, pig livers are very common in markets and constitute a potential reservoir for HEV-3 infections. In this study, we could not retrieve HEV-RNA from any human serum sample collected from individuals continuously exposed to pigs and pork meat and from the controls. In contrast, in a previous study, HEV strains were successfully isolated from 9 of 141 sera from patients with acute sporadic hepatitis in Hanoi, and all of them had sequences closely related to the genotype HEV-4 [42]. Another study also has described a 56-year-old Japanese male who acquired an HEV-4 infection after ingestion of uncooked shellfish while traveling in Vietnam [43]. This and our previous and present findings speculate that HEV genotypes 3 and 4 circulate in Vietnam and may constitute a zoonotic disease risk.
Understanding the molecular epidemiology and transmission route of zoonotic pathogens such as HEV-3 and -4 is important in Vietnam, because the burden of HEV infection and related liver diseases or extrahepatic disorders is still underestimated. Indeed, there have been no studies so far regarding the cases of chronic hepatitis E in immunocompromised individuals and HEV infection-related cases of neurological (eg, neuralgic amyotrophy, Guillain-Barré syndrome), hematological (eg, thrombocytopenia), gastroenterological (eg, acute pancreatitis), and nephrological conditions (eg, glomerulonephritis) [44]. In addition, tools for routine testing for hepatitis E are not available in most medical units and hospitals in Vietnam, and awareness about the occurrence of hepatitis E needs to be raised.
CONCLUSIONS
This study indicates a high prevalence of HEV infection in domestic pigs and individuals particularly exposed to pigs and pork meat, but also among the controls involved. Our study provides insight into HEV transmission dynamics and shows that domestic pigs may be an important zoonotic reservoir for HEV infection in Vietnam. Although information on HEV genotypes infecting humans is still scarce, we could infer that the genotypes HEV-3 and -4 may be the cause of acute sporadic hepatitis E, rather than other genotypes. Further studies on the occurrence of zoonotic hepatitis E in Vietnam are required.
Acknowledgments
We thank all study subjects for their participation.
Author contributions. T. P. V. designed and supervised the study and contributed to the materials and reagents. P. X. H., N. L. T., B. T. S., C.-T. B., and H. V. T. participated in the study design, recruited participants, and collected samples. N. X. H., P. X. H., T. V. S., B. T. S., D. P. G., M. T. B., and D. T. A. performed the experimental procedures. N. X. H. and B. W. performed the statistical and phylogenetic analysis. T. P. V. and P. G. K. contributed to materials and reagents. N. X. H. and T. P. V. wrote the manuscript. H. V. T. and C. G. M. revised the main draft. All authors agreed with the results and conclusions.
Disclaimer. The funder has no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Financial support. This study was supported by Deutscher Akademischer Austauschdienst-Partnerschaften für den Gesundheitssektor in Entwicklungsländern (DAAD-PAGEL) (57140033, 57445019) for student fellowship. We acknowledge the financial support from the Federal Ministry of Education and Research, Germany (Projekt Nr. BMBF01DP19006A; BMBF01DP17047) and from the Ministry of Science and Technology, (MoST) Vietnam (Project ID: HEPNET-010).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.World Health Organization. 2016. Geneva, Switzerland: WHO. Available at: https://www.who.int/news-room/fact-sheets/detail/hepatitis-e [Google Scholar]
- 2. Kamar N, Dalton HR, Abravanel F, Izopet J. Hepatitis E virus infection. Clin Microbiol Rev 2014; 27:116–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arends JE, Ghisetti V, Irving W, et al. . Hepatitis E: an emerging infection in high income countries. J Clin Virol 2014; 59:81–8. [DOI] [PubMed] [Google Scholar]
- 4. Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N Engl J Med 2012; 367:1237–44. [DOI] [PubMed] [Google Scholar]
- 5. Pérez-Gracia MT, Suay-García B, Mateos-Lindemann ML. Hepatitis E and pregnancy: current state. Rev Med Virol 2017; 27:e1929. [DOI] [PubMed] [Google Scholar]
- 6. Kamar N, Selves J, Mansuy JM, et al. . Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 2008; 358:811–7. [DOI] [PubMed] [Google Scholar]
- 7. Ruggeri FM, Di Bartolo I, Ponterio E, et al. . Zoonotic transmission of hepatitis E virus in industrialized countries. New Microbiol 2013; 36:331–44. [PubMed] [Google Scholar]
- 8. Takahashi M, Okamoto H. Features of hepatitis E virus infection in humans and animals in Japan. Hepatol Res 2014; 44:43–58. [DOI] [PubMed] [Google Scholar]
- 9. Tei S, Kitajima N, Takahashi K, Mishiro S. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 2003; 362:371–3. [DOI] [PubMed] [Google Scholar]
- 10. Bouquet J, Cheval J, Rogée S, et al. . Identical consensus sequence and conserved genomic polymorphism of hepatitis E virus during controlled interspecies transmission. J Virol 2012; 86:6238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Schryver A, De Schrijver K, François G, et al. . Hepatitis E virus infection: an emerging occupational risk? Occup Med (Lond) 2015; 65:667–72. [DOI] [PubMed] [Google Scholar]
- 12. Teixeira J, Mesquita JR, Pereira SS, et al. . Prevalence of hepatitis E virus antibodies in workers occupationally exposed to swine in Portugal. Med Microbiol Immunol 2017; 206:77–81. [DOI] [PubMed] [Google Scholar]
- 13. Smith DB, Simmonds P, Jameel S, et al. . Consensus proposals for classification of the family Hepeviridae. J Gen Virol 2014; 95:2223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Tong H, Hoan NX, Wang B, et al. . Hepatitis E virus mutations: functional and clinical relevance. EBioMedicine 2016; 11:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woo PC, Lau SK, Teng JL, et al. . New hepatitis E virus genotype in Bactrian Camels, Xinjiang, China, 2013. Emerg Infect Dis 2016; 22:2219–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kamar N, Bendall R, Legrand-Abravanel F, et al. . Hepatitis E. Lancet 2012; 379:2477–88. [DOI] [PubMed] [Google Scholar]
- 17. Takahashi M, Nishizawa T, Sato H, et al. . Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J Gen Virol 2011; 92:902–8. [DOI] [PubMed] [Google Scholar]
- 18. Sridhar S, Teng JL, Chiu TH, et al. . Hepatitis E virus genotypes and evolution: emergence of camel hepatitis E variants. Int J Mol Sci 2017; 18:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee GH, Tan BH, Teo EC, et al. . Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 2016; 150:355–7-e3. [DOI] [PubMed] [Google Scholar]
- 20. Aggarwal R, Jameel S. Hepatitis E. Hepatology 2011; 54:2218–26. [DOI] [PubMed] [Google Scholar]
- 21. Geng Y, Zhang H, Huang W, et al. . Persistent hepatitis E virus genotype 4 infection in a child with acute lymphoblastic leukemia. Hepat Mon 2014; 14:e15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujiwara S, Yokokawa Y, Morino K, et al. . Chronic hepatitis E: a review of the literature. J Viral Hepat 2014; 21:78–89. [DOI] [PubMed] [Google Scholar]
- 23. Hoan NX, Tong HV, Hecht N, et al. . Hepatitis E virus superinfection and clinical progression in Hepatitis B patients. EBioMedicine 2015; 2:2080–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berto A, Pham HA, Thao TT, et al. . Hepatitis E in southern Vietnam: Seroepidemiology in humans and molecular epidemiology in pigs. Zoonoses Public Health 2018; 65:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lapinski TW, Jaroszewicz J. Hepatitis E virus infection--a new threat for Europe. Przegl Epidemiol 2016; 70:11–4, 103–106. [PubMed] [Google Scholar]
- 26. Hau CH, Hien TT, Tien NT, et al. . Prevalence of enteric hepatitis A and E viruses in the Mekong River delta region of Vietnam. Am J Trop Med Hyg 1999; 60:277–80. [DOI] [PubMed] [Google Scholar]
- 27. Corwin AL, Khiem HB, Clayson ET, et al. . A waterborne outbreak of hepatitis E virus transmission in southwestern Vietnam. Am J Trop Med Hyg 1996; 54:559–62. [DOI] [PubMed] [Google Scholar]
- 28. Tran HT, Ushijima H, Quang VX, et al. . Prevalence of hepatitis virus types B through E and genotypic distribution of HBV and HCV in Ho Chi Minh City, Vietnam. Hepatol Res 2003; 26:275–80. [DOI] [PubMed] [Google Scholar]
- 29. Yue N, Wang Q, Zheng M, et al. . Prevalence of hepatitis E virus infection among people and swine in mainland China: a systematic review and meta-analysis. Zoonoses Public Health 2019; 66:265–75. [DOI] [PubMed] [Google Scholar]
- 30. Abravanel F, Lhomme S, Chapuy-Regaud S, et al. . Hepatitis E virus reinfections in solid-organ-transplant recipients can evolve into chronic infections. J Infect Dis 2014; 209:1900–6. [DOI] [PubMed] [Google Scholar]
- 31. Mansuy JM, Bendall R, Legrand-Abravanel F, et al. . Hepatitis E virus antibodies in blood donors, France. Emerg Infect Dis 2011; 17:2309–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan MC, Kwok K, Hung TN, Chan PK. Molecular epidemiology and strain comparison between Hepatitis E viruses in human Sera and pig livers during 2014 to 2016 in Hong Kong. J Clin Microbiol 2017; 55:1408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yazaki Y, Mizuo H, Takahashi M, et al. . Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J Gen Virol 2003; 84:2351–7. [DOI] [PubMed] [Google Scholar]
- 34. Okano H, Takahashi M, Isono Y, et al. . Characterization of sporadic acute hepatitis E and comparison of hepatitis E virus genomes in acute hepatitis patients and pig liver sold as food in Mie, Japan. Hepatol Res 2014; 44:E63–76. [DOI] [PubMed] [Google Scholar]
- 35. Li W, Shu X, Pu Y, et al. . Seroprevalence and molecular detection of hepatitis E virus in Yunnan Province, China. Arch Virol 2011; 156:1989–95. [DOI] [PubMed] [Google Scholar]
- 36. Li W, She R, Wei H, et al. . Prevalence of hepatitis E virus in swine under different breeding environment and abattoir in Beijing, China. Vet Microbiol 2009; 133:75–83. [DOI] [PubMed] [Google Scholar]
- 37. Intharasongkroh D, Sa-Nguanmoo P, Tuanthap S, et al. . Hepatitis E virus in pork and variety meats sold in fresh markets. Food Environ Virol 2017; 9:45–53. [DOI] [PubMed] [Google Scholar]
- 38. Szabo K, Trojnar E, Anheyer-Behmenburg H, et al. . Detection of hepatitis E virus RNA in raw sausages and liver sausages from retail in Germany using an optimized method. Int J Food Microbiol 2015; 215:149–56. [DOI] [PubMed] [Google Scholar]
- 39. Pavio N, Merbah T, Thébault A. Frequent hepatitis E virus contamination in food containing raw pork liver, France. Emerg Infect Dis 2014; 20:1925–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moor D, Liniger M, Baumgartner A, Felleisen R. Screening of ready-to-eat meat products for hepatitis E virus in Switzerland. Food Environ Virol 2018; 10:263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Forgách P, Nowotny N, Erdélyi K, et al. . Detection of hepatitis E virus in samples of animal origin collected in Hungary. Vet Microbiol 2010; 143:106–16. [DOI] [PubMed] [Google Scholar]
- 42. Hijikata M, Hayashi S, Trinh NT, et al. . Genotyping of hepatitis E virus from Vietnam. Intervirology 2002; 45:101–4. [DOI] [PubMed] [Google Scholar]
- 43. Koizumi Y, Isoda N, Sato Y, et al. . Infection of a Japanese patient by genotype 4 hepatitis e virus while traveling in Vietnam. J Clin Microbiol 2004; 42:3883–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pischke S, Hartl J, Pas SD, et al. . Hepatitis E virus: infection beyond the liver? J Hepatol 2017; 66:1082–95. [DOI] [PubMed] [Google Scholar]