Abstract
The G-quadruplex-forming VEGF-binding aptamer V7t1 was previously found to be highly polymorphic in a K+-containing solution and, to restrict its conformational preferences to a unique, well-defined form, modified nucleotides (LNA and/or UNA) were inserted in its sequence. We here report an in-depth biophysical characterization of V7t1 in a Na+-rich medium, mimicking the extracellular environment in which VEGF targeting should occur, carried out combining several techniques to analyse the conformational behaviour of the aptamer and its binding to the protein. Our results demonstrate that, in the presence of high Na+ concentrations, V7t1 behaves in a very different way if subjected or not to annealing procedures, as evidenced by native gel electrophoresis, size exclusion chromatography and dynamic light scattering analysis. Indeed, not-annealed V7t1 forms both monomeric and dimeric G-quadruplexes, while the annealed oligonucleotide is a monomeric species. Remarkably, only the dimeric aptamer efficiently binds VEGF, showing higher affinity for the protein compared to the monomeric species. These findings provide new precious information for the development of improved V7t1 analogues, allowing more efficient binding to the cancer-related protein and the design of effective biosensors or theranostic devices based on VEGF targeting.
INTRODUCTION
The Vascular Endothelial Growth Factor (VEGF) family comprises several cytokine homodimeric proteins playing essential roles in physiological angiogenesis, lymphangiogenesis and vasculogenesis (1). Besides being involved in physiological processes, VEGF is also implicated in the development and progression of several diseases, such as age-related macular degeneration (2), diabetes (3), rheumatoid arthritis (4) and numerous cancer forms (5–9). Several studies have also suggested its key role in angiogenesis and vascularization of a large number of solid tumours, whereby the serum levels of VEGF are considered a useful tumour marker (10–12). The VEGF family includes six glycoproteins: VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGF-E and the Placenta Growth Factor (PlGF) (13). The most important protein in this family is VEGF-A, which presents five different isoforms consisting of 121, 145, 165, 189 and 206 amino acids (14). In a large variety of biological systems, VEGF165 and VEGF121 represent the most abundant isoforms, with distinct activities in promoting tumour angiogenesis (15,16). The identification of VEGF as the main regulator of angiogenesis has led to recognize it as a major therapeutic target and therefore it is not surprising that most anti-angiogenic strategies are based on VEGF inhibition (17–19). Using SELEX (Systematic Evolution of Ligands by Exponential Enrichment) strategies, various oligonucleotide aptamers have been identified against this target. Among them, pegaptanib sodium, the active component of Macugen, was approved by FDA in 2004 for the treatment of age-related macular degeneration (20).
In 2010, Nonaka et al. (21) identified the DNA aptamer Vap7 by three rounds of SELEX towards the isoform VEGF121, and subsequently evolved a truncated form, named V7t1, able to bind the two most abundant VEGF isoforms, VEGF121 and VEGF165. From the dissociation constant values, determined by Surface Plasmon Resonance (SPR) experiments, they showed that V7t1 has even higher affinity than Vap7 for VEGF165 (Kd = 1.4 versus 20 nM), with comparable affinity for VEGF121 (Kd ∼ 1.1 nM).
V7t1, with the sequence d(TGTGGGGGTGGACGGGCCGGGTAGA), is a 25-mer DNA oligonucleotide presenting four G-rich tracts, respectively consisting of five, two, three and again three consecutive guanine residues, along with two additional isolated guanines, that offer different alternatives for the formation of a G-quadruplex (G4) core.
Plavec et al. analysed this aptamer in K+-containing solutions, evidencing its marked structural polymorphism and, to reduce it, investigated V7t1 analogues containing LNA (locked nucleic acid) and UNA (unlocked nucleic acid) monomers in various positions of its sequence (22). Insertion of three LNA monomers at specific sites of the oligonucleotide sequence provided the so-called RNV66 aptamer, forming a well-defined, parallel G4 structure, with three stacked G-tetrads (22).
Among the studied V7t1 analogues, RNV66 proved to be the best candidate, showing improved antiproliferative activity on human breast cancer cells with respect to the unmodified V7t1 aptamer, also displaying significant antitumoral effects in breast cancer mice models (23). Molecular docking followed by molecular dynamic simulations on RNV66 showed that this aptamer is able to bind to VEGF in the receptor-binding region, thus inhibiting its recognition by VEGFR2 and further confirming its very promising therapeutic potential (23).
Stimulated by the discovery of RNV66, we decided to revisit the unmodified V7t1 aptamer, whose structural features, and particularly bioactive conformation, are still largely unknown. Indeed, Plavec et al. (22) provided only preliminary information on V7t1, proving its ability to form several G4 structures in a K+-containing solution, apparently also stabilized by a few Watson–Crick hydrogen-bonded GC base pairs, as evidenced by the 1H NMR analysis of the imino protons region.
In particular, nothing is known on the structure and properties of V7t1 in solutions with high content of Na+ ions, which represent the saline conditions of the extracellular environment, in which VEGF is most abundant and its recognition should occur.
Taking into account the strong interest for VEGF as an essential therapeutic target and the antiproliferative activity shown by V7t1 on several cancer cell lines (23), a detailed analysis has been here undertaken in order to elucidate the biophysical properties of this oligonucleotide in a Na+-rich buffer. In particular, we have studied the conformational behaviour of V7t1 and the thermal stability of its structures in solution exploiting different techniques, i.e. gel electrophoresis, size exclusion chromatography (SE-HPLC), dynamic light scattering (DLS), circular dichroism (CD), UV spectroscopy and differential scanning calorimetry (DSC), also analysing its resistance to nuclease digestion. A Na+-rich buffered solution (25 mM HEPES, 150 mM NaCl, pH 7.4, here named HEPES/Na+) was selected as a good mimic of the extracellular media.
Additionally, the ability of V7t1 to bind VEGF165 was investigated by electrophoretic mobility shift assay (EMSA), revealing new, unexpected insights on the conformations of V7t1 preferentially recognized by the protein.
MATERIALS AND METHODS
Preparation of the oligonucleotide samples
The sequences of the here studied oligonucleotides are listed in the Supplementary Material and are all purchased from biomers.net GmbH, Germany. Purified and lyophilized V7t1 was dissolved in Milli-Q water and its concentration was determined by UV–vis spectroscopy in a 1 cm path length cuvette, measuring the absorbance at 260 nm (at 90°C) and using the molar extinction coefficient ϵ260 = 281 000 cm−1 M−1. Then, the oligonucleotide was diluted from the initial stock solution (>280 μM) in the selected Na+-rich buffer solution (25 mM HEPES, 150 mM NaCl, pH 7.4, here indicated as HEPES/Na+). Samples of annealed V7t1 were obtained by heating the appropriate aptamer solution at 95°C for 5 min and then leaving it to slowly cool to r.t. overnight, to allow their structuring into the thermodynamically most stable conformations (24). Not-annealed and annealed V7t1 samples were then kept at 4°C until use.
Protein samples
Recombinant human VEGF165 (GenScript) was purchased from TwinHelix srl (Italy) and prepared according to the manufacturer's instructions. Protein concentration was confirmed by Bradford assay (Bio-Rad) using Bovine Gamma Globuline (BGG) as a standard.
Gel electrophoresis
Not-annealed (N.A.) and annealed (A.) V7t1 samples (5 μM), in HEPES/Na+, were mixed with 4× loading buffer (20% glycerol, 0.1% bromophenol blue in 20 mM Tris, pH 6.8) and loaded on 10% polyacrylamide gels with TBE (Tris–borate–EDTA) 0.5× as running buffer. Other oligonucleotides used in PAGE experiments as size references and their loading concentrations on the gels were: the 26-mer tel26 from human telomere (2 μM), known to adopt a monomolecular G4 structure in solution (25,26), the 50-mer human telomeric DNA sequence (0.5 μM) able to form two consecutive G-quadruplex structures (25) and the 42-mer R1.2 (27,28) as a control G-rich oligonucleotide (3 μM). Their sequences are specified in the Supplementary Material. The gels were then run at 100 V for 45 min at r.t., stained with GelGreen Nucleic Acid Stain in 0.1 M NaCl for 30 min and visualized with a UV transilluminator (BioRad ChemiDoc XRS). For the analysis of peak 1 and peak 2 (see below), proper solutions in HEPES/Na+ (4 and 3 μM, respectively), were supplemented with 5% glycerol immediately before their loading on 10% polyacrylamide gel with TBE 1× as running buffer. Then the gel was run at r.t. and 70 V for 2 h, finally stained and visualized as described above.
Size exclusion chromatography
SE-HPLC analyses were performed using an Agilent HPLC system, equipped with a UV/vis detector, on a Yarra 3 μm analytical column (300 × 4.60 mm; Phenomenex). The elution was monitored at λ = 254 nm with 0.2 ml min−1 flow rate. In order to directly compare the elution behaviour of different oligonucleotide systems, the absorbance values at 254 nm were plotted against the relative elution volumes Ve/V0 as x-axis, where Ve is the elution volume and V0 is the dead volume (1.86 ml). The mobile phases used were: (i) HEPES/Na+ (pH 6.8) for not-annealed and annealed V7t1 (2 μM); (ii) 10 mM phosphate buffer/100 mM KCl (pH 6.8), for TBA, TBA–cTBA duplex, tel26 and its dimer (2 μM) and (iii) 5 mM phosphate buffer/100 mM KCl (pH 6.8) for c-kit2 (5 μM). The solutions were chosen so that the reference oligonucleotides adopted a unique, well-defined secondary structure in solution. For the HPLC separation of peak 1 and peak 2, a ca. 70 μM solution of not-annealed V7t1 was used: the concentration of the two species separated by HPLC was determined by UV analysis, as described above.
Dynamic light scattering
DLS measurements of V7t1 in HEPES/Na+ buffer solution at 50 μM concentration were obtained on a Zetasizer Nano ZS (Malvern Instruments) using 12 mm square polystyrene cuvettes (DTS0012) from Malvern Instruments. In detail, 50 nmol of lyophilized V7t1 were dissolved in 1 ml of the selected buffer and then filtered through a 0.2 μm membrane filter (Merck Millipore), so to obtain a 50 μM solution (ca. 0.4 mg/ml, necessary to obtain a good signal/noise ratio). All the measurements were carried out at r.t. with a scattering angle of 173° and an equilibration time of 60 s. The Z-average (radius, expressed in nm) and polydispersity index (PdI) values, with the associated standard deviations of three determinations, were directly obtained from the measurements fitting the correlation functions with the cumulant analysis algorithm as implemented by the Malvern software (ISO 13321).
Circular dichroism (CD) spectroscopy
CD spectra and CD-melting curves were recorded on a Jasco J-715 spectropolarimeter equipped with a Peltier-type temperature control system (model PTC-348WI), using a quartz cuvette with a path length of 1 cm (3 ml internal volume, Hellma). CD parameters for the V7t1 spectra recording were the following: spectral window 220–320 nm, data pitch 1 nm, band width 2 nm, response 4 s, scanning speed 100 nm/min, 3 accumulations. The experiments were performed on not-annealed and annealed samples in HEPES/Na+ buffer at a concentration of 2 μM. Each experiment was performed in duplicate.
The molar ellipticity [θ] (deg cm2 dmol–1) was calculated from the equation [θ] = θobs/10 × l × C, where θobs is the ellipticity (mdeg), C is the oligonucleotide molar concentration and l is the optical path length of the cell (cm).
Melting/annealing curves were recorded following the CD signal (at 263 and/or 296 nm) upon increasing/decreasing the temperature (1°C/min) in the temperature range 20–90°C. The Tm values were determined as the maxima of the first derivative plots of the melting/annealing curves (associated error ± 1°C). Each experiment was performed in duplicate.
UV spectroscopy
The UV spectra and UV-melting curves were obtained on a Cary 5000 UV-Vis-NIR spectrophotometer equipped with a temperature controller system, using 1 cm path length cuvette (1 ml internal volume, Hellma). The UV–vis spectra were recorded in the range 220–320 nm using a scanning speed of 100 nm/min with the appropriate baseline subtracted. The TDS analysis was carried out by subtracting the UV spectrum recorded at a temperature below the Tm (20°C), at which the aptamer is fully structured, from that obtained at a temperature above the Tm (100°C), when the oligonucleotide is fully destructured (29,30). The absorbance versus temperature profiles of not-annealed and annealed V7t1 samples were recorded following the absorbance changes (at 260 and/or 295 nm) in the temperature range 20–90°C or 20–100°C. The Tm values were calculated as the maxima of the first derivative plots of the melting curves (associated error: ±1°C). Each experiment was performed in duplicate. UV and CD melting curves of annealed V7t1 were also modelled according to the van’t Hoff analysis (29,31), allowing the determination of the thermodynamic parameters of the unfolding processes.
Differential scanning calorimetry
Differential scanning measurements were performed on a last generation nano-DSC (TA Instruments). The excess molar heat capacity function ΔCp was obtained after baseline subtraction, assuming that the baseline is given by the linear temperature dependence of the native-state heat capacity (32). A buffer versus buffer scan was subtracted from the sample scan and linear-polynomial baselines were drawn for each scan. The baseline-corrected thermograms were then normalized with respect to the DNA single strand molar concentration to obtain the corresponding molar heat capacity curves. The measurements were performed with a DNA strand concentration of 140 μM in the selected HEPES/Na+ buffer. All the systems were tested for reversibility by running the heating and cooling curves at the scan rate of 1°C min−1 in the 5–115°C temperature range. A constant pressure of 3 atm was employed to prevent bubble formation upon heating and expand the heating temperature range up to 115°C. Model-free enthalpies were obtained by integrating the area under the heat capacity versus the temperature curves. Tm is the temperature corresponding to the maximum of each DSC peak. Only for the reversible transitions, the entropy values were obtained by integrating the curve ΔCp/T versus T (where ΔCp is the molar heat capacity and T is the temperature in Kelvin).
Enzymatic stability assays
The stability of not-annealed and annealed V7t1 in biological media was analysed by incubating the oligonucleotide samples, dissolved in HEPES/Na+ buffer at 10 μM concentration, in 80% fetal bovine serum (FBS) at 37°C. Then, at fixed times, 2 μl aliquots were withdrawn from the mixtures, added to 4 μl of a gel-loading buffer containing formamide (85% formamide, 2% 0.5 M EDTA pH 8.0, 5% H2O, 8% bromophenol blue) to quench the enzymatic reactions, heated at 95°C for 3 min, and finally stored at –20°C until subsequent analysis. Thereafter, all the samples were analysed by electrophoresis on 20% denaturing polyacrylamide gels using 8 M urea in TBE (Tris–borate–EDTA, 1×) as running buffer. The gels were run at r.t., first for 5 min at 100 V and then for 130 min at 210 V, later stained with GelGreen Nucleic Acid Stain for 30 min and finally visualized with a UV transilluminator (BioRad ChemiDoc XRS). PAGE experiments were carried out using two different reference oligonucleotides, i.e. a 22-mer and a 24-mer of the c-myc sequence, as a length control of the partially digested oligonucleotides (see Supplementary Material).
Electrophoresis mobility shift assay (EMSA)
For the electrophoresis mobility shift assays, 30 pmol of V7t1 were incubated with 40 pmol of VEGF165. In the titration experiments, 25 pmol of purified peak 1 were incubated with different equivalents (0.3, 0.5, 0.7, 1.0, 1.5, 2.0, 2.5 eq.) of the protein in 9 μl of HEPES/Na+ buffer at pH 7.4 for 30 min at 4°C. Glycerol was added to all the samples to a final concentration of 5% immediately before their loading on the gel. Electrophoresis was carried out at constant 45 V on 7% polyacrylamide gels in 1× TAE (Tris Acetate EDTA), pH 8.0 (33). The gels were stained for 30 min with GelGreen Nucleic Acid Stain and visualized on a UV transilluminator (BioRad ChemiDoc XRS). After the DNA staining, the gels were washed in water and stained again with Colloidal Coomassie G-250 to visualize the protein.
RESULTS
Biophysical characterisation of V7t1 aptamer in HEPES/Na+ buffer solution
The VEGF-binding aptamer V7t1, dissolved in a K+-rich medium, gave indication of high polymorphism (22). We have investigated the conformational preferences of this oligonucleotide in a Na+-rich medium, which mimics the extracellular environment in which VEGF targeting and/or inhibition should occur. We observed marked differences in the behaviour of not-annealed and annealed V7t1 samples (24).
Gel electrophoresis analysis
Native gel electrophoresis was used to characterize not-annealed and annealed V7t1 samples in terms of number and molecularity of the species present in solution. Appropriate reference oligonucleotides, i.e. a 26-mer, a 42-mer and a 50-mer, were also loaded as standards (see Supplementary Material).
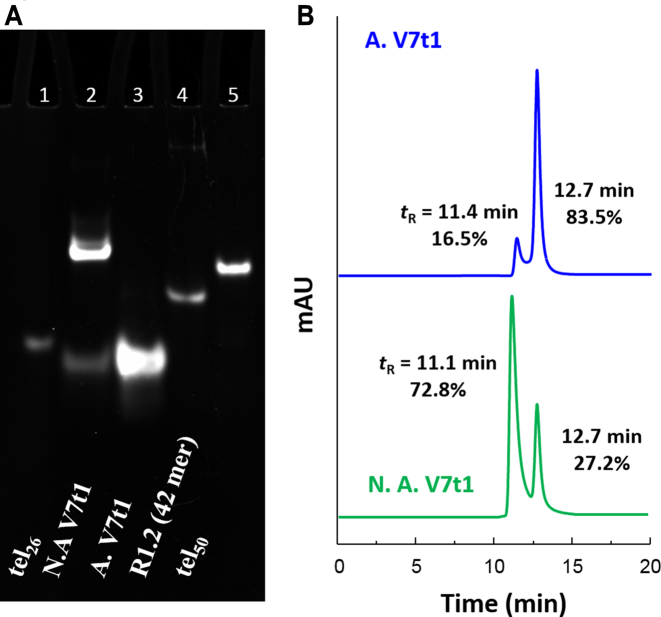
In Figure 1A, a picture of the native 10% polyacrylamide gel is reported. In comparison with the 26-mer tel26 (lane 1), which migrated on the gel as a single band, not-annealed V7t1 showed two bands, with a marked difference in electrophoretic mobility (lane 2), indicative of very different structuring. The band with a gel mobility similar to tel26, differing from V7t1 only for one nucleotide in length, could be attributed to a species with similar overall size and shape. On the other hand, the retarded band showed a gel mobility similar to the 50-mer tel50 (lane 5), indicative of a species with the overall size and charge of a dimeric V7t1 structure. In contrast, annealed V7t1 (lane 3) showed only one band with electrophoretic mobility similar to tel26, compatible with a monomolecular structure.
Figure 1.
(A) 10% polyacrylamide gel electrophoresis analysis under native conditions of V7t1 in the selected HEPES/Na+ buffer, run at 100 V and r.t. for 45 min in TBE 0.5×. Lane 1: tel26 (2 μM); lane 2: not-annealed V7t1 (N.A. V7t1, 5 μM); lane 3: annealed V7t1 (A. V7t1, 5 μM); lane 4: R1.2 (3 μM); lane 5: tel50 (0.5 μM). (B) Size exclusion HPLC analysis of not-annealed and annealed V7t1 (green and blue lines, respectively) at 2 μM concentration in the selected HEPES/Na+ buffer.
These results provided clear evidence that not-annealed V7t1 is present in solution as two different species with a gel mobility similar to tel26 and tel50, respectively, suggesting the coexistence of a monomeric and a dimeric structure. Notably, after annealing, the retarded band disappeared, indicating that the dimeric species did not reform after the heating/cooling cycle.
Size exclusion chromatography analysis
Size exclusion chromatography (SE-HPLC) can be a useful tool for studying the conformational behaviour of polymorphic G4 structures (34,35). Thus, in order to further investigate the number and nature of the species formed by V7t1 in HEPES/Na+ buffer, we exploited also this technique for its analysis, using tel26 as a reference. Indeed, tel26 is similar in size and known to fold in solution as a monomolecular G4 structure, forming, in the presence of Na+ ions, one main species consisting in an antiparallel basket-type G4 structure (26). Accordingly, tel26 gave only one peak on an analytical SEC column (tR = 12.7 min), consistent with a single, monomolecular G4 structure (Supplementary Figure S1).
The analysis of not-annealed V7t1 (Figure 1B, green line) resulted in a main peak with tR = 11.1 min, accompanied by another peak (ca. 27% in area) at tR = 12.7 min. In the chromatogram of annealed V7t1 (Figure 1B, blue line), the relative abundance of the two peaks was approximately inverted: the peak with longer elution times represented ca. 84% of the total area. SE-HPLC data confirmed the electrophoretic results, proving the co-existence of two forms for V7t1 under the studied conditions, plausibly a dimeric and a monomeric structure. In particular, the species with tR = 12.7 min was consistent with a monomeric G4 structure.
To further corroborate the idea that the faster eluting species in the SE-HPLC profile of V7t1 was a dimer, we compared its migration with standard oligonucleotides (whose sequences are listed in Table S1), known to fold either as monomers or dimers. It is in fact known that the retention times of different oligonucleotides depend on their molecularity, shape and length in bases. In particular, we compared monomolecular TBA versus the TBA–cTBA duplex (Supplementary Figure S2-I); monomolecular tel26 vs. tel50, known to form two consecutive G4 structures (25) (Supplementary Figure S2-II) and monomolecular versus dimeric c-kit2 G4 structures (36) (Supplementary Figure S2-III). The experimental differences for all the investigated cases were similar to the one found for the two peaks of V7t1 (Supplementary Figure S2-IV), further supporting the hypothesis that they correspond to monomeric and dimeric species, respectively.
DLS experiments
To analyse the size of the species formed by V7t1, we also performed DLS measurements. Due to the inherent low sensitivity of the technique and the small size of our samples, DLS experiments were performed at 50 μM V7t1 concentration, i.e. more than one order of magnitude higher than in the aforementioned experiments. The DLS profiles of not-annealed and annealed V7t1 samples in HEPES/Na+ buffer are shown in Supplementary Figure S3a, while the Z-average size (radius, expressed in nm) and associated polydispersity index (PdI) values are reported in Supplementary Figure S3b. The DLS profile of annealed V7t1 indicated the presence of one population with hydrodynamic radius (RH) centered at 2.77 nm, reasonably corresponding to a monomolecular G4 (37). In the case of not-annealed V7t1, the DLS measurements showed one main population with hydrodynamic radius (RH) centered at 4.49 nm, compatible with a dimeric species (37,38). In both cases, large aggregates were not found in the studied samples, indicating their high homogeneity within the detection limits of the technique.
DLS results completed the overall picture observed by gel electrophoresis and SE-HPLC analyses, confirming that V7t1 - in the HEPES/Na+ buffer - mainly consisted of dimeric structures, which could be converted to monomers upon annealing, also when analysed at very high concentrations.
Spectroscopic properties of V7t1
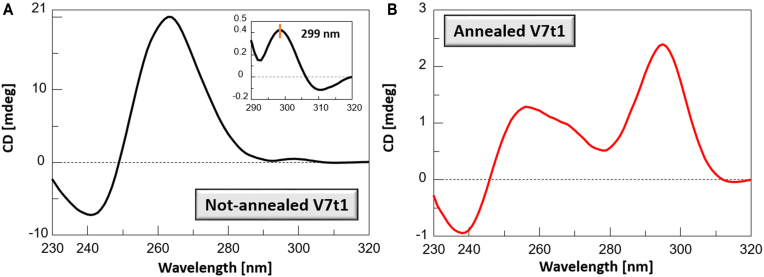
The CD spectrum of the not-annealed V7t1 sample showed two positive bands—the main one, very intense with a maximum centred at 263 nm, and another, very weak, at about 299 nm—together with a negative band with a minimum at 240 nm (Figure 2A). These spectroscopic features resemble the CD spectrum of V7t1 previously reported in a K+-rich buffer (50 mM) (22) and are consistent with the presence in solution of parallel G4 structures (39).
Figure 2.
CD spectra of not-annealed (A) and annealed (B) V7t1 samples recorded at 2 μM concentration in the selected HEPES/Na+ buffer. The inset in A shows the very weak band at 299 nm.
The CD spectrum of the annealed V7t1 sample was dramatically different, showing two positive bands, with maxima centered at 256 and 296 nm, together with a negative band with minimum at 237 nm (Figure 2B). These spectral features—similar to those found by Nonaka et al. in TBSE–KCl buffer (10 mM Tris/HCl, 100 mM NaCl, 50 mM KCl, 0.05 mM EDTA, pH 7.0) (21)—may account for the coexistence in solution of several G-quadruplex structures, essentially of hybrid and antiparallel topologies (40). The different conformational behaviour of V7t1 when in annealed or not-annealed form had indeed been observed also by Plavec et al. (22) in a K+-containing buffer, but not further investigated.
Remarkably, the overall intensity of the CD signals of the annealed sample was almost 10-fold lower than that of the not-annealed one at the same concentration: this so dramatic CD signal intensity difference could be reasonably due to two different contributions. Indeed, the high CD signal intensity of the not-annealed sample may indicate additional G-tetrad stacks compared to the annealed one, as expected for dimeric structures, probably including also intermolecular G4–G4 association, (41,42) which could further reinforce the overall base stacking interactions responsible for the large positive CD band centered at ca. 260 nm. On the other hand, the very weak CD signal intensity of the annealed sample could be the result of the presence of different G4 conformations, with positive and negative bands counterbalancing their contributions.
The samples were also analysed by UV thermal difference spectra (TDS): both not-annealed and annealed samples showed a TDS with a maximum at 273 nm and a minimum at 295 nm, consistent with the typical ‘fingerprint’ of G4 structures (Supplementary Figure S4) (29,30). However, the characteristic minimum at 295 nm was not very pronounced, suggesting that perhaps not all the oligonucleotide is folded into a G4 and other structures can be also present in the investigated samples.
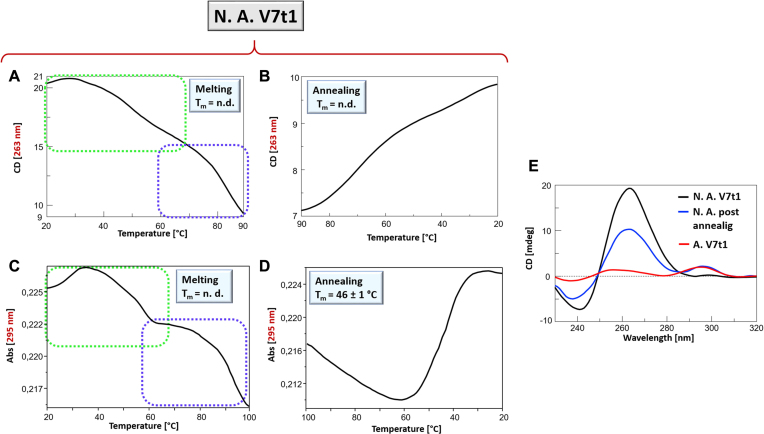
The thermal stability of V7t1 was then analysed by CD- and UV-melting experiments (43). In the case of not-annealed V7t1, both the CD- and UV-melting profiles showed a marked decrease of the signal upon increasing the temperature (Figure 3A–C) (29). Notably, in both cases the profiles did not show the sigmoidal behaviour expected for a unique, cooperative transition, but rather a curve with at least two transitions and apparent Tm values of ca. 50°C and 80°C, respectively. These profiles could be due to the presence of mainly two different species in solution, i.e. monomeric and dimeric G4s, in accordance with the results of the native PAGE and SEC analyses, and/or to multi-process denaturation pathways. Remarkably, the CD- and UV-annealing profiles, recorded upon cooling down the samples from 90 to 20°C, were not superimposable to the corresponding melting profiles. In particular, the CD-annealing profile (Figure 3B) did not show a clearly defined sigmoidal behaviour. In turn, the UV-annealing profile provided a sigmoidal curve in the range 65–20°C, with an apparent Tm value of ca. 46°C (Figure 3D).
Figure 3.
Not-annealed V7t1 sample at 2 μM concentration in the selected HEPES/Na+ buffer analysed by CD- (A, B) and UV-melting experiments (C, D): CD-melting (A) and CD-annealing (B) profiles, recorded at 263 nm; UV-melting (C) and UV-annealing (D) profiles recorded at 295 nm. All the experiments were performed at a scan rate of 1°C/min. n.d. = not determined. (E) overlapped CD spectra at 20°C of V7t1 samples: not-annealed V7t1 (black line), after fast cooling the sample from 90 to 20°C at 1°C/min (blue line) and after slow annealing from 95 to 20°C at ca. 0.3°C/min (red line).
The analysis of the CD spectra of not-annealed V7t1 acquired at different temperatures during the melting process revealed that its denaturation was not complete even at 90°C (Supplementary Figure S5A). Interestingly, in the successive cooling experiment the CD signal at 296 nm was detectably enhanced on decreasing the temperature from 50 to 20°C (Supplementary Figure S5B), suggesting that, after heating, different G4 structures were formed. Particularly, by plotting the CD signal at 296 nm as a function of the temperature, an almost sigmoidal behaviour with an apparent Tm of ca. 50°C was observed (Supplementary Figure S5C), in approximately good accordance with the UV-annealing data, clearly indicating that the thermally-induced reorganization of the systems in solution provided G4 structures with antiparallel or hybrid-type topologies, not initially present in the not-annealed sample. Remarkably, the initial CD signal intensity of not-annealed V7t1 sample at 20°C (Figure 3E, black line) after the cooling process was not restored (Figure 3E, blue line), thus indicating irreversible folding/unfolding processes. In line with this observation, the annealing processes proved to be also affected by the scan rate: notably, the spectrum recorded going back to 20°C after a heating/cooling cycle at 1°C/min (fast annealing, Figure 3E, blue line) was also different from the spectrum obtained for the slowly annealed V7t1 sample (ca. 0.3°C/min, Figure 3E, red line); thus, fast and slow annealing procedures resulted in different refolded G4 topologies, with different contributions to the CD bands at 263 and 295 nm.
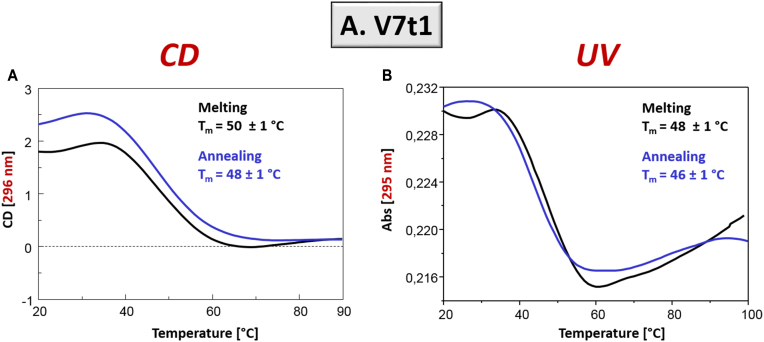
Differently from the not-annealed V7t1 sample, the CD and UV spectroscopic characterization carried out on the annealed V7t1 sample showed in all cases sigmoidal denaturation pathways, with almost superimposable melting/cooling profiles and only limited hysteresis, indicating that under the experimental conditions used (scan rate: 1°C/min), the related processes were essentially reversible (Figure 4).
Figure 4.
Annealed V7t1 sample at 2 μM concentration in the selected HEPES/Na+ buffer: (A) CD-melting and annealing profiles, recorded at 296 nm and (B) UV-melting and annealing profiles recorded at 295 nm. Melting and annealing curves were respectively depicted as black and blue lines. All the experiments were performed at a scan rate of 1°C/min.
Monitoring the CD signal at 296 nm and the UV absorbance at 295 nm, apparent Tm values of 50 and 48°C, and of 48 and 46°C, were respectively found for the heating and cooling profiles (Figure 4, black and blue lines, respectively), indicating the formation of quite stable G4 structures. The analysis of the CD spectra recorded on increasing the temperature showed significant changes, with a shift in the positive band maxima from 296 to 287 nm, on one side, and a rearrangement in the region from 255 to 266 nm, on the other (Supplementary Figure S6A), essentially reversible upon cooling (Supplementary Figure S6B).
Taken together, CD and UV results demonstrated that V7t1 structuring was extremely sensitive to the preparation method and annealing procedures, being the kinetic species (present in the not-annealed sample) very different in terms of G4 folding and stability from the thermodynamically favoured ones (present in the slowly annealed V7t1).
The spectroscopic data obtained by UV and CD experiments on both V7t1 samples were well consistent among them and in line with the previously described gel electrophoresis, DLS and SE-HPLC data. In particular, for the not-annealed sample, complex denaturation pathways were found, confirming the coexistence of different G4 structures in solution; on the contrary, the annealed V7t1 sample mainly denoted reversible G4 foldings, characteristic of monomolecular G4 structures.
Differential scanning calorimetry
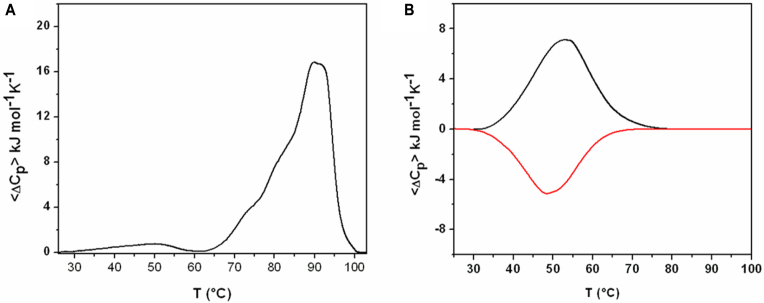
The melting behaviour of not-annealed and annealed V7t1 samples was further investigated by DSC. In order to completely unfold the extremely thermostable structure found in the not-annealed V7t1 sample, an over-pressure of 2 atm was applied to the DSC cell so to expand the heating temperature range up to 115°C. In line with the CD-melting data, the DSC profile (Figure 5A) of the first heating scan was markedly different from the profiles obtained in the successive heating/cooling scans (Figure 5B and Supplementary Figure S7), revealing the presence of kinetically trapped species in the not-annealed sample. Inspection of the heating profile revealed a large multicomponent peak centered at ∼90°C, with a higher temperature shoulder (∼93°C) and two lower temperatures shoulders (∼73 and 80°C), denoting complex denaturation pathways and/or multiple co-existing starting species (Figure 5A). The overall enthalpy change measured by integrating the area under the main DSC peak was about 241 kJ/mol, a value consistent with the presence of at least three G-tetrads per mol of single strand (i.e. six G-tetrads for a dimeric G4) (25). It should be noted that, in the 30–60°C temperature range preceding the main peak, a broad pre-melting transition was present, which, being barely detectable, was neglected in the successive analysis of the DSC profile as the corresponding heat was too small to be accurately determined. As for the CD- and UV-melting experiments, after annealing the sample, the successive heating/cooling profiles were completely superimposable, indicating a reversible unfolding process with a transition peak centered at about 52°C (Figure 5B and Supplementary Figure S7). Due to the reversibility of this transition, a full thermodynamic analysis was possible, giving the corresponding thermodynamic parameters reported in Table 1. The Tm values were in good agreement with those obtained by CD and UV measurements, also considering that the oligonucleotide concentrations used in the DSC analyses were ∼70-fold higher than those used in the CD- and UV-melting experiments.
Figure 5.
(A) Excess heat capacity profile for a not-annealed V7t1 sample (140 μM strand conc.). (B) Excess heat capacity heating (black) and cooling (red) profile for an annealed V7t1 sample (140 μM strand conc.) in the selected HEPES/Na+ buffer.
Table 1.
Thermodynamic parameters obtained: (a) from the DSC analysis for not-annealed and annealed V7t1 samples, and (b) from CD and UV experiments for the unfolding process of annealed V7t1. Errors on UV and CD-derived parameters are within ± 10%.
| (a) DSC analysis | T m (°C) | ΔHm (kJ mol−1) | ΔSm (kJ mol−1K−1) |
| Not-annealed V7t1 | 89.9±0.5 | 241±12 | - |
| Annealed V7t1 | 52.2±0.5 | 131±7 | 0.40±0.02 |
| b) annealed V7t1 | T m (°C) | ΔHv.H (kJ mol−1) | ΔSv.H (kJ mol−1K−1) |
| UV (295 nm) | 48 ± 1 | 130 | 0.40 |
| CD (296 nm) | 50 ± 1 | 133 | 0.41 |
This observation is a further evidence of the monomolecular nature of the observed transition, suggesting that, after annealing, mainly monomolecular G4 species were formed, consistently with the gel electrophoresis, DLS and SE-HPLC data. The overall enthalpy change for the annealed V7t1 was 131 kJ/mol, suggesting the formation of a G4 structure stabilized by two G-tetrads (44,45). These data seem to somehow corroborate Nonaka's structural hypothesis for V7t1 containing only two G-tetrads in TBSE–KCl buffer, which would imply that seven guanine residues are not involved in the G4 core formation and belong to the loop regions (21).
Furthermore, analysis of the enthalpies values reveals that the dimeric structure is stabilized by more favourable interactions than the species formed in the annealed sample. These interactions are likely due to the formation of additional G-tetrads and/or other inter-strand interactions. Overall, the DSC results are consistent with the presence of highly thermostable, dimeric G4 species in the not-annealed V7t1 sample that were kinetically trapped and, once melted, did not reform after cooling. On the other hand, under equilibrium conditions (slowly annealed samples) mainly monomolecular G4 species were formed.
Although thermodynamic parameters for the unfolding process obtained by DSC measurements are more reliable than those extracted by spectroscopic methodologies, the UV- and CD-melting curves of annealed V7t1 sample were studied also by van’t Hoff analysis (Table 1). Notably, the obtained enthalpy and entropy changes, as well as the Tm values, were very close to those determined by DSC analysis.
Stability of V7t1 in fetal bovine serum (FBS)
In order to investigate their enzymatic stability in serum, not-annealed and annealed V7t1 samples were analysed by incubating them in 80% FBS at 37°C. At fixed times, aliquots of the incubation samples were collected and then analysed by 20% denaturing PAGE in comparison with two different reference oligonucleotides, a 22-mer and a 24-mer c-myc sequence.
After 2 h, the not-annealed V7t1 sample was still essentially unaffected by serum nucleases. Only a very limited oligonucleotide fragmentation was observed, with bands having electrophoretic mobility comparable to the 22-mer c-myc, used as length control, suggesting that V7t1 was shortened by three nucleotides as a consequence of the enzymatic digestion (Supplementary Figure S8A). Then, no other degradation product appeared up to 18 h; further digestion, though scarce, became visible only after 2 days of treatment, and the system remained essentially stable for 6 days.
Analogously to the not-annealed sample, also annealed V7t1 proved to be very resistant to nuclease digestion, as expected for a G4-forming oligonucleotide. After 2 h in FBS, it essentially lost only a three-nucleotide fragment. However, after 5 h, the annealed V7t1 samples gave also further degradation products, providing evidence that in this arrangement the sugar-phosphate backbone of V7t1 was somehow more accessible to nucleases (Supplementary Figure S8B).
Electrophoretic analysis of VEGF-V7t1 binding
After investigating the electrophoretic, spectroscopic and thermodynamic features of V7t1, as well as its resistance to nucleases in not-annealed and annealed forms, its interaction and binding with the VEGF protein was analysed by electrophoretic mobility shift assay (EMSA) under non-denaturing conditions, which is a sensitive and widely used method to detect protein–nucleic acid interactions (33). This approach is essentially based on the analysis of the electrophoretic mobility of an oligonucleotide, which is typically high when free in solution, and is significantly retarded when complexed with a protein.
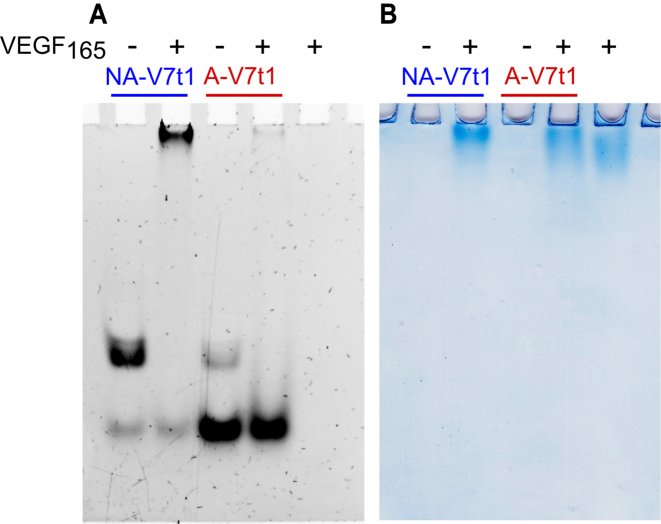
Both not-annealed and annealed V7t1 samples were incubated with ∼1.3 eq. VEGF165 and the resulting mixtures analysed by PAGE using the free oligonucleotides as controls. The gels were subjected to a double staining procedure, using first a staining specific for nucleic acids (GelGreen) and then one specific for proteins (Coomassie). The control aptamers showed the bands expected on the basis of the SE-HPLC profiling of annealed and not annealed V7t1.
Nucleic acids staining showed the appearance of a new retarded band, attributable to the formation of the aptamer-protein complex in the sample containing not-annealed V7t1 and VEGF165, but, surprisingly, only the band corresponding to the dimeric form of the oligonucleotide completely disappeared, whereas neither the mobility nor the intensity of the faster migrating band of Vt71 monomer changed (Figure 6A). On the other hand, upon incubation of VEGF165 with annealed V7t1, the band of the prevailing monomeric form did not disappear. Only the minor amount of dimer still present in this sample did interact with VEGF165, indicating that, even in the presence of high excess of the monomeric form, the binding to dimeric V7t1 was highly preferred (Figure 6A) under the experimental conditions used.
Figure 6.
(A) GelGreen- and (B) Coomassie-stained electrophoretic mobility shift assay (EMSA) of annealed and not annealed V7t1 (30 pmol) incubated with VEGF165 (40 pmol) in HEPES/Na+ buffer.
Coomassie staining confirmed these results. VEGF165 is a highly glycosylated protein, which produces, even in denaturing and reducing SDS-PAGE, smeared bands due to its high intrinsic heterogeneity (Supplementary Figure S9). This smeared migration was indeed also visible in the EMSA experiments (Figure 6B). Nonetheless, a sharper migration profile compared to the free protein was observed when VEGF165 was incubated with the not-annealed V7t1 sample, clearly visible also in the Coomassie-stained gel (Figure 6B) and attributable to the drastic change in the mass-to-charge ratio of the protein upon complex formation.
Separation and characterisation of the two species of not-annealed V7t1
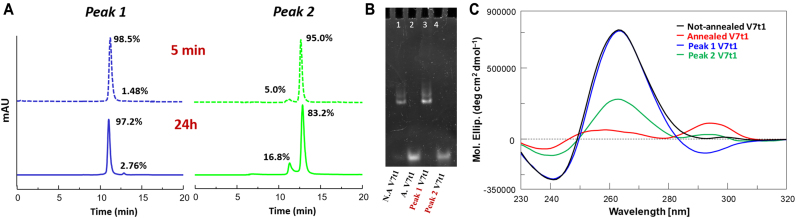
The not-annealed V7t1 sample was chromatographed on a SE-HPLC analytical column in order to isolate its monomeric and dimeric forms (Figure 1b, green line). After the separation, both peaks were then analysed over time for 24 h by re-injecting them on the same column. The main peak at tR = 11.1 min (dimer) and the one at tR = 12.7 min (monomer), when reinjected, showed a single peak with the same retention time as the original collected fractions (Figure 7A). Notably, the re-equilibration between the two forms was very limited: indeed, after 24 h, the separated peak 1 showed only 2.8% of the monomeric form (versus the 25.7% present in the original sample), whereas in the separated peak 2 the dimeric form reappeared only in a modest amount (16.8% versus ca. 73% present in the not-annealed V7t1 sample before HPLC separation). The analysis over time demonstrated that both peak 1 and peak 2 were quite stable over a 24 h period, and thus we could isolate and separately analyse them by electrophoretic and spectroscopic techniques (TDS and CD spectra analysis, CD- and UV-melting experiments). On a 10% polyacrylamide gel, in comparison with not-annealed and annealed V7t1 (Figure 7B, lanes 1 and 2, respectively), peak 1 (lane 3) exclusively showed the retarded band of the not-annealed sample, identified as the dimeric species, whereas peak 2 (lane 4) exhibited only one band with the same electrophoretic mobility as annealed V7t1, i.e. the monomeric aptamer.
Figure 7.
(A) Size exclusion HPLC analysis of isolated peak 1 (tR = 11.1 min) and peak 2 (tR = 12.7 min) reinjected 5 min (dashed blue and green lines, respectively), and 24 h (continuous blue and green lines, respectively) after their separation on the SEC column; (B) 10% polyacrylamide gel electrophoresis under native conditions of V7t1 and its components, as isolated from the not-annealed V7t1 sample in HEPES/Na+ buffer, run at 70 V at r.t. for 2 h in TBE 1× buffer. Lane 1: not-annealed V7t1 (4 μM); lane 2: annealed V7t1 (4 μM); lane 3: peak 1 of V7t1 (4 μM); lane 4: peak 2 of V7t1 (3 μM); (C) overlapped CD spectra of not-annealed V7t1 (2 μM), annealed V7t1 (2 μM), peak 1 (2 μM) and peak 2 (0.4 μM) in the selected HEPES/Na+ buffer (black, red, blue and green lines, respectively).
The CD spectrum of separated peak 1 (Figure 7C, blue line) essentially exhibited the spectral features of not-annealed V7t1, compatible with a G4 parallel topology, except for the appearance of a negative band with a minimum centered at ca. 290 nm. This negative band could be related to the one previously associated with the peculiar stacking interactions between terminal octads, allowing for the head-to-head or tail-to-tail dimerization of parallel G4 monomeric structures (48–50). In our case, it could be hypothesized that the pending extremities of V7t1 not involved in the G4 core formation, plausibly the terminal 3′ TAGA tract, could be faced in a 3′–3′ stacked dimer and further stabilize the whole structure by additional interactions with the adjacent G-tetrads.
Inspection of the CD spectrum of the isolated peak 2 (Figure 7C, green line) showed a negative band at ca. 240 nm accompanied by a large positive band at 263 nm, diagnostic of a parallel G4 structure, in turn accompanied by a weak positive band centered at ca. 295 nm. The absence of the negative band at 290 nm found in peak 1 was consistent with its attribution to structural motifs characteristic of a dimeric structure; in contrast, an additional, weak positive band centered at ca. 295 nm appeared, showing the coexistence in solution also of antiparallel and/or hybrid-type G4 topologies.
The differences observed in the CD amplitudes of the main bands at 263 and 295 nm comparing the CD spectra of peak 2 with that of the annealed V7t1 sample (Figure 7C, green and red lines) essentially reflect the conformational reorganization of the monomeric V7t1 G4 foldings upon annealing, but could also be due to the presence of tiny amounts (ca. 5%, as estimated by SE-HPLC) of peak 1 in the analysed solution.
The TDS—determined by substracting the UV spectra recorded at 20°C from the ones recorded at 100°C for each separated peak—showed a maximum at 273 nm and a minimum at 295 nm, diagnostic of the presence of G4 structures (Supplementary Figure S10) (29,30).
The 263 nm-monitored CD-melting and annealing profiles of isolated peak 1 (Supplementary Figure S11) at 2 μM concentration did not show a sigmoidal behaviour, but essentially two barely hinted transitions, quite similar to those observed in the case of not-annealed V7t1 (Figure 3). Also its UV-melting profile monitored at 295 nm was similar to the one of not-annealed V7t1, not showing the characteristic sigmoidal shape of a unique, cooperative transition (Supplementary Figure S12A). On the contrary, the UV-annealing profile of this sample monitored at 295 nm provided a sigmoidal behaviour with apparent Tm value of 45°C (Supplementary Figure S12B), similarly to the not-annealed V7t1 (Figure 3D). As far as peak 2 is concerned, since the amount of this peak we could recover after multiple injections on the SE-HPLC column was insufficient to get it at high concentration, we could perform only UV-thermal denaturation experiments, in which both the melting and annealing profiles, monitored at 295 nm, provided sigmoidal curves with apparent Tm value of 50°C and 43°C, respectively (Supplementary Figure S12C), showing some hysteresis under the tested experimental conditions. On the contrary, at the same conc. the CD-monitored heating/cooling experiments only gave very noisy profiles (data not shown).
Taken together these results indicated that the conformational behaviour of the separated peak 1 was well correlated to that observed for the not-annealed V7t1 sample, consistently with being its main component. In turn, peak 2, though being a V7t1 monomeric form as is the annealed V7t1 sample, showed somehow different CD spectral features and apparent Tm values, obtained by UV measurements, compared to the annealed V7t1. Also taking into account the tiny contamination of peak 1 possibly contained in peak 2, the different behaviour of the V7t1 monomeric forms present in the annealed and in the not-annealed samples can be essentially related to the structural rearrangements that its mainly parallel G4 structures undergo after thermal treatments, finally resulting in a mixture of hybrid-type and antiparallel G4 conformations.
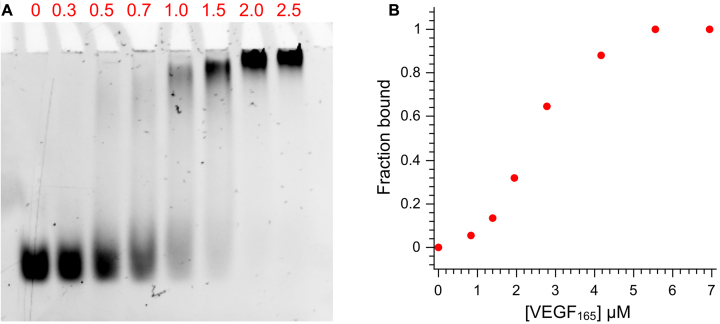
Having analysed the electrophoretic, spectral and thermal stability properties of the different V7t1 species, which only in its dimeric form, under our experimental conditions, proved to bind VEGF165 with high affinity, the EMSA analysis was then repeated on purified peak 1. The peak 1 sample (25 pmol, 2.8 μM) was incubated for 30 min at 4°C with increasing equivalents of VEGF165 and the resulting complexes were analysed on a 7% native polyacrylamide gel. Nucleic acid staining (Figure 8A) confirmed the formation of the peak 1-VEGF165 complex, showing the complete disappearance of the free oligonucleotide band after addition of 2 eq. of protein. The resulting data, plotted as fraction of bound V7t1 versus protein concentration (Figure 8B), evidenced a cooperative binding of the aptamer, which recognized the protein with high affinity, in general overall accordance with the data reported by Nonaka et al. (21) and Lönne et al. (51), reporting Kd values in the nM range, as determined by SPR and MST experiments, respectively.
Figure 8.
(A) EMSA of 2.8 μM peak 1 incubated with different equivalents of VEGF165 (red numbers. Protein concentration in lanes 1–8: 0, 0.8, 1.4, 1.9, 2.8, 4.2, 5.6, 6.9 μM, respectively) in HEPES/Na+ buffer. (B) fraction of bound peak 1 reported as a function of VEGF165 concentration.
DISCUSSION
Being involved in angiogenesis and vascularization of many solid tumours, VEGF is a valuable therapeutic target to develop effective drugs in cancer treatment. The VEGF-binding aptamer V7t1 proved to bind with high affinity the two main isoforms of the target protein and exhibited promising anticancer properties on different cancer cell lines (23). Also, V7t1 showed a remarkable structural polymorphism in a K+-rich environment, being able to fold into several G4 structures (22), but no data have been previously reported on its conformational preferences in Na+-rich solutions.
Considering the great potential of V7t1 as efficient and selective VEGF ligand, herein we have investigated its physico-chemical properties using different techniques, i.e. gel electrophoresis, SE-HPLC, DLS, CD-melting, UV-melting and DSC analysis. In detail, we aimed at obtaining a deeper structural insight on V7t1 structuring in a Na+-rich buffered solution (25 mM HEPES, 150 mM NaCl), which mimics the extracellular media in which VEGF targeting should occur.
Under the studied experimental conditions, the behaviour of the aptamer proved to be dramatically influenced by the preparation method and annealing procedure. Remarkably, our results have demonstrated that not-annealed samples of V7t1 mainly consisted of two different species: a dimer and a monomer. The dimer, which represents the most abundant species in this sample, is probably formed by stacking of two parallel G4 monomers, each stabilized by three G-tetrads, and/or by dimeric interlocked parallel G4 structures stabilized by six G-tetrads, as suggested by the measured enthalpy change values. Upon annealing, this aptamer rearranged to give monomeric G4 structures, probably mixtures of long-looped hybrid and antiparallel conformations, stabilized by two G-tetrads, according to the entalphy change values measured by DSC experiments. This finding is in accordance with studies on other G-rich sequences forming dimeric G-quadruplexes, showing that Na+ ions favour the formation of intermolecular assemblies through kinetically stabilized G-quadruplex intermediates (52,53).
Evidence of the coexistence of monomeric and dimeric G4 forms in V7t1 has been obtained by native PAGE, SE-HPLC and DLS experiments. Noteworthy, not-annealed and annealed V7t1 samples showed dramatically different CD and DSC profiles, indicating the formation of G4 structures differing not only for their molecularity (dimer vs. monomer), but also for the topology and number of consecutive G-tetrads. CD- and UV-melting analysis on not-annealed V7t1 revealed complex thermal denaturation profiles, evidencing the presence of essentially two main transitions in solution, which could be ascribed to the presence of two forms in solution endowed with different thermal stability, but could also account for multiple events on the same G4 structure, e.g. dissociation of the G4 dimer into G4 monomers accompanied by architectural rearrangement and, then, complete unfolding.
In contrast, the annealed V7t1 gave sigmoidal curves, with Tm values derived by CD- and UV-melting experiments in good agreement (ca. 48°C) and no relevant hysteresis on comparing the heating and cooling profiles, thus indicating essentially reversible processes.
Notably, both not-annealed and annealed V7t1 samples proved to be resistant to nuclease digestion, even though with marked differences. In fact, both samples lost only a trinucleotide fragment after ca. 2 h of FBS incubation; however, the annealed V7t1—only present as a monomeric G4—after 5 h showed also other degradation products, whereas the not-annealed sample proved to be much more resistant, consistently with a dimeric structure. Overall, these experiments suggested that both the dimeric and monomeric forms of V7t1 exposed to the solvent a terminal trinucleotide, not involved in the G4 core formation—most plausibly the 5′TGT end, assuming that some similarities with the structure solved for RNV66 (22) can apply – in turn masking the 3′-extremity to nuclease digestion. Since these protection effects were much more marked for the V7t1 dimer than for the monomer, this could indicate that the dimerization process favoured a head-to-head or tail-to-tail stacked dimer—plausibly a 3′–3′ dimer—over a head-to-tail dimer, and/or an interlocked dimer composed of two parallel G4-forming strands with the 3′-ends strongly masked to the solvent.
The presence in solution of a hypothetical head-to-head or tail-to-tail stacked dimer G4 structure is corroborated by the CD spectrum of the isolated dimer (peak 1) which showed a negative band centered at 290 nm, already observed in other 5′–5′ stacked G4 dimers (48–50). However, the available data do not allow unambiguously determining if mainly a stacked or an interlocked dimer formed. High resolution techniques are required to address this issue in a conclusive manner but, most probably, both structures are present in solution (41).
The equilibrium between the monomeric and dimeric forms present in not-annealed V7t1 samples was very slow (only limited interconversion was observed after 24 h), so it was possible to isolate and separately analyse them. The irreversibility of the dimer-monomer interconversion can be explained in terms of hydration, which plays a major role in determining G-quadruplex stability, as well described in the literature (54,55). Upon G-quadruplex formation, and more markedly, multimerization, the whole surface accessible to water is minimized. When thermal treatments disrupt all inter- and intrastrand H-bonds stabilizing the G4 architecture, the resulting unfolded strands are maximally exposed to water and therefore get hydrated. If a cooling process follows, a G4 structural reorganization with the minimum G-tetrads requirement, involving the lowest dehydration cost, is favoured, possibly stabilized also by other structural elements (e.g., hairpins) formed in the loops (56).
Spectroscopic and SE-HPLC analyses clearly indicated that peak 1 essentially behaved as the not-annealed V7t1, as expected being its main component. A central issue of this study has been the analysis of the binding properties of V7t1, evaluated by EMSA. Experiments on not-annealed V7t1 provided clear evidence of the formation of a V7t1–VEGF complex involving only the band attributed to the dimeric species. In fact, neither the intensity nor the mobility of the band attributed to the monomeric V7t1 changed in the presence of the protein, even upon addition of an excess of VEGF165. Remarkably, these results clearly proved that the dimeric form of V7t1 has high avidity for VEGF165, contrarily to its monomeric form, although it cannot be ruled out that, at higher protein concentrations or exploring different solution conditions, also the monomeric V7t1 might interact with the protein.
In conclusion, our investigations revealed interesting and unprecedented results showing that V7t1 is able to form both monomeric and dimeric G-quadruplex structures in a Na+-rich solution; notably, the target protein recognizes with high affinity only the parallel dimeric G4 species, largely predominant when the aptamer does not undergo a slow annealing procedure.
Aiming at a more efficient binding to the VEGF protein so to develop valuable and highly selective targeting agents for both diagnostics and/or therapeutic treatments, these findings provide new, fundamental information for the design of improved VEGF-binding aptamers. Studies to obtain more effective V7t1 analogues, also based on its covalent dimers, are currently underway in our laboratories to produce potential G-quadruplex based drugs (57) and/or biosensors (58) for, respectively, in vivo and ex vivo applications.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research (AIRC) [IG2015 n. 17037 to D.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Shibuya M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: a crucial target for anti- and pro-angiogenic therapies. Genes Cancer. 2011; 2:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jonas J.B., Neumaier M.. Vascular endothelial growth factor and basic fibroblast growth factor in exudative age-related macular degeneration and diffuse diabetic macular edema. Ophthalmic Res. 2007; 39:139–142. [DOI] [PubMed] [Google Scholar]
- 3. Adamis A.P., Miller J.W., Bernal M.T., D’Amico D.J., Folkman J., Yeo T.K., Yeo K.T.. Increased vascular endothelial growth factor levels in the vitreous of eyes with proliferative diabetic retinopathy. Am. J. Ophthalmol. 1994; 118:445–450. [DOI] [PubMed] [Google Scholar]
- 4. Koch A.E., Harlow L.A., Haines G.K., Amento E.P., Unemori E.N., Wong W.L., Pope R.M., Ferrara N.. Vascular endothelial growth factor. A cytokine modulating endothelial function in rheumatoid arthritis. J. Immunol. 1994; 152:4149–4156. [PubMed] [Google Scholar]
- 5. Guo P., Fang Q., Tao H.Q., Schafer C.A., Fenton B.M., Ding I., Hu B., Cheng S.Y.. Overexpression of vascular endothelial growth factor by MCF-7 breast cancer cells promotes estrogen-independent tumor growth in vivo. Cancer Res. 2003; 63:4684–4691. [PubMed] [Google Scholar]
- 6. Shijubo N., Kojima H., Nagata M., Ohchi T., Suzuki A., Abe S., Sato N.. Tumor angiogenesis of non-small cell lung cancer. Microsc. Res. Tech. 2003; 60:186–198. [DOI] [PubMed] [Google Scholar]
- 7. Yamaguchi R., Yano H., Iemura A., Ogasawara S., Haramaki M., Kojiro M.. Expression of vascular endothelial growth factor in human hepatocellular carcinoma. Hepatology. 1998; 28:68–77. [DOI] [PubMed] [Google Scholar]
- 8. Mesiano S., Ferrara N., Jaffe R.B.. Role of vascular endothelial growth factor in ovarian cancer: inhibition of ascites formation by immunoneutralization. Am. J. Pathol. 1998; 153:1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown L.F., Berse B., Jackman R.W., Tognazzi K., Guidi A.J., Dvorak H.F., Senger D.R., Connolly J.L., Schnitt S.J.. Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in breast cancer. Hum. Pathol. 1995; 26:86–91. [DOI] [PubMed] [Google Scholar]
- 10. Tawada K., Ishihara T., Kobayashi A., Yamaguchi T., Tsuyuguchi T., Matsuyama M., Yokosuka O.. Quantitative analysis of vascular endothelial growth factor in liver metastases from pancreatic carcinoma as a predictor of chemotherapeutic effect and prognosis. Clin. Cancer Res. 2008; 14:7438–7443. [DOI] [PubMed] [Google Scholar]
- 11. Konno H., Tanaka T., Baba M., Kanai T., Matsumoto K., Kamiya K., Nakamura S., Baba S.. Quantitative analysis of vascular endothelial growth factor in colon cancer. Eur. Surg. Res. 1998; 30:273–278. [DOI] [PubMed] [Google Scholar]
- 12. Toi M., Kondo S., Suzuki H., Yamamoto Y., Inada K., Imazawa T., Taniguchi T., Tominaga T.. Quantitative analysis of vascular endothelial growth factor in primary breast cancer. Cancer. 1996; 77:1101–1106. [DOI] [PubMed] [Google Scholar]
- 13. Ferrara N., Gerber H.P., LeCouter J.. The biology of VEGF and its receptors. Nat. Med. 2003; 9:669–676. [DOI] [PubMed] [Google Scholar]
- 14. Harper S.J., Bates D.O.. VEGF-A splicing: the key to anti-angiogenic therapeutics?. Nat. Rev. Cancer. 2008; 8:880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Catena R., Muniz-Medina V., Moralejo B., Javierre B., Best C.J.M., Emmert-Buck M.R., Green J.E., Baker C.C., Calvo A.. Increased expression of VEGF121/VEGF165-189 ratio results in a significant enhancement of human prostate tumor angiogenesis. Int. J. Cancer. 2007; 120:2096–2109. [DOI] [PubMed] [Google Scholar]
- 16. Yu J.L., Rak J.W., Klement G., Kerbel R.S.. Vascular endothelial growth factor isoform expression as a determinant of blood vessel patterning in human melanoma xenografts. Cancer Res. 2002; 62:1838–1846. [PubMed] [Google Scholar]
- 17. Torimura T., Iwamoto H., Nakamura T., Abe M., Ikezono Y., Wada F., Sakaue T., Masuda H., Hashimoto O., Koga H. et al.. Antiangiogenic and antitumor activities of Aflibercept, a soluble VEGF receptor-1 and -2, in a mouse model of hepatocellular carcinoma. Neoplasia. 2016; 18:413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meadows K.L., Hurwitz H.I.. Anti-VEGF therapies in the clinic. Cold Spring Harb. Perspect. Med. 2012; 2:a006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferrara N., Hillan K.J., Novotny W.. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun. 2005; 333:328–335. [DOI] [PubMed] [Google Scholar]
- 20. Ng E.W.M., Shima D.T., Calias P., Cunningham E.T., Guyer D.R., Adamis A.P.. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006; 5:123–132. [DOI] [PubMed] [Google Scholar]
- 21. Nonaka Y., Sode K., Ikebukuro K.. Screening and improvement of an anti-VEGF DNA aptamer. Molecules. 2010; 15:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marušič M., Veedu R.N., Wengel J., Plavec J.. G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold. Nucleic Acids Res. 2013; 41:9524–9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edwards S.L., Poongavanam V., Kanwar J.R., Roy K., Hillman K.M., Prasad N., Leth-Larsen R., Petersen M., Marušič M., Plavec J. et al.. Targeting VEGF with LNA-stabilized G-rich oligonucleotide for efficient breast cancer inhibition. Chem. Commun. 2015; 51:9499–9502. [DOI] [PubMed] [Google Scholar]
- 24. Petraccone L., Pagano B., Giancola C.. Studying the effect of crowding and dehydration on DNA G-quadruplexes. Methods. 2012; 57:76–83. [DOI] [PubMed] [Google Scholar]
- 25. Petraccone L., Spink C., Trent J.O., Garbett N.C., Mekmaysy C.S., Giancola C., Chaires J.B.. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011; 133:20951–20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D.. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006; 34:2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moccia F., Platella C., Musumeci D., Batool S., Zumrut H., Bradshaw J., Mallikaratchy P., Montesarchio D.. The role of G-quadruplex structures of LIGS-generated aptamers R1.2 and R1.3 in IgM specific recognition. Int. J. Biol. Macromol. 2019; 133:839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zümrüt H.E., Batool S., Van N., George S., Bhandari S., Mallikaratchy P.. Structural optimization of an aptamer generated from LIgand-Guided Selection (LIGS) resulted in high affinity variant toward mIgM expressed on Burkitt's lymphoma cell lines. Biochim. Biophys. Acta - Gen. Subj. 2017; 1861:1825–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mergny J.-L., Lacroix L.. UV melting of G-quadruplexes. Curr. Protoc. Nucleic Acid Chem. 2009; 37:17.1.1–17.1.15. [DOI] [PubMed] [Google Scholar]
- 30. Mergny J.-L., Li J., Lacroix L., Amrane S., Chaires J.B.. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005; 33:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marky L.A., Breslauer K.J.. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987; 26:1601–1620. [DOI] [PubMed] [Google Scholar]
- 32. Pagano B., Randazzo A., Fotticchia I., Novellino E., Petraccone L., Giancola C.. Differential scanning calorimetry to investigate G-quadruplexes structural stability. Methods. 2013; 64:43–51. [DOI] [PubMed] [Google Scholar]
- 33. Hellman L.M., Fried M.G.. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007; 2:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Largy E., Mergny J.-L.. Shape matters: size-exclusion HPLC for the study of nucleic acid structural polymorphism. Nucleic Acids Res. 2014; 42:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dailey M.M., Clarke Miller M., Bates P.J., Lane A.N., Trent J.O.. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010; 38:4877–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuryavyi V., Phan A.T., Patel D.J.. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010; 38:6757–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Riccardi C., Musumeci D., Russo Krauss I., Piccolo M., Paduano L., Montesarchio D.. Exploring the conformational behaviour and aggregation properties of lipid-conjugated AS1411 aptamers. Int. J. Biol. Macromol. 2018; 118:1384–1399. [DOI] [PubMed] [Google Scholar]
- 38. Meier M., Moya-torres A., Krahn N.J., Mcdougall M.D., Orriss L., Mcrae E.K.S., Booy E.P., Mceleney K., Patel T.R.. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018; 46:5319–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karsisiotis A.I., Hessari N.M.A., Novellino E., Spada G.P., Randazzo A., Webba da Silva M.. Topological characterization of nucleic acid G-Quadruplexes by UV absorption and circular dichroism. Angew. Chem. - Int. Ed. Eng. 2011; 50:10645–10648. [DOI] [PubMed] [Google Scholar]
- 40. Vorlíčková M., Kejnovská I., Sagi J., Renčiuk D., Bednářová K., Motlová J., Kypr J.. Circular dichroism and guanine quadruplexes. Methods. 2012; 57:64–75. [DOI] [PubMed] [Google Scholar]
- 41. Varizhuk A.M., Protopopova A.D., Tsvetkov V.B., Barinov N.A., Podgorsky V. V, Tankevich M. V, Vlasenok M.A., Severov V. V, Smirnov I.P., Dubrovin E. V et al.. Polymorphism of G4 associates: from stacks to wires via interlocks. Nucleic Acids Res. 2018; 46:8978–8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. del Villar-Guerra R., Trent J.O., Chaires J.B.. G-Quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem. - Int. Ed. 2018; 57:7171–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mergny J.-L., Phan A., Lacroix L.. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 44. Olsen C.M., Gmeiner W.H., Marky L.A.. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-quartet stacking. J. Phys. Chem. B. 2006; 110:6962–6969. [DOI] [PubMed] [Google Scholar]
- 45. Petraccone L., Erra E., Esposito V., Randazzo A., Mayol L., Nasti L., Barone G., Giancola C.. Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry. 2004; 43:4877–4884. [DOI] [PubMed] [Google Scholar]
- 46. Stoltenburg R., Krafčiková P., Víglaský V., Strehlitz B.. G-quadruplex aptamer targeting Protein A and its capability to detect Staphylococcus aureus demonstrated by ELONA. Sci. Rep. 2016; 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gray D.M. Berova Woody N, Polavarapu PL, Nakanishi K, Woody RW. Circular dichroism of protein-nucleic acid interactions. Comprehensive Chiroptical Spectroscopy. 2012; 2:615–633. [Google Scholar]
- 48. Nici F., Oliviero G., Falanga A.P., D’Errico S., Marzano M., Musumeci D., Montesarchio D., Noppen S., Pannecouque C., Piccialli G. et al.. Anti-HIV activity of new higher order G-quadruplex aptamers obtained from tetra-end-linked oligonucleotides. Org. Biomol. Chem. 2018; 16:2349–2355. [DOI] [PubMed] [Google Scholar]
- 49. D’Atri V., Borbone N., Amato J., Gabelica V., D’Errico S., Piccialli G., Mayol L., Oliviero G.. DNA-based nanostructures: the effect of the base sequence on octamer formation from d(XGGYGGT) tetramolecular G-quadruplexes. Biochimie. 2014; 99:119–128. [DOI] [PubMed] [Google Scholar]
- 50. Borbone N., Amato J., Oliviero G., D’Atri V., Gabelica V., De Pauw E., Piccialli G., Mayol L.. d(CGGTGGT) forms an octameric parallel G-quadruplex via stacking of unusual G(:C):G(:C):G(:C):G(:C) octads. Nucleic Acids Res. 2011; 39:7848–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lönne M., Bolten S., Lavrentieva A., Stahl F., Scheper T., Walter J.G.. Development of an aptamer-based affinity purification method for vascular endothelial growth factor. Biotechnol. Rep. 2015; 8:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trajkovski M., Webba Da Silva M., Plavec J.. Unique structural features of interconverting monomeric and dimeric G-quadruplexes adopted by a sequence from the intron of the N-myc gene. J. Am. Chem. Soc. 2012; 134:4132–4141. [DOI] [PubMed] [Google Scholar]
- 53. Smargiasso N., Rosu F., Hsia W., Colsom P., Baker E.S., Bowers M.T., De Pauw E., Gabelica V.. G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008; 130:10208–10216. [DOI] [PubMed] [Google Scholar]
- 54. Miller M.C., Buscaglia R., Chaires J.B., Lane A.N., Trent J.O.. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG 3(TTAG3)3). J. Am. Chem. Soc. 2010; 132:17105–17107. [DOI] [PubMed] [Google Scholar]
- 55. Miyoshi D., Karimata H., Sugimoto N.. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006; 128:7957–7963. [DOI] [PubMed] [Google Scholar]
- 56. Fujimoto T., Nakano S.I., Sugimoto N., Miyoshi D.. Thermodynamics-hydration relationships within loops that affect G-quadruplexes under molecular crowding conditions. J. Phys. Chem. B. 2013; 117:963–972. [DOI] [PubMed] [Google Scholar]
- 57. Platella C., Riccardi C., Montesarchio D., Roviello G.N., Musumeci D.. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. BBA - Gen. Subj. 2017; 1861:1429–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Musumeci D., Platella C., Riccardi C., Moccia F., Montesarchio D.. Fluorescence sensing using DNA aptamers in cancer research and clinical diagnostics. Cancers. 2017; 9:174–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.