Abstract
MicroRNAs (miRNAs) are short, noncoding RNAs that regulate gene expression by suppressing mRNA translation and reducing mRNA stability. A miRNA can potentially bind many mRNAs, thereby affecting the expression of oncogenes and tumor suppressor genes as well as the activity of whole pathways. The promise of miRNA therapeutics in cancer is to harness this evolutionarily conserved mechanism for the coordinated regulation of gene expression, and thus restoring a normal cell phenotype. However, the promiscuous binding of miRNAs can provoke unwanted off-target effects, which are usually caused by high-dose single-miRNA treatments. Thus, it is desirable to develop miRNA therapeutics with increased specificity and efficacy. To achieve that, we propose the concept of miRNA cooperativity in order to exert synergistic repression on target genes, thus lowering the required total amount of miRNAs. We first review miRNA therapies in clinical application. Next, we summarize the knowledge on the molecular mechanism and biological function of miRNA cooperativity and discuss its application in cancer therapies. We then propose and discuss a systems biology approach to investigate miRNA cooperativity for the clinical setting. Altogether, we point out the potential of miRNA cooperativity to reduce off-target effects and to complement conventional, targeted, or immune-based therapies for cancer.
MicroRNA THERAPEUTICS IN CANCER
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNAs, which negatively regulate gene expression by inducing translational repression or mRNA decay (1,2). In animals, miRNAs are primarily encoded in the genome as individual genes or as clusters (3). miRNA biogenesis starts with transcription of primary miRNAs that are processed by Drosha and Dicer to generate mature miRNAs. Subsequently, the mature miRNAs are bound by Argonaute protein to form RNA-induced silencing complexes that can recognize complementary mRNAs to repress target gene expression (3). Since the discovery of the first miRNA, the field of miRNA biology has expanded considerably (1,4–6). Because each miRNA regulates the expression of multiple genes, miRNAs can efficiently regulate and coordinate multiple cellular pathways and processes, like the ones involved in cellular growth and proliferation (6,7). Insights into the roles of miRNAs in the onset, progression and dissemination of cancers have made them attractive tools and targets for novel therapeutic approaches (8–10). Dysregulation of miRNAs can contribute to cancer establishment and progression, with miRNAs acting as oncogenes (oncomiRs) (11–13) or tumor suppressors (14–17). Hence, miRNA-based therapies fall into two different approaches: (i) the inhibition of oncomiRs by miRNA antagonists, such as antisense oligonucleotides, antagomirs and miRNA sponges, and (ii) augmenting the expression of tumor suppressor miRNAs using miRNA mimics, like double-stranded synthetic miRNAs and miRNA expression vectors. The choice of miRNA therapies depends on the disease mechanism and on whether the intended outcome is the gain or loss of gene function. miRNA antagonists designed with complementary sequences to oncomiRs prevent them from being loaded into Argonaute proteins, thereby reducing suppression of their tumor suppressor gene targets (18,19). miRNA mimics are used to restore lost or diminished function of tumor suppressor miRNAs whose downregulation leads to the activation of oncogenic pathways (18,20).
Like other RNA therapies, a key obstacle for miRNA therapeutics is the development of efficient delivery systems that facilitate safe and effective application (21–23). As small RNAs are prone to degradation by RNases in serum or in the endocytic compartment of cells, two different strategies have been investigated. One is to alter anti-miRNA oligonucleotides through chemical modification, yielding locked nucleic acids (LNAs) or a phosphorothioate-modified RNA backbone(24,25). For example, safety, tolerability, pharmacokinetics and potential efficacy of MRG-106, a LNA-modified antisense inhibitor of oncomiR miR-155, is currently tested in an ongoing clinical trial (NCT02580552) for treating patients with cutaneous T-cell lymphoma. A parallel effort goes into development of delivery vehicles, such as viral vectors, lipid-based systems and polymeric vectors, to encapsulate miRNAs for protection and allow endosomal escape (24–26). For instance, a lipid-formulated miR-34 mimic (MRX34) is the first miRNA drug that entered a phase I trial (NCT01829971) and was later tested in patients with primary liver cancer or metastatic cancer that has spread to the liver. In a more recent clinical trial (NCT02369198), miR-16 mimics packaged by mini-cells were intravenously administered in patients with mesothelioma or non-small cell lung cancer (27). Besides cancer, miRNA therapeutics have also been tested in other diseases like hepatitis C (28) and type 2 diabetes (29), and some of them have already succeeded in phase II trials (10). Interestingly, increasing evidence shows that miRNAs can cooperate to more efficiently regulate the expression of their target genes (42,43). Since this finding can improve and expand current miRNA-based therapies, we elaborate on it and discuss its potential clinical application in the following section.
MicroRNA COOPERATIVITY IN CANCER: MECHANISMS, FUNCTIONS AND POTENTIAL THERAPIES
Although the use of miRNA mimics and miRNA inhibitors as therapeutics seems promising, only a small number of miRNA therapeutics has so far progressed into clinical development. One major challenge is the identification of the best miRNA candidates or miRNA targets for different types of cancers. An intuitive strategy is to combine gene expression analysis with miRNA target prediction algorithms, such as TargetScan (30). However, this strategy is compromised by indirect effects and complex gene regulation involving different molecular species and their dynamical interactions. Thus, biochemical techniques based on Argonaute and miRNA immunoprecipitation (e.g. HITS-CLIP and PAR-CLIP) have been developed to experimentally identify miRNA–target interactions at a transcriptome-wide scale (31). Another challenge for miRNA therapeutics is to avoid or minimize toxicity and off-target effects. As a single miRNA often represses its targets quite weakly (21,32), high doses of effective miRNA mimics are usually required to achieve the expected effect. However, high doses also provoke undesired consequences, including unintended targeting by the administered miRNAs. For example, the MRX34 clinical trial had to be terminated due to immune-related adverse events involving patient deaths. Such failures may be prevented by lowering the dosage of the miRNA mimic and consequently reducing off-target effects, but therapeutic benefits could dwindle equally. To overcome this problem, a reasonable approach would be to use lower-dose combinations of miRNAs that synergistically regulate the expression of a shared target. In this context, we expect a reduction or avoidance of undesired events in patients when multiple miRNAs are co-administered at lower levels compared to an individual high-dose miRNA treatment.
Cooperative and synergistic miRNA regulation is an intriguing yet poorly explored mechanism. Different miRNAs can for example cooperate by regulating multiple, complementary targets in a pathway (Figure 1). There are a few experimentally validated examples where miRNAs exert synergistic effects in cancer (Table 1). Co-transfection of miR-34a and miR-15a/16 led to increased cell cycle arrest in non-small cell lung cancer cells due to the fact that miR-15a and miR-16 specifically downregulate CCNE1 and CCND3. Such gene regulation exerts a complementary effect to cell-cycle regulation by miR-34a (33). Pencheva et al. identified that miR-1908, miR-199a-5p and miR-199a-3p jointly target ApoE signaling in melanoma. LNA-mediated inhibition of these miRNAs strongly suppressed melanoma metastasis (34). In comparison to single-miRNA treatment in acute lymphoblastic leukemia cells of children, co-expression of miR-125b, miR-100 and miR-99a resulted in downregulation of multiple targets that is causally linked to the resistance to the chemotherapeutic agent vincristine (35). Through the investigation of nine miRNA pairs in glioma cells, Zhao et al. found that extensive synergy occurred among upregulated miRNAs. They showed that the highest synergistic effect increasing apoptosis of glioma cells is achieved through simultaneous inhibition of miR-20a and miR-21 (36). Derepression of tumor suppressor genes (PDCD4, BTG2 and NEDD4L) by inhibiting miR-21, miR-23a and miR-27a showed synergistic effects toward reducing pancreatic tumor growth and progression (37). Furthermore, considering multiple targeting of miRNAs, researchers proposed miRNAs as adjuvants in conjunction with available cancer therapies (38). For instance, miRNA modulation can be used to increase the efficiency of small-molecule inhibitors targeting oncogenes (39) and to lower effective doses of chemotherapies (40). We have demonstrated how a potential therapeutic gain can be achieved by utilizing miR-205 and miR-342 as co-adjuvants to sensitize tumor cells to a genotoxic anti-cancer drug (41).
Figure 1.
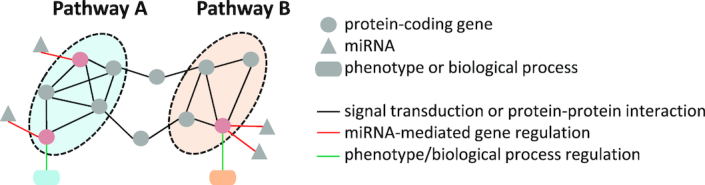
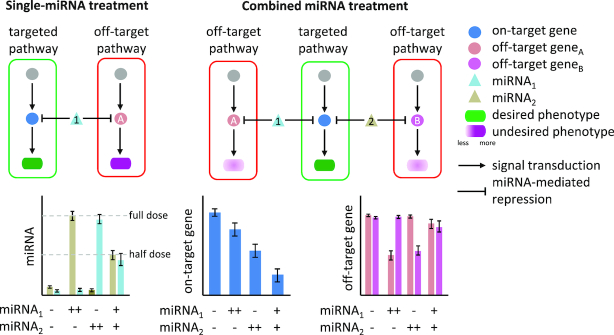
Implementation of miRNA cooperativity through targeting of a shared pathway or of a shared protein-coding gene. Targeting of several interlinked protein-coding genes by multiple miRNAs leads to the regulation of a pathway and thereby modulation of the phenotypic outcome (pathway A). Concerted targeting of a protein-coding gene by two miRNAs can induce efficient regulation of a biological process that is controlled by the gene (pathway B). miRNA targets are highlighted in red.
Table 1.
Experimentally verified cooperative miRNAs
| Target gene | Cooperating miRNAs | Reagents | Effect | Phenotype | Cancer | Reference |
|---|---|---|---|---|---|---|
| CCNE1 and other genes | miR-34a, miR-15/16 | Precursor | Synergistic | Cell cycle arrest | NSCLC (in vitro) | (33) |
| APOE, DNAJA4 | miR-1908, miR-199a, miR-199a | Inhibitors | Synergistic | Metastasis | Melanoma (in vitro, in vivo) | (34) |
| Multiple genes | miR-125b, miR-100, miR-99a | Precursor | Synergistic | Chemoresistance | ALL (in vitro) | (35) |
| n/a | miR-20a, miR-21 | Inhibitors | Synergistic | Apoptosis | Glioma (in vitro) | (36) |
| PDCD4, BTG2*, NEDD4L | miR-21, miR-23a, miR-27a | Inhibitors | Synergistic | Tumor growth | PDAC (in vitro, in vivo) | (37) |
| E2F1* | miR-205, miR-342 | Mimics | Synergistic | Chemoresistance | Melanoma NSCLC (in vitro) | (41) |
| CDKN1A* | miR-572, miR-93 | Mimics | Synergistic | n/a | Melanoma (in vitro) | (49) |
| TGFBR2 | miR-9, miR-130b | Mimics | Additive | n/a | NSCLC (in vitro) | (50) |
| DMPK* | miR-206, miR-148a | Precursor | Synergistic | n/a | n/a | (51) |
| RASA1, SPRED1 | miR-21, miR-206 | Mimics | Synergistic | Apoptosis | TNBC (in vitro) | (52) |
| RUNX3 | miR-130a, miR-495 | Mimics, inhibitors | Synergistic | Apoptosis Angiogenesis | GC (in vitro, in vivo) | (53) |
| PDCD4, TPM1, RhoC, HoxD10, EGFR, MMP2 | miR-21, miR-10b | Inhibitors | Synergistic | Chemoresistance Tumor proliferation and invasion | Glioma (in vitro, in vivo) | (40,100) |
The table lists experimentally verified cooperative miRNAs that regulate the expression of a gene or a phenotype in a concerted manner. The genes with adjacent (13–35 nts) miRNA binding sites are highlighted by asterisks. The regulatory effect is derived from a quantitative analysis of gene or phenotype regulation by the specified miRNAs. The effect is additive when any amount of one miRNA can be substituted with the same amount of the other miRNA without increasing or decreasing the effect of the treatment; the effect is synergistic when the combined treatment leads to a significantly stronger effect than a treatment with the same total amount of either miRNA. The effects of gene regulation by cooperative miRNAs on cell phenotypes are also given if confirmed in the study, and the category of the experiments (in vitro or in vivo) is specified. Non-small cell lung cancer (NSCLC); acute lymphoblastic leukemia (ALL); pancreatic ductal adenocarcinoma (PDAC); triple-negative breast cancer (TNBC); gastric cancer (GC); not available (n/a)
On the other hand, miRNA cooperativity can be implemented through efficient downregulation of targets common to multiple miRNAs (Figure 1). Specifically, early genome-wide studies indicated that miRNAs with binding sites located in close proximity in the 3′ untranslated region (UTR) of a shared target are more likely to induce stronger gene repression (42–44). Using this criterion, we developed a computational model and published a database containing thousands of predictions of such cooperative miRNA pairs (45) (https://triplexrna.org). The model can be improved by considering the interplay between mRNAs, miRNA-induced silencing complexes and RNA-binding proteins that can result in multiple, distinct mechanisms of miRNA cooperation (46,47). It is also worth noting that endogenous miRNA expression levels and the abundance of miRNA target sites may impact the magnitude of miRNA cooperativity (48).
The notion of cooperative gene regulation by miRNAs is supported by experimental evidence. We have shown in melanoma cells that miR-572 and miR-93 cooperatively repress CDKN1A protein synthesis during the DNA damage response (49). Using a luciferase reporter system, Mitra et al. demonstrated that miR-9–5p and miR-130b-3p cooperatively regulate TGFBR2 expression by binding to its 3′ UTR (50). Similarly, Koscianska et al. validated the cooperative regulation of DMPK by miR-206 and miR-148a (51). Furthermore, some studies have demonstrated the potential of using antagomirs or mimics of cooperative miRNAs as cancer therapies. Knockdown of miR-21 and miR-206 restored the expression of their common targets RASA1 and SPRED1, which inhibit in vivo tumor initiation by suppressing the activity of the RAS-ERK signaling pathway (52). Inhibition of miR-130a and miR-495 significantly reduced cell proliferation and angiogenesis in gastric cancer cells both in vitro and in vivo through the recovery of RUNX3 expression under hypoxic conditions (53). More recently, we have demonstrated the cooperative repression of E2F1 by miR-205 and miR-342 in aggressive melanoma and lung cancer cell lines, and found that overexpression of both miRNAs reduced the cells’ chemoresistance (41). Of note, due to the promiscuous binding of miRNAs, a miRNA can play contradictory roles by targeting different genes (54). Thus, it is reasonable to speculate that cooperative miRNAs targeting the same gene or pathway may oppose each other's direction of regulation in other circumstances. Consequently, the action of miRNAs depends on the cellular environment.
Moreover, increasing evidence has indicated that miRNA-mediated gene regulatory networks are critical for inferring cooperative miRNA regulation in cancer. The complexity of such networks results from the multiplicity of miRNA–target interactions, which often engage in reciprocal feedback and feedforward loops (55,56). Hence, network approaches have been applied to infer cooperative regulation among miRNAs (57,58). Xu et al. identified cancer-specific cooperative miRNAs using networks that are reconstructed using miRNA target information, the biological functions of the miRNA targets and their regulatory interactions (59). Li et al. developed an algorithm to detect synergistic miRNA regulatory modules in ovarian, breast and thyroid cancers (60). Shao et al. investigated miRNA cooperativity in pan-cancer networks and identified similar network structures and expression profiles of cooperative miRNA pairs for cancers with similar tissue origins (61).
In addition, it is important to investigate target hubs subject to coordinated regulation by dozens of miRNAs, as these are often key regulators of biological processes (62). We have shown that tight control of CDKN1A expression by multiple and cooperative miRNAs is important for cell cycle regulation (49). Similarly, hub miRNAs that target many genes play a dominant role in regulatory networks and thus bear potential as prognostic markers and therapeutic targets. Yang et al. identified several important hub miRNAs that are involved in a gene regulatory network in serous ovarian cancer and demonstrated in vivo the ability of miR-506 to reduce tumor growth by targeting SNAI2 (63). Li et al. identified miR-524 and miR-628 as preeminent regulators of malignant transformation in glioma and also showed their ability to individually predict the survival of patients with different glioma grades (64). Su et al. identified five hub miRNAs (miR-16–2-3p, miR-890, miR-3201, miR-602, and miR-877) as potential diagnostic and prognostic biomarkers for pancreatic ductal adenocarcinoma (65).
Taken together, the above studies demonstrate the power of combining computational and experimental methods in advancing our understanding of gene regulation by cooperative miRNAs and suggest that miRNA cooperativity should be considered to enhance efficacy and reduce toxicity of miRNA-based therapies for cancer.
A ROAD MAP TOWARD USING SYSTEMS BIOLOGY APPROACHES TO INVESTIGATE microRNA COOPERATIVITY IN ANTI-CANCER THERAPEUTICS
As mentioned before, a challenge in miRNA therapeutics is the potential toxicity linked to off-target effects. To overcome this problem, we propose to reduce off-target effects by using lower doses of cooperative miRNAs that synergistically repress common targets (Figure 2), with candidate selection implemented in a systems biology approach. This approach, combining computational and experimental methods, has become a staple in advancing our understanding of gene regulation by miRNAs (31,56,66–67) and also holds promise for biomedical application (41,68–69). We believe these methods can be re-purposed and combined into integrative workflows to exploit miRNA cooperativity in anti-cancer therapeutics. In particular, we are interested in miRNA cooperativity that is facilitated by adjacent miRNA target sites in the human transcriptome (44,45), as such cooperating miRNAs may exert synergistic effects on repressing their common targets (42). By definition, the synergistic action of cooperative miRNAs requires a lower total dose to achieve the same degree of repression as a single-miRNA treatment; yet the actual amounts and ratios for in vitro or in vivo deployments depend on the specific binding partners and also on the cellular context.
Figure 2.
Exploiting miRNA cooperativity in cancer therapeutics. Single-miRNA treatment requires high-dose administration of a specific miRNA to obtain a desired phenotype. This could lead to unwanted regulation of off-targets and thus to undesired phenotypes. Such effects can be reduced or avoided by combining miRNAs that jointly repress one gene, which not only reduces the amount of miRNAs required but also decreases off-target effects and thus the risk of toxicity. The bar plots illustrate the expression profiles of miRNAs (left), on-target genes (middle) and off-target genes (right) in biological scenarios where single or combined miRNA treatment is applied. Compared to the single treatments (++), the combined miRNA treatment (+) at lower dose is more efficient in repressing the on-target gene. The experimental design and expected observation are adapted from a study, in which the synergetic repression of E2F1 using low doses of miR-205 and miR-342 was validated in vitro (41). Due to the reduction of miRNA doses, we also expect less impact on the off-target genes by the combined miRNA treatment. Bar colors correspond to node colors in the upper diagram.
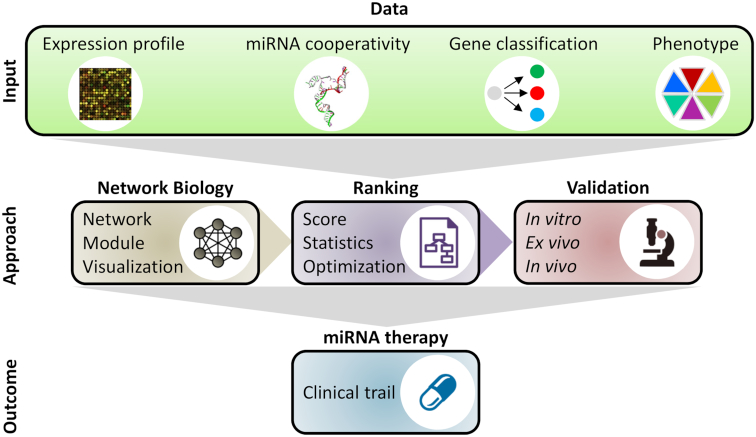
In the following, we describe an exemplary workflow that integrates existing methods to systematically identify cooperative miRNA pairs targeting cancer genes in the context of a given metastatic cancer (Figure 3). The workflow consists of five steps: (i) reconstruction of a gene regulatory network containing molecular species, their interactions as well as their association to phenotypes and clinical features of the investigated tumor entity; (ii) extraction of a core regulatory network that contains key molecules and interactions of biomedical interest; (iii) addition of predicted cooperating miRNA pairs to the core network; (iv) ranking of miRNA candidates for therapeutic intervention; and (v) at last, validation of the identified miRNA candidates using in vitro and in vivo experiments, with clinical tests and trials initiated in the case of success. In the following, we describe and discuss each step of the workflow in detail.
Figure 3.
A systems biology approach to investigate the application of miRNA cooperativity in cancer therapy. The approach feeds biological data of various sources into computational methods that predict cooperative miRNAs regulating the expression of cancer genes. Experimental validation of the computational results followed by clinical trials may ultimately lead to the discovery of novel miRNA therapies for cancer.
Reconstruction of gene regulatory networks
Information about molecular species and their interactions related to the tumor entity and the investigated therapeutic scenario can be collected and curated from the scientific literature and databases. This information is integrated into a network in which the nodes account for molecules (e.g. signaling proteins, transcription factors and miRNAs) and the edges for their interactions (e.g. activation and inhibition). Methods, tools and databases for network reconstruction with an emphasis on miRNAs have been reviewed by others (31,70).
In knowledge-driven studies, researchers reconstruct a gene regulatory network from a few core genes of interest. For instance, to investigate the role of the E2F1 signaling axis in the context of chemotherapy for melanoma, Vera et al. reconstructed a network with the core genes E2F1, p73/DNp73 and miR-205 and expanded it using databases on transcriptional and post-transcriptional gene regulation and protein–protein interactions (69). Using the network, the authors showed that the core genes can regulate the expression of pro- and anti-apoptotic genes (i.e. Bax, Hrk and BCL2) through an incoherent feedforward loop. This regulation of apoptotic genes by the signaling axis influences the sensitivity of tumor cells to genotoxic drugs.
In discovery-driven studies, researchers usually reconstruct a large network consisting of all genes and their interactions with relevance for the disease. Dreyer et al. developed a comprehensive network containing signaling pathways that are crucial for the development and progression of cutaneous melanoma and enriched it with melanoma-associated miRNAs (71). Using the network, the authors analyzed patient data on responsiveness to immunotherapy in melanoma and identified a subnetwork linked to epithelial-to-mesenchymal transition that is regulated by miR-34a and miR-18a. Sumazin et al. reconstructed an extensive miRNA-mediated network in glioblastoma, in which more than 7000 protein-coding genes are regulated by miRNAs. Follow-up analyses showed that miRNA-mediated interactions regulate driver genes of tumor initiation and subtype differentiation and thus may enable crosstalk between canonical oncogenic pathways underlying glioblastoma (68).
Extraction of core regulatory networks
Due to a large number of participating molecules, their heterogeneity and the complex interactions among them, the behavior of comprehensive regulatory networks is difficult to simulate and to connect with specific observable disease outcomes. Thus, we need to modularize the network to understand general principles that govern the structure and behavior of a biological system (72). Toward this end, one can integrate multi-omics and clinical data with network properties to identify a so-called core regulatory network, a module that is central in the tumor of interest and also relevant in the therapeutic context. The influence that a core regulatory network wields is reflected in the properties of its components. For example, hub genes connect the core to the rest of the network, while mutation or aberrant expression of core molecules makes them good candidates for therapeutic manipulation. Sun et al. reconstructed a comprehensive glioblastoma-specific regulatory network composed of miRNAs and transcription factors (73). By filtering the network components using connectivity metrics (i.e. node degree), the authors obtained a core regulatory network for glioblastoma and identified several miRNAs that may regulate the disease via the Notch signaling pathway. Similarly, Sadeghi et al. identified key regulatory interactions responsible for the primary tumor to metastasis transition in prostate cancer (74). Using a network inference approach that integrates patient-derived transcriptomic data, the authors reconstructed a comprehensive regulatory network including protein-coding genes, miRNAs and transcription factors. Key transcription factors and miRNAs were determined for this network using node degree and betweenness centrality measures whereby an integrative TF-miRNA regulatory core that drives prostate cancer progression was identified.
Integration of miRNA cooperativity information
There are a few computational tools that predict gene regulation by cooperative miRNAs, ranging from distance-defined cooperative regulation to structural modeling. The former predicts miRNA cooperativity based on the distance between predicted or validated miRNA binding sites (44). Structural modeling, on the other hand, predicts cooperative miRNA pairs by simulating molecular interactions of RNA-induced silencing complexes, miRNAs and mRNA targets. This approach better resembles the molecular processes underlying miRNA cooperativity but comes at a high computational cost (47). The TriplexRNA database combines miRNA target prediction algorithms, sequence and structural analyses, and kinetic modeling to predict and decipher the synergism that cooperative miRNAs exert on their targets; it not only increases the precision and accuracy of distance-based predictions but also makes the genome-wide identification of cooperative miRNAs computationally affordable (45).
By following the three steps described above, we reconstructed a network accounting for metastatic melanoma in the context of anti-PD1 immunotherapy (Figure 4; see Supplementary Figure S1 and Supplementary Materials for details). Precisely, we made use of availiable comprehensive networks accounting for key pathways in cutaneous melanoma (71) and for cell-cycle regulation linked to the emergence of cancer-associated phenotypes (75). We derived a core regulatory network by combining network motifs, network topology and gene expression profiles that were measured in melanoma biopsies from patients who received anti-PD1 immunotherapy (76). Network motifs are small structures like feedback and feedforward loops that are frequently recurring in large networks. These motifs encode relevant regulatory features and are often deregulated in cancer (56,77–78). Next, we characterized types of cancer-related genes (i.e. oncogenes and tumor suppressors) using the UniProtKB database (79) and annotated miRNA-target interactions with experimental validation using miRTarBase (80) and starBase (81). We annotated the network with the hallmarks of cancer that are linked to protein-coding genes involved in relevant biological functions or signaling pathways (82). Then, we included miRNA pairs that cooperate in the regulation of protein-coding genes in the core network by utilizing the TriplexRNA database (45). To filter the miRNAs in the core network, we used datasets that characterize miRNA expression profiles in three human cell lines (83), namely, one melanocyte cell line, one melanoma cell line with a weak metastatic phenotype (A375), and one with a strong metastatic phenotype (A2058). By integrating all data, we aimed to identify cooperative miRNA pairs that show abnormal expression levels in melanoma cells, making them potential targets for manipulating the expression of oncogenic and tumor suppressor genes.
Figure 4.
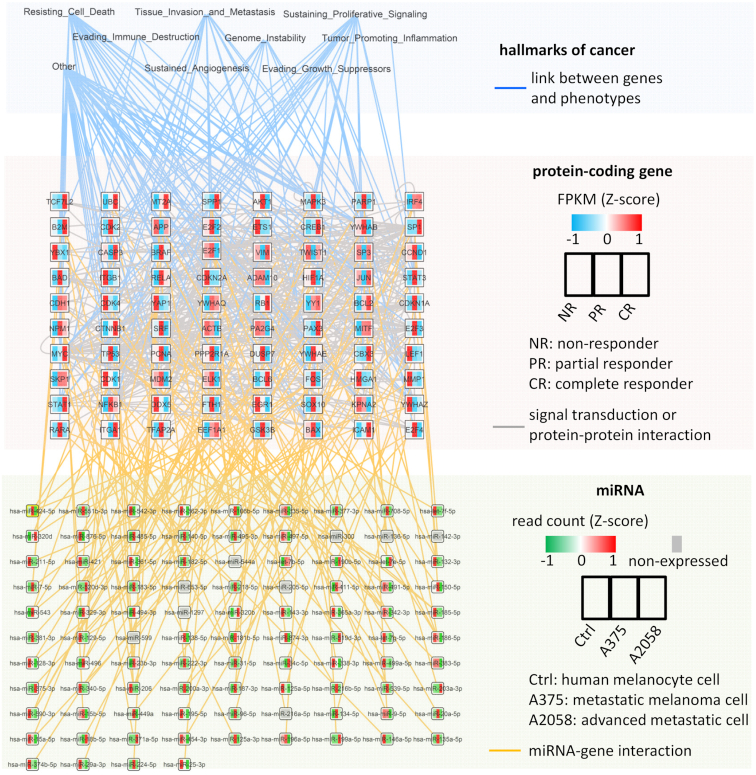
Overview of miRNA cooperativity in the melanoma network. The network contains hallmarks of cancer (top layer), protein-coding genes (middle layer), miRNAs (bottom layer) and their interactions. If a protein-coding gene is connected to two miRNAs (orange lines), it means its mRNA is predicted to form a triplex with the miRNAs. We show only the triplex with the highest score for a protein-coding gene (Supplementary Table S1). All identified cooperative miRNAs can be found in Supplementary Table S2 or at www.synmirapy.net. The data used to reconstruct the network can be found in Supplementary Table S3.
As a result, we obtained a comprehensive network built from protein–protein interactions, cooperative miRNA-mediated gene regulation, and hallmarks-of-cancer associations (Figure 4). This kind of network can be used to visualize and mine high-throughput data generated from in vitro or in vivo experiments, or from patient-derived samples. For example, the color of protein-coding genes in Figure 4 indicates their expression levels in melanoma patient groups according to their classification into non-responders (NR), partial responders (PR) and complete responders (CR) to anti-PD1 therapy in Hugo et al. (76).
Ranking miRNA candidates for therapeutic intervention
While a core regulatory network is less complex, it is enriched with potential therapeutic targets and thus allows for more targeted downstream experimental investigations. Additional criteria can be used to select the most promising candidates for experimental validation, making experimental validation even more cost-effective in terms of time and resources. Often knowledge and hypothesis-driven inspection of computational results (e.g. a table of differentially expressed miRNAs and their putative targets) is used to select relevant and promising candidates for experimental validation (84,85). Alternatively, one can combine biological knowledge, quantitative data and mathematical modeling to derive a systematic and unbiased ranking of candidate genes. For instance, Dhawan et al. used a multi-variable linear model to identify miRNAs common to hallmark gene signatures across different cancers and ranked the identified miRNAs based on their strength of association to these signatures (denoted by the estimated model coefficients) (86). The analysis prioritized putative associations between miRNAs and hallmark phenotypes of cancer. By ranking putative miRNA binding sites, Mavrakis et al. identified five miRNAs that can produce cooperative effects on tumor suppressor genes implicated in the pathogenesis of T-cell acute lymphoblastic leukemia (87).
In this study, we ranked triplexes formed by target mRNAs and the predicted cooperative miRNAs using an equation that involves the computed equilibrium concentration and the minimum free energy of the triplexes (see Supplementary Materials for details). We interpret the former as the likelihood of triplex formation and the latter as the stability of the triplex. The equation is formulated based on the assumption that stability is secondary to the likelihood of triplex formation, and we hypothesized that triplexes with higher scores are more likely to represent true positives (Supplementary Table S1). It is also worth noting that molecular dynamics simulations can be employed to substantiate the predicted triplexes through analyzing their structural and thermodynamic properties (41,45). All identified triplexes for the melanoma network are accessible at an interactive website, allowing further investigation (www.synmirapy.net).
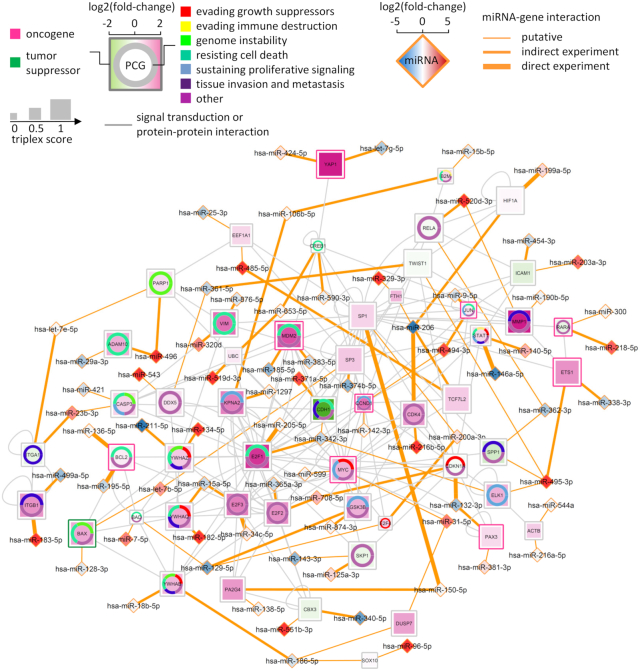
Furthermore, we built an interaction network from the selected miRNAs and their targets and enriched it with multiple layers of information, creating a powerful tool to select miRNA candidates for experimental validation. More specifically, we included: (i) the classification of genes as an oncogene or a tumor suppressor gene; (ii) the association of protein-coding genes with the hallmarks of cancer; (iii) the fold-change of protein-coding gene expression levels for the comparison between non-responders and complete responders to anti-PD1 treatment from the dataset (76); (iv) the fold-change of miRNA expression levels for the comparison between melanoma and melanocyte cell lines; (v) existing experimental evidence for the miRNA–target interactions; and (vi) the score for the likelihood of protein-coding genes to be cooperatively repressed by its targeting miRNAs. The information visualized and aggregated in the network provides us with a tool to identify cancer genes whose expression could be modulated by cooperative miRNAs (Figure 5 and Table 2). In particular, we predicted therapeutic strategies that deploy either miRNA mimics to suppress oncogenes or miRNA inhibitors to upregulate the expression of tumor suppressor genes. For instance, we found that the expression of the oncogene BCL2, which is a regulator of apoptosis (88), is increased in melanoma patients that do not respond to anti-PD1 treatment. Both BCL2-targeting miRNAs (miR-195 and miR-136) are downregulated in melanoma cells and their abilities to repress BCL2 were also validated at the mRNA and protein levels (miRTarBase entry: MIRT006868 and MIRT005362). This suggests that simultaneous upregulation of both miRNAs constitutes an efficient treatment option to repress BCL2. Such a miRNA treatment might be an alternative to BCL2 antisense therapies for melanoma (89,90). A similar treatment strategy could also be employed for the other identified oncogenes as their expression is upregulated in non-responding patients and their cooperative miRNA regulators are either simultaneously downregulated or differentially modulated (i.e. one miRNA is upregulated and the other is downregulated; Table 2). The tumor suppressor BAX is a pro-apoptotic gene whose activation leads to the release of cytochrome c and consequently cell death (91). The data showed that BAX expression is upregulated in non-responding patients while the expression of its cooperating miRNAs is differentially modulated (i.e. let-7b is upregulated and miR-128 is downregulated) in tumor cells. Hence, targeting let-7b with a specific inhibitor might augment the expression level of the tumor suppressor.
Figure 5.
Predictive miRNA treatments for melanoma. The network shows protein-coding genes (PCG; squares) that could be targeted by cooperative miRNAs (diamonds) for therapy based on their expression profiles in the context of melanoma. The associations between protein-coding genes and cancer hallmarks are denoted by colors in the ring chart. The gene classification (oncogene or tumor suppressor) is denoted by the color of the square border. The color of protein-coding gene indicates the fold-change of its expression levels for the comparison between patients undergoing anti-PD1 treatment (non-responders versus complete responders). Two miRNAs connecting to a protein-coding gene (orange lines) were predicted to form the highest-scoring triplex with the mRNA of the protein-coding gene. The size of protein-coding genes increases with the score assigned to their triplexes. The color of the miRNA indicates the fold change of its expression level for the comparison between melanoma cells and human melanocytes (A375 + A2058 versus control). The thickness of the orange lines is determined by whether or not the putative miRNA–gene interactions have been validated by experiments. The validation data are from miRTarBase r2018 and starBase v2.0. As direct experiments we consider reporter assays, qRT-PCR and Western blots. Indirect experiments are high-throughput experiments, such as microarray, RNA sequencing, HITS-CLIP and PAR-CLIP. The data used to visualize the network can be found in Supplementary Table S4.
Table 2.
Cooperative miRNAs targeting cancer genes
| Target gene | Cooperative miRNAs | Direct therapy | Role in cancer | Reference |
|---|---|---|---|---|
| MDM2 | miR-185-5p* (D) miR-383-5p* (D) | Small molecule (101) | Overexpressed in cancers; Negative regulator of TP53 | (101,102) |
| ETS1 | miR-338-3p* (D) miR-495-3p* (U) | RNA interference (103) | Overexpressed in cancer cells; Chemoresistance Regulate the RAS/MAPK pathway | (103–105) |
| YAP1 | let-7g-5p* (D) miR-424-5p* (U) | Small molecule (106) | Activated in a broad number of solid tumors Involved in tumor initiation, growth, metastasis and chemoresistance; Regulate the Hippo pathway | (106–108) |
| MYC | miR-31-5p (U) miR-599 (N) | Small molecule (109) | Deregulated in more than half of human cancers; Regulate pathways underlying cell growth, cell-cycle progression, metabolism, and survival | (109,110) |
| BCL2 | miR-136-5p# (U) miR-195-5p# (D) | Small molecule (88) Oligonucleotides (89) RNA interference (90) | Regulate mitochondrial integrity, subsequent caspase activation, and apoptosis | (17,88–90,111) |
| PAX3 | miR-132-3p* (D) miR-381-3p (U) | n/a | Contribute to cell survival in melanoma; Drive oncogenic transformation; Enhance cell invasion and migration | (112–114) |
| CCND1 | miR-142-3p (N) miR-494-3p (U) | n/a | Sometimes amplified in melanoma cell-cycle regulator | (115,116) |
| RARA | miR-218-5p* (U) miR-300 (N) | Natural and synthetic retinoids (117) | Modulate cell growth and differentiation in response to retinoids; Induce chemoresistance | (117–120) |
| JUN | miR-9-5p* (D) miR-494-3p* (U) | Small molecule (121) | Overexpressed in a large fraction of human melanoma samples; Promote cell proliferation and tumor progression; Regulate the MAPK and JNK/JUN pathway | (121–123) |
| BAX | let-7b-5p (U) miR-128-3p (D) | Small molecule (17) mRNA lipofection (124) Rrg-Gly-Asp-based peptides (125) | Promote apoptosis; Regulate the mitochondrial apoptosis pathway | (17,91,111,124,125) |
The table lists identified oncogenes (regular font) and one tumor suppressor gene (in italics) that can be targeted by cooperative miRNAs. The texts in parentheses indicate the regulation of miRNAs in melanoma cells (D: downregulated, U: upregulated, N: non-expressed). The superscripts indicate whether or not the miRNA–gene interactions have been validated by direct (#) or indirect (*) experiments. The third and fourth columns show available therapies for the target genes and their known role in cancer, respectively. n/a: not available.
Experimental validation of candidate miRNAs
Ultimately, the computational methods described previously are utilized to support and accelerate experimental findings. At the same time, the computationally predicted cooperative miRNA target interactions have to be validated by in vitro and in vivo experiments prior to considering their use in clinical trials. There are numerous examples where putative miRNA-mediated gene regulation in cancer has been validated experimentally (92,93). For instance, using target prediction algorithms, Lal et al. predicted a large number of candidate target genes for miR-24 that are associated with cell proliferation. The authors experimentally confirmed that more than a hundred of the putative targets are directly downregulated by overexpressed miR-24 and that miR-24 plays a role in regulating cell-cycle progression and DNA repair (94). We predicted the cooperative regulation of CDKN1A by miR-572 and miR-93 and validated that both miRNAs can consistently repress the expression of CDKN1A after provoked genotoxic stress in melanoma cells (49).
Previous systems biology-driven studies showed that upregulation of E2F1 can undermine the effect of anti-PD1 treatment through negative regulation of apoptosis and promotion of processes associated to cancer aggressiveness, such as epithelial-to-mesenchymal transition (71,75). Our analysis suggests that abnormal expression of E2F1 in melanoma may be caused by the loss or downregulation of miRNAs such as miR-205 and miR-342 (Figure 5). Specifically, the data showed that miR-205 is endogenously expressed in neither melanocytes nor melanoma cells, while miR-342 expression is downregulated in melanoma cells. Thus, one therapeutic strategy complementing the anti-PD1 immunotherapy could be augmenting the expression of miR-205 and miR-342 to cooperatively downregulate E2F1 in malignant melanoma. In this line, we combined sequence alignment, molecular dynamics simulations, and kinetic modeling to investigate cooperative miRNAs targeting E2F1 (41). We identified and validated that simultaneous upregulation of miR-205 and miR-342 is an efficient way to achieve E2F1 repression using in vitro experiments. The combined treatment using low doses of both miRNAs enhanced E2F1 repression significantly and also increased the tumor cells’ sensitivity to conventional chemotherapy. Taken together, these results show how computational methods like the ones reviewed and discussed here can be used to prioritize miRNA candidates for anti-cancer therapies and also guide experimental validation.
DISCUSSION AND CONCLUSION
Despite the rapid development of miRNA therapeutics for cancer, there are still concerns related to the effects of miRNA modulation. These concerns include the lack of target specificity, the chemical design of anti-miRNA and miRNA mimics, as well as the efficiency of miRNA delivery systems to specifically reach tumor cells (25,39). Here, we discussed a therapeutic strategy that combines two cooperative miRNAs to increase efficacy and specificity in gene regulation, and thus significantly reduce the total dose of miRNAs needed to achieve a desired effect. Our previous work showed that cooperative miRNAs used in reduced doses increase the sensitivity of tumor cells to chemotherapy through enhanced repression of the intended target (41). It has also been reported that pooling of small interfering RNAs (siRNAs) can eliminate off-target effects, which is achieved by lowering the concentration of individual siRNAs (95). Interestingly, siRNAs designed to mimic miRNA features, with reduced sequence complementarity to their designated target sites, show improved efficacy and fewer off-target effects (96). Consequently, one can speculate that lower doses of multiple miRNAs which act in a cooperative manner may also reduce off-target effects that are the cause for toxicity in miRNA treatments (97). Of note, it has been shown that a reduction in off-target effects may not be achieved when using low-complexity pools of a few siRNAs (95). Although miRNAs as natural cellular products may be more efficient than siRNAs in repressing target genes, it is still required to thoroughly their assess off-target effects (96).
We proposed a systems biology approach that integrates computational methods with experiments to identify and validate cooperative miRNA pairs that can synergistically modulate the expression of cancer genes and also be used as therapeutic targets for complementing cancer therapies. While the approach has been demonstrated to be successful in vitro, it is necessary to expand and upgrade it before applying newly found miRNA therapies in a clinical setting. This is because miRNA interactions are highly complex since not only mRNAs but also pseudogenes, long ncRNAs and circular RNAs are among their potential off-targets. One mechanism whereby these RNAs can interfere with miRNA function is by miRNA sponging, sequestering them away from their intended targets (98,99). To prevent titration effects by such decoys, an obvious strategy is to avoid selecting cooperating miRNAs that have a higher affinity to bind to competing RNAs than to their intended targets. This requires expression profiling of RNA competitors and also the analysis of properties of corresponding miRNA binding sites.
Furthermore, targeted delivery of cooperating miRNAs is crucial to achieve therapeutic effects. However, there is a lack of means to ensure precise transmission of miRNA-targeting reagents such as miRNA mimics and inhibitors (22). Thus, further work is required in the design of vehicles for their efficient delivery in vivo, especially for tissues where natural uptake of miRNAs is modest and it is necessary to use carriers to transfect miRNA reagents. For instance, one possible strategy is to coat miRNA-containing particles with ligands, peptides or antibodies that can recognize and interact with specific receptors expressed on the surface of tumor cells. Other potential issues, such as how to ensure an effective and sufficient miRNA dose in the appropriate target cells in order to achieve therapeutic effects, are also under investigation (24).
Despite many challenges in the clinical application of miRNAs, we believe that the advancement of computational models and biotechnology will allow miRNA cooperativity to enhance and diversify miRNA therapeutics, and thus provide additional treatments against cancer.
DATA AVAILABILITY
The data presented in the article is available at www.synmirapy.net.
Supplementary Material
ACKNOWLEDGEMENTS
X.L. and J.V. conceived the study. X.L., M.E. and J.V. developed the method. X.L. and M.E. analyzed the data. X.L. prepared the figures and wrote the software. X.L. and J.V. wrote the original manuscirpt. All authors reviewed and edited the manuscirpt and approved its final version.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Federal Ministry of Education and Research (BMBF) [eBio:MelEVIR 031L0073A to J.V.]; Staedler Stiftung [ww/eh 30/16 to J.V.]; Cancer Institute of NSW Fellowship to U.S.; ELAN-fund of Universitätsklinikum Erlangen [16-08-16-1-Lai to X.L.]. We also acknowledge support by Deutsche Forschungsgemeinschaft and Friedrich-Alexander-Universität Erlangen-Nürnberg within the funding program Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 2. Iwakawa H.-O., Tomari Y.. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015; 25:651–665. [DOI] [PubMed] [Google Scholar]
- 3. Treiber T., Treiber N., Meister G.. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019; 20:5–20. [DOI] [PubMed] [Google Scholar]
- 4. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gebert L.F.R., MacRae I.J.. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019; 20:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel D.P. Metazoan microRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ebert M.S., Sharp P.A.. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012; 149:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kasinski A.L., Slack F.J.. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer. 2011; 11:849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayes J., Peruzzi P.P., Lawler S.. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014; 20:460–469. [DOI] [PubMed] [Google Scholar]
- 10. Rupaimoole R., Slack F.J.. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017; 16:203–222. [DOI] [PubMed] [Google Scholar]
- 11. Hirono T., Jingushi K., Nagata T., Sato M., Minami K., Aoki M., Takeda A.H., Umehara T., Egawa H., Nakatsuji Y. et al.. MicroRNA-130b functions as an oncomiRNA in non-small cell lung cancer by targeting tissue inhibitor of metalloproteinase-2. Sci. Rep. 2019; 9:6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu J., Xu C., Ruan L., Wu J., Li Y., Zhang X.. MicroRNA-146b overexpression promotes human bladder cancer invasion via enhancing ETS2-Mediated mmp2 mRNA transcription. Mol. Ther. Nucleic Acids. 2019; 16:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao W., Wang X., Wang T., Xing J.. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging (Albany NY). 2019; 11:615–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Svoronos A.A., Engelman D.M., Slack F.J.. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016; 76:3666–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mansoori B., Mohammadi A., Ghasabi M., Shirjang S., Dehghan R., Montazeri V., Holmskov U., Kazemi T., Duijf P., Gjerstorff M. et al.. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J. Cell Physiol. 2019; 234:9816–9825. [DOI] [PubMed] [Google Scholar]
- 16. Wach S., Brandl M., Borchardt H., Weigelt K., Lukat S., Nolte E., Al-Janabi O., Hart M., Grässer F., Giedl J. et al.. Exploring the MIR143-UPAR axis for the inhibition of human prostate cancer cells in vitro and in vivo. Mol. Ther. Nucleic Acids. 2019; 16:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh R., Letai A., Sarosiek K.. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019; 20:175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bader A.G., Brown D., Winkler M.. The promise of microRNA replacement therapy. Cancer Res. 2010; 70:7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M.. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005; 438:685–689. [DOI] [PubMed] [Google Scholar]
- 20. Jin H.Y., Gonzalez-Martin A., Miletic A.V., Lai M., Knight S., Sabouri-Ghomi M., Head S.R., Macauley M.S., Rickert R.C., Xiao C.. Transfection of microRNA mimics should be used with caution. Front. Genet. 2015; 6:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y., Gao D.-Y., Huang L.. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 2015; 81:128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levin A.A. Treating disease at the RNA level with oligonucleotides. N. Engl. J. Med. 2019; 380:57–70. [DOI] [PubMed] [Google Scholar]
- 23. Stylianopoulos T., Jain R.K.. Combining two strategies to improve perfusion and drug delivery in solid tumors. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:18632–18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z., Rana T.M.. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 2014; 13:622–638. [DOI] [PubMed] [Google Scholar]
- 25. Slaby O., Laga R., Sedlacek O.. Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 2017; 474:4219–4251. [DOI] [PubMed] [Google Scholar]
- 26. Castelli D.D., Terreno E., Cabella C., Chaabane L., Lanzardo S., Tei L., Visigalli M., Aime S.. Evidence for in vivo macrophage mediated tumor uptake of paramagnetic/fluorescent liposomes. NMR Biomed. 2009; 22:1084–1092. [DOI] [PubMed] [Google Scholar]
- 27. van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L. et al.. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017; 18:1386–1396. [DOI] [PubMed] [Google Scholar]
- 28. Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjärn M., Hansen J.B., Hansen H.F., Straarup E.M. et al.. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008; 36:1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M.. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011; 474:649–653. [DOI] [PubMed] [Google Scholar]
- 30. Agarwal V., Bell G.W., Nam J.-W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bracken C.P., Scott H.S., Goodall G.J.. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016; 17:719–732. [DOI] [PubMed] [Google Scholar]
- 32. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N.. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008; 455:58–63. [DOI] [PubMed] [Google Scholar]
- 33. Bandi N., Vassella E.. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol. Cancer. 2011; 10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pencheva N., Tran H., Buss C., Huh D., Drobnjak M., Busam K., Tavazoie S.F.. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012; 151:1068–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akbari Moqadam F., Lange-Turenhout E.A.M., Ariës I.M., Pieters R., den Boer M.L.. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk. Res. 2013; 37:1315–1321. [DOI] [PubMed] [Google Scholar]
- 36. Zhao Y., Cui X., Zhu W., Chen X., Shen C., Liu Z., Yang G., Liu Y., Zhao S.. Synergistic regulatory effects of microRNAs on brain glioma cells. Mol. Med. Rep. 2017; 16:1409–1416. [DOI] [PubMed] [Google Scholar]
- 37. Frampton A.E., Castellano L., Colombo T., Giovannetti E., Krell J., Jacob J., Pellegrino L., Roca-Alonso L., Funel N., Gall T.M.H. et al.. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology. 2014; 146:268–277. [DOI] [PubMed] [Google Scholar]
- 38. Sotillo E., Thomas-Tikhonenko A.. Shielding the messenger (RNA): microRNA-based anticancer therapies. Pharmacol. Ther. 2011; 131:18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gulei D., Berindan-Neagoe I.. Combined therapy in cancer: the non-coding approach. Mol. Ther. Nucleic Acids. 2018; 12:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malhotra M., Sekar T.V., Ananta J.S., Devulapally R., Afjei R., Babikir H.A., Paulmurugan R., Massoud T.F.. Targeted nanoparticle delivery of therapeutic antisense microRNAs presensitizes glioblastoma cells to lower effective doses of temozolomide in vitro and in a mouse model. Oncotarget. 2018; 9:21478–21494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lai X., Gupta S.K., Schmitz U., Marquardt S., Knoll S., Spitschak A., Wolkenhauer O., Pützer B.M., Vera J.. MiR-205-5p and miR-342-3p cooperate in the repression of the E2F1 transcription factor in the context of anticancer chemotherapy resistance. Theranostics. 2018; 8:1106–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saetrom P., Heale B.S.E., Snøve O., Aagaard L., Alluin J., Rossi J.J.. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007; 35:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rinck A., Preusse M., Laggerbauer B., Lickert H., Engelhardt S., Theis F.J.. The human transcriptome is enriched for miRNA-binding sites located in cooperativity-permitting distance. RNA Biol. 2013; 10:1125–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schmitz U., Lai X., Winter F., Wolkenhauer O., Vera J., Gupta S.K.. Cooperative gene regulation by microRNA pairs and their identification using a computational workflow. Nucleic Acids Res. 2014; 42:7539–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Broderick J.A., Salomon W.E., Ryder S.P., Aronin N., Zamore P.D.. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011; 17:1858–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Flamand M.N., Gan H.H., Mayya V.K., Gunsalus K.C., Duchaine T.F.. A non-canonical site reveals the cooperative mechanisms of microRNA-mediated silencing. Nucleic Acids Res. 2017; 45:7212–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M.. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell. 2016; 64:565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lai X., Schmitz U., Gupta S.K., Bhattacharya A., Kunz M., Wolkenhauer O., Vera J.. Computational analysis of target hub gene repression regulated by multiple and cooperative miRNAs. Nucleic Acids Res. 2012; 40:8818–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mitra R., Edmonds M.D., Sun J., Zhao M., Yu H., Eischen C.M., Zhao Z.. Reproducible combinatorial regulatory networks elucidate novel oncogenic microRNAs in non-small cell lung cancer. RNA. 2014; 20:1356–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koscianska E., Witkos T.M., Kozlowska E., Wojciechowska M., Krzyzosiak W.J.. Cooperation meets competition in microRNA-mediated DMPK transcript regulation. Nucleic Acids Res. 2015; 43:9500–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sharma S.B., Lin C.-C., Farrugia M.K., McLaughlin S.L., Ellis E.J., Brundage K.M., Salkeni M.A., Ruppert J.M.. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol. Cell Biol. 2014; 34:4143–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee S.H., Jung Y.D., Choi Y.S., Lee Y.M.. Targeting of RUNX3 by miR-130a and miR-495 cooperatively increases cell proliferation and tumor angiogenesis in gastric cancer cells. Oncotarget. 2015; 6:33269–33278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Banzhaf-Strathmann J., Edbauer D.. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun. Signal. 2014; 12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vera J., Lai X., Schmitz U., Wolkenhauer O.. MicroRNA-regulated networks: the perfect storm for classical molecular biology, the ideal scenario for systems biology. Adv. Exp. Med. Biol. 2013; 774:55–76. [DOI] [PubMed] [Google Scholar]
- 56. Lai X., Wolkenhauer O., Vera J.. Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res. 2016; 44:6019–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu B., Li J., Cairns M.J.. Identifying miRNAs, targets and functions. Brief. Bioinformatics. 2014; 15:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang J., Pham V.V.H., Liu L., Xu T., Truong B., Li J., Le T.D.. Identifying miRNA synergism using multiple-intervention causal inference. 2019; bioRxiv doi: 10.1101/652180, 29 May 2019, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- 59. Xu J., Li C.-X., Li Y.-S., Lv J.-Y., Ma Y., Shao T.-T., Xu L.-D., Wang Y.-Y., Du L., Zhang Y.-P. et al.. MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res. 2011; 39:825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li Y., Liang C., Wong K.-C., Luo J., Zhang Z.. Mirsynergy: detecting synergistic miRNA regulatory modules by overlapping neighbourhood expansion. Bioinformatics. 2014; 30:2627–2635. [DOI] [PubMed] [Google Scholar]
- 61. Shao T., Wang G., Chen H., Xie Y., Jin X., Bai J., Xu J., Li X., Huang J., Jin Y. et al.. Survey of miRNA-miRNA cooperative regulation principles across cancer types. Brief. Bioinformatics. 2018; doi:10.1093/bib/bby038. [DOI] [PubMed] [Google Scholar]
- 62. Shalgi R., Lieber D., Oren M., Pilpel Y.. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput. Biol. 2007; 3:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang D., Sun Y., Hu L., Zheng H., Ji P., Pecot C.V., Zhao Y., Reynolds S., Cheng H., Rupaimoole R. et al.. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013; 23:186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li Y., Xu J., Chen H., Bai J., Li S., Zhao Z., Shao T., Jiang T., Ren H., Kang C. et al.. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic Acids Res. 2013; 41:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Su Q., Zhu E.C., Qu Y.-L., Wang D.-Y., Qu W.-W., Zhang C.-G., Wu T., Gao Z.-H.. Serum level of co-expressed hub miRNAs as diagnostic and prognostic biomarkers for pancreatic ductal adenocarcinoma. J. Cancer. 2018; 9:3991–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Steinkraus B.R., Toegel M., Fulga T.A.. Tiny giants of gene regulation: experimental strategies for microRNA functional studies. Wiley Interdiscip. Rev. Dev. Biol. 2016; 5:311–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Napolitano M., Comegna M., Succoio M., Leggiero E., Pastore L., Faraonio R., Cimino F., Passaro F.. Comparative analysis of gene expression data reveals novel targets of senescence-associated microRNAs. PLoS One. 2014; 9:e98669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sumazin P., Yang X., Chiu H.-S., Chung W.-J., Iyer A., Llobet-Navas D., Rajbhandari P., Bansal M., Guarnieri P., Silva J. et al.. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011; 147:370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vera J., Schmitz U., Lai X., Engelmann D., Khan F.M., Wolkenhauer O., Pützer B.M.. Kinetic modeling-based detection of genetic signatures that provide chemoresistance via the E2F1-p73/DNp73-miR-205 network. Cancer Res. 2013; 73:3511–3524. [DOI] [PubMed] [Google Scholar]
- 70. Alshalalfa M. miRNA regulation in the context of functional protein networks: principles and applications. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014; 6:189–199. [DOI] [PubMed] [Google Scholar]
- 71. Dreyer F.S., Cantone M., Eberhardt M., Jaitly T., Walter L., Wittmann J., Gupta S.K., Khan F.M., Wolkenhauer O., Pützer B.M. et al.. A web platform for the network analysis of high-throughput data in melanoma and its use to investigate mechanisms of resistance to anti-PD1 immunotherapy. Biochim. Biophys. Acta. 2018; 1864:2315–2328. [DOI] [PubMed] [Google Scholar]
- 72. Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W.. From molecular to modular cell biology. Nature. 1999; 402:C47–C52. [DOI] [PubMed] [Google Scholar]
- 73. Sun J., Gong X., Purow B., Zhao Z.. Uncovering MicroRNA and transcription factor mediated regulatory networks in glioblastoma. PLoS Comput. Biol. 2012; 8:e1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sadeghi M., Ranjbar B., Ganjalikhany M.R., Khan M, Schmitz F., Wolkenhauer U., Gupta S.K.. MicroRNA and transcription factor gene regulatory network analysis reveals key regulatory elements associated with prostate cancer progression. PLoS One. 2016; 11:e0168760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Khan F.M., Marquardt S., Gupta S.K., Knoll S., Schmitz U., Spitschak A., Engelmann D., Vera J., Wolkenhauer O., Pützer B.M.. Unraveling a tumor type-specific regulatory core underlying E2F1-mediated epithelial-mesenchymal transition to predict receptor protein signatures. Nat. Commun. 2017; 8:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G. et al.. Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell. 2016; 1:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007; 8:450–461. [DOI] [PubMed] [Google Scholar]
- 78. Tsang J., Zhu J., van Oudenaarden A.. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007; 26:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chou C.-H., Shrestha S., Yang C.-D., Chang N.-W., Lin Y.-L., Liao K.-W., Huang W.-C., Sun T.-H., Tu S.-J., Lee W.-H. et al.. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018; 46:D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li J.-H., Liu S., Zhou H., Qu L.-H., Yang J.-H.. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014; 42:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Knijnenburg T.A., Bismeijer T., Wessels L.F.A., Shmulevich I.. A multilevel pan-cancer map links gene mutations to cancer hallmarks. Chin. J. Cancer. 2015; 34:439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ding N., Wang S., Yang Q., Li Y., Cheng H., Wang J., Wang D., Deng Y., Yang Y., Hu S. et al.. Deep sequencing analysis of microRNA expression in human melanocyte and melanoma cell lines. Gene. 2015; 572:135–145. [DOI] [PubMed] [Google Scholar]
- 84. Iliopoulos D., Polytarchou C., Hatziapostolou M., Kottakis F., Maroulakou I.G., Struhl K., Tsichlis P.N.. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci. Signal. 2009; 2:ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nolte E., Wach S., Silva I.T., Lukat S., Ekici A.B., Munkert J., Müller-Uri F., Kreis W., Oliveira Simões C.M., Vera J. et al.. A new semisynthetic cardenolide analog 3β-[2-(1-amantadine)- 1-on-ethylamine]-digitoxigenin (AMANTADIG) affects G2/M cell cycle arrest and miRNA expression profiles and enhances proapoptotic survivin-2B expression in renal cell carcinoma cell lines. Oncotarget. 2017; 8:11676–11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dhawan A., Scott J.G., Harris A.L., Buffa F.M.. Pan-cancer characterisation of microRNA across cancer hallmarks reveals microRNA-mediated downregulation of tumour suppressors. Nat. Commun. 2018; 9:5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mavrakis K.J., Van Der Meulen J., Wolfe A.L., Liu X., Mets E., Taghon T., Khan A.A., Setty M., Rondou P., Vandenberghe P. et al.. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat. Genet. 2011; 43:673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Anvekar R.A., Asciolla J.J., Missert D.J., Chipuk J.E.. Born to be alive: a role for the BCL-2 family in melanoma tumor cell survival, apoptosis, and treatment. Front. Oncol. 2011; 1:fonc.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jansen B., Wacheck V., Heere-Ress E., Schlagbauer-Wadl H., Hoeller C., Lucas T., Hoermann M., Hollenstein U., Wolff K., Pehamberger H.. Chemosensitisation of malignant melanoma by BCL2 antisense therapy. Lancet. 2000; 356:1728–1733. [DOI] [PubMed] [Google Scholar]
- 90. Reddy T.L., Garikapati K.R., Reddy S.G., Reddy B.V.S., Yadav J.S., Bhadra U., Bhadra M.P.. Simultaneous delivery of Paclitaxel and Bcl-2 siRNA via pH-Sensitive liposomal nanocarrier for the synergistic treatment of melanoma. Sci. Rep. 2016; 6:35223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu Z., Ding Y., Ye N., Wild C., Chen H., Zhou J.. Direct activation of bax protein for cancer therapy. Med. Res. Rev. 2016; 36:313–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Thomas M., Lieberman J., Lal A.. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 2010; 17:1169–1174. [DOI] [PubMed] [Google Scholar]
- 93. Thomson D.W., Bracken C.P., Goodall G.J.. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011; 39:6845–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lal A., Navarro F., Maher C.A., Maliszewski L.E., Yan N., O’Day E., Chowdhury D., Dykxhoorn D.M., Tsai P., Hofmann O. et al.. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to ‘seedless’ 3′UTR microRNA recognition elements. Mol. Cell. 2009; 35:610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hannus M., Beitzinger M., Engelmann J.C., Weickert M.-T., Spang R., Hannus S., Meister G.. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014; 42:8049–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wu H., Ma H., Ye C., Ramirez D., Chen S., Montoya J., Shankar P., Wang X.A., Manjunath N.. Improved siRNA/shRNA functionality by mismatched duplex. PLoS One. 2011; 6:e28580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. van Rooij E., Purcell A.L., Levin A.A.. Developing microRNA therapeutics. Circ. Res. 2012; 110:496–507. [DOI] [PubMed] [Google Scholar]
- 98. Yoon J.-H., Abdelmohsen K., Gorospe M.. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014; 34:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hausser J., Zavolan M.. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat. Rev. Genet. 2014; 15:599–612. [DOI] [PubMed] [Google Scholar]
- 100. Dong C.G., Wu W.K.K., Feng S.Y., Wang X.J., Shao J.F., Qiao J.. Co-inhibition of microRNA-10b and microRNA-21 exerts synergistic inhibition on the proliferation and invasion of human glioma cells. Int. J. Oncol. 2012; 41:1005–1012. [DOI] [PubMed] [Google Scholar]
- 101. Burgess A., Chia K.M., Haupt S., Thomas D., Haupt Y., Lim E.. Clinical overview of MDM2/X-Targeted therapies. Front. Oncol. 2016; 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Vassilev L.T. MDM2 inhibitors for cancer therapy. Trends Mol. Med. 2007; 13:23–31. [DOI] [PubMed] [Google Scholar]
- 103. Wu M., Liu X., Jin W., Li Y., Li Y., Hu Q., Chu P.K., Tang G., Ping Y.. Targeting ETS1 with RNAi-based supramolecular nanoassemblies for multidrug-resistant breast cancer therapy. J. Control Release. 2017; 253:110–121. [DOI] [PubMed] [Google Scholar]
- 104. Garrett-Sinha L.A. Review of Ets1 structure, function, and roles in immunity. Cell Mol. Life Sci. 2013; 70:3375–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tetsu O., McCormick F.. ETS-targeted therapy: can it substitute for MEK inhibitors. Clin. Transl. Med. 2017; 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Oku Y., Nishiya N., Shito T., Yamamoto R., Yamamoto Y., Oyama C., Uehara Y.. Small molecules inhibiting the nuclear localization of YAP/TAZ for chemotherapeutics and chemosensitizers against breast cancers. FEBS Open Bio. 2015; 5:542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Felley-Bosco E., Stahel R.. Hippo/YAP pathway for targeted therapy. Transl. Lung Cancer Res. 2014; 3:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zanconato F., Battilana G., Cordenonsi M., Piccolo S.. YAP/TAZ as therapeutic targets in cancer. Curr. Opin. Pharmacol. 2016; 29:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wang H., Hammoudeh D.I., Follis A.V., Reese B.E., Lazo J.S., Metallo S.J., Prochownik E.V.. Improved low molecular weight Myc-Max inhibitors. Mol. Cancer Ther. 2007; 6:2399–2408. [DOI] [PubMed] [Google Scholar]
- 110. Chen H., Liu H., Qing G.. Targeting oncogenic Myc as a strategy for cancer treatment. Signal. Transduct. Target Ther. 2018; 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yip K.W., Reed J.C.. Bcl-2 family proteins and cancer. Oncogene. 2008; 27:6398–6406. [DOI] [PubMed] [Google Scholar]
- 112. Scholl F.A., Kamarashev J., Murmann O.V., Geertsen R., Dummer R., Schäfer B.W.. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001; 61:823–826. [PubMed] [Google Scholar]
- 113. Nguyen T.H., Barr F.G.. Therapeutic approaches targeting PAX3-FOXO1 and its regulatory and transcriptional pathways in rhabdomyosarcoma. Molecules. 2018; 23:E2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wachtel M., Schäfer B.W.. PAX3-FOXO1: zooming in on an ‘undruggable’ target. Semin. Cancer Biol. 2018; 50:115–123. [DOI] [PubMed] [Google Scholar]
- 115. Fu M., Wang C., Li Z., Sakamaki T., Pestell R.G.. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004; 145:5439–5447. [DOI] [PubMed] [Google Scholar]
- 116. Flaherty K.T., Hodi F.S., Fisher D.E.. From genes to drugs: targeted strategies for melanoma. Nat. Rev. Cancer. 2012; 12:349–361. [DOI] [PubMed] [Google Scholar]
- 117. Soprano K.J., Soprano D.R.. Retinoic acid receptors and cancer. J. Nutr. 2002; 132:3809S–3813S. [DOI] [PubMed] [Google Scholar]
- 118. Demary K., Wong L., Liou J.S., Faller D.V., Spanjaard R.A.. Redox control of retinoic acid receptor activity: a novel mechanism for retinoic acid resistance in melanoma cells. Endocrinology. 2001; 142:2600–2605. [DOI] [PubMed] [Google Scholar]
- 119. Johansson H.J., Sanchez B.C., Mundt F., Forshed J., Kovacs A., Panizza E., Hultin-Rosenberg L., Lundgren B., Martens U., Máthé G. et al.. Retinoic acid receptor alpha is associated with tamoxifen resistance in breast cancer. Nat. Commun. 2013; 4:2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang X., Dasari S., Nowakowski G.S., Lazaridis K.N., Wieben E.D., Kadin M.E., Feldman A.L., Boddicker R.L.. Retinoic acid receptor alpha drives cell cycle progression and is associated with increased sensitivity to retinoids in T-cell lymphoma. Oncotarget. 2017; 8:26245–26255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bennett B.L., Sasaki D.T., Murray B.W., O’Leary E.C., Sakata S.T., Xu W., Leisten J.C., Motiwala A., Pierce S., Satoh Y. et al.. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gurzov E.N., Bakiri L., Alfaro J.M., Wagner E.F., Izquierdo M.. Targeting c-Jun and JunB proteins as potential anticancer cell therapy. Oncogene. 2008; 27:641–652. [DOI] [PubMed] [Google Scholar]
- 123. Wagner E.F., Nebreda A.R.. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 2009; 9:537–549. [DOI] [PubMed] [Google Scholar]
- 124. Okumura K., Nakase M., Inui M., Nakamura S., Watanabe Y., Tagawa T.. Bax mRNA therapy using cationic liposomes for human malignant melanoma. J. Gene Med. 2008; 10:910–917. [DOI] [PubMed] [Google Scholar]
- 125. Karageorgis A., Claron M., Jugé R., Aspord C., Thoreau F., Leloup C., Kucharczak J., Plumas J., Henry M., Hurbin A. et al.. Systemic delivery of tumor-targeted Bax-derived membrane-active peptides for the treatment of melanoma tumors in a humanized SCID mouse model. Mol. Ther. 2017; 25:534–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the article is available at www.synmirapy.net.