Abstract
Background
Transmission of human hepegivirus 1 (HHpgV-1), a novel human pegivirus, is closely associated with hepatitis C virus (HCV). The impact of HHpgV-1 viremia on HCV infection is unknown. This study aimed to (a) evaluate the impact of HHpgV-1 viremia on HCV viral load and liver injury and (b) elucidate the clinical and molecular epidemiology of HHpgV-1 infection.
Methods
Individuals with HHpgV-1 viremia (cases) were identified by screening plasma from 655 HCV-infected adults. HHpgV-1 isolates were sequenced for phylogenetic analysis, and viral load was quantified. Cases were age- and sex-matched to HCV-infected individuals without HHpgV-1 viremia (controls) in a 1:3 ratio. A retrospective case–control analysis was performed to identify differences in HCV viral load and parameters of liver injury.
Results
Among HCV-infected adults, 16/655 (2.4%) had HHpgV-1 viremia. Risk groups for HHpgV-1 infection included intravenous drug users, blood product recipients, tattoo recipients, and men who have sex with men. Viral sequences clustered into 2 distinct HHpgV-1 genogroups. Cases had a higher mean HCV viral load than controls, with difference between means of 0.58 log10 IU/mL (P = .009). Cases were more likely to have an HCV viral load >5 log10 IU/mL (P = .028). Multiple regression demonstrated the impact of HHpgV-1 viral load and infection status on HCV viral load. HHpgV-1 infection was not associated with higher liver function tests, fibrosis scores, or imaging abnormalities.
Conclusions
HHpgV-1 viremia is associated with a higher HCV viral load in co-infected patients. HHpgV-1 infection does not affect progression of HCV-related liver disease.
Keywords: hepatitis C virus, human hepegivirus 1, hepatitis
Human hepegivirus 1 (HHpgV-1), also known as human pegivirus 2 (HPgV-2), is a recently discovered RNA virus belonging to species H of the Pegivirus genus within the family Flaviviridae [1]. Although HHpgV-1 is phylogenetically closer to human pegivirus 1 (also known as GBV-C), it shares features with hepatitis C virus (HCV) such as a type IV internal ribosomal entry site in the 5’-untranslated region and several potential glycosylation sites in the envelope protein [2].
HHpgV-1 was independently discovered by 2 groups among blood product recipients and HCV-infected patients [2, 3]. Since then, HHpgV-1 viremia has been documented in populations in Cameroon, China, Iran, the United Kingdom, the United States, and Vietnam [4–8]. The virus appears to have a particular predilection for co-infection with HCV among persons who inject drugs and hemophilia patients [5, 9]. This had led to a postulation of blood-borne transmission. Among HCV-infected individuals, the prevalence of HHpgV-1 viremia ranges from 0.3% to 1.6% [5, 10]. In individuals co-infected with HCV and HIV, the prevalence rises to 3.4%–10.9% [4, 9, 10].
HHpgV-1 viremia appears to persist in approximately 30% of infected individuals, with spontaneous clearance in the remainder [10]. Although HHpgV-1 infection is highly linked to concomitant HCV infection, individuals with isolated HHpgV-1 infections have been described. This excludes a “helper virus” relationship akin to hepatitis delta virus and hepatitis B virus (HBV) [9].
The effect of HHpgV-1 infection on HCV-related liver damage is uncertain because studies to date have either been uncontrolled or have only included a small number of HHpgV-1 cases [4, 6, 9, 10]. In this study, we aimed to measure the prevalence and epidemiology of HHpgV-1 viremia in the HCV-infected population in Hong Kong. Furthermore, we investigated the impact of active HHpgV-1 infection on HCV viral loads and related liver damage by comparing HHpgV-1 viremic HCV carriers with HCV-infected comparators without HHpgV-1 viremia.
METHODS
Study Setting and Samples
The study was conducted in the microbiology laboratory of Queen Mary Hospital, Hong Kong, between August 31, 2017, and January 31, 2019. Archived HCV RNA–positive plasma samples from 655 adults with chronic HCV infection were retrieved for HHpgV-1 virus RNA detection. Some individuals had more than 1 sample sent for HCV viral load detection during the study period. These samples were also included in the study to investigate incidence of acute HHpgV-1 acquisition and spontaneous viral clearance during the study period. Nucleic acid was extracted from 730 samples from 655 adults, followed by HHpgV-1 RNA detection by quantitative real time RT-PCR (qRT-PCR) using the methodology described below. The study received ethics approval from the institutional review board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW 14–249).
HHpgV-1 qRT-PCR
Aliquots of each plasma sample (20 µL) were combined together into mini-pools (9 to 10 plasma aliquots per mini-pool). Nucleic acid was extracted from plasma mini-pools using the EZ1 Virus Mini Kit v2.0 (Qiagen, Hilden, Germany) and eluted into a final volume of 60 µL. The mini-pools were screened for HHpgV-1 RNA using qRT-PCR and a LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland). qRT-PCR was performed as described by Frankel et al., except that we used the reverse complement of the original probe sequence to avoid quenching the fluorescence of the reporter dye by the guanosine base at the 5’-end of the probe sequence [11]. Primer and probe sequences were as follows: HPgV2_5UTR_F (5’-CGCTGATCGTGCAAAGGGATG-3’), HPgV2_5UTR_R (5’-GCTCCACGGACGTCACACTGG-3’), and HPgV2_5UTR_Probe (5’HEX-ATCAGGCTGTACGGAGTGGTG-IABkFQ3’). Each 20-µL reaction mix contained 1X QuantiNova Probe RT-PCR Master Mix, 1X QN Probe RT-Mix, 0.8 µM of forward and reverse primers, a 0.2-µM probe, and a 5-µL template. Reactions were incubated at 45oC for 10 minutes and 95oC for 5 minutes, followed by 50 cycles at 95oC for 5 seconds and 55oC for 30 seconds. For mini-pools testing positive for HHpgV-1 (Cp < 40), individual plasma samples constituting the mini-pool were extracted and underwent HHpgV-1 qRT-PCR to identify positive plasma within the mini-pool and quantify HHpgV-1 RNA. Plasmid concentrations ranging from 102 to 106 copies/reaction were used to generate standard curves for the qRT-PCR run.
HHpgV-1 NS5B Gene Sequencing and Phylogenetic Analysis
Reverse transcription was performed using the SuperScript IV kit (Invitrogen, Carlsbad, CA). Conventional sequencing of a 549-nucleotide fragment of the HHpgV-1 NS5B gene was done on cDNA derived from HHpgV-1-positive plasma samples using primers HPgV2_8614F_NS5B: 5’-GGTACGCTTACAACAAGCAGTG-3’ and HPgV2_9235R_NS5B: 5’-TCAACCCTCTCATGCCAAAG-3’. PCR was performed by preparing a PCR mixture (25 µL) containing 1 µL of cDNA, 1X PCR buffer II (10 mM Tris-HCl [pH 8.3], 50 mM KCl) 2mM of MgCl2, 200 µM of each dNTP, 1 µM of forward and reverse primers, and 1.0 U of Taq polymerase (Applied Biosystems, Foster City, CA). The mixtures were then incubated in an automated thermocycler with a hot start at 95oC for 10 minutes, followed by 50 cycles of 94oC for 1 minute, 55oC for 1 minute, 72oC for 1 minute, and a final extension at 72oC for 10 minutes. All PCR products were gel-purified using the QIAquick gel extraction kit (Qiagen). Both strands of the PCR products were sequenced with the 3730 DNA Analyzer (Applied Biosystems) using PCR primers. Sequences were assembled and manually edited to produce final sequences of the partial HHpgV-1 NS5B gene by BioEdit, version 7.2.5 (NC State University, Raleigh, NC). Final sequences were aligned with other HHpgV-1 sequences downloaded from GenBank. Phylogenetic trees were constructed using MEGA7, and sequence identity matrices were generated using MatGAT [12, 13].
Cases and Controls
HCV-infected individuals with circulating HHpgV-1 RNA in blood were designated as cases. Controls were HCV-infected individuals without HHpgV-1 viremia who were sex-matched and age-matched (+/- 2 years) to individual cases manually by an investigator who was blinded to the clinical details of cases and controls. Controls were recruited from within the same sample pool and individually tested negative for HHpgV-1 RNA. The case:control ratio was 1:3, as determined by the sample size formula of Kelsey et al. assuming a 2-sided significance level of 95% and study power of 80% [14].
Demographic, clinical, and laboratory data of cases and controls were retrieved from the electronic patient record. These included HCV viral load, genotype, route of HCV infection, time since HCV diagnosis, hepatitis B surface antigen (HBsAg), and HIV infection status. Alanine aminotransferase (ALT), total bilirubin, platelet count, and prothrombin time collected concomitantly with the HCV viral load were recorded. Fibrosis scores, measured by transient elastography (FibroScan) and liver imaging reports, were retrieved. Modalities of liver imaging included ultrasound and contrast-enhanced computed tomography. Radiographer comments of chronic parenchymal changes, fatty infiltration, cirrhosis, and hepatocellular carcinoma were considered imaging abnormalities.
HCV Viral Load and Genotyping
Before July 2018, HCV viral load measurement and genotyping were performed using the Abbott RealTime HCV viral load assay and Genotype II kit, respectively (Abbott, Des Plaines, IL), as previously described [15]. After July 2018, HCV viral load measurement and genotyping were performed using the Roche Cobas 4800 system (Roche), which had excellent correlation with the Abbott kit based on extensive in-house verification (verification data are available upon request). HCV viral loads were expressed in IU/mL to further ensure interassay reproducibility in reporting.
Statistical Analysis
Statistical analysis was performed using the Student t test, Welch’s t test for independent samples with unequal variances, the Fisher exact test, the chi-square test, simple and multiple linear regression using XLSTAT (Addinsoft, Long Island, NY), and the Microsoft Office Excel Analysis ToolPak add-in program. Scatter column graphs and scatter plots were produced using GraphPad Prism, version 8.1 (GraphPad Software, La Jolla, CA).
RESULTS
Prevalence and Epidemiology of HHpgV-1 Infection
The samples were combined into 75 mini-pools for HHpgV-1 screening; 12/75 (16%) mini-pools were positive. Independent samples constituting each mini-pool were tested for HHpgV-1, and 16 positive samples were found. Therefore, the final prevalence of HHpgV-1 viremia in the HCV-infected population was 16/655 (2.4%). None of the individuals with multiple samples collected during the study period tested positive for HHpgV-1. The median age of patients was 54.5 years, and most were male (81.2%). We retrieved risk factors for HCV infection in each patient as this would be an indicator of the route of acquisition of HHpgV-1 infection. Intravenous drug use was the most common route of HCV infection (8/16, 50%). Only 1 of the 8 patients was still using intravenous drugs at the time of the study; the other 7 patients had already abstained for several years. The second most common risk factor for HCV acquisition was via contaminated blood products, accounting for 4 patients (4/16, 25%): 2 of these patients suffered from hemophilia and had probably received non–virally inactivated clotting factor concentrates in early life, whereas the other 2 had received blood transfusions before 1991, when routine HCV screening of blood products was not yet introduced in Hong Kong. In addition, we identified an HHpgV-1-infected individual with no risk factors for HCV apart from receiving a tattoo. Another patient reported having sex with men, with no other risk factors for HCV. No identifiable risk factor for blood-borne virus infections could be identified in 2 female patients. All 16 patients were chronically infected with HCV and had been followed up for a median duration (interquartile range) of 6 (2.75–9.25) years. Half the patients were infected with genotype 1 HCV, and 5/16 (31.25%) patients were infected with genotype 6, which is the second most common HCV genotype in Hong Kong [16]. HBsAg positivity was documented in 4/16 (25%) cases, whereas 1 patient was known to be co-infected with HIV. None of the cases had received anti-HCV direct-acting antivirals (DAAs) at the time of HCV viral load and liver function test (LFT) measurement. The demographic and virological details of the 16 cases are summarized in Table 1.
Table 1.
Baseline Characteristics of Cases and Controls
| Parameters | Cases (n = 16) | Controls (n = 48) | P |
|---|---|---|---|
| Median age (interquartile range), y | 54.5 (46.8–65) | 54.5 (46.8–64.8) | .996 |
| Sex, No. (%) | |||
| Male | 13 (81.2) | 39 (81.2) | 1.000 |
| Female | 3 (18.8) | 9 (18.8) | |
| Ethnicity, No. (%) | |||
| Chinese | 15 (93.7) | 44 (91.7) | 1.000 |
| Others | 1 (6.3) | 4 (8.3) | |
| Route of HCV infection, No. (%) | |||
| Intravenous drug use | 8 (50.0) | 25 (52.1) | .107 |
| Blood transfusion before 1991 | 2 (12.5) | 11 (22.9) | |
| Hemophilia | 2 (12.5) | 0 (0.0) | |
| Men who have sex with men | 1 (6.25) | 3 (6.25) | |
| Tattoo | 1 (6.25) | 0 (0.0) | |
| Unknown | 2 (12.5) | 9 (18.75) | |
| Infecting HCV genotype, No. (%) | |||
| Genotype 1: 1a or 1b | 8 (50.0) | 18 (37.5) | .485 |
| Genotype 6 | 5 (31.25) | 19 (39.6) | |
| Other genotypes | 0 (0.0) | 5 (10.4) | |
| Unknown/indeterminate genotype | 3 (18.75) | 6 (12.5) | |
| Known HBsAg positivity, No. (%) | 4 (25.0) | 5 (10.4) | .210 |
| Known HIV positivity, No. (%) | 1 (6.25) | 2 (4.2) | 1.000 |
Abbreviations: HBsAg, hepatitis B surface antigen; HCV, hepatitis C virus.
HHpgV-1 Viral Load and Sequence Analysis
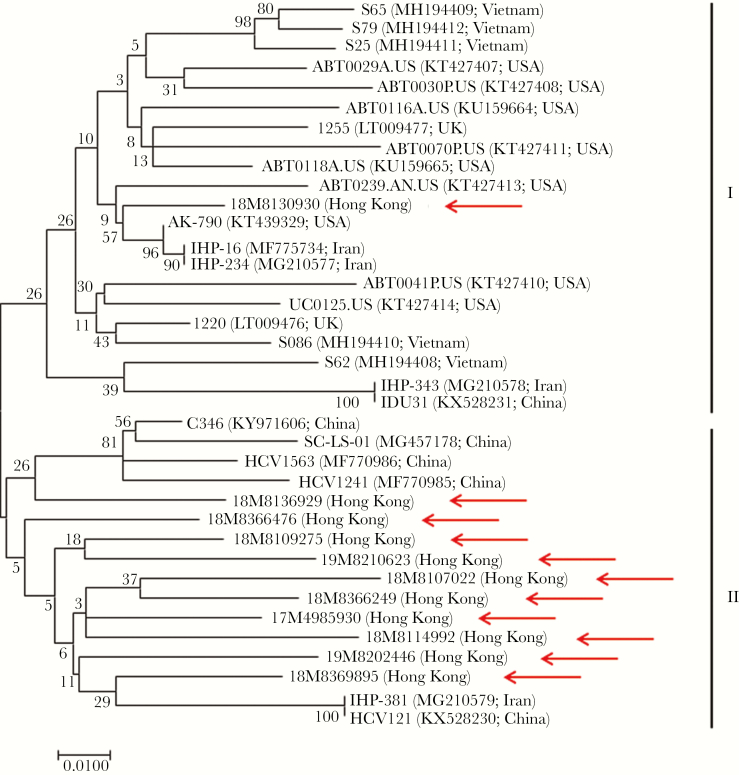
HHpgV-1 viral loads of the 16 cases ranged from 3.7 to 6.5 log10 copies/mL, with a mean of 5.5 log10 copies/mL. Sequencing of a near-complete fragment of NS5B was possible for 11/16 isolates with adequate viral loads. Phylogenetic analysis showed that HHpgV-1 sequences from our study and GenBank clustered into 2 distinct genogroups, designated I and II (Figure 1). These 2 groups shared a nucleotide identity of 90.3%–94.7%. Genogroup I mostly included isolates from Iran, the United Kingdom, the United States, and Vietnam. One of the HHpgV-1 isolates from our study (18M8130930) clustered with Iranian and American hemophilia patients. The patient from whom this sequence was derived also suffered from hemophilia, indicating transmission through non–virally inactivated plasma-derived clotting factor concentrates. Most of the isolates from our study clustered in genogroup II with isolates from China.
Figure 1.
Phylogenetic analysis of the partial NS5B region of the 11 HHpgV-1 strains detected in Hong Kong and other strains available in GenBank. The tree was constructed by the maximum-likelihood method using the Kimura 2-parameter model with gamma distributed with invariant sites (G + I). The bootstrap analysis was performed with 1000 replicates. The analysis included 549 nucleotide positions. GenBank accession numbers and country/city are shown in parentheses. The scale bar indicates the estimated number of substitutions per site. Arrows indicate the strains detected in this study.
Selection and Characteristics of Controls
Each case was matched with 3 HCV-infected control individuals on the basis of age and sex. Control plasma samples were individually tested for HHpgV-1 RNA and confirmed to be negative. None of the 48 control patients had ever received DAAs at the time of viral load and LFT measurement. Background features of cases and controls, including ethnicity, route of HCV infection, infecting HCV genotypes, HBsAg, and HIV co-infection rates, were congruent (Table 1).
Comparison of Cases and Controls
Cases and controls were compared for hepatitis C viral load and various parameters of liver injury. As depicted in Figure 2A, cases had a mean HCV viral load of 6.21 log10 IU/mL, whereas the mean HCV viral load of controls was 5.63 log10 IU/mL (difference between means, 0.58 log10 IU/mL; 95% confidence interval [CI], 0.15–1.01 log10 IU/mL). This difference achieved statistical significance (P = .009 by Welch t test). All cases had an HCV viral load >5 log10 IU/mL, compared with 36/48 (75%) controls (P = .028). Among cases, HHpgV-1 viral load and HCV viral load were not significantly correlated. The correlation coefficient (r) was 0.257 (95% CI, –0.274 to 0.667; P = .337) (Supplementary Figure 1a).
Figure 2.
Scatter column graphs comparing (A) hepatitis C virus viral load, (B) alanine aminotransferase, (C) bilirubin, (D) prothrombin time, (E) platelet count, and (F) fibrosis scores between cases and controls. The number of cases is 16 and the number of controls is 48, unless stated otherwise. Error bars represent mean and standard error of the mean. Asterisk indicates statistical difference by Welch’s t test.
To further analyze the link between HCV viral load and HHpgV-1 status, we constructed a multiple regression model to detect variables that may have an effect on HCV viral load. Variables included infecting genotype (genotype 1 or genotype 6), HBsAg status, ALT levels, and HHpgV-1 infection status (as a categorical variable). The model was constructed using data from 55 patients (13 cases, 42 controls) who had complete data for all variables using the backward selection approach. A P value of .15 was used as the cutoff for entry and elimination for backward stepwise regression. Although the model could not completely predict HCV variability, backward stepwise regression produced a final model with ALT, infecting genotype, and HHpgV-1 status as significant explanatory variables (P < .15; adjusted R2 = .103).
When HHpgV-1 status was substituted with the actual HHpgV-1 viral load in a multiple regression model involving the 13 cases for whom data for all explanatory variables were available, HHpgV-1 viral load was the only variable selected in the final model (P = .142 by backward stepwise regression; adjusted R2 = .112).
Cases and controls were then compared for liver injury parameters including liver function test parameters, fibrosis scores, and imaging abnormalities. The chi-square and Fisher exact tests were used as appropriate to compare proportions of cases and controls with abnormal imaging findings and liver function/fibrosis scores above the upper limit of normal. Cases tended to have higher risk (relative risk > 1) of having ALT and bilirubin above the upper limit of normal and also tended to be at higher risk of having fibrosis scores >7 kPa and imaging abnormalities, but these results did not achieve statistical significance (Table 2). ALT, bilirubin, platelet counts, prothrombin time, and liver stiffness were then analyzed as continuous variables. Cases and controls did not show any significant difference in these parameters by Welch’s t test (Figures 2B–F). As shown in Supplementary Figure 1b, ALT was not significantly correlated with HHpgV-1 viral load (r = –.389; P = .136).
Table 2.
Comparison of HCV Viral Load and Parameters of Liver Damage Between Cases and Controls
| Outcome | Cases | Controls | P | RR (95% CI) | ||
|---|---|---|---|---|---|---|
| No/Totala | % | No/Totala | % | |||
| ALT ≥58 U/L | 9/16 | 56.25 | 21/48 | 43.75 | .386 | 1.28 (0.75–2.20) |
| Total bilirubin ≥23 µmol/L | 3/16 | 18.75 | 3/48 | 6.25 | .159 | 3.00 (0.67–13.40) |
| Prothrombin time ≥13.6 sec | 1/14 | 7.14 | 9/38 | 23.68 | .254 | 0.30 (0.04–2.17) |
| Platelet count ≤154 ×109/L | 7/16 | 43.75 | 12/48 | 25.00 | .264 | 1.75 (0.83–3.67) |
| Fibrosis score ≥7 kPa | 6/7 | 85.71 | 15/23 | 65.22 | .393 | 1.31 (0.86–2.01) |
| Any imaging abnormality | 8/14 | 57.14 | 12/39 | 30.77 | .081 | 1.86 (0.97–3.57) |
| Parenchymal changes | 4/14 | 28.57 | 7/39 | 17.94 | .453 | 1.59 (0.55–4.62) |
| Fatty infiltration | 1/14 | 7.14 | 3/39 | 7.69 | 1.000 | 0.93 (0.11–8.21) |
| Cirrhosis | 2/14 | 14.28 | 0/39 | 0 | .081 | 13.33 (0.68–261.95)b |
| Hepatocellular carcinoma | 1/14 | 7.14 | 2/39 | 5.13 | 1.000 | 1.39 (0.14–14.19) |
Abbreviations: ALT, alanine aminotransferase; CI, confidence interval; RR, relative risk.
aTotal may be <16 (for cases) or <48 (for controls) due to missing data.
b0.5 added to all cells to avoid 0 error in relative risk calculation.
DISCUSSION
The advent of next-generation sequencing has accelerated virus discovery by allowing unbiased screening for novel viruses in various samples [17]. However, many of these new agents are “orphan viruses” of uncertain clinical significance. There is a need for controlled clinical studies to systematically demonstrate the clinical significance (or lack thereof) of these newly discovered agents.
One such orphan virus is HHpgV-1, the second human pegivirus, which was discovered in 2015 [2, 3]. Research to date shows that infection by this virus is closely related to concomitant HCV infection; both transient and persistent infections have been documented [10]. Given its predilection to infect intravenous drug users and hemophiliacs, HHpgV-1 is likely to be a blood-borne virus [6, 9]. In this study, the prevalence of HHpgV-1 viremia among HCV-infected persons in Hong Kong was 16/650 (2.4%), which is consistent with studies in similar populations in the United Kingdom and United States [3, 5]. We further analyzed these 16 cases, which constitute one of the largest cohorts of HHpgV-1 viremic individuals in the literature. Their demographic characteristics revealed novel potential routes of acquisition such as receiving a tattoo or having sex with men, both recognized risk factors for blood-borne viruses [18, 19].
Phylogenetic analysis revealed 2 distinct genogroups with nucleotide identity of 90.3%–94.7% between the 2 groups. Most local isolates clustered in genogroup II along with isolates from Chinese patients in previous studies. One of the HHpgV-1 isolates from our study (18M8130930) clustered in genogroup 1 along with isolates from Iranian and American hemophilia patients. The patient from whom this sequence was derived also suffered from hemophilia, suggesting transmission through commercially available plasma-derived clotting factor concentrates before pathogen reduction therapy was routinely implemented.
The clinical significance of HHpgV-1 infection is far from certain. Studies to date have only involved small numbers of cases and have only analyzed a few parameters of liver injury [9, 10]. These analyses indicated a neutral effect of HHpgV-1 viremia on ALT levels, HCV viral load, and liver stiffness. The 16 cases identified in our study were compared with a matched control group (HCV-infected individuals without HHpgV-1 viremia) in a 1:3 ratio to evaluate effects of active HHpgV-1 infection on HCV viral load and liver damage. The most significant finding was the impact of HHpgV-1 status on HCV viral load. On average, cases were found to have 0.5 log10-IU/mL higher HCV viral load than controls. Although HCV viral load variability is notoriously difficult to model [20, 21], our study showed that HHpgV-1 infection status and viral load were one of few potentially significant explanatory variables. Concomitant infection with multiple hepatotropic viruses can result in intricate interviral interactions, which have been best described for HBV–HCV co-infection [22]. We speculate that certain HHpgV-1 nonstructural proteins may boost HCV production in co-infected hepatocytes. A reciprocal positive effect of HCV viral load on HHpgV-1 infection also cannot be excluded. Cell culture models for HHpgV-1 and HCV-HHpgV-1 co-infection are urgently required to elucidate this possibility.
The impact of HCV viral load on disease progression and treatment response is controversial. Some studies have associated a higher HCV viral load with hepatitis, fibrosis, and hepatocellular carcinoma [23–25]. HCV viral load is a predictor of sustained virologic response to PEGylated interferon and ribavirin [26]. In the DAA era, pretreatment HCV viral load is less important for clinical decision-making. However, pretreatment viral load <106 IU/mL is a predictor of sustained virologic response when using 8-week regimens of sofosbuvir/ledipasvir [27]. The impact of HHpgV-1 infection on HCV viral loads found in our study is unlikely to have a major impact on HCV treatment planning, but the adverse effect of a higher HCV viral load on disease progression must be considered. We found that HHpgV-1-infected individuals tended to have higher relative risks of abnormal LFTs, imaging findings, and fibrosis scores. However, these analyses did not reach statistical significance. Future studies with larger numbers of HHpgV-1 viremic cases would be required to delineate this impact.
Our study had several limitations. Being a retrospective study, it is not possible to state definitively whether the HHpgV-1 infections were persistent or transient. As we did not perform HHpgV-1 serologic testing, we cannot exclude that some of the controls had also been infected with HHpgV-1 in the past and had spontaneously cleared the infection. However, we believe this would not significantly impact the results of our study, as we were interested in the effect of active HHpgV-1 viremia on HCV infection. Another limitation was that we only examined HCV viral loads and liver function at a single time point, so survival analysis was not possible. Furthermore, none of the cases had undergone liver biopsy, which is the gold standard for detecting necroinflammation and fibrosis. Manual matching of cases and controls could have led to bias, although we tried to minimize this by blinding the investigator to clinical details of cases and controls during the matching process. Despite these limitations, we believe that the data presented in the study provide new impetus to research on HHpgV-1 infection.
In summary, HHpgV-1 infection is present in 2.4% of HCV-infected individuals and may be acquired via a variety of high-risk behaviors including blood-borne and sexual exposures. Although HHpgV-1 infection does not appear to impact liver function, it may be associated with higher HCV viral loads in co-infected persons.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. This study was partly supported by the donation of Michael Seak-Kan Tong and funding by the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for Department of Health of the Hong Kong Special Administrative Region; the Sanming Project of Medicine in Shenzhen; the High Level Hospital-Summit Program in Guangdong, The University of Hong Kong-Shenzhen Hospital; and the Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, the Ministry of Education of China. The sponsors had no role in the design and conduct of the study, in the collection, analysis and interpretation of data, or in the preparation, review or approval of the manuscript.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. S.S. conceived the study, performed statistical analysis, and drafted the manuscript; C.C.Y.Y. designed primers and probes and performed phylogenetic analysis on sequences; N.F.S.C., S.W., and K.H.L. performed the PCR and sequencing experiments; J.F.W.C. and V.C.C.C. critically reviewed the manuscript; K.Y.Y. supervised the study.
References
- 1. Smith DB, Becher P, Bukh J, et al. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol 2016; 97:2894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kapoor A, Kumar A, Simmonds P, et al. Virome analysis of transfusion recipients reveals a novel human virus that shares genomic features with hepaciviruses and pegiviruses. MBio 2015; 6:e01466–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berg MG, Lee D, Coller K, et al. Discovery of a novel human pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog 2015; 11:e1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anh NT, Hong NTT, Nhu LNT, et al. ; Southeast Asia Infectious Disease Clinical Research Network Detection and characterization of human pegivirus 2, Vietnam. Emerg Infect Dis 2018; 24:2063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonsall D, Gregory WF, Ip CL, et al. Evaluation of viremia frequencies of a novel human pegivirus by using bioinformatic screening and PCR. Emerg Infect Dis 2016; 22:671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bijvand Y, Aghasadeghi MR, Sakhaee F, et al. First detection of human hepegivirus-1 (HHpgV-1) in Iranian patients with hemophilia. Sci Rep 2018; 8:5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H, Wan Z, Sun Q, et al. Second human pegivirus in hepatitis C virus-infected and hepatitis C virus/HIV-1-co-infected persons who inject drugs, China. Emerg Infect Dis 2018; 24:908–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodgers MA, Holzmayer V, Vallari A, et al. Hepatitis C virus surveillance and identification of human pegivirus 2 in a large Cameroonian cohort. J Viral Hepat 2019; 26:30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kandathil AJ, Breitwieser FP, Sachithanandham J, et al. Presence of human hepegivirus-1 in a cohort of people who inject drugs. Ann Intern Med 2017; 167:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang H, Wan Z, Xu R, et al. A novel human pegivirus, HPgV-2 (HHpgV-1), is tightly associated with hepatitis C virus (HCV) infection and HCV/human immunodeficiency virus type 1 coinfection. Clin Infect Dis 2018; 66:29–35. [DOI] [PubMed] [Google Scholar]
- 11. Frankel M, Forberg K, Coller KE, et al. Development of a high-throughput multiplexed real time RT-PCR assay for detection of human pegivirus 1 and 2. J Virol Methods 2017; 241:34–40. [DOI] [PubMed] [Google Scholar]
- 12. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campanella JJ, Bitincka L, Smalley J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 2003; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelsey JL, Whitemore AS, Evans AS, et al. Methods in Observational Epidemiology. 2nd ed Oxord: Oxford University Press; 1996. [Google Scholar]
- 15. Sridhar S, Yip CCY, Chan JFW, et al. Impact of inter-genotypic recombination and probe cross-reactivity on the performance of the Abbott RealTime HCV Genotype II assay for hepatitis C genotyping. Diagn Microbiol Infect Dis 2018; 91:34–7. [DOI] [PubMed] [Google Scholar]
- 16. Hui YT, Wong GLH, Fung JYY, et al. Territory-wide population-based study of chronic hepatitis C infection and implications for hepatitis elimination in Hong Kong. Liver Int 2018; 38:1911–9. [DOI] [PubMed] [Google Scholar]
- 17. Sridhar S, To KK, Chan JF, et al. A systematic approach to novel virus discovery in emerging infectious disease outbreaks. J Mol Diagn 2015; 17:230–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan DP, Sun HY, Wong HT, et al. Sexually acquired hepatitis C virus infection: a review. Int J Infect Dis 2016; 49:47–58. [DOI] [PubMed] [Google Scholar]
- 19. Crowley D, Lambert JS, Betts-Symonds G, et al. The seroprevalence of untreated chronic hepatitis C virus (HCV) infection and associated risk factors in male Irish prisoners: a cross-sectional study, 2017. Euro Surveill 2019; 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomas DL, Astemborski J, Vlahov D, et al. Determinants of the quantity of hepatitis C virus RNA. J Infect Dis 2000; 181:844–51. [DOI] [PubMed] [Google Scholar]
- 21. Ticehurst JR, Hamzeh FM, Thomas DL. Factors affecting serum concentrations of hepatitis C virus (HCV) RNA in HCV genotype 1-infected patients with chronic hepatitis. J Clin Microbiol 2007; 45:2426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Konstantinou D, Deutsch M. The spectrum of HBV/HCV coinfection: epidemiology, clinical characteristics, viral interactions and management. Ann Gastroenterol 2015; 28:221–8. [PMC free article] [PubMed] [Google Scholar]
- 23. Heller T, Seeff LB. Viral load as a predictor of progression of chronic hepatitis C? Hepatology 2005; 42:1261–3. [DOI] [PubMed] [Google Scholar]
- 24. Noh R, Lee DH, Kwon BW, et al. Clinical impact of viral load on the development of hepatocellular carcinoma and liver-related mortality in patients with hepatitis C virus infection. Gastroenterol Res Pract 2016; 2016:7476231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hisada M, Chatterjee N, Kalaylioglu Z, et al. Hepatitis C virus load and survival among injection drug users in the United States. Hepatology 2005; 42:1446–52. [DOI] [PubMed] [Google Scholar]
- 26. Bruno G, Fasano M, Saracino A, et al. Real life experience in treatment of HIV-1/HCV-coinfected patients with pegylated interferon alpha and ribavirin: predictors of SVR. New Microbiol 2015; 38:21–7. [PubMed] [Google Scholar]
- 27. Kowdley KV, Gordon SC, Reddy KR, et al. ; ION-3 Investigators Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med 2014; 370:1879–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.