Abstract
eIF3 is a large multiprotein complex serving as an essential scaffold promoting binding of other eIFs to the 40S subunit, where it coordinates their actions during translation initiation. Perhaps due to a high degree of flexibility of multiple eIF3 subunits, a high-resolution structure of free eIF3 from any organism has never been solved. Employing genetics and biochemistry, we previously built a 2D interaction map of all five yeast eIF3 subunits. Here we further improved the previously reported in vitro reconstitution protocol of yeast eIF3, which we cross-linked and trypsin-digested to determine its overall shape in 3D by advanced mass-spectrometry. The obtained cross-links support our 2D subunit interaction map and reveal that eIF3 is tightly packed with its WD40 and RRM domains exposed. This contrasts with reported cryo-EM structures depicting eIF3 as a molecular embracer of the 40S subunit. Since the binding of eIF1 and eIF5 further fortified the compact architecture of eIF3, we suggest that its initial contact with the 40S solvent-exposed side makes eIF3 to open up and wrap around the 40S head with its extended arms. In addition, we mapped the position of eIF5 to the region below the P- and E-sites of the 40S subunit.
INTRODUCTION
Gene expression is regulated at multiple levels, including the translation of mRNAs into proteins. It can be divided into four phases: initiation, elongation, termination and ribosome recycling. In eukaryotes, translation initiation starts with the assembly of the 43S preinitiation complex (PIC), which is composed of the 40S ribosomal subunit, eukaryotic initiation factors eIF1, eIF1A, eIF3 and eIF5, and the eIF2–GTP–Met-tRNAiMet ternary complex (TC). The eIF4F cap-binding complex then brings mRNA to the 43S PIC to form the 48S PIC. Subsequently, the 48S PIC with the anticodon of Met-tRNAiMet in the ribosomal P-site scans the mRNA leader sequence for the proper initiation site, which is in majority of cases the AUG triplet. Upon AUG recognition, accompanied by an irreversible hydrolysis of the GTP molecule on eIF2, series of intricate events and conformational changes in the 48S PIC result in a complete accommodation of Met-tRNAiMet of the ribosomal P-site, closure of the 40S mRNA binding channel and ejection of most of eIFs from the scanning-arrested 48S PIC. eIF5B bound by GTP then mediates the 60S subunit joining step to form, upon its departure triggered by GTP hydrolysis, the 80S initiation complex poised for elongation (reviewed in (1–5)).
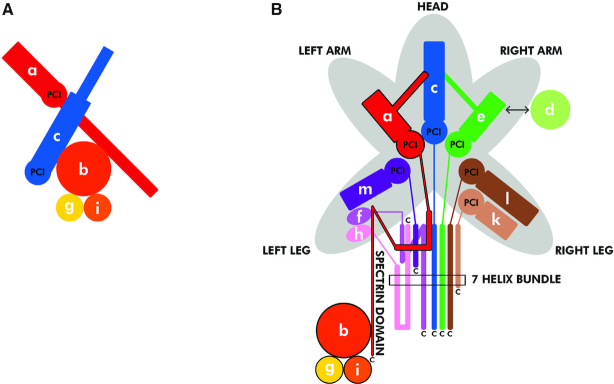
eIF3 is a multiprotein complex comprising 5 core subunits in the budding yeast (a/Tif32, b/Prt1, c/Nip1, i/Tif34, g/Tif35; Figure 1A and Supplementary Figure S1), whereas 12 core subunits constitute eIF3 in mammals (a, b, c, d, e, f, g, h, i, k, l, m; Figure 1B) (reviewed in (3,4)). eIF3 is one of the key players in the entire translation pathway. By definition it serves as an ‘orchestra conductor’ promoting or fine tuning virtually all initiation steps (3). In addition to that, eIF3 was also shown to: (i) stay present on early elongating ribosomes (6,7) in order to stimulate reinitiation on downstream cistrons (8–13), (ii) control translation termination in vivo (14), (iii) promote ribosomal recycling in vitro (15,16) and last but not least (iv) critically promote programmed stop codon readthrough (17,18). Owing to the multitude of roles that eIF3 has been implicated in, it is not surprising that deregulated eIF3 expression is associated with different pathological conditions, including neurodegenerative disorders and cancer (reviewed in (4,19–22)).
Figure 1.
Schematic models depicting structural organization of the yeast and mammalian eIF3 complexes. (A and B) In Saccharomyces cerevisiae, the eIF3 complex is formed by 5 subunits (A), whereas in mammals it consists of 12 subunits (B). These schematics illustrate both similarities as well as differences between budding yeast and mammalian eIF3. One of the main structural domains shared by several eIF3 subunits—the PCI (for Proteasome, COP9, Initiation factor 3) domain—is shown in bold in both panels; adopted from (71).
As can be seen, each eIF3 subunit contains at least one well-defined domain implicated in mediating either protein–protein or protein–RNA interactions (Figure 2A and B). In particular, both a/Tif32 and c/Nip1 contain the proteasome-COP9-eIF3 (PCI) domain, which is characteristic for proteins occurring in multimeric protein complexes. By mediating PCI-to-PCI protein–protein interactions, these domains were shown to directly build and stabilize complexes like proteasome and COP9 signalosome, as well as the 12-subunit mammalian eIF3 complex (23). A substantial inner part of b/Prt1 and basically the whole i/Tif34 are formed by the 9- and 7-bladed WD40 beta-propellers, respectively (24–26). In analogy to the PCI domain, this fold also serves as a mediator of complex protein–protein interactions. At last, the extreme N-terminus of b/Prt1 and the C-terminal half of g/Tif35 contain the RNA recognition motif (RRM), typically mediating protein-RNA interactions, rarely, like in case of b/Prt1, mediating also protein–protein interactions (27). Interestingly, both the c/Nip1-PCI and a/TIF32-PCI domains are on the other hand unexpectedly capable of making protein-RNA interactions (28,29). All these domains are not only crucial for the eIF3 integrity, as described below, they also mediate important eIF3 interactions with the small ribosomal subunit and protein factors associated with individual phases of translation (reviewed in (4)).
Figure 2.
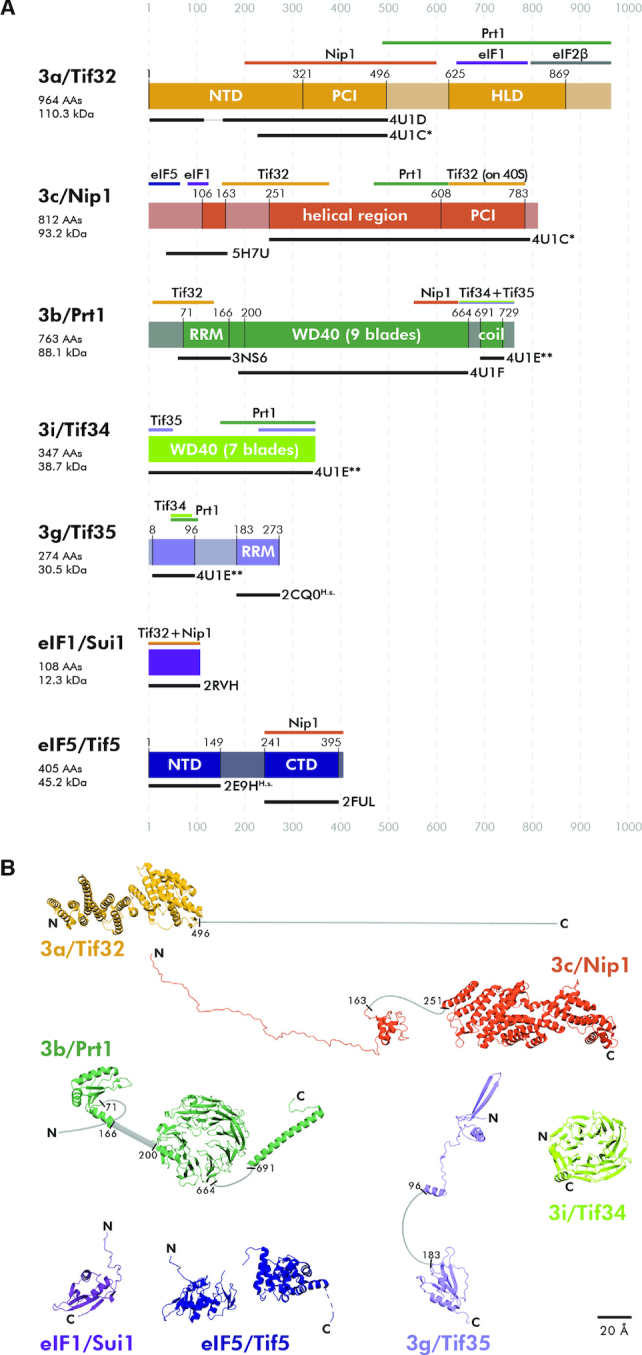
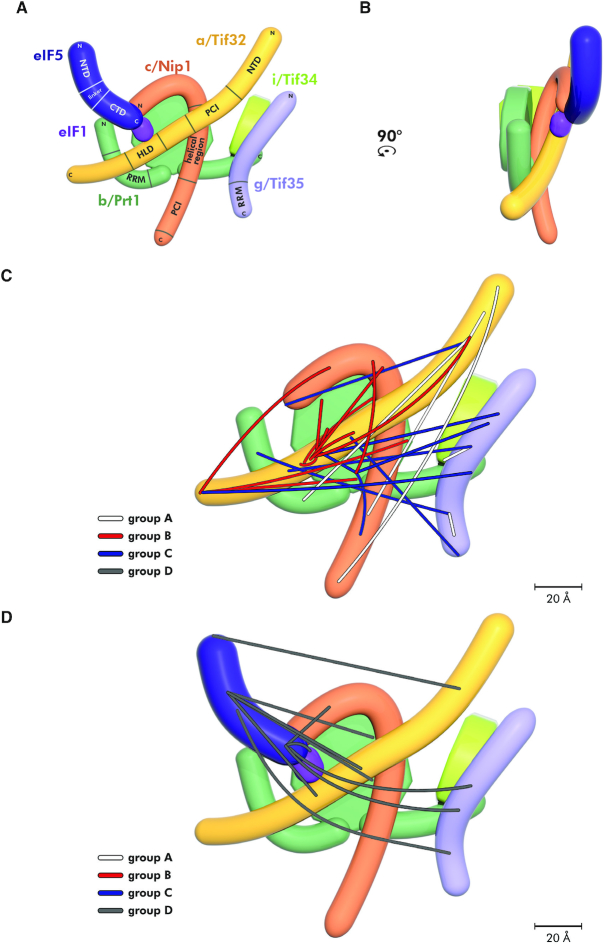
The overview of specific domains, available structures, and interacting partners of eIF1, eIF5 and individual eIF3 subunits. (A) Schematic representations of all eIF3 subunits, eIF1 and eIF5 with indicated structural motifs set in the context of the full length proteins. Color bars above each protein delineate binding regions of its interacting partner(s). Black bars under each protein indicate its parts with solved and published 3D structures including the respective PDB accession numbers. Lighter shades represent regions with either unknown structure or predicted to be unstructured. Please note that PDB code: 4U1C* contains a heterodimer of a/Tif32 and c/Nip1, whereas PDB code: 4U1E** contains a trimeric complex of b/Prt1-g/Tif35-i/Tif34. Structures 2CQ0 and 2E9H are human variants of the corresponding yeast proteins. PCI: Proteasome, COP9, Initiation factor 3 domain; HLD: HCR1-like domain; RRM: RNA recognition motif; WD40: beta propeller domain; NTD: N-terminal domain; CTD: C-terminal domain. (B) Summary of all known 3D structures of the eIF3 subunits/domains set in the context of the full length proteins. Unknown or unstructured parts are depicted in light gray.
The Saccharomyces cerevisiae eIF3 complex is formed by a triangle of three large mutually interconnected subunits (Supplementary Figure S1). The a/Tif32 and b/Prt1 subunits interact with each other via the C-terminal HCR1-like domain (HLD) of a/Tif32 and the N-terminal RRM of b/Prt1 (27,30,31), whereas the c/Nip1 subunit is anchored to this trimeric core via its central helical region making separate contacts with the PCI domain of a/Tif32 (c/Nip1 residues 157–370; (30,31)), and the C-terminal end of the WD40 domain of b/Prt1 (c/Nip1 residues 371–570; (27,30)). Based on the crystal structure of the isolated segments of a/Tif32 (residues 225–500) and c/Nip1 (residues 250–800) it may seem that these two proteins interact with each other via their PCI domains (26), as could be expected. However, our data presented below indicate that there are two modes of the a/TIF32–c/Nip1 interaction—one which does not involve the PCI domain of c/Nip1 occurring in the ribosome-free eIF3 complex as clearly demonstrated before (30), and the other (PCI-to-PCI) being adopted upon eIF3 binding to the PIC. At last, the extended C-terminal alpha-helix of b/Prt1 recruits the WD40 subunit i/Tif34 and g/Tif35 via its N-terminal domain containing a Zn-finger motif (25,26,32,33).
Despite the known subunit-interaction map and high resolution structures of several eIF3 domains (Figure 2B) (reviewed in (4)), the structure of the whole eIF3 complex free in solution from any organism has remained elusive. Semi-successful achievements were made by analyzing the overall structural arrangement of both yeast and mammalian eIF3 bound to the 40S subunit in various PICs, where several well-resolved but otherwise discontinued densities attributed to various eIF3 modules were described. Namely, reconstruction and intensive integrative modeling of the yeast 40S–eIF1–eIF1A–eIF3 complex stabilized by cross-linking revealed that the a/Tif32-c/Nip1 PCI heterodimer sits stably near the mRNA exit channel (26,34). In fact, functional placement of the a/Tif32 N-terminal half (including its PCI domain) near the mRNA exit channel was predicted by a number of earlier genetic and biochemical studies (10,11,35–38). The b/Prt1-i/Tif34 subcomplex, represented by the RRM and both beta-propellers, was conversely shown to be in contact with the mRNA entry channel. In accord with the earlier work identifying contacts between yeast g/TIF35 and RPS3/uS3 and RPS20/uS10 (33), the unresolved g/Tif35-NTD was predicted to lie between the i/Tif34 beta-propeller and the 40S body (26,34), while the proper position of the g/Tif35-RRM has remained unknown. The a-c/PCI and b-g-i/WD40-RRM modules are flexibly connected by the extended C-terminal half of a/Tif32 stretching over ribosomal proteins RPS2/uS5 and RPS3/uS3, and helices 16–18 of 18S rRNA (35,39), most of which, however, has not been resolved yet. In any case, the b-g-i module seems well positioned to interact with incoming mRNA by modulating the mRNA entry channel (26) (even in mammals (40,41)), and thus to control the rate and processivity of scanning for AUG recognition as demonstrated before (25,33,38,39,42–44), while the a-c module extends the mRNA exit channel and seems to stabilize mRNA in the PIC (29,37,38). Interestingly, based on the most recent yeast cryo-EM structures (45,46), the eIF3b–g–i module seems to be rather mobile. The current view predicts that upon mRNA recruitment to and its stabilization at the 48S PICs, this module transposes itself—most probably thanks to the C-terminal domain of eIF3a that seems to operate as a controllable mechanical arm—to the interface side, with the WD40 domain of eIF3b residing in the vicinity of RPS23/uS12, h44 and the eIF2γ subunit, and the eIF3b-RRM motif directly contacting eIF1 (4,45,47). This eIF3b rearrangement with eIF3i-g subunits still attached to it then supposedly enables the onset of scanning.
One of the most important contacts that eIF3 makes with other eIFs is that with eIF5 (48–51). eIF5 has three major roles: (i) during the scanning process it stimulates hydrolysis of GTP in TC acting as an eIF2-specific GTPase activating protein (GAP); (ii) it also promotes stringent AUG selection (2); and (iii) it additionally regulates GDP to GTP exchange on eIF2 by competing with eIF2B (eIF2-specific Guanine nucleotide exchange factor) for binding to eIF2-GDP (52). eIF5 consists of an N-terminal domain (NTD, residues 1–149 containing the GAP function) (53) and a C-terminal domain (CTD, residues 241–395) (54), connected by a long flexible linker (residues 150–240). They both perform critical functions the molecular details of which are unclear due to a poorly described location of eIF5 within the PIC. For example, it was proposed that movement of the eIF5–NTD and the eIF1A C-terminal tail toward one another within the PIC upon AUG recognition represents one of the hallmarks capturing this intricate process (55). The eIF5–CTD was in turn shown to stimulate assembly of the PIC and control start codon selection by promoting the dissociation of eIF1 from the scanning-arrested PICs (55). In a partial 48S pre-initiation complex from yeast (py48S), the eIF5–CTD was tentatively assigned to a low-resolution density near eIF2γ (56); however, its true location remains to be unambiguously determined. While this manuscript was in preparation, the eIF5–NTD in the py48S–eIF5N complex was mapped near the P-site at essentially the same position where eIF1 binds in the open/POUT state (46), which is characterized by only partially accommodated Met-tRNAiMet. In this most recent structure, the Met-tRNAiMet with the eIF5–NTD replacing eIF1 seems to be conversely fully accommodated in the P-site, tilted toward the 40S body, apparently setting the stage for its interaction with eIF5B. Hence it was proposed that the eIF5–NTD stabilizes the codon–anticodon interaction and the closed/PIN state of the PIC, prevents eIF1 rebinding, and promotes a conformation of the 48S PIC compatible with eIF5B-mediated subunit joining.
An unprecedented complexity of the web of eIF3 interactions among its own subunits, as well as with other eIFs like eIF5 and the ribosome ensuring the multitude of its roles has pressingly called for: (i) resolving its structure as a free holocomplex or when in complex with eIF5 and another important binding partner eIF1; (ii) and for describing the conformational changes that eIF3 and its associated eIFs undergo prior to their binding to the 40S subunit. Here we attempted to achieve this long-standing goal by improving the original yeast eIF3 reconstitution protocol (31) and subjecting the in vitro assembled eIF3 in a free form or bound to the 40S or 80S ribosomes to (i) X-ray crystallography, (ii) cryo-electron microscopy (cryo-EM) and (iii) chemical cross-linking and advanced Mass spectrometry. Using the latter approach, as the only feasible, we mapped distances between the solvent-exposed parts of the free eIF3 complex and revealed that it has a rather globular 3D arrangement. Employing the same method we modeled the 3D structure of the eIF3–eIF1–eIF5 assembly and described a rather dramatic rearrangement that eIF3 must undergo when contacting the small ribosomal subunit. Finally, this approach also allowed us to map the binding site of both N- and C-terminal domains of eIF5 in the PIC.
MATERIALS AND METHODS
Chemicals
Cross-linkers disuccinimidyl glutarate (DSG), disuccinimidyl suberate (DSS), bis(sulfosuccinimidyl) glutarate (BS2G), bis(sulfosucdinimidyl) suberate (BS3) and their four times (d4) deuterated analogs were purchased from Proteochem (USA). Sequencing grade modified trypsin was obtained from Promega (USA). MS2 cleavable cross-linker disuccinimidyl dibutyric urea (DSBU), dimethyl sulfoxide (DMSO), water and HPLC solvents were LC/MS grade purity and were obtained from Thermo Scientific (USA). Other chemicals at the highest available purity were purchased from Sigma-Aldrich (USA).
Plasmids and bacterial strains
The lists and descriptions of plasmids and bacterial strains used throughout this study can be found in Table 1.
Table 1.
Plasmids used in this study and relevant bacterial strains used for protein expression
| Plasmid | Description | Bacterial strain used for protein expression | Source of reference |
|---|---|---|---|
| pGEX-6P-1-GST-Nip1 | GST-Nip1 fusion inserted into pGEX-6P-1 | BL21 (DE3) | (57) |
| pET22b-Prt1-His | Prt1 inserted into pET22b, generating a C-terminally His6 Tag. | Rosetta 2 (DE3) | this study |
| pET28b-Tif32-His | Tif32 inserted into pET28b, generating a C-terminally His6 Tag. | Rosetta 2 (DE3) | this study |
| pET22b-Tif34-Tif35-His | Tif34-Tif35 fusion protein inserted into pET22b, generating a C-terminally His6 Tag. An HRV 3C protease cleavage site is inserted between Tif34 and Tif35. | Rosetta 2 (DE3) | this study |
| pTYB2-eIF1 | fusion of the C-terminus of eIF1 to the N-terminus of the intein/chitin binding domain inserted into pTYB2 | Rosetta 2 (DE3) | (57) |
| pTYB2-eIF5 | fusion of the C-terminus of the eIF5 to the N-terminus of the intein/chitin binding domain inserted into pTYB2 | Rosetta 2 (DE3) | (57) |
Modifications to the protocol for the recombinant eIF3 preparation
Yeast eIF3 subunits were expressed in Escherichia coli (Table 1) and purified using the modified protocol to that employed before (31) as outlined in Results. In addition, we replaced 2 mM dithiothreitol (DTT) in the K-200 buffer used for the eIF3 reconstitution with 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP); the new K-200 buffer (200 mM KCl, 20 mM HEPES/KOH (pH 7.5), 10% glycerol, 2 mM TCEP).
Purification of eIF1, eIF5 and the 40S ribosomal subunit
eIF1 and eIF5 were purified as described elsewhere (Acker et al., (57)) except that the Enzyme storage buffer contained 2 mM TCEP instead of 2 mM DTT; the new Enzyme storage buffer (20 mM HEPES/KOH (pH 7.4), 1 M KOAc, 10% glycerol, 2 mM TCEP).
The 40S ribosomal subunits were purified as described before (57).
Assembly of the protein complexes of interest
The eIF3–eIF1–eIF5 complex (11.5 μM) was assembled by mixing the reconstituted eIF3 complex with the purified proteins in a molar ratio of 1:1:1 in the Protein binding buffer (25 mM HEPES/KOH (pH 7.5), 15 mM MgCl2, 100 mM KCl and 2 mM TCEP).
The eIF3–40S complex (2 μM) was assembled by mixing the reconstituted eIF3 complex with the purified 40S subunits in a molar ratio of 1.2:1 in the Protein binding buffer.
The eIF5–40S complex (2 μM) was assembled by mixing eIF5 with the purified 40S subunits in a molar ratio of 1.2:1 in the Protein binding buffer.
Chemical cross-linking of protein complexes
The eIF3 complex (1.42 μM) was cross-linked using disuccinimidyl suberate (DSS) or disuccinimidyl glutarate (DSG) in 100 and 200 molar excess to complex.
The eIF3–eIF1–eIF5 complex (2.78 μM) was cross-linked using DSS, DSG bis(sulfosuccinimidyl)suberate (BS3) or bis(sulfosuccinimidyl)glutarate (BS2G), respectively. DSG and DSS were dissolved in DMSO and used in 100 and 200 molar excess to complex whereas BS2G a BS3 were dissolved in protein binding buffer and used in 500, 1000 and 2000 molar excess.
The eIF3–40S and eIF5–40S complexes (2 μM) were cross-linked using BS3 or disuccinimidyl dibutyric urea (DSBU) in 2000 molar excess to both complexes.
All cross-linking agents excluding DSBU were used as equimolar mixture of non-deuterated and four–times deuterated form (DSG d0/d4, DSSd0/d4, BS2G d0/d4 and BS3d0/d4) in all above described cross-linking experiments. Reaction mixtures were incubated for 30 min at room temperature, quenched by ethanolamine to a final concentration of 1mM for eIF3 complex or 5 mM for eIF3–eIF1–eIF5 complex, eIF3–40S and eIF5–40S complexes and analyzed in gel and in solution.
Analysis of cross-linked eIF3–40S complexes using sucrose gradient centrifugation and dot blot
To determine the composition of the cross-linked eIF3–40S complexes, 23 μg of the sample after cross-linking was analyzed using 7.5-30% sucrose gradient centrifugation as described in (58). Altogether 23 fractions were collected in 33 s intervals (corresponding to the volume of ∼500 μl). Fractions containing the 40S subunits (10 - 13) were subjected to dot blotting (10 μl aliquots of each fraction were loaded) using rabbit anti-Tif32 (1:1000) and anti-Tif35 (1:2000) primary antibodies. A control dot blot contained 10 μl of the binding buffer as a negative control, 4 μl of the purified 40S subunits [8 μM] and 2 μl of reconstituted eIF3 complex [19 microM].
Protein electrophoresis to purify cross-linked complexes to be analyzed by mass spectrometry
The cross-linking reaction mixture was mixed with the 6× concentrated Sample loading buffer (330 mM Tris, 60% glycerol, 416 mM SDS). Samples were incubated for 5 min at 90°C and then loaded onto the Criterion TGX 4–20% Tris-Glycine gel (133 × 87 × 1 mm, 12+2 wells, BIORAD). Separation was performed in the Tris-glycine running buffer at 200 V. After separation, the gels were stained by GelCode™ Blue Stain Reagent (Thermo Scientific) and destained with distilled water.
Enzymatic digestion
For in-gel digestion, the bands of interest were excised and destained. The disulfide bonds were reduced with 20 mM TCEP in 50 mM ethylmorpholine buffer (pH 8.5) for 10 min at 50°C and free cysteines were alkylated with 20 mM iodacetamide in 50 mM ethylmorpholine buffer (pH 8.5) for 20 min at room temperature in the dark. The gel pieces were covered with trypsin solution (trypsin in 100 mM ethylmorpholine buffer (pH 8.5) with 10% AcN, enzyme:protein ratio 1:20) and incubated at 37°C overnight. The digestion was stopped by adding trifluoracetic acid to 0.1% and resulting peptide mixture was dried by SpeedVac (Eppendorf, Germany).
For in-solution digestion, the cross-linking reaction mixture was diluted 1:1 (protein:buffer) with 100mM ethylmorpholine buffer (pH 8.5). The disulfide bonds were reduced with 20 mM TCEP in 50 mM ethylmorpholine buffer (pH 8.5) for 10 min at 50°C and free cysteines were alkylated with 20 mM iodacetamide in 50 mM ethylmorpholine buffer (pH 8.5) for 20 min at room temperature in the dark. Acetonitrile was added to the mixture to 10% before digestion. Trypsin in 100 mM ethylmorpholine buffer (pH 8.5) with 10% AcN was added to give a final concentration 1:20 (w/w) trypsin:protein and digestion was carried out at 37°C overnight. The digestion was stopped by adding trifluoracetic acid to 0.1% and resulting peptide mixture was dried by SpeedVac (Eppendorf, Germany).
Mass spectrometry analysis
For the eIF3 and eIF3–eIF1–eIF5 complexes cross-linking experiments, dried peptides were resuspended in 20 μl of solvent A (0.1% formic acid in 2% AcN) for further processing. A 1 μl of each peptide mixture was injected onto a reversed-phase trap column (Zorbax 300SB-C18 5 μm, 0.3 × 5 mm, Agilent Technologies, USA). Eluting peptides were subsequently separated on a 60°C-heated reversed-phase analytical column (Zorbax 300SB-C18 3.5 μm, 0.3 × 15 mm, Agilent Technologies, USA) using Agilent 1200 series System (Agilent Technologies, USA) at a flow rate of 10 μl/min under the following gradient conditions: 5–40% solvent B (0.1% formic acid in 98% AcN) in 35 min, 40–95% B in 3 min, 3 min in 95% B, 95–2% B in 1 min and 10 min in 2% B. The HPLC system was coupled to ESI-Spray which directly introduces ions into a solariX XR FT-ICR mass spectrometer (Bruker Daltonics, Germany) equipped with 15 T superconducting magnet. The instrument was calibrated on-line by using The ESI-TOF tuning mix (Agilent Technologies, USA). Mass spectral data was acquired in positive broadband mode over the m/z range 250–2500, with 1M data points transient, 0.2 s ion accumulation and four scans were accumulated per spectrum. Data acquisition was performed using solariXControl FTMS control application software.
For the eIF3–40S and eIF5–40S complexes, samples were separated identically as eIF3 complex. However, peptide mixtures were analyzed by MS/MS analysis in data independent mode with broad isolation window (1000 m/z). Mass spectral data was acquired in positive broadband mode over the m/z range 250–2500, with 1M data points transient, 15 eV collision voltage, 0.2 s ion accumulation, 0.8 s MS/MS accumulation and two scans were accumulated per spectrum.
Cross-links identification
For identification of cross-links from LC-MS analysis, we used in house developed software (LinX) based on the Links algorithm, previously described as Automated Spectrum Assignment Program (ASAP) (59,60). Because it requires input in the format of tab separated m/z and intensity values, it was necessary to program a data output script using the internal scripting language (Visual Basic) of the DataAnalysis 4.4 software suite (Bruker Daltonics, Germany). This script uses the DataAnalysis implementation of the SNAP 2.0 algorithm to generate deconvoluted spectra and then exports a file containing the sorted monoisotopic masses and their corresponding intensities (and other related data). LinX software identifies cross-linked peptides by matching experimental data to a theoretical library generated based on the protein sequence, protease specificity, cross-linker reactivity and composition and protein chemical modification. The parameters for the search were defined as follows: enzyme–trypsin (specificity—cleavage after lysine and arginine, not cleaved after modification, three missed cleavages); variable modification—oxidation on methionine, fixed modification—carbamidation on cysteines, cross-linker—Type 0,1 and 2 products for each cross-linker (specificity to N-terminus, lysine and tyrosine); mass error—2 ppm.
Cross-links analyzed by LCMS/MS were identified using StavroX (61) or MeroX (62) (DSBU) with similar parameters as for LinX and mass error for MS/MS fragments—2 ppm and FDR < 5%.
All cross-links identified by the Links, StavroX and MeroX were manually confirmed by examination of the raw data.
3D modeling
The 3D models were made using 3Ds Max 2019 (Autodesk) and PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.)
RESULTS AND DISCUSSION
Optimization of the yeast eIF3 purification and reconstitution protocol
Since native eIF3 purified directly from yeast cells in large scale often contains other ‘contaminating’ proteins, predominantly the three major eIF3 interacting partners, Hcr1, eIF1 and eIF5 (27,48,63,64), the in vitro E. coli expression/reconstitution protocol was developed by the Ficner's lab for the entire eIF3 complex (31). To obtain higher yields of proteins without truncated variants for our downstream structural applications, we set out to optimize this protocol as follows.
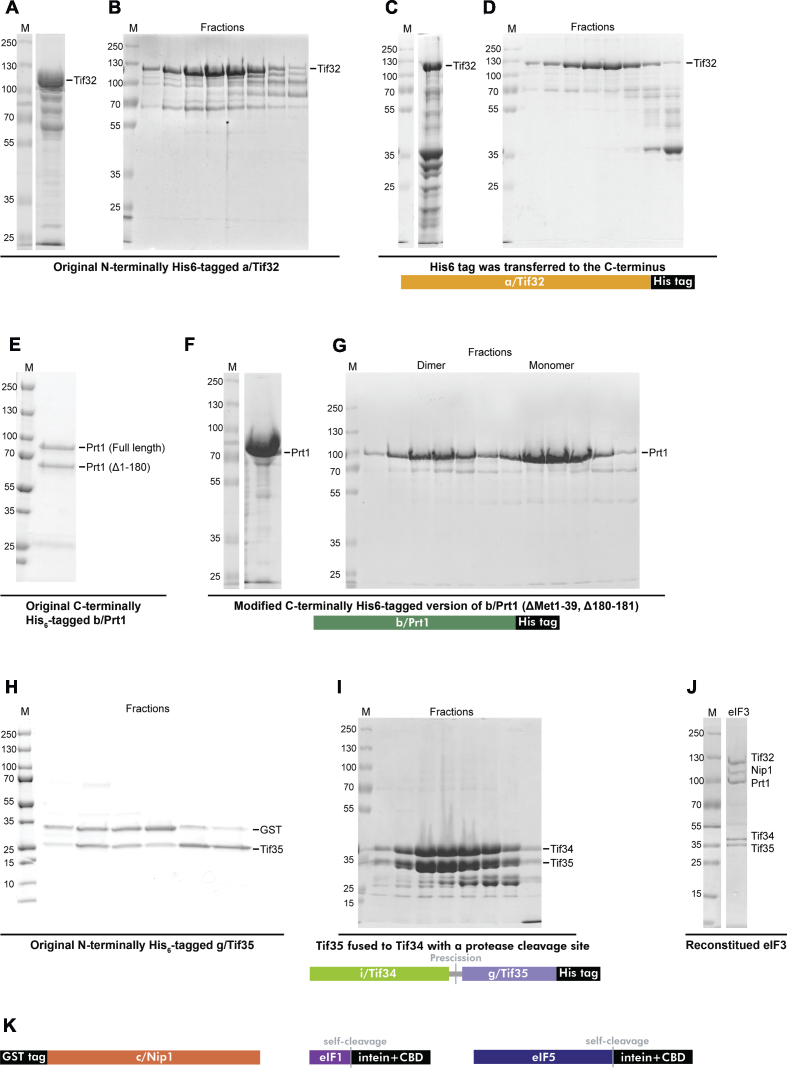
In the original plasmid, TIF32 was N-terminally His-tagged (31), and its expression led to a presence of the C-terminally truncated protein variants (Figure 3A). Therefore, it was difficult to separate the full-length Tif32 from the truncated derivatives because they also carried an N-terminal His6 tag and the size of the truncated variants was similar to that of the full-length protein (Figure 3B). To circumvent this, the His6 tag was relocated to the C-terminus by subcloning the TIF32 gene into the pET28b vector (Figure 3C and D). The resulting construct was expressed in E. coli and the Tif32-His6 protein was purified by the two-step column chromatography. After the Ni charged Immobilized metal affinity chromatography (IMAC), the full-length and N-terminally truncated Tif32 proteins were eluted (Figure 3C) and subsequently subjected to size-exclusion chromatography (SEC), where it was finally possible to separate the full-length Tif32-His6 from its much smaller N-terminally truncated variants (Figure 3D). The typical yield was ∼1.5 mg per liter of culture.
Figure 3.
The refined eIF3 reconstitution protocol. (A and B) The CBB stained SDS-PAGE gels showing the original N-terminally His6-tagged a/Tif32 eluted from the IMAC column (A), which was subsequently subjected to gel filtration chromatography—only the peak fractions are displayed (B). (C and D) Same as in (A and B) only the reworked C-terminally His6-tagged a/Tif32-His6 is shown including its schematic. (E) The original C-terminally His6-tagged b/Prt1 purified via IMAC is shown; the full-length and truncated (Δ1-180) b/Prt1 variants were detected. (F and G) The modified C-terminally His6-tagged version of b/Prt1 (Δ1-39,180-181) eluted via IMAC (F) and further subjected to gel filtration chromatography (only the peak fractions are displayed) is shown including its schematic (G). Please note that two peaks corresponding to the dimeric and monomeric b/Prt1 proteins were observed (the Saccharomyces cerevisiae b/Prt1 reportedly forms a homodimer when it is concentrated (26)). (H) The CBB stained SDS-PAGE gels showing the original g/Tif35 subunit expressed as a fusion protein with GST that was subjected to gel filtration chromatography; peak fractions were detected after the glutathione column purification and HRV 3C Protease cleavage. (I) The i/Tif34 and g/Tif35 subunits expressed as the C-terminally His6-tagged fusion i/Tif34-g/Tif35 protein (see the schematics below) were first purified via IMAC, followed by the HRV 3C Protease cleavage (the protease was eliminated using the glutathione column) and the resulting eluate was then subjected to gel filtration chromatography; the peak fractions contained both proteins. (J) An example of the eIF3 reconstitution according to the modified protocol. (K) Schematics of c/NIP1, eIF1 and eIF5 fusion proteins; all purified according to original protocols (31,57).
The PRT1 gene was in the original protocol subcloned into the pET22b vector with a C-terminal His6 tag and the expressed protein was purified by column chromatography starting with IMAC followed by SEC. It was previously shown that expression of S. cerevisiae Prt1 does not start from the first Met occurring in its 5′ UTR (Met1 according to (31)) but from Met40 (27); in accord, the putative amino acid sequence preceding Met40 is not conserved with any other organism. Since the original eIF3 reconstitution protocol developed by (31) did not reflect this fact, we first re-subcloned the PRT1 gene into the pET22b vector to begin with ‘Met40’ (henceforth Met1*). In addition, with the original purification procedure, about 50% of the typical yield contained N-terminally truncated (Δ1-180) Prt1 (31) (Figure 3E). To avoid this problem, a deletion mutant lacking Met140* and Pro141* (Met180 and Pro181 according to (31)) was generated to remove this protease cleavage site. Neither Met140* and Pro141* are conserved (Figure 3F and G). Therefore we think we have a good reason to believe that the Prt1ΔM140,P141* derivative is fully functional; it certainly does not affect the eIF3 complex assembly (Figure 3J). Applying the same purification protocol as used by (31), we greatly diminished the appearance of the originally observed truncation (Figure 3F and G). The yield was ∼15 mg per liter of culture.
The NIP1 gene was originally subcloned into the pGEX-6P-1 vector with an N-terminal GST tag (31). The protein was first purified using a glutathione column, followed by an HRV 3C Protease cleavage and SEC. The yield was ∼1 mg per liter of culture, but ∼50% of the purified Nip1 protein unexpectedly did not assemble into the eIF3 complex, which significantly reduced the overall reconstitution yield. Plasmids with an N-terminal His6 and MBP tags, respectively, were also tested, but neither the final purification yields nor the Nip1 assembly into the rest of the complex improved (data not shown).
The TIF35 gene was originally cloned into the pGEX6P-1 vector with an N-terminal GST tag (31). As in case of Nip1, the Tif35 protein was purified by a glutathione column, followed by an HRV 3C Protease cleavage and SEC. However, Tif35 non-specifically associates with the GST moiety even after the cleavage and thus cannot be easily separated by SEC (Figure 3H). This problem indicates that Tif35 might need a binding partner that could prevent its non-specific interaction with GST and increase its stability. Therefore, the TIF35 gene was fused with the gene encoding its native binding partner Tif34 by a short linker encoding an HRV 3C Protease cleavage site. This fusion TIF35-TIF34 DNA fragment was subcloned into pET22b with a C-terminal His6 tag (Figure 3I). The resulting fusion protein was first purified by a Ni column, followed by HRV 3C Protease cleavage and the protease was eliminated using the glutathione column. The resulting eluate was then subjected to SEC and as can be seen in Figure 3I, both Tif34 and Tif35 proteins eluted in the same peak further underscoring their stable and strong binding (Figure 3I). The apparent appearance of degradation products and/or other proteins in the corresponding Tif34-Tif35 elution peaks did not hamper the completion of this improved eIF3 expression protocol because these were completely eliminated by SEC that captures the entire protocol of the eIF3 reconstitution (Figure 3J, see also below). The total yield was ∼30 mg per liter of culture.
The recombinant eIF3 was reconstituted by mixing the purified Tif32, Prt1, Nip1, and Tif35 and Tif34 subunits in 1:1.5:1.5:2.25:2.25 molar ratio in a given order. The reconstitution mixture was incubated for 15 min at 4°C and subjected to one final SEC to yield highly pure, 5-subunit eIF3 in a perfect stoichiometry (Figure 3J).
Taken together, we succeeded in optimization of the existing protocol for expression and purification of eIF3 subunits. Problems with truncated variants of full-length proteins contaminating the purified samples were eliminated and, in addition, we can now obtain higher yields of individual subunits (except for c/Nip1). Thus this protocol should be more suitable for x-ray crystallography and cryo-EM sample preparation. Owing to the fact that the expression of soluble c/Nip1 is much lower than those of the other subunits, the quality and yield of the reconstituted eIF3 is still dominated by the c/Nip1 preparation. Nevertheless, introduced modifications of the a/Tif32, b/Prt1, g/Tif35 and i/Tif34 purification schemes significantly simplified their preparations.
Free yeast eIF3 complex adopts a globular 3D architecture
Numerous laboratories have attempted to solve the structure of yeast, as well as mammalian eIF3 complex when free in solution. With respect to yeast eIF3 only very low-resolution single particle electron microscopy (EM) images (2D class averages) of negatively stained free yeast eIF3 are available (31). Since our own attempts using the in vitro reconstituted eIF3 complexes also failed, we decided to apply chemical cross-linking coupled with advanced mass spectrometry on our highly pure complexes in perfect stoichiometry in effort to reveal the overall shape and geometry of free yeast eIF3. We employed two different cross-linkers containing an amine-reactive N-hydroxysuccinimide (NHS) ester at each end of either a 5-carbon (DSG or water-soluble variant BS2G; a minimal/maximal length of the Cα-Cα cross-link is 20.5/26.4 Å) or an 8-carbon (DSS or water-soluble variant BS3; a minimal/maximal length of the Cα-Cα cross-link is 24/30.2 Å) spacer arm, respectively (65,66). According to previously published observations (67), the eIF3 complex was cross-linked using either 100- or 200-fold molar excess of the cross-linker over the protein complex to avoid perturbation of protein structure. This experiment was repeated twice with each cross-linker, hence altogether we obtained eight unique datasets. As a negative control, an untreated samples (buffer only) were always run in parallel and, as expected, no signals corresponding to cross-linked peptides were detected (data not shown). To minimize false positive assignments, only experimental values differing from theoretical ones less than 2 ppm for the both isotopes were considered and multiple assignments were discarded. Such strict criteria prohibit the misinterpretation of mass spectrometric data.
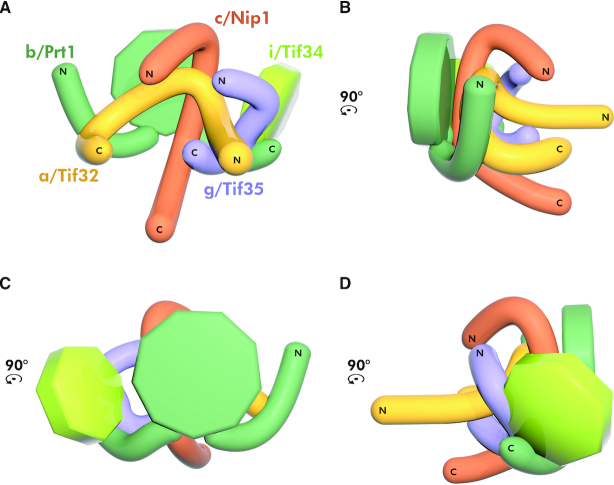
To be able to precisely and synoptically map all cross-links between surface-exposed lysine residues of individual eIF3 domains, we first refined our previously published 2D model of the ribosome-free eIF3 complex (30) (Supplementary Figure S1) using the Autodesk 3Ds MAX software (Figure 4A–D). Please note that this schematic 2D model: (i) reflects as accurately as possible all contact points among individual eIF3 subunits that have been mapped so far using various in vivo and in vitro protein binding assays and, at the same time, (ii) purposefully disregards all contacts that eIF3 subunits may establish among each other when bound to the ribosome (reviewed in (3,4); for further details please see below).
Figure 4.
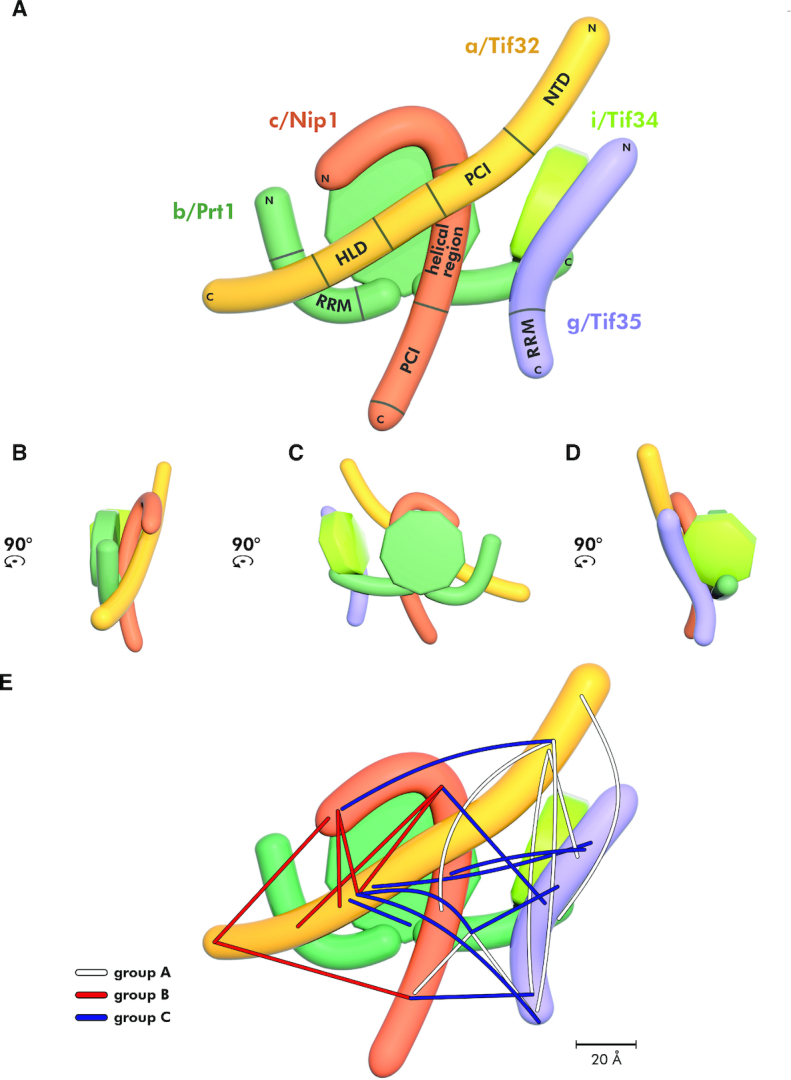
The refined 2D model of the budding yeast eIF3 complex with the indicated placement of the obtained cross-links. (A–D) Planar projections of the eIF3 complex with known contact points between its individual subunits/defined domains in all four views rotated by 90°CCW. (E) The summary of all cross-links obtained with the reconstituted eIF3 divided into three color-coded groups based on their positions and directions.
In total, we obtained 22 unique cross-links (Figure 4E). Two cross-links out of those 22 (between the c/Nip1 middle helical region and the g/Tif35-NTD, and the a/Tif32-HLD and the g/Tif35-RRM) were obtained in all eight datasets, eight were found in both DSG and DSS, eight or three only in DSG or DSS, respectively, and five in both replicates with at least one of the cross-linkers (for the overview and count of the occurrence of individual cross-links obtained in given experimental conditions specified above please see Supplementary Figure S2 and Table S1). Interestingly, three main directions/groups can be clearly distinguished with respect to the spatial arrangement of these cross-links (Figure 4E). The first group (A) runs roughly vertically and includes cross-links of the N-terminal domain of a/Tif32 with both halves of g/Tif35, the middle region of c/Nip1, and the extreme CTD of b/Prt1, as well as those of the b/Prt1-CTD with the c/Nip1-PCI domain and the tip of the g/Tif35-RRM (Figure 4E). The second group (B) emanates from the very C-terminus of a/Tif32 and contacts the very N-terminus of c/Nip1, as well as its C-terminal PCI domain; it also includes cross-links between the first N-terminal 1/3 of c/Nip1 and the a/Tif32-HLD. The third group (C) runs roughly horizontally across the entire complex from the a/Tif32-HLD toward both ends of g/Tif35 and the WD40 and CTD of b/Prt1, as well as from the first N-terminal 1/3 of c/Nip1 toward g/Tif35 and the a/Tif32-NTD; it also includes cross-links between the middle helical region of c/Nip1 and the g/Tif35-NTD, the c/Nip1-PCI and the g/Tif35-RRM and at last between the b/Prt1-CTD and g/Tif35 (Figure 4E). Taking into account the average lengths of the generated cross-links by the selected cross-linkers (see above) that we set as delimiting parameters in the aforementioned software, we accommodated the individual parts of the 2D model to fit into our cross-linking map and thus build the 3D model of eIF3 (Figure 5A–D).
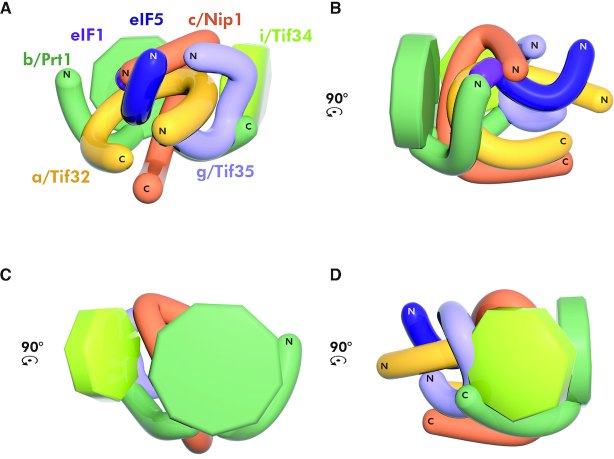
Figure 5.
The compact 3D architecture of the yeast eIF3 complex free in solution modeled based on the obtained cross-links. (A–D) The 3D architecture of yeast eIF3 free in solution modeled using restraints from all cross-links depicted in Figure 4 viewed from all four sides rotated by 90°CCW. Color-coding of individual eIF3 subunits corresponds to Figure 4; for details see the main text.
The three cross-linking directions described above clearly suggest that free eIF3 adopts a tightly packed conformation with both of its WD40 domains (b/Prt1 and i/Tif34), as well as the RRM domains (b/Prt1 and g/Tif35) exposed to the surface (Figure 5). The other exposed domains are the extreme ends of a/Tif32 (consistent with the fact that the extreme CTD binds eIF2 off the ribosome (30) and the NTD contacts uS2/Rps0A (35,36)), as well as those of c/Nip1 (the NTD associates with eIFs 1 and 5 (50,63) and the CTD contacts the small ribosomal protein RACK1/Asc1 (28)).
It is noteworthy that in neither of our models of the ribosome-free eIF3 (Figure 4A and Supplementary Figure S1) the PCI domains of a/Tif32 and c/Nip1 subunits interact with each other, which is in contrast to the PCI-to-PCI interaction resolved in the reported cryo-EM structures of PICs (34,45). The reason is that with the exception of the crystal structure obtained with the N-terminal 2/3 of c/Nip1 in complex with the second half of the a/Tif32 (Figure 2A; PDB code: 4U1C) (26), neither of in vitro or in vivo biochemical experiments carried out with either individual subunits or the entire eIF3 (wild-type or mutant) free of the ribosome confirmed the existence of the PCI-to-PCI interaction between these two proteins (27,30,31). In fact, we showed earlier that the c/Nip1 residues 157–370 are both necessary and sufficient for binding to a/Tif32 in vitro and in vivo (30,31). Hence, as discussed below, we propose that the establishment of the PCI-to-PCI interaction materializes only upon eIF3 binding to the 40S ribosome. In favor of our model, no specific cross-links between both PCI domains were seen, and the group A cross-link detected between the a/Tif32-NTD and the helical region of c/Nip1 could not be formed in the prospective PCI-to-PCI arrangement (26,45) simply because the distance between the two lysine residues involved in this cross-link exceeds the maximal length by over 2-fold (Supplementary Figure S4A). There are two other cross-links that further support this rationale (see the next chapter).
Binding of eIF1 and eIF5 further compacts the globular 3D geometry of eIF3
As mentioned above, yeast eIF3 has two prominent interacting partners—eIF1 and eIF5 (Figure 6A and B) and it was proposed that eIF3 actually promotes their association with the small ribosomal subunit during formation of the 43S PIC in vivo (reviewed in (3)). Whereas eIF5 interacts with the tip of the N-terminal domain of c/Nip1 (51), eIF1 contacts the region further downstream but still within the extreme N-terminus of c/Nip1 (51). In fact, eIF1 was proposed to possess more than one binding site within the c/Nip1-NTD (68) and, in addition, it was also shown to contact the a/Tif32-HLD (30). Taking into account the dramatic differences in the conformation of free versus 40S-bound eIF3, we were wondering whether perhaps eIF1 and eIF5 association with eIF3 serves as a trigger for its structural rearrangement prior to its binding to the 40S subunit. To test that, we purified the eIF1 and eIF5 proteins and formed a trimeric eIF3-1-5 complex in vitro that we subjected to the cross-linking protocol using selected cross-linkers (DSS, BS3, DSG and BS2G). We unambiguously identified 36 intermolecular cross-links (Supplementary Table S2). To simplify the cross-linking map, we split these 36 cross-links into two classes: class (i) features 25 cross-links that were detected among the eIF3 subunits and illustrates the conformational alterations that binding of eIFs 1 and 5 imposes on the eIF3 complex (compare Figure 6C versus 4E); class (ii) depicts nine cross-links between eIF5 and eIF3 subunits indicating the eIF5 placement on eIF3 (Figure 6D). Interestingly, we found only one cross-link between eIF5 and eIF1 (Figure 6D) and no cross-links between eIF1 and eIF3 subunits (most probably due to the fact that eIF1 is a small protein with only four lysines that are likely occluded by its interacting partners; namely the c/Nip1-NTD, the a/Tif32-HLD, and the eIF5–CTD).
Figure 6.
The refined 2D model of budding yeast eIF3 in complex with eIFs 1 and 5 with the indicated placement of the obtained cross-links. (A and B) Planar projections of the eIF3-1-5 ‘supercomplex’ with known contact points among individual proteins/defined domains in all four views rotated by 90°CCW. (C) The summary of all cross-links among individual eIF3 subunits (deliberately excluding cross-links with eIF1 and eIF5) obtained in the eIF3-1-5 ‘supercomplex’ that were divided into three color-coded groups (A–C) based on their positions and directions. (D) The summary of all cross-links between eIF5 and other proteins (deliberately excluding cross-links given in (C)) in the eIF3-1-5 ‘supercomplex’ (group D).
As can be seen in Figure 6C, Supplementary S3A and Table S2, even though some eIF3-free-specific cross-links seen in Figure 4E disappeared, three major directions of cross-links are still clearly discernible. Hence the major change between free eIF3 and eIF3 complexed with eIF1 and eIF5 is that the latter shows an elaborate net of cross-links among the inner parts of its subunits, including the β-propeller of b/Prt1. In addition to that, we observed cross-links of the a/Tif32 terminal domains with the c/Nip1-CTD, as well as with the entire length of g/Tif35. These findings strongly suggest that eIF3 complexed with eIFs 1 and 5 is packed even more tightly than when in its free form. In agreement, eIF5 shows the same classification of cross-links: (i) with the inner parts of eIF3 subunits; (ii) with the NTDs of a/Tif32 and c/Nip1, (iii) as well as with nearly the entire length of g/Tif35 (Figure 6D, Supplementary Figure S3B-C and Table S2).
Based on these data we modified the 3D model of eIF3 (Figure 5A–D) to reflect its nearly perfectly globular character when bound by eIFs 1 and 5 (Figure 7A–D). Both WD40 domains (b/Prt1 and i/Tif34), the RRM domains (b/Prt1 and g/Tif35), as well as the extreme ends of a/Tif32 and c/Nip1 (except for the NTD of the latter which is occluded by eIF5) are still exposed to the surface but in a surprisingly compact array.
Figure 7.
The globular 3D architecture of the eIF3-1-5 ‘supercomplex’ free in solution modeled based on the obtained cross-links. (A–D) The 3D architecture of the yeast eIF3-1-5 ‘supercomplex’ modeled using restraints from all cross-links depicted in Figure 6 viewed from all four sides rotated by 90°CCW. Color-coding of individual eIF3 subunits corresponds to Figure 6; for details see the main text.
In further support of our model of non-interacting PCI domains of a/Tif32 and c/Nip1, no specific cross-links between both PCI domains were detected also in this complex, and two cross-links that we detected between the a/Tif32-NTD and the helical region of c/Nip1 do not fit with the published arrangement observed in the cryo-EM structures (26,45) due to their distance restrains (Supplementary Figure S4B).
It is also noteworthy that we detected one intrasubunit cross-link within the RRM domain of g/Tif35 (Supplementary Figure S4C). The structure of yeast g/Tif35-RRM has not been solved yet, hence Supplementary Figure S4C depicts the structure of the human eIF3g-RRM that is well-conserved with its yeast counterpart (33). It is the only cross-link in all our datasets occurring in the solved structure; it connects one of the RRM β-sheets (β1) with the loop between two other β sheets. Since the distance between the corresponding cross-linked lysines in this crystal structure (∼20.7 Å) fits well with the maximal length of the employed cross-linking agent (24 Å), this result serves as a proof of principle of the meaningfulness of our cross-linking approach.
The a/Tif32 and c/Nip1 subunits must restructure in order to adopt their extended conformations on the 40S ribosomal surface
Our compactly packed 3D cross-linking model of eIF3 in complex with eIFs 1 and 5 is in sharp contrast to all available cryo-EM structures of eIF3 in complex with the 40S ribosome (26,34,45–46), where a/Tif32 and c/Nip1 are heterodimerized through their PCI domains, sitting near the mRNA exit pore with the c/Nip1-NTD and a/Tif32-CTD running in the opposite direction –in the anthropomorphic terms representing the eIF3 arms embracing the 40S head from the opposite sides. To reconcile these findings, we hypothesize that eIF3 must go through a series of conformational changes upon its initial association with the small ribosomal subunit that completely reshape its arrangement in 3D.
To examine that, we purified 40S ribosomal subunits and formed the eIF3–40S complexes that were cross-linked with DSBU; i.e. a mass-spectrometry-cleavable cross-linker that contains an amine-reactive NHS ester at each end of an 11-atom spacer arm (69). DSBU has similar reactivity to DSS but contains an urea linker that can be cleaved using collision-induced dissociation (CID); an approximate length of the Cα-Cα cross-link is 25.3 Å. The cleavable cross-linker enables desirable generation of more fragments and was instrumental in overcoming the expected technical difficulties associated with the high complexity of the eIF3–40S samples.
We first tested whether the chosen DSBU concentrations did not result in a massive over-cross-linking of the sample that could generate two or more eIF3–40S complexes joined together. Hence we subjected our samples to high velocity sucrose gradient centrifugation and collected all fractions carrying obvious A260 peaks. As can be seen in Supplementary Figure S5, the only dominant peak (across fractions 10–11) was observed and found to correspond to that section of the gradient where the free 40S subunits and 43–48S PICs migrate (58,70). Interestingly, the cross-linker generated also a minor, faster migrating peak that we attribute to a 40S dimer. The collected fractions were subsequently spotted onto a nitrocellulose membrane and subjected to Western blotting using anti-g/Tif35 and anti-a/Tif32 polyclonal antisera (Supplementary Figure S5). Only the fraction 10 representing the major peak gave a specific signal with both antibodies, whereas the minor peak occurring in the fraction 13 did not carry any eIF3 complexes.
Running MeroX (DSBU) and StavroX (BS3) algorithms we obtained 81 high confidence inter-molecular cross-links, out of which 11 were connecting various eIF3 subunits and the rest bridged some eIF3 subunit and some Rps (for the overview please see Supplementary Table S3 and Document S1 containing all assigned fragment spectra). Based on this web of cross-links we constructed a 3D map of the eIF3–40S complex. After accommodation of all cross-links (taking into account their average lengths as precisely as possible) two pictures emerge. One most probably depicts the initial docking of eIF3 onto the ribosomal surface (Figure 8A and B; see below for details), and the other closely resembles previously published Cryo-EM structures of not only yeast but also mammalian eIF3–40S complexes (Figure 8C–H); underscoring the validity and precision of our cross-linking approach. To synoptically visualize all obtained cross-links, we decided to divide them into the following groups: panels A and B show specific Lysine-to-Lysine cross-links of all subunits with the designated small ribosomal proteins; panel C shows cross-links of the a/Tif32 subunit with small ribosomal proteins occurring on the solvent-exposed side of the 40S; panel D shows cross-links of b/Prt1 and g/Tif35 subunits with small ribosomal proteins in the same place; panel E of c/Nip1 and i/Tif34 with small ribosomal proteins in the same place; panel F illustrates all cross-links that are made with the beak side of the 40S; panel G depicts cross-links of c/Nip1 with the interface side of the 40S; and panel H all cross-links occurring on the platform side.
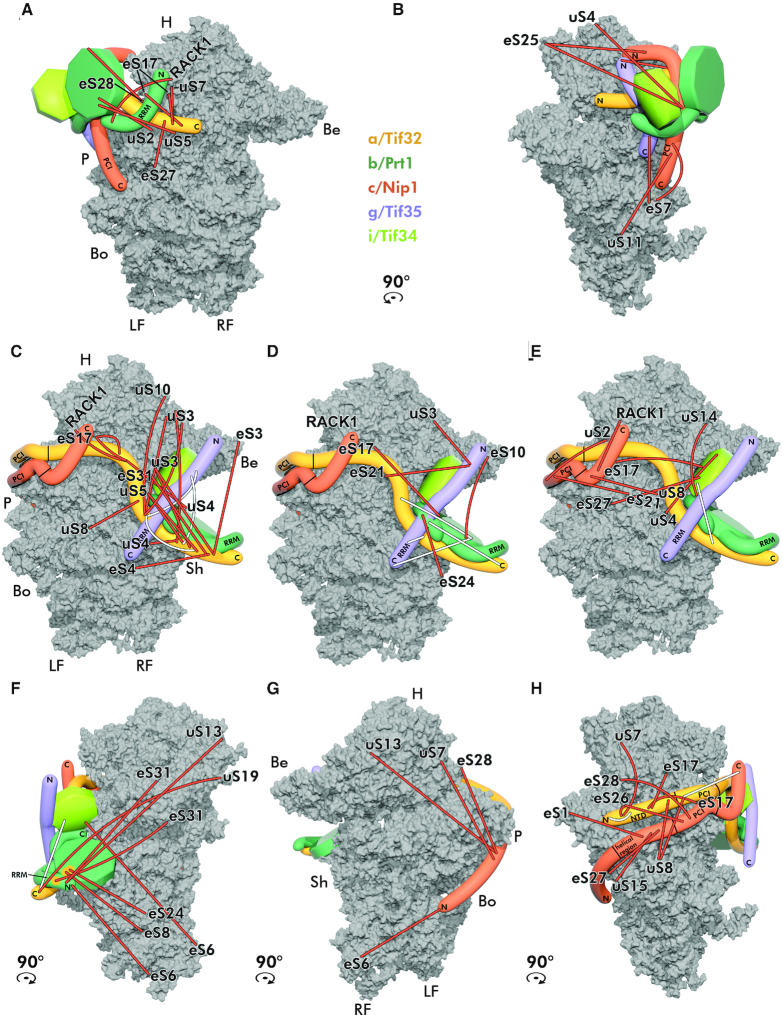
Figure 8.
The eIF3 complex undergoes a dramatic structural rearangement upon its binding to the 40S subunit. (A–H) Placement of eIF3 on the 40S ribosomal subunit modeled based on the obtained cross-links. (A) Position of eIF3 prior to the major rearangement of individual subunits with corresponding crosslinks shown from the solvent-exposed side. (B) The same as A but only cross-links mapping to the platform side are shown. (C) The eIF3 complex after the structural rearangement on the 40S subunit. Only cross-links between a/Tif32 and ribosomal proteins are shown in the solvent-exposed view of the 40S subunit (PDB code: 3JAM). (D) Only cross-links between c/Nip1 and i/Tif34 and ribosomal proteins are shown in the solvent-exposed view of the 40S subunit. (E) Only cross-links between b/Prt1 and g/Tif35 and ribosomal proteins are shown in the solvent-exposed view of the 40S subunit. (F) Only cross-links mapping to the beak side are shown; yellow circles indicate three cross-links emanating from the C-terminal domain of a/Tif32 in contact with b/Prt1. (G) Only cross-links mapping to the intersubunit side are shown. (H) Only cross-links mapping to the platform side are shown. Cross-links between ribosomal proteins and all eIF3 subunits are depicted in orange, intramolecular cross-links between eIF3 subunits in white; identities of small ribosomal proteins are indicated at the end of each cross-link. Color-coding of individual eIF3 subunits corresponds to previous figures. H–head; P–platform; Be–beak; Bo–body; Sh–shoulder; RF–right foot; LF–left foot. For details see the main text.
As can be seen, the N-terminal half of a/Tif32 and the C-terminal 2/3 of c/Nip1 cross-link with small ribosomal proteins on the platform side of the 40S subunit (Figure 8C, E, G and H; orange sticks) strongly supporting both yeast and mammalian cryo-EM studies showing that these two eIF3 domains are firmly attached to the region around the mRNA exit channel (reviewed in (4)). On the other hand, the Rps cross-links obtained with the C-terminal half of a/Tif32 and the remaining three eIF3 subunits strongly suggest existence of at least two different conformations. (i) Cross-links obtained with uS7/Rps5, eS17/Rps17, eS27/Rps27, uS2/Rps0, eS7/Rps7, uS11/Rps14 and RACK1 indicate that the entire b/Prt1-g/Tif35-i/Tif34-a/Tif32-CTD module is closely attached to the c/Nip1-a/Tif32-NTD module at the entry channel as when free in solution (Figure 8A and B); (ii) cross-links detected with uS3/Rps3, uS10/Rps20, eS31/Rps31, uS5/Rps2, eS4/Rps4, uS4/Rps9, uS14/Rps29, eS10/Rps10 and eS24/Rps24 support the extended conformation as seen in the available cryo-EM studies (Figure 8C–E). Since practically all eIF3–eIF3 cross-links were obtained within but not between these two modules (Figure 8C–H; white sticks), we think that the latter conformation is more prevalent in our samples and thus—as expected—more stable.
Taking into account: (i) the eIF3 domains that are (based on our cross-linking analysis) exposed to the surface in the ribosome-free form of eIF3, together with (ii) the well-known bridges between eIF3 and the 40 subunit, we speculate that the initial eIF3 docking site(s) on the 40S ribosome could be represented by one (or more) of the following interactions: (a) a/Tif32-NTD–uS2/Rps0A (36); (b) a/Tif32-CTD–uS5/Rps2 and/or –uS3/Rps3 (39); (c) c/Nip1-CTD–RACK1/ASC1 (28); (d) g/Tif35-NTD–uS10/Rps20 and/or –uS3/Rps3 (33); and at last (e) b/Prt1-WD40– uS4/Rps9 (24). Importantly, earlier we demonstrated that the internal deletion of the RRM domain from b/Prt1 produces a trimeric complex composed of the Δ100-b/prt1 mutant protein associated with only g/Tif35 and i/Tif34 that completely fails to interact with the 40S subunit in vivo (27). We also showed that the deletion of the C-terminal 1/3 of a/Tif32 (in a/Tif32-Δ5) generates a complex containing the N-terminal 2/3 of a/Tif32, c/Nip1 and eIF5, called the minimal 40S-binding unit (MBU), which is sufficient for 40S binding in vivo and in vitro (35). The N and C termini of c/Nip1 and the a/Tif32-NTD were found to be required for 40S binding by the eIF3–eIF1–eIF2–eIF5 complex in vivo, whereas eIF5 was necessary for binding only when the a/Tif32-CTD was absent. Therefore, we suggest that the most probable mediators of the initial contact between eIF3 and the 40S are the a/Tif32-NTD–uS2/Rps0A and c/Nip1-CTD–RACK1/ASC1 interactions (Figure 8A and B). This is consistent with the fact that the a/Tif32-PCI–c/Nip1-PCI heterodimer is considered to be a stable anchor of eIF3 on the ribosomal surface, as mentioned above, whereas the a/Tif32-CTD–b/Prt1–g/Tif35–i/Tif34 module can shuffle from the solvent-exposed region near the mRNA entry channel to the interface side and back (reviewed in (4)).
Hence, we propose that upon the initial eIF3–40S contact mediated by the a/Tif32-NTD and c/Nip1-CTD (Figure 8A and B), these two subunits undergo a conformational switch from their ribosome-free mode into their PCI-to-PCI heterodimer mode of binding (Figure 8C–H), so that (i) the c/Nip1–b/Prt1 contact is severed allowing the tightly packed eIF3 to unfold, (ii) the a/Tif32-CTD adopts its extended form enabling the b/Prt1–g/Tif35–i/Tif34 module associated with it to settle near the mRNA entry channel and (iii) the c/Nip1-NTD (restricted in its movement in the free form) can wrap around the platform and reach the 40S P-site to which it could deliver eIFs 1 and 5 during the 43S PIC formation, as suggested earlier (51,68).
The cross-links obtained among small ribosomal proteins and the a/Tif32-CTD–b/Prt1–g/Tif35–i/Tif34 module on the 40S beak side (Figure 8F) do support the flexibility of this module consistent with its shuffling ability. Similarly, cross-links obtained between the c/Nip1-NTD and small ribosomal proteins residing at the 40S head versus right foot (Figure 8G) illustrate the flexibility of the c/Nip1-NTD that is most probably instrumental during AUG selection process (51,68). Consistently, earlier cross-linking mass spectrometry data predicted an interaction of the N-terminal segment of c/Nip1 with eIF1 bound to the 40S platform (26), and a density presumably corresponding to the eIF3c residues 115–220 was identified not too far away from the P-site-bound eIF1 in both the py48S-closed/open complexes (45).
To conclude, the fact that eIF3 is tightly packed in the ribosome-free form suggests that this arrangement could have a regulatory role; i.e. to expose only the 40S- and eIF1- and eIF5-contact domains while preventing the hidden domains from making functional interactions until they become accessible on the ribosomal surface for binding to other factors.
eIF5 occupies the region below the 40S P- and E-sites
As described above, eIF5 plays a critical role in the AUG recognition process, yet its precise position within the 43–48S PICs has remained elusive. Having purified the eIF5 protein, we attempted to form the eIF5–40S complex that could be subjected to our DSBU cross-linking approach (using the same concentration as with the eIF3–40S complex ensuring that the complex did not get over-crosslinked (Supplementary Figure S6)) in order to reveal the placement of both, functionally relatively independent NTD and CTD domains of eIF5 on the 40S ribosome. In total, we obtained 33 high-confidence cross-links and built the 3D model of the eIF5–40S complex (Figure 9A–C). Since we were interested in the position of eIF5 in PICs, we disregarded 12 cross-links the positions of which would collide with those of eIF3 subunits (a few of them mapped onto the solvent-exposed side of 40S ribosome). Out of the remaining 21 cross-links, we detected 18 cross-links between eIF5 and some ribosomal protein (three cross-links emanate from the linker region) and three cross-links within the eIF5 protein itself (Figure 9A—in white); for the overview of cross-links please see Supplementary Table S4 and Document S2 containing all considered collision spectra.
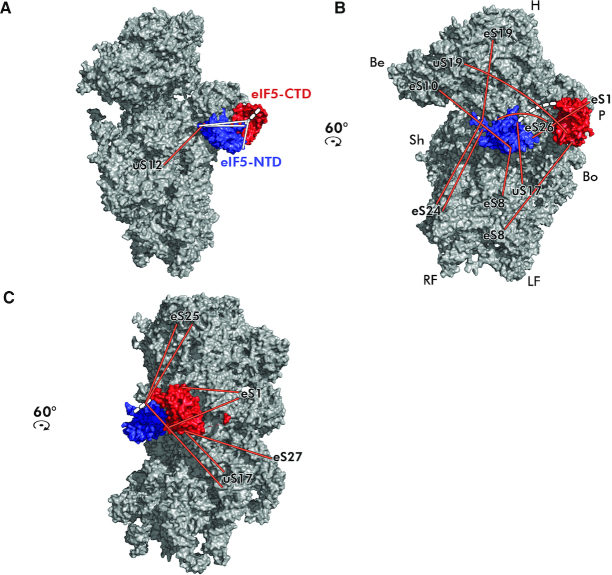
Figure 9.
Placement of both terminal domains of eIF5 on the 40S ribosomal subunit modeled based on the obtained cross-links. (A–C) The N-terminal (PDB code: 2E9H—in blue) and the C-terminal (PDB code: 2FUL—in red) domains of eIF5 were modeled onto the intersubunit side of the 40S subunit (PDB code: 3JAM) based on the obtained cross-links. The linker region, unresolved in the available 3D structures, is depicted as a dashed white line. Cross-links between ribosomal proteins and eIF5 are depicted in orange, intramolecular cross-links between the eIF5 halves in white; identities of small ribosomal proteins are indicated at the end of each cross-link. For details see the main text.
The very tip of the eIF5–NTD (residues 1–149, containing the GAP function) was cross-linked with uS12/Rps23, eS19/Rps19 and eS24/Rps24 (Figure 9A and B). The other half of the eIF5–NTD was cross-linked with eS10/Rps10, eS26/Rps26, eS8/Rps8, uS17/Rps11 and eS1/Rps1 (Figure 9B), while the linker region was cross-linked with eS25/Rps25 and uS17/Rps11 (Figure 9C). The latter ribosomal protein also cross-linked to the eIF5–CTD together with eS1/Rps1 and eS27/Rps27 (Figure 9C). Taken together, these cross-links place the eIF5–NTD near the P-site where eIF1 binds in the open/POUT state, in agreement with the only recently published work by others (46), whereas the eIF5–CTD seems to reside closer to the platform, near the E-site (Figure 10). In fact, in this arrangement the eIF5–CTD occurs next to its binding partner c/Nip1-NTD in eIF3, suggesting that the c/Nip1–NTD–eIF5–CTD interaction is instrumental for the eIF3-mediated delivery of eIF5 (and most likely also of eIF1) to its operational site on the ribosomal interphase as proposed earlier (50,51,68). In addition, in this arrangement the eIF5–CTD is also ideally suited to control the start codon selection by promoting the dissociation of eIF1 from the scanning-arrested PICs as shown in (55).
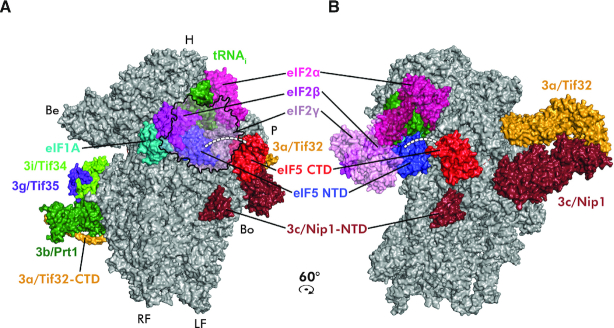
Figure 10.
Model of the eIF5-containing PIC at the late initiation stage. The partial yeast 48S PIC with the eIF5 N-terminal domain is shown (adopted from (46); PDB code: 6FYX) containing the 40S subunit (gray), eIF5–NTD (blue), eIF1A (teal), eIF2α (hotpink), eIF2β (magenta), eIF2γ (pink), a/Tif32 (orange), b/Prt1 (forest green), c/Nip1 (ruby), i/Tif34 (chartreuse), g/Tif35 (purple blue) and Met-tRNAiMet. (green). Placement of the eIF5–CTD (PDB code: 2FUL) was modeled based on the obtained cross-links shown if Figure 9. The eIF5 linker region is depicted as a dashed white line. The density corresponding to the c/Nip1-NTD is indicated. (A) View from the intersubunit side; eIF2β is partially transparent and eIF2γ is depicted merely as an outline to visualize the eIF5–NTD. (B) View from the platform side rotated by 60°CW.
Despite the obvious limitations of this approach (for example the studied eIF5–40S complex might never assemble in the cell without other eIFs), we believe that our data are relevant and, in simple terms of the molecular functioning of eIF5 in the context of the 48S PIC, entitle us to propose the following (Figure 10). Upon the initial docking of eIF1 and eIF5 (via its CTD) to their places underneath the P- and E-sites by the c/Nip1-NTD, respectively, and formation of the 48S PIC, the 40S ribosome starts scanning for AUG. The position of the arginine finger-containing eIF5–NTD at this stage remains to be determined, yet we think that it must be situated close to the GTP-binding pocket of eIF2γ. Selection of AUG, triggering the irreversible GTP hydrolysis, then spurs the eIF5–CTD to promote eIF1 dissociation or replacement from its ‘P-site’ binding spot to allow the relocation of the eIF5–NTD to this very spot in a synchronous movement with the eIF1A C-terminal tail toward one another. This way the eIF5–NTD can stabilize the codon-anticodon interaction and thus lock the 48S PIC in the closed/PIN state to enable eIF5B-mediated subunits joining as recently proposed (46).
Concluding remarks
Here we mastered the chemical cross-linking methodology combined with advanced mass spectrometry to map the 3D architecture and relative positions of all eIF3 subunits when (i) free in solution, (ii) in complex with other eIFs or (iii) when bound to the 40S ribosomal subunit, based on which we expanded our knowledge on molecular mechanics of the general initiation pathway. Together with other studies of a similar kind on different complexes, we think that our approach can be viewed as the detailed instruction manual for other researchers working with multiprotein assemblies, the crystal or cryo-EM structures of which are very hard to obtain.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Ralf Ficner, Jon R. Lorsch and Hüseyin Besir for material and advice. We are indebted to Vojtěch Spiwok and Dalibor Trapl for their help with modeling of protein complexes. At last, we are very thankful to Olga Krýdová, Vladimíra Matoušková and Veronika Kovaničová for technical and administrative assistance. We acknowledge the Centre of molecular structure Core Facility at BIOCEV.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation Grants [GA17-06238S to L.S.V., GA16-24309S to P.N.]; European Regional Development Funds [CZ.1.05/1.1.00/02.0109 BIOCEV]; The Czech Infrastructure for Integrative Structural Biology, MEYS CR [LM2015043 CIISB for CMS BIOCEV; LTC17065]; European Commission H2020 (European Proteomics Infrastructure Consortium providing Access—EPIC-XS Project [823839]. Y.I. was supported by the Overseas Research Fellowships of JSPS and the Uehara Memorial Foundation. Funding for open access charge: Czech Science Foundation Grant [GA17-06238S].
Conflict of interest statement. None declared.
REFERENCES
- 1. Frank J. The mechanism of translation [version 1; peer review: 3 approved]. F1000Res. 2017; 6:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hinnebusch A.G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 2017; 42:589–611. [DOI] [PubMed] [Google Scholar]
- 3. Valášek L.S. ‘Ribozoomin’—translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Pept. Sci. 2012; 13:305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valasek L.S., Zeman J., Wagner S., Beznoskova P., Pavlikova Z., Mohammad M.P., Hronova V., Herrmannova A., Hashem Y., Gunisova S.. Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res. 2017; 45:10948–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hershey J.W.B., Sonenberg N., Mathews M.B.. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 2018; 4:a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park H.S., Himmelbach A., Browning K.S., Hohn T., Ryabova L.A.. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001; 106:723–733. [DOI] [PubMed] [Google Scholar]
- 7. Mohammad M.P., Munzarova Pondelickova V., Zeman J., Gunisova S., Valasek L.S.. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res. 2017; 45:2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thiebeauld O., Schepetilnikov M., Park H.S., Geldreich A., Kobayashi K., Keller M., Hohn T., Ryabova L.A.. A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J. 2009; 28:3171–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schepetilnikov M., Dimitrova M., Mancera-Martinez E., Geldreich A., Keller M., Ryabova L.A.. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013; 32:1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szamecz B., Rutkai E., Cuchalova L., Munzarova V., Herrmannova A., Nielsen K.H., Burela L., Hinnebusch A.G., Valášek L.. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008; 22:2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munzarová V., Pánek J., Gunišová S., Dányi I., Szamecz B., Valášek L.S.. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-Acting mRNA elements preceding short uORFs. PLoS Genet. 2011; 7:e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gunisova S., Valasek L.S.. Fail-safe mechanism of GCN4 translational control-uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res. 2014; 42:5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gunisova S., Hronova V., Mohammad M.P., Hinnebusch A.G., Valasek L.S.. Please do not recycle! translation reinitiation in microbes and higher eukaryotes. FEMS Microbiol. Rev. 2018; 42:165–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beznosková P., Cuchalová L., Wagner S., Shoemaker C.J., Gunišová S., Von der Haar T., Valášek L.S.. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS Genet. 2013; 9:e1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pisarev A.V., Hellen C.U.T., Pestova T.V.. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007; 131:286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pisarev A.V., Skabkin M.A., Pisareva V.P., Skabkina O.V., Rakotondrafara A.M., Hentze M.W., Hellen C.U.T., Pestova T.V.. The Role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010; 37:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beznoskova P., Wagner S., Jansen M.E., von der Haar T., Valasek L.S.. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015; 43:5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beznoskova P., Gunisova S., Valasek L.S.. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA. 2016; 22:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gomes-Duarte A., Lacerda R., Menezes J., Romao L.. eIF3: a factor for human health and disease. RNA Biol. 2018; 15:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hershey J.W. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta. 2015; 1849:792–800. [DOI] [PubMed] [Google Scholar]
- 21. Robichaud N., Sonenberg N.. Translational control and the cancer cell response to stress. Curr. Opin. Cell Biol. 2017; 45:102–109. [DOI] [PubMed] [Google Scholar]
- 22. Scheper G.C., van der Knaap M.S., Proud C.G.. Translation matters: protein synthesis defects in inherited disease. Nat. Rev. Genet. 2007; 8:711–723. [DOI] [PubMed] [Google Scholar]
- 23. Pick E., Hofmann K., Glickman M.H.. PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell. 2009; 35:260–264. [DOI] [PubMed] [Google Scholar]
- 24. Liu Y., Neumann P., Kuhle B., Monecke T., Schell S., Chari A., Ficner R.. Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit. Structure. 2014; 22:923–930. [DOI] [PubMed] [Google Scholar]
- 25. Herrmannová A., Daujotyte D., Yang J.C., Cuchalová L., Gorrec F., Wagner S., Danyi I., Lukavsky P.J., Valášek L.S.. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-Initiation complex assembly. Nucleic Acids Res. 2012; 40:2294–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erzberger J.P., Stengel F., Pellarin R., Zhang S., Schaefer T., Aylett C.H., Cimermancic P., Boehringer D., Sali A., Aebersold R. et al.. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell. 2014; 158:1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valášek L., Phan L., Schoenfeld L.W., Valášková V., Hinnebusch A.G.. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recoginition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001; 20:891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kouba T., Rutkai E., Karasková M., Valášek L.S.. The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of the pre-initiation complexes. Nucleic Acids Res. 2012; 40:2683–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khoshnevis S., Gunišová S., Vlčková V., Kouba T., Neumann P., Beznosková P., Ficner R., Valášek L.S.. Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes. Nucleic Acids Res. 2014; 42:4123–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valášek L., Nielsen K.H., Hinnebusch A.G.. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002; 21:5886–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khoshnevis S., Hauer F., Milon P., Stark H., Ficner R.. Novel insights into the architecture and protein interaction network of yeast eIF3. RNA. 2012; 18:2306–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Asano K., Phan L., Anderson J., Hinnebusch A.G.. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998; 273:18573–18585. [DOI] [PubMed] [Google Scholar]
- 33. Cuchalová L., Kouba T., Herrmannová A., Danyi I., Chiu W.-L., Valášek L.. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell Biol. 2010; 30:4671–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aylett C.H., Boehringer D., Erzberger J.P., Schaefer T., Ban N.. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat. Struct. Mol. Biol. 2015; 22:269–271. [DOI] [PubMed] [Google Scholar]
- 35. Valášek L., Mathew A., Shin B.S., Nielsen K.H., Szamecz B., Hinnebusch A.G.. The yeast eIF3 subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003; 17:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kouba T., Danyi I., Gunišová S., Munzarová V., Vlčková V., Cuchalová L., Neueder A., Milkereit P., Valášek L.S.. Small ribosomal protein RPS0 stimulates translation initiation by mediating 40S-binding of eIF3 via its direct contact with the eIF3a/TIF32 subunit. PLoS One. 2012; 7:e40464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pisarev A.V., Kolupaeva V.G., Yusupov M.M., Hellen C.U.T., Pestova T.V.. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008; 27:1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aitken C.E., Beznoskova P., Vlckova V., Chiu W.L., Zhou F., Valasek L.S., Hinnebusch A.G., Lorsch J.R.. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. Elife. 2016; 5:e20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chiu W.-L., Wagner S., Herrmannová A., Burela L., Zhang F., Saini A.K., Valášek L., Hinnebusch A.G.. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell Biol. 2010; 30:4415–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Hellen C.U., Pestova T.V., Frank J.. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013; 153:1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. des Georges A., Dhote V., Kuhn L., Hellen C.U., Pestova T.V., Frank J., Hashem Y.. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015; 525:491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. ElAntak L., Wagner S., Herrmannová A., Karásková M., Rutkai E., Lukavsky P.J., Valášek L.. The indispensable N-terminal half of eIF3j co-operates with its structurally conserved binding partner eIF3b-RRM and eIF1A in stringent AUG selection. J. Mol. Biol. 2010; 396:1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nielsen K.H., Szamecz B., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G.. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004; 23:1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nielsen K.H., Valášek L., Sykes C., Jivotovskaya A., Hinnebusch A.G.. Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol. Cell Biol. 2006; 26:2984–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Llacer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell. 2015; 59:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Llacer J.L., Hussain T., Saini A.K., Nanda J.S., Kaur S., Gordiyenko Y., Kumar R., Hinnebusch A.G., Lorsch J.R., Ramakrishnan V.. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. Elife. 2018; 7:e39273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simonetti A., Brito Querido J., Myasnikov A.G., Mancera-Martinez E., Renaud A., Kuhn L., Hashem Y.. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol. Cell. 2016; 63:206–217. [DOI] [PubMed] [Google Scholar]
- 48. Phan L., Zhang X., Asano K., Anderson J., Vornlocher H.P., Greenberg J.R., Qin J., Hinnebusch A.G.. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell Biol. 1998; 18:4935–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Asano K., Phan L., Valasek L., Schoenfeld L.W., Shalev A., Clayton J., Nielsen K., Donahue T.F., Hinnebusch A.G.. A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNA(i)Met promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition. Cold Spring Harb. Symp. Quant. Biol. 2001; 66:403–415. [DOI] [PubMed] [Google Scholar]
- 50. Valášek L., Nielsen K.H., Zhang F., Fekete C.A., Hinnebusch A.G.. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell Biol. 2004; 24:9437–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Karaskova M., Gunisova S., Herrmannova A., Wagner S., Munzarova V., Valasek L.S.. Functional characterization of the role of the N-terminal domain of the c/Nip1 subunit of eukaryotic initiation factor 3 (eIF3) in AUG recognition. J. Biol. Chem. 2012; 287:28420–28434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jennings M.D., Zhou Y., Mohammad-Qureshi S.S., Bennett D., Pavitt G.D.. eIF2B promotes eIF5 dissociation from eIF2•GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev. 2013; 27:2696–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Conte M.R., Kelly G., Babon J., Sanfelice D., Youell J., Smerdon S.J., Proud C.G.. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry. 2006; 45:4550–4558. [DOI] [PubMed] [Google Scholar]
- 54. Wei Z., Xue Y., Xu H., Gong W.. Crystal structure of the C-terminal domain of S.cerevisiae eIF5. J. Mol. Biol. 2006; 359:1–9. [DOI] [PubMed] [Google Scholar]
- 55. Nanda J.S., Saini A.K., Munoz A.M., Hinnebusch A.G., Lorsch J.R.. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J. Biol. Chem. 2013; 288:5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hussain T., Llacer J.L., Fernandez I.S., Munoz A., Martin-Marcos P., Savva C.G., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014; 159:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Acker M.G., Kolitz S.E., Mitchell S.F., Nanda J.S., Lorsch J.R.. Reconstitution of yeast translation initiation. Methods Enzymol. 2007; 430:111–145. [DOI] [PubMed] [Google Scholar]
- 58. Valášek L., Szamecz B., Hinnebusch A.G., Nielsen K.H.. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007; 429:163–183. [DOI] [PubMed] [Google Scholar]
- 59. Schilling B., Row R.H., Gibson B.W., Guo X., Young M.M.. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 2003; 14:834–850. [DOI] [PubMed] [Google Scholar]
- 60. Young M.M., Tang N., Hempel J.C., Oshiro C.M., Taylor E.W., Kuntz I.D., Gibson B.W., Dollinger G.. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5802–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gotze M., Pettelkau J., Schaks S., Bosse K., Ihling C.H., Krauth F., Fritzsche R., Kuhn U., Sinz A.. StavroX–a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 2012; 23:76–87. [DOI] [PubMed] [Google Scholar]
- 62. Gotze M., Pettelkau J., Fritzsche R., Ihling C.H., Schafer M., Sinz A.. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass Spectrom. 2015; 26:83–97. [DOI] [PubMed] [Google Scholar]
- 63. Phan L., Schoenfeld L.W., Valášek L., Nielsen K.H., Hinnebusch A.G.. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNAiMet. EMBO J. 2001; 20:2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Algire M.A., Maag D., Savio P., Acker M.G., Tarun S.Z. Jr, Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L. et al.. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002; 8:382–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schneider M., Belsom A., Rappsilber J.. Protein tertiary structure by crosslinking/mass spectrometry. Trends Biochem. Sci. 2018; 43:157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R.. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell Proteomics. 2010; 9:1634–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rozbesky D., Rosulek M., Kukacka Z., Chmelik J., Man P., Novak P.. Impact of chemical cross-linking on protein structure and function. Anal. Chem. 2018; 90:1104–1113. [DOI] [PubMed] [Google Scholar]
- 68. Obayashi E., Luna R.E., Nagata T., Martin-Marcos P., Hiraishi H., Singh C.R., Erzberger J.P., Zhang F., Arthanari H., Morris J. et al.. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: structural role for eIF3c and its control by eIF5. Cell Rep. 2017; 18:2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Muller M.Q., Dreiocker F., Ihling C.H., Schafer M., Sinz A.. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal. Chem. 2010; 82:6958–6968. [DOI] [PubMed] [Google Scholar]
- 70. Pospisek M., Valasek L.. Polysome profile analysis–yeast. Methods Enzymol. 2013; 530:173–181. [DOI] [PubMed] [Google Scholar]
- 71. Wagner S., Herrmannova A., Sikrova D., Valasek L.S.. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Res. 2016; 44:10772–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.