Abstract
Viruses have developed different mechanisms to manipulate their hosts, including the process of viral mimicry in which viruses express important host proteins. Until recently, examples of viral mimicry were limited to mimics of growth factors and immunomodulatory proteins. Using a comprehensive bioinformatics approach, we have shown that viruses possess the DNA/RNA with potential to encode 16 different peptides with high sequence similarity to human peptide hormones and metabolically important regulatory proteins. We have characterized one of these families, the viral insulin/IGF-1–like peptides (VILPs), which we identified in four members of the Iridoviridae family. VILPs can bind to human insulin and IGF-1 receptors and stimulate classic postreceptor signaling pathways. Moreover, VILPs can stimulate glucose uptake in vitro and in vivo and stimulate DNA synthesis. DNA sequences of some VILP-carrying viruses have been identified in the human enteric virome. In addition to VILPs, sequences with homology to 15 other peptide hormones or cytokines can be identified in viral DNA/RNA sequences, some with a very high identity to hormones. Recent data by others has identified a peptide that resembles and mimics α-melanocyte-stimulating hormone’s anti-inflammatory effects in in vitro and in vivo models. Taken together, these studies reveal novel mechanisms of viral and bacterial pathogenesis in which the microbe can directly target or mimic the host endocrine system. These findings also introduce the concept of a system of microbial hormones that provides new insights into the evolution of peptide hormones, as well as potential new roles of microbial hormones in health and disease.
Through millions of years of coevolution, viruses have developed elegant survival strategies to manipulate their host and avoid immune recognition. These strategies include antigenic variation used by RNA viruses (1, 2) and molecular mimicry by large DNA viruses (3–5). Mimicry is a common evolutionary phenomenon that occurs when an organism or cell imitates another to gain an advantage in competing for resources, protection, or survival. Viral mimicry is a mechanism employed by viruses to generate molecules that resemble host growth factors or immune response regulators such as cytokines, chemokines, and their receptors for the benefit of the virus. In some cases, this may be to either co-opt or disrupt host immune function to obtain an advantage, but whether this is always true is not known.
One mechanism of viral mimicry uses divergent evolution involving the transfer of a gene from a host genome to a viral genome via horizontal gene transfer. This subtype of evolution usually results in changes in the molecule, but with sufficient similarity to be recognized as similar to the original (6)—that is, the mimic sequence encodes a protein that is highly similar to the host protein being mimicked.
Another viral mimicry mechanism acts through convergent evolution where random genetic mutations of viral sequences create factors that independently mimic host features (7, 8). These types of viral mimics ordinarily have no sequence similarity with the host proteins being mimicked, but instead they have structural or functional similarities. Although horizontal transfer is known to occur in bacterial and other microbes, this later mechanism is also responsible for producing host protein mimetics (see below). Mimics that favor pathogen fitness and are evolutionarily advantageous for survival will be selected through evolutionary processes, such as natural selection.
Until our recent discovery of viral insulin/IGF-1–like peptides (VILPs) (9), knowledge on viral mimicry mechanisms was limited to mimics of growth factors and immunomodulatory proteins. Viruses need actively dividing cells to replicate their genetic material and promote infection. To do this, viruses can mimic several growth factors and cell cycle modulators, such as cyclins. Vaccinia growth factor was the first growth factor mimic identified in vaccinia virus. It is the homolog of epidermal growth factor and can stimulate cell proliferation (10). Other growth factor mimics include the simian sarcoma virus–derived platelet-derived growth factor (7) and the epidermal and TGF-like molecules of vaccinia virus (8, 9). Likewise, Kaposi sarcoma–associated herpesvirus encodes v-cyclins that mimic cyclin D (11), a putative viral oncogene. U197 protein of cytomegalovirus mimics a host kinase (cyclin-dependent kinase) to directly promote cell proliferation (12). The phosphatidylinositol 3-kinase (PI3K) pathway, which plays a critical role in regulating growth, differentiation, and a number of metabolic processes, is also targeted by viruses (13). Indeed, the AKT gene was first identified as a cellular homolog of the retroviral oncogene v-Akt in the AKT8 mouse-transforming retrovirus (14) and thus named c-AKT (15).
Another category of viral mimics is those that can affect the host immune system by imitating the function of cytokines, chemokines, and their corresponding receptors. Viral homologs of IL-6, IL-10, and IL-17 have been identified and found to regulate host cellular immune responses (16–19). Mimicking host cytokines is proven to be an effective strategy to reduce proinflammatory cytokine production. Furthermore, mimicking host cytokine receptors to neutralize cytokine activity is another way to suppress host proinflammatory responses. In some cases, these viral proteins mimic the extracellular binding domains of host cytokine receptors, but they lack transmembrane and intracellular signaling domains (e.g., soluble viral TNF receptor, IL-1β receptor, and IFN-γ receptor) (20–25). Viral chemokine mimics can act as either agonists or antagonists of host chemokine receptors and help facilitate virus dissemination and growth (26–31). Soluble chemokine binding mimicry has also been identified as soluble receptor and nonhomologous soluble chemokine-binding mimicry. Viral chemokine receptors potentially play a role in the regulation of the physiological state of the host cells and/or the ability of cells to respond to chemotactic signals involved in cell proliferation or cell reprogramming (32–37). Some chemokine binding viral mimics have little or no similarity in their primary sequence to host chemokine receptors, but they can function as chemokine inhibitors by binding and neutralizing host chemokines (38–40). Viral mimicry of other immune modulators has also been reported in orthopoxvirus, retrovirus, and herpesvirus. Some of these take advantage of the host cell’s complement response by producing mimics of complement activation regulators (41, 42). Still, others mimic major histocompatibility complex proteins and help viruses evade detection and destruction by natural killer cells (43).
One of the main strategies of infected cells to stop infection is undergoing apoptosis. Several viruses, including African swine fever virus and bovine herpesvirus-4, express Bcl family homologs that protect host cells from apoptosis (44, 45). It is also reported that enveloped viruses (alphavirus, Ebola virus, and dengue virus) use a strategy of apoptotic mimicry to promote virus entry, enhance virus binding to host cells, or dampen the host immune response (46–49).
Discovery of Viral Hormones
Peptide hormones are secreted by endocrine cells into the circulation and regulate a variety of physiological functions, including metabolism, growth, and development. These act by binding and activating specific receptors on target cells. Blood carries a multitude of signaling and nonsignaling molecules (e.g., metabolites, proteins, vitamins). To eliminate nonspecific stimulation of downstream signaling, the peptide hormone and its cognate receptor have evolved highly specific structures, requiring the conservation of certain binding residues and structures. In some cases, the ligand is a small unstructured peptide, but in others, the interaction between the hormone and its receptor depends on a complex three-dimensional structure of the receptor and ligand. In the case of insulin, its structure arises due to the hormone’s two disulfide-linked peptide chains. Based on the abundance and diversity of viruses, we hypothesized that viruses might encode proteins that can mimic human peptide hormones and manipulate host physiology by stimulating or inhibiting these human hormone receptors.
Using a comprehensive bioinformatics approach, we searched all available viral genomes in the viral/viroid genome database at the National Center for Biotechnology Information for sequences with significant similarity to all known precursors and mature sequences of 62 human peptide hormones and metabolism-related cytokines. In total, we identified 16 viral homologs or peptides of which 14 have significant sequence similarity to different hormone families, including insulin, IGF-1 and IGF-2, endothelin-1 (ET1) endothelin-2, TGF-β1 and TGF-β2, fibroblast growth factors 19 and 21, inhibin, adiponectin, resistin, adipsin, and irisin in various viruses (Table 1) (9). In addition to these hormones, we also confirmed two regulatory cytokines, TNF (50) and IL-6 (51). In some cases, such as for insulin/IGFs and endothelins, there was not only significant similarity in the primary sequence, but also conservation of cysteine residues with identical spacing to the human hormones, thus allowing critical disulfide bond formation. In other cases, such as adiponectin, the homology was limited to a repetitive sequence, which may or may not be important in adiponectin function. All of these hormones have unique physiological functions, creating the possibility for many viral effects in regulating the host functions (Table 1).
Table 1.
Human Hormones and Regulatory Proteins Having Homolog Sequences in Viral Genomes
| Human Hormone or Regulatory Protein Sequences Used for the Search | Viruses Carrying the Viral Mimic of the Protein | Physiological Function in Humans |
|---|---|---|
| Insulin: 28–57 (B-chain) and 88–110 (A-chain) | Lymphocystis disease virus Sa | Glucose, fat, and protein metabolism |
| Lymphocystis disease virus 1 | Adipocyte development | |
| Arterial muscle tone | ||
| Cognition | ||
| Inflammation | ||
| Fertility | ||
| IGF-1: 49–118 | Lymphocystis disease virus Sa | Cell growth/maturation/proliferation/differentiation |
| Singapore grouper iridovirus | Glucose metabolism | |
| Lymphocystis disease virus 1 | Aging | |
| Neuropathy | ||
| Osteogenic | ||
| Tumor growth | ||
| IGF-2: 25–91 | Singapore grouper iridovirus | Cell growth/maturation/proliferation/migration/differentiation |
| Lymphocystis disease virus Sa | Fetoplacental development | |
| Atherosclerosis | ||
| Glucose metabolism | ||
| Tumor growth | ||
| Reproduction | ||
| Endothelin-1: 53–73 and 53–90 | Deerpox virus | Vasoconstriction |
| Proinflammation | ||
| Profibrotic | ||
| Tumorigenesis | ||
| Proangiogenesis | ||
| Mitogen | ||
| Proapoptosis | ||
| Endothelin-2: 49–69 | Deerpox virus | Vasoconstriction |
| Ovulation | ||
| Proinflammation | ||
| Profibrotic | ||
| Antiangiogenesis | ||
| Pro-tumor invasion and cancer metastatic | ||
| Fibroblast growth factor-21: 29–209 | Singapore grouper iridovirus | Brown fat thermogenesis |
| Orgyia pseudotsugata MNPV | Fatty acid oxidation | |
| Choristoneura rosaceana alphabaculovirus | Ketosis | |
| Lymantria xylina MNPV | White fat glucose metabolism | |
| White fat browning | ||
| Anti-inflammation | ||
| Nutrient restriction | ||
| Fibroblast growth factor-19: 25–216 | Singapore grouper iridovirus | Nutrient metabolism |
| Bile acid metabolism | ||
| Embryonic development | ||
| Tissue morphogenesis | ||
| Tumor invasion and growth | ||
| Irisin (fibronectin type III domain-containing protein 5): 32–212 and 32–143 | Bacteriophage SPP1 | White fat browning |
| Inflammatory markers for T2DM and complications | ||
| Hippocampal neurogenesis | ||
| Cognition | ||
| Anti-aging | ||
| Nutrient metabolism marker | ||
| TGF-β1: 279–390 | Deerpox virus | Inflammatory cytokine |
| TGF-β2: 303–414 | Pigeonpox virus | Tumorigenesis |
| Penguinpox virus | Tumor suppressor | |
| Fowlpox virus | Antimitogenic, | |
| Canarypox virus | T-cell (development, differentiation, homeostasis, and tolerance) | |
| Turkeypox virusa | Bone formation | |
| Angiogenesis | ||
| Hematopoiesis | ||
| Neuroprotection | ||
| Resistin: 19–108 | Gordonia phage ClubL | Resist insulin action |
| Endothelial dysfunction | ||
| Proinflammation Proangiogenesis | ||
| Provascular smooth muscle migration and proliferation | ||
| Cardiovascular disease marker Proatherosclerosis | ||
| Inhibin β A-chain: 311–426 | Deerpox virus | Pituitary hormone secretion and hypothalamic secretion |
| Turkeypox virus | Gonadal hormone secretion | |
| Canarypox virus | Insulin secretion | |
| Penguinpox virus | Germ cell development and maturation | |
| Pigeonpox virus | Erythroid differentiation | |
| Fowlpox virus | Embryonic axial development | |
| Bone growth | ||
| Neuronal cell survival | ||
| IL-6: 30-212b,c | Human herpesvirus 8 | Fat metabolism |
| Promyeloma and plasmacytoma | ||
| Neuronal cell differentiation | ||
| TNF: 57–233b,c | Pteropox virus | Fat metabolism |
| Induce insulin resistance in adipocyte | ||
| Adipocyte proliferation and remodeling |
The identical regions are listed following the name of hormones as residue numbers of the prepropeptides.
Abbreviation: T2DM, type 2 diabetes mellitus.
Turkeypox virus mimic is identified for just TGF-β2 and not TGF-β1.
Only metabolism-related functions of these cytokines are shown. Because many hits are identified for adiponectin, mainly based on the collagen domain, it was not added to this table.
Viral homology sequence was identified earlier by others.
Discovery of these viral hormones indicates a new potential mechanism of viral pathogenesis. For example, apoptosis is an important innate immune defense mechanism against viral infections. Insulin/IGF-1 are well known to suppress apoptosis and promote cell growth and proliferation (52–55). Thus, viruses that express these insulin/IGF-1 molecules have the potential to inhibit apoptosis and facilitate growth and proliferation of the infected host cells. This may also benefit the virus in terms of survival, propagation, and replication. More importantly, it adds a new dimension to the concept of viral mimicry and host–pathogen interaction. Thus, understanding the biology of these viral hormones has the potential to provide new targets for prevention and therapy of human disease.
Characterization of VILPs
The insulin superfamily of peptide hormones has been extensively studied in vertebrates and some invertebrates. Although there have been previous suggestions of insulin-like molecules in bacteria (56), these have not been widely accepted, because DNA sequence analysis has not identified any bacterial, archaeal, or fungal potential insulin-like sequence. In most mammalian species, there are separate insulin and IGF-1 receptors (i.e., IR and IGF1R) and one insulin and two IGFs that interact with them. Invertebrates, such as Drosophila and Caenorhabditis elegans, alternatively, have a single insulin/IGF receptor, tyrosine kinase receptor, capable of interacting with multiple insulin-like peptides (57–59). In these organisms, the single receptor predominantly regulates longevity and stress resistance (60). In mammals, however, there are separate receptors for insulin and IGF-1. The IR primarily controls metabolism, whereas the IGF1R controls growth (55). Despite the diversity in primary sequences of insulin, IGFs, and their receptors across species, most of the critical features are conserved. This conservation allows for allowing structural and function similarities across species. As a result, insulins and IGFs are able to bind to each other’s receptors, as do insulins and IGFs of different species, albeit with varying affinities, resulting in differences in biological potency (61). Indeed, it was because of the ability of many peptide hormones to be active across species lines that people with diabetes were able to initially be treated with insulins extracted from porcine and bovine pancreas, and when these were not available insulins from other mammals and even fish. During the last four decades, multiple laboratories have studied how insulin and IGF-1 bind to their receptors and signal, how these ligands control growth and metabolism (62–64), and how insulin signaling is altered in diabetes, obesity, and other insulin-resistant states (55).
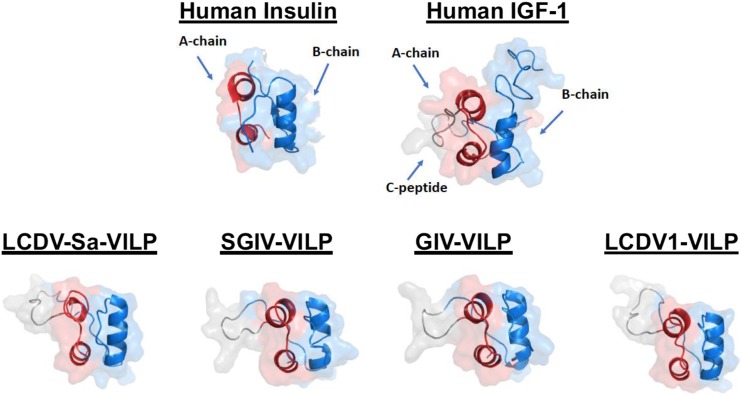
Using homology search of all viral sequences focusing on both primary sequence and conservation of the cysteine residues critical for intrachain and interchain disulfide bond formation, we identified a family of four VILPs, each encoded by a different member of the Iridoviridae family: (i) Lymphocystis disease virus (LCDV)-1, (ii) LCDV-Sa, (iii) Grouper iridovirus (GIV), and (iv) Singapore GIV (SGIV). Although these viruses were initially isolated from fish, we and others have identified the DNA sequences of these viruses in the human gut (9, 65, 66). The LCDV VILPs are ∼50% and SGIV/GIV VILPs are ∼40% identical to the A- and B-chain of human insulin and IGF-1. These VILPs were also predicted to have the structure of other members of the insulin/IGF-1 superfamily structure via the presence of highly conserved cysteine residues, which create two interchain and one intrachain disulfide bonds critical to formation of an insulin-like tertiary structure (Fig. 1). All of the VILPs also have potential signal peptide sequences that might allow secretion from infected cells.
Figure 1.
Predicted tertiary structure of VILPs and comparison with insulin and IGF-1. The structure is very well conserved among different VILPs. The A-chain is red; the B-chain is blue; the C-peptide is gray. The C-peptide of human IGF-1 is not cleaved, shown in gray. PyMOL (v1.8.6.2; Schrodinger, Inc., New York, NY) was used to make the illustrations. Predicted VILP structures were obtained using I-TASSER (Zhang Lab, University of Michigan, Ann Arbor, MI). For human insulin, see Protein Data Bank ID 2KQP; for human IGF-1, see Protein Data Bank ID 3LRI. T2D, type 2 diabetes.
Although both insulin and IGF-1 are synthesized as single-chain peptides, one of the main differences is the length and processing of their respective C-peptides. Insulin is synthesized as preproinsulin (110 amino acids) containing both the signal peptide and a C-peptide of 33 to 35 amino acids depending on the species. Following cleavage of the signal peptide, proinsulin is processed within the β-cell secretory granules by sequence cleavage of the C-peptide at both its N and C termini followed by removal of residual amino acids of the A- and B-chain to form mature insulin. In humans and other vertebrates, the mature hormone is a double-chained molecule composed of a 21–amino acid A-chain and a 30–amino acid B-chain that are held together with disulfide bonds. For IGF-1 and IGF-2, in contrast, the C-peptide is shorter (12 and 8 amino acids, respectively) and not cleaved, resulting in a single-chain molecule with three intrachain disulfide bonds. Additionally, both IGF-1 and IGF-2 have C-terminal extensions on the B-chain referred to as the D-domain and E-domain. All four VILPs have short C-peptides (10 amino acids for LCDV VILPs and 7 amino acids for GIV and SGIV), resembling IGFs. They also lack the typical conserved dibasic residues found at the ends of insulin C-peptide, suggesting that VILPs form a single-chain IGF-1–like structure, although this remains to be proven, because there are some potential basic amino acid cleavage sites in all VILPS. Although LCDV-1 and LCDV-Sa VILPs are composed of 80–amino acid residues resembling insulin’s domain structure, both GIV and SGIV VILPs also have the potential to encode C-terminal extensions similar to, or even longer than, the D and E regions in IGF-1; however, these show no sequence homology to those in human IGF-1.
For initial functional characterization, we chose to chemically synthesize these VILPs as single-chain peptides with the C-peptide intact and used murine preadipocyte cell lines in which the endogenous IR and IGF1R genes had been inactivated and the human IR or human IGF1R (hIGF1R) was expressed by stable transfection. Using these cells in a binding competition assay, we demonstrated that at least two of these VILPs can bind to hIGF1R with a better affinity than human insulin but with a lesser affinity than human IGF-1. The VILPs also competed for the human IR but were much weaker than human insulin and even tended to be weaker than human IGF-1 for this receptor. Consistent with this, all VILPs tested stimulated receptor autophosphorylation with higher potency on hIGF1R than on the human IR.
The PI3K/Akt pathway and the Ras-MAPK pathway are two major pathways downstream of IR and IGF1R, with the former regulating mainly metabolic effects and the latter regulating cell growth and differentiation (55, 67). Consistent with the binding data, all VILPs acting via hIGF1R strongly stimulated AKT and ERK phosphorylation. Some, when analyzed at a higher concentration and at later time points, also stimulated AKT and ERK phosphorylation in cells expressing only the inulin receptor, suggesting that VILPs may act through the IR but with delayed kinetics and at high concentrations compared with native insulin.
We also determined the ability of VILPs to increase glucose uptake using the differentiated murine 3T3-L1 white adipocyte cell line. All VILPs stimulated glucose uptake, albeit at lower potencies (twofold to threefold less) than insulin and IGF-1. Thus, despite the low binding affinity to the IR (∼3 orders of magnitude less than the human insulin), VILPs are able to stimulate biological function. This suggests that these peptides may bind to the receptors in some unique way compared with the mammalian hormones. This could also be due to 3T3-L1 cells responding through the IGF-1 receptor, because these cells express both insulin and IGF-1 receptors. Whether this occurs with normal fat cells remains to be determined.
Using human fibroblasts, we showed that GIV and LCDV-1 VILPs are strong mitogens, whereas SGIV was not able to stimulate DNA synthesis at the concentrations tested. Transfection of a cDNA for LCDV-1 viral insulin to the AML-12 mouse hepatocyte cell line also resulted in stimulation of the insulin/IGF-1 signaling pathways and increased DNA synthesis, suggesting that these cells can express, secrete, and respond to this viral ligand. Finally, we tested the in vivo function of VILPs by IP injection of LCDV-1 and SGIV VILPs into healthy male mice. Under these conditions, LCDV-1 significantly lowered blood glucose. The time course, however, appears slower and more prolonged (9), suggesting a long-lasting effect.
VILPs: Their Potential Roles for the Virus and in Infected Fish
All four VILPs identified thus far are in four different fish viruses (LCDV-1, LCDV-Sa, GIV, and SGIV), and all belong to the Iridoviridae family. Iridoviridae family viruses are large double-stranded DNA viruses that are known to infect poikilothermic vertebrates, such as reptiles, amphibians, and fish, as well as insects (68). Iridoviridae infections are common in saltwater commercially and ecologically important species of fish. In these species, these viruses target several organs, resulting in high levels of morbidity and mortality, and from a fish-farming perspective significant economic losses (69). The role of VILPs during the Iridoviridae infection remains unclear. Fish insulins are among the first vertebrate insulins isolated, with various forms encoded by multiple insulin genes and secreted by pancreas and extrapancreatic tissues such as adipose tissue, brain, gastrointestinal tract, and pituitary (70–72). The function of these insulins is mediated through the IR and involves multiple aspects of development, metabolism, growth, and feeding (73).
It has been reported that SGIV VILP is an early-transcribed gene during viral infection. SGIV VILP facilitates growth of grouper embryonic cells by promoting G1/S transition. This is associated with increases in SGIV replication (74). The IR is one of the most common receptors on the cell surface and a great viral target for attachment. Whether VILPs are on the surface of the virus and could bind to these receptors, however, remains unknown.
Micropinocytosis-induced macropinosome formation is driven by actin rearrangement, which is regulated by the PI3K/Akt/protein kinase C signaling pathway (75, 76). Fish insulins acting on IRs linked to the PI3K/Akt signaling pathway are not only essential for the metabolic and mitogenic action of insulin, but they may also stimulate the process of micropinocytosis (77, 78). SGIV VILP might hijack this endocytic pathway to facilitate its entry into grouper cells. GIV was implicated to make use of the Bcl-2 protein GIV66 to inhibit premature host cell apoptosis for its own proliferation and survival (79). IGF-1 has been reported to block cell apoptosis via inactivating Bcl-2–interacting mediator of cell death protein (80). GIV VILP might play a role similar to IGF-1 as an apoptosis inhibitor to facilitate GIV survival in host cells. It has also been reported that SGIV entered host grouper cells via clathrin-mediated endocytosis and micropinocytosis (81). Furthermore, LCDV-caused fish lymphocystis is characterized by hypertrophied cells and macroscopic nodules primarily on the epidermis (fins and skin), which may well be an IGF-1–like response. Insulin/IGF-1 signaling is well documented to control interfollicular morphogenesis and determine epidermal proliferative potential (82, 83). LCDV insulin/IGF-1 mimicry could promote fish epidermal cell hypertrophy through activation of insulin/IGF-1 signaling in host cells.
VILPs and Their Potential Link to Human Disease
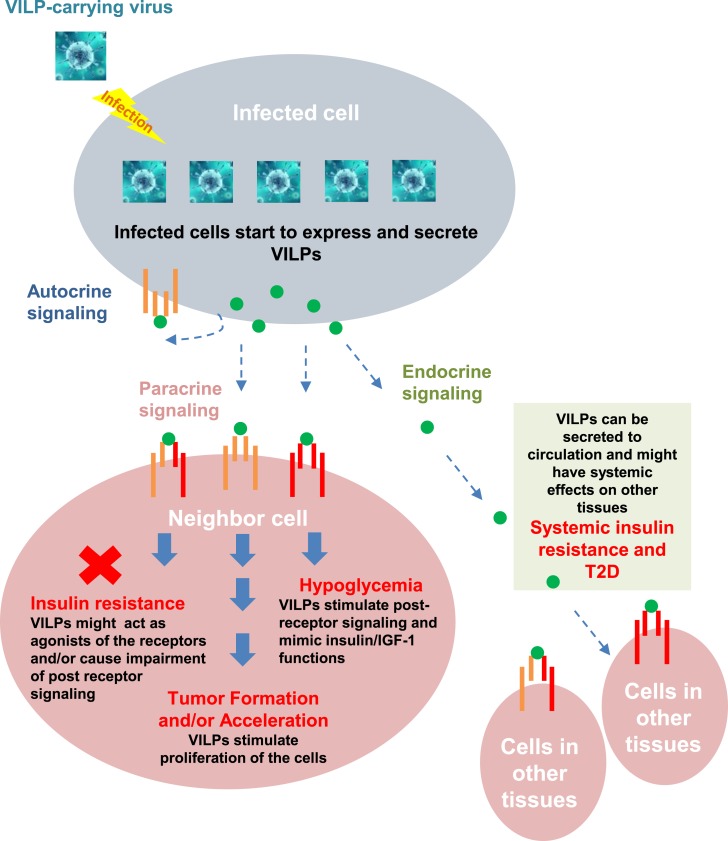
Although there are no reports on human infection by Iridoviridae members, reanalysis of published data on the human gut virome shows the presence of viral sequences of Iridoviridae known to have VILP sequences (9). Thus, two recent studies of the human fecal virome identified DNA sequences belonging to LCDV-1 and SGIV (65, 66). Moreover, LCDV-1 sequences have been identified in samples of human blood (84). These data support our hypothesis that humans are exposed to VILP-containing viruses. Because VILPs mimic human insulin/IGF-1 function and structure, they have potential to play an important role in human health and disease.
It is well established that insulin and IGF-1 are critical for maintaining metabolic homeostasis and normal development. Impaired insulin and IGF-1 secretion or action are important pathogenetic components of several human diseases. Insulin levels are elevated in insulin-resistant states, such as in type 2 diabetes and obesity (85), whereas insulin levels are remarkably decreased in type 1 diabetes (T1D) (86). Given the ability of VILPs to bind to insulin/IGF-1 receptors, VILPs could act as IR/IGFR agonists or, perhaps under some circumstances, antagonists. They can also compete with endogenous insulin/IGF-1 to bind the IR or IGF1R, and in the case of the former, could be involved in the pathogenesis of type 2 diabetes or hypoglycemia. The VILPs characterized in our study could also modify endogenous IGF-1 action, either stimulating cell growth or inhibiting IGF-1 and IGF-2 growth-promoting effects. Although much less potent than native insulin or IGF-1, it is possible that infected cells have very high concentrations of VILPs, which could act locally or at a distance. It is also possible that other viruses will be found to contain even more potent VILPs.
Another potential role of VILP-containing viruses in humans could include involvement in the pathogenesis of T1D. T1D is an autoimmune disease characterized by the immune-mediated destruction of the insulin-secreting β-cells in the pancreas. The development of T1D is preceded by months to years of lymphocytic infiltration of islets and T-cell–mediated β-cell destruction. CD8+ T-cells are the most abundant immune cell type in insulitis (87). Various environmental factors have been considered as the potential trigger of the autoimmunity, but the exact factor or factors remain unclear. Insulin is one of the earliest and main targets of the autoreactive T-cells in both nonobese diabetic mice and humans with T1D (86, 88). Indeed, >90% of the anti-insulin CD4 T-cell clones target amino acids 9 to 23 of the human insulin B-chain (B:9-23) in nonobese diabetic mice. B:9-23–specific T-cells are also present in patients with T1D (89). Proinsulin-specific (90, 91) CD8+ T-cells and CD4+ T-cells recognizing fusion proteins containing sequences of insulin and other pancreatic peptides (92) have also been identified and shown to be cytotoxic using in vitro models.
In terms of humoral immune response, an early marker of T1D autoimmunity is the development of four types of islet autoantibodies. Among them, insulin autoantibodies (IAAs) are the only autoantibodies specific to β-cells, and they are usually the first to be detected (93, 94). In humans, IAAs develop months to years before the onset of overt diabetes (95), and there is a significant correlation between IAA concentrations and the rate of progression to T1D (96). Molecular mimicry—that is, when the immune response to a foreign antigen targets a similar epitope as a host protein—is a potential cause of autoimmunity (97–99). This mechanism is based on the potential for some degree of degeneracy of T-cell recognition (100, 101). This could be either pathogenic (102, 103) or protective (104, 105). The molecular mimicry hypothesis gained ground in T1D research during the 1990s (106, 107). However, today there are very few studies investigating this concept. Our preliminary data and the literature (108) suggest that Iridoviridae members can infect mammalian cells and that humans are exposed to these viruses. Based on this evidence, we speculate that Iridoviridae or other viruses that produce VILPs can act as insulin mimics. These viruses could potentially stimulate or desensitize T-cells and thus contribute to the pathogenesis of T1D by triggering T-cell cross-reactions with endogenous insulin or protect against the disease. We are currently exploring these hypotheses.
In addition to mimicking or blocking insulin action, VILPs could act through IGF-1 receptors. IGF-1 and IGF-2 are mediators of GH action and are central in normal growth and development. IGFs may also act as paracrine or autocrine factors and affect tumor development through stimulating cell proliferation and inhibiting apoptosis (109). Endometrial cancer, pancreatic cancer, and colon cancer are among the cancers most strongly related to a history of diabetes and hyperinsulinemia (110). Elevated IGF-1 has also been shown to be related to prostate (111) and colorectal cancers (112). In addition to these correlations, in vitro and animal experiments show that IGF-1 may have a direct effect on tumor development by stimulating cellular proliferation and by inhibiting apoptosis (113, 114). Viruses play a key role in several human cancers, including liver cancer (hepatitis B and C viruses) and cervical cancer (human papillomavirus) (115, 116). Note that one of the most important signs of Iridoviridae infection in fish is the appearance of wart-like growths on the fins, skin, or gills that could be related to an IGF-1–like effect of VILPs on cell proliferation (117). In vitro VILPs stimulate DNA synthesis and growth of murine and human cell lines, demonstrating that VILPs are able to promote cell proliferation (9). Although we do not have any experimental evidence in cancer, our preliminary data suggest that VILPs are mitogens and may accelerate or stimulate tumor formation and growth in the infected region (Fig. 2).
Figure 2.
VILPs and their potential role in diabetes and cancer. VILPs bind to IR (red), IGF1R (orange), and hybrid receptors (red and orange) of targeted cells via autocrine, paracrine, endocrine, and other signaling to affect the pathogenesis of diabetes and cancer.
Other Viral Hormones: ET1
VILPs are only 1 of the 14 viral hormone–like peptides that we have identified through bioinformatics searches. The functions of most of the others have not been studied, and their roles in both viruses and humans remain largely unknown. For example, our analysis identified viral endothelin-1 in three viruses: two Deerpox viruses (W-848-83, W-1170-84) (118) and one Eptesipox virus (119). Poxviruses are known to encode peptides able to mimic host factors. They also infect a wide range of hosts, including primates (e.g., monkeypox) (120). Deerpox virus belongs to the Poxviridae family, and in deer causes keratoconjunctivitis, proliferative-ulcerative skin lesions, and dermatitis on facial and foot surfaces (121). Deer-associated parapoxviruses have also recently been shown to infect humans (122).
Human ET1 is a potent vasoconstrictor primarily secreted by endothelial cells. ET1 receptors are primarily found in smooth muscle and endothelial cells. ET1 binds to its receptors, leading to vasoconstriction and proinflammatory effects via cytokine production and recruitment of inflammatory cells in vascular smooth cells. In endothelial cells, ET1 regulates nitric oxide release and inhibits apoptosis (123, 124). Therefore, in humans, high ET1 levels are associated with pulmonary vascular, cardiovascular, airway disorders (i.e., hypertension, artery stiffness, asthma) and acute and chronic inflammatory diseases, including inflammatory lung disease (125). Viral endothelin-like molecules may play a role in these human diseases by functioning as an ET1 agonist or antagonist, interfering with normal host ET function. This remains to be determined.
Bacterial Hormone Mimetics
As early as the 1970s, Dr. Jesse Roth and colleagues suggested the presence of hormone-like molecules in bacteria (56). Although the initial studies suggested an insulin-like molecule in Escherichia coli, no DNA sequences resembling insulin have been found in these bacteria. Nonetheless, it remains possible that some molecule of a different structure could be produced in bacteria that could either mimic insulin or act as an antigen in the pathogenesis of T1D. Indeed, in recent work, Qiang et al. (126) have discovered a bacterial peptide in Escherichia coli that can mimic human melanocortin hormone and termed it melanocortin-like peptide of E. coli (MECO-1). MECO-1 is a 33–amino acid peptide present in the C terminus of E. coli elongation factor G. Although it has some sequence resemblance to human α-melanocyte-stimulating hormone (α-MSH) and ACTH, the similarity at the primary sequence level is <25%. Nonetheless, Qiang et al. (126) showed that MECO-1 exerts α-MSH’s anti-inflammatory function through the α-MSH receptor. Stimulation of downstream signaling inhibited the release of cytokines, and the anti-inflammatory effects of MECO-1 are similar to α-MSH both in vitro and in vivo. On cultured RAW 264.7 macrophages, MECO-1 can prevent TNF-α release in response to a wide range of proinflammatory agents, including lipopolysaccharide, Toll-like 3 receptor proinflammatory agent (poly:C), peptidoglycan (Toll-like 2 receptor activator), and high-mobility group protein B1 (HMGB-1; Toll-like 2 receptor and Toll-like 4 receptor activator). Anti-inflammatory effects of MECO-1 were also observed in human peripheral blood mononuclear cells. In this model, HMGB-1–induced TNF-α release was attenuated by MECO-1.
In vivo studies confirmed these in vitro observations and demonstrated that MECO-1 mimics α-MSH function. MECO-1 can rescue mice from death as caused by lipopolysaccharide administration or cecal ligation and puncture–induced polymicrobial sepsis. In a dextran-induced colitis mouse model, IP injection of MECO-1 was also shown to improve severe body weight loss in colitis mice, whereas administration of anti–MECO-1 antibody exacerbated colitis in mice. Interestingly, MECO-1–like structures were also identified in the C terminus of elongation factor G of other gut microbes, such as Bacteroides thetaiotamicron and Bacteroides fragilis. The gut microbiome is known for regulating inflammation (127), and the MECO-1–like peptides might contribute to anti-intestinal inflammation. As in the case of VILPs, the discovery of MECO-1 in E. coli raises several new questions regarding host–microbe interactions based on microbial hormones and indicates an unexplored niche in human gut microbiome research.
Concluding Remarks and Future Perspectives
The discovery and characterization of the first viral hormones expands our understanding of host–microbe interactions based on a new microbial mechanism: manipulation of the host endocrine system. In terms of viral hormone discovery, this may be the tip of the iceberg because the number of available viral genomes is still limited to ∼3% of predicted mammalian viruses (8852 viruses in the National Center for Biotechnology Information as of June 2019 out of an estimated 300,000 mammalian viruses) (128). Indeed, when we first searched for insulin mimics in 2015, we identified only three viruses (LCDV-1, GIV, and SGIV), and as the number of completed viral genome projects increased, we identified a fourth VILP in LCDV-Sa in 2016. As the body of virome research increases, along with the advancement of sequencing technology (particularly metagenomic sequencing), the decrease in sequencing costs, improvement in virome database annotation, and discovery of novel viruses, hormone-like molecules will be identified in more viruses. The genetic diversity and abundance of viruses and their ability to mimic human hormones suggest that humans are already exposed to unidentified viruses manipulating our endocrine system, and the link to human disease and health will be discovered in the future.
Thus far, our approach to identifying hormone mimics in viruses has been limited to a primary sequence search for a significant alignment (divergent evolution). Alternatively, viral (or bacterial) hormone-like peptides may also have evolved de novo from unrelated sequences in viral genomes with a structural or functional convergence with human hormones. In this case, the amino acid sequence homology between the viral mimic and the host peptide may be low, but structural and functional mimicry may persist. Recently, a bioinformatics analysis of the human gut metagenome revealed human gastrointestinal bacteria N-acryl amides structurally resembling ligands of G protein–coupled receptors that affect such receptors involved in GLP-1 secretion and glucose homeostasis (129). Moreover, using a structural bioinformatics approach, Drayman et al. (130) demonstrated that both viral and bacterial pathogens use ligand mimicry to recognize host ligand receptors. The primary sequence similarity of these microbial ligands and host ligands are low, but these microbial proteins structurally mimic host ligands and engage with their cognate host receptors. In total, four viral and two bacterial surface proteins were identified as ligand mimics of the different host receptors. Using polyomavirus SV40 capsid protein VP1 and the host ligand, the authors showed an SV40–receptor tyrosine kinase Axl interaction. This study is one of the first examples of structural screening for ligand mimics of pathogens to identify host–pathogen receptor interactions and suggests that there may be several other viral hormone mimics that will be missed in a primary sequence-based homology analysis.
In the past, endocrinologists had a very human-centric approach to studying human physiology. The new-generation data produced in our and other studies suggests that hormone ligands could also be found in “unexpected species.” In addition to the discovery of microbial hormones, a novel evolutionary approach in endocrinology can benefit from new discoveries and fuel drug development. The discovery of insulin-like molecules in fish-hunting cone snails (Conus geographus), as well as the use of these venomous insulins for capture of prey, is a good example of how this novel interspecies evolutionary approach can be used. Cone snail insulins were shown to act on the human IR, activate downstream insulin signaling, and reverse blood glucose levels in both zebrafish and mouse diabetes models. Most importantly, cone snail insulins lack the terminal octopeptide of the B-chain and do not form hexamers, as insulin does. Therefore, the new insulin is absorbed faster from subcutaneous injection and could give clues for new fast-acting insulins for diabetes treatment (131–133). This has some analogy to the discovery of exendin-4 in Gila monster (Heloderma suspectum) venom. Exendin-4 is a potent binder of the human GLP-1 receptor and mimics GLP-1 function on increasing insulin production, reducing blood glucose, and improving β-cell glucose sensitivity (134, 135). Importantly, unlike human GLP-1, which has a very short half-life (∼2 minutes) in plasma, exendin-4 has a half-life of ∼2.4 hours, and clinically its effect can last for up to 8 hours (136). Therefore, exendin-4 became the first GLP-1 analog used as an antidiabetic therapy (135). The above examples suggest that evolution uses hormones for different goals, and continuing to discover hormone ligands in “unexpected” species could help move development of therapeutic drugs forward.
Viruses and bacteria have long been viewed as pathogens. The development of virome and microbiome research furthers our understanding of microbe–human interactions and shows that microbiota are able to impact host immunity, metabolism, and behavior via modulation of host hormonal activities and signaling (137–140). The use of fecal microbiota transplantation is becoming a novel approach to cure metabolic syndrome. For instance, gut microbial transplantation from lean donors to recipient patients with metabolic disorders are being explored as a novel method to improve insulin sensitivity or induce weight loss (141). An interesting example of gut microbiome–endocrine crosstalk was recently reported using germ-free mice colonized with conventional specific pathogen-free gut microbiota. Yan et al. (142) showed that serum IGF-1 levels are increased with colonization in germ-free mice as a result of IGF-1 production in liver and adipose tissue. Likewise, vancomycin treatment of conventional mice reduced serum IGF-1 levels, suggesting that the gut microbiome regulates IGF-1 production. If this relates to a bacterially produced stimulator of IGF-1 or to more general effects of nutrition on IGF-1 secretion remains to be determined.
Among all of the research studying the correlation between microbiota and host hormone activities, the underlying mechanisms remain largely unknown. As our understanding deepens regarding the role that the human microbiota plays in health and disease, alongside discoveries of novel microbial hormone mimics (e.g., VILPs, MECO-1), a window is opening into novel mechanisms of host–microbiome crosstalk and their effects on the endocrine system. Further studies are required to determine the biological activities of the hormone-like mimetics and their roles in human health and disease. These studies will provide new targets for human disease prevention and treatment. Studying the functions of these diseases will further advance our understanding of the physiological relevance of viral mimics in human health and disease.
Acknowledgments
We thank Dr. Marie Holm Solheim for help with the initial structural modeling of VILPs and Fig. 1.
Financial Support: This work was supported by National Institutes of Health Grants 1K01DK117967-01 (to E.A.) and R01DK031026 and R01DK033201 (to C.R.K.).
Glossary
Abbreviations:
- ET1
endothelin-1
- GIV
Grouper iridovirus
- hIGF1R
human IGF-1 receptor
- HMGB-1
high-mobility group protein B1
- IAA
insulin autoantibody
- IGF1R
IGF-1 receptor
- IR
insulin receptor
- LCDV
Lymphocystis disease virus
- MECO-1
melanocortin-like peptide of E. coli
- PI3K
phosphatidylinositol 3-kinase
- SGIV
Singapore grouper iridovirus
- T1D
type 1 diabetes
- VILP
viral insulin/IGF-1–like peptide
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data Availability:
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References and Notes
- 1. Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. 2000;18(1):861–926. [DOI] [PubMed] [Google Scholar]
- 2. Nash P, Barrett J, Cao JX, Hota-Mitchell S, Lalani AS, Everett H, Xu XM, Robichaud J, Hnatiuk S, Ainslie C, Seet BT, McFadden G. Immunomodulation by viruses: the myxoma virus story. Immunol Rev. 1999;168(1):103–120. [DOI] [PubMed] [Google Scholar]
- 3. Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. 2003;3(1):36–50. [DOI] [PubMed] [Google Scholar]
- 4. Amara A, Mercer J. Viral apoptotic mimicry. Nat Rev Microbiol. 2015;13(8):461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat Rev Microbiol. 2009;7(11):787–797. [DOI] [PubMed] [Google Scholar]
- 6. Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457(7228):485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franzosa EA, Xia Y. Structural principles within the human-virus protein-protein interaction network. Proc Natl Acad Sci USA. 2011;108(26):10538–10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kropp KA, Angulo A, Ghazal P. Viral enhancer mimicry of host innate-immune promoters. PLoS Pathog. 2014;10(2):e1003804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Altindis E, Cai W, Sakaguchi M, Zhang F, GuoXiao W, Liu F, De Meyts P, Gelfanov V, Pan H, DiMarchi R, Kahn CR. Viral insulin-like peptides activate human insulin and IGF-1 receptor signaling: a paradigm shift for host–microbe interactions. Proc Natl Acad Sci USA. 2018;115(10):2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buller RM, Chakrabarti S, Cooper JA, Twardzik DR, Moss B. Deletion of the vaccinia virus growth factor gene reduces virus virulence. J Virol. 1988;62(3):866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Verschuren EW, Jones N, Evan GI. The cell cycle and how it is steered by Kaposi’s sarcoma-associated herpesvirus cyclin. J Gen Virol. 2004;85(Pt 6):1347–1361. [DOI] [PubMed] [Google Scholar]
- 12. Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320(5877):797–799. [DOI] [PubMed] [Google Scholar]
- 13. Dunn EF, Connor JH. HijAkt: the PI3K/Akt pathway in virus replication and pathogenesis. Prog Mol Biol Transl Sci. 2012;106:223–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bellacosa A, Testa JR, Staal SP, Tsichlis PN. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science. 1991;254(5029):274–277. [DOI] [PubMed] [Google Scholar]
- 15. Bellacosa A, Franke TF, Gonzalez-Portal ME, Datta K, Taguchi T, Gardner J, Cheng JQ, Testa JR, Tsichlis PN. Structure, expression and chromosomal mapping of c-akt: relationship to v-akt and its implications. Oncogene. 1993;8(3):745–754. [PubMed] [Google Scholar]
- 16. Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci USA. 2000;97(4):1695–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spencer JV, Lockridge KM, Barry PA, Lin G, Tsang M, Penfold ME, Schall TJ. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J Virol. 2002;76(3):1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aoki Y, Jaffe ES, Chang Y, Jones K, Teruya-Feldstein J, Moore PS, Tosato G. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93(12):4034–4043. [PubMed] [Google Scholar]
- 19. Yao Z, Fanslow WC, Seldin MF, Rousseau AM, Painter SL, Comeau MR, Cohen JI, Spriggs MK. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3(6):811–821. [DOI] [PubMed] [Google Scholar]
- 20. Smith CA, Davis T, Wignall JM, Din WS, Farrah T, Upton C, McFadden G, Goodwin RG. T2 open reading frame from the Shope fibroma virus encodes a soluble form of the TNF receptor. Biochem Biophys Res Commun. 1991;176(1):335–342. [DOI] [PubMed] [Google Scholar]
- 21. Benedict CA, Butrovich KD, Lurain NS, Corbeil J, Rooney I, Schneider P, Tschopp J, Ware CF. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J Immunol. 1999;162(12):6967–6970. [PubMed] [Google Scholar]
- 22. Tidona CA, Darai G. The complete DNA sequence of lymphocystis disease virus. Virology. 1997;230(2):207–216. [DOI] [PubMed] [Google Scholar]
- 23. Alcamí A, Smith GL. A soluble receptor for interleukin-1β encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell. 1992;71(1):153–167. [DOI] [PubMed] [Google Scholar]
- 24. Spriggs MK, Hruby DE, Maliszewski CR, Pickup DJ, Sims JE, Buller RM, VanSlyke J. Vaccinia and cowpox viruses encode a novel secreted interleukin-1-binding protein. Cell. 1992;71(1):145–152. [DOI] [PubMed] [Google Scholar]
- 25. Upton C, Mossman K, McFadden G. Encoding of a homolog of the IFN-gamma receptor by myxoma virus. Science. 1992;258(5086):1369–1372. [DOI] [PubMed] [Google Scholar]
- 26. Boshoff C, Endo Y, Collins PD, Takeuchi Y, Reeves JD, Schweickart VL, Siani MA, Sasaki T, Williams TJ, Gray PW, Moore PS, Chang Y, Weiss RA. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278(5336):290–294. [DOI] [PubMed] [Google Scholar]
- 27. Kledal TN, Rosenkilde MM, Coulin F, Simmons G, Johnsen AH, Alouani S, Power CA, Lüttichau HR, Gerstoft J, Clapham PR, Clark-Lewis I, Wells TN, Schwartz TW. A broad-spectrum chemokine antagonist encoded by Kaposi’s sarcoma-associated herpesvirus. Science. 1997;277(5332):1656–1659. [DOI] [PubMed] [Google Scholar]
- 28. Zou P, Isegawa Y, Nakano K, Haque M, Horiguchi Y, Yamanishi K. Human herpesvirus 6 open reading frame U83 encodes a functional chemokine. J Virol. 1999;73(7):5926–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lüttichau HR, Stine J, Boesen TP, Johnsen AH, Chantry D, Gerstoft J, Schwartz TW. A highly selective CC chemokine receptor (CCR)8 antagonist encoded by the poxvirus molluscum contagiosum. J Exp Med. 2000;191(1):171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saederup N, Lin YC, Dairaghi DJ, Schall TJ, Mocarski ES. Cytomegalovirus-encoded β chemokine promotes monocyte-associated viremia in the host. Proc Natl Acad Sci USA. 1999;96(19):10881–10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Penfold ME, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. Cytomegalovirus encodes a potent α chemokine. Proc Natl Acad Sci USA. 1999;96(17):9839–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn MC, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385(6614):347–350. [DOI] [PubMed] [Google Scholar]
- 33. Waldhoer M, Kledal TN, Farrell H, Schwartz TW. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J Virol. 2002;76(16):8161–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lalani AS, Barrett JW, McFadden G. Modulating chemokines: more lessons from viruses. Immunol Today. 2000;21(2):100–106. [DOI] [PubMed] [Google Scholar]
- 35. Milne RS, Mattick C, Nicholson L, Devaraj P, Alcami A, Gompels UA. RANTES binding and down-regulation by a novel human herpesvirus-6β chemokine receptor. J Immunol. 2000;164(5):2396–2404. [DOI] [PubMed] [Google Scholar]
- 36. Murphy PM. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat Immunol. 2001;2(2):116–122. [DOI] [PubMed] [Google Scholar]
- 37. Tulman ER, Afonso CL, Lu Z, Zsak L, Kutish GF, Rock DL. Genome of lumpy skin disease virus. J Virol. 2001;75(15):7122–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mossman K, Nation P, Macen J, Garbutt M, Lucas A, McFadden G. Myxoma virus M-T7, a secreted homolog of the interferon-γ receptor, is a critical virulence factor for the development of myxomatosis in European rabbits. Virology. 1996;215(1):17–30. [DOI] [PubMed] [Google Scholar]
- 39. Graham KA, Lalani AS, Macen JL, Ness TL, Barry M, Liu LY, Lucas A, Clark-Lewis I, Moyer RW, McFadden G. The T1/35kDa family of poxvirus-secreted proteins bind chemokines and modulate leukocyte influx into virus-infected tissues. Virology. 1997;229(1):12–24. [DOI] [PubMed] [Google Scholar]
- 40. van Berkel V, Barrett J, Tiffany HL, Fremont DH, Murphy PM, McFadden G, Speck SH, Virgin HW IV. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J Virol. 2000;74(15):6741–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kotwal GJ, Isaacs SN, McKenzie R, Frank MM, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250(4982):827–830. [DOI] [PubMed] [Google Scholar]
- 42. McKenzie R, Kotwal GJ, Moss B, Hammer CH, Frank MM. Regulation of complement activity by vaccinia virus complement-control protein. J Infect Dis. 1992;166(6):1245–1250. [DOI] [PubMed] [Google Scholar]
- 43. Senkevich TG, Bugert JJ, Sisler JR, Koonin EV, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273(5276):813–816. [DOI] [PubMed] [Google Scholar]
- 44. Cheng EH, Nicholas J, Bellows DS, Hayward GS, Guo HG, Reitz MS, Hardwick JMA. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94(2):690–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Telford EA, Watson MS, Aird HC, Perry J, Davison AJ. The DNA sequence of equine herpesvirus 2. J Mol Biol. 1995;249(3):520–528. [DOI] [PubMed] [Google Scholar]
- 46. Bhattacharyya S, Zagórska A, Lew ED, Shrestha B, Rothlin CV, Naughton J, Diamond MS, Lemke G, Young JA. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe. 2013;14(2):136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jemielity S, Wang JJ, Chan YK, Ahmed AA, Li W, Monahan S, Bu X, Farzan M, Freeman GJ, Umetsu DT, Dekruyff RH, Choe H. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 2013;9(3):e1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sandersfeld LM, Quinn K, Weller M, McCray PB Jr, Chiorini J, Maury W. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci USA. 2011;108(20):8426–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moller-Tank S, Kondratowicz AS, Davey RA, Rennert PD, Maury W. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J Virol. 2013;87(15):8327–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen X, Xun K, Chen L, Wang Y. TNF-α, a potent lipid metabolism regulator. Cell Biochem Funct. 2009;27(7):407–416. [DOI] [PubMed] [Google Scholar]
- 51. Ghanemi A, St-Amand J. Interleukin-6 as a “metabolic hormone.” Cytokine. 2018;112:132–136. [DOI] [PubMed] [Google Scholar]
- 52. Párrizas M, Saltiel AR, LeRoith D. Insulin-like growth factor 1 inhibits apoptosis using the phosphatidylinositol 3′-kinase and mitogen-activated protein kinase pathways. J Biol Chem. 1997;272(1):154–161. [DOI] [PubMed] [Google Scholar]
- 53. Kooijman R. Regulation of apoptosis by insulin-like growth factor (IGF)-I. Cytokine Growth Factor Rev. 2006;17(4):305–323. [DOI] [PubMed] [Google Scholar]
- 54. Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6(1):a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Le Roith D, Shiloach J, Heffron R, Rubinovitz C, Tanenbaum R, Roth J. Insulin-related material in microbes: similarities and differences from mammalian insulins. Can J Biochem Cell Biol. 1985;63(8):839–849. [DOI] [PubMed] [Google Scholar]
- 57. Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 1995;14(14):3373–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nagasawa H, Kataoka H, Isogai A, Tamura S, Suzuki A, Ishizaki H, Mizoguchi A, Fujiwara Y, Suzuki A. Amino-terminal amino acid sequence of the silkworm prothoracicotropic hormone: homology with insulin. Science. 1984;226(4680):1344–1345. [DOI] [PubMed] [Google Scholar]
- 59. Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, Liu LX, Doberstein SK, Ruvkun G. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15(6):672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van Heemst D. Insulin, IGF-1 and longevity. Aging Dis. 2010;1(2):147–157. [PMC free article] [PubMed] [Google Scholar]
- 61. Muggeo M, Ginsberg BH, Roth J, Neville DM Jr, De Meyts P, Kahn CR. The insulin receptor in vertebrates is functionally more conserved during evolution than insulin itself. Endocrinology. 1979;104(5):1393–1402. [DOI] [PubMed] [Google Scholar]
- 62. Boucher J, Charalambous M, Zarse K, Mori MA, Kleinridders A, Ristow M, Ferguson-Smith AC, Kahn CR. Insulin and insulin-like growth factor 1 receptors are required for normal expression of imprinted genes. Proc Natl Acad Sci USA. 2014;111(40):14512–14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. O’Neill BT, Lauritzen HP, Hirshman MF, Smyth G, Goodyear LJ, Kahn CR. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell Reports. 2015;11(8):1220–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emanuelli B, Vienberg SG, Smyth G, Cheng C, Stanford KI, Arumugam M, Michael MD, Adams AC, Kharitonenkov A, Kahn CR. Interplay between FGF21 and insulin action in the liver regulates metabolism. J Clin Invest. 2014;124(2):515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, Rodriguez-Brito B, Salamon P, Rohwer F. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159(5):367–373. [DOI] [PubMed] [Google Scholar]
- 67. Boucher J, Tseng YH, Kahn CR. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J Biol Chem. 2010;285(22):17235–17245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chinchar VG, Hick P, Ince IA, Jancovich JK, Marschang R, Qin Q, Subramaniam K, Waltzek TB, Whittington R, Williams T, Zhang QY; Ictv Report Consortium. ICTV virus taxonomy profile: Iridoviridae. J Gen Virol. 2017;98(5):890–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chinchar VG, Waltzek TB, Subramaniam K. Ranaviruses and other members of the family Iridoviridae: their place in the virosphere. Virology. 2017;511:259–271. [DOI] [PubMed] [Google Scholar]
- 70. Youson JH, Al-Mahrouki AA. Ontogenetic and phylogenetic development of the endocrine pancreas (islet organ) in fish. Gen Comp Endocrinol. 1999;116(3):303–335. [DOI] [PubMed] [Google Scholar]
- 71. Caruso MA, Blaufuss PC, Kittilson JD, Raine J, Sheridan MA. Isolation and characterization of a mRNA encoding a novel insulin receptor (IR) subtype, IR2, from rainbow trout (Oncorhynchus mykiss) and patterns of expression of the four IR subtypes, IR1–IR4, in tissues and during embryonic development. Gen Comp Endocrinol. 2010;169(3):258–268. [DOI] [PubMed] [Google Scholar]
- 72. Hrytsenko O, Wright JR Jr, Morrison CM, Pohajdak B. Insulin expression in the brain and pituitary cells of tilapia (Oreochromis niloticus). Brain Res. 2007;1135(1):31–40. [DOI] [PubMed] [Google Scholar]
- 73. Hernández-Sánchez C, Mansilla A, de Pablo F, Zardoya R. Evolution of the insulin receptor family and receptor isoform expression in vertebrates. Mol Biol Evol. 2008;25(6):1043–1053. [DOI] [PubMed] [Google Scholar]
- 74. Yan Y, Cui H, Guo C, Li J, Huang X, Wei J, Qin Q. An insulin-like growth factor homologue of Singapore grouper iridovirus modulates cell proliferation, apoptosis and enhances viral replication. J Gen Virol. 2013;94(Pt 12):2759–2770. [DOI] [PubMed] [Google Scholar]
- 75. Haga Y, Miwa N, Jahangeer S, Okada T, Nakamura S. CtBP1/BARS is an activator of phospholipase D1 necessary for agonist-induced macropinocytosis. EMBO J. 2009;28(9):1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6(9):e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cuatrecasas P. Interaction of insulin with the cell membrane: the primary action of insulin. Proc Natl Acad Sci USA. 1969;63(2):450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bickel PE. Lipid rafts and insulin signaling. Am J Physiol Endocrinol Metab. 2002;282(1):E1–E10. [DOI] [PubMed] [Google Scholar]
- 79. Banjara S, Mao J, Ryan TM, Caria S, Kvansakul M. Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim. J Biol Chem. 2018;293(15):5464–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Linseman DA, Phelps RA, Bouchard RJ, Le SS, Laessig TA, McClure ML, Heidenreich KA. Insulin-like growth factor-I blocks Bcl-2 interacting mediator of cell death (Bim) induction and intrinsic death signaling in cerebellar granule neurons. J Neurosci. 2002;22(21):9287–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang S, Huang X, Huang Y, Hao X, Xu H, Cai M, Wang H, Qin Q. Entry of a novel marine DNA virus, Singapore grouper iridovirus, into host cells occurs via clathrin-mediated endocytosis and macropinocytosis in a pH-dependent manner. J Virol. 2014;88(22):13047–13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stachelscheid H, Ibrahim H, Koch L, Schmitz A, Tscharntke M, Wunderlich FT, Scott J, Michels C, Wickenhauser C, Haase I, Brüning JC, Niessen CM. Epidermal insulin/IGF-1 signalling control interfollicular morphogenesis and proliferative potential through Rac activation. EMBO J. 2008;27(15):2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Edmondson SR, Thumiger SP, Werther GA, Wraight CJ. Epidermal homeostasis: the role of the growth hormone and insulin-like growth factor systems. Endocr Rev. 2003;24(6):737–764. [DOI] [PubMed] [Google Scholar]
- 84. Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS One. 2014;9(8):e105221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. [DOI] [PubMed] [Google Scholar]
- 86. Nakayama M. Insulin as a key autoantigen in the development of type 1 diabetes. Diabetes Metab Res Rev. 2011;27(8):773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pugliese A. Autoreactive T cells in type 1 diabetes. J Clin Invest. 2017;127(8):2881–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jasinski JM, Eisenbarth GS. Insulin as a primary autoantigen for type 1A diabetes. Clin Dev Immunol. 2005;12(3):181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Michels AW, Landry LG, McDaniel KA, Yu L, Campbell-Thompson M, Kwok WW, Jones KL, Gottlieb PA, Kappler JW, Tang Q, Roep BO, Atkinson MA, Mathews CE, Nakayama M. Islet-Derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2017;66(3):722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kronenberg D, Knight RR, Estorninho M, Ellis RJ, Kester MG, de Ru A, Eichmann M, Huang GC, Powrie J, Dayan CM, Skowera A, van Veelen PA, Peakman M. Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill β-cells. Diabetes. 2012;61(7):1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Abreu JR, Martina S, Verrijn Stuart AA, Fillié YE, Franken KL, Drijfhout JW, Roep BO. CD8 T cell autoreactivity to preproinsulin epitopes with very low human leucocyte antigen class I binding affinity. Clin Exp Immunol. 2012;170(1):57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Delong T, Wiles TA, Baker RL, Bradley B, Barbour G, Reisdorph R, Armstrong M, Powell RL, Reisdorph N, Kumar N, Elso CM, DeNicola M, Bottino R, Powers AC, Harlan DM, Kent SC, Mannering SI, Haskins K. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351(6274):711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang L, Nakayama M, Eisenbarth GS. Insulin as an autoantigen in NOD/human diabetes. Curr Opin Immunol. 2008;20(1):111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Alleva DG, Crowe PD, Jin L, Kwok WW, Ling N, Gottschalk M, Conlon PJ, Gottlieb PA, Putnam AL, Gaur A. A disease-associated cellular immune response in type 1 diabetics to an immunodominant epitope of insulin. J Clin Invest. 2001;107(2):173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Palmer JP, Asplin CM, Clemons P, Lyen K, Tatpati O, Raghu PK, Paquette TL. Insulin antibodies in insulin-dependent diabetics before insulin treatment. Science. 1983;222(4630):1337–1339. [DOI] [PubMed] [Google Scholar]
- 96. Steck AK, Johnson K, Barriga KJ, Miao D, Yu L, Hutton JC, Eisenbarth GS, Rewers MJ. Age of islet autoantibody appearance and mean levels of insulin, but not GAD or IA-2 autoantibodies, predict age of diagnosis of type 1 diabetes: diabetes autoimmunity study in the young. Diabetes Care. 2011;34(6):1397–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Christen U, Hintermann E, Holdener M, von Herrath MG. Viral triggers for autoimmunity: is the “glass of molecular mimicry” half full or half empty? J Autoimmun. 2010;34(1):38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cusick MF, Libbey JE, Fujinami RS. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol. 2012;42(1):102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cunningham MW, Antone SM, Smart M, Liu R, Kosanke S. Molecular analysis of human cardiac myosin-cross-reactive B- and T-cell epitopes of the group A streptococcal M5 protein. Infect Immun. 1997;65(9):3913–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today. 1998;19(9):395–404. [DOI] [PubMed] [Google Scholar]
- 101. Calis JJ, de Boer RJ, Keşmir C. Degenerate T-cell recognition of peptides on MHC molecules creates large holes in the T-cell repertoire. PLOS Comput Biol. 2012;8(3):e1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gautam AM, Liblau R, Chelvanayagam G, Steinman L, Boston T. A viral peptide with limited homology to a self peptide can induce clinical signs of experimental autoimmune encephalomyelitis. J Immunol. 1998;161(1):60–64. [PubMed] [Google Scholar]
- 103. Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80(5):695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ruiz PJ, Garren H, Hirschberg DL, Langer-Gould AM, Levite M, Karpuj MV, Southwood S, Sette A, Conlon P, Steinman L. Microbial epitopes act as altered peptide ligands to prevent experimental autoimmune encephalomyelitis. J Exp Med. 1999;189(8):1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mañá P, Goodyear M, Bernard C, Tomioka R, Freire-Garabal M, Liñares D. Tolerance induction by molecular mimicry: prevention and suppression of experimental autoimmune encephalomyelitis with the milk protein butyrophilin. Int Immunol. 2004;16(3):489–499. [DOI] [PubMed] [Google Scholar]
- 106. Atkinson MA, Bowman MA, Campbell L, Darrow BL, Kaufman DL, Maclaren NK. Cellular immunity to a determinant common to glutamate decarboxylase and coxsackie virus in insulin-dependent diabetes. J Clin Invest. 1994;94(5):2125–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kaufman DL, Erlander MG, Clare-Salzler M, Atkinson MA, Maclaren NK, Tobin AJ. Autoimmunity to two forms of glutamate decarboxylase in insulin-dependent diabetes mellitus. J Clin Invest. 1992;89(1):283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ahlers LR, Bastos RG, Hiroyasu A, Goodman AG. Invertebrate iridescent virus 6, a DNA virus, stimulates a mammalian innate immune response through RIG-I-like receptors. PLoS One. 2016;11(11):e0166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Moschos SJ, Mantzoros CS. The role of the IGF system in cancer: from basic to clinical studies and clinical applications. Oncology. 2002;63(4):317–332. [DOI] [PubMed] [Google Scholar]
- 110. Coughlin SS, Calle EE, Teras LR, Petrelli J, Thun MJ. Diabetes mellitus as a predictor of cancer mortality in a large cohort of US adults. Am J Epidemiol. 2004;159(12):1160–1167. [DOI] [PubMed] [Google Scholar]
- 111. Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, Hennekens CH, Pollak M. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279(5350):563–566. [DOI] [PubMed] [Google Scholar]
- 112. Giovannucci E. Insulin, insulin-like growth factors and colon cancer: a review of the evidence. J Nutr. 2001;131(11Suppl):3109S–3120S. [DOI] [PubMed] [Google Scholar]
- 113. Warren RS, Yuan H, Matli MR, Ferrara N, Donner DB. Induction of vascular endothelial growth factor by insulin-like growth factor 1 in colorectal carcinoma. J Biol Chem. 1996;271(46):29483–29488. [DOI] [PubMed] [Google Scholar]
- 114. Brahmkhatri VP, Prasanna C, Atreya HS. Insulin-like growth factor system in cancer: novel targeted therapies. BioMed Res Int. 2015;2015:538019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fallot G, Neuveut C, Buendia MA. Diverse roles of hepatitis B virus in liver cancer. Curr Opin Virol. 2012;2(4):467–473. [DOI] [PubMed] [Google Scholar]
- 116. Lowy DR, Schiller JT. Reducing HPV-associated cancer globally. Cancer Prev Res (Phila). 2012;5(1):18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Williams T, Barbosa-Solomieu V, Chinchar VG. A decade of advances in iridovirus research. Adv Virus Res. 2005;65:173–248. [DOI] [PubMed] [Google Scholar]
- 118. Afonso CL, Delhon G, Tulman ER, Lu Z, Zsak A, Becerra VM, Zsak L, Kutish GF, Rock DL. Genome of deerpox virus. J Virol. 2005;79(2):966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tu SL, Nakazawa Y, Gao J, Wilkins K, Gallardo-Romero N, Li Y, Emerson GL, Carroll DS, Upton C. Characterization of Eptesipoxvirus, a novel poxvirus from a microchiropteran bat. Virus Genes. 2017;53(6):856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Oliveira GP, Rodrigues RAL, Lima MT, Drumond BP, Abrahão JS. Poxvirus host range genes and virus–host spectrum: a critical review. Viruses. 2017;9(11):E331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Williams ES, Becerra VM, Thorne ET, Graham TJ, Owens MJ, Nunamaker CE. Spontaneous poxviral dermatitis and keratoconjunctivitis in free-ranging mule deer (Odocoileus hemionus) in Wyoming. J Wildl Dis. 1985;21(4):430–433. [DOI] [PubMed] [Google Scholar]
- 122. Roess AA, Galan A, Kitces E, Li Y, Zhao H, Paddock CD, Adem P, Goldsmith CS, Miller D, Reynolds MG, Zaki SR, Damon IK. Novel deer-associated parapoxvirus infection in deer hunters. N Engl J Med. 2010;363(27):2621–2627. [DOI] [PubMed] [Google Scholar]
- 123. Alonso D, Radomski MW. The nitric oxide-endothelin-1 connection. Heart Fail Rev. 2003;8(1):107–115. [DOI] [PubMed] [Google Scholar]
- 124. Davenport AP, Hyndman KA, Dhaun N, Southan C, Kohan DE, Pollock JS, Pollock DM, Webb DJ, Maguire JJ. Endothelin. Pharmacol Rev. 2016;68(2):357–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Goraca A. New views on the role of endothelin (minireview). Endocr Regul. 2002;36(4):161–167. [PubMed] [Google Scholar]
- 126. Qiang X, Liotta AS, Shiloach J, Gutierrez JC, Wang H, Ochani M, Ochani K, Yang H, Rabin A, LeRoith D, Lesniak MA, Böhm M, Maaser C, Kannengiesser K, Donowitz M, Rabizadeh S, Czura CJ, Tracey KJ, Westlake M, Zarfeshani A, Mehdi SF, Danoff A, Ge X, Sanyal S, Schwartz GJ, Roth J. New melanocortin-like peptide of E. coli can suppress inflammation via the mammalian melanocortin-1 receptor (MC1R): possible endocrine-like function for microbes of the gut. NPJ Biofilms Microbiomes. 2017;3(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JA, Daszak P, Lipkin WI. A strategy to estimate unknown viral diversity in mammals. MBio. 2013;4(5):e00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, Chu J, Vila-Farres X, Kaplitt J, Rogoz A, Calle PY, Hunter C, Bitok JK, Brady SF. Commensal bacteria make GPCR ligands that mimic human signalling molecules [published correction appears in Nature. 2018;556(7699):135]. Nature. 2017;549(7670):48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Drayman N, Glick Y, Ben-nun-shaul O, Zer H, Zlotnick A, Gerber D, Schueler-Furman O, Oppenheim A. Pathogens use structural mimicry of native host ligands as a mechanism for host receptor engagement. Cell Host Microbe. 2013;14(1):63–73. [DOI] [PubMed] [Google Scholar]
- 131. Ahorukomeye P, Disotuar MM, Gajewiak J, Karanth S, Watkins M, Robinson SD, Flórez Salcedo P, Smith NA, Smith BJ, Schlegel A, Forbes BE, Olivera B, Hung-Chieh Chou D, Safavi-Hemami H. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. eLife. 2019;8:e41574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Robinson SD, Safavi-Hemami H. Insulin as a weapon. Toxicon. 2016;123:56–61. [DOI] [PubMed] [Google Scholar]
- 133. Menting JG, Gajewiak J, MacRaild CA, Chou DH, Disotuar MM, Smith NA, Miller C, Erchegyi J, Rivier JE, Olivera BM, Forbes BE, Smith BJ, Norton RS, Safavi-Hemami H, Lawrence MC. A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin. Nat Struct Mol Biol. 2016;23(10):916–920. [DOI] [PubMed] [Google Scholar]
- 134. Göke R, Fehmann HC, Linn T, Schmidt H, Krause M, Eng J, Göke B. Exendin-4 is a high potency agonist and truncated exendin-(9–39)-amide an antagonist at the glucagon-like peptide 1-(7–36)-amide receptor of insulin-secreting beta-cells. J Biol Chem. 1993;268(26):19650–19655. [PubMed] [Google Scholar]
- 135. Thorens B, Porret A, Bühler L, Deng SP, Morel P, Widmann C. Cloning and functional expression of the human islet GLP-1 receptor. Demonstration that exendin-4 is an agonist and exendin-(9–39) an antagonist of the receptor. Diabetes. 1993;42(11):1678–1682. [DOI] [PubMed] [Google Scholar]
- 136. Kim D, MacConell L, Zhuang D, Kothare PA, Trautmann M, Fineman M, Taylor K. Effects of once-weekly dosing of a long-acting release formulation of exenatide on glucose control and body weight in subjects with type 2 diabetes. Diabetes Care. 2007;30(6):1487–1493. [DOI] [PubMed] [Google Scholar]
- 137. Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev. 2015;39(4):509–521. [DOI] [PubMed] [Google Scholar]
- 138. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. [DOI] [PubMed] [Google Scholar]
- 139. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. [DOI] [PubMed] [Google Scholar]
- 140. Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome–gut–brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18(6):666–673. [DOI] [PubMed] [Google Scholar]
- 141. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916.e7. [DOI] [PubMed] [Google Scholar]
- 142. Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, Sartor BR, Aliprantis AO, Charles JF. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci USA. 2016;113(47):E7554–E7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.