ABSTRACT
Background
Changes in gut microbiota are associated with cardiometabolic disorders and are influenced by diet. Almonds are a rich source of fiber, unsaturated fats, and polyphenols, all nutrients that can favorably alter the gut microbiome.
Objectives
The aim of this study was to examine the effects of 8 wk of almond snacking on the gut (fecal) microbiome diversity and abundance compared with an isocaloric snack of graham crackers in college freshmen.
Methods
A randomized, controlled, parallel-arm, 8-wk intervention in 73 college freshmen (age: 18–19 y; 41 women and 32 men; BMI: 18–41 kg/m2) with no cardiometabolic disorders was conducted. Participants were randomly allocated to either an almond snack group (56.7 g/d; 364 kcal; n = 38) or graham cracker control group (77.5 g/d; 338 kcal/d; n = 35). Stool samples were collected at baseline and 8 wk after the intervention to assess primary microbiome outcomes, that is, gut microbiome diversity and abundance.
Results
Almond snacking resulted in 3% greater quantitative alpha-diversity (Shannon index) and 8% greater qualitative alpha-diversity (Chao1 index) than the cracker group after the intervention (P < 0.05). Moreover, almond snacking for 8 wk decreased the abundance of the pathogenic bacterium Bacteroides fragilis by 48% (overall relative abundance, P < 0.05). Permutational multivariate ANOVA showed significant time effects for the unweighted UniFrac distance and Bray–Curtis beta-diversity methods (P < 0.05; R2 ≤ 3.1%). The dietary and clinical variables that best correlated with the underlying bacterial community structure at week 8 of the intervention included dietary carbohydrate (percentage energy), dietary fiber (g), and fasting total and HDL cholesterol (model Spearman rho = 0.16; P = 0.01).
Conclusions
Almond snacking for 8 wk improved alpha-diversity compared with cracker snacking. Incorporating a morning snack in the dietary regimen of predominantly breakfast-skipping college freshmen improved the diversity and composition of the gut microbiome. This trial was registered at clinicaltrials.gov as NCT03084003.
Keywords: adolescence, amplicon sequence variants, ANCOM, cardiovascular, functional foods, gut, metabolism, minority, nutrients, nuts
Introduction
Specific gastrointestinal (gut) microbiome profiles are associated with obesity and cardiometabolic disorders (1). For example, an overweight phenotype and glucose intolerance are characterized by an increase in pathogenic bacteria and a decrease in anti-inflammatory and butyrate-producing bacteria (1). In addition, low gut microbiome diversity indicated by the number of operational taxonomic units (OTUs) is associated with greater adiposity (2), insulin resistance (2), dyslipidemia (increase in triglycerides and decrease in HDL cholesterol) (3), and a proinflammatory phenotype (2) compared with high gut microbiome diversity. Positive dietary modulation of the gut microbiome by consuming diets rich in prebiotics (1), probiotics (1), and plant-based foods (4) can improve biomarkers of cardiometabolic health (5).
Nuts such as almonds are rich sources of fiber, unsaturated fats, and polyphenols (6), all nutrients that can favorably alter the gut microbiome (7–9). Differences in relative abundance of specific bacterial taxa have been observed with varying doses of almonds and pistachios (42 g or 84 g) (10), different almond processed forms (42 g) (11), and walnut (42 g) (12) consumption over short periods (<3 wk). Furthermore, whole almonds (56 g) and almond skin (10 g) supplementation for 6 wk increased abundance of targeted bacterial species such as Bifidobacterium spp. and Lactobacillus spp. (13). In addition, walnut (43 g) consumption for 8 wk increased the abundance of butyric acid–producing species (14). However, little is known about the effects of longer-term almond consumption in young adults.
We have previously demonstrated the effects of almond snacking on cardiometabolic profiles in college freshmen (15). As an extension of that study, we evaluated the impact of 8 wk of almond snacking (57 g/d) on the gut microbiome abundance and diversity compared with an isocaloric snack of graham crackers in college freshmen. A secondary analysis examined the association of dietary and clinical (anthropometric, glucoregulatory, and cardiovascular) variables with the microbiome community structure. We also highlight issues with the use of common statistical frameworks in nutrition research that do not account for compositional constraints while analyzing differential abundance of microbiome data.
Methods
Participants
Seventy-three (41 women and 32 men) young adults (aged 18–19 y; BMI: 18–41 kg/m2) participating in a snacking intervention were recruited (15). The eligibility criteria were as follows: 1) aged 18–21 y; 2) newly enrolled, first-year college students with no nut allergies; 3) nonsmokers; and 4) no diagnosed endocrine or cardiometabolic disorders. Participants were recruited via public advertisements. Participants who met eligibility criteria provided written, informed consent prior to commencement of study visits. All procedures involving human subjects were approved by the University of California Merced Institutional Review Board. The study is registered on clinicaltrials.gov (registration number: NCT03084003).
Study design and protocol
The primary study was an 8-wk randomized, controlled, parallel-arm intervention examining the effects on glucoregulatory profiles of almonds compared with cracker snacking (15). The sample size calculations for the primary study were based on glucose and insulin profiles at the end of the 8-wk intervention (15). For the present analysis, outcomes related to the gut microbiome were examined. In brief, participants were assigned into 1 of 2 study arms. Participants in the almond group (n = 38) consumed 57 g/d [2 oz; 327 kcal; 14% carbohydrate (8 g fiber), 74% fat, 13% protein] of whole, dry-roasted almonds. Participants in the cracker group (n = 35) consumed 5 sheets (77.5 g/d) of graham crackers [325 kcal; 74% carbohydrate (2.5 g fiber), 20% fat, 6% protein] and were asked to avoid all nuts, seeds, and nut-containing products. Stool samples were collected in sterilized collection containers at baseline and 8 wk after the intervention and stored at −80°C. Anthropometric, biochemical, and dietary data were collected and analyzed as described previously (15).
DNA extraction and sequencing
DNA from homogenized stool samples was extracted using the MoBio power soil DNA isolation kit (MoBio Laboratories, Inc). The quality and quantity of the DNA was confirmed using a Nanodrop 1000 (Thermo Fisher Scientific). The 16S rRNA gene V4/V5 variable region PCR primers 530F/926R, GTGCCAGCMGCNGCGG/CCGTCAATTYYTTTRAGTTT, with barcode on the forward primer, were used in a 28-cycle PCR using the HotStarTaq Plus Master Mix Kit (Qiagen) under the following conditions: 94°C for 3 min, followed by 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, after which a final elongation step at 72°C for 5 min was performed. After amplification, PCR products were checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands. Sequencing was performed at MR DNA (www.mrdnalab.com) on a MiSeq (Illumina).
Sequence quality control
The raw sequence data from the Illumina platform were converted into forward and reverse read files using the FASTQ processor (www.mrdnalab.com), which were then imported to QIIME 2, an open-source microbiome analysis platform (16) for further analysis. The paired-end sequences were demultiplexed, and then denoised, dereplicated, and merged with the DADA2 quality control package (17) in QIIME2. The amplicon sequence variants (ASVs: features and associated representative sequences) resulting from DADA2, are the final products used in all downstream analyses. ASVs are higher-resolution artifacts than commonly used operational taxonomic units (OTUs) (18). A total of 3259 ASVs were detected after demultiplexing and DADA2 quality control.
Taxonomic analysis
The ASVs were assigned taxonomy in QIIME2 using a naive Bayes classifier, which was trained on the 530–926 region of the Greengenes 13_8 database reference sequences clustered at 99% sequence similarity (19). The differential abundance of the ASVs at the different taxonomic levels, namely, phylum, class, family, order, genus, and species, was analyzed using the analysis of composition of microbiomes (ANCOM) statistical framework in R (version 3.5.2) (20). The ANCOM framework uses standard statistical tests to compare Aitchison log-ratios of observed abundances of a taxon relative to a predefined taxon with adjustment for multiple testing (here the Benjamini–Hochberg procedure) (20). For example, testing the effect of snack group on a specific feature, when a total of 5 features are observed, would involve computation of 4 log-ratios for that feature, and the snack effect P value for each ratio would need to be adjusted for multiple testing of 4 ratios. The overall statistical significance of the snack effect would then be determined by the number of subhypotheses (per ratio) rejected compared with a cutoff value (set at 0.6). We included the following statistical tests within the flexible ANCOM framework: 1) linear mixed model analysis with time and group as fixed factors, and participant as a random effect; and 2) nonparametric analysis of time and group effects using the nparLD package in R (21). Because both the parametric and nonparametric tests gave similar results, we have reported only the parametric results. The ANCOM framework was also modified to test for effects of BMI category and sex, as well as pairwise comparisons for significant interaction effects with adjustment for multiple comparisons using the Bonferroni procedure. Only taxa prevalent in at least 25% of the samples were included in the analysis to avoid confounding results due to low-frequency taxa (20).
Alpha- and beta-diversity analysis
Alpha-diversity measures were assessed on the raw abundances and abundances of sequences rarefied to an even sequence depth of 8268 sequences per sample in QIIME2 (Supplemental Figure 1). The following alpha-diversity measures were assessed: 1) Chao1 index, a measure of species richness that is particularly useful for low-abundance datasets (22); 2) observed OTUs, a measure of the number of distinct features; 3) Shannon index, a measure of richness and evenness (23); and 4) Simpson evenness measure, a measure of how well represented a species is (24). The alpha-diversity measures were analyzed in R using linear mixed-model analysis with time and snack as fixed factors and participant as a random effect. Analyses were adjusted for baseline when baseline values had a significant effect on the model.
Phylogenetic beta-diversity measures such as weighted UniFrac (quantitative, i.e., weighs branches of phylogenetic tree based on abundance) and unweighted UniFrac (qualitative, i.e., fraction of unique branches in phylogenetic tree) (25), and nonphylogenetic Bray–Curtis dissimilarity, which quantifies differences between samples based on abundance per count (26), were assessed on the raw, rarefied, and cumulative-sum–scaled (CSS) datasets. The ordinations of the matrices obtained from the raw, rarefied, and CSS datasets were compared using Procrustes analysis in R (27). The effects of time and time × snack on the distance/dissimilarity matrices were analyzed by the permutational multivariate ANOVA (PERMANOVA) (28,29) framework in the vegan package of R (27).
Taxa and environmental associations with community structure
The structural redundancy in community composition was quantified through the BVSTEP (30) analysis in the sinkr package of R (31). The input dataset for the taxa variables consisted of the ASV abundances (at the class level). The best set of environmental (i.e., clinical and dietary) variables associated with the community structure (i.e., ASVs abundance) was deduced by computing the maximum (rank) correlation of all environmental variables with Bray–Curtis dissimilarities using the BIOENV (32) analysis in the vegan package of R (27). The significance for the BVSTEP and BIOENV procedures was assessed using the Mantel test (27). The input dataset for the environmental variables comprised: snack group (week 8 only), sex; body mass; BMI; fat mass percentage; waist circumference; systolic and diastolic blood pressures; fasting total, HDL, and LDL cholesterol; fasting triglycerides; fasting insulin; fasting glucose; total energy intake; dietary carbohydrate percentage; dietary fat percentage; dietary protein percentage; dietary fiber; total saturated fat; MUFAs; PUFAs; oleic acid; linoleic acid; and α-tocopherol.
The best subsets of taxa and environmental variables at baseline and 8 wk after the intervention were plotted as vectors along nonmetric, multidimensional scaling (NMDS) plots, and the significance of the individual variables on the 2-dimensional ordinations was assessed using vegan function envfit (27). It is important to note that the results of envfit do not supersede the results of the BVSTEP and BIOENV analyses, but instead describe the individual contribution of the best model variables to the 2-dimensional ordinations.
Results
Participant characteristics and findings from parent study
The participants’ demographic, clinical, and dietary characteristics have been described in detail previously (15). The parent study exploring the glucoregulatory and cardiometabolic outcomes demonstrated a smaller decline in HDL cholesterol but similar reductions in fasting glucose and LDL cholesterol, and greater postprandial insulin sensitivity during the oral-glucose-tolerance test following almond consumption for 8 wk compared with cracker consumption (15).
ANCOM results of selected taxa prevalent in at least 25% of the samples
Firmicutes (64%), Bacteroidetes (29%), and Actinobacteria (4%) were the most dominant bacterial phyla in this study population at baseline. The ANCOM results depict an overall significant time effect (P < 0.05) indicating an increase over 8 wk for order RF39 (phylum Tenericutes) and decrease for family S24.7 (phylum Bacteroidetes), and genera Alistipes, Butyricimonas, and Odoribacter (all in phylum Bacteroidetes), and an increase for genus Lachnospira (phylum Firmicutes) (Table 1). In addition, Bacteroides fragilis decreased significantly (P < 0.05) in the almond group over the 8-wk intervention (time × snack effect, P < 0.05; Table 1). The relative abundance percentages of all selected taxa at the different taxonomic levels are shown in Supplemental Table 1.
TABLE 1.
ANCOM results of selected taxa prevalent in at least 25% of the samples obtained from college freshmen in the almond and cracker groups at baseline and 8 wk after the intervention
| Relative abundance (%)1 | |||||||
|---|---|---|---|---|---|---|---|
| Baseline | Week 8 | W-taxa2 | |||||
| Taxa | Almond (n = 38) | Cracker (n = 35) | Almond (n = 38) | Cracker (n = 35) | Time | Snack | Time × snack |
| p__Tenericutes.c__Mollicutes.o__RF39 | 0.35 ± 1.35 | 0.49 ± 2.35 | 0.54 ± 1.35 | 0.87 ± 2.92 | 10* | 0 | 0 |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__S24.7 | 0.06 ± 0.2 | 0.11 ± 0.35 | 0.08 ± 0.25 | 0.04 ± 0.08 | 26* | 0 | 0 |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__Rikenellaceae.g__Alistipes | 4.33 ± 5.22 | 2.63 ± 3.4 | 2.41 ± 3.09 | 2.07 ± 3.19 | 36* | 0 | 0 |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__Odoribacteraceae.g__Butyricimonas | 0.08 ± 0.19 | 0.12 ± 0.48 | 0.06 ± 0.16 | 0.03 ± 0.1 | 37* | 0 | 0 |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__Odoribacteraceae.g__Odoribacter | 0.25 ± 0.36 | 0.2 ± 0.34 | 0.09 ± 0.14 | 0.11 ± 0.19 | 39* | 0 | 0 |
| p__Firmicutes.c__Clostridia.o__Clostridiales.f__Lachnospiraceae.g__Lachnospira | 0.08 ± 0.12 | 0.1 ± 0.18 | 0.16 ± 0.17 | 0.1 ± 0.19 | 45* | 0 | 0 |
| p__Proteobacteria.c__Betaproteobacteria.o__Burkholderiales.f__Alcaligenaceae.g__Sutterella | 0.19 ± 0.37 | 0.08 ± 0.15 | 0.71 ± 1.61 | 0.13 ± 0.33 | 4 | 38* | 0 |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__Bacteroidaceae.g__Bacteroides.s__fragilis | 0.66 ± 1.22** | 0.66 ± 1.6 | 0.34 ± 0.76 | 0.76 ± 1.61 | 3 | 0 | 68* |
Values presented are means ± SDs of relative abundance percentages of participants in the almond and cracker groups at baseline and week 8 of the intervention. **Significantly different from week 8. ANCOM, analysis of composition of microbiomes; c, class; f, family; g, genus; o, order; p, phylum; s, species.
W-taxa represent the number of significant log-ratios for that taxon at the time, snack, and time × snack levels. *Denotes overall significance at a 60% cutoff, that is, at least 60% of the log-ratios were significant (i.e., P < 0.05) after adjusting for multiple testing of log-ratios using the Benjamini–Hochberg procedure. Analysis was a linear mixed model analysis within the ANCOM framework.
Almond group had greater alpha-diversity at week 8 compared with the cracker group
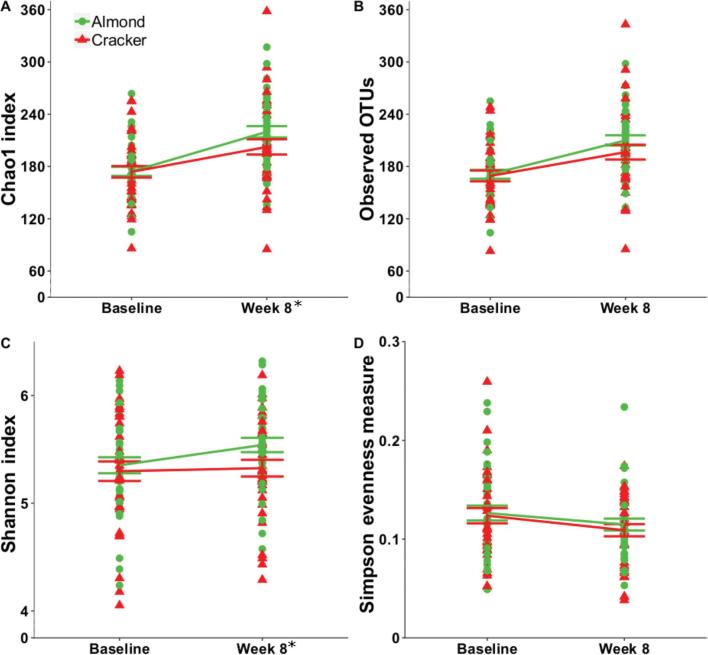
Chao1 index, observed OTUs, and Shannon index measures for the raw and rarefied abundances significantly increased (time effect, P < 0.05), whereas the Simpson evenness measure for raw abundances significantly decreased (time effect, P < 0.05) over the 8-wk intervention (Figure 1, Supplemental Table 2). The almond group had significantly greater baseline-adjusted Chao1 index and Shannon index (P < 0.05) for raw and rarefied abundances and observed OTUs (P < 0.05) measures for rarefied abundances at week 8 (Figure 1, Supplemental Table 2). The rarefaction curves for Chao1 index, observed OTUs, and Shannon index (Supplemental Figures 2–5) confirm the greater diversity, at different sequencing depths, in the almond group compared with the cracker group at week 8 of the intervention.
FIGURE 1.
Alpha-diversity indices (calculated from raw abundances). (A) Chao1 index, (B) observed OTUs, (C) Shannon index, and (D) Simpson evenness measure of college freshmen in the almond and cracker groups at baseline and week 8 of the intervention. Values are individual data points representing each participant at baseline and week 8. Means ± SDs of the 2 snack groups at baseline and week 8 are also plotted. Analyses were conducted using 1) linear mixed model with snack (almond or cracker) as between-subject factor, time (baseline or week 8) as within-subject factor, and participant as random factor; and 2) analysis of covariance with baseline value as covariate and snack as between-subject factor. *Snack effect (adjusting for baseline value), P < 0.05. Almond: n = 38, cracker: n = 35. OTU, operational taxonomic unit.
Procrustes analysis indicated that the raw and rarefied datasets were highly correlated
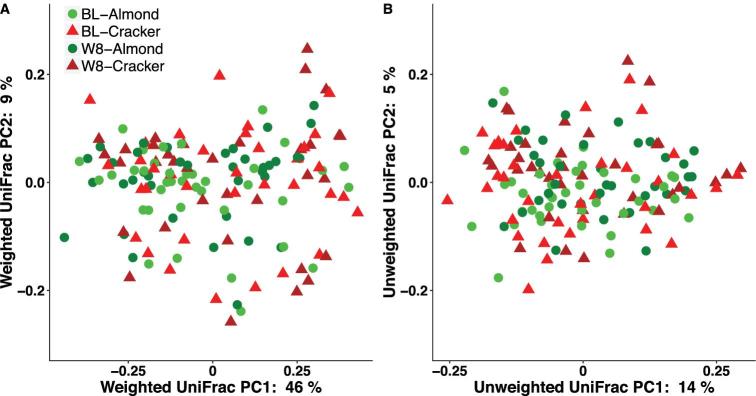
The principal coordinates analysis (PCoA) plots of the weighted and unweighted UniFrac measures of the raw data at baseline and week 8 of the intervention are depicted in Figure 2. Procrustes analysis found that the raw compared with rarefied ordinations of weighted and unweighted UniFrac measures and Bray–Curtis dissimilarities were highly correlated (Procrustes correlation coefficient >0.94; P < 0.01; Supplemental Table 3) indicating that the raw and rarefied matrices are similar.
FIGURE 2.
Principal coordinates analysis plots of beta-diversity measures (calculated from raw abundances). (A) Weighted UniFrac, and (B) unweighted UniFrac of college freshmen in the almond and cracker groups at baseline and week 8 of the intervention. Almond: n = 38, cracker: n = 35. BL, baseline; PC, principal component; W8, week 8.
PERMANOVA analysis indicated differences in beta-diversity over time for the raw and rarefied datasets
PERMANOVA analyses showed significant, albeit small, time effects for the unweighted UniFrac distance and Bray–Curtis dissimilarity matrices of the raw and rarefied datasets (P < 0.05; R2 ≤3.1%; Supplemental Table 4), which are not visible in the 2-dimensional PCoA plots (Figure 2). There were no significant effects of snack and time × snack on any distance/dissimilarity matrices. The best subset of taxa contributing to the community structure at baseline and week 8 is explored in the section below.
The structural redundancy in the community structure was characterized by 40–55% of the identified microbial classes
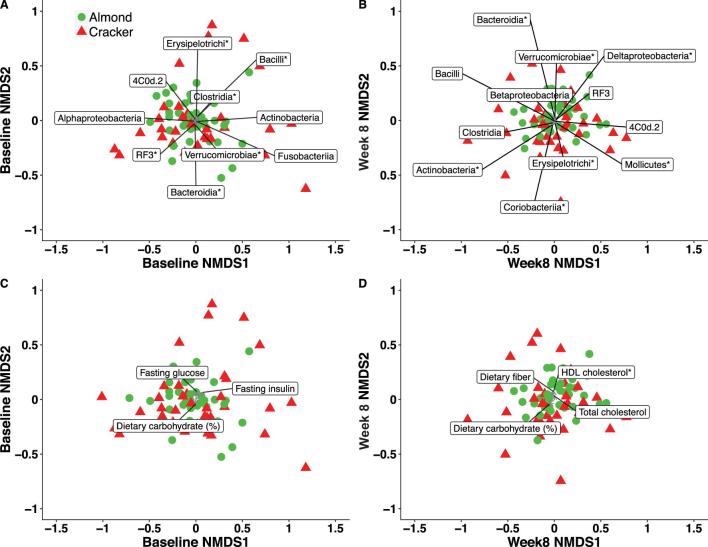
The best correlated subset of taxa variables included 10 of the 24 identified microbial classes at baseline (model Pearson correlation coefficient = 0.96; P = 0.001; Figure 3A) and 13 of 24 classes at week 8 (model Pearson correlation coefficient = 0.96; P = 0.001, Figure 3B). Of notable interest are the absence of Fusobacteria and Alphaproteobacteria from the week 8 model, and the presence of Coriobacteria and Mollicutes in the week 8 model. The 2-dimensional NMDS ordination at baseline was (individually) significantly associated with the Bacteroidia (phylum Bacteroidetes), Bacilli, Clostridia, and Erysipelotrichi (phylum Firmicutes), RF3 (phylum Tenericutes), and Verrucomicrobiae (phylum Verrucomicrobia) classes (P < 0.05 envfit test; Figure 3A, Supplemental Table 5). The 2-dimensional NMDS ordination at week 8 of the intervention was (individually) significantly associated with the Bacteroidia (phylum Bacteroidetes), Erysipelotrichi (phylum Firmicutes), Coriobacteria and Actinobacteria (phylum Actinobacteria), Mollicutes (phylum Tenericutes), Verrucomicrobiae (phylum Verrucomicrobia), and Deltaproteobacteria (phylum Proteobacteria) classes (P < 0.05 envfit test; Figure 3B, Supplemental Table 5).
FIGURE 3.
Nonmetric multidimensional scaling (NMDS) plot of the Bray–Curtis dissimilarities of college freshmen with best set of (A) taxa variables at baseline, (B) taxa variables at week 8, (C) environmental variables at baseline, and (D) environmental variables at week 8. NMDS baseline stress = 0.176, NMDS week 8 stress = 0.198. *P < 0.05 from envfit. The black line shows the direction of the (increasing) gradient, and the length of the line is proportional to the correlation between the variable and the NMDS score. Line lengths should not be compared across plots. Almond: n = 38; cracker: n = 35.
Dietary and clinical variables contributed to a small amount of variation in the community structure
The best correlated subset of environmental variables at baseline (model Spearman rho = 0.17; P = 0.014) and week 8 (model Spearman rho = 0.16; P = 0.01) is depicted in Figure 3C,D. The subset of environmental variables with the highest correlation with the community structure comprised dietary carbohydrate (%), fasting glucose, and fasting insulin at baseline (Figure 3C, Supplemental Table 5) and dietary carbohydrate (%), dietary fiber, and fasting total and HDL cholesterol at week 8 (Figure 3D, Supplemental Table 5). Moreover, fasting HDL cholesterol demonstrated a statistically significant association with the 2-dimensional NMDS ordination at week 8 of the intervention (R2 = 10%; P < 0.05 envfit test; Supplemental Table 5).
Discussion
Almond snacking (57 g/d) for 8 wk resulted in 3% greater quantitative alpha-diversity (Shannon index) and 8% greater qualitative alpha-diversity (Chao1 index) compared with isocaloric cracker snacking at the end of the 8-wk intervention. In addition, almond snacking decreased overall B. fragilis relative abundance by 48%. The snacking intervention increased alpha-diversity over time, but the unique nutrient profile of almonds had a greater impact on alpha-diversity than graham crackers. Increased bacterial richness is associated with favorable health outcomes (2). In this same cohort of study participants, glucose tolerance and postprandial insulin sensitivity were improved with almond snacking (15), suggesting that improved gut microbiome and carbohydrate metabolism could be associated. The fiber, monounsaturated fat, and polyphenol content of almonds are likely responsible for the greater alpha-diversity. The beneficial effects of dietary fiber on modulating the gut microbiome diversity are well characterized (33). Monounsaturated fats (compared with saturated fats) are associated with promoting a positive gut microbiome profile as well (34). Polyphenols in almond skins are partially bioavailable (35), and the unabsorbed polyphenols are metabolized by specific colon microbiota into absorbable metabolites (8). Sustained consumption of polyphenol-rich diets can stimulate the growth of beneficial bacteria (8), thereby promoting greater diversity as well.
Other nut studies have either not assessed (13) or have not detected any significant differences in alpha-diversity (10–12, 14). Three of those studies were short-term (<3 wk) (10–12), suggesting that the intervention duration was not sufficient to induce significant changes in species richness as seen in other short-term dietary interventions (36, 37). Conversely, an 8-wk walnut intervention did not alter alpha-diversity; however, phylogenetic beta-diversity (between-sample diversity) was different by 5% between the walnut and control diets (14). In the present study, the 8-wk intervention can only explain, at most, 3% of the variance in beta-diversity with no differential effect of snack group. However, the beta-diversity was positively correlated with alpha-diversity (P < 0.01, data not shown) suggesting that beta-diversity increased over time as well. Increased microbiome diversity could promote greater stability of the microbiome in the long term, thereby contributing to functional resilience against extreme stress and perturbations as purported by the classical ecological resilience theory (38, 39).
Another important finding of the present study was the decrease in B. fragilis with almond consumption. B. fragilis is an anaerobic pathogen that is most frequently isolated from clinical specimens and is considered the most virulent Bacteroides species because of its: 1) adhesion properties that facilitate adherence to host tissues, 2) lipopolysaccharide capsule that protects it from the host's immune system, and 3) histolytic enzyme activity (40). Although enterotoxins produced by specific B. fragilis strains can cause gastrointestinal inflammation (41), other reports indicate that colonization of germ-free mice with polysaccharide-producing B. fragilis can contribute to the maturation of the immune system (42). We did not assess inflammatory profiles or immune system markers in this study; however, Sugizaki and Naves (6) propose that the dietary fiber and polyphenol component of nuts can promote a homeostatic intestinal state via host–microbe interactions.
Over 90% of the variation in the bacterial community structure pre- and post-intervention was explained by ∼40–50% of taxa at the class level. The taxa characterizing the structural redundancy were mostly similar at both time points of the intervention with the notable contributions of classes Mollicutes (phylum Tenericutes), which was predominantly composed of the order RF39, and Coriobacteriia (phylum Actinobacteria), which was mostly composed of the family Coriobacteriaceae at week 8 of the intervention. The relation between bacteria in the order RF39 and the host's metabolism is not defined. However, twin studies demonstrated that an increased abundance of RF39 was associated positively with a lean phenotype (43) and negatively with metabolic syndrome (44). The Coriobacteriaceae in the gut could contribute to the modulation of bile acid, steroid, and dietary polyphenol metabolism (45).
Other favorable changes in the gut microbiome profile over the 8-wk intervention involved a decrease in Alistipes (phylum Bacteroidetes), which is indicative of a shift away from an animal-based diet, and increase in Lachnospira (phylum Firmicutes), which are pectin degraders (46). Additionally, a decrease in the butyric acid producers, Odoribacter and Butyricimonas (Bacteroidetes genera), was observed over 8 wk. Although a decrease in butyric acid–producing potential is typically associated with cardiometabolic disorders (47), an increase in butyric acid producers of the order Bacteroidales was reported in murine models of colitis (48, 49). As noted before, in times of physiological stress such as inflammation, there can be an increased reliance of colonocytes on butyric acid for energy, hence promoting the growth of butyric acid producers (48). Thus, it can be postulated that a decrease in stress can reduce the reliance on butyric acid producers. In addition, the abundance of uncultured Bacteroidetes family S24-7 decreased over 8 wk, but this has not been well studied in humans. However, murine studies report conflicting effects of diets on S24-7 in obese and diabetes models (50–52).
Dietary components such as carbohydrate and fiber, and cardiometabolic markers such as total and HDL cholesterol, explained <3% of the variation in the bacterial community composition at week 8 of the intervention in this cohort. The association of carbohydrate and fiber intake with the underlying community structure is not surprising because these are the principal energy sources for bacteria (53). Additionally, we have demonstrated that incorporating a morning snack into the diet of these predominantly breakfast-skipping college freshmen improved fasting glucose, and total and LDL cholesterol, with a protective effect of almond consumption on HDL cholesterol (15). Hence, changes in cholesterol profiles could partly be related to the gut microbiome composition. In some studies, intrinsic factors outweighed dietary intervention and drove community structure (54), whereas in others, dietary factors that were significantly associated with interindividual variation of gut microbiome outnumbered other intrinsic factors (55). The present study population was homogeneous in terms of age (18–19 y), predominantly belonged to ethnic/racial minority groups, and was at low to moderate cardiometabolic risk (15). Hence, intra- and inter-individual factors such as genetics and environment (43) could have a greater influence on the gut microbiome composition.
Developing statistical models for analyzing differential abundance of taxa is an active area of research. Compositional data such as relative abundances exist in the simplex rather than Euclidean space, that is, relative abundances across all taxa sum to 1 within a given sample. Because standard statistical tests such as t test, ANOVA, linear regression, and others do not account for the simplex nature of compositional data, they should not be directly used for analyzing relative abundance data. The ANCOM method is a highly sensitive statistical framework that uses Aitchison log-ratios to test for differences in the mean abundances of log-transformed taxa, which can be used to draw inferences regarding relative abundance differences in the ecosystem (20) (represented by the gut here). The ANCOM method controls the false discovery rate, while maintaining high power (20, 56), and can incorporate standard statistical tests within the framework.
Strengths of the study included: 1) excellent participant compliance (15), 2) the use of high-resolution ASVs, and 3) the incorporation of the sensitive and powerful ANCOM method for analyzing differential abundance. Limitations included: 1) the lack of a “no snack” group to capture changes in the gut microbial profile in response to the students’ regular diet, and 2) the use of 16S rRNA profiling instead of shotgun sequencing. However, the 16S taxonomic resolution concerns are partly mitigated by sequencing ASVs (57) of the V4 and V5 variable regions.
In conclusion, almond snacking for 8 wk led to greater alpha-diversity than the isocaloric cracker group in college freshmen. In addition, almond snacking decreased the relative abundance of B. fragilis, a commonly isolated pathogenic bacterium. In general, incorporating a morning snack in the dietary regimen of predominantly breakfast-skipping college freshmen improved the diversity and composition of the gut microbiome.
Supplementary Material
Acknowledgements
The authors’ responsibilities were as follows—JD, RMO: designed and conducted the study, analyzed data, and wrote the paper; ZL: provided study design and sample processing support; JD, ZL, RMO: shared equal responsibility for writing the manuscript and its final content; and all authors: read and approved the final manuscript.
Notes
The present study was supported by the Almond Board of California (principal investigator: RMO). JD is supported by the National Institute on Minority Health and Health Disparities of the NIH under award number K99MD012815. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders. The funders had no role in the study conception, design and implementation, data collection, data analysis, or interpretation of results.
Author disclosures: RMO discloses grants and nonfinancial support from the Almond Board of California for conducting the study; JD and ZL, no conflicts of interest.
Supplemental Figures 1–5 and Supplemental Tables 1–5 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/cdn.
Abbreviations used: ANCOM, analysis of composition of microbiomes; ASV, amplicon sequence variant; CSS, cumulative-sum–scaled; NMDS, nonmetric, multidimensional scaling; OTU, operational taxonomic unit; PCoA, principal coordinates analysis; PERMANOVA, permutational multivariate ANOVA.
References
- 1. Festi D, Schiumerini R, Eusebi LH, Marasco G, Taddia M, Colecchia A. Gut microbiota and metabolic syndrome. World J Gastroenterol 2014;20:16079–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S et al.. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541–6. [DOI] [PubMed] [Google Scholar]
- 3. Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JAM, Brandsma E, Marczynska J, Imhann F, Weersma RK et al.. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ Res 2015;117:817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeffery IB, O'Toole PW. Diet-microbiota interactions and their implications for healthy living. Nutrients 2013;5:234–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brahe LK, Astrup A, Larsen LH. Can we prevent obesity-related metabolic diseases by dietary modulation of the gut microbiota?1. Adv Nutr 2016;7:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sugizaki CSA, Naves MMV. Potential prebiotic properties of nuts and edible seeds and their relationship to obesity. Nutrients. [Internet]. 2018;10(11):1645 [cited January 31, 2019]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6266159/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang BG, Hur KY, Lee MS. Alterations in gut microbiota and immunity by dietary fat. Yonsei Med J 2017;58:1083–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem 2013;24:1415–22. [DOI] [PubMed] [Google Scholar]
- 9. Graf D, Di Cagno R, Fåk F, Flint HJ, Nyman M, Saarela M, Watzl B. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. [Internet]. 2015;26 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4318938/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ukhanova M, Wang X, Baer DJ, Novotny JA, Fredborg M, Mai V. Effects of almond and pistachio consumption on gut microbiota composition in a randomised cross-over human feeding study. Br J Nutr 2014;111:2146–52. [DOI] [PubMed] [Google Scholar]
- 11. Holscher HD, Taylor AM, Swanson KS, Novotny JA, Baer DJ. Almond consumption and processing affects the composition of the gastrointestinal microbiota of healthy adult men and women: a randomized controlled trial. Nutrients 2018;10:E126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holscher HD, Guetterman HM, Swanson KS, An R, Matthan NR, Lichtenstein AH, Novotny JA, Baer DJ. Walnut consumption alters the gastrointestinal microbiota, microbially derived secondary bile acids, and health markers in healthy adults: a randomized controlled trial. J Nutr 2018;148:861–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Z, Lin X, Huang G, Zhang W, Rao P, Ni L. Prebiotic effects of almonds and almond skins on intestinal microbiota in healthy adult humans. Anaerobe 2014;26:1–6. [DOI] [PubMed] [Google Scholar]
- 14. Bamberger C, Rossmeier A, Lechner K, Wu L, Waldmann E, Fischer S, Stark RG, Altenhofer J, Henze K, Parhofer KG. A walnut-enriched diet affects gut microbiome in healthy caucasian subjects: a randomized, controlled trial. Nutrients 2018;10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dhillon J, Thorwald M, De La Cruz N, Vu E, Asghar SA, Kuse Q, Diaz Rios LK, Ortiz RM. Glucoregulatory and cardiometabolic profiles of almond vs. cracker snacking for 8 weeks in young adults: a randomized controlled trial. Nutrients 2018;10:960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F et al.. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. [Internet] 2019. Available from: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 2017;11:2639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Env Microbiol 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. [Internet]. 2015;26 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4450248/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noguchi K, Gel YR, Brunner E, Konietschke F. nparLD: an R software package for the nonparametric analysis of longitudinal data in factorial experiments. J Stat Softw. [Internet]. 2012;50 [cited September 24, 2018]. Available from: https://www.jstatsoft.org/article/view/v050i12. [Google Scholar]
- 22. Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat 1984;11:265–70. [Google Scholar]
- 23. Shannon C, Weaver W. The mathematical theory of communication. Univ Ill Press: 131 New York, NY, USA: Association for Computing Machinery (ACM); 2001. [Google Scholar]
- 24. Simpson EH. Measurement of diversity. Nature 1949;163:688. [Google Scholar]
- 25. Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Env Microbiol 2007;73:1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sørensen TJ. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. København: I kommission hos E. Munksgaard; 1948. [Google Scholar]
- 27. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P et al.. vegan: community ecology package. [Internet]. [cited March 24, 2019]. Available from: https://CRAN.R-project.org/package=vegan.
- 28. Anderson MJ. A new method for non-parametric multivariate analysis of variance: Non-Parametric Manova For Ecology. Austral Ecol 2001;26:32–46. [Google Scholar]
- 29. Anderson MJ. Permutational multivariate analysis of variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online; [Internet]. American Cancer Society; 2017. [cited March 24, 2019]. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118445112.stat07841. [Google Scholar]
- 30. Clarke KR, Warwick RM. Quantifying structural redundancy in ecological communities. Oecologia 1998;113:278–89. [DOI] [PubMed] [Google Scholar]
- 31. Taylor M. sinkr: a collection of functions with emphasis in multivariate data analysis. [Internet]. 2017. [cited March 24, 2019]. Available from: https://github.com/marchtaylor/sinkr. [Google Scholar]
- 32. Clarke R, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol 1993;92:205–19. [Google Scholar]
- 33. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 2018;23:705–15. [DOI] [PubMed] [Google Scholar]
- 34. Cândido FG, Valente FX, Grześkowiak ŁM, Moreira APB, Rocha DMUP, Alfenas R de CG. Impact of dietary fat on gut microbiota and low-grade systemic inflammation: mechanisms and clinical implications on obesity. Int J Food Sci Nutr 2018;69:125–43. [DOI] [PubMed] [Google Scholar]
- 35. Mandalari G, Tomaino A, Rich GT, Lo Curto R, Arcoraci T, Martorana M, Bisignano C, Saija A, Parker ML, Waldron KW et al.. Polyphenol and nutrient release from skin of almonds during simulated human digestion. Food Chem 2010;122:1083–8. [Google Scholar]
- 36. Kaczmarek JL, Liu X, Charron CS, Novotny JA, Jeffery EH, Seifried HE, Ross SA, Miller MJ, Swanson KS, Holscher HD. Broccoli consumption affects the human gastrointestinal microbiota. J Nutr Biochem 2018;63:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al.. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levine JM, D'Antonio CM. Elton revisited: a review of evidence linking diversity and invasibility. Oikos 1999;87:15–26. [Google Scholar]
- 39. Yachi S, Loreau M. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci U S A 1999;96:1463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 2007;20:593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu S, Powell J, Mathioudakis N, Kane S, Fernandez E, Sears CL. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infect Immun 2004;72:5832–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005;122:107–18. [DOI] [PubMed] [Google Scholar]
- 43. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT et al.. Human genetics shape the gut microbiome. Cell 2014;159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lim MY, You HJ, Yoon HS, Kwon B, Lee JY, Lee S, Song Y-M, Lee K, Sung J, Ko G. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 2017;66:1031–8. [DOI] [PubMed] [Google Scholar]
- 45. Clavel T, Lepage P, Charrier C. The family Coriobacteriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: Actinobacteria. [Internet]. [cited February 10, 2019] Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. p. 201–38. Available from: 10.1007/978-3-642-30138-4_343. [DOI] [Google Scholar]
- 46. Bang S-J, Kim G, Lim MY, Song E-J, Jung D-H, Kum J-S, Nam Y-D, Park C-S, Seo D-H. The influence of in vitro pectin fermentation on the human fecal microbiome. AMB Express. [Internet] 2018;8:98 [cited 2019 Feb 10]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6004267/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. [Internet]. 2017;2:e00130–17.[cited February 16, 2019]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5715108/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hamilton MK, Boudry G, Lemay DG, Raybould HE. Changes in intestinal barrier function and gut microbiota in high-fat diet-fed rats are dynamic and region dependent. Am J Physiol Gastrointest Liver Physiol 2015;308:G840–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samanta AK, Torok VA, Percy NJ, Abimosleh SM, Howarth GS. Microbial fingerprinting detects unique bacterial communities in the faecal microbiota of rats with experimentally-induced colitis. J Microbiol 2012;50:218–25. [DOI] [PubMed] [Google Scholar]
- 50. Ormerod KL, Wood DLA, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal'Molin CGO, Palfreyman RW, Nielsen LK, Cooper MA et al.. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 2016;4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, Waget A, Klopp P, Iacovoni J, Klopp C et al.. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 2012;61:543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V et al.. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One 2014;9:e92193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2014;7:17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lang JM, Pan C, Cantor RM, Tang WHW, Garcia-Garcia JC, Kurtz I, Hazen SL, Bergeron N, Krauss RM, Lusis AJ. Impact of individual traits, saturated fat, and protein source on the gut microbiome. mBio. [Internet] 2018;9 [cited January 31, 2019]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6299478/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S et al.. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016;352:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A et al.. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Edgar RC. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 2018;34:2371–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.