Abstract
The spindle assembly checkpoint (SAC) ensures the fidelity of chromosome segregation during mitosis. Here, we show that ULK1, a serine/threonine kinase that plays a key role in initiation of autophagy, also has an important function in the activation of SAC. ULK1 phosphorylates the SAC protein Mad1 at Ser546 to recruit Mad1 to kinetochores. Furthermore, Rod/ZW10/Zwilch (RZZ) complex may serve as a receptor for phos-Ser546-Mad1 at kinetochore, since phosphorylation of Mad1 by ULK1 strengthens the interaction between Mad1 and RZZ complex. In addition, deletion of ULK1 increases chromosome instability and cytotoxicity of paclitaxel, resulting in significant impairment of cancer cell growth. These findings highlight the role of ULK1 as a protein kinase controlling the fidelity of chromosome segregation and cell-cycle progression.
INTRODUCTION
The spindle assembly checkpoint (SAC) ensures the accurate segregation of chromosome by delaying anaphase until all sister chromatid pairs make proper bipolar attachments to the mitotic spindle. The SAC includes mitotic arrest deficient (MAD) and budding uninhibited by benzimidazole (BUB) genes (1). At unattached kinetochores, Mad1–Mad2 complex temporarily sequester Cdc20, an activator of the anaphase-promoting complex/cyclosome (APC/C) (2,3). This enables the formation of mitotic checkpoint complex (MCC) that prevents the activation of APC/C, which initiates anaphase by targeting securin and cyclin B for proteasome-mediated degradation. Thus, the kinetochore-bound Mad1–Mad2 complex plays important roles in MCC assembly and SAC activity regulation (4). The Mad1/Mad2 complex’ crystal structure reveals a 2:2 tetramer (5,6). Mad2 is recruited to the kinetochore via its interaction with Mad1 (7).
In human cells, there are two kinetochore recruitment pathways for Mad1. One is the Knl1/Bub3/Bub1 pathway. The MELT motif of kinetochore protein KNL1 is phosphorylated by Mps1 kinase to target Bub1:Bub3 to kinetochores (8–10). Then, Mps1 phosphorylation of conserved domain 1 (CD1) in Bub1 promotes Bub1:Mad1 interaction (11–13). On the other hand, a second pathway that complemented the Knl1/Bub3/Bub1 pathway for Mad1 recruitment depends on the Rod/ZW10/Zwilch (RZZ) kinetochore complex (14). Once Mad1 is retained, two different outcomes are produced based on the function of the two distinct complexes mentioned above. While RZZ is responsible for Mad1/2 tethering and kinetochore expansion, the KBB pathway is critical for the generation of the wait anaphase signal (14,15). A recent study indicated that RZZ is responsible for keeping Mad1 stable on the kinetochore, although both Bub1 and RZZ contribute to the localization of MAD1 (16). Molecular mechanisms of Mad1 recruitment to kinetochores are of broad interest, since this event is known to be important for mitotic progression and error-free chromosome segregation.
Unc-51-like kinases 1/2 (ULK1/2) is a serine/threonine protein kinase that plays important roles in autophagy initiation (17–25). During autophagy process, the activated ULK1/2 phosphorylates Atg13, Fip200 and Beclin-1 leading to autophagy induction (26–28). Besides autophagy-related protein substrates, several autophagy-unrelated ULK1 substrates were found. For example, during deprivation of amino acid and growth factors, ULK1/2 directly phosphorylates key glycolytic enzymes to sustain glycolysis (29). Stimulator of interferon genes (STING) is also phosphorylated by ULK1 to prevent the persistent transcription of innate immune genes (30). ULK1/2 phosphorylates SEC16A and regulates endoplasmic reticulum (ER) export that is essential for cellular homeostasis (31). Our previous study also identified cochaperone Cdc37 as a substrate of ULK1 that disrupted its ability to coordinate Hsp90 for maintaining the stability and functions of protein kinases (32).
Here in this study, we show that ULK1 phosphorylates the spindle checkpoint protein Mad1 at Ser546. This phosphorylation is required for Mad1 recruitment to kinetochores, proper mitotic progression, faithful chromosome alignment and segregation. Furthermore, deletion of ULK1 in cancer cells increases chromosome instability and cytotoxicity of paclitaxel, resulting in significant impairment of tumor cell growth.
MATERIALS AND METHODS
Cell culture and plasmid transfection
Cells were grown in DMEM with 10% (v/v) fetal bovine serum and the appropriate amount of penicillin/streptomycin in a 37°C incubator with a humidified 5% CO2 atmosphere. Transient and stable transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.
Plasmids and siRNA
cDNA of MAD1 was amplified and cloned into p3xFLAG-CMV-10, pEGFP-C1 and pET28a vectors. ΔN (Δ1-485a.a.), ΔMIM (Δ485-584a.a.) and ΔC (Δ584-718a.a.) of MAD1 were amplified and cloned into pEGFP-C1 vector. ΔN (Δ1-278a.a.), ΔST (Δ278-828a.a.) and ΔC (Δ828-1051a.a.) of ULK1 were amplified and cloned into p3xFLAG-CMV-10 vector. For expression of MAD1–MAD2, cDNA of MAD1 and MAD2 was amplified and cloned into pETDuet-1 vectors. pETDuet-1 vector was kindly provided by Dr Caihong Yun (Peking University Health Science Center, China). MAD1 and ULK1 mutation constructs were generated with a Fast mutagenesis kit (Vazyme).
The sense-strand sequence of negative control siRNA was 5′-UUCUCCGAACGUGUCACGU-3′. The other siRNA sequences were as follows:
ULK1: 5′-CACTGACCTGCTCCTTAA-3′ (#1) and 5′-GGAGAAAACTTGTAGGTGT-3′ (#2).
ZW10: 5′-UGAUCAAUGUGCUGUUCAA-3′ (#1) and 5′-AAGGGTGAGGTGTGCAATATG-3′ (#2).
Knl1:5′-GGAAUCCAAUGCUUUGAGA-3′ (#1) and 5′-GCAUGUAUCUCUUAAGGAA-3′ (#2).
MAD1:5′-CAGGCAGUGUCAGCAGAAC-3′ (#1) and 5′-CCACAGGGCAGCAGCAUGA-3′ (#2).
For MAD1 knockdown, siRNA was transfected twice in 2 days. All RNAi oligonucleotides were purchased from Shanghai GenePharma Company.
Generation of CRISPR/Cas9 KO cell lines
The Cas9 knockout cell lines were generated using CRISPR–Cas9 methods in HCT116 cells. We used the SpCas9-2A-Puro vector purchased from Addgene (#48139; deposited by Feng Zhang). The sgRNA was designed by online software (http://crispr.mit.edu), and the sgRNA sequences were as follows:
ULK1 sgRNA sequence 5′-CGAAGGCGCCGTGGCCGATC-3′ (#1) and 5′-AGCAGATCGCGGGCGCCATG-3′ (#2);
Atg3 sgRNA sequence 5′-GTGAAGGCATACCTACCAAC-3′;
Atg13 sgRNA sequence 5′-GAATGGACACATTACCTTGA-3′;
Atg7 sgRNA sequence 5′- GAAGCTGAACGAGTATCGGC-3′;
FIP200 sgRNA sequence 5′-CACCTGAAGATCGGCTCTACGCCC-3′.
The plasmids were transfected into HCT116 cells and selected with 2.5 μg/ml puromycin.
Antibodies and reagents
The antibodies used were anti-MAD1 (proteintech, #18322-1-AP; Santa Cruz, #sc-47746), anti-CENP-B (Santa Cruz, #sc-376283; #sc-22788), anti CDC20 (Santa Cruz, #sc-5296), anti-MAD2 (proteintech, #10337-1-AP; Covance, #PRB-452C-200), anti-BUBR1 (proteintech, #11504-2-AP), anti-cyclinB1 (abclonal, #A2056), anti-ZW10 (abcam, #ab21582), anti-Flag-tag (Sigma-Aldrich, #F1804), anti-ULK1 (Santa Cruz, #sc-33182; proteintech, #20986-1-AP), anti-Atg13 (CST, #13468S), anti-FIP200 (proteintech, #17250-1-AP), anti-Atg3 (MBL, #M133-3), anti-GFP-tag (Santa Cruz, #sc-9996), anti-His-tag (MBL, #D291-3), anti-BUB1 (proteintech, #13330-1-AP), anti-BUB3 (proteintech, #27073-1-AP), anti-H3S10 (abcam, #ab32017), anti-Knl1 (abclonal, A13108), anti-ROD (abclonal, A13064), anti-ZWILCH (proteintech, #14281-1-AP), anti-α-tubulin (CST, #3873T) and anti-actin (Santa Cruz, #sc-7210). A phospho (ph)-MAD1-S546 specific antibody was generated by Beijing Biodragon Immunotechnologies Co. Ltd.
Treatments included thymidine (Sigma, T9250), RO3306 (Selleck, S7747), Taxol (Santa Cruz, sc-201439), nocodazole (Selleck, S2775), vinblastine (Selleck, S4505).
Chromosome spread and Karyotype analysis
Cells in 10-cm dishes were accumulated in mitosis by treatment with 100 ng/ml colcemid for 3–5 h. Cells in the media, as well as adherent cells, were collected by centrifugation and resuspended in 75 mM KCl for 9 min at room temperature. Three drops of freshly made fixative agent, consisting of methanol and acetic acid at a 3:1 ratio, were added to cells, which were then collected by centrifugation. Cells were resuspended in a residual volume by flicking. A total of 4 ml of fresh fixative agent was added dropwise while mixing cells after every 0.5 ml. Cells were washed again with fresh fixative agent twice more before being resuspended in ∼400 μl fixative agent. A total of 70 μl of cells were dropped onto precleaned microscope slides from a height of 2 ft. Slides were dried slowly in a fume hood for 10 s and then dried quickly on a hot plate at 74°C for 30 s. Slides were stained with 1 μg/ml 4',6-diamidino-2-phenylindole (DAPI), washed with PBS solution, mounted using Vectashield and sealed with nail polish. Chromosome spreads were imaged by using a 100×, 1.4-NA objective. At least 150 metaphase spreads were counted for each cell line using CytoVision®.
Enrichment of cells in mitosis (nocodazole and ‘shake-off’)
Cells were treated with thymidine for 24 h and then released into 200 nM nocodazole for 16 h. Mitotic cells were collected by shake-off. Microscopic examination had shown ∼70–80% of cells were in prometaphase after this treatment compared with 2–3% in untreated cells.
RO3306 washout experiments
For RO3306 washout experiments, cells were initially treated with 2 mM thymidine for 24 h, washed twice with PBS and released in fresh medium for 4 h prior to treatment with 9 μM RO3306. After 8 h, the cells were washed twice with PBS and incubated with fresh medium for different times.
Western blotting
Equal amounts of proteins (20–50 μg) were size fractionated by 6–15% SDS polyacrylamide gel electrophoresis.
Co-immunoprecipitation
Cells were harvested and lysed for 30–45 min at 4°C in buffer A (50 mM Tris–HCl at pH 7.5, 0.5 mM phenylmethanesulfonyl fluoride (PMSF), 0.5 mM benzamidine, 5 μM leupeptin, 1 mM dithiothreitol, 20 mM NaF), supplemented with 0.5% Triton X-100. Centrifugation of the lysates (10 min at 1800 g) yielded a supernatant (S) and pellet (P). The pellets were washed twice with buffer A. For the immunoprecipitation assays, the washed chromatin pellets were resuspended in 50 mM Tris–HCl at pH 8.0, supplemented with 1.5 mM CaCl2 and 20 mM NaF, and treated with 30 U of micrococcal nuclease for 30 min at 37°C. The soluble and insoluble fractions were separated by centrifugation (2 min at 664 g). About 2 g of the indicated antibody was added into the supernatant and incubated at 4°C overnight. Then, 30 μl of protein G Sepharose slurry (GE healthcare) was added and incubated for 2 h at 4°C. The beads were washed three times. The precipitated components were analyzed by western blotting.
Protein purification
Recombinant His-tagged proteins were expressed in and purified from Escherichia coli by Ni2+-Sepharose affinity (GE Healthcare), and the bound protein was eluted with 250 mM imidazole in PBS and desalted by buffer exchange with PBS.
In Vitro MAD1 phosphorylation assay
HCT116 cells were grown, and each dish was transfected with 8 μg of Flag-ULK1. After 48 h post-transfection, cells were lysed in MLB (10 mM Tris at pH 7.5, 2 mM EDTA, 100 mM NaCl, 1% Nonidet P-40, 50 mM NaF, 1 mM Na3VO4 and 1% EDTA-free protease and phosphatase inhibitor cocktails (Roche Applied Science)). ULK1 proteins were immunoprecipitated with anti-Flag-tag (Sigma) antibodies and then washed with MLB once and radioimmune precipitation assay buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 50 mM NaF, 1 mM EDTA, 1 mM EGTA, 0.05% SDS, 1% Triton X-100, and 0.5% deoxycholate) twice followed by washing with kinase assay buffer containing 20 mM HEPES at pH 7.4, 1 mM EGTA, 0.4 mM EDTA, 5 mM MgCl2 and 0.05 mM dithiothreitol. His-MAD1 and His-MAD1-S546A were bacterially purified. The kinase reaction was performed at 37°C for 30 min, and the reaction was terminated by adding SDS sample buffer and subjected to SDS-PAGE.
In Vitro BUB1 phosphorylation assay
Purified human FLAG-BUB1-(437-509) traps were incubated for 60 min at 30°C with bacterially expressed and purified His-tagged MPS1-(519-808) and/or CDK2/cyclin A and in buffer containing 20 mM Tris/HCl at pH 7.5, 1 mM DTT, 0.1 mg/ml BSA, 2 mM MgAc and 1 mM ATP. Subsequently, the traps were used for pull-down with recombinant His-MAD1 and His-MAD1-S546A.
Immunofluorescence
Cells were cultured in confocal dishes to ∼60% confluence. After treatment, cells were fixed with 4% paraformaldehyde in PHEM buffer (60 mM PIPES, 25 mM HEPES, pH 6.9, 10 mM EGTA, 4 mM MgSO4) for 20 min at room temperature. Fixed cells were extracted with 0.5% Triton X-100 in PHEM buffer for 10 min. The dishes were incubated in blocking solution (0.8% BSA in PBS) and exposed overnight to primary antibody (1:100 dilution for all antibodies) at 4°C. The cells were then washed three times with blocking solution and then exposed to a secondary antibody (1:100 dilution) conjugated to FITC/TRITC. Cells were observed and analyzed under a confocal microscope (Zeiss LSM880).
Live cell microscopy
For determination of mitotic index, nocodazole (3.3 μM) was added to media. After 4, 8, 12 or 16 h, cells were observed using a 10×, 0.25-NA objective and counted the proportion of cells in mitosis to generate a mitotic index graph.
To measure mitotic timing, cells were grown in 384-well plate. After treatment, images were acquired at 5- or 30-min intervals on a high content screening system (Perkin Elmer) with a heated chamber and an automated stage at 37°C. Images were acquired using a 60×, 0.95-NA objective and perfect focus.
Chromosome segregation was observed in cells stably expressing histone H2B-RFP or H2B-mCherry grown in 35-mm dishes with glass bottoms in CO2-independent media. Image were acquired every 2 min by using a 63×, 1.4-NA oil-immersion objective (Zeiss LSM880). Maximum projections of in-focus planes were assembled in Elements, exported as jpg files, and converted to move files in QuickTime with a play rate of 5 frames/s.
Cell growth curve
Cells treated with taxol (5 nM) were seeded at a concentration of 5 × 104 cells/well in 12-well plates. After 24, 48, 72 and 96 h, the cells were harvested, diluted with a Trypan blue working solution, and counted to generate a growth curve.
Colony formation
Cells treated with taxol (5 nM) for 7 days were washed in PBS three times, digested and then plated in 6-well plates. After culture in a drug-free medium for 2 weeks, the cells were fixed using methanol and stained with Crystal Violet to identify visible colonies. The number of colonies was calculated using ImageJ (Image Processing and Analysis in Java).
Statistical analysis
Statistical analysis was performed by using the SPSS statistical software package (standard version 20; SPSS Inc., Chicago, IL, USA). For two groups statistical analyses, unpaired Student’s t-test was used; N.S.: non-significant, *: P < 0.05, **: P < 0.01, ***: P < 0.001. Error bars represent standard error of standard deviation (SD), as indicated in the legends. For more than two groups statistical analyses, the Dunnett test and the Tukey test were used; N.S.: non-significant, *: P < 0.05, **: P < 0.01, ***: P < 0.001.
RESULTS
ULK1 preserves chromosome integrity
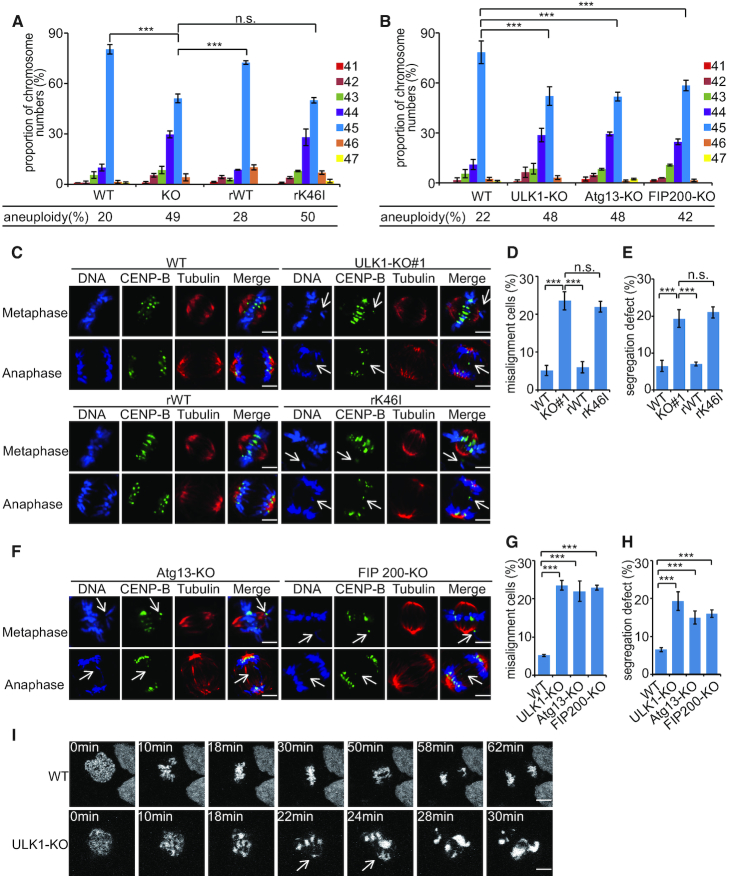
Autophagy deficiency promotes chromosomal instability, such as increased DNA damage, gene amplification and aneuploidy (33). Since ULK1 plays an important role in autophagy, we examined the effect of ULK1 on chromosome stability by generated ULK1 knocked-out HCT116 and DLD1 cells using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology (Supplementary Figure S1A and B). The control cell lines showed a relatively stable karyotype (about 80% normal karyotype with 45 chromosomes in HCT116 cells, about 74% normal karyotype with 46 chromosomes in DLD1 cells), while ULK1-KO cells were found to be highly aneuploid (Figure 1A and Supplementary Figure S1C). This alteration was partially rescued by re-expression of WT ULK1 but not kinase-impaired K46I ULK1 (Figure 1A), suggesting that ULK1 function in chromosome stability is dependent on its kinase activity. In autophagy induction process, the kinase activity of ULK1 is highly dependent on its binding partner, for example Atg13 and FIP200 (21,27,34). We therefore knocked-out Atg13 or FIP200 in HCT116 cells and then observed similar effect that caused by depletion of ULK1 (Figure 1B; Supplementary Figure S1D and E), pointing to a crucial role of ULK1 complex in sustaining chromosome stability.
Figure 1.
ULK1 preserves chromosome integrity. (A) A set of Cas9-resistant rescue forms of ULK1 plasmids (ULK1-rWT or rK46I) were stable transfected into HCT116 ULK1-KO cells. Cells were used for chromosome spread analyses and individual chromosomes were counted. The distribution of chromosome numbers is shown and the percentage of aneuploidy was calculated (45 chromosomes represent the relative euploid karyotype of HCT116 cells). At least 150 metaphase spreads were counted for each cell line. Data represent the mean ± SD of three independent experiments. Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (B) HCT116 WT cells, ULK1-KO cells (sgRNA#1), Atg13-KO cells and FIP200-KO cells were used for chromosome spread analyses and individual chromosomes were counted. Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (C) HCT116 WT cells, ULK1-KO cells, ULK1-rWT cells and ULK1-rK46I cells synchronized by thymidine (2 mM) double block were released for the indicated periods of time. These cells were immunostained with the indicated antibodies. The white arrows point to the fragmented chromatin or lagging chromosomes; scale bars: 5 μM. (D and E) Quantification of the misalignment and segregation defect cells in panel (C). One hundred cells in each indicated phase of mitosis were analyzed. Data represent the mean ± SD of three independent experiments. Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (F) HCT116 Atg13-KO cells and FIP200-KO cells synchronized by thymidine (2 mM) double block were released for the indicated periods of time. These cells were immunostained with the indicated antibodies. The white arrows point to the fragmented chromatin or lagging chromosomes; scale bars: 5 μM. (G and H) Quantification of the misalignment and segregation defect cells in panel (F). One hundred cells in each indicated phase of mitosis were analyzed. Data represent the mean ± SD of three independent experiments. Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (I) HeLa WT cells and ULK1-KO cells expressing mCherry-Histone H2B (for chromosome staining) synchronized by thymidine (2 mM) double block were released for 9 h. Live-cell confocal time-lapse images were taken at the indicated time points. Arrows indicate unaligned chromosomes and lagging chromosome/chromatid; scale bars: 10 μM.
To determine whether chromosome integrity is related with inaccurate chromosome segregation, abnormal chromosome incidences were visualized by immunofluorescence. Depletion of ULK1 complex resulted in chromosome misalignment in metaphase (Figure 1C–H and Supplementary Figure S1F–K) and abnormal chromosome segregation and lagging chromatids in anaphase (Figure 1C–H and Supplementary Figure S1F–K). In addition, live-cell microscopy of HeLa cells stably expressing the histone marker H2B–mCherry revealed that ULK1-depleted cells show defects of chromosome alignment and chromosome segregation (Figure 1I; Supplementary Figure S1L, and Supplementary Videos S1 and S2). These results indicate that ULK1 is necessary for fidelity of chromosome segregation.
ULK1 is required to maintain sufficient activity of the spindle assembly checkpoint
Spindle checkpoint ensures accurate chromosome segregation; therefore, we next tested the role of ULK1 in spindle assembly checkpoint (SAC). Live-cell imaging assay was performed by using HeLa cells stably expressing H2B-mCherry. ULK1 depletion yielded a significant decrease in mitosis duration (12.5 min faster than control cells, Figure 2A), indicating that ULK1 has a contribution to SAC strength during unperturbed mitosis. Then, we measured SAC strength by quantifying the amount of MAD2 that co-immunoprecipitates with CDC20. After RO3306 washout, CDC20:MAD2 interaction steadily increased for 40 min and subsequently decreased (Figure 2B). On the contrary, in ULK1-depleted cells, the interaction of CDC20:MAD2 was less pronounced, suggesting that ULK1 regulated CDC20:MAD2 interaction in early mitosis.
Figure 2.
ULK1 is required to maintain sufficient activity of the spindle assembly checkpoint. (A) Time-lapse analysis of the duration of mitosis from nuclear envelope breakdown to anaphase onset. WT and ULK1-KO Hela cell lines expressing mCherry-H2B were synchronized with thymidine for 24 h and released into fresh medium for 6 h. n indicates number of cells, m is the median duration of mitosis. (B) HCT116 WT cells and ULK1-KO cells were synchronized by sequential thymidine-RO3306 treatment and released into fresh medium prior to CDC20 immunoprecipitation. (C) HeLa cells were transfected with negative siRNA (siNC) or ULK1 siRNA (siULK1). After knockdown of endogenous ULK1, ULK1-rWT or ULK1-rK46I was transfected into HeLa cells. These cells were treated with 3.3 μM nocodazole. n = 300 cells per time point and per treatment, from three independent experiments. (D) HCT116 WT cells, ULK1-KO cells, ULK1-rWT cells and ULK1-rK46I cells were treated with thymidine for 24 h and then released into 200 nM nocodazole 16 h before immunoblotting. Mitotic cells were collected by shake-off. (E) Time-lapse analysis of the duration of mitosis. WT and ULK1-KO Hela cell lines expressing mCherry-H2B were synchronized with thymidine for 24 h and released into fresh medium for 6 h prior to treatment with the indicated drugs. n indicates number of cells, m is the median duration of mitosis. (F) Time-lapse analysis of the duration of mitosis. HeLa ULK1-KO cell lines expressing mCherry-H2B were transfected with GFP-ULK1-WT or GFP-ULK1-K46I. Indicated cells were treated with thymidine for 24 h and released into fresh medium for 6 h prior to treatment with 200 nM nocodazole. n indicates number of cells, m is the median duration of mitosis. Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (G) Time-lapse analysis of the duration of mitosis. HCT116 WT cells, ULK1-KO cells, Atg3-KO cells, Atg7-KO cells, ULK1/Atg3-KO cells and ULK1/Atg7-KO cells expressing RFP-H2B were treated with thymidine for 24 h and released into fresh medium for 6 h prior to treatment with or without 200 nM nocodazole. n indicates number of cells, m is the median duration of mitosis. Asterisks indicate significance (the Dunnett test; ***: P < 0.001).
The effect of ULK1 on SAC was also determined under mitotic poisons treatment. As shown in Figure 2C and Supplementary Figure S2A–C, ULK1-knockdown cells exhibited reduced mitotic index after nocodazole treatment compared with wild-type cells. In addition, Cyclin B1 levels were reduced in ULK1-KO cells after nocodazole treatment, consistent with an early mitotic exit due to a SAC defect (Figure 2D). This effect was largely rescued by expression of the wild-type ULK1. To further validate the SAC data in single cells, we measure the duration of mitosis in cells that had been treated with taxol, vinblastine or nocodazole. ULK1 depletion reduced the mitotic duration of mitotic poisons-arrested cells, and this phenotype was rescued by the expression of WT-ULK1, but not by kinase-impaired K46I ULK1 (Figure 2E–F).
Because ULK1 plays important roles in autophagy initiation (17–23), we then tested whether ULK1-mediated autophagy is related with effect of ULK1 on SAC. By using the autophagy-deficient Atg3/Atg7 knockout HCT116 cell line (Supplementary Figure S2D), we found that ULK1 was still required for maintaining the duration of mitosis (Figure 2G and Supplementary Figure S2E), supporting the autophagy-independent role of ULK1 in regulating the activity of SAC.
ULK1 is involved in Mad1 recruitment to the kinetochores
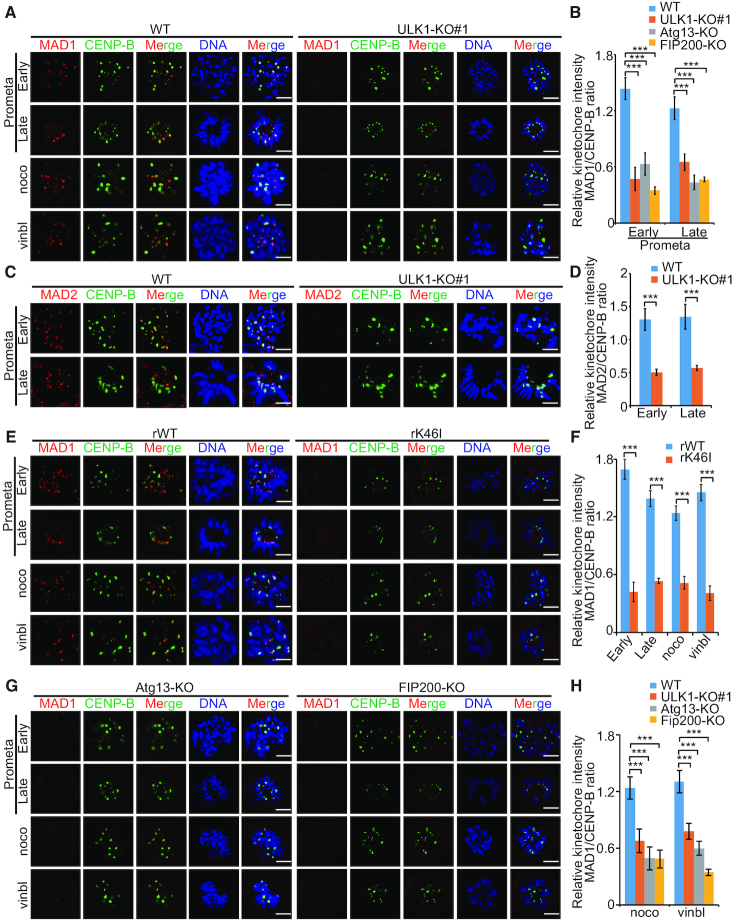
We next investigated whether ULK1 affect the accumulation of SAC effectors at the kinetochores. We found that ULK1 depletion reduced the recruitment of Mad1 and Mad2 to kinetochores in prometaphase and in cells that had been blocked with spindle poisons (Figure 3A–F and Supplementary Figure S3A–D). In contrast, the recruitment of other SAC component proteins (such as Bub1, Bub3 and BubR1) to kinetochore regions was not affected (Supplementary Figure S4A–D). Similar results were also obtained in Atg13-KO and FIP200-KO HCT116 cells (Figure 3G and H) but not Atg3-KO cells (Supplementary Figure S4E–F), indicating that ULK1 complex regulated Mad1 recruitment to the kinetochores also in an autophagy-independent manner. Since RZZ complex is particularly important for Mad1 kinetochore localization (14,35), we also tested ZW10’s location and found that the recruitment of ZW10 to kinetochores was not affected by ULK1 (Supplementary Figure S4G–H).
Figure 3.
ULK1 is involved in mad1 recruitment to the kinetochores. (A) HCT116 WT cells and ULK1-KO (#1) cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h, or into medium containing nocodazole (330 nM) or vinblastine (4.4 μM) for 10 h; scale bars, 5 μM. (B) Quantification of the kinetochore MAD1/CENP-B ratio in (A). Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (C) HCT116 WT cells and ULK1-KO (#1) cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h; scale bars: 5 μM. (D) Quantification of the kinetochore MAD2/CENP-B ratio in (C). (E) A set of Cas9-resistant rescue forms of ULK1 plasmids (ULK1-rWT or rK46I) were stable transfected into HCT116 ULK1-KO (#1) cells. ULK1-rWT- or ULK1-rK46I-expressing HCT116 cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h, or into medium containing nocodazole (330 nM) or vinblastine (4.4 μM) for 10 h; scale bars: 5 μM. (F) Quantification of the kinetochore MAD1/CENP-B ratio in (E). (G) Atg13-KO cells and FIP200-KO HCT116 cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h, or into medium containing nocodazole (330 nM) or vinblastine (4.4 μM) for 10 h; scale bars: 5 μM. (H) Quantification of the kinetochore MAD1/CENP-B ratio in (G). Asterisks indicate significance (the Dunnett test; ***: P < 0.001).
Mad1 is a substrate of ULK1
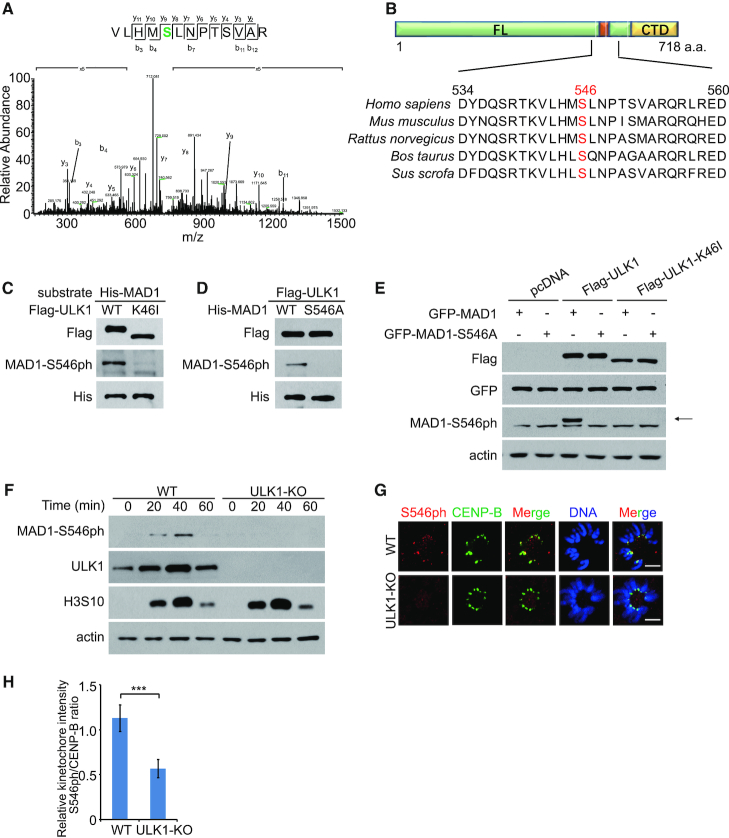
ULK1 is a well-known serine–threonine kinase, we therefore tested whether ULK1-dependent Mad1 recruitment is related with its phosphorylation. Exogenous ULK1 and MAD1 can bind with each other, and the interaction between ULK1 and Mad1 is not affected by ULK1’s kinase activity (Supplementary Figure S5A and B). In addition, Mad1’s C-terminal is both necessary and sufficient to mediate interaction with ULK1’s C-terminal (Supplementary Figure S5C and D). Mass spectrometry analysis indicated that highly conserved site S546 of Mad1 was phosphorylated by ULK1 (Figure 4A and B). To further confirm these sites, Mad1-WT and Mad1-S546A His-fusion protein were purified, and an in vitro phosphorylation assay was performed. By using an antibody for the S546-phosphorylated peptide of Mad1, we found that His-Mad1-WT was phosphorylated by wild-type ULK1, but not the kinase impaired K46I ULK1 mutant (Figure 4C and D).
Figure 4.
Mad1 is a substrate of ULK1. (A) Identification of S546 phosphorylation on His-MAD1 incubated in vitro with ULK1 by using mass spectrometry analysis. (B) Schematic of the phosphorylation sites Ser546 in human and other species MAD1. (C) Flag-ULK1 or Flag-ULK1-K46I proteins were immunopurified from transfected HEK293 cells. Then in vitro phosphorylation assays were performed. (D) Flag-ULK1 protein was immunopurified from transfected HEK293 cells. In vitro phosphorylation assays were then performed in the presence of His-MAD1 or His-MAD1-S546A proteins. (E) An empty plasmid and an expression plasmid for Flag-ULK1 or Flag-ULK1-K46I were co-transfected with GFP-MAD1 or GFP-MAD1-S546A into HCT116 cells for 48 h. Proteins were then extracted for western blotting. (F) HCT116 WT cells and ULK1-KO cells were synchronized by sequential thymidine-RO3306 treatment and released into fresh medium. Proteins were then extracted for western blotting. (G) HCT116 WT cells and ULK1-KO cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h; scale bars: 5 μM. (H) Quantification of the kinetochore S546ph/CENP-B ratio in (G). Asterisks indicate significance (the Dunnett test; ***: P < 0.001).
Next, we tried to determine whether Mad1-S546 is phosphorylated by ULK1 in vivo. A significant increase of phosphorylated S546-Mad1 was detected in ULK1-overexpressed HCT116 cells (Figure 4E), suggesting that ULK1 plays a critical role in regulating Mad1 phosphorylation. In addition, we also synchronized HCT116 cells at the G2/M phase border with RO3306 and then released the block by removing RO3306 for different times. Immunoblotting studies revealed a significant increase in phosphorylated Mad1 that was detected in mitotic HCT116 cells, but not in ULK1-KO HCT116 cells (Figure 4F). Anti-phos-Mad1 (Ser546) antibody immunostaining was also performed and analyzed in WT and ULK1-KO cells. As shown in Figure 4G and H, p-Mad1 (Ser546) is targeted to kinetochore in prometaphase only in WT-HCT116 cells, suggesting that ULK1 plays a critical role in regulating Mad1 phosphorylation. Since the S546 located near the Mad2-interacting motif (MIM) (541 aa to 545 aa), the interaction between Mad1-S546A and Mad2 was then detected. We found that the Mad1–Mad2 binding was not affected by S546 mutation (Supplementary Figure S5E), indicating that S546 phosphorylation is not involved in Mad1–Mad2 complex formation.
Mad1-Ser546 phosphorylation is involved in Mad1 kinetochore location
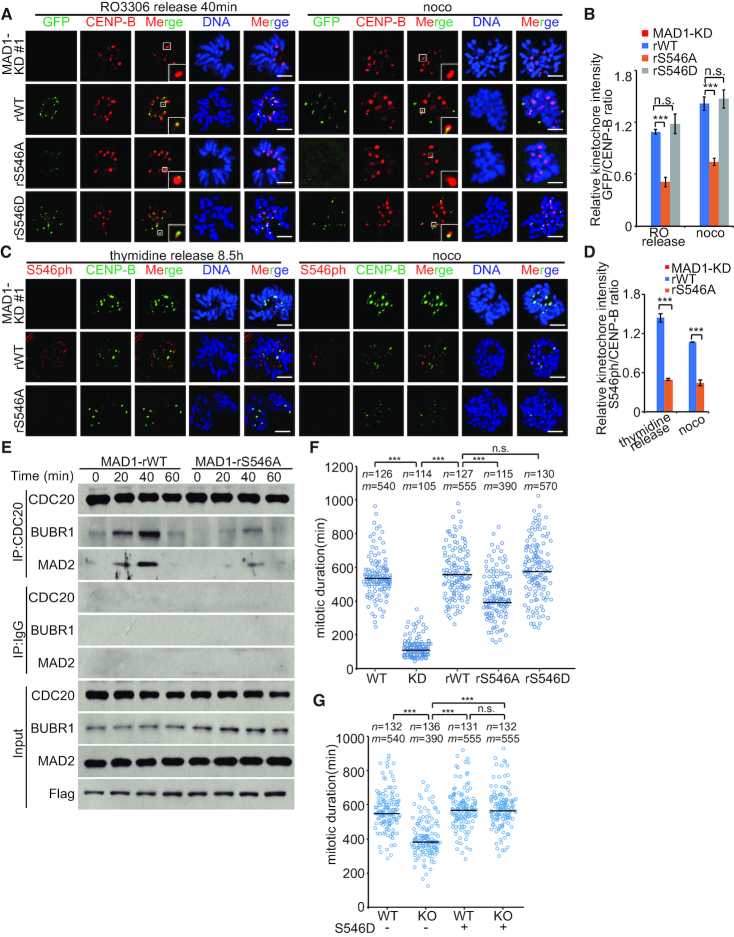
To detect the involvement of S546 phosphorylation in the kinetochore targeting of Mad1, we next transfected Mad1-WT, Mad1-S546A or Mad1-S546D plasmid into HCT116 Mad1-KD cells (Supplementary Figure S6A). We found that Mad1 kinetochore-localization was compromised when Ser546 was mutated to alanine (Figure 5A and B; Supplementary Figure S6B and C). Anti-phos-Mad1 (Ser546) antibody immunostaining was also performed and analyzed in Mad1-WT- and Mad1-S546A-expressing cells. As shown in Figure 5C and D, p-Mad1 (Ser546) is targeted to kinetochore in prometaphase, suggesting that kinetochore-located Mad1 is phosphorylated during mitosis. In addition, we also created Mad1-5A plasmid which mutates the MIM motif (541 aa to 545 aa) to alanines. As shown in Supplementary Figure S6D–F, Mad1-5A-S546A has ∼37% of the kinetochore-binding activity of Mad1-5A, suggesting the role of Ser546 in the kinetochore recruitment of Mad1.
Figure 5.
Mad1-Ser546 phosphorylation is involved in Mad1 kinetochore location. (A) GFP-MAD1-WT, GFP-MAD1-S546A or GFP-MAD1-S546D was transfected into stable MAD1-RNAi (#1) HCT116 cells. Cells were synchronized by sequential thymidine-RO3306 treatment and released into fresh medium for 40 min or thymidine-nocodazole treatment for 10 h; scale bars: 5 μM. (B) Quantification of the kinetochore GFP/CENP-B ratio in (A). Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (C) Flag-MAD1-WT or Flag-MAD1-S546A was transfected into HCT116 MAD1-KD cells. Cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h, or into medium containing nocodazole (330 nM) for 10 h; scale bars: 5 μM. (D) Quantification of the kinetochore S546ph/CENP-B ratio in (C). (E) HCT116 WT cells were transfected with MAD1 siRNA (siMAD1). At 48 h after transfection, MAD1-rWT or MAD1-rS546A was transfected into HCT116 cells. Cells were synchronized by sequential thymidine-RO3306 treatment and released into fresh medium prior to CDC20 immunoprecipitation. (F) Time-lapse analysis of the duration of mitosis. MAD1-KD cell lines expressing RFP-H2B stably were transfected with GFP-MAD1-WT, GFP-MAD1-S546A or GFP-MAD1-S546D. Indicated cells were treated with thymidine for 24 h and released into fresh medium for 6 h prior to treatment with 200 nM nocodazole. n indicates number of cells, m is the median duration of mitosis. Asterisks indicate significance (the Tukey test; ***: P < 0.001). (G) Time-lapse analysis of the duration of mitosis. HCT116 WT cells and ULK1-KO cells expressing RFP-H2B stably were transfected with GFP-MAD1-S546D. Indicated cells were treated with thymidine for 24 h and released into fresh medium for 6 h prior to treatment with 200 nM nocodazole. n indicates number of cells, m is the median duration of mitosis. Asterisks indicate significance (the Tukey test; ***: P < 0.001).
Next, to test the role of Mad1-Ser546 in SAC regulation, we measured SAC strength by quantifying the amount of MAD2 that co-immunoprecipitates with CDC20. In Mad1-S546A-expressing cells, the interaction of CDC20:MAD2 was less pronounced than that in Mad1-WT-expressing cells, suggesting that Mad1-S546 phosphorylation regulated CDC20:MAD2 interaction in mitosis (Figure 5E). Next, we used Mad1-WT-, Mad1-S546A- and Mad1-S546D-expressing cell lines to observe the duration of mitosis in single cells (Figure 5F). Upon nocodazole treatment, the expression of Mad1-S546A considerably reduced the duration of mitosis compared with Mad1-WT-expressing cells (Figure 5F), furthering confirming the role of Mad1-Ser546 phosphorylation in SAC regulation. Moreover, to investigate the function of ULK1 in the activation of spindle assembly is indeed dependent on the phosphorylation of MAD1, we transfected Mad1-S546D into ULK1 KO cells. As shown in Figure 5G, Mad1-S546D partly rescued the duration of mitosis, supporting the role of Mad1 phosphorylation in the ULK1-regulated SAC.
RZZ complex may serve as a receptor for phos-Ser546-Mad1 at Kinetochore
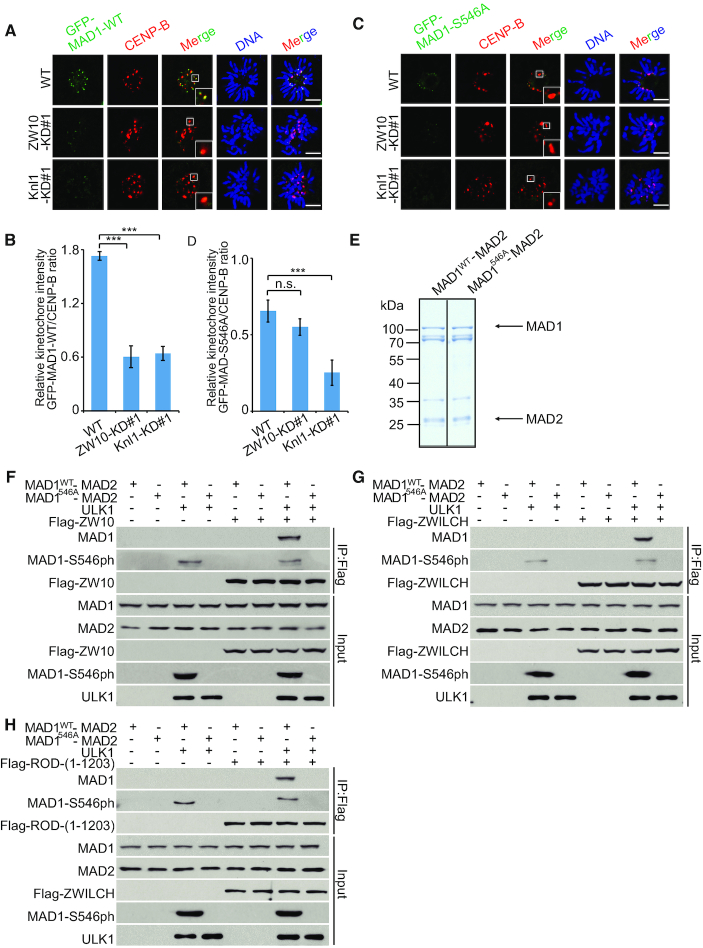
Since RZZ and Knl1/Bub3/Bub1 (KBB) have distinct roles in recruiting Mad1 at kinetochores, we then generate Knl1-knockdown and ZW10-knockdown cells to observe which pathway contributed for phos-S546-Mad1 recruitment (Supplementary Figure S7A and B). As shown in Figure 6A–D and Supplementary Figure S7C–F, Knl1 depletions reduce the amount of kinetochore-bound Mad1-S546A by 62%, whereas ZW10 depletion only reducing Mad1-S546A levels by 17%. We next focused our attention on potential mechanisms of RZZ complex-mediated phos-S546-Mad1 kinetochore recruitment. How RZZ might help recruit Mad1:Mad2 is unclear, as no physical interaction between endogenous Mad1 and RZZ has been reported in mammalian cells. In Drosophila, only a small fraction of overexpressed RZZ and Mad1 co-precipitate (36). Consistent with previous reports, we did not observe the interaction between endogenous RZZ and WT-Mad1/S546A-Mad1 either. Then we purified Mad1 His-fusion protein and performed an in vitro ULK1-phosphorylation assay to obtain phos-S546-Mad1 protein. After incubating with flag-Rod-/flag-Zw10-/flag-Zwilch-expressing cell lysates, wild-type Mad1, but not phosphorylation-incompetent S546A-Mad1, was detected in IPs of cell lysates (Supplementary Figure S7G–I), indicating that phosphorylation of Mad1 by ULK1 can promote the interaction between Mad1 and RZZ complex. Since Mad1 is tightly bound to Mad2 in cells, we also tested the interaction between RZZ and Mad1–Mad2 complex. By using reconstituted Mad1–Mad2 complex (Figure 6E), we obtained similar results and further confirmed that Mad1 phosphorylated at S546 could interact with the RZZ complex (Figure 6F–H).
Figure 6.
RZZ complex may serve as a receptor for phos-Ser546-Mad1 at kinetochore. (A) HCT116 WT cells, ZW10-KD (#1) cells and Knl1-KD (#1) cells were transfected with GFP-MAD1-WT. Cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h; scale bars: 5 μM. (B) Quantification of the kinetochore GFP-MAD1-WT/CENP-B ratio in (A). Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (C) HCT116 WT cells, ZW10-KD (#1) cells and Knl1-KD (#1) cells were transfected with GFP-MAD1-S546A. Cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 8.5 h; scale bars: 5 μM. (D) Quantification of the kinetochore GFP-MAD1-S546A/CENP-B ratio in (C). Asterisks indicate significance (the Dunnett test; ***: P < 0.001). (E) MAD1–MAD2 recombinant proteins used for the in vitro phosphorylation assays. Relevant protein bands were indicated by arrows. (F andH) HCT116 cells were transfected with or without the flag-ZW10 (F), flag-ZWILCH (G) or flag-Rod (H) plasmid. Flag protein was extracted on the beads as described. In vitro phosphorylated MAD1–MAD2 by ULK1 was then incubated with the beads. Western blotting was performed by using indicated antibodies.
Inhibition of ULK1 increases the cancer cells sensitivity to taxol
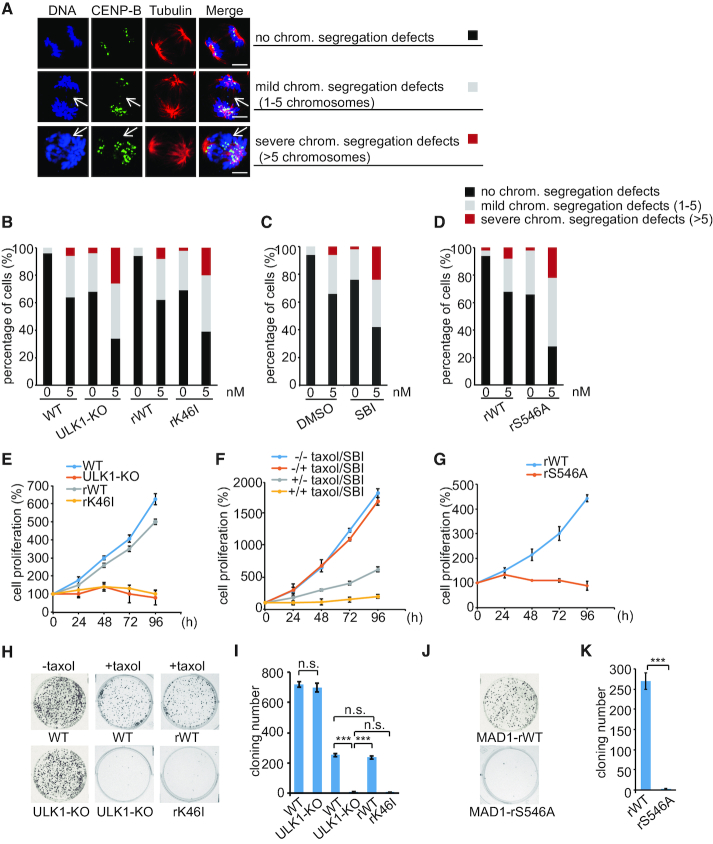
A functional mitotic checkpoint has been reported by numerous groups to be required for sensitivity to microtubule poisons (37–40). In contrast, cells with weakened mitotic checkpoint have also been reported to be sensitive to mitotic poisons (41–43). We then tried to investigate the role of ULK1-mediated Mad1 phosphorylation in regulating the sensitivity of cells to microtubule poisons. Paclitaxel (taxol), a microtubule-stabilizing antineoplastic agent, has been expected to exert its anti-tumor effects by inducing mitosis arrest for decades. However, intratumoral concentrations of taxol are lower than that necessary to affect mitotic arrest (44). Therefore, we next tested the effect of ULK1 on the fidelity of chromosome segregations in response to low-dose taxol. We found that upon 5 nM taxol treatment, the frequency and severity of chromosome segregation errors was increased in ULK1-KO cells or in cells treated with SBI-0206965, a highly selective ULK1 inhibitor (45) (Figure 7A–C; Supplementary Figure S8A and B). Furthermore, compared with that in Mad1-WT-expressing cells, the percentage of severely mis-segregated chromosomes in Mad1-S546A-expressing cells increased from 8% to 22% after taxol treatment (Figure 7D). These data suggested that inhibition of ULK1-induced Mad1 phosphorylation leads to higher chromosomal instability (CIN) upon low-dose taxol treatment. Then, we tested whether that high CIN could reduce cell proliferation. As shown in Figure 7E and F, and Supplementary Figure S8C and D, ULK1-inibibited cells are much more sensitive to taxol than wild-type cells with normal ULK1 level or activity. A rescue experiment in Mad1-KD cells yielded similar results, with ∼400% proliferation rate in cells with Mad1-WT treated with taxol for 4 days, but only 90% proliferation rate in cells expressing Mad1-S546A (Figure 7G). Additionally, we also tested the colony formation of the ULK1-KO and wild-type cell lines under the treatment of 5 nM taxol. We found that colony formation capacity of ULK1-KO cells or Mad1-S546A-expressing cells was drastically reduced by taxol compared with that of control cells, suggesting that ULK1 inhibition sensitizes tumor cells to clinically relevant doses of taxol. (Figure 7H–K).
Figure 7.
Inhibition of ULK1 increases the cancer cells sensitivity to taxol. (A) Immunofluorescence images of cells with different chromosome segregation defects. Cells were synchronized by thymidine (2 mM) double block and released into fresh medium for 9 h. ‘No chrom. segregation defects’, ‘Mild chrom. segregation defects’ or ‘Severe chrom. segregation defects’ indicate cells with 0, 1–5, or more than 5 chromosomes segregation defects in anaphase, respectively. (B) HCT116 WT cells, ULK1-KO cells, rWT cells and rK46I cells were treated with thymidine for 24 h and then released into medium with or without 5 nM taxol for 9 h before staining for DNA and quantification. (C) HCT116 WT cells were treated with or without 2 μM SBI throughout the course of the experiment. Cells were treated with thymidine for 24 h and then released into fresh medium or into medium containing 5 nM taxol for 9 h before staining for DNA and quantification. (D) A set of RNAi-resistant rescue forms of MAD1 plasmids (MAD1-rWT or rS546A) were stably transfected into HCT116 MAD1-KD cells. Cells were treated with thymidine for 24 h and then released into fresh medium or into medium containing 5 nM taxol for 8.5 h before staining for DNA and quantification. (E) HCT116 WT cells, ULK1-KO cells, ULK1-rWT cells and ULK1-rK46I cells were treated with taxol (5 nM). After 24, 48, 72 and 96 h, the cells were harvested and counted to generate a growth curve. (F) HCT116 WT cells were treated with or without 2 μM SBI throughout the course of the experiment. Cells were treated with taxol (5 nM). After 24, 48, 72 and 96 h, the cells were harvested and counted to generate a growth curve. (G) A set of RNAi-resistant rescue forms of MAD1 plasmids (MAD1-rWT or rS546A) were stably transfected into HCT116 MAD1-KD cells. Cells were treated with taxol (5 nM) for different times, then harvested and counted to generate a growth curve. (H) HCT116 WT cells, ULK1-KO cells, ULK1-rWT cells and ULK1-rK46I cells were treated with or without taxol (5 nM). After 2 weeks, the cloning formation was analyzed with Crystal Violet staining. (I) Quantification of the colony numbers in (H). Asterisks indicate significance (the Tukey test; ***: P < 0.001). (J) Colony formations of indicated treatment in (G). (K) Quantification of the colony numbers in (J).
DISCUSSION
In yeast, Atg1 is a central autophagy regulator required for bulk autophagy (46). In mammalian cells, Atg1 has two homologs known as ULK1 and ULK2. ULK1/2 forms a complex with Atg13, FIP200 and Atg101. Distinct from most other types of Atg-deficient mice, FIP200-deficient mice (47) or Atg13-deficient mice (48) die in utero. Mice lacking either ULK1 or ULK2 are viable, but ULK1/2 double-knockout mice die within 24 h of birth (49), indicating that ULK1/2–Atg13–FIP200 complex have additional functions in pathways other than autophagy. Here in our study, we found that, under non-stress conditions, ULK1 complex also plays important roles in SAC regulation. Moreover, we demonstrate that ULK1 phosphorylates Mad1 S546, which leads to the recruitment of Mad1 to kinetochores and precise control of SAC to guarantee accurate chromosome segregation. Several reports have shown that Mad1 phosphorylation contributes to its function regulation. For example, phosphorylated Mad1 at Thr716 directly binds to Cdc20 and inhibits APC/CCdc20 presumably through stimulating MCC assembly (50). Thr680 phosphorylation is important for the kinetochore localization of Mad1 and its SAC function (51). ATM-mediated S214 phosphorylation regulates Mad1 dimerization as well as heterodimerization with Mad2, and contributes to activation of the SAC and the maintenance of chromosomal stability (52). Here our results show that ULK1 is also an important regulator for Mad1 phosphorylation and its subsequent localization to kinetochore. After phosphorylation on Ser546, Mad1 is prone to localize to the kinetochore and recruit Mad2 to form mitotic checkpoint complex. Substitution of Ser546 in Mad1 with alanine (Ser546A) diminishes the capacity of Mad1 kinetochore targeting and SAC activation (Figure 5A and B). These data support the notion that ULK1-dependent phosphorylation of Mad1 on Ser546 plays a significant role in SAC and faithful chromosome segregation.
In human cells, there are two kinetochore recruitment pathways for Mad1:Mad2. One is the Knl1/Bub3/Bub1 (KBB) pathway and the other is the Rod/ZW10/Zwilch (RZZ) kinetochore complex. In the KBB pathway, Bub1 kinase is required for Mad1–Mad2 kinetochore recruitment (53,54), and protein phosphorylation modification plays important roles in this process. For example, Mps1 phosphorylation of conserved domain 1 (CD1) in Bub1 promotes Bub1:Mad1 interaction (11–13). In our study, we tried to investigate whether the interaction between Mad1 and Bub1 was effected by Mad1 S546 phosphorylation. However, no obvious change was observed for the binding of Bub1 with wild-type Mad1 and phosphorylated Mad1 by ULK1 (Supplementary Figure S7J), indicating that phos-S546-Mad1 kinetochore recruitment may be independent on KBB pathway. On the other hand, through a poorly understood mechanism, RZZ pathway stabilizes kinetochore localization of the Mad1:Mad2 complex (14,35,55–58). How RZZ might help recruit Mad1:Mad2 is unclear, as no physical interaction between Mad1 and RZZ has been reported. In Drosophila, only a small fraction of RZZ and Mad1 co-precipitate (36). In our study, by using in vitro purified Mad1 protein, we found that wild-type Mad1, but not S546A-Mad1, was able to interact with RZZ complex that is overexpressed in HCT116 cells (Figure 6F–H). In addition, Knl1 but not ZW10 depletions reduce the amount of kinetochore-bound Mad1-S546A and mitosis duration in Mad1-S546A-expressing cells (Figure 6C and D). Therefore, we hypothesize that RZZ pathway is more important for phosphorylated Mad1 recruitment to kinetochore. The specific relationship between Mad1 and this pathway will be the subject of our future work.
Previous study showed that partial reduction of essential mitotic checkpoint components in tumor cells caused mild chromosome mis-segregations, but no lethality (59). In our study, we found that inhibition of ULK1-induced Mad1 phosphorylation also leads to an increase in mild chromosome segregation errors (Figure 7A–D). While combining ULK1 inhibition with non-lethal dose of taxol increases the severity of segregation errors and reduces the proliferation rate of tumor cells (Figure 7E–F). A growing body of evidence suggests that high rates of CIN cause cell death and tumor suppression (60). It has been reported that ULK1 inhibitor SBI-0206965 greatly synergized with mTOR inhibitor to kill tumor cells (45). In our study, we found that ULK1 inhibition also enhanced cancer cells sensitivity to taxol, thus providing a strong rationale for their combined use in the clinic.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFA0503900]; National Natural Science Foundation of China [81472581, 81672712, 81874145, 81621063, 91319302, 31261140372]. Funding for open access charge: National Key R&D Program of China [2017YFA0503900].
Conflict of interest statement. None declared.
REFERENCES
- 1. Varetti G., Musacchio A.. The spindle assembly checkpoint. Curr. Biol. 2008; 18:R591–R595. [DOI] [PubMed] [Google Scholar]
- 2. De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D. et al.. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005; 15:214–225. [DOI] [PubMed] [Google Scholar]
- 3. Primorac I., Musacchio A.. Panta rhei: the APC/C at steady state. J. Cell Biol. 2013; 201:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maldonado M., Kapoor T.M.. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 2011; 13:475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sironi L., Melixetian M., Faretta M., Prosperini E., Helin K., Musacchio A.. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001; 20:6371–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K.T., Musacchio A.. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002; 21:2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen R.H., Shevchenko A., Mann M., Murray A.W.. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 1998; 143:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. London N., Ceto S., Ranish J.A., Biggins S.. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 2012; 22:900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., Musacchio A.. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife. 2013; 2:e01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., Millar J.B.. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 2012; 22:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brady D.M., Hardwick K.G.. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000; 10:675–678. [DOI] [PubMed] [Google Scholar]
- 12. London N., Biggins S.. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 2014; 28:140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mora-Santos M.D., Hervas-Aguilar A., Sewart K., Lancaster T.C., Meadows J.C., Millar J.B.. Bub3-Bub1 Binding to Spc7/KNL1 Toggles the Spindle Checkpoint Switch by Licensing the Interaction of Bub1 with Mad1-Mad2. Curr. Biol. 2016; 26:2642–2650. [DOI] [PubMed] [Google Scholar]
- 14. Silio V., McAinsh A.D., Millar J.B.. KNL1-Bubs and RZZ provide two separable pathways for checkpoint activation at human kinetochores. Dev. Cell. 2015; 35:600–613. [DOI] [PubMed] [Google Scholar]
- 15. Rodriguez-Rodriguez J.A., Lewis C., McKinley K.L., Sikirzhytski V., Corona J., Maciejowski J., Khodjakov A., Cheeseman I.M., Jallepalli P.V.. Distinct Roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol. 2018; 28:3422–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang G., Kruse T., Guasch Boldu C., Garvanska D.H., Coscia F., Mann M., Barisic M., Nilsson J.. Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J. 2019; 38:e100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan J., Kuroyanagi H., Kuroiwa A., Matsuda Y., Tokumitsu H., Tomoda T., Shirasawa T., Muramatsu M.. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem. Biophys. Res. Commun. 1998; 246:222–227. [DOI] [PubMed] [Google Scholar]
- 18. Yan J., Kuroyanagi H., Tomemori T., Okazaki N., Asato K., Matsuda Y., Suzuki Y., Ohshima Y., Mitani S., Masuho Y. et al.. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999; 18:5850–5859. [DOI] [PubMed] [Google Scholar]
- 19. Chan E.Y., Longatti A., McKnight N.C., Tooze S.A.. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell Biol. 2009; 29:157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan E.Y., Kir S., Tooze S.A.. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 2007; 282:25464–25474. [DOI] [PubMed] [Google Scholar]
- 21. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., Mizushima N.. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008; 181:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Young A.R., Chan E.Y., Hu X.W., Kochl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A.. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006; 119:3888–3900. [DOI] [PubMed] [Google Scholar]
- 23. Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010; 22:132–139. [DOI] [PubMed] [Google Scholar]
- 24. Guo Y., Peng D., Zhou J., Lin S., Wang C., Ning W., Xu H., Deng W., Xue Y.. iEKPD 2.0: An update with rich annotations for eukaryotic protein kinases, protein phosphatases and proteins containing phosphoprotein-binding domains. Nucleic Acids Res. 2019; 47:D344–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deng W., Ma L., Zhang Y., Zhou J., Wang Y., Liu Z., Xue Y.. THANATOS: an integrative data resource of proteins and post-translational modifications in the regulation of autophagy. Autophagy. 2018; 14:296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.Y., Kim J., Kim H., Neufeld T.P., Dillin A., Guan K.L.. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013; 15:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N. et al.. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009; 20:1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H.. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009; 20:1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li T.Y., Sun Y., Liang Y., Liu Q., Shi Y., Zhang C.S., Zhang C., Song L., Zhang P., Zhang X. et al.. ULK1/2 Constitute a bifurcate node controlling glucose metabolic fluxes in addition to autophagy. Mol. Cell. 2016; 62:359–370. [DOI] [PubMed] [Google Scholar]
- 30. Konno H., Konno K., Barber G.N.. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013; 155:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joo J.H., Wang B., Frankel E., Ge L., Xu L., Iyengar R., Li-Harms X., Wright C., Shaw T.I., Lindsten T. et al.. The noncanonical role of ULK/ATG1 in ER-to-golgi trafficking is essential for cellular homeostasis. Mol. Cell. 2016; 62:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li R., Yuan F., Fu W., Zhang L., Zhang N., Wang Y., Ma K., Li X., Wang L., Zhu W.G. et al.. Serine/threonine kinase Unc-51-like Kinase-1 (Ulk1) phosphorylates the Co-chaperone Cell Division Cycle Protein 37 (Cdc37) and thereby disrupts the stability of Cdc37 Client Proteins. J. Biol. Chem. 2017; 292:2830–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathew R., Kongara S., Beaudoin B., Karp C.M., Bray K., Degenhardt K., Chen G., Jin S., White E.. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007; 21:1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ganley I.G., Lam du H., Wang J., Ding X., Chen S., Jiang X.. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009; 284:12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caldas G.V., Lynch T.R., Anderson R., Afreen S., Varma D., DeLuca J.G.. The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells. Open Biol. 2015; 5:150160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Defachelles L., Raich N., Terracol R., Baudin X., Williams B., Goldberg M., Karess R.E.. RZZ and Mad1 dynamics in Drosophila mitosis. Chromosome Res. 2015; 23:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kasai T., Iwanaga Y., Iha H., Jeang K.T.. Prevalent loss of mitotic spindle checkpoint in adult T-cell leukemia confers resistance to microtubule inhibitors. J. Biol. Chem. 2002; 277:5187–5193. [DOI] [PubMed] [Google Scholar]
- 38. Sudo T., Nitta M., Saya H., Ueno N.T.. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 2004; 64:2502–2508. [DOI] [PubMed] [Google Scholar]
- 39. Kops G.J., Foltz D.R., Cleveland D.W.. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:8699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swanton C., Marani M., Pardo O., Warne P.H., Kelly G., Sahai E., Elustondo F., Chang J., Temple J., Ahmed A.A. et al.. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007; 11:498–512. [DOI] [PubMed] [Google Scholar]
- 41. Lee E.A., Keutmann M.K., Dowling M.L., Harris E., Chan G., Kao G.D.. Inactivation of the mitotic checkpoint as a determinant of the efficacy of microtubule-targeted drugs in killing human cancer cells. Mol. Cancer Ther. 2004; 3:661–669. [PubMed] [Google Scholar]
- 42. Sihn C.R., Suh E.J., Lee K.H., Kim T.Y., Kim S.H.. p55CDC/hCDC20 mutant induces mitotic catastrophe by inhibiting the MAD2-dependent spindle checkpoint activity in tumor cells. Cancer Lett. 2003; 201:203–210. [DOI] [PubMed] [Google Scholar]
- 43. Zhang Y., Lees E.. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: Model for spindle checkpoint regulation. Mol. Cell Biol. 2001; 21:5190–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zasadil L.M., Andersen K.A., Yeum D., Rocque G.B., Wilke L.G., Tevaarwerk A.J., Raines R.T., Burkard M.E., Weaver B.A.. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl. Med. 2014; 6:229ra243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Egan D.F., Chun M.G., Vamos M., Zou H., Rong J., Miller C.J., Lou H.J., Raveendra-Panickar D., Yang C.C., Sheffler D.J. et al.. Small molecule inhibition of the autophagy kinase ULK1 and Identification of ULK1 substrates. Mol. Cell. 2015; 59:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsukada M., Ohsumi Y.. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993; 333:169–174. [DOI] [PubMed] [Google Scholar]
- 47. Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J.L.. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 2006; 175:121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaizuka T., Mizushima N.. Atg13 is essential for autophagy and cardiac development in mice. Mol. Cell Biol. 2015; 36:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheong H., Wu J., Gonzales L.K., Guttentag S.H., Thompson C.B., Lindsten T.. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy. 2014; 10:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ji Z., Gao H., Jia L., Li B., Yu H.. A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. Elife. 2017; 6:e22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chi Y.H., Haller K., Ward M.D., Semmes O.J., Li Y., Jeang K.T.. Requirements for protein phosphorylation and the kinase activity of polo-like kinase 1 (Plk1) for the kinetochore function of mitotic arrest deficiency protein 1 (Mad1). J. Biol. Chem. 2008; 283:35834–35844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang C., Hao J., Kong D., Cui X., Zhang W., Wang H., Guo X., Ma S., Liu X., Pu P. et al.. ATM-mediated Mad1 Serine 214 phosphorylation regulates Mad1 dimerization and the spindle assembly checkpoint. Carcinogenesis. 2014; 35:2007–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sharp-Baker H., Chen R.H.. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 2001; 153:1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gillett E.S., Espelin C.W., Sorger P.K.. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004; 164:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E.. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 2005; 15:856–861. [DOI] [PubMed] [Google Scholar]
- 56. Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R. 3rd, Tagaya M., Cleveland D.W.. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005; 169:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matson D.R., Stukenberg P.T.. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J. Cell Biol. 2014; 205:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang G., Lischetti T., Hayward D.G., Nilsson J.. Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat. Commun. 2015; 6:7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Janssen A., Kops G.J., Medema R.H.. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19108–19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Funk L.C., Zasadil L.M., Weaver B.A.. Living in CIN: Mitotic infidelity and its consequences for tumor promotion and suppression. Dev. Cell. 2016; 39:638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.