Abstract
The intron-lariat spliceosome (ILS) complex is highly conserved among eukaryotes, and its disassembly marks the end of a canonical splicing cycle. In this study, we show that two conserved disassembly factors of the ILS complex, Increased Level of Polyploidy1-1D (ILP1) and NTC-Related protein 1 (NTR1), positively regulate microRNA (miRNA) biogenesis by facilitating transcriptional elongation of MIRNA (MIR) genes in Arabidopsis thaliana. ILP1 and NTR1 formed a stable complex and co-regulated alternative splicing of more than a hundred genes across the Arabidopsis genome, including some primary transcripts of miRNAs (pri-miRNAs). Intriguingly, pri-miRNAs, regardless of having introns or not, were globally down-regulated when the ILP1 or NTR1 function was compromised. ILP1 and NTR1 interacted with core miRNA processing proteins Dicer-like 1 and Serrate, and were required for proper RNA polymerase II occupancy at elongated regions of MIR chromatin, without affecting either MIR promoter activity or pri-miRNA decay. Our results provide further insights into the regulatory role of spliceosomal machineries in the biogenesis of miRNAs.
INTRODUCTION
MiRNAs are a class of endogenous small non-coding RNAs that predominantly mediate post-transcriptional gene silencing (PTGS). Similar to messenger RNA (mRNA), canonical MIR genes are transcribed by DNA-dependent RNA polymerase II (Pol II), and undergo a series of co-transcriptional modifications including 5′ capping and 3′ polyadenylation and/or splicing (1). Primary transcripts of miRNAs (pri-miRNAs) are folded into stem-loop structures with imperfect complementarity at the stem regions, features that are recognized by the RNase III enzyme DICER in animals and Dicer-like 1 (DCL1) in plants (2). Plant miRNA/miRNA* duplexes are methylated by HUA Enhancer 1 (HEN1), and one of the strands is selectively incorporated into Argonaute (AGO) proteins to guide PTGS (3,4).
Steady levels of intracellular miRNAs are controlled by multiple mechanisms including transcription, processing and stability (5). In addition to the core components of miRNA metabolic and action pathways (Pol II, DCL1, HEN1 and AGO1), more than a dozen accessory/regulatory proteins affecting miRNA abundance have been identified over the last decade, revealing the complexity of miRNA homeostasis (6). Importantly, several proteins show dual effects on MIR gene transcription and pri-miRNA processing, suggesting a tight coupling between the two processes. In fact, DCL1 is recruited to MIR loci in a pri-miRNA dependent manner (7). Moreover, a number of genes linked to spliceosomal function affect miRNA biogenesis, including Tough (TGH), Stabilized 1 (STA1), Thoc 2 (THO2), Sickle (SIC), Debranching RNA Lariats 1 (DBR1), Glycine-rich RNA-binding Protein 7 (GRP7), Modifier of snc1;2 (MOS2), High Osmotic Stress Gene Expression 5 (HOS5), Small 1 (SMA1) (8–16), as well as multiple members of the MOS4-associated complex (MAC, also known as the Nineteen Complex [NTC]) (17–20). Moreover, alternative splicing (AS) of pri-miRNAs has been reported to affect DCL1 processing efficiency (21,22). Yet, our knowledge of the regulation of miRNA biogenesis by spliceosome-associated proteins remains limited and ambiguous.
The intron-lariat spliceosome (ILS) complex is evolutionarily conserved among eukaryotes (23). In the last stage of a canonical spliceosome cycle, the ILS complex is dismantled by the DEAH-box-containing RNA helicase PRP43, which facilitates the recycling of splicing factors and degradation of intron lariats (24,25). The Arabidopsis genome encodes two putative PRP43 proteins, At3g62310 and At2g47250, hereafter referred to as PRP43a and PRP43b, respectively. In budding yeast (Saccharomyces cerevisiae), NTC-related protein 1 (NTR1) and NTR2 function as key co-factors of PRP43 (26). While NTR1 is highly conserved among eukaryotes, NTR2 appears absent in mammals and plants. Instead, the human C2ORF3 protein interacts with both hPRP43 and NTR1/TFIP11, and depletion of C2ORF3 causes splicing defects (27). However, little is known about the plant ILS complex. In Arabidopsis, the C2ORF3 homolog, Increased Level of Polyploidy1-1D (ILP1), interacts with NTR1/Spliceosomal Timekeeper Locus 1 (STIPL1) and regulates AS of a subset of genes (28–30).
Through a candidate gene-based approach, here we show that ILP1 dysfunction results in reduced levels of miRNAs, trans-acting small interfering RNAs (ta-siRNAs) and a subset of heterochromatin-associated siRNAs (hc-siRNAs). Our data suggest that ILP1, NTR1, PRP8 and PRP43 are likely components of the ∼660 kDa plant ILS complex. The ilp1-1 and ntr1-1 mutants had many shared defects in plant morphology, gene expression, splicing, and small RNA biogenesis. RNA-seq data showed that pri-miRNAs, regardless of having introns or not, were globally decreased in abundance in ilp1-1 and ntr1-1. In contrast, ILP1 and NTR1 did not affect the transcription levels of protein coding genes with splicing defects in cognate mutant. Mechanistically, ILP1 and NTR1 interacted with DCL1 and SE, and promoted transcriptional elongation of MIR genes without affecting transcription initiation. These results suggest dual functions of ILP1 and NTR1 in regulating MIR gene transcription and splicing.
MATERIALS AND METHODS
Plant materials
All the plant materials used in this study were in the Columbia (Col) background except for ago4-1, which was in the Landsberg erecta-0 (Ler-0) background. The T-DNA insertion mutants including ilp1-1 (SALK_030650C), ntr1-1 (SALK_073187C), hyl1-2 (SALK_064863), se-2 (SAIL_44_G12), dcl1-9 (CS3828), ago4-1 (CS6364), ago1-36 (Salk_087076) and prr9-1 (SALK_007551C) were obtained from the Arabidopsis Biological Resources Center (ABRC) (https://abrc.osu.edu). Additionally, mutants toc1-2 (31) and cca1-1 lhy-20 (32), and transgenic reporter lines p35S::MIR172b (33), pMIR167a::GUS (20), pMIR172b::GUS (19), pCCA1::LUC and pTOC1::LUC (34) were used in this study. Other transgenic plants, including p35S::NTR1-10xMYC, pILP1::ILP1-GFP, p35S::PRP43b-3xFLAG and p35S::amiR-LUC + p35S::LUC, were obtained by the floral dip method (35). Double mutants and introduction of transgene reporters into respective genotypes were generated by genetic crossing and characterized by genotyping in the F2 or F3 generation.
Antibodies and DNA/LNA oligos
All antibodies, polymerase chain reaction (PCR) primers and Northern probes used in this study are listed in Supplementary Table S1.
Plasmid construction
To construct pILP1::ILP1-GFP, the ILP1 gene fragment (without stop codon) along with ∼1.5 kb of the upstream sequence were amplified and sub-cloned into the pENTR-D/TOPO entry vector (Thermo Fisher Scientific Inc.). The resultant plasmids were cloned into the pMDC204 destination vector via LR recombination. To construct p35S::PRP43b-3xFLAG, the PRP43b coding sequence (CDS) (without stop codon) was sub-cloned into the pENTR1A, and then transferred to p35SC3F (a modified pEarleygate100 vector with 3× FLAG tag downstream of the Gateway cassette) via LR recombination. To construct p35S::NTR1-10xMYC, the NTR1 CDS (without stop codon) was PCR amplified and directly cloned into pGWB520 using the EcoR V and Pac I restriction sites. To construct p35S::amiR-LUC + p35S::LUC, the luciferase (LUC) CDS was amplified from pGreenII 0800–LUC and cloned into pCAMBIA1301 vector using the restriction sites Nco I and BstP I to produce pCAMBIA1301-LUC. The amiR-LUC + tNOS (NOS Terminator, tNOS) sequence was synthesized de novo and cloned into pCAMBIA1301-LUC using the Kpn I and Hind III sites. At last, the whole amiR-LUC-tNOS + p35S::LUC fragment was digested with Kpn I and Sal I restriction endonucleases, and inserted into pCHF3 to produce p35S::amiR-LUC + p35S::LUC. To construct bimolecular fluorescence complementation (BiFC) vectors, the respective CDS sequences were PCR amplified and cloned into pXY103 (containing N-terminal half of yellow fluorescent protein [nYFP]) and/or pXY104 (containing C-terminal half of YFP [cYFP]) using Kpn I and Sal I (for ILP1 and NTR1) or Kpn I and Xba I (for DCL1, HYL1, and SE) sites. To construct PRP43b-cYFP, cYFP was PCR amplified and digested using Pac I and Sac I restriction endonucleases, and then inserted into pGWB520 to generate pGWB-cYFP. Then, pENTR-PRP43b was transferred into pGWB-cYFP via LR recombination.
GUS staining and LUC detection
Three-week-old transgenic seedlings in Col-0 or the ilp1-1 mutant background were subjected to β-glucuronidase (GUS) staining, as described previously (36). To determine LUC activity, seedlings were sprayed with a solution containing 300 μg/ml luciferin and 0.1% Triton X-100, and incubated in the dark for 10 min. Luminescence was monitored by the Tanon 5200 chemiluminescent imaging system (Tanon Science & Technology Co Ltd., Shanghai, China). To analyze circadian rhythms, seedlings were entrained under long-day conditions for 6 days and then transferred to constant light for luminescence measurements according to previous work (37).
RNA analysis
Quantitative real-time PCR (qPCR), small RNA Northern blot hybridization and RNA immunoprecipitation assays were performed as described previously (8). RNA half-life analysis was performed according to Lidder et al. (38). Briefly, the fifth to eighth leaves harvested from 1-month-old dcl1-7 and dcl1-7 ilp1-1 plants were transferred to a culture dish containing half-strength Murashige and Skoog (1/2 MS) medium. After a 30-min incubation, 3′-deoxyadenosine (Cordycepin, Sigma) was added to a final concentration of 0.6 mM. Samples were collected at indicated time points, and the decay rate of pri-miRNAs and mRNAs was analyzed by qPCR.
Deep sequencing
To sequence small RNAs, total RNA was extracted from inflorescence tissues. Small RNA libraries were constructed using the TruSeq Small RNA Library Preparation Kit, and single-end sequencing was performed to produce 50-bp reads (SE50) using the Illumina Hiseq 2500 platform (Novogene, Beijing, China). DEGseq was used for differential miRNA expression based on a modified miRNA annotation file, which combined miRNAs with identical sequences (39). The accuracy of miRNA processing was determined as described previously (40). Determination of 24-nt hc-siRNA clusters and expression analysis were performed as previously described (41). For mRNA sequencing, total RNA was isolated from 7-day-old seedlings grown under constant white light. Oligo(dT) purification was performed to yield poly(A)+ RNA, which was subject to strand-specific mRNA library construction. Paired-end sequencing was performed to produce 2 × 150-bp reads using the Illumina Hiseq X Ten platform (Novogene, Beijing, China). Differential gene expression was performed using the DESeq2 software, and AS events were detected with rMATS (42,43). Nucleotides frequency analysis was calculated by the WebLogo application (http://weblogo.threeplusone.com) (44). Sequences around 5′ and 3′ splice sites were extracted from the TAIR10 genome, and those around branch-point sites were obtained from the ERISdb database (http://lemur.amu.edu.pl/share/ERISdb/home.html) (45). Intergenic pri-miRNAs with FPKM≥1 in at least one sample were used for differential expression analysis (46).
Gel filtration
For gel filtration, 500 μl of soluble proteins extracted from inflorescence tissues using the protein extraction buffer (10% glycerol, 50 mM Tris–HCl [pH 8.0], 5 mM MgCl2, 150 mM NaCl, 0.25% NP-40 and 2 mM Dithiothreitol [DTT]) were loaded on a Superdex 200 10/300 GL column (GE Healthcare). Gel filtration was performed on an ÄKTA system at a rate of 0.5 ml/min. Samples were collected at 1 min intervals and divided into two aliquots for protein and RNA assays.
Protein–protein interactions
Co-immunoprecipitation (co-IP) assays were performed using either stably transformed Arabidopsis plants or transiently transformed Nicotiana benthamiana leaves expressing the respective proteins, as described previously (8). Protein extracts prepared from pistils (In our hands, endogenous DCL1 is hardly to be detected in other tissues but can be robustly detected in pistils by Western blot.) of pILP1::ILP1-GFP or p35S::GFP transgenic plants were subjected to mass spectrometry (Mass-Spec) to identify ILP1-interacting proteins and to determine interactions between ILP1 and core processing proteins (DCL1, SE and HYL1). Inflorescence tissues were used for testing the interaction among ILS proteins. For Mass-Spec identification of ILP1 interacting proteins, ILP1-GFP and GFP control proteins were immunoprecipitated by GFP-trap beads. The immunoprecipitated proteins were subjected to in-gel digestion with trypsin. The resulting peptides were extracted twice with 30% acetonitrile, vacuum-dried and dissolved in 1% formic acid prior to Mass-Spec analysis. Mass-Spec analysis and raw data processing were performed as described previously (47). For BiFC assays, paired cYFP and nYFP fusion proteins were co-expressed in N. benthamiana leaves or Arabidopsis protoplasts (8,48). BiFC signals were excited at 488 nm and detected with a narrow barrier (493–525 nm; BA493–525) under a confocal microscope (Leica SP8).
Chromatin immunoprecipitation (ChIP) assay
The 3-week-old seedlings from indicated genotypes were subjected to ChIP as previously described (49). Precipitated DNAs were used for qPCR analysis, and Pol II occupancy was calculated as percentage of input.
RESULTS
ILP1 promotes both miRNA and siRNA accumulation
To identify novel components in miRNA biogenesis in Arabidopsis, we systematically mined candidate genes based on a number of selection criteria, including phylogenetic conservation, nuclear localization and/or mutant phenotype. Homozygous genotypes of T-DNA insertion mutants obtained from ABRC were identified by genotyping. Mutants showing multiple developmental defects, which are common and necessary, though not sufficient, features of miRNA-pathway defective mutants (50), were further screened by small RNA Northern blot analysis using miR167 and miR159 as dual reporters. A mutant (SALK_030650C / ilp1-1, defective in the ILP1 gene) showing a significant reduction in the accumulation of both reporter miRNAs was studied in detail.
Morphological analysis revealed that the ilp1-1 mutant exhibited pleiotropic phenotypes including short root, delayed growth, altered leaf shape, late flowering, abnormal silique phyllotaxy and abscisic acid (ABA) hypersensitivity (Figure 1A and Supplementary Figure S1A–C). Examination of additional miRNAs in inflorescences and leaves confirmed a role of ILP1 in the miRNA pathway (Figure 1B and Supplementary Figure S1D). Since target cleavage is a major mode of miRNA action in plants (51), we monitored the transcript levels of several miRNA targets by qPCR using primers spanning cleavage sites. The levels of TasiRNA precursor RNA 2 (TAS2), Phavoluta (PHV), Heme Activator Protein Homolog 2B (HAP2B), Target Of EAT 2 (TOE2) and TOE3 were moderately increased in ilp1-1 compared with the wild-type plant (WT; Col-0) (Supplementary Figure S1E). Next, we conducted a small RNA-seq assay using ilp1-1 and Col-0 inflorescences (two biological replicates), and detected a global reduction of miRNAs in ilp1-1 relative to WT (Figure 1C and Supplementary Table S2). Also of note was that ILP1 ablation did not affect the accuracy of miRNA processing (Supplementary Figure S1F).
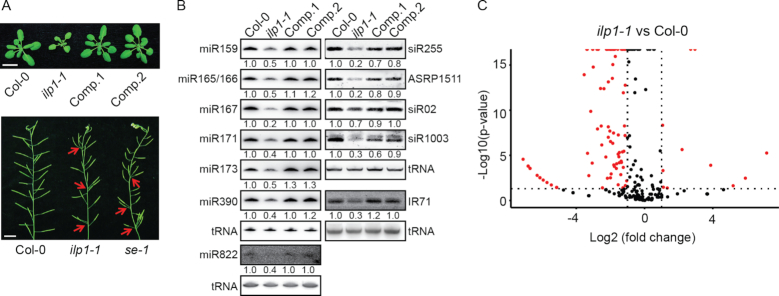
Figure 1.
ILP1 is involved in small RNA biogenesis. (A) Phenotypic characterization of the ilp1-1 mutant. Arrows indicate clustered siliques. Scale bar = 1 cm. Comp. 1 and Comp. 2 are two independent ilp1-1+pILP1::ILP1-GFP complementation lines. (B) Northern blot analysis of small RNA abundance in inflorescence tissues of different genotypes. Numerals indicate relative abundance. Values in Col-0 were arbitrarily set to 1. Transfer RNA (tRNA) served as a loading control. (C) Volcano plot showing differential expression of miRNAs between the ilp1-1 mutant and Col-0. Two biological replicates from mixed stages of inflorescence tissues were subjected to small RNA-seq.
Additionally, the amount of miR822, which depends on DCL4 (52), was decreased in ilp1-1 (Figure 1B), suggesting that ILP1 affects multiple small RNA pathways. We thus investigated the effect of ilp1-1 on the accumulation of other types of siRNAs, including a DCL2-dependent siRNA (IR71), two DCL3-dependent hc-siRNAs (siR02 and siR1003), and two DCL1-/DCL4- dependent ta-siRNAs (siR255 and ASRP1511) (8). Results showed that the accumulation of all examined siRNAs was greatly reduced in the ilp1-1 mutant plants, except for siR02, which only showed a moderate reduction (Figure 1B). Introduction of ILP1 fused to GFP under the control of its native promoter (pILP1::ILP1-GFP) into ilp1-1 fully rescued the developmental defects and restored the expression of small RNAs (Figure 1A and B). Nevertheless, small RNA-seq data revealed that 24-nt hc-siRNAs are less significantly affected by the ILP1 dysfunction as compared to miRNAs (Supplementary Figure S1G and Table S2). We thus chose to focus on miRNA in this study.
ILP1 interacts with the disassembly factors of the ILS complex, NTR1 and PRP43
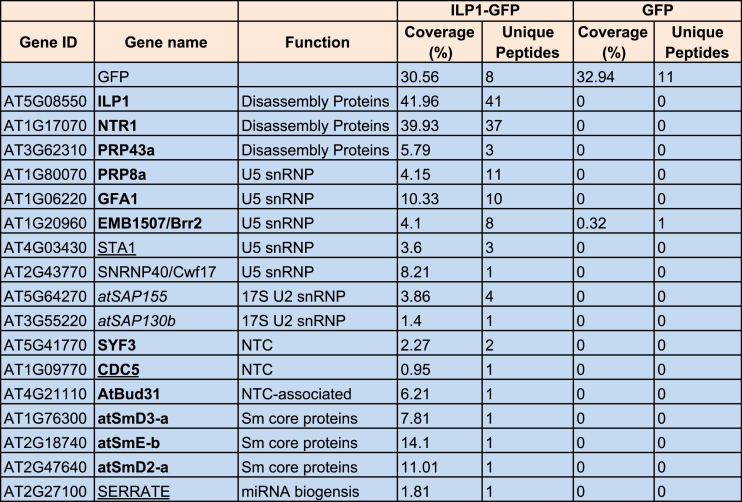
To investigate how ILP1 regulates miRNA biogenesis and plant development, we immunoprecipitated ILP1-GFP from the ilp1-1+pILP1::ILP1-GFP transgenic plants, followed by Mass-spec analysis. Consistent with recent published data (29), NTR1 was the strongest hit among all candidate interacting proteins (Table 1). Notably, PRP43a and additional spliceosome components, especially several U5 small nuclear ribonucleoproteins (snRNPs), were also retrieved from our Mass-Spec assay (Table 1). To validate these data, we introduced the p35S::PRP43b-3xFLAG construct into the pILP1::ILP1-GFP transgenic background. Transgenic plants effectively expressing both PRP43b-3xFLAG and ILP1-GFP were selected and crossed with a p35S::NTR-10xMYC transgenic plant. The resultant F1 plants were used for size exclusion chromatography and in vivo co-IP assays. Proteins including ILP1-GFP, NTR1-10xMYC and endogenous PRP8, a central component of U5 snRNP, co-migrated at ∼660 kDa (Figure 2A). By contrast, PRP43b-3xFLAG not only existed in the ∼660 kDa region, but was also present in lower molecular weight regions. Additionally, U5 and U6 small nuclear RNAs (snRNAs), but not U1 snRNA, peaked at ∼660 kDa regions, consistent with the fact that U5 and U6 are components of the ILS complex (53). Moreover, the abundance of U6, but not that of U4 and U5 snRNAs, was increased in the ilp1-1 and ntr1-1 mutants, possibly due to defects in snRNA recycling (Supplementary Figure S2C). Nevertheless, it has been reported that NTR1 interacts with U1 and U4, in addition to U2, U5 and U6, as revealed by RNA immunoprecipitation followed by reverse-transcription PCR (RT-PCR) detection (29), implicating that NTR1 and ILP1 may have broader functions during splicing.
Table 1.
List of potential ILP1 interacting proteins related to splicing as identified by Mass-Spec analysis
|
ILP1-GFP and GFP were used as bait proteins. Known ILS components, the hPRP43-interacting proteins (27) and, known miRNA pathway proteins are indicated in bold, italic and underlined font, respectively.
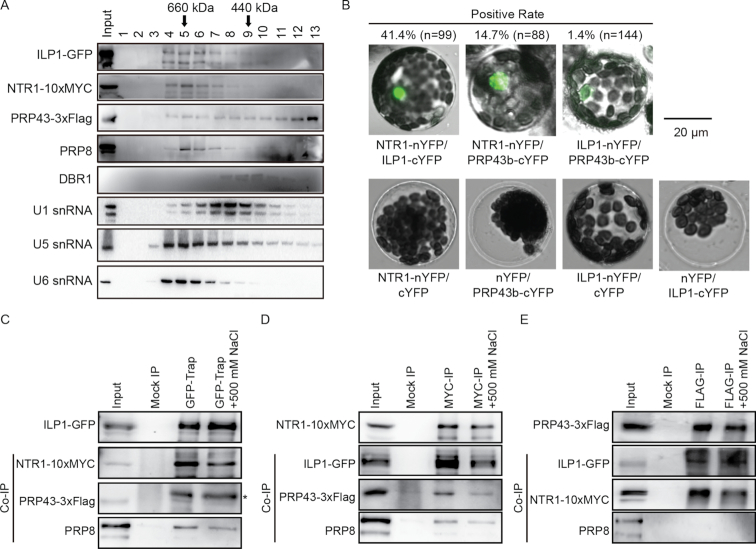
Figure 2.
Characterization of the Arabidopsis ILS complex. (A) Size exclusion chromatography analysis of soluble proteins extracted from inflorescence tissues of plants simultaneously expressing pILP1::ILP1-GFP, p35S::NTR1-10xMYC and p35S::PRP43b-3xFLAG. Proteins were analyzed by Western blot, whereas U1, U5 and U6 RNAs were checked by Northern blot analysis. (B) Reciprocal bimolecular fluorescence complementation (BiFC) analyses among ILP1, NTR1 and PRP43b. Scale bar = 20 μm. Paired constructs were transiently expressed in Arabidopsis leaf protoplasts, and fluorescence was measured 24 h after transfection. Percentage means positive BiFC ratio for corresponding protein pairs. Values (n) indicate the number of counted protoplasts. See also Supplementary Figure S2A for results at lower magnification. (C–E) Reciprocal co-immunoprecipitation (co-IP) assays among ILP1-GFP, NTR1-10xMYC and PRP43b-3xFLAG using ILP1-GFP (C), NTR1-10xMYC (D) and PRP43b-3xFLAG (E) as a bait. Endogenous PRP8 was analyzed using a commercial anti-PRP8 antibody. Asterisk indicates unknown bands. Input = 25% for IPs and 1% for co-IPs, except for the interactions between ILP1-GFP and NTR1-10xMYC, where input was 5% for co-IPs.
Next, we investigated their interactions by reciprocal immunoprecipitation. As shown in Figure 2C and D, both NTR1 and ILP1 effectively captured each other as well as PRP43b and PRP8. By contrast, PRP43b successfully captured NTR1 but barely ILP1 and PRP8 (Figure 2E). Importantly, a high salt (500 mM) wash only slightly weakened these interactions but did not abolish them completely, indicating a stable association among these proteins. Considering a high affinity between NTR1 and ILP1 and a broader distribution of PRP43b (Table 1 and Figure 2A), it is possible that NTR1 bridges the interaction between ILP1 and PRP43, while only a sub-proportion of PRP43 is in complex with NTR1 and ILP1. To test this possibility, spatial proximities among these proteins were tested by BiFC assays via transient expression of paired split-YFP fusion proteins in Arabidopsis protoplasts. LUC was co-expressed as a control of transfection efficiency. We found that the NTR1-nYFP/ILP1-cYFP pair produced both the brightest fluorescence and the highest positive rate (41.4%, n = 99 protoplasts). In sharp contrast, in the ILP1-nYFP/PRP43b-cYFP pair, only few protoplasts (1.4%, n = 144) gave weak fluorescence. The NTR1–nYFP/PRP43b-cYFP pair was between the two in terms of both fluorescence intensity and positive rate (14.7%, n = 88) (Figure 2B and Supplementary Figure S2A). Similar results were obtained when co-expressed in N. benthamiana leaves (Supplementary Figure S2B). Taken together, these data suggested that ILP1 and PRP43b may be relatively distant in space, and/or that their interaction is transient.
ILP1 and NTR1 act in the same or similar biological pathway(s)
Previously, examination of 144 selected AS events has shown that ILP1 and NTR1 affect the splicing of a subset of AS sites (29). To gain a global view of how ILP1 and NTR1 regulate gene expression and splicing, we conducted a strand-specific RNA-seq analysis. To avoid potential effects of altered circadian rhythm (Figure 4C, see below), we used 7-day-old seedlings of Col-0, ilp1-1 and ntr1-1 grown under constant white light. Principal components analysis (PCA) did not separate ilp1-1 and ntr1-1 in the first component, which explained 61.84% of the variation, suggesting similar changes in global gene expression in ilp1-1 and ntr1-1 (Figure 3A). Compared with the WT, 1249 and 865 differentially expressed genes (DEGs; |log2FC| ≥ 1, P < 0.05, negative binomial distribution model test with DESeq2) were identified in the ilp1-1 and ntr1-1 mutants, respectively. Among these genes, 496 (P < 2.2 × 10−16; Fisher's exact test) were either up- or down-regulated in both mutants, with only three genes up-regulated in ntr1-1 but down-regulated in ilp1-1 and none vice versa (Figure 3B and C; Supplementary Tables S3 and 4). Next, we identified AS events by rMATS (42), and determined that retained introns (RI) were the most frequently affected AS type in both mutants (Supplementary Figure S3A-B and Table S5). Approximately 50% of the significantly affected RI events (and 43% for all AS types) were shared between ilp1-1 and ntr1-1 (P < 2.2 × 10−16; Fisher's exact test) (Figure 3D; Supplementary Tables S6 and 7). The above data suggested that ILP1 and NTR1 probably function in the same complex. If so, we predicted that for those RI sites with significant defects in only one mutant (i.e. significant in ilp1-1 but not in ntr1-1, or vice versa), they should be more affected in the other mutant than all RI controls. Indeed, for RI introns only significantly affected in ntr1-1, we observed a remarkable trend toward having lower FDR scores (for false discovery rates) in ilp1-1 (Supplementary Figure S3C). The same held true for vice versa (Supplementary Figure S3D). Taken together, our data supported the interaction between ILP1 and NTR1 and their shared function in regulation of AS. Representative loci representing different types of AS, including a core circadian gene LHY, were validated by RT-PCR (Figure 3E and F). Results showed a tendency of combined AS types (e.g. retained introns plus alternative 3′ splice sites [RI+3′SS] or retained introns plus alternative 5′ splice sites [RI+5′SS]) affected by ILP1 and NTR1 (Supplementary Figure S3E). Notably, NTR1 and ILP1 also affected the splicing of some pri-miRNAs but not that of Serrate (SE) dependent pre-mRNAs (Figure 3G and Supplementary Figure S3F) (54).
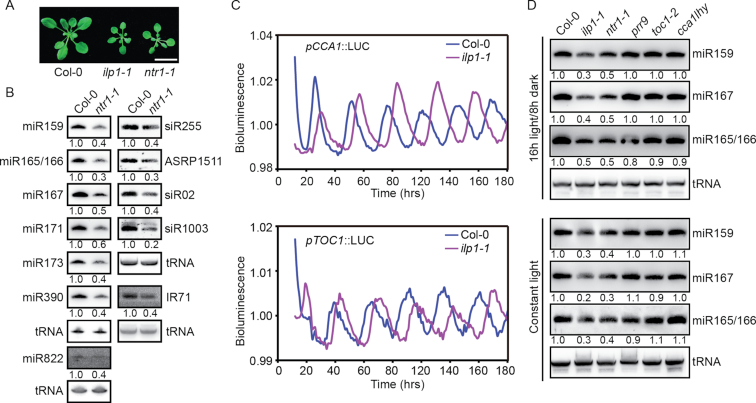
Figure 4.
NTR1 and ILP1 have dual functions in small RNA biogenesis and circadian clock regulation. (A) Three-week-old plants of Col-0, ilp1-1 and ntr1-1. Scale bar = 1 cm. (B) Northern blot analysis of small RNA abundance in inflorescence tissues of Col-0 and ntr1-1 plants. Numerals indicate relative abundance. Values of Col-0 were arbitrarily set to 1. tRNA served as a loading control. (C) Bioluminescence of pCCA1::LUC and pTOC1::LUC in Col-0 and ilp1-1 plants. (D) Northern blot analysis of small RNA abundance in 7-day-old seedlings of different genotypes. Plants were grown on  MS plates under either 16 h light/8 h dark photoperiod (sampled at Zeitgeber time 2) or constant light conditions. Numerals indicate relative abundance. Values of Col-0 were arbitrarily set to 1.
MS plates under either 16 h light/8 h dark photoperiod (sampled at Zeitgeber time 2) or constant light conditions. Numerals indicate relative abundance. Values of Col-0 were arbitrarily set to 1.
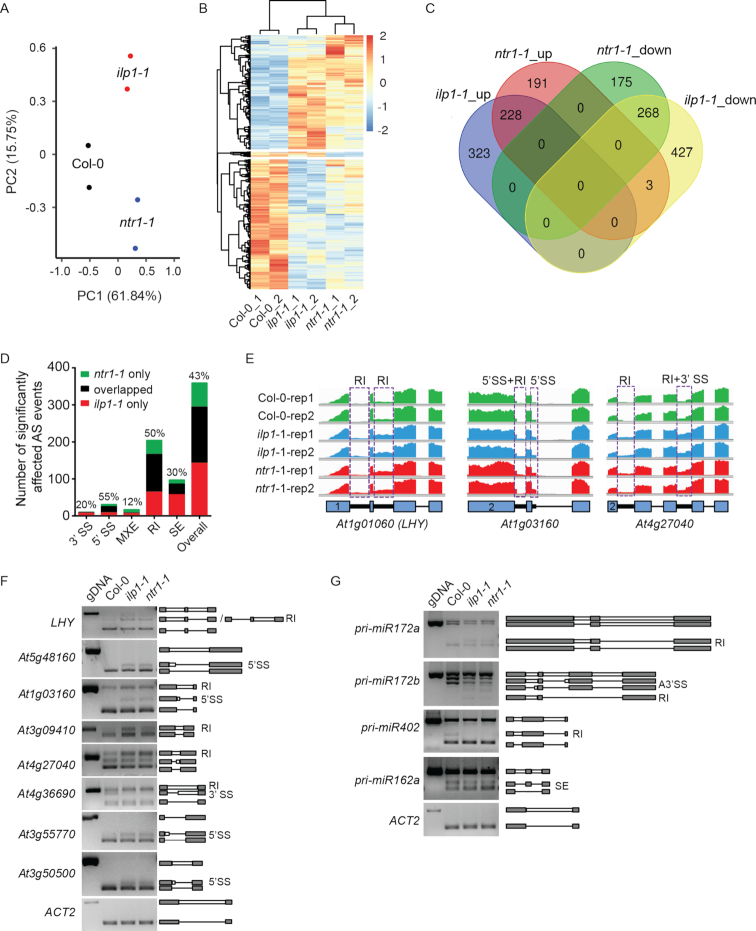
Figure 3.
Comparison of differential gene expression and AS among Col-0, ilp1-1 and ntr1-1 plants. (A) Principal component analysis of RNA-seq data. (B) Clustering analysis of six samples with significantly differentially expressed genes (DEGs) in either ilp1-1 or ntr1-1 plants. (C) Venn diagram showing overlaps of significantly differential expressed genes among different categories. (D) Bar charts showing the overlap of significantly affected alternative splice sites between ilp1-1 and ntr1-1. Values above each bar represent the percentage of overlapped sites. A3SS, alternative 3′ splice sites; A5SS, alternative 5′ splice sites; MXE, mutually exclusive exons; RI, retained introns; SE, skipped exons. (E) Representative examples of affected AS events in ilp1-1 and ntr1-1 plants. Numerals in the box indicate the sequence number of exons. (F) Validation of affected AS events by reverse-transcription PCR (RT-PCR) analysis. (G) RT-PCR analysis of AS of pri-miRNAs. Gray boxes, lines and white boxes indicate exons, spliced introns and retained introns, respectively.
In general, introns are either constitutively or alternatively spliced in wild-type plants. We found that all ILP1 and NTR1 dependent introns were AS introns according to the TAIR10 annotation. To further explore whether ILP1 and NTR1 dependent introns share any common features, we focused on the analysis of those with intron retention defects in both mutants (i.e. ILP1 and NTR1-dependent RI introns). We found that these RI introns used essentially the same set of cis-elements as non-affected RI introns (Supplementary Figure S3G), had no preference on neither the locations within host genes, nor the selection of host genes in terms of the total number of introns (Supplementary Figure S3H and I). Although ILP1 and NTR1 dependent RI introns were markedly longer than all introns and all AS introns, they did not differ from all RI introns characterized by rMATS (Supplementary Figure S3J).
NTR1 is involved in small RNA biogenesis
If the ILS complex plays a role in small RNA biogenesis, we expected that the loss of function of NTR1 should also reduce the steady levels of small RNAs. We identified a T-DNA insertion mutant of NTR1 (SALK_073187C, ntr1-1), which morphologically resembled ilp1-1 (Figure 4A; Supplementary Figures S1C and 4B). The ntr1-1 mutant showed decreased expression of miRNAs, ta-siRNAs and hc-siRNAs comparable to those observed in ilp1-1 (Figure 4B). Introduction of p35S::NTR1-10xMYC into ntr1-1 fully rescued the morphological phenotype and miRNA levels (Supplementary Figure S4A and C). Strikingly, the ilp1-1 ntr1-1 double mutant appeared embryonic lethal (Supplementary Figure S4D), indicating a synergistic interaction between these two proteins, albeit that ILP1 and NTR1 probably worked together as a complex.
Since the ntr1-1 mutant has a long circadian period (30), we examined whether ILP1 ablation causes altered circadian rhythm. We introduced the pCCA1::LUC and pTOC1::LUC reporter constructs into the ilp1-1 mutant. Similar to ntr1-1, ilp1-1 showed a longer circadian period than the WT control (Figure 4C). Since both ilp1-1 and ntr1-1 exhibited altered circadian rhythms, we next investigated whether disruptions of normal circadian rhythm affect miRNA biogenesis. As shown in Figure 4D, the ilp1-1 and ntr1-1 mutants consistently showed reduced miRNA levels under both constant light and 16-hr light, whereas mutations in the core circadian pathway genes (PRR9, TOC1, CCA1 and LHY) (55) did not cause obvious changes in miRNA abundance. Thus, ILP1 and NTR1 regulated miRNA biogenesis independent of their roles in circadian rhythm regulation.
ILP1 and NTR1 promote pri-miRNA accumulation
To determine which step(s) of the small RNA pathways is ILP1/NTR1 involved in, we first investigated whether ILP1 and NTR modulate the expression and/or splicing of genes involved in the small RNA biogenesis and action pathways. RNA-seq data suggested that neither the expression levels nor the splicing patterns of known small RNA pathway genes were significantly affected in ilp1-1 or ntr1-1, except for AGO10, which had intron retention defects (Supplementary Figure S5A–C, Supplementary Tables S5 and 8). However, global reduction of miRNAs in ilp1-1 is unlikely due to mis-splicing of AGO10, because AGO10 is specifically associated with miR165/166 and promotes their degradation (56). Transcript levels of eight miRNA biogenesis pathway genes (DCL1, Hyponastic Leaves 1/HYL1, SE, SIC, TGH, STA1, Dawdle/DDL and HEN1) were validated by qPCR (Supplementary Figure S5D). We also monitored the protein levels of three core pri-miRNA processing members (DCL1, HYL1 and SE) and two small RNA effector proteins (AGO1 and AGO4). The result showed that all were unaffected in either mutant, except for AGO1, which showed a slight reduction (Supplementary Figure S5E). Reduced AGO1 abundance is consistent with reduced miRNA levels in ilp1-1 and ntr1-1, as AGO1 and miRNAs are known to be co-stabilized (57,58).
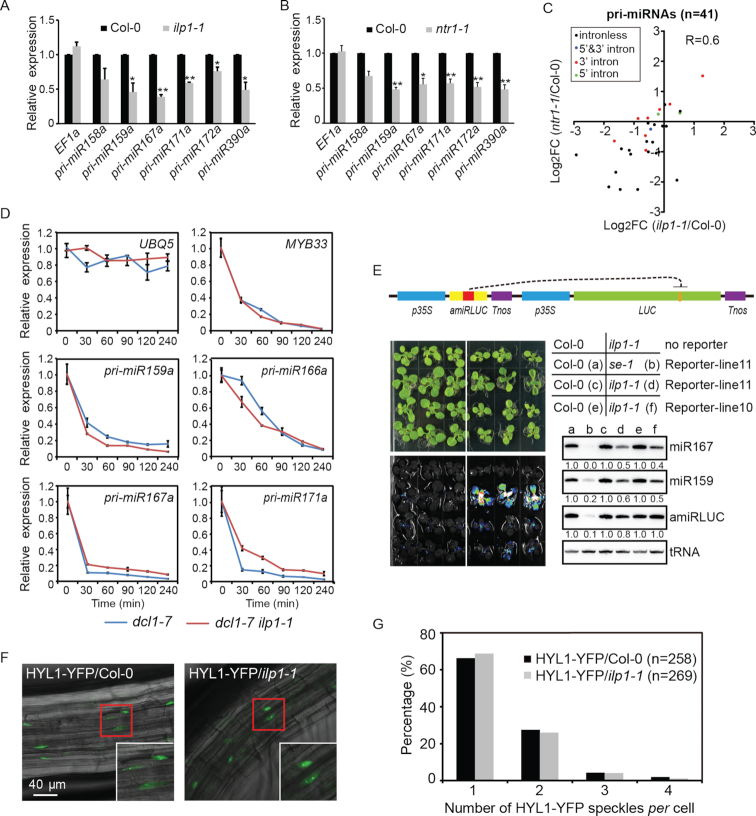
Mature miRNAs are mainly processed from pri-miRNAs by DCL1. In principle, the levels of pri-miRNAs would be reduced in mutants defective in miRNA transcription but increased in mutants defective in miRNA processing. Results of qPCR analysis revealed a significant reduction of all six tested pri-miRNAs in both ilp1-1 and ntr1-1 mutants (Figure 5A and B). To minimize the effect of processing, we crossed ilp1-1 with dcl1-7 ; dcl1-7 is a hypomorphic allele of DCL1 characterized by impaired miRNA processing (59). Since homozygous dcl1-7 plants are female sterile (60), we generated the dcl1-7 ilp1-1 double mutant in the F2 siblings by genotyping. dcl1-7 ilp1-1 was morphologically similar to dcl1-7 , but with a more severe phenotype (Supplementary Figure S5F). Although the impairment of miRNA processing in dcl1-7 resulted in a dramatic over-accumulation of five tested pri-miRNAs, their expression levels were consistently lower in dcl1-7 ilp1-1 than those in dcl1-7 (Supplementary Figure S5G).
Figure 5.
ILP1 and/or NTR1 promotes MIR gene expression without affecting pri-miRNA stability, processing and dicing body (D-body) formation. (A and B) Quantification of pri-miRNAs in Col-0, ilp1-1 and ntr1-1 plants by quantitative real-time PCR (qPCR). Data were normalized to GAPDH. Expression of EF1a was analyzed in parallel as a control. Significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01; paired t-test; n = 3 biological replicates). (C) Scatter plot showing the pri-miRNAs expression in ilp1-1 and ntr1-1. Intergenic pri-miRNAs with FPKM≥1 in at least one biological samples were selected for analysis (n = 41). 5′ and 3′ indicate intron positions relative to the pre-miRNA stem loop. (D) qPCR analysis of pri-miRNAs decay at different time intervals after cordycepin treatment. Data were normalized to GAPDH. UBQ5 and MYB33 served as long and short half-life RNA controls, respectively. Error bars indicate standard deviation (SD; n = 3 technical replicates). The experiment was repeated once with similar results. (E) Effect of ilp1-1 on p35S::amiR-LUC + p35S::LUC reporter as indicated by bioluminescence and Northern blot assays. Plants were grown on  MS plates for 3 weeks and analyzed. Lines 10 and 11 represent two independent transgenic events. For Northern blot hybridization, tRNA was used as a loading control. For each comparison group, values in Col-0 background were arbitrarily set to 1. (F and G) Effect of ilp1-1 on D-body formation as indicated by HYL1-YFP speckles. Root tissues from HYL1-YFP/Col-0 and HYL1-YFP/ilp1-1 backgrounds were examined using an upright fluorescent microscope (Zeiss Axio Observer A1). Values in parentheses indicate numbers of counted nuclei.
MS plates for 3 weeks and analyzed. Lines 10 and 11 represent two independent transgenic events. For Northern blot hybridization, tRNA was used as a loading control. For each comparison group, values in Col-0 background were arbitrarily set to 1. (F and G) Effect of ilp1-1 on D-body formation as indicated by HYL1-YFP speckles. Root tissues from HYL1-YFP/Col-0 and HYL1-YFP/ilp1-1 backgrounds were examined using an upright fluorescent microscope (Zeiss Axio Observer A1). Values in parentheses indicate numbers of counted nuclei.
To gain a genome-wide view of ILP1 and NTR1 in regulating pri-miRNA expression, we analyzed the expression levels of 41 intergenic pri-miRNAs with FPKM ≥ 1 in at least one biological samples in our RNA-seq data. The results showed that pri-miRNAs were globally down-regulated in ilp1-1 and ntr1-1, with a significant correlation between the two mutants (Figure 5C and Supplementary Table S9). A number of pri-miRNAs are known to have one or more introns, and these introns may impact transcription elongation and/or DCL1 processing (21,22,61). Although ILP1 and NTR1 affected the splicing of some pri-miRNAs, no obvious differences were detected between intron-containing and intronless pri-miRNAs (Figure 5C). In contrast, we found five out of seven expressed TAS genes were moderately up-regulated in both ilp1-1 and ntr1-1 (Supplementary Figure S5H and Table S10). Thus, decreased ta-siRNA levels in ilp1-1 and ntr1-1 are not due to reduced TAS gene expression, but more likely a consequence of compromised miRNA biogenesis (Figure 1B and Figure 4B). Elevated expression of TAS genes are consistent with their roles as miRNA targets. However, we didn’t see a trend toward up-regulation of miRNA targets at global level (Supplementary Figures S5I and Table S11). NTR1 was previously reported to promote transcription elongation of protein coding genes harboring NTR1-dependent splicing sites (61). To test this, 141 genes with shared AS defects in both ilp1-1 and ntr1-1 were selected for analysis. To our surprise, no significant changes in gene expression were observed in either mutant (Supplementary Figure S5J), implicating that there might be no correlation between ILP1/NTR1 dependent splicing and gene expression.
Reduced accumulation of pri-miRNAs in ilp1-1 and ntr1-1 could also be due to accelerated degradation, as reported in ddl and prl1 mutants (20,62). To test whether ILP1 and NTR1 influence the stability of pri-miRNAs, we evaluated the effect of ILP1 or NTR1 dysfunction on the half-lives of pri-miRNAs. The fifth and eighth leaves were collected from 1-month-old dcl1-7 and dcl1-7 ilp1-1 (or dcl1-7 ntr1-1) plants and transferred to 1/2 MS medium supplemented with 0.6 mM cordycepin, a general transcription inhibitor used widely in Arabidopsis research (38). Tissues were sampled at the indicated time points, and levels of pri-miRNAs and control mRNAs were monitored by qPCR. The results showed that the decay rates of pri-miRNAs in dcl1-7 ilp1-1 and dcl1-7 ntr1-1 were comparable to those in dcl1-7 (Figure 5D and Supplementary Figure S5K), indicating that ILP1 and NTR1 do not affect pri-miRNA decay.
The above results point to the likelihood that ILP1 and NTR1 promote MIR gene expression. However, it is also possible that they play a role in pri-miRNA processing. To test this possibility, a LUC-based artificial miRNA reporter line (p35S::amiR-LUC + p35S::LUC) was generated, based on a published design (63). In this reporter system, the expression of amiR-LUC silences the expression of LUC, resulting in weak luminescence. Two independent single-copy transgenic lines were selected in the T2 generation and crossed with ilp1-1 or se-1, which served as controls for impaired pri-miRNA processing. As expected, se-1 greatly diminished amiR-LUC expression and produced bright luminescence (Figure 5E). In sharp contrast, only weak luminescence recovery and a slight reduction, if any, in amiR-LUC expression was observed in ilp1-1 (Figure 5E).
We also tested whether ILP1 is required for dicing body (D-body) formation (64,65). We introduced the D-body reporter pHYL1::HYL1-YFP into ilp1-1 by crossing, and D-body numbers per cell were counted under a fluorescence microscope. As shown in Figure 5F and G, the distribution of D-body number was comparable between ilp1-1 and WT plants. Based on these observations, we conclude that ILP1 promotes MIR gene expression with little effect on either processing or decay of pri-miRNAs.
ILP1 and NTR1 may facilitate Pol II transcription elongation at MIR loci
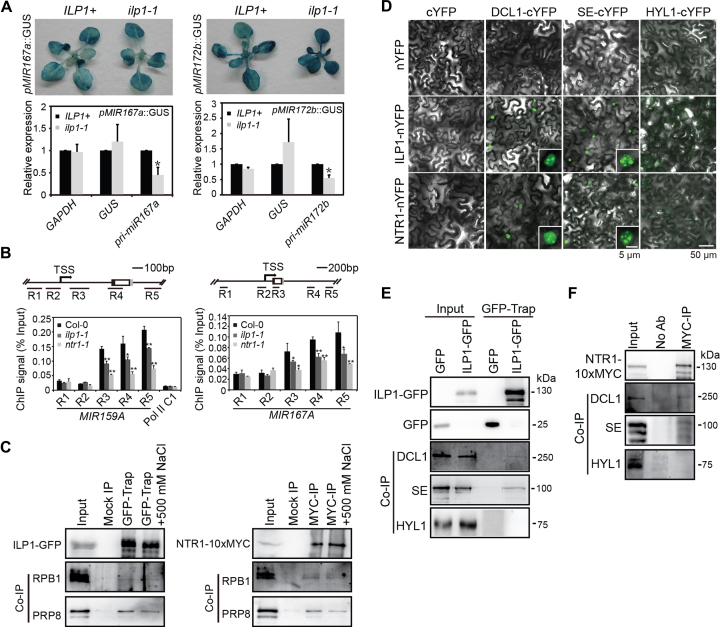
Given that pri-miRNA levels were globally reduced in ilp1-1 and ntr1-1, we surmised that ILP1 and NTR1 promote MIR gene expression. Two MIR promoter reporter lines (pMIR167a::GUS and pMIR172b::GUS) (19,20) were individually crossed with ilp1-1. The ilp1-1 and ILP1+/+ progenies harboring the respective reporter transgene were identified in the F2 generation. Lines homozygous for either transgene were confirmed in the F3 generation and subjected to GUS staining assay. Strikingly, neither promoter reduced the expression of GUS in ilp1-1 as compared with WT. For the pMIR172b::GUS reporter, the GUS expression was increased when ILP1 was compromised (Figure 6A). We next measured the Pol II occupancies at the MIR loci by ChIP-PCR using an antibody directed against the second largest subunit of Pol II, with Pol II_C1, an intergenic locus with no Pol II occupancy, as a background control (19). While no changes were detected at the promoter regions and transcription start sites, significantly reduced occupancies of Pol II was observed at the downstream regions in the ilp1-1 and ntr1-1 mutants, suggesting that ILP1 and NTR1 may affect elongation rather than initiation of MIR gene transcription (Figure 6B and Supplementary Figure S6A). We also measured Pol II occupancies on GUS genes (pMIR167a::GUS) and several coding genes harboring ILP1/NTR1-dependent AS introns. As shown in Supplementary Figure S6B and C, ILP1 dysfunction did not affect Pol II occupancies in both upstream and downstream regions of these genes.
Figure 6.
ILP1 and NTR1 promote transcription elongation of MIR genes. (A) Expression of pMIR167a::GUS and pMIR172b::GUS in ilp1-1 and ILP1+ genetic backgrounds. Top panels, GUS staining. Bottom panels, qPCR quantification of GUS transcripts. Data were normalized relative to EF1a. Endogenous pri-miR167a and pri-miR172b were analyzed in parallel. Significant differences are indicated by asterisks (*, P < 0.05; paired t-test; n = 3 biological replicates). (B) Chromatin immunoprecipitation (ChIP)-qPCR analysis of RPB1 occupancies across MIR159A and MIR167A loci. Black and gray boxes indicate miR-5p and miR-3p positions, respectively. Pol II C1, an intergenic fragment, served as a negative control. Significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01; paired t-test; n = 3 biological replicates). (C) Immunoprecipitation of ILP1-GFP and NTR1-10xMYC, followed by Western blot analysis of RPB1 and PRP8. Inflorescence tissues of pILP1::ILP1-GFP and p35S::NTR1-10xMYC transgenic plants were used. Input = 25% for IP samples and 0.7% for co-IP samples. (D) BIFC to test the interactions between ILS proteins and core pri-miRNA processing proteins. Insets showing zoom-in views of single nuclei. (E and F) Immunoprecipitation of ILP1-GFP and NTR1-10xMYC, followed by Western blot analysis of DCL1, HYL1 and SE. Input = 40% for IP and 0.7% for co-IP, respectively.
If ILP1 facilitated Pol II elongation at MIR genes, it should also affect the elongation process of artificially introduced miRNA driven by the constitutive 35S promoter. We surmised that the weak effect of ILP1 mutation on the p35S::amiR-LUC + p35S::LUC reporter might be due to the use of a short hairpin construct, which diminished the influence of transcription elongation. We then introduced a p35S::MIR172b transgene, which contained a long MIR172b precursor RNA with three introns (33), into the ilp1-1 mutant background. We monitored the expression of transgene-derived miR172 at young seedling stages, when endogenous miR172 was expressed at non-detectable levels (Supplementary Figure S6D) (66,67). The result showed that transgene-derived miR172b was expressed at lower levels in ilp1-1 than that in the WT background (Supplementary Figure S6D and E).
To test whether ILP1 and/or NTR interacts with Pol II, we extracted proteins from inflorescence tissues of ilp1-1+pILP1::ILP1-GFP and ntr1-1+p35S::NTR1-10xMYC transgenic plants, and performed co-IP using the GFP-trap and MYC-agarose beads, respectively. PRP8 (positive control) was readily detected in ILP1-GFP and NTR1-10xMYC immunoprecipitates, and RPB1 co-immunoprecipitated with ILP1-GFP and NTR1-10xMYC, albeit at low affinities (Figure 6C).
ILP1 and NTR1 interact with pri-miRNA processing machinery
We next investigated if ILP1 and NTR1 were associated with the dicing machinery. BiFC assays in the tobacco transient expression system revealed that ILP1 and NTR1 were in close proximity to DCL1 and SE, but not to HYL1, at sub-nuclear foci (Figure 6D). Taking advantage of the available antibodies for DCL1, HYL1 and SE, we performed in vivo co-IP assays using pistil tissues from ilp1+pILP1:: ILP1-GFP and ntr1-1+p35S::NTR1-10xMYC complementation plants. Consistent with the BiFC results, DCL1 and SE, but not HYL1, were co-immunoprecipitated with both ILP1-GFP and NTR1-10xMYC (Figure 6E and F). However, we were unable to detect a robust association of ILP1-GFP or NTR1-10xMYC with MIR chromatin, which could be due to a transient interaction between ILP1-GFP/NTR1-10xMYC and MIR chromatins (Supplementary Figure S6F).
DISCUSSION
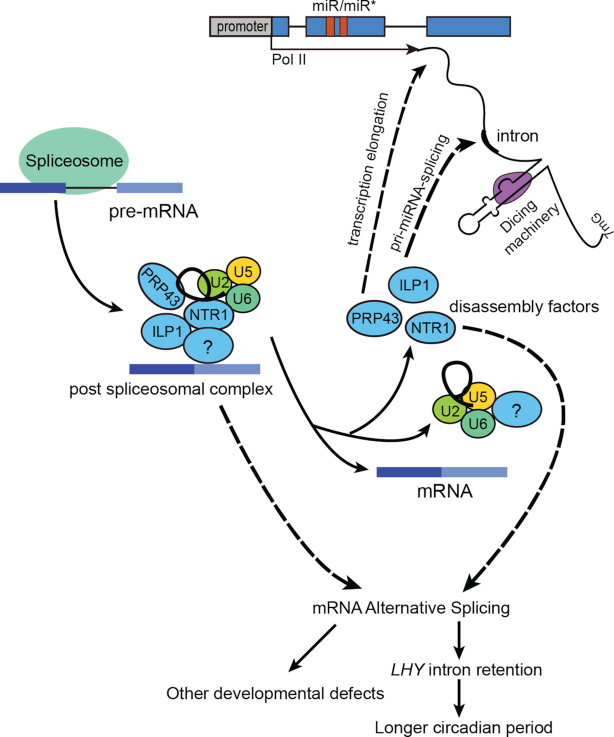
In this study, we showed that ILP1 and NTR1, two key co-factors of the ILS disassembly complex, positively regulated miRNA biogenesis. Based on our current data, we favor a model in which ILP1 and NTR1 regulate miRNA accumulation mainly by facilitating transcription elongation (Figure 7). This model is supported by several lines of evidence presented in this study and a previous study (28). First, ILP1 and/or NTR1 ablation reduced pri-miRNA levels without changing their promoter activities and half-lives (Figure 5A–D and Figure 6A). Second, ILP1 and NTR1 directly or indirectly interacted with Pol II (Figure 6C). Third, ILP1 and NTR1 were required for proper Pol II occupancies at downstream regions but not promoter regions of MIR genes (Figure 6B and Supplementary Figure S6A–C). At last, NTR1 has been shown to colocalize with Ser2-phosphorylated Pol II (29). Consistent with the model, several metazoan splicing factors, including SC35, TAT-SF1 and SKIP, have been reported to stimulate transcription elongation through their interactions with PTEF-b, a key elongation factor (68,69,70,71). Strikingly, for protein-coding genes with AS defects in ilp1-1 and ntr1-1, no differential expression was detected in either mutant (Supplementary Figure S5J), indicating that splicing and gene expression may be uncoupled activities of ILP1 and NTR1 for at least coding genes, or that the effect of splicing alteration on coding gene expression is too weak to be observed. This observation is in contrast to a previous study that NTR1 is required for proper transcription elongation at alternative exons (61). Future studies with more careful investigations will be required to clarify these discrepant observations. ILP1 and NTR1 also regulate AS of several pri-miRNAs (Figure 3G). Considering that splicing affects pri-miRNA processing and ILP1/NTR1 interact with DCL1 and SE (Figure 6D–F), it is possible that ILP1 and NTR1 also affect pri-miRNA processing. However, we believe that this should not be the main underlying mechanism because pri-miRNA levels were globally reduced in ilp1-1 and ntr1-1 mutants, and ILP1 had little effect on amiR-LUC expression under the control of the constitutive 35S promoter (Figure 5A–C and E).
Figure 7.
Working model of the spliceosome disassembly factors. Dismantling of the post-spliceosomal complex is mediated by spliceosome disassembly factors: ILP1, NTR1 and PRP43. Both ILP1 and NTR1 regulate plant development via miRNA dependent and independent pathways. On one hand, ILP1 and NTR1 regulate some developmental processes, including circadian rhythm, via proper splicing of a subset of precursor mRNAs (pre-mRNAs). On the other hand, ILP1 and NTR1 modulate miRNA abundance through promoting Pol II transcription elongation at MIR genes and/or pri-miRNA splicing.
Not all pri-miRNAs have detectable introns, which raises the question of how ILP1 and NTR1 globally regulate miRNA biogenesis. It is possible that ILP1 and NTR1 are recruited to MIR regions via interactions with miRNA-specific factors such as DCL1 and SE. Consistent with this, DCL1 is reported to be associated with MIR chromatin (7). Nevertheless, we only detect weak effect of ILP1 on the p35S::amiR-LUC reporter. We hypothesized that the use of a short hairpin construct may diminish the influence of ILP1 on transcription elongation. Indeed, ILP1 reduced the expression of exogenously introduced miR172b reporter, which harbors a much longer precursor (p35S::MIR172b) (33). Alternatively, pri-miRNAs splicing could be underestimated due to their low expression levels, and the total number of intron-containing pri-miRNAs might be much more than previously thought. In addition, it is possible that spliceosomal factors may interact with weak intron sites even without splicing. Consistent with this, it has been reported that long non-coding RNAs are less efficiently spliced than coding genes (72).
Several miRNA targets were moderately but significantly up-regulated in both mutants by qPCR and five out of seven TAS genes were up-regulated by RNA-seq (Supplementary Figures S1E and 5H). However, we didn’t see any significant changes of miRNA targets at global level (Supplementary Figure S5I). In a recently published study, there were also only few miRNA targets that were significantly up-regulated in pp4r3a, a mutant with reduced miRNA levels comparable to ILP1 or NTR1 dysfunction (46). In fact, even in the core miRNA biogenesis and action pathway mutants background (e.g. dcl1, ago1, hyl1 and hen1), only a subset of miRNA targets were significantly up-regulated, with the majority showing little to no changes (73,74). Considering many miRNAs are differentially expressed in differently cell types, future investigations at single-cell level may be applied to more accurately examine the impact of miRNAs on their target genes’ expression.
Dismantling of the ILS complex, catalyzed by PRP43 and its cofactors, plays an essential role in recycling snRNAs and splicing factors. We showed that ILP1 and NTR1 were in complex with PRP43. While PRP43 and NTR1 are conserved among plants, animals and yeast, the ILP1 homolog is absent in yeast (26,27,53). In budding yeast, NTR2 is another key co-factor of PRP43 (26). It will be interesting to test whether ILP1 is functionally equivalent to the yeast NTR2. The ILP1 and NTR1 proteins regulated AS of only a few hundreds of AS introns, with no obvious effect on the splicing of constitutively spliced introns (Figure 3D–G). This may be due to slightly weakened function of the disassembly complex harboring single mutations in ILP1 and NTR1. Surprisingly, the ilp1-1 ntr1-1 double knockout mutant showed embryonic lethality (Supplementary Figure S4D), suggesting a cooperative role of ILP1 and NTR1 either in maintaining the functional integrity of the disassembly complex or in other unknown synergistic functions.
Splicing of protein-coding genes occurs in all eukaryotes and exhibits pathway conservation across kingdoms (75). By contrast, much less is known about the significance of spliceosome machinery in regulating the biogenesis and functions of non-coding RNAs. The current study, as well as previous studies, has uncovered a large group of splicing-related proteins that regulate plant miRNA biogenesis (8–20). However, it is unclear whether these proteins share common mechanisms or perform distinct functions. Future studies are needed to reveal the relationships among these proteins and their mechanisms of action.
DATA AVAILABILITY
Raw sequence reads from small RNA-seq and RNA-seq data were deposited in the NCBI Sequence Read Archive database under the accession numbers SRP162482 and SRP162483, respectively. Raw data for Mass-Spec analysis are available via ProteomeXchange with identifier PXD011542.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Shuxin Zhang for providing the pMIR167::GUS line, Xiaodong Xu for the pCCA1::LUC and the pTOC1::LUC reporter lines, Yuda Fang for the pHYL1:: HYL1-GFP seeds and Xumin Zhang for Mass-Spec analysis. We also thank Xiaofei Yu for critical reading of the manuscript.
Authors’ contribution: Designed research: G.R., J.W. and S.C.; Performed research: J.W., S.C., X.W., Z.L., X.L., Y.Y. and C.Y.; Contributed new reagents/analytic tools: G.R. B.Z., H.L., T.N., L.L. and J.M.; Analyzed data: G.R., N.J. J.W., S.C. and N.L.; Wrote the paper: G.R. and J.W.
Notes
Present address: Xiaoyan Wang, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen 518060, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [91740101, 31622009, 31471221, 31771480]; National Key R&D Program of China [2016YFA0503200]. Funding for open access charge: National Natural Science Foundation of China [91740101].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rogers K., Chen X.M.. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013; 25:2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P.. MicroRNAs in plants. Gene Dev. 2002; 16:1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu B., Yang Z.Y., Li J.J., Minakhina S., Yang M.C., Padgett R.W., Steward R., Chen X.M.. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005; 307:932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mi S.J., Cai T., Hu Y.G., Chen Y., Hodges E., Ni F.R., Wu L., Li S., Zhou H., Long C.Z. et al.. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell. 2008; 133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J.L., Mei J., Ren G.D.. Plant microRNAs: biogenesis, homeostasis, and degradation. Front. Plant Sci. 2019; 10:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu Y., Jia T.R., Chen X.M.. The “how" and “where" of plant microRNAs. New Phytol. 2017; 216:1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fang X.F., Cui Y.W., Li Y.X., Qi Y.J.. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants. 2015; 1:15075. [DOI] [PubMed] [Google Scholar]
- 8. Ren G.D., Xie M., Dou Y.C., Zhang S.X., Zhang C., Yu B.. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:12817–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ben Chaabane S., Liu R.Y., Chinnusamy V., Kwon Y., Park J.H., Kim S.Y., Zhu J.K., Yang S.W., Lee B.H.. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2013; 41:1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhan X.Q., Wang B.S., Li H.J., Liu R.Y., Kalia R.K., Zhu J.K., Chinnusamy V.. Arabidopsis proline- rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:18198–18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu X.Y., Shi Y.P., Li J.R., Xu L., Fang Y.D., Li X., Qi Y.J.. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013; 23:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koster T., Meyer K., Weinholdt C., Smith L.M., Lummer M., Speth C., Grosse I., Weigel D., Staiger D.. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 2014; 42:9925–9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen T., Cui P., Xiong L.M.. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015; 43:8283–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Francisco-Mangilet A.G., Karlsson P., Kim M.H., Eo H.J., Oh S.A., Kim J.H., Kulcheski F.R., Park S.K., Manavella P.A.. THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 2015; 82:1018–1029. [DOI] [PubMed] [Google Scholar]
- 15. Li Z.W., Wang S.P., Cheng J.P., Su C.B., Zhong S.X., Liu Q., Fang Y.D., Yu Y., Lv H., Zheng Y. et al.. Intron lariat RNA inhibits MicroRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLos Genet. 2016; 12:e1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li S., Xu R., Li A., Liu K., Gu L., Li M., Zhang H., Zhang Y., Zhuang S., Wang Q. et al.. SMA1, a homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2018; 46:9148–9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li S.J., Liu K., Zhou B.J., Li M., Zhang S.X., Zeng L.R., Zhang C., Yu B.. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell. 2018; 30:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia T.R., Zhang B.L., You C.J., Zhang Y., Zeng L., Li S.P., Johnson K.C.M., Yu B., Li X., Chen X.M.. The arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell. 2017; 29:2626–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang S.X., Xie M., Ren G.D., Yu B.. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang S.X., Liu Y.H., Yu B.. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLos Genet. 2014; 10:e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bielewicz D., Kalak M., Kalyna M., Windels D., Barta A., Vazquez F., Szweykowska-Kulinska Z., Jarmolowski A.. Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 2013; 14:622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwab R., Speth C., Laubinger S., Voinnet O.. Enhanced microRNA accumulation through stemloop-adjacent introns. EMBO Rep. 2013; 14:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wan R.X., Yan C.Y., Bai R., Lei J.L., Shi Y.G.. Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae. Cell. 2017; 171:120–132. [DOI] [PubMed] [Google Scholar]
- 24. Martin A., Schneider S., Schwer B.. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002; 277:17743–17750. [DOI] [PubMed] [Google Scholar]
- 25. Arenas J.E., Abelson J.N.. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsai R.T., Fu R.H., Yeh F.L., Tseng C.K., Lin Y.C., Huang Y.H., Cheng S.C.. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Gene Dev. 2005; 19:2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoshimoto R., Okawa K., Yoshida M., Ohno M., Kataoka N.. Identification of a novel component C2ORF3 in the lariat-intron complex: lack of C2ORF3 interferes with pre-mRNA splicing via intron turnover pathway. Genes Cells. 2014; 19:78–87. [DOI] [PubMed] [Google Scholar]
- 28. Yoshizumi T., Tsumoto Y., Takiguchi T., Nagata N., Yamamoto Y.Y., Kawashima M., Ichikawa T., Nakazawa M., Yamamoto N., Matsui M.. Increased level of polyploidy1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell. 2006; 18:2452–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dolata J., Guo Y.W., Kolowerzo A., Smolinski D., Brzyzek G., Jarmolowski A., Swiezewski S.. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 2015; 34:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones M.A., Williams B.A., McNicol J., Simpson C.G., Brown J.W.S., Harmer S.L.. Mutation of arabidopsis spliceosomal timekeeper locus1 causes circadian clock defects. Plant Cell. 2012; 24:4066–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Mas P., Panda S., Kreps J.A., Kay S.A.. Cloning of the ARABIDOPSIS CLOCK CONE TOC1, an autoregulatory response regulator homolog. Science. 2000; 289:768–771. [DOI] [PubMed] [Google Scholar]
- 32. Song Y., Jiang Y.P., Kuai B.K., Li L.. CIRCADIAN CLOCK-ASSOCIATED 1 Inhibits Leaf Senescence in Arabidopsis. Front. Plant Sci. 2018; 9:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen X.M. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004; 303:2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie Q.G., Wang P., Liu X., Yuan L., Wang L.B., Zhang C.G., Li Y., Xing H.Y., Zhi L.Y., Yue Z.L. et al.. LNK1 and LNK2 Are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell. 2014; 26:2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clough S.J., Bent A.F.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 36. Weigel D., Glazebrook J.. Arabidopsis: a Laboratory Manual. 2002; Cold Spring Harbor Press Laboratory Press, NY. [Google Scholar]
- 37. Li X., Ma D.B., Lu S.X., Hu X.Y., Huang R.F., Liang T., Xu T.D., Tobin E.M., Liu H.T.. Blue light- and low temperature-regulated COR27 and COR28 play roles in the arabidopsis circadian clock. Plant Cell. 2016; 28:2755–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lidder P., Gutierrez R.A., Salome P.A., McClung C.R., Green P.J.. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 2005; 138:2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L.K., Feng Z.X., Wang X., Wang X.W., Zhang X.G.. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010; 26:136–138. [DOI] [PubMed] [Google Scholar]
- 40. Liu C.G., Axtell M.J., Fedoroff N.V.. The Helicase and RNaseIIIa Domains of Arabidopsis Dicer-Like1 Modulate Catalytic Parameters during MicroRNA Biogenesis. Plant Physiol. 2012; 159:748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M., Strahl B.D., Patel D.J., Jacobsen S.E.. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013; 498:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shen S.H., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Szczesniak M.W., Kabza M., Pokrzywa R., Gudys A., Makalowska I.. ERISdb: a database of plant splice sites and splicing signals. Plant Cell Physiol. 2013; 54:E10. [DOI] [PubMed] [Google Scholar]
- 46. Wang S., Quan L., Li S., You C., Zhang Y., Gao L., Zeng L., Liu L., Qi Y., Mo B. et al.. The PROTEIN PHOSPHATASE4 complex promotes transcription and processing of primary microRNAs in Arabidopsis. Plant Cell. 2019; 31:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu Z., Huang J.C., Huang J.N., Li Q.Q., Zhang X.M.. Lys-C/Arg-C, a more specific and efficient digestion approach for proteomics studies. Anal. Chem. 2018; 90:9700–9707. [DOI] [PubMed] [Google Scholar]
- 48. Yoo S.D., Cho Y.H., Sheen J.. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007; 2:1565–1572. [DOI] [PubMed] [Google Scholar]
- 49. Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J.. Chromatin techniques for plant cells. Plant J. 2004; 39:776–789. [DOI] [PubMed] [Google Scholar]
- 50. Chen X.M. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009; 25:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carbonell A., Fahlgren N., Garcia-Ruiz H., Gilbert K.B., Montgomery T.A., Nguyen T., Cuperus J.T., Carrington J.C.. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012; 24:3613–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P.. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gene Dev. 2006; 20:3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen W.J., Shulha H.P., Ashar-Patel A., Yan J., Green K.M., Query C.C., Rhind N., Weng Z.P., Moore M.J.. Endogenous U2.U5.U6 snRNA complexes in S. pombe are intron lariat spliceosomes. RNA. 2014; 20:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rascht G., Weigel D.. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nagel D.H., Kay S.A.. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012; 22:R648–R657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu Y., Ji L.J., Le B.H., Zhai J.X., Chen J.Y., Luscher E., Gao L., Liu C.Y., Cao X.F., Mo B.X. et al.. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017; 15:e2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vaucheret H., Vazquez F., Crete P., Bartel D.P.. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004; 18:1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P.. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schauer S.E., Jacobsen S.E., Meinke D.W., Ray A.. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002; 7:487–491. [DOI] [PubMed] [Google Scholar]
- 60. Robinsonbeers K., Pruitt R.E., Gasser C.S.. Ovule development in wild-type Arabidopsis and 2 female-sterile mutants. Plant Cell. 1992; 4:1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dolata J., Guo Y.W., Kolowerzo A., Smolinski D., Brzyzek G., Jarmolowski A., Swiezewski S.. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 2015; 34:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu B., Bi L., Zheng B.L., Ji L.J., Chevalier D., Agarwal M., Ramachandran V., Li W.X., Lagrange T., Walker J.C. et al.. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10073–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Manavella P.A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., Weigel D.. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012; 151:859–870. [DOI] [PubMed] [Google Scholar]
- 64. Fang Y.D., Spector D.L.. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007; 17:818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fujioka Y., Utsumi M., Ohba Y., Watanabe Y.. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in arabidopsis. Plant Cell Physiol. 2007; 48:1243–1253. [DOI] [PubMed] [Google Scholar]
- 66. Aukerman M.J., Sakai H.. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003; 15:2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M.. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007; 19:2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bres V., Gomes N., Pickle L., Jones K.A.. A human splicing factor,SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Gene Dev. 2005; 19:1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Saldi T., Cortazar M.A., Sheridan R.M., Bentley D.L.. Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 2016; 428:2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lin S.R., Coutinho-Mansfield G., Wang D., Pandit S., Fu X.D.. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008; 15:819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fong Y.W., Zhou Q.. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001; 414:929–933. [DOI] [PubMed] [Google Scholar]
- 72. Tilgner H., Knowles D.G., Johnson R., Davis C.A., Chakrabortty S., Djebali S., Curado J., Snyder M., Gingeras T.R., Guigo R.. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for IncRNAs. Genome Res. 2012; 22:1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Allen E., Xie Z., Gustafson A.M., Carrington J.C.. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005; 121:207–221. [DOI] [PubMed] [Google Scholar]
- 74. Ronemus M., Vaughn M.W., Martienssen R.A.. MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by Argonaute, Dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell. 2006; 18:1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Keren H., Lev-Maor G., Ast G.. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010; 11:345–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads from small RNA-seq and RNA-seq data were deposited in the NCBI Sequence Read Archive database under the accession numbers SRP162482 and SRP162483, respectively. Raw data for Mass-Spec analysis are available via ProteomeXchange with identifier PXD011542.