Abstract
Background
Predictive molecular biomarkers to select optimal treatment for patients with glioblastoma and other cancers are lacking. New strategies are needed when large randomized trials with correlative molecular data are not feasible.
Methods
Gene signatures (GS) were developed from 31 orthotopic glioblastoma patient-derived xenografts (PDXs), treated with standard therapies, to predict benefit from radiotherapy (RT-GS), temozolomide (Chemo-GS), or the combination (ChemoRT-GS). Independent validation was performed in a heterogeneously treated clinical cohort of 502 glioblastoma patients with overall survival as the primary endpoint. Multivariate Cox analysis was used to adjust for confounding variables and evaluate interactions between signatures and treatment.
Results
PDX models recapitulated the clinical heterogeneity of glioblastoma patients. RT-GS, Chemo-GS, and ChemoRT-GS were correlated with benefit from treatment in the PDX models. In independent clinical validation, higher RT-GS scores were associated with increased survival only in patients receiving RT (P = 0.0031, hazard ratio [HR] = 0.78 [0.66–0.92]), higher Chemo-GS scores were associated with increased survival only in patients receiving chemotherapy (P < 0.0001, HR = 0.66 [0.55–0.8]), and higher ChemoRT-GS scores were associated with increased survival only in patients receiving ChemoRT (P = 0.0001, HR = 0.54 [0.4–0.74]). RT-GS and ChemoRT-GS had significant interactions with treatment on multivariate analysis (P = 0.0009 and 0.02, respectively), indicating that they are bona fide predictive biomarkers.
Conclusions
Using a novel PDX-driven methodology, we developed and validated 3 platform-independent molecular signatures that predict benefit from standard of care therapies for glioblastoma. These signatures may be useful to personalize glioblastoma treatment in the clinic and this approach may be a generalizable method to identify predictive biomarkers without resource-intensive randomized trials.
Keywords: biomarker, Glioblastoma, patient-derived xenograft, radiation, signature, temozolomide
Key Points.
1. Predictive biomarkers for glioblastoma are lacking.
2. We developed a novel patient-derived xenograft approach.
3. We developed biomarkers for response to standard glioblastoma treatments.
Importance of the Study.
Developing predictive genomic biomarkers to guide cancer therapy typically requires large randomized trials with correlative molecular data. We developed a novel alternative approach utilizing a panel of orthotopic patient-derived glioblastoma xenografts to identify gene signatures that predict benefit from radiotherapy, temozolomide, or the combination of the two. We then independently validated these signatures in a clinical cohort of 502 glioblastoma patients. These signatures may be useful to select the optimal treatment for patients with glioblastoma or to select patients for clinical trials. Furthermore, this approach represents a potentially generalizable method to develop predictive biomarkers without resource-intensive randomized trials.
Following surgery, most patients with glioblastoma (GBM) are treated with a combination of temozolomide (TMZ) and radiotherapy (RT), while some receive either RT or TMZ alone. Molecular information to select from these treatment options is lacking. While promoter methylation of the gene encoding O6-methylguanine-DNA methyltransferase (MGMT) may predict for treatment benefit from TMZ, there are no biomarkers to inform response to RT or multi-modality therapy.1–4
To date, few predictive genomic biomarkers have been developed to guide treatment decisions for patients with cancer. A notable exception is breast cancer, where a 21-gene expression recurrence score guides decision making regarding adjuvant cytotoxic chemotherapy.5 Similar efforts are ongoing in prostate cancer to determine which men should receive adjuvant hormonal therapy6 or radiation following surgery.7 These efforts to personalize treatment have benefited from the high incidence of breast and prostate cancers, which facilitates the large number of patients and randomized trials necessary for predictive biomarker development.8 Rarer cancers, such as GBM, have more limited patient numbers and randomized trial data, and require alternative approaches to develop predictive biomarkers.
Orthotopic patient-derived xenografts (PDXs), in which tumor tissue directly from patients is implanted into the relevant body site in mice, recapitulate much of the biology of human tumors, including the microenvironment, intratumoral heterogeneity, and, in GBM, the blood–brain barrier.9 Orthotopic PDXs typically maintain the treatment responsiveness of their founder tumors10 and can be used to assess individual biomarkers.11–13 However, to our knowledge, there are no reported studies utilizing large numbers of PDXs combined with high-throughput gene expression profiling as a strategy to identify predictive biomarkers for treatment response.
In the first study of its kind, we performed RNA sequencing on a large cohort of GBM PDXs at baseline. We treated these PDXs with RT, TMZ, or RT+TMZ and developed gene signatures (GS) predicting treatment response (termed RT-GS, Chemo-GS, and ChemoRT-GS). We then independently validated the gene signatures in a clinical GBM cohort to assess the performance of the GS as predictive biomarkers, and compared our results with MGMT promoter methylation and gene expression.
Methods
Patient-Derived Xenografts
Data on 31 orthotopic GBM PDXs with baseline RNAseq data were obtained from the Mayo Clinic PDX National Resource (https://www.mayo.edu/research/labs/translational-neuro-oncology/mayo-clinic-brain-tumor-patient-derived-xenograft-national-resource. Accessed June 4, 2019). Clinical characteristics and genomic data on these PDXs are summarized in Supplementary Table 1 and are publicly available at (http://www.mayo.edu/research/documents/7-mayo-pdx-clinical-data/doc-20339608. Accessed June 4, 2019) and (http://www.mayo.edu/research/documents/1-mayo-pdx-genotype/doc-20339599. Accessed June 4, 2019). These PDX models have been used to evaluate various treatments, including RT (n = 67 experiments on 31 PDXs), TMZ (n = 137 experiments on 31 PDXs), and RT+TMZ (n = 79 experiments on 29 PDXs), as previously described.14,15 Gene expression was log2 transformed, then centered and scaled using the “scale” function in R. Additional details on the analyses in the PDXs can be found in the Supplementary Methods.
Clinical Validation
Independent clinical validation was performed in The Cancer Genome Atlas (TCGA)16 GBM cohort because of the large numbers of patients and the availability of gene expression, treatment data, and clinical outcomes. While other published GBM cohorts exist, they lack one or more of these required data.17,18 Gene expression data were downloaded from the University of California Santa Cruz cancer browser.19 Affymetrix U133A microarray data were selected for analysis, rather than RNAseq data, as microarray data were available on more samples. Expression, treatment, and outcomes were available for 502 patients. Gene expression was centered and scaled as above. Patients were classified as having received chemotherapy if they had received TMZ or other alkylating chemotherapy during their treatment course and were classified as having received ChemoRT if they had received both modalities of chemotherapy and radiation. Additional details on the analyses in TCGA can be found in the Supplementary Methods.
Gene Signature Development
The primary endpoint for each PDX experiment was the ratio of survival time with treatment relative to survival time without treatment. Spearman’s correlation was calculated for each gene to this ratio. Gene expression signatures for treatment response were developed using the genes with the highest absolute correlation coefficients. A score was created from the top genes by averaging20,21 their expression. For any gene selected for signature development with a negative correlation coefficient, the expression was multiplied by −1 such that a higher value always corresponded with increased benefit from treatment. All model development was performed exclusively in the PDXs. To identify biological pathways associated with treatment response, we used gene set enrichment analysis (GSEA).22 Additional details on the PDXs, RNAseq, formulas for the signatures, and GSEA can be found in the Supplementary Methods.
Statistical Analysis
The primary endpoint for clinical validation was overall survival. Once signatures were defined in the PDX data, independent clinical validation was performed without further modification. To assess for predictive potential, Cox regression was performed to test the interaction between the signatures and treatment.23 Multivariate analysis (MVA) of interactions was used to adjust for treatment selection bias as previously described.24 Gene signatures, MGMT promoter methylation, and gene expression were treated as continuous variables in Cox regression. This allowed the results to be comparable to each other, and is suggested by Janes et al23 for treatment selection biomarker evaluation. Therefore, all statistical inference was performed using gene signatures as continuous variables. Continuous variables are categorized into tertiles within Kaplan–Meier curves only for the purposes of visualization within the main text. The pre-specified analyses were the assessments of the 3 treatment signatures, MGMT promoter methylation, and MGMT expression for treatment benefit. P-values <0.05 were considered significant.
Results
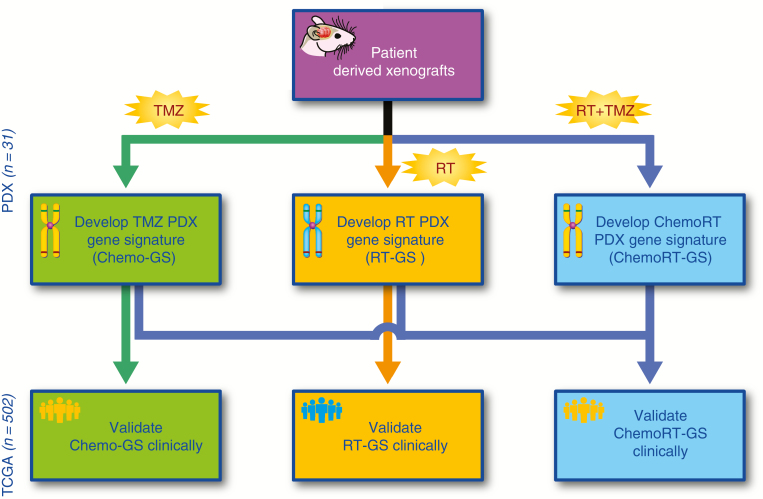
The overall study schema is depicted in Fig. 1. We utilized the gene expression and treatment response data from the PDXs to develop gene signatures for RT, chemotherapy, and ChemoRT response, which were then independently validated in a clinical cohort.
Fig. 1.
Flow diagram detailing the analysis pipeline.
Patient-Derived Xenografts
The PDXs recapitulated the clinical heterogeneity of human GBMs. Sixty-one percent were from male patients and 39% from females. The median age at diagnosis was 63 years, with a range from 38 to 83 years. MGMT promoter methylation occurred in 45% of samples.1 All PDXs were isocitrate dehydrogenase 1 (IDH1) wild-type (ie, derived from primary GBMs), but mutations in EGFR, PTEN, and TP53 were all present.16 Clinical and molecular characteristics are further summarized in Supplementary Table 1, with all data publicly available online at the Mayo Clinic PDX National Resource website. Treatment benefit was greatest with TMZ + RT combined, followed by TMZ, and then by RT (Supplementary Fig. 1). There was limited overlap between the top 100 genes that were positively and negatively correlated with response to RT, TMZ, and RT+TMZ (Supplementary Fig. 2, Supplementary Table 2). Within the top 10 pathways correlated with resistance to RT, GSEA revealed that several were related to epithelial-mesenchymal transition and extracellular matrix interactions (Supplementary Fig. 3A). RAS signaling pathways were represented in the top pathways correlated with TMZ resistance (Supplementary Fig. 3B). For pathways correlated with resistance to RT+TMZ, 9 out of the top 10 pathways were associated with DNA replication (Supplementary Fig. 3C).
Clinical Validation
Patients in the clinical validation cohort (Supplementary Table 3, N = 502) were treated with combined ChemoRT (65%), RT alone (16%), or chemotherapy alone (3%) or received no treatment (16%). Patients treated with ChemoRT had the best outcomes, followed by single modality treatment (RT or chemotherapy), and patients receiving no treatment had the worst outcomes (Supplementary Fig. 4). MGMT promoter methylation was highly inversely correlated with MGMT gene expression (Spearman’s rho = −0.54, P < 0.0001), as expected, since promoter methylation silences MGMT, and is consistent with the literature.25
Alkylating Chemotherapy Response Signature
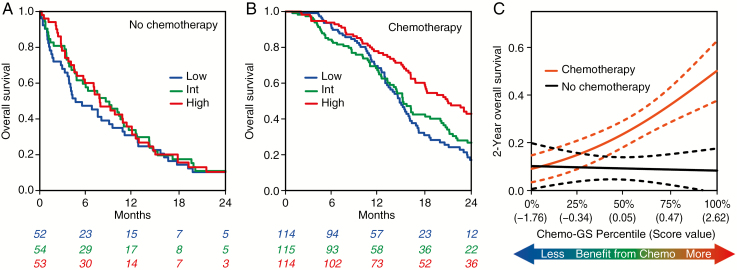
We rank genes for correlation to TMZ response in the PDX models and found that MGMT had the second-highest ranked absolute correlation coefficient (Spearman’s rho: −0.47). Because of the known biology of MGMT promoter methylation and increased sensitivity to alkylating chemotherapy,26 the fact that it was highly correlated with TMZ response in the PDXs supported the validity of our methodology. Therefore, we hypothesized that a gene signature consisting of the average of MGMT and the only gene ranked higher GPRASP1, Spearman’s rho = −0.48) would predict response to chemotherapy. The absolute correlation of the 2-gene Chemo-GS was higher than MGMT alone (Spearman’s rho: −0.53) supporting the addition of GPRASP1. To validate clinically, we compared patients who received chemotherapy (with or without RT) to patients who did not (RT alone or no treatment). The Chemo-GS was associated with improved survival only in patients who received chemotherapy (P < 0.0001, HR = 0.66 [0.55–0.8]), but not in those who did not (P = 0.14, HR = 0.81 [0.62–1.07]; Fig. 2A, B). Higher Chemo-GS indicated an increased benefit from chemotherapy (Fig. 2C). MGMT promoter methylation was borderline associated with survival in patients who received chemotherapy (P = 0.065, HR = 0.86 [0.74–1.01]) and not associated in patients who did not receive chemotherapy (P = 0.96, HR = 1.01 [0.78–1.30]; Supplementary Fig. 5A, B). MGMT gene expression was borderline associated with survival in the chemotherapy treated patients (P = 0.085, HR = 1.1 [0.99–1.23]) and not associated in patients who did not receive chemotherapy (P = 0.48, HR = 1.07 [0.89–1.28]; Supplementary Fig. 5C, D). The MVA interactions were not significant for Chemo-GS (Table 1), MGMT promoter methylation (P = 0.64), or MGMT expression (P = 0.25).
Fig. 2.
(A-C) Assessment of performance of Chemo-GS in independent clinical validation. Chemotherapy is defined as alkylating chemotherapy with or without RT. No chemotherapy is defined as RT alone or no treatment. For visualization purposes, continuous scores are divided into tertiles (low, intermediate, high). (A) Kaplan–Meier curves showing that Chemo-GS is not prognostic in patients who did not receive chemotherapy. (B) Kaplan–Meier curves showing that Chemo-GS is prognostic in patients who received chemotherapy. (C) Interaction plot showing logistic regression models fitting Chemo-GS to 2-year overall survival. Patients with a higher Chemo-GS score derived more benefit from chemotherapy.
Table 1 .
Interaction MVA of each signature in the clinical validation cohort
| Chemo-GS1 | RT-GS2 | ChemoRT-GS3 | ||||
|---|---|---|---|---|---|---|
| P-value | HR | P-value | HR | P-value | HR | |
| Chemo-GS:Chemo | 0.8934 | 0.98 [0.72–1.33] | Not included | Not included | ||
| Chemo-GS | 0.0284 | 0.76 [0.6–0.97] | ||||
| RT-GS:RT | Not included | 0.0009 | 0.4 [0.23–0.69] | Not included | ||
| RT-GS | 0.0019 | 2.26 [1.35–3.77] | ||||
| ChemoRT-GS:ChemoRT | Not included | Not included | 0.0204 | 0.56 [0.34–0.91] | ||
| ChemoRT-GS | 0.5467 | 1.13 [0.76–1.68] | ||||
| Chemo | 0.2085 | 0.68 [0.37–1.24] | 0.0608 | 0.56 [0.3–1.03] | 0.1876 | 0.67 [0.36–1.22] |
| RT | <0.0001 | 0.41 [0.29–0.57] | <0.0001 | 0.36 [0.26–0.51] | <0.0001 | 0.43 [0.3–0.6] |
| ChemoRT | 0.8923 | 0.96 [0.49–1.85] | 0.7482 | 1.12 [0.57–2.18] | 0.8708 | 0.95 [0.49–1.84] |
| Prior treatment | 0.1815 | 0.71 [0.42–1.18] | 0.3877 | 0.8 [0.48–1.33] | 0.2474 | 0.74 [0.45–1.23] |
| Resection | 0.2021 | 1.21 [0.9–1.63] | 0.3255 | 1.16 [0.87–1.55] | 0.2664 | 1.18 [0.88–1.58] |
| Male vs Female | 0.2974 | 1.12 [0.91–1.37] | 0.2673 | 1.12 [0.91–1.38] | 0.2486 | 1.13 [0.92–1.4] |
| Age | <0.0001 | 1.03 [1.02–1.04] | <0.0001 | 1.03 [1.02–1.04] | <0.0001 | 1.03 [1.02–1.04] |
1Left: interaction MVA for Chemo-GS (as a continuous variable) with alkylating chemotherapy.
2Middle: interaction MVA for RT-GS (as a continuous variable) with RT.
3Right: interaction MVA for ChemoRT-GS (as a continuous variable) with alkylating chemotherapy and RT.
Radiation Response Signature
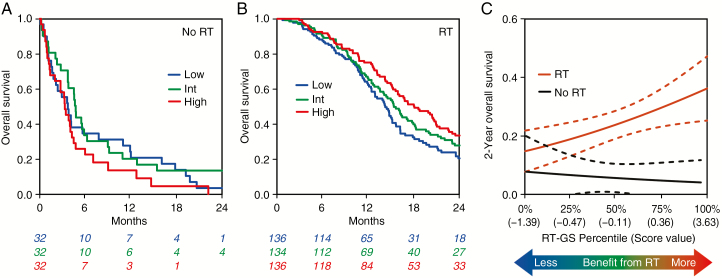
We next examined RT response, for which there are no clinically utilized predictive markers in GBM. Applying the exact same methodology used to generate Chemo-GS, we integrated the top 2 most correlated genes from the PDXs into RT-GS (average of CHGA, MAPK8, Spearman’s rho = 0.47, 0.41, respectively). In clinical validation, we compared patients who received RT (with or without chemotherapy) with those who did not (chemotherapy alone or no treatment). The 2-gene RT-GS was associated with improved survival only in the patients who received RT (P = 0.0031, HR = 0.78 [0.66–0.92]) and not in patients who did not receive RT (P = 0.28, HR = 1.28 [0.82–2.0]; Fig. 3A, B). Higher RT-GS scores indicated more of a benefit from RT (Fig. 3C). On interaction MVA, the RT-GS:RT treatment interaction term was highly significant (P = 0.0009; Table 1), indicating that RT-GS is a predictive biomarker for response to radiation.
Fig. 3.
(A–C) Assessment of performance of RT-GS in independent clinical validation. RT is defined as RT with or without chemotherapy. No RT is defined as chemotherapy alone or no treatment. For visualization purposes, continuous scores are divided into tertiles (low, intermediate, high). (A) Kaplan–Meier curves showing that RT-GS is not prognostic in patients who did not receive RT. (B) Kaplan–Meier curves showing that RT-GS is prognostic in patients who received RT. (C) Interaction plot showing logistic regression models fitting RT-GS to 2-year overall survival. Patients with a higher RT-GS score derived more benefit from RT.
Chemotherapy and Radiation Response Signature
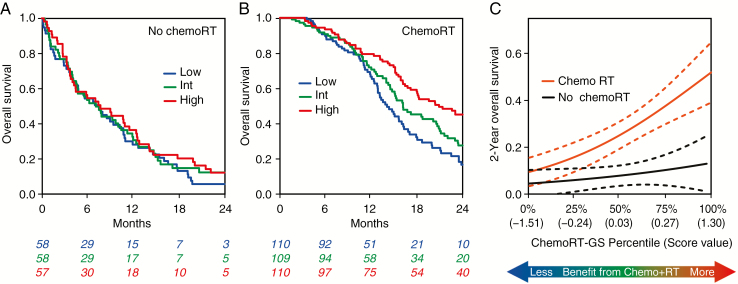
We next examined response to combined modality therapy, for which there is also no clinically utilized predictive marker. Since chemotherapy and RT response may independently contribute to ChemoRT response, we incorporated the individual chemotherapy and RT response signatures from above into the combined response score. To account for interactions between RT and chemotherapy (such as the generation of complex DNA damage), we also used the top 2 genes specifically correlated with RT+TMZ treatment in the PDXs (ATP6V0A2, FGF7, Spearman’s rho = −0.7, −0.69, respectively) to develop a 6-gene ChemoRT-GS. We then compared patients who received ChemoRT with those who had received single modality treatment or no treatment. As with the other 2 signatures, ChemoRT-GS was associated with improved survival only in patients treated with ChemoRT (P = 0.0001, HR = 0.54 [0.4–0.74]) and not in those not treated with ChemoRT (P = 0.26, HR = 0.8 [0.54–1.18]; Fig. 4A, B). Higher ChemoRT-GS scores indicated more of a benefit from ChemoRT (Fig. 4C). The multivariate interaction term was significant (P = 0.02; Table 1), indicating that ChemoRT-GS is a predictive biomarker for response to dual therapy with ChemoRT. MGMT promoter methylation was associated with survival in patients who received ChemoRT (P = 0.033, HR = 0.84 [0.71–0.99]) and not associated in patients who did not receive ChemoRT (P = 0.79, HR = 1.03 [0.82–1.31]; Supplementary Fig. 6A, B), with a non-significant MVA interaction (P = 0.55). Similarly, MGMT gene expression was borderline associated with survival in patients who received ChemoRT (P = 0.057, HR = 1.11 [1.00-0.25]) and not associated in patients who did not receive ChemoRT (P = 0.45, HR = 1.07 [0.9–1.27]; Supplementary Fig. 6C, D), with a non-significant MVA interaction (P = 0.77).
Fig. 4.
(A–C) Assessment of performance of ChemoRT-GS in independent clinical validation. ChemoRT is defined as alkylating chemotherapy with RT. No ChemoRT is defined as chemo alone, RT alone, or no treatment. For visualization purposes, continuous scores are divided into tertiles (low, intermediate, high). (A) Kaplan–Meier curves showing that ChemoRT-GS is not prognostic in patients who did not receive ChemoRT. (B) Kaplan–Meier curves showing that ChemoRT-GS is prognostic in patients who received ChemoRT. (C) Interaction plot showing logistic regression models fitting ChemoRT-GS to 2-year overall survival. Patients with a higher ChemoRT-GS score derived more benefit from ChemoRT.
Clinical and Molecular Associations
Associations between the three signatures and clinical and molecular variables are presented in Supplementary Tables 4–6. Of note, Chemo-GS was associated with MGMT promoter methylation as expected, since MGMT gene expression is part of the signature. Chemo-GS was also associated with age at diagnosis. Higher RT-GS scores were also associated with younger age at diagnosis, consistent with the observation that younger patients may benefit more from RT.27 Similarly, ChemoRT-GS was associated with both MGMT promoter methylation and age, which is expected, as both Chemo-GS and RT-GS are components of ChemoRT-GS. Scores of all 3 signatures were higher in IDH1-mutant tumors, suggesting that patients whose tumors harbor the IDH1 mutation may derive increased benefit from multiple therapies. When we include IDH1 mutation as a covariate in the MVA interaction, the signature:treatment interactions remain significant for both RT-GS (P = 0.025) and ChemoRT-GS (P = 0.042), suggesting that the IDH1 mutation is not exclusively responsible for the predictive nature of these signatures.
Discussion
In the first study of its kind, we have utilized a PDX-based approach to develop 3 different gene signatures to predict GBM responsiveness to chemotherapy, radiation, and the combination. We independently validated these signatures in a clinical cohort of GBM patients. Each signature was prognostic only in patients receiving the signature-associated treatment. RT-GS and ChemoRT-GS represent the first molecular predictors of RT and ChemoRT response in GBM. The significant interaction between signatures and treatments indicate that they predict response to therapy rather than simply being prognostic.
The pathways associated with treatment resistance are consistent with known biology. MGMT, which predicts for TMZ resistance in patients and laboratory models of GBM,26 was the second most highly correlated gene with TMZ resistance across our PDX cohort. GSEA also revealed biologically relevant pathways associated with therapy resistance. Pathways involved with epithelial-to-mesenchymal transition were associated with GBM PDX radioresistance. This finding is in agreement with literature reports in GBM and other cancers and suggests that therapeutic approaches targeting this phenotype should be explored in combination with radiotherapy in GBM.28–30 Increased expression of RAS signaling pathways was associated with TMZ resistance, which could be due to the role of RAS/mitogen-activated protein kinase signaling in cell survival.31 Numerous pathways related to DNA elongation and replication were associated with resistance to combined TMZ and radiation treatment, perhaps indicating that this machinery allows GBMs to detoxify the complex DNA damage that forms when radiation is combined with alkylating chemotherapy.32
This xenograft-driven methodology may be especially beneficial for rare tumors. Conventional approaches to predictive biomarker development require molecular profiling of large numbers of patient samples from patients treated with control or the treatment of interest in order to account for population-level genomic heterogeneity. This has been most successful in cancers with high incidences such as breast and prostate but is less feasible in rarer cancers.5,7 In our PDX-driven approach, genomically identical PDXs are treated with placebo or the treatment of interest, which allows for the quantification of gene-level effects on treatment benefit without large numbers of profiled samples. While the signatures developed from PDXs must still be validated on samples from patients, this methodology greatly increases the feasibility of biomarker development in cancers where patient numbers or clinical trial samples are limiting.
This methodology is also versatile. A single, simple approach led to predictive biomarkers of 3 standard-of-care treatments for GBM. Because PDX cohorts are also amenable to high-throughput testing of novel therapies,13 we anticipate that this methodology could be used during drug development to identify the patients most likely to benefit from a given therapy and potentially guide initial clinical trial design. The versatility of this methodology is also enhanced by its platform independence. Because these biomarkers of response can be applied to tumors whose transcript levels have been quantified by quantitative PCR, microarray, or RNAseq, they do not require that a specific commercial test be performed. This platform independence should facilitate future validation of these signatures using clinical trial specimens.
These gene signatures have potential clinical utility. Outside of a clinical trial, we expect that most patients will continue to receive combined chemoradiation regardless of their gene score. However, in patients able to tolerate only a single treatment modality, the RT-GS and Chemo-GS scores could be used to select RT or TMZ with more precision than the currently used MGMT promoter methylation assay.3,4 In the setting of a clinical trial, patients with a high ChemoRT-GS score should strongly consider treatment regimens involving both radiation and chemotherapy, whereas patients with a low score could be offered trials with novel therapy strategies. Patients with high RT-GS scores but low RT-Chemo and ChemoRT-GS scores may be excellent candidates for trials involving standard radiation but novel systemic therapy and/or novel radiosensitizing strategies.
Our work has limitations. PDX RNAseq data were obtained at a single timepoint, while treatment response experiments were performed numerous times over several years, during which mouse-specific evolution could have occurred.33 Experimental number also varied between PDXs, which could bias our results toward PDXs with more replicates. Despite these limitations, MGMT was the second most correlated gene with TMZ response, which underscores the validity of this model. While we believe this approach may be generalizable to develop predictive biomarkers for other therapeutics, care should be taken regarding immune-oncology agents, as PDXs are typically grown in immunocompromised mice. The validation dataset is not a randomized control trial and therefore has treatment selection bias, unaccounted confounders, and imbalances in the numbers of patients receiving different treatment modalities. We attempted to adjust for this by including potential confounders in our MVA,24 but such corrections are likely imperfect and additional validation is needed prior to using these tools in the clinic. Clinical validation would ideally be in prospective trials, with arms containing RT or TMZ monotherapy and/or observation. We also noted that TMZ monotherapy outperformed RT monotherapy in PDX models, but RT monotherapy was more efficacious in TCGA. We believe that this finding is due to the lower absolute dose of RT (20 Gy in 10 fractions) typically used in the PDX experiments.
As oncology moves toward the molecular classification of tumors, there is a strong need for molecular signatures that not only risk-stratify patients (prognostic biomarkers) but can also guide treatment decisions (predictive biomarkers). These PDX-derived signatures may be useful to personalize GBM treatment in the clinic. Furthermore, this PDX approach may be a generalizable method to identify predictive biomarkers, which is especially useful in rarer cancers.
Funding
DRW is funded by a University of Michigan Cancer Center Core grant (P30CA046592), as well as grants from the American Cancer Society, the Forbes Institute for Cancer Discovery, and the Jones Family Foundation Fund within the Chad Carr Pediatric Brain Tumor Center. JNS and the Mayo GBM Xenograft National Resource are supported by grants from the NCI including R24NS092940 and P50CA108961. MY is funded by a Patient-Centered Outcomes Research Institute (PCORI) award (ME-1409–21219). The views in this publication are solely the responsibility of the authors and do not necessarily represent the views of the PCORI, its Board of Governors or Methodology Committee.
Conflict of interest statement. A provisional patent application on this work has been filed. Unrelated to the content of this manuscript, SGZ has received travel expenses from GenomeDx Biosciences Inc, a genitourinary cancer molecular diagnostics company, and has patent applications on gene signatures not related to this work. SLC is an employee of and FYF and CWS have an ownership interest in PFS Genomics Inc, a breast cancer molecular diagnostics company.
Authorship statement: Conception and design: SGZ, JNS, DRW. Acquisition of data: BLC, ACM, TLS, JNS, DRW. Analysis and interpretation of data: SGZ, MY, DES, SLC, FYF, MMK, CWS. Drafting the article or revising it critically for important intellectual content: All authors. Final approval of the version to be published: All authors.
Supplementary Material
Acknowledgments
We would like to acknowledge the graphical artist assistance of Steven Kronenberg.
References
- 1. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 2. Perry JR, Laperriere N, O’Callaghan CJ, et al. ; Trial Investigators Short-Course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med. 2017;376(11):1027–1037. [DOI] [PubMed] [Google Scholar]
- 3. Wick W, Platten M, Meisner C, et al. ; NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. [DOI] [PubMed] [Google Scholar]
- 4. Malmström A, Grønberg BH, Marosi C, et al. ; Nordic Clinical Brain Tumour Study Group (NCBTSG) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. [DOI] [PubMed] [Google Scholar]
- 5. Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao SG, Chang SL, Erho N, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 2017;3(12):1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. Lancet Oncol. 2016;17(11):1612–1620. [DOI] [PubMed] [Google Scholar]
- 8. Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J Clin Oncol. 2009;27(24):4027–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JY, Kim SY, Park C, et al. Patient-derived cell models as preclinical tools for genome-directed targeted therapy. Oncotarget. 2015;6(28):25619–25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stewart EL, Mascaux C, Pham NA, et al. Clinical utility of patient-derived xenografts to determine biomarkers of prognosis and map resistance pathways in EGFR-mutant lung adenocarcinoma. J Clin Oncol. 2015;33(22):2472–2480. [DOI] [PubMed] [Google Scholar]
- 11. Rubio-Viqueira B, Jimeno A, Cusatis G, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12(15):4652–4661. [DOI] [PubMed] [Google Scholar]
- 12. Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1(6):508–523. [DOI] [PubMed] [Google Scholar]
- 13. Gao H, Korn JM, Ferretti S, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–1325. [DOI] [PubMed] [Google Scholar]
- 14. Carlson BL, Grogan PT, Mladek AC, et al. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int J Radiat Oncol Biol Phys. 2009;75(1):212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitange GJ, Mladek AC, Schroeder MA, et al. Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep. 2016;14(11):2587–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Puchalski RB, Shah N, Miller J, et al. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360(6389):660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldman M, Craft B, Swatloski T, et al. The UCSC cancer genomics browser: update 2015. Nucleic Acids Res. 2015;43(Database issue):D812–D817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yau C, Sninsky J, Kwok S, et al. An optimized five-gene multi-platform predictor of hormone receptor negative and triple negative breast cancer metastatic risk. Breast Cancer Res. 2013;15(5):R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tutt A, Wang A, Rowland C, et al. Risk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signature. BMC Cancer. 2008;8:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Janes H, Pepe MS, Bossuyt PM, Barlow WE. Measuring the performance of markers for guiding treatment decisions. Ann Intern Med. 2011;154(4):253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. White NM, Zhao SG, Zhang J, et al. Multi-institutional analysis shows that low PCAT-14 expression associates with poor outcomes in prostate cancer. Eur Urol. 2017;71(2):257–266. [DOI] [PubMed] [Google Scholar]
- 25. Everhard S, Tost J, El Abdalaoui H, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11(4):348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6(1):39–51. [DOI] [PubMed] [Google Scholar]
- 27. Barker FG 2nd, Chang SM, Larson DA, et al. Age and radiation response in glioblastoma multiforme. Neurosurgery. 2001; 49(6):1288–1297; discussion 1297–1298. [DOI] [PubMed] [Google Scholar]
- 28. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang L, Graham PH, Hao J, et al. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013;4:e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davis FM, Stewart TA, Thompson EW, Monteith GR. Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci. 2014;35(9):479–488. [DOI] [PubMed] [Google Scholar]
- 31. Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286(5443):1358–1362. [DOI] [PubMed] [Google Scholar]
- 32. Morgan MA, Parsels LA, Maybaum J, Lawrence TS. Improving the efficacy of chemoradiation with targeted agents. Cancer Discov. 2014;4(3):280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ben-David U, Ha G, Tseng YY, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49(11):1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.