Abstract
Nitric oxide (NO) is now established as an important signalling molecule in plants where it influences growth, development, and responses to stress. Despite extensive research, the most appropriate methods to measure and localize these signalling radicals are debated and still need investigation. Many confounding factors such as the presence of other reactive intermediates, scavenging enzymes, and compartmentation influence how accurately each can be measured. Further, these signalling radicals have short half-lives ranging from seconds to minutes based on the cellular redox condition. Hence, it is necessary to use sensitive and specific methods in order to understand the contribution of each signalling molecule to various biological processes. In this review, we summarize the current knowledge on NO measurement in plant samples, via various methods. We also discuss advantages, limitations, and wider applications of each method.
Keywords: Chemiluminiscence, haemoglobin, mitochondria, nitric oxide, quantum cascade laser, redox
This review summarizes current approaches to detect nitric oxide and discuss advantages and disadvantages of each method.
Introduction
The gaseous free radical nitric oxide (NO) emerged as an important signalling molecule in plants. This versatile molecule plays various roles in plants during various developmental stages (seed germination, root growth, flowering, growth of pollen tube, and leaf senescence), stem cell regulation, stomatal movement, regulation of primary metabolism, and protection against biotic/abiotic stresses (Besson-Bard et al., 2008b; Wilson et al., 2008; Yu et al., 2014; Domingos et al., 2015; Fancy et al., 2017).
In the case of animals, NO acts as a potent endogenous vasodilator and plays important roles in inflammation, inhibition of platelet aggregation, regulation of immune responses, cGMP signalling, and protection during ischaemia and reperfusion injury (Schmidt and Walter, 1994; Phillips et al., 2009). In animals, the majority of NO is produced by a family of nitric oxide synthases (NOSs). NOS catalyses deamination of arginine to citrulline. Three NOS enzymes are characterized in animal systems, namely endothelial nitric oxide synthase (eNOS), inducible nitric oxide synthase (iNOS), and neuronal nitric oxide synthase (nNOS). NOS has homology to P450 cytochrome c reductases. NOS activity requires various substrates and cofactors such as NADPH, FMN, FAD, tetrahydrobiopterin (BH4), and calcium/calmodulin. In addition to NOS, mitochondria of animal system are also known to produce NO via a nitrite-dependent pathway (Shiva, 2010).
Comparison of NO signalling in animals and plants revealed several commonalities (Wendehenne et al., 2001). Plant NO signalling operates via the S-nitrosylation process and also via activation of calcium, protein kinase cGMP signalling, and reactive oxygen species (ROS) (Wendehenne et al., 2001; Gaupels et al., 2011). Despite several attempts to identify NOS, a clear picture of its existence in land plants has not been obtained so far. However, the existence of NOS-like activity has been shown in various species and also in response to stress (reviewed in Corpas et al., 2009). The conclusions on NOS activity are mainly based on two methods, one is the inhibition of NO production in response to l-arginine analogues such as AET [2-(2-aminoethyl)isothiourea], PBITU [S,S′-1,3-phenylene-bis(1,2-ethanediyl)-bis-isothiourea], l-NIL [N6-(1-iminoethyl)- l-lysine], l-NAME (NG-nitro-l-arginine methyl ester), and l-NMMA (NG-monomethyl-l-arginine), and the second is the arginine/citrulline assay. The latter has recently been questioned because argininosuccinate, synthesized as a side reaction from arginine and fumarate by argininosuccinate lyase, can give false-positive results (Tischner et al., 2007). Recent evidence suggests that a few algal species such as Ostreococcus tauri possess NOS (Foresi et al., 2010; Santolini et al., 2017).
NO is also produced via nitrate reductase (NR) by utilizing nitrite as an ultimate substrate in an NAD(P)H-dependent reaction (Rockel et al., 2002; Planchet et al., 2005). NR contains molybdenum in its active site, and the enzyme activity is inhibited by tungstate which is an antagonist of molybdate (Bright et al., 2006). In Arabidopsis, NR is encoded by two genes NIA1 and NIA2. Deletion of NIA2 in Arabidopsis resulted in only 10% NR activity compared with wild-type plants (Wilkinson and Crawford, 1993). NR-dependent NO increases in light and dark with anoxia treatment (Planchet et al., 2005). In addition, mitochondria can reduce nitrite to NO under low oxygen conditions using nitrite as substrate (Gupta et al., 2005; Vishwakarma et al., 2018); this occurs at the site of complex III, IV, and alternative oxide (AOX) (Gupta et al., 2018). In addition, NO may also be synthesized by a plasma membrane-bound nitrite-NO oxidoreductase (Ni-NOR). However, it was found to be root specific and acts as a sensor of nitrate availability in the soil (Stohr and Stremlau, 2006). However, other reductive pathways for NO synthesis operating via xanthine dehydrogenase/oxidase where nitrite acts as a substrate (Godber et al., 2000) are also present. Plants also produce NO via polyamine- and hydroxylamine-mediated pathways. These pathways are still poorly characterized and need further investigation (Tun et al., 2006; Rumer et al., 2009).
Crucially, NO rapidly reacts with O2−, leading to formation of ONOO−. The production of ONOO− takes place in close proximity to the sites of superoxide generation. Due to the negative charge of O2·− and its shorter half-life compared with NO (Vranova et al., 2002), ONOO− production is more dependent on O2·− levels. Peroxynitrite can undergo homolytic cleavage to give HO· but its main biological function may be to react with tyrosine residues within proteins to form 3-nitrotyrosine. This occurs via addition of a nitro group in the ortho position of the aromatic ring in tyrosine (Kolbert et al., 2017). The formation of nitrotyrosine represents an irreversible oxidative post-translational modification that is accepted as a biomarker of nitrosative stress. The incorporation of a nitro group into protein tyrosine can significantly change protein structure with consequent functional defects (e.g. enzyme inactivation). In plants, so far only loss of function by nitration has been reported (Begara-Morales et al., 2013; Kolbert et al., 2017). Nitration is a mechanism distinct from S-nitrosation where NO can covalently but reversibly oxidize the thiols groups of reduced reactive cysteine residues in proteins. S-Nitrosylation can activate or repress protein function (Gupta, 2011) and can be assessed using biotin-switch technology (BST). BST is currently the most commonly used method used for identification of nitrosylated proteins (Jaffrey and Snyder, 2001; Palmieri et al., 2010) which is so well established that it will not be considered in this review. An excellent consideration of BST can be found in many papers; for example, Forrester et al. (2009).
Given the importance of NO, and its interaction in plant physiology, this review will critically consider current methods to measure NO. Measuring poses particular problems which will be discussed. In doing so, we will attempt to highlight the best practices so that the functions of NO can be assessed.
Assays for nitric oxide
NO quantification in tissues can be difficult due to the potential interference of various metabolites and its half-life, which can be extremely short, ranging from a few seconds to minutes. Therefore, it is necessary to use sensitive methods to detect NO. Further, as each method has advantages and disadvantages, it is recommended to use at least two methods for NO quantification (Gupta and Igamberdiev, 2013). There are several methods which are being used widely throughout plant and animal systems for the measurement of NO, such as fluorescent probes, the Griess reaction, EPR, chemiluminescence, oxyhaemoglobin (Hb-O2) assay, laser photoacoustics, membrane inlet-mass spectrometry, and amperometric methods with NO-specific electrodes. These methods either measure free NO or oxidized products of NO such as NO2− or NO3−.
DAF or MNIP-Cu fluorescent probes
One frequently used method to detect NO is based on the fluorophore diaminofluorescein (DAF) in cell-permeable forms (DAF-2DA or DAF-FM DA) as developed by Kojima et al. (1998). The utility of these dyes have been demonstrated in various species, tissues types, and responses. Thus, DAF dyes have been used for live imaging of NO in cell suspensions (Planchet and Kaiser, 2006), tobacco leaves (Cvetkovska and Vanlerberghe, 2012), lateral root primordia of tomato (Correa-Aragunde et al., 2004), and differentiating xylem of Zinnia elegans vascular bundles (Gabaldon et al., 2005). Other examples include legumes, such as Medicago truncatula suspension cells and leaves (Kepczynski and Cembrowska-Lech, 2018), protoplasts of soybean roots treated with indole-3-acetic acid (IAA) (Hu et al., 2005), and in vascular tissue (xylem), epidermal cells and root hairs of pea seedlings (Corpas et al., 2006). DAF dyes have also been used to provide spatial information on NO generation patterns in response to various stresses (Arita et al., 2006; Planchet and Kaiser, 2006; Neill et al., 2008; De Michele et al., 2009; Mur et al., 2011; Wany et al., 2017, 2019; Vishwakarma et al., 2018). Examples include tobacco epidermal (abaxial) sections treated with cryptogein, a fungal elicitor from Phytophthora cryptogea (Foissner et al., 2000), or within developing B. graminis fungal sp. hordei appressoria (Prats et al., 2008).
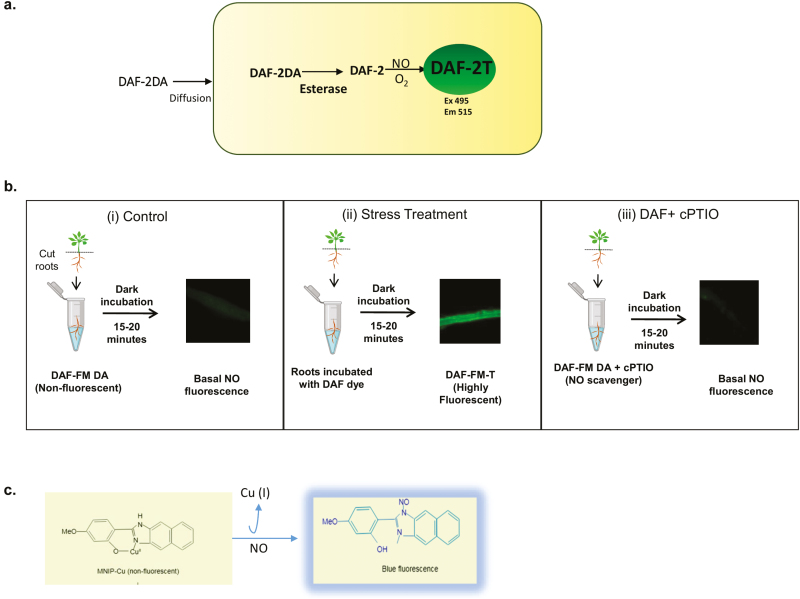
The reaction mechanism of DAF-2DA, is shown in Fig. 1(a). Once the DAF dyes enter the cell, ester bonds are hydrolysed by cellular esterases and DAF-2 is released. The resulting DAF-2 is N-nitrosated, forming the highly fluorescent triazolofluorescein (DAF-2T) which can be easily detected by fluorescence microscopy or fluorimetry. The advantage of DAF dyes is that they are highly sensitive (detection limit in the nanomolar range), non-invasive, readily commercially available, cost-effective, and easy to handle. It should be noted that DAF-2 does not directly react with NO but rather with N2O3 (Kojima et al., 1998), hence DAF fluorescence depends on auto-oxidation of NO, which in turn is influenced by the cellular redox status (Planchet and Kaiser, 2006). However, NO2−, NO3−, ONOO−, and H2O2 do not cause DAF-2DA to fluoresce (Kojima et al., 1998). This specificity of DAF dyes was questioned when it was found that DAF-2 reacts with dehydroascorbic acid (DHA) and ascorbic acid to produce compounds that fluoresce within the range of DAF-2T. To overcome this, Ye et al. (2004), devised a method of assaying NO by freezing DAF-2 on dry ice. The NO present in the assay diffuses and reacts with frozen DAF-2, but the other less mobile compounds such as DHA would not be able to react (Zhang et al., 2002). DAF-2DA was later replaced by 4-amino-5-methylamino-2',7'-difluorofluorescein diacetate (DAF-FM-DA) that possesses greater photostability and sensitivity towards NO (~3 nM in comparison with 5 nM of DAF-2DA) (Kojima et al., 1998). However, DAF-FM-DA has limited use, due to its pH sensitivity (Vitecek et al., 2008), which means that it exhibits different pH values in plant cell compartments during different stresses, such as hypoxia, pathogen attack, and osmotic stress, which can lead to artefactual ‘NO’ measurements.
Fig. 1.
In vivo NO fluorescence visualized by using DAF fluorescent dyes. (a) Reaction mechanisms of DAF-2DA. The diacetate groups of DAF dyes are removed by intracellular esterases, the diffused DAF-2 in the presence of NO (and O2) forms highly fluorescent DAF-2T. (b) Suggested use of DAF dyes for measurement of NO under stress conditions. (i) Shows DAF fluorescence in roots of control seedlings incubated in DAF-FM-DA (10 μM in 50 mM HEPES buffer, pH 7.2) for 15 min in the dark. (ii) NO production under stress conditions; after incubation, roots shows intense DAF-FM fluorescence. (iii) Similarly, roots from stress-induced plants display basal NO fluorescence when cPTIO (an NO scavenger) is used along with DAF-FM under the same incubating conditions. (c) MNIP-Cu probe reacts directly with NO and gives blue fluorescence at 420 nm.
This method requires verification using the NO scavenger, 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO). To unequivocally demonstrate that NO is being measured using the DAF dyes, the signal needs to be reduced through the application of cPTIO. This molecule reacts with NO to form the NO2· radical and cPTI (Akaike and Maeda, 1996).
To aid investigators who may be new to the use of DAF dyes, we present a suggested approach in Fig. 1(b).
This is a fluorescence-based method which can be observed using confocal or fluorescence microscopy. MNIP-Cu is a cell-permeable fluorescent dye which forms a complex by direct reaction with NO and gives blue fluorescence (Fig. 1c). Its excitation and emission are in the ranges 330–385 nm and 420–500 nm, respectively. Dye is prepared by mixing MNIP with CuSO4. This probe appears to be more specific for NO detection and it can be a better alternative to DAF dyes. MNIP-Cu has an extra advantage as it is sensitive under both anoxic and oxygen-rich conditions (Keisham et al., 2019). Yadav et al. (2013) demonstrated the auxin-induced NO detection in the hypocotyls of sunflower using MNIP-Cu. MNIP-Cu was also used for a localization study in whole seedlings, hypocotyl segments, stigma, and protoplasts of sunflower (Helianthus annuus L.) (Jain et al., 2016; Keisham et al., 2019).
Griess reaction
Plant researchers have used the Griess reagent to measure the nitrite level during low oxygen, seed germination, and plant–pathogen interactions (Doherty and Cohn, 2000; Liu et al., 2007; Vandelle and Delledonne, 2008; Vitecek et al., 2008; Wany et al., 2017; Vishwakarma et al., 2018). In addition, Griess reagent is also used in measurement of NR activity via change in nitrite concentration in a time course. In this method, NR activity is measured as nitrate to nitrite reduction coupled with NAD(P)H (Planchet et al., 2005; Wany et al., 2017).
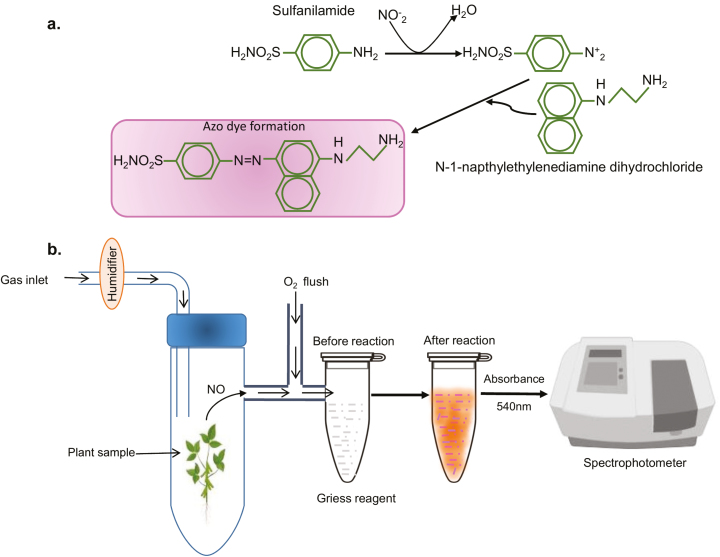
This method was developed by a German chemist, Johann Peter Griess, in 1879. In this assay, nitrite is detected by reaction with sulfanilic acid and α-naphthylamine under acidic conditions to produce azo dye which is magenta in colour, which can be measured in a spectrophotometer at 540 nm. In subsequent developments of the assay, sulfanilic acid and α-naphthylamine were replaced by sulfanilamide and N-(1-napthyl) ethylenediamine (NED), respectively. The Griess reaction is actually an indirect measurement of NO. The available NO in the sample is oxidized to NO2− (as shown below in the reaction) and followed by reaction with Griess reagent which produces a magenta colour (Fig. 2a).
Fig. 2.
Detection of gaseous nitric oxide in plants through the Griess assay. (a) Reaction mechanism which shows that NO is first oxidized to nitrite and reacts with sulfanilamide to give a diazonium salt. The diazonium salt then reacts with N -(1-naphthyl) ethylenediamine (NED) and forms a magenta coloured azo compound showing absorbance at a maximum of 540 nm. (b) Set-up for measurement of NO using the Gas Phase Griess reagent.
2NO+O2→2NO2
NO+NO2→N2O3
N2O3+H2O→2NO2−+2H+
The nitrite levels in unknown samples are calculated with a standard curve of nitrite (1–10 µM range). The advantage of this method is that it is quick, inexpensive, and can be used in any molecular biology laboratory. Further, a great advantage of this Griess reagent assay is that it can also be used for measurement of NO emitted from samples via the gas phase. Here, released NO is oxidized and reacts with sulfanilamide and NED to yield azo dye (Wany et al., 2017). This simple set-up is described in Fig. 2(b).
The exact methodology used in the Griess assay can vary between research groups. In some studies, sulfanilamide and NED (prepared in acidic medium: HCl/H3PO4) are added together to the nitrite sample (Planchet and Kaiser, 2006; García-Robledo et al., 2014; Vishwakarma et al., 2018). Alternatively, sulfanilamide is first allowed to react with nitrite, followed by NED after a 10 min incubation (Wany et al., 2017).
EPR spin trap
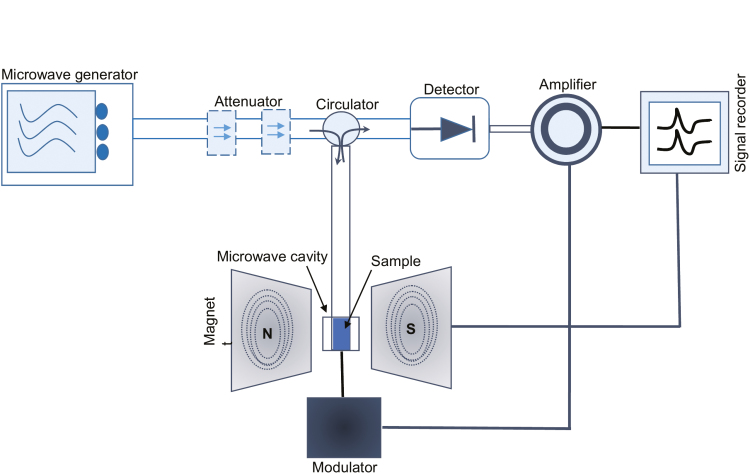
EPR spectroscopy is a reliable method to detect the short-lived NO radical. EPR is based on detection of unpaired electrons in magnetic fields within a microwave cavity which display a ‘resonance’ between parallel and an antiparallel electron spin orientations (Kleschyov et al., 2007) (Fig. 3). EPR is aided by the use of a chemical ‘spin trap’ that produces persistent paramagnetic adducts which are stable for minutes to hours to allow their detection (Steffen-Heins and Steffens, 2015). Use of a spin trap is essential for NO detection, given its very low concentration and short half-life. The principle of a spin trap is as follows:
Fig. 3.
A simple illustration of EPR spectroscopy employing magnetic field modulation. The microwave bridge typically operates at 9.5 GHz, with modulation and demodulation circuits at 100 kHz. Magnetic scanning is done very slowly (i.e. 10–100 mHz).
R*+ST→R-SA
where R* is the radical intermediate, ST is a spin trap, and R-SA is the radical spin trap adduct that is detected by EPR spectroscopy.
EPR sensitivity in detecting NO and other radicals depends on various factors such as the spin trap used, transient radical concentration, the reaction kinetics of the radicals to form adducts, and their stability (Steffen-Heins and Steffens, 2015). If used correctly, this method can detect pmoles of NO (Weaver et al., 2005).
NO spin-trapping agents can be endogenous in origin such as Hb or other metalloproteins, or exogenous compounds such as diethyldithiocarbamate (DETC) and N-methyl-d-glucaminedithiocarbamate (MGD). Hbs represent an attractive option for use as an NO spin trap agent. They can react with NO very rapidly in both oxy- and deoxy-Hb forms. Met-Hb can also be used to detect NO by this method. The interaction of NO and Hb leads to formation of paramagnetic Hb/NO derivatives which can be detected by EPR (Kosaka, 1999).
Use of iron-dithiocarbamates in EPR exploits the high affinity of NO for iron. Iron-dithiocarbamate ST [Fe(S2CN-R R′)2] side groups include methyl-, ethyl-, glucamine, sarcosine, or amino acids. This R side group variation can be useful for targeting NO (Kleschyov et al., 2007). 2-Phenyl-4,4,5,5-tetramethylimidazole-1-oxy-3-oxide (PTIO) or cPTIO (a class of imidazolineoxy-N-oxides) can be used as spin traps, and these compounds react with rate constants of the order of 104 mol−1 s−1 with concomitant generation of NO2−/NO3−. This method has some limitation as the cPTIO (NNO) produced in the reaction is degraded within a few minutes. INO, a compound, generated by a reaction between cPTIO and NO, has not been detected in Arabidopsis cell cultures (D’Alessandro et al., 2013).
Example studies which used EPR to include the measurement of NO in wounded Arabidopsis thaliana leaves using [Fe(II)(DETC)2], linked NO to jasmonic acid signalling (Huang et al., 2004), metal-induced programmed cell death (De Michele et al., 2009), innate immunity (Zeidler et al., 2004), and its effect on chloroplastic lipids and protein (Jasid et al., 2006). Bright et al. (2009) reported that all hydrated pollen produce NO, which might have a possible link to allergic responses such as rhinitis (hay fever) in humans.
The EPR approach can also be elaborated through the use of 14N and 15N labels in experiments so that both NO and its enzymatic source can be measured (Maia and Moura, 2016). EPR spectroscopy has been used to show the NO2−-dependent NO synthesis upon the Pseudomonas syringae-elicited hypersensitive response, in NR-deficient A. thaliana (Modoloa et al., 2006). In this method, the sample extract was incubated with labelled 15NO2−, 15NO3−, or l-(15N2-guanidineimino) arginine and the spin trap (MGD)2Fe(II). The NO production was quantified by measuring the levels of labelled nitrosyl complex (MGD)2Fe(II)15NO.
In considering whether to use EPR spectroscopy, its advantages and disadvantages need to be weighed up. EPR advantages include direct assay, high specificity due to use of NO spin traps, no interference with NO2− and NO3−, and compatibility for nitrogen isotope substitution. However, its disadvantages are considerable, particularly its cost. It is also semi-quantitative and requires special expertise to analyse the derived data. In addition, the method may generate and artificially detect the NO from HNO, nitrite, and S-nitrosothiols (Bellavia et al., 2015; Csonka et al., 2015).
Chemiluminescence method
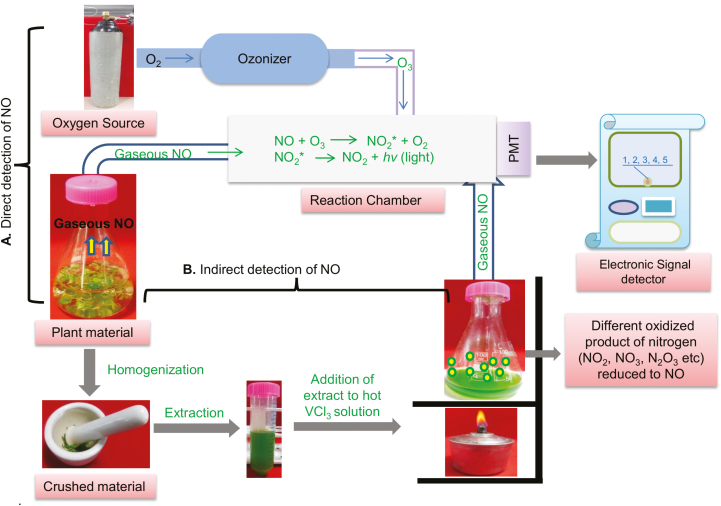
Of all of the various methods developed for detecting NO, chemiluminescence is one of the most robust and widely used. This method is based on the reaction of NO with ozone (O3), which emits a quantum of light. The light produced is directly proportional to the amount of NO produced in the biological system in a given time (Gupta et al., 2005; Planchet et al., 2005).
The constituent reactions are the reaction of NO with O3 to produce nitrogen dioxide in the excited state,
NO+O3→NO2*+O2
In the second step, NO2* returns to its ground state which leads to emission of photons (hν),
NO2*→NO2+hν
The photons emitted can be counted in a photomultiplier tube (Gupta et al., 2005; Planchet et al., 2005).
The chemiluminescent assay is exploited with often commercially available equipment with different parts (Fig. 4a). The first part is the sample chamber whose dimensions can be changed depending on sample number and size. For instance, a chamber can be designed to measure NO from a whole plant or it can be a very small (2–5 ml) beaker that can be used to assess cells and organelles such as mitochondria. Secondly, a constant source of gas flow (air or N2) with an adjusted flow rate (1 or 1.5 l min−1) is maintained inside the chamber by flow controllers. The chamber is connected to the chemiluminescence detector which is also linked to an ozone generator. The NO signal can be calibrated through the use of NO-free air.
Fig. 4.
Chemiluminescence detection of nitric oxide from plant samples. (a) Direct detection of chemiluminescence by passing released NO from plant material to the reaction chamber containing ozone. NO reacts with ozone, leading to formation of NO2* which then jumps to the ground state emitting light which is proportional to the amount of NO produced. (b) Indirect detection of chemiluminescence by injecting plant extract into hot acidic VCl3 where released NO passes into the reaction chamber containing ozone and then signals are detected.
The chemiluminescence method is highly sensitive, with an NO detection limit down to a pmole range (Planchet et al., 2005). There are many example studies where the chemiluminescence method was used to measure, from cell suspensions, root and leaf tissues, organelles of various species of plants (reviewed in Wany et al., 2016). This method has a significant advantage over other methods as it allows NO measurements from both gaseous and aqueous samples. If the biological sample is aqueous, NO can be driven to gas phase by bubbling the solution with an inert gas or shaking the sample (Gupta et al., 2005; Planchet et al., 2005).
A potential disadvantage is that chemiluminiscence only measures gaseous NO and no other forms of nitrogen or reactive nitrogen intermediates (Planchet and Kaiser, 2006). Thus, the considerable levels of NO that are oxidized in living tissues cannot be easily detected by using this method (Table 1). However, by using an indirect chemiluminiscence method, the oxidized forms of NO such as NO2− and NO3− can be reduced back to NO and therefore detected. For this, sample extracts must be injected into heated (95 °C) acidic vanadium chloride (VCl3); subsequently, it generates chemiluminescence, the NO signal, which is recorded in the signal detector sensitive enough to trap parts per billion (ppb) amounts of NO (Fig. 4b) (Rumer et al., 2009).
Table 1.
Advantages and disadvantages of NO methods
| No | Method | Advantages | Disadvantages |
|---|---|---|---|
| 1 | DAF-FM, DAF-FM-DA, and DAF-2DA |
(i) Commercially available in market (ii) Highly sensitive; measures NO levels at nanomolar concentrations (iii) Allows cellular localization studies |
(i) pH sensitive (ii) Requires oxygen (iii) DAF-2 does not directly react with NO (iv) The uptake of DAF-2DA varies with tissue type and organelle (v) Forms an N2O3 intermediate in the reaction (vi) Ascorbic and dehydroascorbic acid can react with DAF (vii) Accurate quantitative measurement is not possible |
| MNIP-Cu fluorescent probes | (i) Cheaper than DAF (ii) Reacts directly with NO (iii) MNIP-Cu probe can measure NO at anoxic, hypoxic, and normoxic conditions |
(i) Precise quantitative measurement is not possible | |
| 2 | Griess reaction | (i) Rapid and inexpensive (ii) Possible to measure NO in the gas phase |
(i) Poor sensitivity compared with other methods (ii) Quantification can be difficult (iii) Not applicable to cellular localization studies |
| 3 | EPR spin trap | (i) NO can be measured down to picomolar concentrations (ii) Used properly, EPR spectroscopy is highly specific to NO |
(i) A semi-quantitative method requiring complex methodology to acquire and analyse the results (ii) Online measurement of NO is difficult (iii) Spin traps can be expensive (iv) Equipment is expensive and requires specialist expertise to be used properly (v) Not applicable to cellular localization studies (vi). It cannot measure NO emitted into the gas phase |
| 4 | Chemiluminescence | (i) Measures NO in gas phase (ii) NO measurement in the ppb range is possible (iii) Highly reliable (iv) Can be used to measure NO from in vitro enzymatic reactions, tissues, and organelles |
(i) Difficult to measure oxidized forms of NO (ii) Equipment is expensive and requires specialist expertise to be used properly (iii) Not applicable to cellular localization studies |
| 5 | Oxyhaemoglobin assay | (i) A sensitive method, measures NO levels in the range of 1.3–2.8 nM (ii) Specialized equipment is not required |
(i) Intracellular ROS can interfere with the formation of methaemoglobin complex leading to false-positive results. (ii) Rapid pH and temperature changes in the cellular environment can affect the method’s sensitivity (iii) Several haem-containing proteins fall within the range of the same absorbance (iv) Not applicable to cellular localization studies |
| 6 | Mass spectroscopy | (i) Quantification of NO can be done efficiently by using 15N isotopes (ii) The combination MIMS/RIMS allows the simultaneous detection of NO, O2, and CO2 |
(i) Equipment is expensive and requires specialist expertise to be used properly (i) Not applicable to cellular localization studies (iii) Expensive and expertise is needed |
| 7 | Quantum cascade laser | (i) A very sensitive method, measuring NO down to ppb range (ii) Allows online, in vivo NO production to be determined |
(i) Equipment is expensive and requires specialist expertise to be used properly (ii) Requires interdisciplinary approaches to analyse the results (iii) Can only measure NO in the gas phase (iv) Localization is not possible |
Oxyhaemoglobin assay
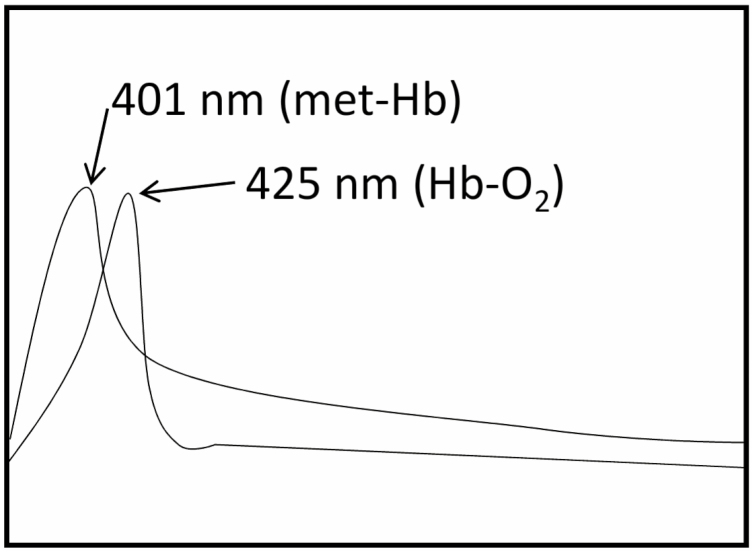
During various physiochemical reactions occurring inside the cell, NO can interact with the Hb moiety in several ways such as occupying Hb’s vacant haems in a cooperative manner such as nitrosylation of Hb thiols or by reacting with superoxide produced during biochemical reactions (Gow et al., 1999). This oxidative reaction between NO and HbO2 generates Met-Hb and nitrate, which results in a very simple and reliable method for estimating NO produced in living systems, known as the oxyhaemoglobin assay (Feelisch et al., 1996). This quantitative assay for NO estimation using a spectrophotometer was first described by Feelisch and Noack (1987). The reaction mechanism involves the shift in absorbance from 415 nm which belongs to HbO2 to 401 nm due to formation of Met-Hb (Fig. 5) (Larfars and Gyllenhammar, 1995). The available NO can be quantified by measuring the difference in absorbance at 401 nm and 421 nm (Murphy and Noack, 1994). The HbO2 assay can detect NO in the range of 1.3–2.8 nM in a short time, so it is a very fast and sensitive method (Murphy and Noack, 1994). HbO2 assay has been used widely in many plant studies related to NO signalling. Orozco-Cardenas and Ryan (2002) reported that NO negatively modulates wound signalling in tomato plants. They used excised leaves of wounded or non-wounded tomato and homogenized them in 1 ml of ice-chilled buffer containing 0.1 M sodium acetate, 1 M NaCl, and 1% (w/v) ascorbic acid, pH 6.0. Supernatant collected after centrifugation at 10 000 g for 20 min at 4 °C, passed through 0.8×4 cm columns in 1×8 resin, was used for spectrophotometric analysis by measuring the conversion of HbO2 to Met-Hb. The reaction mechanism is as follows:
Fig. 5.
A simple diagram shows the absorbance peak of Met-Hb and HbO2. The reaction of NO with Met-Hb causes a shift in the peak from 401 nm to 411 nm which indicates the oxidation of Met-Hb into HbO2.
HbO2+NO→MetHb+NO3−
A fresh calibration is required before each experiment. Calibration is performed by oxidation of HbO2 with potassium ferricyanide, with an NO donor (SNP, SNAP, and GSNO) that generates a known concentration of NO (Hetrick and Schoenfisch, 2009). Ghafourifar et al. (2005) used this method for the measurement of mitochondrial NOS activity. However, this method is not in regular use for several reasons. The first reason is that ROS produced inside the cell can also oxidize HbO2 that can give false readings. This can be prevented by addition of superoxide dismutase and catalase during measurement (Ma et al., 2016). Secondly, variation in pH, which is a common phenomenon occurring inside a plant cell, can affect the sensitivity of this assay (Mur et al., 2011). In addition, temperature change, and the presence of haem-containing proteins can also interfere with the assay as these parameters influence the absorbance spectrum of Met-Hb (Noack et al., 1992).
Mass spectrometry
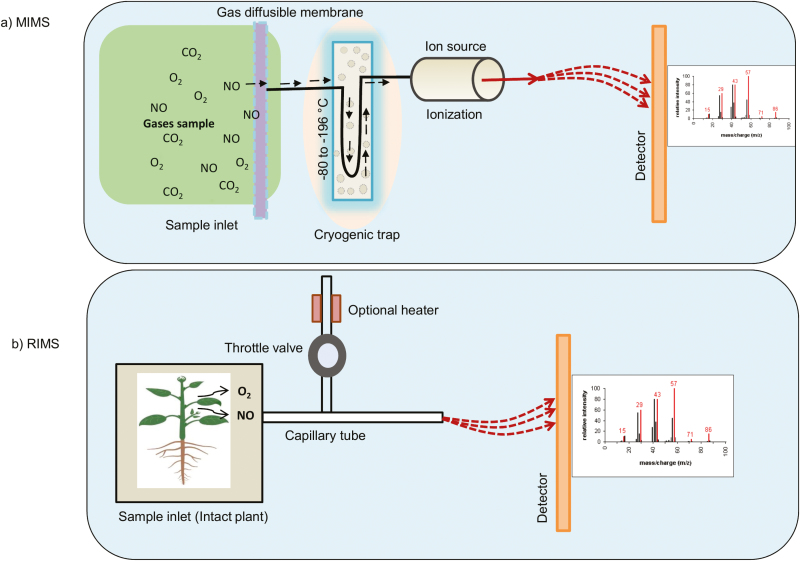
Conrath et al. (2004) demonstrated the use of membrane inlet mass spectrometry (MIMS) to detect NO by a non-invasive isotope pairing technique. MIMS can quantify NO in aqueous solution, but the RIMS (restriction capillary inlet mass spectrometry) approach can measure NO from the gas phase. The MIMS set-up comprises a semi-permeable membrane as an inlet which is connected to a mass spectrometer (Fig. 6a). Samples such as cell suspensions should be stirred and passed through a Teflon membrane (50 µm) that is permeable to NO, following which it is subsequently analysed by MS. In RIMS, NO passes directly through the samples, such as leaves, to the mass spectrometer (Fig. 6b).
Fig. 6.
Schematic view of the experimental set-up of MIMS- and RIMS-based NO measurement. (a) In MIMS, a cell suspension in an 8–10 ml reaction chamber is circulated over a thin Teflon membrane by a magnetic stirrer. The dissolved gases diffuse through the membrane and pass through the ionization chamber of the mass spectrometer. (b) In RIMS, the sample can be a leaf of a small plant or root tissue cells which fit into the leaf cuvette. The produced gases pass through the capillary tube before entering the ionization chamber of the mass spectrometer. A throttle valve connected to a capillary tube regulates the flow of gases to the ionization chamber.
The advantage of this method is that it has the ability to distinguish different isotopic forms of 15N-labelled nitrate, nitrite, arginine, and hydroxylamines. N2 isotopes can also be calculated; thus quantification can be achieved by this method. A combination of MIMS/RIMS can allow the fast, specific, and simultaneous detection of NO, O2, and CO2 from the same sample (Conrath et al., 2004). Indeed, this was demonstrated following the addition of nitrate to mouse macrophages, plant tissue cultures (tobacco, parsley, and soybean), tobacco leaves, whole Arabidopsis plants, green algae, fungi such as Pythium sp., Botrytis sp., and Fusarium sp., and some species of cyanobacteria (Conrath et al., 2004). In all these assays, oxygen concentration was maintained at <1%. However, this technique did not detect NO release when a plant or its leaves is sprayed with equimolar concentrations of phosphate or even water. The technique can be extended to detect S-nitrosothiols coupled with LC or GC (Palmieri et al., 2010).
This superficially attractive method is, however, not often used, most probably due to the expensive equipment required (Table 1).
Quantum cascade laser
Quantum cascade laser (QCL) is one of the most advanced methods for NO detection. A QCL-based detector works at ambient temperature and is highly applicable for gas sensing due to its high sensitivity (parts per billion), compact size, excellent spectral quality, and low power requirements. NO measurements using QCLs have recently been reviewed by us (Montilla-Bascon et al., 2018), so are only briefly considered here.
For NO detection, a QCL emitting light at 5.3 µm (1850 cm−1) is required. There are two different lasers currently being used, namely AllnAs/GalnAs- and GaAs/AlGaAs-based materials. A gas line from the sample being analysed is passed into a gas cell which has mirrors at both ends. This allows multiple reflections of the laser light inside the cell so that a cell that could be 30 cm long has an effective path length of 76 m. This explains the term ‘multi-pass gas cell’ in QCL detection. The NO concentration is determined from comparison with a gas mixture of known NO concentrations in nitrogen. For long-term reproducibility of sample measurement, the optical set-up is placed in a temperature controlled box.
One advantage of this QCL-based NO detector over chemiluminescence is the possibility of long-term measurements due to a low ventilation rate over the sample (usually 1–1.5 l h–1) so that the sample can be affected by dehydration. However, the disadvantages are significant as the QCL approach requires considerable specialist equipment and expertise. Significantly, most NO measurement studies using QCL have been based on collaborations between biologists and physicists. An example of this is a comparison of the sensitivity of the QCL and chemiluminescence approach in tomato plants infected with Botrytis cinerea. Data obtained from both methods showed identical NO detection levels (Mur et al., 2011). There is the potential for QCL miniaturization to make it more amenable for general laboratory use, but currently interdisciplinary collaboration remains the best means to access and exploit this power approach to NO detection.
NO electrodes
Measurement of NO by electrodes is one of the cheapest and alternative ways to measure NO. In a typical NO electrode, the Teflon/platinum serves as the main electrode and Ag/AgCl2 as the reference electrode (Besson-Bard et al., 2008a). The main and reference electrodes are enclosed in a glass or steel tube filled with a solution of 0 mM NaCl/0.3 nM HCl. The open end is covered with an NO-permeable membrane. In this method, NO can be detected based on its oxidation at +0.8 V to +0.9 V in comparison with the reference electrode (Shibuki, 1990). These can be constructed in a less expensive way (Besson-Bard et al., 2008a), cell death in tobacco BY2 cells (Ma et al., 2010), and cryptogein-induced cell death (Besson-Bard et al., 2008a). The disadvantage is that NO emitted into the gas stream cannot be measured by using this method.
Conclusions
In conclusion, in this review we have considered various means of NO mneasurement. The list is not exhaustive, but reflects the experiences of the authors. In each case, we recommend care being used to confirm that the signals obtained truly reflect the radical molecule being assessed, ideally through the use of scavengers, inhibitors, mutants, or overexpressing lines. If possible, more than one assay should be used to corroborate the results, although for practical reasons this may be not possible.
Acknowledgements
This work is funded by a Ramalingaswami Fellowship and IYBA, SERB ECR to KJG.
References
- Akaike T, Maeda H. 1996. Quantitation of nitric oxide using 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO). Methods in Enzymology 268, 211–221. [DOI] [PubMed] [Google Scholar]
- Arita NO, Cohen MF, Tokuda G, Yamasaki H. 2006. Fluorometric detection of nitric oxide with diaminofluoresceins (DAFs): applications and limitations for plant NO research. In: Lamattina L, Polacco JC, eds. Nitric oxide in plant growth, development and stress physiology. Berlin Heidelberg: Springer, 269–280. [Google Scholar]
- Begara-Morales JC, Chaki M, Sánchez-Calvo B, Mata-Pérez C, Leterrier M, Palma JM, Barroso JB, Corpas FJ. 2013. Protein tyrosine nitration in pea roots during development and senescence. Journal of Experimental Botany 64, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia L, Kim-Shapiro DB, King SB. 2015. Detecting and monitoring NO, SNO and nitrite in vivo. Future Science OA 1, FSO36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Griveau S, Bedioui F, Wendehenne D. 2008a.Real-time electrochemical detection of extracellular nitric oxide in tobacco cells exposed to cryptogein, an elicitor of defence responses. Journal of Experimental Botany 59, 3407–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. 2008b.New insights into nitric oxide signaling in plants. Annual Review of Plant Biology 59, 21–39. [DOI] [PubMed] [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. 2006. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. The Plant Journal 45, 113–122. [DOI] [PubMed] [Google Scholar]
- Bright J, Hiscock SJ, James PE, Hancock JT. 2009. Pollen generates nitric oxide and nitrite: a possible link to pollen-induced allergic responses. Plant Physiology and Biochemistry 47, 49–55. [DOI] [PubMed] [Google Scholar]
- Conrath U, Amoroso G, Köhle H, Sültemeyer DF. 2004. Non-invasive online detection of nitric oxide from plants and some other organisms by mass spectrometry. The Plant Journal 38, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, Valderrama R, Palma JM, León AM, Sandalio LM, del Río LA. 2006. Constitutive arginine-dependent nitric oxide synthase activity in different organs of pea seedlings during plant development. Planta 224, 246–254. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM, del Río LA, Barroso JB. 2009. Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytologist 184, 9–14. [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L. 2004. Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218, 900–905. [DOI] [PubMed] [Google Scholar]
- Csonka C, Páli T, Bencsik P, Görbe A, Ferdinandy P, Csont T. 2015. Measurement of NO in biological samples. British Journal of Pharmacology 172, 1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC. 2012. Coordination of a mitochondrial superoxide burst during the hypersensitive response to bacterial pathogen in Nicotiana tabacum. Plant, Cell & Environment 35, 1121–1136. [DOI] [PubMed] [Google Scholar]
- D’Alessandro S, Posocco B, Costa A, Zahariou G, Schiavo FL, Carbonera D, Zottini M. 2013. Limits in the use of cPTIO as nitric oxide scavenger and EPR probe in plant cells and seedlings. Frontiers in Plant Science 4, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michele R, Vurro E, Rigo C, Costa A, Elviri L, Di Valentin M, Careri M, Zottini M, Sanità di Toppi L, Lo Schiavo F. 2009. Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiology 150, 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty LC, Cohn MA. 2000. Seed dormancy in red rice (Oryza sativa). XI. Commercial liquid smoke elicits germination. Seed Science Research 10, 7. [Google Scholar]
- Domingos P, Prado AM, Wong A, Gehring C, Feijo JA. 2015. Nitric oxide: a multitasked signaling gas in plants. Molecular Plant 8, 506–520. [DOI] [PubMed] [Google Scholar]
- Fancy NN, Bahlmann AK, Loake GJ. 2017. Nitric oxide function in plant abiotic stress. Plant, Cell & Environment 40, 462–472. [DOI] [PubMed] [Google Scholar]
- Feelisch M, Kubitzek D, Werringloer J. 1996. The oxyhaemoglobin assay. In: Feelisch M, Stamler JS, eds. Methods in nitric oxide research. New York: Wiley, 472–473. [Google Scholar]
- Feelisch M, Noack EA. 1987. Correlation between nitric oxide formation during degradation of organic nitrates and activation of guanylate cyclase. European Journal of Pharmacology 139, 19–30. [DOI] [PubMed] [Google Scholar]
- Foissner I, Wendehenne D, Langebartels C, Durner J. 2000. In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. The Plant Journal 23, 817–824. [DOI] [PubMed] [Google Scholar]
- Foresi N, Correa-Aragunde N, Parisi G, Caló G, Salerno G, Lamattina L. 2010. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. The Plant Cell 22, 3816–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Foster MW, Benhar M, Stamler JS. 2009. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radical Biology & Medicine 46, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón C, Gómez Ros LV, Pedreño MA, Ros Barceló A. 2005. Nitric oxide production by the differentiating xylem of Zinnia elegans. New Phytologist 165, 121–130. [DOI] [PubMed] [Google Scholar]
- García-Robledo E, Corzo A, Papaspyrou S. 2014. A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Marine Chemistry 162, 30–36. [Google Scholar]
- Gaupels F, Kuruthukulangarakoola GT, Durner J. 2011. Upstream and downstream signals of nitric oxide in pathogen defence. Current Opinion in Plant Biology 14, 707–714. [DOI] [PubMed] [Google Scholar]
- Ghafourifar P, Asbury ML, Joshi SS, Kincaid ED. 2005. Determination of mitochondrial nitric oxide synthase activity. Methods in Enzymology 396, 424–444. [DOI] [PubMed] [Google Scholar]
- Godber BL, Doel JJ, Sapkota GP, Blake DR, Stevens CR, Eisenthal R, Harrison R. 2000. Reduction of nitrite to nitric oxide catalyzed by xanthine oxidoreductase. Journal of Biological Chemistry 275, 7757–7763. [DOI] [PubMed] [Google Scholar]
- Gow AJ, Luchsinger BP, Pawloski JR, Singel DJ, Stamler JS. 1999. The oxyhemoglobin reaction of nitric oxide. Proceedings of the National Academy of Sciences, USA 96, 9027–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ. 2011. Protein S-nitrosylation in plants: photorespiratory metabolism and NO signaling. Science Signaling 4, jc1. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Igamberdiev AU. 2013. Recommendations of using at least two different methods for measuring NO. Frontiers in Plant Science 4, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Kumari A, Florez-Sarasa I, Fernie AR, Igamberdiev AU. 2018. Interaction of nitric oxide with the components of the plant mitochondrial electron transport chain. Journal of Experimental Botany 69, 3413–3424. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Stoimenova M, Kaiser WM. 2005. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. Journal of Experimental Botany 56, 2601–2609. [DOI] [PubMed] [Google Scholar]
- Hetrick EM, Schoenfisch MH. 2009. Analytical chemistry of nitric oxide. Annual Review of Analytical Chemistry 2, 409–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Neill SJ, Tang Z, Cai W. 2005. Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiology 137, 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Stettmaier K, Michel C, Hutzler P, Mueller MJ, Durner J. 2004. Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218, 938–946. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. 2001. The biotin switch method for the detection of S-nitrosylated proteins. Science Signaling 2001, pl1. [DOI] [PubMed] [Google Scholar]
- Jain P, David A, Bhatla SC. 2016. A novel protocol for detection of nitric oxide in plants. Methods in Molecular Biology 1424, 69–79. [DOI] [PubMed] [Google Scholar]
- Jasid S, Simontacchi M, Bartoli CG, Puntarulo S. 2006. Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiology 142, 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kępczyński J, Cembrowska-Lech D. 2018. Application of flow cytometry with a fluorescent dye to measurement of intracellular nitric oxide in plant cells. Planta 248, 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleschyov AL, Wenzel P, Munzel T. 2007. Electron paramagnetic resonance (EPR) spin trapping of biological nitric oxide. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences 851, 12–20. [DOI] [PubMed] [Google Scholar]
- Keisham M, Jain P, Singh N, von Toerne C, Bhatla SC, Lindermayr C. 2019. Deciphering the nitric oxide, cyanide and iron-mediated actions of sodium nitroprusside in cotyledons of salt stressed sunflower seedlings. Nitric Oxide 88, 10–26. [DOI] [PubMed] [Google Scholar]
- Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. 1998. Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Analytical Chemistry 70, 2446–2453. [DOI] [PubMed] [Google Scholar]
- Kolbert Z, Feigl G, Bordé Á, Molnár Á, Erdei L. 2017. Protein tyrosine nitration in plants: present knowledge, computational prediction and future perspectives. Plant Physiology and Biochemistry 113, 56–63. [DOI] [PubMed] [Google Scholar]
- Kosaka H. 1999. Nitric oxide and hemoglobin interactions in the vasculature. Biochimica et Biophysica Acta 1411, 370–377. [DOI] [PubMed] [Google Scholar]
- Lärfars G, Gyllenhammar H. 1995. Measurement of methemoglobin formation from oxyhemoglobin. A real-time, continuous assay of nitric oxide release by human polymorphonuclear leukocytes. Journal of Immunological Methods 184, 53–62. [DOI] [PubMed] [Google Scholar]
- Liu HY, Yu X, Cui DY, Sun MH, Sun WN, Tang ZC, Kwak SS, Su WA. 2007. The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Research 17, 638–649. [DOI] [PubMed] [Google Scholar]
- Ma Z, Marsolais F, Bykova NV, Igamberdiev AU. 2016. Nitric oxide and reactive oxygen species mediate metabolic changes in barley seed embryo during germination. Frontiers in Plant Science 7, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Xu W, Xu H, Chen Y, He Z, Ma M. 2010. Nitric oxide modulates cadmium influx during cadmium-induced programmed cell death in tobacco BY-2 cells. Planta 232, 325–335. [DOI] [PubMed] [Google Scholar]
- Maia LB, Moura JJ. 2016. Detection of nitric oxide by electron paramagnetic resonance spectroscopy: spin-trapping with iron-dithiocarbamates. Methods in Molecular Biology 1424, 81–102. [DOI] [PubMed] [Google Scholar]
- Modoloa LV, Augusto O, Almeida IMG, Pinto-Maglio CAFP, Oliveira HC, Seligman K, Salgado I. 2006. Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Science 171, 34–40. [Google Scholar]
- Montilla-Bascón G, Mandon J, Harren FJM, Mur LAJ, Cristescu SM, Prats E. 2018. Quantum cascade lasers-based detection of nitric oxide. Methods in Molecular Biology 1747, 49–57. [DOI] [PubMed] [Google Scholar]
- Mur LA, Mandon J, Cristescu SM, Harren FJ, Prats E. 2011. Methods of nitric oxide detection in plants: a commentary. Plant Science 181, 509–519. [DOI] [PubMed] [Google Scholar]
- Murphy ME, Noack E. 1994. Nitric oxide assay using hemoglobin method. Methods in Enzymology 233, 240–250. [DOI] [PubMed] [Google Scholar]
- Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, Morris P, Ribeiro D, Wilson I. 2008. Nitric oxide, stomatal closure, and abiotic stress. Journal of Experimental Botany 59, 165–176. [DOI] [PubMed] [Google Scholar]
- Noack E, Kubitzek D, Kojda G. 1992. Spectrophotometric determination of nitric oxide using hemoglobin. Neuroprotocols 1, 133–139. [Google Scholar]
- Orozco-Cárdenas ML, Ryan CA. 2002. Nitric oxide negatively modulates wound signaling in tomato plants. Plant Physiology 130, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri MC, Lindermayr C, Bauwe H, Steinhauser C, Durner J. 2010. Regulation of plant glycine decarboxylase by S-nitrosylation and glutathionylation. Plant Physiology 152, 1514–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips L, Toledo AH, Lopez-Neblina F, Anaya-Prado R, Toledo-Pereyra LH. 2009. Nitric oxide mechanism of protection in ischemia and reperfusion injury. Journal of Investigative Surgery 22, 46–55. [DOI] [PubMed] [Google Scholar]
- Planchet E, Jagadis Gupta K, Sonoda M, Kaiser WM. 2005. Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. The Plant Journal 41, 732–743. [DOI] [PubMed] [Google Scholar]
- Planchet E, Kaiser WM. 2006. Nitric oxide (NO) detection by DAF fluorescence and chemiluminescence: a comparison using abiotic and biotic NO sources. Journal of Experimental Botany 57, 3043–3055. [DOI] [PubMed] [Google Scholar]
- Prats E, Carver TL, Mur LA. 2008. Pathogen-derived nitric oxide influences formation of the appressorium infection structure in the phytopathogenic fungus Blumeria graminis. Research in Microbiology 159, 476–480. [DOI] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM. 2002. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. Journal of Experimental Botany 53, 103–110. [PubMed] [Google Scholar]
- Rümer S, Gupta KJ, Kaiser WM. 2009. Plant cells oxidize hydroxylamines to NO. Journal of Experimental Botany 60, 2065–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini J, André F, Jeandroz S, Wendehenne D. 2017. Nitric oxide synthase in plants: where do we stand? Nitric Oxide 63, 30–38. [DOI] [PubMed] [Google Scholar]
- Schmidt HH, Walter U. 1994. NO at work. Cell 78, 919–925. [DOI] [PubMed] [Google Scholar]
- Shiva S. 2010. Mitochondria as metabolizers and targets of nitrite. Nitric Oxide 22, 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen-Heins A, Steffens B. 2015. EPR spectroscopy and its use in planta—a promising technique to disentangle the origin of specific ROS. Frontiers in Environmental Science 3, 15. [Google Scholar]
- Stöhr C, Stremlau S. 2006. Formation and possible roles of nitric oxide in plant roots. Journal of Experimental Botany 57, 463–470. [DOI] [PubMed] [Google Scholar]
- Shibuki K. 1990. An electrochemical microprobe for detecting nitric oxide release in brain tissue. Neuroscience Research 9, 69–76. [DOI] [PubMed] [Google Scholar]
- Tischner R, Galli M, Heimer YM, Bielefeld S, Okamoto M, Mack A, Crawford NM. 2007. Interference with the citrulline-based nitric oxide synthase assay by argininosuccinate lyase activity in Arabidopsis extracts. FEBS Journal 274, 4238–4245. [DOI] [PubMed] [Google Scholar]
- Tun NN, Santa-Catarina C, Begum T, Silveira V, Handro W, Floh EI, Scherer GF. 2006. Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant & Cell Physiology 47, 346–354. [DOI] [PubMed] [Google Scholar]
- Vandelle E, Delledonne M. 2008. Methods for nitric oxide detection during plant–pathogen interactions. Methods in Enzymology 437, 575–594. [DOI] [PubMed] [Google Scholar]
- Vishwakarma A, Kumari A, Mur LAJ, Gupta KJ. 2018. A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radical Biology & Medicine 122, 40–51. [DOI] [PubMed] [Google Scholar]
- Vitecek J, Reinohl V, Jones RL. 2008. Measuring NO production by plant tissues and suspension cultured cells. Molecular Plant 1, 270–284. [DOI] [PubMed] [Google Scholar]
- Vranová E, Inzé D, Van Breusegem F. 2002. Signal transduction during oxidative stress. Journal of Experimental Botany 53, 1227–1236. [PubMed] [Google Scholar]
- Wany A, Gupta AK, Kumari A, Gupta S, Mishra S, Jaintu R, Pathak PK, Gupta KJ. 2016. Chemiluminescence detection of nitric oxide from roots, leaves, and root mitochondria. Methods in Molecular Biology 1424, 15–29. [DOI] [PubMed] [Google Scholar]
- Wany A, Gupta AK, Kumari A, Mishra S, Singh N, Pandey S, Vanvari R, Igamberdiev AU, Fernie AR, Gupta KJ. 2019. Nitrate nutrition influences multiple factors in order to increase energy efficiency under hypoxia in Arabidopsis. Annals of Botany 123, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wany A, Kumari A, Gupta KJ. 2017. Nitric oxide is essential for the development of aerenchyma in wheat roots under hypoxic stress. Plant, Cell & Environment 40, 3002–3017. [DOI] [PubMed] [Google Scholar]
- Weaver J, Porasuphatana S, Tsai P, Budzichowski T, Rosen GM. 2005. Spin trapping nitric oxide from neuronal nitric oxide synthase: a look at several iron-dithiocarbamate complexes. Free Radical Research 39, 1027–1033. [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J. 2001. Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends in Plant Science 6, 177–183. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM. 1993. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Molecular & General Genetics 239, 289–297. [DOI] [PubMed] [Google Scholar]
- Wilson ID, Neill SJ, Hancock JT. 2008. Nitric oxide synthesis and signalling in plants. Plant, Cell & Environment 31, 622–631. [DOI] [PubMed] [Google Scholar]
- Yadav S, David A, Baluška F, Bhatla SC. 2013. Rapid auxin-induced nitric oxide accumulation and subsequent tyrosine nitration of proteins during adventitious root formation in sunflower hypocotyls. Plant Signaling & Behavior 8, e23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Kim WS, Rubakhin SS, Sweedler JV. 2004. Measurement of nitric oxide by 4,5-diaminofluorescein without interferences. The Analyst 129, 1200–1205. [DOI] [PubMed] [Google Scholar]
- Yu M, Lamattina L, Spoel SH, Loake GJ. 2014. Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytologist 202, 1142–1156. [DOI] [PubMed] [Google Scholar]
- Zeidler D, Zahringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J. 2004. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proceedings of the National Academy of Sciences, USA 101, 15811–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kim WS, Hatcher N, Potgieter K, Moroz LL, Gillette R, Sweedler JV. 2002. Interfering with nitric oxide measurements. 4,5-Diaminofluorescein reacts with dehydroascorbic acid and ascorbic acid. Journal of Biological Chemistry 277, 48472–48478. [DOI] [PubMed] [Google Scholar]