OBJECTIVES:
The composition of the small intestinal microbiota has not yet been characterized thoroughly using culture-independent techniques. We compared small intestinal microbial communities in patients with and without small intestinal bacterial overgrowth (SIBO) using culture-dependent and culture-independent bacterial identification approaches.
METHODS:
Small bowel aspirate and mucosal samples were collected from patients with suspected SIBO. The aspirates were cultured to diagnose SIBO, defined as ≥104 colony-forming units/mL coliform or ≥105 colony-forming units/mL upper aerodigestive tract bacteria. Bacteria in the aspirates and mucosa were identified using 16S rRNA gene sequencing. We compared small intestinal microbiome composition between groups with and without a culture-based SIBO diagnosis.
RESULTS:
Analysis of the aspirate and mucosal microbial communities from 36 patients revealed decreased α-diversity but no differences in β-diversity in patients with SIBO compared with those without SIBO. There were no significant differences in the relative abundance of individual taxa from the aspirates or mucosa after adjustment for false discovery rate between patients with and without SIBO. Subgroup analysis revealed significant differences in mucosal β-diversity between the coliform and upper aerodigestive tract subgroups. Relative abundances of a mucosal Clostridium spp. (P = 0.05) and an aspirate Granulicatella spp. (P = 0.02) were higher in coliform SIBO vs non-SIBO subgroups. The microbial composition and relative abundance of multiple taxa significantly differed in the mucosal and aspirate specimens.
DISCUSSION:
Culture-based results of small bowel aspirates do not correspond to aspirate microbiota composition but may be associated with species richness of the mucosal microbiota.
INTRODUCTION
Small intestinal bacterial overgrowth (SIBO) has historically been described in the context of maldigestion and malabsorption in patients with anatomic abnormalities (e.g., fistulae, strictures, bowel reconstruction, bowel obstruction), underlying dysmotility secondary to diabetes or scleroderma, and hypochlorhydria (1). This syndrome has also been implicated in the pathophysiology of numerous gastrointestinal and hepatobiliary diseases including common conditions such as irritable bowel syndrome (IBS) (2) and functional dyspepsia (3). Typical SIBO-associated symptoms include bloating, diarrhea, abdominal pain, and gas. SIBO symptoms may overlap with those of functional gastrointestinal disorders. However, the role of SIBO in the pathogenesis of functional gastrointestinal disorders remains controversial given that its definition relies on tests, including breath test and culture of small bowel aspirates, which have not been appropriately validated (4). Indirect techniques such as breath testing have important limitations related to heterogeneous testing methods and lack of standardization in interpretation. To address these limitations, guidelines for breath testing indications, performance, and interpretation were recently published in a North American consensus document (5). Yet, there remain important knowledge gaps in how these tests reflect the microbial landscape of the human small intestine.
Small bowel aspirates and culture were previously considered the gold standard for the diagnosis of SIBO. SIBO has traditionally been defined by abnormally high amounts of bacteria in the small intestine and more precisely by >105 colony-forming units (CFU)/mL of colonic-type bacteria from jejunal aspirates (6). However, the validity of this approach has been questioned as the proposed cutoff of 105 seemed more predictive of postsurgical patients in a previous systematic review (7). Others have suggested that lower cutoff values of 103 CFU/mL be considered as diagnostic of SIBO (5), especially if coliforms are present (8). Standard culture-based techniques are limited in their ability to characterize fully the complex diversity of the small intestinal microbiome, particularly as a large proportion of bacterial species remain uncultured (9). In light of increasing evidence supporting the hypothesis that the intestinal microbiome serves a key role in host physiology and gastrointestinal disease, better characterization of the small intestinal microbiome will be a critical step in developing novel diagnostic and therapeutic strategies in a number of human diseases and disorders.
In the recent years, the introduction of culture-independent techniques has dramatically expanded our ability to characterize the complexity of the intestinal microbiota. It also has increased our knowledge of the culture conditions needed to detect and identify previously difficult to culture organisms (10). Integration of rapid molecular approaches (11) for the assessment of SIBO in conditions such as IBS has yielded promising data including evidence for decreased microbial diversity from duodenal aspirates in patients with IBS compared with control subjects (12). However, these findings are preliminary. Overall, there are limited data reporting detailed characterizations of the human small intestinal microbiome. It may be hypothesized that alterations in the small intestinal microbiome are associated with culture-proven SIBO, which in turn could suggest that although imperfect, culture-proven SIBO is at least a useful surrogate marker of small intestinal dysbiosis. However, further investigation is needed to (i) explore the role of microbial diversity in SIBO using culture-independent approaches and (ii) describe the interactions between the host and small intestinal microbiome in the context of gastrointestinal disease. This study characterizes small intestinal microbial composition of small bowel aspirates and mucosal specimens in patients with and without a diagnosis of SIBO defined by qualitative and quantitative culture of proximal small bowel aspirates.
METHODS
Study design and participants
We conducted a cross-sectional study comparing microbial profiles of small bowel specimens in patients with and without a culture-based diagnosis of SIBO presenting to the Motility and Neurogastroenterology Clinic at Indiana University from March 2013 to November 2015. The study protocol was approved by the Indiana University School of Medicine Institutional Review Board. Participants were prospectively enrolled after signing informed consent at the time of small bowel enteroscopy. All patients with suspected SIBO based on clinical symptoms and undergoing small bowel enteroscopy for diagnostic evaluation were eligible for inclusion regardless of clinical history. We excluded patients using antibiotics, probiotics, or colon cleansing preparations in the last 30 days before enteroscopy and those who were pregnant or could not provide informed consent.
Variables
Demographic data (age, gender, race/ethnicity) and anthropometric assessments (body mass index [BMI], kg/m2) were collected at the time of enrollment. Clinical history was reviewed to assess for the presence of abdominal distension, diarrhea, and the risk factors for SIBO including conditions (e.g., proton pump inhibitor use) and surgeries associated with decreased acid production (e.g., Billroth I or II, vagotomy, gastric bypass for obesity) (13). Clinical symptoms were further assessed using the validated Patient Assessment of Gastrointestinal Disorder-Symptoms Severity Index questionnaire (14). All demographic and clinical data were collected on enrollment and before the assessment of SIBO status to minimize potential for information bias. Outcomes of interest were (i) quantitative and qualitative cultures of jejunal aspirates and (ii) microbial composition of jejunal aspirates and mucosal biopsy specimens.
Study procedures
Study participants underwent upper enteroscopy using a pediatric colonoscope (11.3 mm diameter) or a small caliber upper enteroscope (9.2 mm diameter) for the collection of luminal aspirate and mucosal biopsy specimens within the proximal jejunum using a standardized protocol as previously detailed (13). Procedure details are outlined in the supplementary material (see Table, Supplemental Digital Content, http://links.lww.com/CTG/A89), which outlines the procedures for enteroscopy. The enteroscope was advanced past the ligament of Treitz to the maximum extent of reach into the jejunum, and an aspiration catheter was introduced through the working channel to collect at least 2 mL of luminal fluid. After the collection of luminal aspirates, 2 mucosal biopsies were obtained from the proximal jejunum on withdrawal of the endoscope. Mucosal biopsy samples were stored at −80 °C for subsequent 16S rRNA analysis. Jejunal aspirate samples were split at the time of collection to be sent for quantitative and qualitative bacterial cultures and to be stored at −80 °C for subsequent polymerase chain reaction analysis.
Quantitative culture of jejunal aspirates
Small bowel aspirates were cultured for both aerobic and anaerobic bacteria using standard techniques. Aspirate specimens were plated on blood agar, MacConkey agar, chocolate agar, and colistin and nalidixic acid agar plates, and incubated for a minimum of 48 hours. Bacterial isolates were identified by species and quantified as CFU/mL. Given the lack of a universally accepted cutoff for defining SIBO at the time this study was conducted, we used the following thresholds to define coliform and upper aerodigestive tract (UAT) SIBO, respectively: (i) presence of ≥104 CFU/mL Gram-negative aerobic and anaerobic colonic-type bacteria (e.g., Escherichia coli spp, Klebsiella spp, Proteus mirabilis, Acinetobacter spp, Enterobacter spp, Citrobacter spp, Bacteroides spp, or Clostridium spp) and (ii) presence of ≥105 CFU/mL Gram-positive aerobes or facultative anaerobes or other bacteria characteristic of the proximal gut and oropharynx (e.g., Streptococcus spp, Staphylococcus spp, Enterococcus spp, Lactobacillus spp, Fusobacterium spp, or Peptostreptococcus spp) (4,15,16). Participants were classified as having SIBO if the culture results revealed evidence of either UAT or coliform SIBO.
Molecular analysis of jejunal aspirates and mucosal biopsy specimens
Luminal aspirate samples and mucosal biopsy samples were thawed on ice. Total nucleic acids were extracted using the DNeasy tissue kit (Qiagen, Venlo, The Netherlands), and the eluted DNA was stored at −20 °C. Gel electrophoresis and fluorescent assay (Qubit assay kit; Life Technologies, Carlsbad, CA) were used to determine the quality and quantity of isolated genomic DNA (gDNA). gDNA specimens were stored at 4 °C for immediate use in 16S rRNA sequencing. Polymerase chain reaction amplification of V1–V3 regions of 16S rRNA alleles was performed using degenerate 16S rRNA primers with a barcoding strategy previously validated by our laboratory. Reagent controls were processed in parallel to monitor for contamination. Normalized amplicon libraries were pooled and sequenced using the Illumina MiSeq sequencer (Illumina, San Diego, CA) using paired 300 bp reads to generate 4,765,249 total reads. 16S sequences were processed and fed into the Mothur package (v1.37.4) to generate lists of microbial taxa and relative loads. Subsampling at a depth of 1,563 total reads was performed for each sample to correct for biases caused by differential sequencing depth. Consensus sequences or contigs were identified by overlapping paired end reads. The standard operating procedure of Mothur for MiSeq data processing was used to trim primers and barcodes from each read and to remove chimeric and low-quality sequences. Operational taxonomic units (OTUs) were identified with a sequence similarity cutoff at 0.97, and the representative OTUs were taxonomy classified using the RDP classifier (v2.11) (17) at the genus level and Bayesian Lowest Common Ancestor method (18) at the species level. The R software environment (https://www.r-project.org/) was used to calculate standard ecological α-diversity (Observed taxa, Chao1 metrics; S. ACE; Shannon; Simpson; Pielou evenness) with vegan package. Principal coordinate analysis (ape package in R) (19) and nonmetric dimensional scaling method were applied to cluster samples based on Bray-Curtis dissimilarities (ecodist package in R) (20) in anticipation of comparative analyses of β-diversity.
Study endpoints
The primary study endpoint was overall microbial diversity. Secondary endpoints were relative abundance and prevalence of specific bacterial taxa.
Study size
Sample size was determined by the number of available cases during the study period. No formal a priori calculation for sample size was performed for this pilot investigation.
Statistical analysis
Pairwise comparisons of α-diversity indexes between cohorts were performed using the Wilcoxon rank sum test. Principal coordinate analysis and nonmetric dimensional scaling were used to visualize and compare Bray-Curtis dissimilarity, and comparisons between groups were performed using the PERMANOVA approach adjusting for BMI. We also uncovered associations between clinical cohorts and specific bacterial taxa by comparing relative taxa abundance between groups using a linear model adjusting for BMI. A negative binomial regression with mixed model for repeat measurements was applied to analyze the differences in relative abundance of specific bacterial taxa between the groups. Differences in community composition across sampling sites (aspirate vs mucosa) were examined by Bray-Curtis dissimilarities using the PERMANOVA approach. To explore potential heterogeneity related to cultured bacterial types, we also conducted predefined subgroup analyses comparing patients with a diagnosis of coliform SIBO, UAT SIBO, and no SIBO.
Participants with missing data were excluded from the analyses; all statistical tests were performed using the R software environment (http://www.r-project.org). Analyses were confined to taxa with relative abundance of >0.1% in any one of the cohort groups. Two-sided P-values < 0.05 were regarded as significant. Reported P values were corrected for multiple testing using the Benjamini-Hochberg procedure. All sequences and associated metadata were deposited to the NCBI Sequence Read Archive under the BioProject ID PRJNA472002.
RESULTS
Patient characteristics
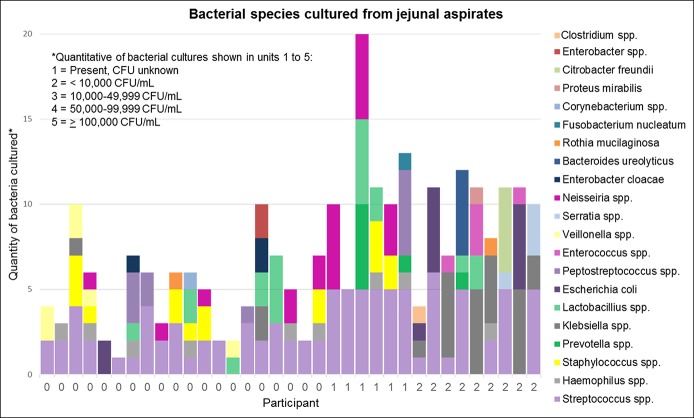
Among the 86 patients who signed informed consent, 76 completed upper enteroscopy with luminal aspiration. 16S sequencing data from either luminal aspirates or biopsy specimens were available in 36 of the 76 patients, of whom 15 had a diagnosis of SIBO based on the culture of jejunal aspirates. 16S sequencing data were not available in 40 patients due to factors related to specimen processing and storage and laboratory handling. Bacterial species cultured from jejunal aspirates of all participants are shown in Figure 1. For the overall cohort, mean BMI (kg/m2) ± SD was 26.5 ± 7.1 and mean age (years) ± SD was 50.1 ± 12.7. There were 33 female patients, and 91.7% were white (Table 1). There were no significant differences in age, race, gender, or BMI in between patients with and without SIBO. Although conditions (e.g., proton pump inhibitor use) and surgeries associated with decreased gastric acid production were more common in patients with SIBO than in patients without SIBO, these differences were not statistically significant. No patients had evidence of mucosal atrophy based on routine histologic evaluation. Comparison of clinical symptoms between patients with and without SIBO showed no significant differences in the proportion of patients reporting diarrhea or daily abdominal distention (Table 1) or in Patient Assessment of Gastrointestinal Disorder-Symptoms Severity Index scores (14). Aspirate and biopsy specimens for 16S sequencing were obtained in 32 and 34 patients, respectively. In some cases, sequencing data were available from only the aspirate or the biopsy specimen but not both. We were unable to amplify bacterial 16S sequences from 2 of the 32 aspirate specimens and 1 of the 34 biopsy specimens.
Figure 1.
Results of quantitative bacterial cultures from jejunal aspirates from individual participants. Quantitative bacterial cultures are shown in units of 1–5 (1 = present, 2 = <10,000 CFU/mL, 3 = 10,000–49,999 CFU/mL, 4 = 50,000–99,999 CFU/mL, 5 = ≥100,000 CFU/mL). Participants along the X‐axis are labeled by final SIBO diagnosis as defined by culture results (0 = no SIBO, 1 = UAT SIBO, 2 = coliform SIBO). CFU, colony-forming unit; SIBO, small intestinal bacterial overgrowth; UAT, upper aerodigestive tract.
Table 1.
Demographics and baseline characteristics among patients with and without SIBO by the culture of jejunal aspirates
Associations between overall microbial composition and SIBO status by culture
Analysis of small intestinal microbial diversity revealed significantly lower mucosal α-diversity (Chao1, ACE) at the genus level (Figure 2) in patients with SIBO compared with patients without SIBO. There was no significant difference in luminal aspirate α-diversity (Chao1, ACE, Shannon, Simpson, Pielou) between patients with and without SIBO. After adjusting for BMI, no significant differences were observed in the aspirate or mucosal microbiota β-diversity (Bray-Curtis dissimilarity) between patients with and without SIBO.
Figure 2.
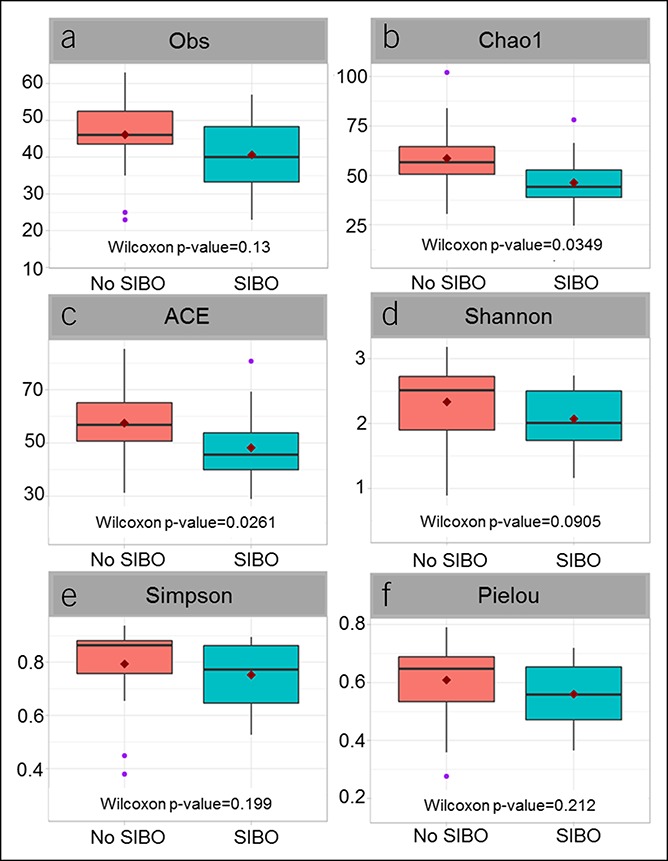
Small intestinal mucosal microbiota demonstrates lower α-diversity in patients with SIBO vs patients without SIBO. SIBO, small intestinal bacterial overgrowth. Alpha diversity determined using Obs (a), Chao1 (b), ACE (c), Shannon (d), Simpson (e), and Pielou (f) indices.
Single taxon-based analyses
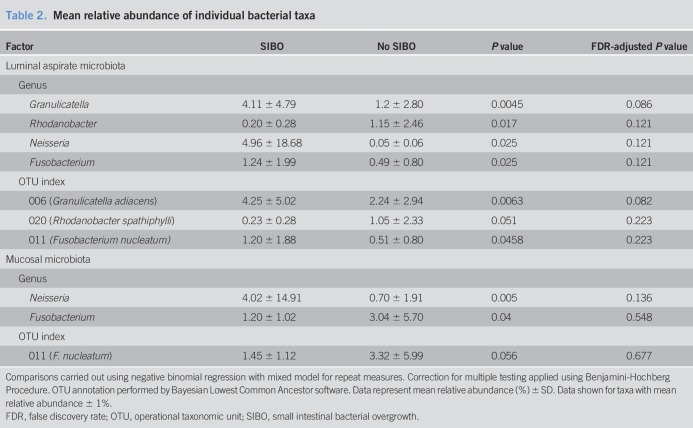
Relative abundance of multiple individual taxa (Table 2) from the aspirates and mucosa was significantly associated with SIBO diagnosis before but not after adjustment for false discovery rate (FDR). Prevalence of individual bacterial taxa in the aspirates and mucosa was not significantly associated with SIBO diagnosis. However, there was a trend toward an increased prevalence of an unassigned OTU (OTU 065) in those with SIBO compared with those without SIBO (P = 0.047).
Table 2.
Mean relative abundance of individual bacterial taxa
Comparison of community composition across sampling sites
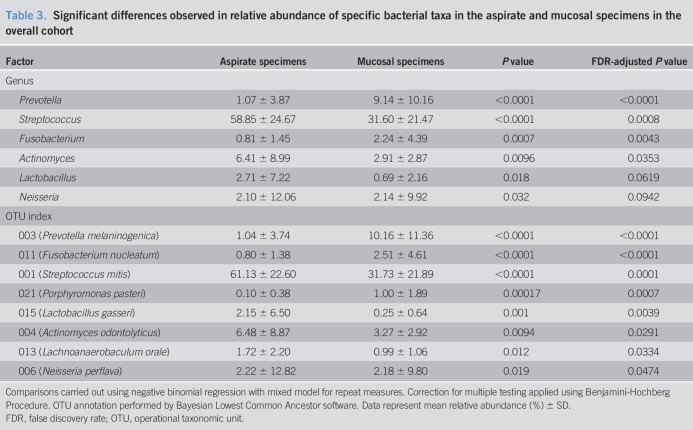
Significant differences in β-diversity (Bray-Curtis dissimilarity) were observed between the aspirate and mucosal microbiota (P = 0.001) at the genus level. These differences remained significant after including subject and BMI as confounding factors. Significant differences were also observed in the relative abundance of multiple individual taxa (Table 3) between the aspirates and mucosa at both genus and OTU levels.
Table 3.
Significant differences observed in relative abundance of specific bacterial taxa in the aspirate and mucosal specimens in the overall cohort
Subgroup analysis of SIBO cohorts by culture type
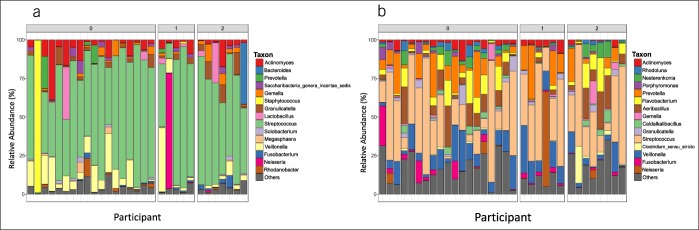
Significant differences in mucosal microbiota β-diversity were observed between patients with coliform and UAT SIBO (Figure 3). There were no differences in mucosal microbiota α-diversity among subgroups (no SIBO, UAT SIBO, and coliform SIBO). Comparisons of taxonomic-level datasets (Table 4) showed an increase in the relative abundance of a Clostridium spp. in mucosal specimens of patients with coliform SIBO vs patients without SIBO (P = 0.05) after correcting for FDR. Granulicatella spp. was also enriched in the aspirates of patients with coliform SIBO vs patients without SIBO (P = 0.02) after adjusting for BMI and correcting for FDR. OTU level analyses showed a higher abundance of a Clostridium perfringens-assigned OTU in the mucosa of patients with coliform SIBO compared with patients without SIBO before (P = 0.001) and after (P = 0.035) adjusting for BMI and correcting for FDR. A Granulicatella-assigned OTU was enriched in the aspirates of patients with coliform SIBO vs patients without SIBO after adjusting for BMI and correcting for FDR (P = 0.01). Prevalence of individual bacterial taxa from the mucosal specimens was not significantly associated with culture diagnosis. Prevalence of Clostridium sensu stricto in the luminal aspirates of patients with coliform SIBO (24%) was higher (P = 0.04) than in patients without SIBO (0.01%) or UAT SIBO (0%). There were no significant differences in the relative abundance of individual taxa in the aspirates or mucosa between patients with UAT SIBO and those with coliform SIBO nor between patients with UAT SIBO and patients without SIBO at either taxonomic level. Relative abundances of individual bacterial taxa in the aspirates and mucosa from each individual are shown in Figure 4.
Figure 3.
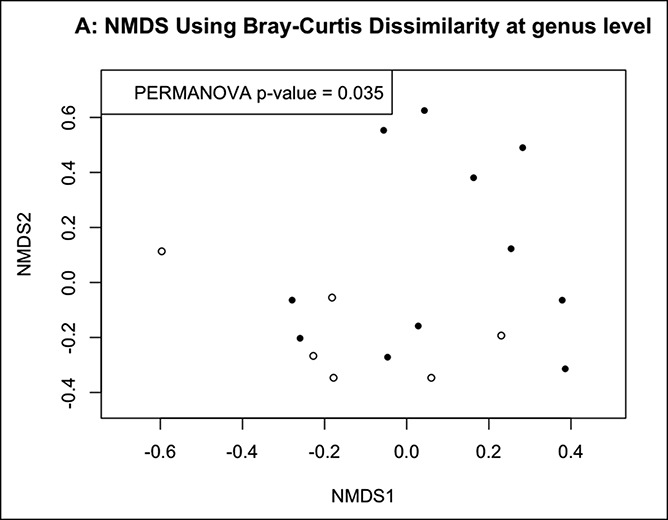
Significant differences in mucosal microbiota β-diversity between patients with coliform SIBO (black circles) and those with UAT SIBO (white circles) at both (a) genus level and (b) OTU levels. SIBO, small intestinal bacterial overgrowth; UAT, upper aerodigestive tract.
Table 4.
Significant differences observed in relative abundance of specific bacterial taxa in the aspirate and mucosal specimens in coliform SIBO vs no SIBO subgroups
Figure 4.
Relative abundances of individual bacterial taxa at the genus level in (a) aspirates and (b) mucosa from each participant. Participants along the X-axis are labeled by final SIBO diagnosis as defined by culture results (0 = no SIBO, 1 = upper aerodigestive tract SIBO, 2 = coliform SIBO). SIBO, small intestinal bacterial overgrowth.
DISCUSSION
In this pilot investigation, we compared luminal aspirate and mucosal microbial communities from patients with and without a culture-based diagnosis of SIBO of the proximal gastrointestinal tract. To date, there are only a handful of studies that have examined the small intestinal microbiome. Relatively little is known regarding its role in symptom generation, although recent data have suggested that the small intestinal microbiome, but not SIBO based on quantitative culture of small bowel aspirates, is associated with gastrointestinal symptoms (21). In our study, we made several interesting observations. First, comparison of microbial communities revealed lower α-diversity (Chao1, ACE index) at the genus level in the mucosal microbiome of patients with SIBO compared with patients without SIBO, but no significant differences in aspirate microbiota α- or β-diversity or in the mucosal microbiota β-diversity were noticed. Findings suggest that whereas mucosal microbial richness (i.e., α-diversity) is decreased in culture-diagnosed SIBO, there are no clear differences at the community level (i.e., β-diversity) or in the luminal aspirate microbiota composition. Our findings are consistent with a recently published report that found a lack of correlation between aspirate microbial composition and duodenal aspirate cultures in SIBO (21). These findings may imply that although small bowel cultures may not reflect luminal aspirate microbial composition, the overgrowth of luminal microbes may be associated with changes in the mucosal microbiota.
Second, analysis by subgroup went on to reveal significant differences in mucosal microbiota β-diversity between patients with coliform SIBO and those with UAT SIBO, suggesting that there may be differences in the community composition of the mucosal microbiota between these 2 SIBO subgroups. These observations may further suggest that the overall SIBO cohort encompasses 2 heterogeneous groups (coliform and UAT SIBO). By combining these groups using broad culture-based cutoffs, we may be diluting our ability to appreciate important differences in the community composition of the luminal and mucosal microbiota between patients with and without bacterial overgrowth.
Single taxon-based testing suggested that the relative abundance of multiple taxa in the aspirate and mucosa of patients with SIBO differed from those of patients without SIBO before but not after adjustment for FDR. Overall results support the longstanding concern that cultivating bacteria in the laboratory does not clearly represent pathologic shifts in the members of the microbial community and is largely insufficient for characterizing the complex microbial composition of the gut.
Subgroup analyses further demonstrated enrichment of Clostridium spp. and a C. perfringens-assigned OTU in the mucosa of patients with coliform SIBO compared with those without SIBO as well as enrichment of Granulicatella spp. and a Granulicatella-assigned OTU in the luminal aspirates of patients with coliform SIBO compared with those without SIBO. Enterotoxinogenic C. perfringens is an important cause of food-borne illnesses and other gastrointestinal diseases such as antibiotic-associated diarrhea (22,23). Higher levels of fecal C. perfringens have been described in elderly individuals residing in long-term care. The presence of C. perfringens in these individuals was associated with significant changes in microbiota composition including decreased levels of Bifidobacterium spp., suggesting that C. perfringens recovery in the stool may reflect a less healthy microbiota (24). Granulicatella spp. are Gram-positive facultative anaerobes that are considered a normal part of the oral bacteriome but have been shown to be increased in patients with cirrhosis with hepatic encephalopathy (25) compared with patients with cirrhosis without encephalopathy. Granulicatella spp. have also been shown to coaggregate with other bacterial species implicated in dental plaque biofilm formations, such as Aggregatibacter actinomycetemcomitans and Fusobacterium nucleatum (26). Although preliminary, our findings may suggest increased relative abundance of mucosal Clostridium spp. and/or luminal Granulicatella spp. may serve as a microbial marker for disturbances in the small intestinal microbiota in patients with coliform SIBO. Findings could also indicate that patients with coliform SIBO may have increased susceptibility to infection by pathogenic microorganisms or abnormal commensal expansion. Lack of significant differences with single-taxon analyses between patients with UAT and coliform SIBO or between patients with UAT and no SIBO may suggest considerable overlap between these particular culture-based diagnostic subgroups.
Last, comparison of small intestinal microbiota composition from the luminal aspirates and mucosal biopsies showed significant differences in β-diversity (Bray-Curtis dissimilarity) and relative abundance of multiple bacterial taxa. Higher abundances of members belonging to Prevotella and Fusobacterium were observed in the mucosal microbiota compared with the luminal microbiota, whereas higher abundances of the genera Streptococcus and Actinomyces and the Lactobacillus gasseri-assigned OTU were observed in the luminal aspirate microbiota. These differences emphasize the distinct stratification of microbial communities within the lumen and mucosa, which have previously been described by others (27,28). It should be noted that there remains a paucity of data describing the changes in microbial composition from the lumen to the mucosa within the human small intestine, which may be directly influenced by aspects of perfusion and oxygen diffusion that are unique to this site (29). The relative importance of the luminal microbiota compared with the mucosa-associated microbiota in disease pathogenesis and the interaction between these 2 communities will require further study.
This study has several strengths including prospective enrollment of study participants with careful assessment of several factors associated with the gut microbiota (age, gender, BMI), use of a standardized technique for the collection of study specimens by 2 experienced endoscopists, and concomitant application of both traditional culture-based methods and culture-independent molecular approaches for the characterization of the small intestinal bacteria. The proportion of patients meeting diagnostic criteria for SIBO in this study was high, which was not unexpected given that our site serves as a tertiary referral center and participants were recruited from among those in whom a diagnosis of SIBO was suspected based on the clinical history and symptoms. Study limitations include the lack of nonmicrobial-defined patient groups and the inability to study SIBO of the mid- and distal gastrointestinal tract. It is also possible that culture techniques were limited in their ability to facilitate the growth of anaerobic organisms due to potential exposure of specimens to air and may, in some cases, have been inadequate for the assessment of SIBO. Because the participants were recruited among patients receiving clinical care at the gastrointestinal motility clinic, study physicians could not be blinded to the clinical history or results of previous testing, as this information was pertinent to patient care. However, patient groups were defined based on objective laboratory testing rather than a clinical diagnosis, and investigators sought to enroll patients both with and without a culture-based diagnosis of SIBO. Furthermore, outcome assessments (i.e., laboratory-based culture results and small intestinal microbiota based on culture-independent techniques) were performed by study team members and by members of the microbiology laboratory who were not directly involved with patient care. Therefore, the impact of a potential selection bias on study recruitment and outcomes due to lack of blinding is overall felt to be minimal. Interpretation of findings are also limited by small sample size and limited generalizability given that our institution serves a tertiary referral center. We did not assess for methanogenic archaea, which may be pathogenic contributors to SIBO (30). However, post hoc analysis did not reveal any significant differences in the relative abundance of methanogenic organisms such as Methanobrevibacter spp. between those with and without SIBO. This is not unexpected as comparison groups were defined based on standard culture results and not on the results of breath testing for methane.
In conclusion, our study suggests that in patients with evidence of proximal SIBO based on culture of small bowel aspirates, luminal overgrowth is not correlated with luminal microbiota composition. Instead, patients with a culture-based diagnosis of proximal SIBO exhibit decreased mucosal microbial richness (e.g., α-diversity). Additionally, mucosal community diversity (e.g., β-diversity) differs between patients with coliform SIBO vs those with UAT SIBO by culture, suggesting that they are distinct microbiologic groups that should be examined separately to better define the clinical utility of culture-based testing. Further prospective investigations that examine the correlation between symptom profiles and the small intestinal aspirate and mucosal microbiota, incorporate deep sequencing techniques, and consider functional and metabolomics profiling will be required to better characterize the relevant microbial communities of the small intestine and to identify future microbial targets for improved diagnosis and management of SIBO.
CONFLICTS OF INTEREST
Guarantor of the article: Andrea Shin, MD, MSc.
Specific author contributions: Andrea Shin, MD, MSc, and Xiang Gao, PhD, MS share co-first authorship. Andrea S. Shin, MD, MSc and Qunfeng Dong, PhD share co-corresponding authorship. A.S.S.: co-investigator, preparation of biologic specimens for molecular analysis, and writing of the manuscript. X.G.: senior biostatistician, data analysis, and writing of the manuscript. M.B.: co-investigator, recruitment, collection of biologic specimens, and writing of the manuscript. A.G.: study coordinator, recruitment, and data collection. D.N.: co-investigator, molecular analysis of biologic specimens, and writing of the manuscript. H.L.: biostatistician, data analysis, and writing of the manuscript. Q.D.: bioinformatician, data analysis, and writing of the manuscript. E.T.: co-investigator and preparation of biologic specimens for molecular analysis. S.T.: co-investigator, fellow, data collection, and editing of the manuscript. R.S.: co-investigator, recruitment, and editing of the manuscript. J.W.: principal investigator, study conceptualization, writing of the protocol, recruitment, and writing of the manuscript.
Financial support: None.
Potential competing interests: None.
Study Highlights.
WHAT IS KNOWN
✓ SIBO remains a poorly defined clinical syndrome, and the validity of currently available techniques to evaluate for abnormal bacterial expansion in the small intestine has been questioned.
✓ The complexity and composition of the small intestinal microbiome are incompletely understood.
WHAT IS NEW HERE
✓ Patients with a culture-based diagnosis of SIBO show no differences in aspirate microbiota composition but exhibit decreased mucosal alpha-diversity, suggesting that decreased mucosal microbial richness is associated with luminal bacterial overgrowth.
✓ Comparisons of patients with a culture-based diagnosis of coliform SIBO and UAT SIBO suggest that the types of bacterial species cultured are associated with distinct changes in the community composition of the mucosal microbiome.
✓ Differences in the relative abundance of individual taxa between patients with coliform and those with UAT SIBO suggest that there may be abnormal expansion of specific bacteria SIBO which are not directly identified by culture-based testing.
TRANSLATIONAL IMPACT
✓ Small intestinal mucosal microbiota and aspirate microbiota are distinct in both community profile and relative abundance of individual taxa, and studying the role of both will be important in understanding the relative contribution of these 2 communities to clinical symptoms.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A89
REFERENCES
- 1.King CE, Toskes PP. Small intestine bacterial overgrowth. Gastroenterology 1979;76:1035–55. [PubMed] [Google Scholar]
- 2.Ford AC, Spiegel BM, Talley NJ, et al. Small intestinal bacterial overgrowth in irritable bowel syndrome: Systematic review and meta-analysis. Clin Gastroenterol Hepatol 2009;7:1279–86. [DOI] [PubMed] [Google Scholar]
- 3.Tziatzios G, Giamarellos-Bourboulis EJ, Papanikolaou IS, et al. Is small intestinal bacterial overgrowth involved in the pathogenesis of functional dyspepsia? Med Hypotheses 2017;106:26–32. [DOI] [PubMed] [Google Scholar]
- 4.Aziz I, Tornblom H, Simren M. Small intestinal bacterial overgrowth as a cause for irritable bowel syndrome: Guilty or not guilty? Curr Opin Gastroenterol 2017;33:196–202. [DOI] [PubMed] [Google Scholar]
- 5.Rezaie A, Buresi M, Lembo A, et al. Hydrogen and methane-based breath testing in gastrointestinal disorders: The North American consensus. Am J Gastroenterol 2017;112:775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simrén M, Stotzer PO. Use and abuse of hydrogen breath tests. Gut 2006;55:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoshini R, Dai SC, Lezcano S, et al. A systematic review of diagnostic tests for small intestinal bacterial overgrowth. Dig Dis Sci 2008;53:1443–54. [DOI] [PubMed] [Google Scholar]
- 8.Ghoshal UC, Ghoshal U. Small intestinal bacterial overgrowth and other intestinal disorders. Gastroenterol Clin North Am 2017;46:103–20. [DOI] [PubMed] [Google Scholar]
- 9.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaughan EE, Schut F, Heilig HG, et al. A molecular view of the intestinal ecosystem. Curr Issues Intest Microbiol 2000;1:1–12. [PubMed] [Google Scholar]
- 11.Kerckhoffs AP, Samsom M, van der Rest ME, et al. Lower bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol 2009;15:2887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giamarellos-Bourboulis E, Tang J, Pyleris E, et al. Molecular assessment of differences in the duodenal microbiome in subjects with irritable bowel syndrome. Scand J Gastroenterol 2015;50:1076–87. [DOI] [PubMed] [Google Scholar]
- 13.Bohm M, Shin A, Teagarden S, et al. Risk factors associated with upper aerodigestive tract or coliform bacterial overgrowth of the small intestine in symptomatic patients. J Clin Gastroenterol 2018. [Epub ahead of print December 19, 2018.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rentz AM, Kahrilas P, Stanghellini V, et al. Development and psychometric evaluation of the patient assessment of upper gastrointestinal symptom severity index (PAGI-SYM) in patients with upper gastrointestinal disorders. Qual Life Res 2004;13:1737–49. [DOI] [PubMed] [Google Scholar]
- 15.Riordan SM, McIver CJ, Wakefield D, et al. Small intestinal mucosal immunity and morphometry in luminal overgrowth of indigenous gut flora. Am J Gastroenterol 2001;96:494–500. [DOI] [PubMed] [Google Scholar]
- 16.Quigley EM, Quera R. Small intestinal bacterial overgrowth: Roles of antibiotics, prebiotics, and probiotics. Gastroenterology 2006;130:S78–90. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Lin H, Revanna K, et al. A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinformatics 2017;18:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004;20:289–90. [DOI] [PubMed] [Google Scholar]
- 20.Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 2007;22:1–19. [Google Scholar]
- 21.Saffouri GB, Shields-Cutler RR, Chen J, et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun 2019;10:2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindström M, Heikinheimo A, Lahti P, et al. Novel insights into the epidemiology of Clostridium perfringens type A food poisoning. Food Microbiol 2011;28:192–8. [DOI] [PubMed] [Google Scholar]
- 23.Sparks SG, Carman RJ, Sarker MR, et al. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J Clin Microbiol 2001;39:883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakshminarayanan B, Harris HM, Coakley M, et al. Prevalence and characterization of Clostridium perfringens from the faecal microbiota of elderly Irish subjects. J Med Microbiol 2013;62:457–66. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs JP, Dong TS, Agopian V, et al. Microbiome and bile acid profiles in duodenal aspirates from patients with liver cirrhosis: The Microbiome, Microbial Markers and Liver Disease Study. Hepatol Res 2018;48:1108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karched M, Bhardwaj RG, Asikainen SE. Coaggregation and biofilm growth of Granulicatella spp. with Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans. BMC Microbiol 2015;15:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albenberg L, Esipova TV, Judge CP, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014;147:1055–63 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parthasarathy G, Chen J, Chen X, et al. Relationship between microbiota of the colonic mucosa vs feces and symptoms, colonic transit, and methane production in female patients with chronic constipation. Gastroenterology 2016;150:367–79 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L, Kelly CJ, Colgan SP. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: Cellular responses to hypoxia. Am J Physiol Cell Physiol 2015;309:C350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahakian AB, Jee SR, Pimentel M. Methane and the gastrointestinal tract. Dig Dis Sci 2010;55:2135–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.