Abstract
To substantiate cross-protection reported across AS04-adjuvanted bivalent human papillomavirus (HPV) vaccine (2vHPV) studies, we reevaluated vaccine effectiveness against type-specific HPV positivity as a function of phylogenetic distance to vaccine target types HPV-16 and -18. We provide evidence of sustained cross-protection up to 8 years postvaccination in a high-risk population in the Netherlands. Moreover, our findings suggest that genomic distance better explains cross-protection than distance measures based on capsid antigens only. Taken together, 2vHPV is predicted to provide partial cross-protection against HPV-31, -33, -35, -45, -52, and possibly -58, that is, acknowledged oncogenic types with close phylogenetic relationships to HPV-16 or -18.
Keywords: HPV, papillomavirus, phylogeny, bivalent vaccine, cross-protection
Ongoing surveillance in Dutch sexual health centers demonstrates sustained effectiveness of the bivalent human papillomavirus (HPV) vaccine against vaccine-targeted types and nontargeted but phylogenetically related types, supporting the hypothesis that vaccine-induced cross-protection is due to genomic similarities between HPV genotypes.
The sexually transmitted human papillomavirus (HPV) is considered a necessary factor for development of cervical cancer and is linked to other anogenital and oropharyngeal carcinomas [1]. Papillomaviruses are characterized by genotype, defined as >10% DNA sequence divergence from other known genotypes (generally termed “types”) in the L1 capsid gene [2]. Most HPV-related malignancies are attributable to types 16 and 18. Consequently, first-generation vaccines, based on recombinant expression of L1 in systems yielding virus-like particles (VLPs), focused on HPV-16 and -18, with the quadrivalent vaccine (4vHPV) also containing L1 VLPs of HPV-6 and -11, primarily associated with anogenital warts. As up to 30% of cervical cancer is attributed to oncogenic types other than 16 or 18, achieving broader protection through cross-reactivity or expansion of the range of VLP types is desirable.
Endeavors to expand the range of VLP types have resulted in the second-generation nonavalent vaccine (9vHPV), containing L1 VLPs from those already contained in 4vHPV plus the 5 next most common types in cervical cancer: HPV types 31, 33, 45, 52, and 58. Alternatively, the minor capsid protein L2, though less immunogenic than L1, is potentially an effective target for prophylaxis, as several subdominant protective epitopes of L2 are well conserved between types and broadly cross-protective in animal models. By contrast, the protection elicited by L1 VLPs is generally taken to be type-restricted (ie, reactive with the homologous type) [1].
First-generation HPV vaccines have shown durable type-specific protection for at least a decade [3]. Importantly, this protection is not absolutely type-restricted, because significant cross-protection has been observed against several nonvaccine types, particularly for the AS04-adjuvanted bivalent vaccine (2vHPV) containing L1 VLPs of HPV-16 and -18 only. In the largest phase 3 trial of 2vHPV, cross-protection was described against persistent HPV-6, -31, -33, -45, -51, and -52 infections, and against incident HPV-35 infection. However, findings with regard to nonvaccine types are equivocal, as the 2vHPV trial from Costa Rica reported significant protection against HPV-31, -45, and -52, insignificant protection against HPV-33, and no effect on HPV-51 [4].
Recent population-based studies from the United Kingdom and the Netherlands confirm some cross-protection from 2vHPV in postvaccine surveillance. In Scotland, a decrease in the prevalence of HPV-31, -33, and -45 was observed among women who underwent their first cervical screening within 7 years after initiating a 2vHPV vaccination program [5]. In the Netherlands, significant cross-protection was estimated against HPV-31, -35, -45, and -52 among female visitors to sexually transmitted infection (STI) clinics who reported to be vaccinated, relative to vaccine-eligible controls [6]. Cross-protection from 2vHPV against HPV-6 or -11 has not been replicated in postvaccine surveillance, neither in England [7] nor in the Netherlands [8].
To reconcile the inconsistencies in cross-protection reported across 2vHPV studies and to assess the type-restricted nature of the protection elicited by L1 VLPs, we reevaluated vaccine effectiveness (VE) against type-specific HPV positivity among STI clinic visitors up to 8 years after vaccination as a function of phylogenetic distance to L1 capsid antigens contained in 2vHPV.
METHODS
We estimated VE from the Papillomavirus Surveillance Among STI Clinic Youngsters in the Netherlands (PASSYON) study, a biennial cross-sectional survey in the Netherlands, as described before [6], but now with an extra study round and including all genotypes in the SPF10-LiPA25 assay (DDL Diagnostics Laboratory). In brief, women aged 16–24 years, who had been eligible for HPV vaccination since 2009 and visited the STI clinic between 2011 and 2017, provided a vaginal swab that was analyzed using a polymerase chain reaction (PCR)–based assay able to detect 25 HPV types, including 12 acknowledged and 3 possibly oncogenic types (Table 1). We compared type-specific HPV positivity between 1305 self-reported vaccinated (≥1 dose) and 799 unvaccinated women. The self-reported vaccination status was validated by serology among those who also provided blood.
Table 1.
Bivalent Human Papillomavirus (HPV) Vaccine Effectiveness Against Type-Specific HPV Positivity
| HPV Typea | VE (95% CI)b | No., Total (No. Vaccinated/No. Nonvaccinated)c | Reference Genomed | Hamming Distancee |
|---|---|---|---|---|
| High-risk | ||||
| 16 | 0.92 (.86–.96) | 100 (13/87) | gi|333031|lcl|HPV16REF.1 | 37 |
| 18 | 0.89 (.78–.94) | 63 (11/52) | gi|60975|lcl|HPV18REF.1 | 41 |
| 31 | 0.66 (.51–.77) | 129 (50/79) | gi|333048|lcl|HPV31REF.1 | 102 |
| 33 | 0.41 (.05–.63) | 73 (37/36) | gi|333049|lcl|HPV33REF.1 | 111 |
| 35 | 0.40 (−.03 to .65) | 55 (28/27) | gi|396997|lcl|HPV35REF.1 | 105 |
| 39 | 0.15 (−.19 to .39) | 165 (98/67) | gi|333245|lcl|HPV39REF.1 | 128 |
| 45 | 0.81 (.55–.92) | 28 (7/21) | gi|397022|lcl|HPV45REF.1 | 91 |
| 51 | −0.24 (−.54 to .01) | 522 (345/177) | gi|333087|lcl|HPV51REF.1 | 180 |
| 52 | 0.36 (.19–.50) | 342 (185/157) | gi|397038|lcl|HPV52REF.1 | 117 |
| 56 | −0.17 (−.59 to .14) | 220 (145/75) | gi|397053|lcl|HPV56REF.1 | 173 |
| 58 | 0.30 (−.06 to .54) | 95 (52/43) | gi|222386|lcl|HPV58REF.1 | 113 |
| 59 | −0.95 (−2.17 to −.20) | 96 (73/23) | gi|557236|lcl|HPV59REF.1 | 125 |
| Probable high-risk | ||||
| 53 | 0.26 (.05–.43) | 332 (189/143) | gi|9627377|lcl|HPV53REF.1 | 173 |
| 66 | 0.02 (−.26 to .24) | 340 (212/128) | gi|1020290|lcl|HPV66REF.1 | 174 |
| 68/73/97 | −0.08 (−.63 to .28) | 110 (71/39) | gi|71726685|lcl|HPV68REF.1 | 139 |
| gi|1491692|lcl|HPV73REF.1 | 150 | |||
| gi|71726694|lcl|HPV97REF.1 | 89 | |||
| Low-risk | ||||
| 6 | −0.15 (−.52 to .14) | 263 (172/91) | gi|60955|lcl|HPV6REF.1 | 161 |
| 11 | −0.07 (−1.00 to .42) | 45 (29/16) | gi|333026|lcl|HPV11REF.1 | 162 |
| 34 | 0.00 (−1.57 to .61) | 19 (12/7) | gi|9627334|lcl|HPV34REF.1 | 154 |
| 40 | 0.08 (−.68 to .50) | 46 (28/18) | gi|397014|lcl|HPV40REF.1 | 163 |
| 42 | −1.27 (−3.34 to −.19) | 57 (45/12) | gi|333211|lcl|HPV42REF.1 | 160 |
| 43 | −0.78 (−2.13 to −.01) | 67 (50/17) | gi|40804474|lcl|HPV43REF.1 | 176 |
| 44 | −0.32 (−1.30 to .24) | 61 (42/19) | gi|1020242|lcl|HPV44REF.1 | 162 |
| 54 | −0.34 (−.95 to .08) | 141 (97/44) | gi|9628437|lcl|HPV54REF.1 | 143 |
| 70 | 0.02 (−1.03 to .52) | 32 (20/12) | gi|1173493|lcl|HPV70REF.1 | 126 |
| 74 | 0.26 (−.19 to .54) | 75 (42/33) | gi|27462483|lcl|HPV74REF.1 | 168 |
Abbreviations: CI, confidence interval; HPV, human papillomavirus.
aHPV genotypes in the SPF10-LiPA25 assay, with 68 being indistinguishable from 73 and 97.
bVE (with 95% CI) was calculated as 1 minus the adjusted odds ratio from a logistic mixed model described in [6].
cNo. of positive test results (among 1305 vaccinated + 799 nonvaccinated women) used in VE estimation.
dWhole-genome reference DNA sequences obtained from the papillomavirus genome database (https://pave.niaid.nih.gov/).
eMinimum number of different amino acids between aligned L1 sequences of reference types and virus-like particles in bivalent vaccine.
Phylogenetic distance of each genotype to the 2vHPV types used for construction of VLPs was calculated from reference DNA sequences obtained via the papillomavirus episteme, a database of curated papillomavirus genomic sequences [9]. We performed a phylogenetic analysis on L1 amino acid composition using a general Dayhoff matrix for evolutionary change in L1 protein with standard codon model and multiple sequence alignment (www.ebi.ac.uk/Tools/msa/muscle/). In addition, we constructed phylogenetic trees directly from DNA sequences on the basis of L1 capsid gene, and on the basis of whole-genome sequences (WGS). Unrooted evolutionary trees from L1 protein or DNA sequences were constructed by maximum likelihood with substitution model selection using IQ-TREE version 1.6.0 software (www.iqtree.org/). Phylogenetic distance was calculated from the consensus tree constructed from 1000 ultra-fast bootstrap trees [10]. Finally, we compared dependence on phylogenetic distance to the Hamming distance from aligned L1 sequences, that is, the number of positions at which the corresponding L1 proteins of reference types are different from the amino acids expressed by VLPs in 2vHPV.
We assessed VE as a function of minimum distance to VLP amino acid composition in L1 protein analysis and as a function of minimum distance to HPV-16 or 18 reference sequences in DNA analyses. Because the LiPA25 assay cannot distinguish between types 68, 73, and 97, we omitted these types from statistical analysis. We also omitted HPV-59, as the estimate of cross-protection against this type is potentially hampered by technical issues in the assay [6]. For the remaining types, we fitted a penalized regression spline to the estimates from the logistic mixed model, weighted by the square root of the number of positive test results used in VE estimation (Table 1), as a function of phylogenetic distance. The smoothness of the function was determined by general cross-validation, and confidence intervals (CIs) were obtained through Bayesian approximation. In addition, we performed weighted covariance analyses on the rank values of the various distance measures, stratified by (putative) oncogenicity of HPV types. Statistical analyses were performed with R version 3.5.1.
RESULTS
Type-specific VE estimates are provided in Table 1. The pooled VE against the 2 vaccine types was 91.0% (95% CI, 86.0%–94.2%). Pooled VE against all (possibly) oncogenic types was 25.8% (95% CI, 17.7%–33.2%), whereas pooled VE against nononcogenic types included in the assay was −4.9% (95% CI, −20.7% to 8.8%). We found no indications for dependency of VE on time since vaccination in stratified analyses, comparing women who were offered vaccination <5 years ago, 5–6 years ago, or 7–8 years ago (Supplementary Table 1).
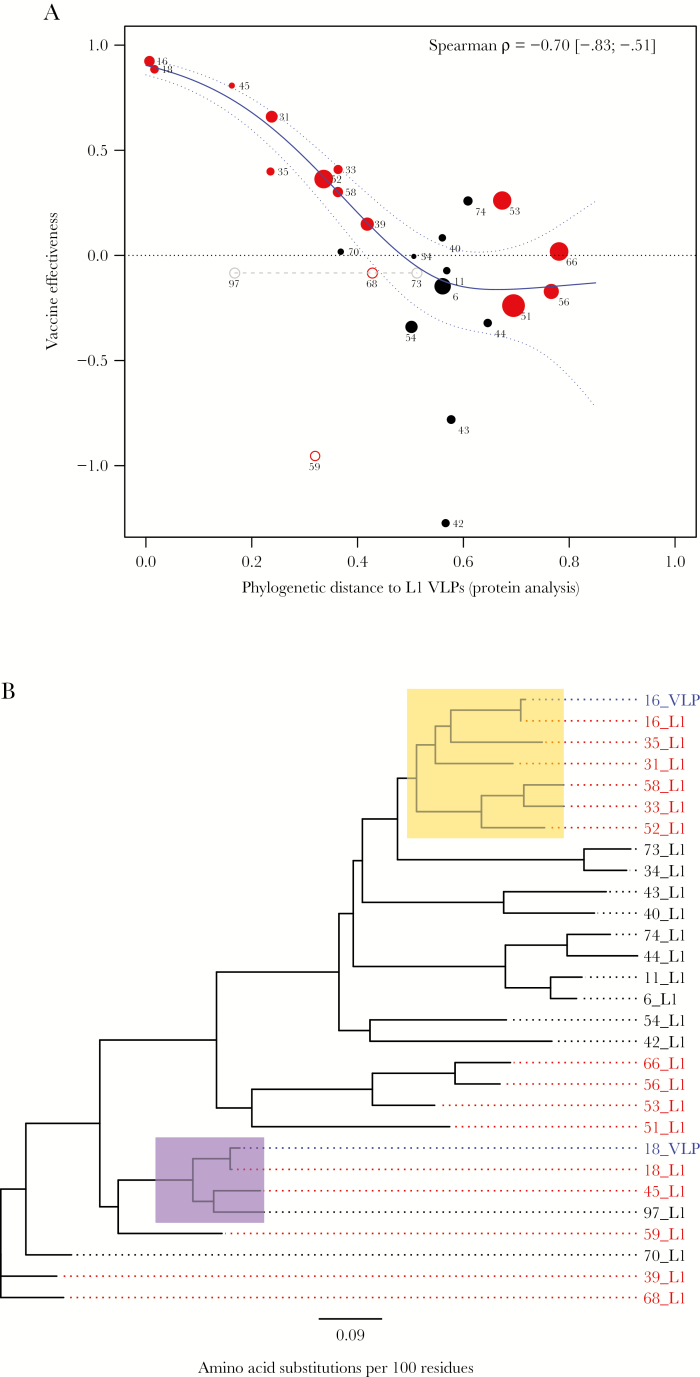
Overall, there was a clear relationship between VE and phylogenetic distance in L1 protein analysis (Figure 1A). The consistently high cross-protection reported for HPV-45 is due to its close relationship to HPV-18 (Figure 1B). The cross-protection of around 50% against HPV-31 and -35 fits their almost equidistant relationship to HPV-16. Likewise, the cross-protection of around 35% against HPV-33, -52, and -58 is in line with these types having approximately similar phylogenetic distance to HPV-16. Our analysis further supports the notion that the estimate of VE against HPV-59 appears to be an outlier. Of the acknowledged oncogenic types, HPV-51, -56, and -66 are most distantly related to either L1 VLP and least likely to be affected by cross-neutralizing antibodies induced by 2vHPV (Supplementary Figure 1A). The significant rank correlation (ρ = −0.70 [95% CI, −.83 to −.51) between VE and phylogenetic distance to L1 VLP in protein analysis was entirely explained by oncogenic types (ρ = −0.93 [95% CI, −.95 to −.89]), as no significant correlation was observed for nononcogenic types (Supplementary Figure 1B).
Figure 1.
Bivalent human papillomavirus (HPV) vaccine effectiveness (VE) as a function of phylogenetic distance to L1 virus-like particles (VLPs). The VE was calculated from cross-sectional prevalence data [6] for all genotypes in the SPF10-LiPA25 assay. Phylogenetic distance to L1 VLPs was calculated from reference DNA sequences, using the Dayhoff model for evolutionary change in L1 protein. Substitution rate heterogeneity among alignment sites was incorporated by assuming γ-distributed rates plus a fraction of invariable sites. A, Red data points denote (possibly) oncogenic types, with HPV-68 being indistinguishable from HPV-73 and -97 (in gray). The size of each data point is plotted proportional to the number of positive test results (n) used in VE estimation. Spearman rank correlation (with 95% confidence limits in brackets) was calculated from all data points weighted by √n, excluding HPV-59 and -68 (open circles). The estimated spline function (in blue) is shown with 95% credible intervals (dotted lines). B, Phylogenetic tree based on L1 protein, with blue tips denoting reference sequences used for construction of L1 VLPs in the bivalent HPV vaccine and red tips denoting (possibly) oncogenic types in the SPF10-LiPA25 assay. The yellow and purple clades highlight types that are close enough to L1 VLPs to benefit (in principle) from cross-protection, according to predictions with 95% confidence from the weighted penalized regression spline in (A).
Analysis based on Hamming distance toward L1 VLPs yielded similar results as L1 phylogenetic analysis (Supplementary Figure 2A). Phylogenetic analysis based on the L1 capsid gene and WGS yielded slightly different depictions but was still comparable to those for L1 protein (Supplementary Figure 2B and 2C). To express the specific association between VE and each phylogenetic distance while controlling for the effect of other measures, we computed their partial rank correlations from the inverse weighted covariance matrix. Apparently, WGS phylogenetic distance to HPV-16 or -18 was the strongest independent determinant of VE (ρpartial = −0.53, P < .01), with HPV-51, -53, -56, and -66 located around the threshold genomic distance still informative for VE. The nononcogenic types were further distanced from vaccine target types in WGS analysis than in analyses based on capsid antigen only, and the partial rank correlations between VE and L1 distance measures were no longer significant when corrected for genomic distance (Supplementary Table 2).
DISCUSSION
This study provides evidence of sustained cross-protection from 2vHPV up to 8 years postvaccination in a high-risk population. Taken together, 2vHPV is predicted to provide partial cross-protection against HPV types 31, 33, 35, 45, 52, and possibly 58—that is, high-risk types belonging to HPV α-7 (including HPV-18) or α-9 (including HPV-16) species [2]. Of those, HPV-35 and -58 are not frequently reported among the cross-protective types, which may be due to their relative rarity as compared to other cross-protective types. Likewise, VE against HPV-35 and -58 was not significantly different from zero in this study, and regression analyses based on phylogenetic distance predicted only small to moderate effect size. Of the other (possibly) oncogenic types, cross-protection may extend to HPV-39 but is unlikely for HPV-51, -53, -56, and -66 (ie, high-risk types belonging to HPV α-5 or α-10 species) [2]. Similarly, cross-protection against low-risk types was not observed in our study [8] and is not to be expected on the basis of phylogenetic analyses.
Although there has been concern about the durability of cross-reactivity, so far there is no evidence for the waning of cross-protection from 2vHPV in women who have been vaccinated 3 times, as per initial recommendation [5, 6, 11]. Moreover, although we have previously shown reliable reporting of vaccination status in our study [6], our VE estimates and their relation with phylogenetic distance may be underestimated by nondifferential misclassification with regard to self-reported vaccination status. It remains to be seen whether cross-reactivity of 2vHPV following vaccination with <3 doses induces similar, long-lasting cross-protection. Analysis of the impact of 2vHPV among female teenagers in the United Kingdom shows evidence of type-specific protection, but not cross-protection following a single dose of vaccine [12].
One possibility as to why cross-protection is better explained by genomic distance than by measures based on the L1 capsid protein is that L1 VLP may induce cross-neutralizing antibodies to L2 that are critical in preventing viral entry into the host cell [1]. Moreover, the adjuvant AS04 in 2vHPV has been suggested to induce a T-cell response that enhances local innate control and provides help for subsequent adaptive immunity [13]. Although the mechanisms of adaptive immunity are still ambiguous, it is worthwhile to point out that vaccination with 2vHPV results not only in reduced incidence rates, but also in reduced viral load in breakthrough infections [14], suggesting that VE extends to control of infection postacquisition. AS04 is particularly effective in activating antigen-presenting cells, inducing cytokines and a T-helper 1–type response, leading to inhibition of viral transcription or translation [1]. Such features could contribute to cross-protective humoral and cellular control of HPV infections, and may be boosted by natural exposure to nonvaccine HPV types.
Cross-protection can be expected to mitigate the potential for type replacement by acting as a substitute of latent competitive pressures induced by vaccine types. Thus, oncogenic types that do not benefit from cross-reactivity should be considered foremost in evaluating type replacement in the wake of vaccination. In this context, the negative VE against HPV-59 might stem from differential sensitivity of the SPF10-LiPA25 assay in vaccinees relative to nonvaccinated controls [6], and not from type replacement. A Finnish community randomized trial on the population effects of 2vHPV, using a different PCR-based assay than ours, found no indications for type replacement by HPV-59 [15]. Instead, HPV-39 and HPV-51 were marked as potential culprits for an increased postvaccination occurrence.
To summarize, our analysis indicates that cross-protection from 2vHPV is sustained up to 8 years postvaccination and that the level of protection correlates with genomic distance to HPV-16 or -18. This suggests that the benefits of 2vHPV vaccination may extend to clinically relevant nonvaccine types, given that oncogenic potential of papillomaviruses itself has a phylogenetic basis [1–3]. Further studies will reveal to what extent cross-protection induced by the bivalent vaccine will contribute to HPV-related disease prevention.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank all participants and personnel within the Medical Microbiological Laboratories and the Public Health Services involved in the PASSYON study for the use of valuable data.
Disclaimer. The funders had no role in study design, data collection and analysis, interpretation of data, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the Dutch Ministry of Health, Welfare and Sport, and by the Strategic Programme from the National Institute for Public Health and the Environment, through grant number S/113005/01/PT (Prometheus project).
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Roden RBS, Stern PL. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat Rev Cancer 2018; 18:240–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Villiers EM. Cross-roads in the classification of papillomaviruses. Virology 2013; 445:2–10. [DOI] [PubMed] [Google Scholar]
- 3. Harper DM, DeMars LR. HPV vaccines—a review of the first decade. Gynecol Oncol 2017; 146:196–204. [DOI] [PubMed] [Google Scholar]
- 4. Skinner SR, Apter D, De Carvalho N, et al. . Human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine for the prevention of cervical cancer and HPV-related diseases. Expert Rev Vaccines 2016; 15:367–87. [DOI] [PubMed] [Google Scholar]
- 5. Kavanagh K, Pollock KG, Cuschieri K, et al. . Changes in the prevalence of human papillomavirus following a national bivalent human papillomavirus vaccination programme in Scotland: a 7-year cross-sectional study. Lancet Infect Dis 2017; 17:1293–302. [DOI] [PubMed] [Google Scholar]
- 6. Woestenberg PJ, King AJ, van Benthem BHB, et al. . Medical Microbiological Laboratories and the Public Health Services Bivalent vaccine effectiveness against type-specific HPV positivity: evidence for cross-protection against oncogenic types among Dutch STI clinic visitors. J Infect Dis 2018; 217:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mesher D, Panwar K, Thomas SL, Beddows S, Soldan K. Continuing reductions in HPV 16/18 in a population with high coverage of bivalent HPV vaccination in England: an ongoing cross-sectional study. BMJ Open 2016; 6:e009915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woestenberg PJ, King AJ, van der Sande MA, et al. . Medical Microbiological Laboratories; Public Health Services No evidence for cross-protection of the HPV-16/18 vaccine against HPV-6/11 positivity in female STI clinic visitors. J Infect 2017; 74:393–400. [DOI] [PubMed] [Google Scholar]
- 9. Van Doorslaer K, Li Z, Xirasagar S, et al. . The papillomavirus episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res 2017; 45:D499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 2013; 30:1188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donken R, King AJ, Bogaards JA, Woestenberg PJ, Meijer CJLM, de Melker HE. High effectiveness of the bivalent human papillomavirus (HPV) vaccine against incident and persistent HPV infections up to 6 years after vaccination in young Dutch women. J Infect Dis 2018; 217:1579–89. [DOI] [PubMed] [Google Scholar]
- 12. Cuschieri K, Kavanagh K, Moore C, Bhatia R, Love J, Pollock KG. Impact of partial bivalent HPV vaccination on vaccine-type infection: a population-based analysis. Br J Cancer 2016; 114:1261–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Didierlaurent AM, Morel S, Lockman L, et al. . AS04, an aluminum salt- and TLR4 agonist-based adjuvant system, induces a transient localized innate immune response leading to enhanced adaptive immunity. J Immunol 2009; 183:6186–97. [DOI] [PubMed] [Google Scholar]
- 14. van der Weele P, Breeuwsma M, Donken R, et al. . Effect of the bivalent HPV vaccine on viral load of vaccine and non-vaccine HPV types in incident clearing and persistent infections in young Dutch females. PLoS One 2019; 14:e0212927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gray P, Palmroth J, Luostarinen T, et al. . Evaluation of HPV type-replacement in unvaccinated and vaccinated adolescent females—post-hoc analysis of a community-randomized clinical trial (II). Int J Cancer 2018; 142:2491–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.