ABSTRACT
Tuberculosis (TB), caused by the intracellular pathogen Mycobacterium tuberculosis, remains one of the leading causes of mortality across the world. There is an urgent requirement to build a robust arsenal of effective antimicrobials, targeting novel molecular mechanisms to overcome the challenges posed by the increase of antibiotic resistance in TB. Mycobacterium tuberculosis has a unique cell envelope structure and composition, containing a peptidoglycan layer that is essential for maintaining cellular integrity and for virulence. The enzymes involved in the biosynthesis, degradation, remodelling and recycling of peptidoglycan have resurfaced as attractive targets for anti-infective drug discovery. Here, we review the importance of peptidoglycan, including the structure, function and regulation of key enzymes involved in its metabolism. We also discuss known inhibitors of ATP-dependent Mur ligases, and discuss the potential for the development of pan-enzyme inhibitors targeting multiple Mur ligases.
Keywords: Tuberculosis (TB), cell envelope, peptidoglycan, metabolism, drug-target validation, antibiotic resistance, antibacterial
A comprehensive, investigative review that highlights key enzymes involved in mycobacterial cell wall peptidoglycan metabolism as novel anti-infective drug targets.
INTRODUCTION
Tuberculosis (TB) is a leading cause of mortality in the world today and is caused by the bacterial pathogen Mycobacterium tuberculosis. Despite innovations in diagnostics and improved access to health care, the global burden of TB remains substantial with around 10 million new cases of infection and 1.6 million deaths reported due to TB in 2017 alone (WHO 2018). An estimated 457 000 cases reported in 2017 presently harbour multi-drug resistant TB (MDR-TB), 8.5% of which are expected to be extensively drug resistant TB (XDR-TB) (WHO 2018), reflecting the urgent need to design and develop novel drugs to treat TB.
The success of M. tuberculosis as a pathogen and its innate resistance to many antimicrobial drugs can be attributed in part to its unique cell wall structure, which has low permeability for many drugs and possesses a large number of efflux pumps (Jarlier and Nikaido 1994, Brennan and Nikaido 1995). The cell wall is a defining characteristic of all bacteria. Amongst the many purposes it serves, maintaining the cell-shape and withstanding turgor are key. The varying properties of the bacterial cell wall, especially the thickness of the peptidoglycan (PG), impart different stain-retention properties to the bacterial cell and enable us to categorise most bacteria into Gram-positive, Gram-negative and acid-fast. The presence of PG across nearly all bacteria indicates that it was likely to have been present in their last common ancestor (Errington 2013). Importantly, PG is essential for bacterial cell survival in most environments, thus making it a good target for anti-infective therapy.
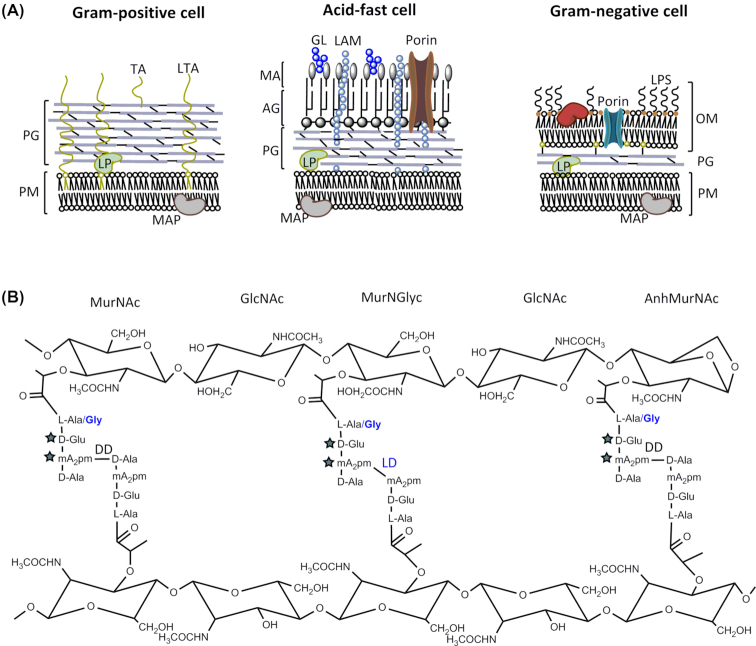
Mycobacteria belong to the diverse family of Actinobacteria. The main components of the mycobacterial cell wall are the PG layer, mycolic acid (MA) and arabinogalactan (AG). The mycobacterial cell wall resembles both the Gram-positive and Gram-negative cell envelope by having a PG layer nearly as thick as the former and an outer, waxy layer mimicking the outer membrane of the latter (Fig. 1A). The cell wall of mycobacteria plays a key role in intrinsic antibiotic resistance and virulence (Forrellad et al. 2013; Becker and Sander 2016) but it is unclear why and how its complex structure evolved. Vincent et al. recently proposed that the ‘mycomembrane’ evolved by successive horizontal acquisition of genes; explaining the narrow distribution of the AG and MA biosynthetic genes in some lineages of the Actinobacteria that evolved later, while the PG genes are broadly distributed (Vincent et al. 2018).
Figure 1.
(A), Differences in the cell envelope architecture of Gram-positive, acid-fast and Gram-negative bacteria. AG, arabinogalactan; GL, glycolipid; LAM, lipoarabinomannan; LP, lipoprotein; LPS, lipopolysaccharide; LTA, lipoteichoic acid; MA, mycolic acid; MAP, membrane-associated protein; OM, outer membrane; PG, peptidoglycan; PM, plasma membrane; TA, teichoic acid . (B), PG structure revealing the unique features present in mycobacterial cells highlighted in blue. The stars depict residues that undergo amidation.
PG is a dynamic structure but the factors that regulate its composition and biogenesis in the slow-growing intra-cellular pathogen M. tuberculosis are not well understood. The mycobacterial PG plays a key role in the cell's growth, cell–cell communication and in the initiation of the host immune response. The cell envelope of some model bacteria such as Escherichia coli, have long been the focus of extensive research; however, these organisms do not serve as appropriate models when studying the unique physiology and biochemistry of M. tuberculosis. Here, we review the different stages of PG metabolism in M. tuberculosis, its importance for survival of the bacilli and the potential therapeutic targets it offers for the design of new anti-infective drugs.
Modifications of cell wall PG in M. tuberculosis
Lying outside the cytoplasmic membrane of M. tuberculosis, the PG layer is covalently attached to AG which itself serves as an attachment site for unique MAs (Fig. 1A). This is referred to as the cell wall core, or as the MA-AG-PG complex (MAPc) (Brennan 2003). Beyond and interspersed within the MAPc are free lipids, phosphatidyl inositol mannosides (PIM), lipomannans (LM) and lipoarabinomannans (LAM). As observed in other microorganisms (Jankute et al. 2015), the disruption of PG synthesis or digestion of the PG layer leads to the formation of L-form bacilli that display a variety of cell shapes and morphology in mycobacteria (Udou, Ogawa and Mizuguchi 1982).
PG is made of glycan strands of alternating, β(1→4)-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues. Adjacent glycan strands are connected by short peptide stems with the sequence l-alanyl-γ-d-isoglutamyl-meso-diaminopimelate-d-alanyl-d-alanine and linked to the d-lactoyl residue of each MurNAc. Peptides protruding from adjacent glycan strands can be cross-linked between the carboxyl group of d-Ala at the fourth position and the ε-amino group of the meso-diaminopimelate (mDAP) in the third position.
While the basic structure of PG is conserved, it is important to note that the PG in mycobacteria undergoes several modifications (Fig. 1B). PG isolated from M. tuberculosis and M. smegmatis contains both N-acetylmuramic acid (MurNAc) and N-glycolylmuramic acid (MurNGlyc), whereas most other bacteria, including M. leprae, exclusively contain MurNAc (Mahapatra et al. 2005a, Petit et al. 1969; Mahapatra et al. 2008). The presence of MurNGlyc increases the resistance of mycobacterial PG to lysozyme (Raymond et al. 2005). Modifications such as N-deacetylation or O-acetylation of GlcNAc and MurNAc respectively, as observed in other pathogenic bacteria (Vollmer 2008) have not been reported in mycobacteria. Consequently, the deletion of the hydroxylase responsible for N-glycolylation, namH, increases the sensitivity towards β-lactams and lysozyme in M. smegmatis (Raymond et al. 2005). Glycolylated PG fragments are more potent at inducing NOD2-mediated host responses such as production of tumour necrosis factor α (Hansen et al. 2014), which may appear to be counter-productive for the success of an intracellular pathogen. However, the enhanced stimulation of the host might enable M. tuberculosis to recruit phagocytic cells, its ecological niche within the host and facilitate transmission to a new host.
The peptide stems in PG undergo modifications such as amidation of the α-carboxylic group of d-isoglutamate (d-iGlu) and the ε-carboxylic group of the mDAP residues (Kotani et al. 1970; Mahapatra et al. 2008). d-iGlu amidation is required for the functioning of PG transpeptidases that cross-link the peptides to form a robust PG (Figueiredo et al. 2012; Zapun et al. 2013) whereas mDAP amidation is involved in modulating PG hydrolysis thereby maintaining the integrity of the cell wall (Nakel, Ghuysen and Kandler 1971; Dajkovic et al. 2017). These modifications reduce the net negative charge of the cell wall, which impairs the efficacy of antimicrobial factors such as lysozyme, antimicrobial peptides and cell wall acting antibiotics (Girardin et al. 2003b; Roychowdhury, Wolfert and Boons 2005). In the PG of M. leprae, Gly replaces l-Ala in position 1 of the peptide stem, and an additional Gly residue is attached to mDAP (Mahapatra et al. 2008).
The PG of M. tuberculosis contains a particularly high percentage (70%–80%) of cross-linked peptides (Wietzerbin et al. 1974), compared to 40%–50% in E. coli (Glauner, Holtje and Schwarz 1988). One-third of the peptide cross-links are DD-(or 4→3) bonds between d-Ala and mDAP, and about two-thirds are LD-(or 3→3) bonds linking two mDAP residues (Wietzerbin et al. 1974; Hett and Rubin 2008) (Fig. 1B). Although LD-crosslinks are relatively rare in many other bacterial species, recent reports have estimated them to account for 80% of the peptide cross-links in the PG of stationary phase or dormant cells of M. tuberculosis (Lavollay et al. 2008). The LD-cross-links are also enhanced to 15%–20% of all cross-links in the PG of stationary phase cells of E. coli, but are significantly reduced (5% of all cross-links) in exponentially growing cells (Pisabarro, de Pedro and Vazquez 1985; Glauner, Holtje and Schwarz 1988; Templin, Ursinus and Holtje 1999).
Interestingly, the pattern of PG incorporation into the cell wall of M. tuberculosis is different compared to the PG of other rod-shaped bacteria (Daniel and Errington 2003; Hett and Rubin 2008). Most rod-shaped bacilli such as E. coli and Bacillus subtilis elongate by inserting nascent PG along the lateral sides of the cell (den Blaauwen et al. 2008). An actin-like protein, MreB, positions the PG-elongation machinery along the short axis of the cell so that glycan strands are inserted circumferentially thus maintaining the rod-shape (Domínguez-Escobar et al. 2011). In contrast, mycobacteria lack MreB and other proteins important for lateral wall elongation and instead incorporate nascent PG at both poles of the cell (Thanky, Young and Robertson 2007; Kieser and Rubin 2014). Mycobacteria exhibit differential incorporation of PG at the two cell poles (old versus new cell pole) at certain stages of the cell cycle (before/after cytokinesis); this allows for asymmetric cell division and results in a heterogeneous population of cells (Kieser and Rubin 2014; Botella et al.2017b). Baranowski et al. reported that the ability of the mycobacterial cell to generate LD-cross-links (discussed later in this review) is essential in maintaining the rod-shape (Baranowski et al. 2018). However, whether this serves as the sole mechanism regulating cell shape is yet to be confirmed.
Imaging PG
The recent application of fluorescence and atomic force microscopy techniques together with new probes for labelling have significantly advanced our understanding of the dynamics of PG synthesis (Radkov et al. 2018). Localisation studies using GFP-labelled PG biosynthetic proteins (Joyce et al. 2012; Grzegorzewicz et al. 2016), fluorescent d-amino acids (Siegrist et al. 2013; Meniche et al. 2014; Boutte et al. 2016; Schubert et al. 2017; Botella et al. 2017b; Rodriguez‐Rivera et al. 2018) and cell wall targeting antibiotics (Thanky, Young and Robertson 2007; Kastrinsky and Barry 2010) have further emphasized the importance of PG remodelling in cell elongation, septum formation and cell division. Aldridge et al. used microfluidics-based live-cell imaging technique to show that the heterogeneous daughter cell populations differ in their susceptibility to antibiotics (Aldridge et al. 2012). Botella et al. compared the spatiotemporal dynamics of PG synthesis in M. smegmatis and M. tuberculosis using super-resolution microscopy combined with fluorescent d-alanine analogues (FDAAs) (Botella et al. 2017b). They reported that FDAAs are predominantly incorporated at one of the two poles in M. smegmatis, whereas M. tuberculosis shows variation in polar dominance depending on the stage in cell cycle. FDAAs are also incorporated along the lateral wall upon damage due to muramidase activity (Garcia-Heredia et al. 2018). Furthermore, fluorescent intracellular membrane domain (IMD)-associated protein reporters revealed that mycobacteria can spatiotemporally coordinate its membrane domain in response to metabolic requirements under different growth conditions (Hayashi et al. 2018). These recent reports on membrane compartmentalisation of PG synthesis and its metabolism during the cell cycle in M. tuberculosis will help to understand mechanisms allowing bacteria to escape host defence systems and to characterise drug targeting steps that have remained elusive before.
Biosynthesis and maturation of PG
The nucleotide precursors of PG were first isolated from Staphylococcus aureus in 1952 (Park 1952). Since then the various steps involved in the biosynthesis of PG have been extensively studied in a number of species. The PG biosynthetic pathway in M. tuberculosis, however, remains largely uncharacterised. In this work we follow the generally accepted assumption that mycobacteria synthesise PG precursors in a similar way to the model bacterium E. coli using enzymes that are homologous to the ones found in the latter. However, as a slow-growing intra-cellular pathogen with varied physiological states, mycobacteria have PG enzymes that differ with respect to structure and regulation compared to their counterparts in E. coli. This review attempts to highlight these critical features. Details of the enzymes discussed throughout the review have been listed in Table S1 (Supporting Information).
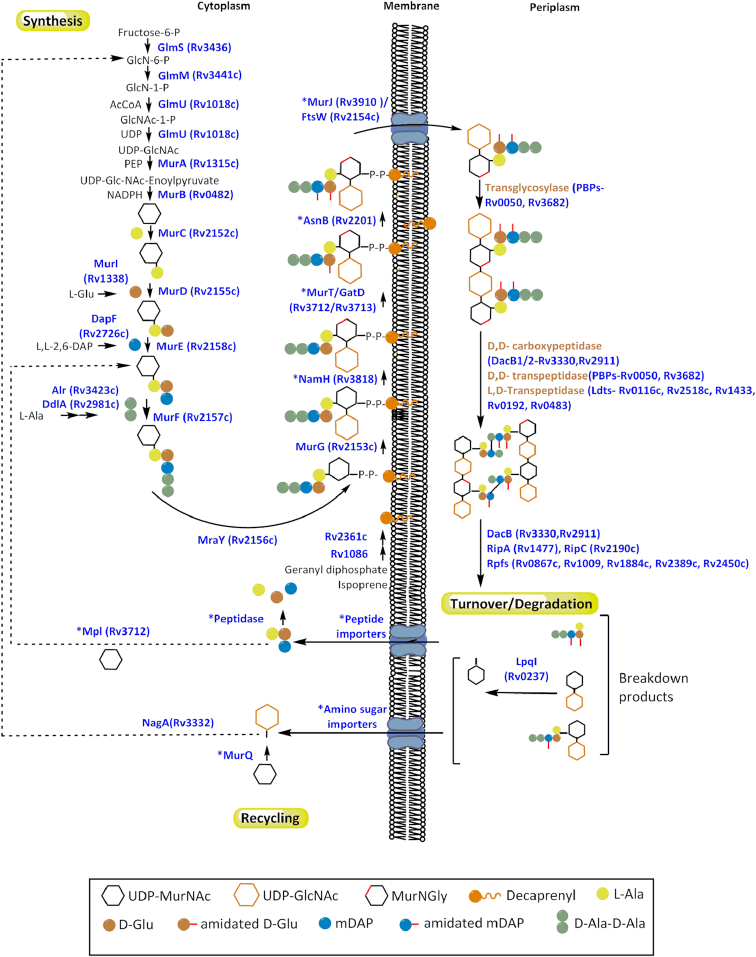
PG biosynthesis: the cytoplasmic steps
PG biosynthesis begins with the conversion of fructose-6-phosphate to UDP-N-acetylglucosamine (UDP-GlcNAc), which is catalysed by the enzymes GlmS, GlmM and GlmU (van Heijenoort 2001a) (Fig. 2). Of these three enzymes, only GlmU has been characterised in M. tuberculosis (Zhang et al. 2009; Jagtap et al. 2012). GlmU is a bifunctional acetyltransferase/uridyltransferase, which converts glucosamine-1-phosphate (GlcN-1-P) to N-acetylglucosamine-1-phosphate (GlcNAc-1-P) and subsequently catalyses the formation of UDP-GlcNAc from GlcNAc-1-P and uridine triphosphate (Zhang et al. 2009; Jagtap et al. 2012; Li et al. 2012). The reaction mechanism of the acetyl transfer by GlmU provides useful insights for drug design, such as the conformational changes of the protein structure upon interaction with the ligand (Craggs et al. 2018). Mycobacterium tuberculosis GlmU is trimeric in solution, whereby each monomer folds into two distinct domains. The N-terminal domain has a typical uridyltransferase fold based on a dinucleotide-binding Rossmann fold and is similar to that observed in the reported structure for GlmU from Streptococcus pneumoniae (Sulzenbacher et al. 2001). The C-terminal domain has a left-handed parallel β-helix fold forming the acetyltransferase active site containing contributions from all three subunits as is characteristic of other acetyltransferase enzymes. However, the GlmU from M. tuberculosis is 6–8 fold less active than that of GlmU from E. coli. Additionally, as M. tuberculosis GlmU lacks free cysteines in the acetyl-CoA binding site or any solvent-accessible cysteines elsewhere, it retains its acetyltransferase activity even in the absence of reducing agents and in the presence of a thiol-reactive reagent; both of which render the E. coli enzyme inactive (Pompeo, van Heijenoort and Mengin-Lecreulx 1998; Zhang et al. 2009). The crystal structure of M. tuberculosis GlmU reveals a unique 30-residue extension which forms a short helix at the C-terminus and is involved in substrate binding (Jagtap et al. 2012). The C-terminus of the enzyme is also involved in auto-regulation through phosphorylation by the serine/threonine protein kinase PknB (Parikh et al. 2009). Dziadek et al. recently reported that GlmU interacts with host immune factor IL-8, thereby increasing mycobacterial attachment to neutrophils and facilitating infection (Dziadek et al. 2016). How a cytosolic bacterial protein serves as a receptor binding entity still requires clarification, however, the enzyme has garnered interest for drug development. The enzyme is essential for M. tuberculosis and it depletion results in severe growth defects in vitro and reduced bacillary loads in mice models (Soni et al. 2015). Further experiments showed that the uridyl- and acetyl-transferase activities are individually essential for the pathogen thus inhibitors designed for either active site would be effective. Also, as the synthesis of UDP-GlcNAc occurs via a different enzymatic route in eukaryotes compared to prokaryotes, any treatment designed against the latter would be expected to have little effect on the former.
Figure 2.
An overview of PG biosynthesis, degradation and recycling pathways representing information currently known in Mycobacterium species. While there has been plethora of information on the PG metabolism in other bacteria, identification of mycobacterial proteins involved in this process has been limited so far. ‘*’ marks enzymes that have not been experimentally established. This includes proteins that are putative or completely unknown and uncertainty on when the enzymatic step occurs in the pathway. The redundancy in the PG hydrolases makes it difficult to assemble a comprehensive list within a figure and only the selected enzymes discussed in this article are highlighted. Recycling of PG in mycobacteria is an especially underexplored area compared to other bacteria and may offer interesting insight into homeostasis mechanisms within the cell.
MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) and MurB (UDP-N-acetylenolpyruvyl-glucosamine reductase) catalyse the conversion of UDP-N-acetylglucosamine (UDP-GlcNAc) to UDP-N-acetylmuramate (UDP-MurNAc). MurA catalyses the transfer of the enolpyruvyl moiety of phosphoenolpyruvate (PEP) to UDP-GlcNAc (De Smet et al. 1999; Xu et al. 2014). MurB (Benson et al. 1993) is an NADPH dependent oxidoreductase that reduces the enolpyruvate moiety to d-lactate resulting in the formation of UDP-MurNAc. While mycobacteria and Gram-negative bacteria like E. coli have a single copy of the murA gene (Brown et al. 1995), a second transferase gene, murA2 occurs as a duplicate copy in low G + C Gram-positive organisms such as S. pneumoniae (Du et al. 2000). Interestingly, both copies perform the same function and can substitute for one another. The purpose for such redundancy at the first committed step of PG biosynthesis may be to accommodate differential regulation (Du et al. 2000). UDP-MurNAc binds to E. coli MurA with high affinity, suggesting an inhibitory or regulatory role in PG biosynthesis (Mizyed et al. 2005). CwlM, an essential, cytoplasmic protein and hypothetical PG amidase, is another regulator of MurA (Boutte et al. 2016). Phophorylated CwlM increases the activity of MurA nearly 30 fold. CwlM is phosphorylated only in the presence of surplus nutrients, thereby revealing a possible mechanism by which the pathogen correlates PG synthesis with the availability of nutrients. Reducing the PG synthesis rate also has implications for drug susceptibility, therefore mechanisms which can activate CwlM and MurA may help to target the ‘persister’ population of M. tuberculosis and reduce treatment duration, one of the main causes of patient non-compliance. The regulatory role of CwlM has since been investigated further and its influences at later stages of PG synthesis have also been revealed and will be discussed in the following section.
Homology modelling, molecular dynamics and molecular docking studies on the mycobacterial MurA and MurB enzymes revealed active site residues and helped identifying potential inhibitors (Babajan et al. 2011; Kumar et al. 2011). The crystal structure of M. tuberculosis MurB with FAD as the prosthetic group was recently reported (Eniyan et al. 2018). MurB is a type I UDP-GlcNAcEP reductase and shares conserved domains with MurB from E. coli and Pseudomonas aeruginosa. The nicotinamide and the enol pyruvyl moieties aligned well in M. tuberculosis MurB upon superimposition with the other MurB structures and show domain III to adopt a more open conformation on binding to the substrates. There are known inhibitors of MurB from Gram-positive and Gram-negative bacteria, however, whether these are active against M. tuberculosis MurB has not been investigated (Bronson et al. ; Yang et al. ).
The assembly of the peptide stem of the PG monomer unit (UDP-MurNGlyc or UDP-MurNAc-pentapeptide) is catalysed by a series of four essential and structurally related ATP-dependent Mur ligases MurC, MurD, MurE and MurF which, in M. tuberculosis, catalyse the addition of l-Ala, d-Glu, mDAP and d-Ala- d-Ala, respectively, with the concomitant hydrolysis of ATP to ADP and inorganic phosphate (Fig. 2) (Basavannacharya et al. 2010a; Basavannacharya et al. 2010b; Munshi et al. 2013; Eniyan et al. 2016). The mycobacterial Mur ligases catalyse amide bond formation via a similar reaction mechanism to that seen in the E. coli enzymes, i.e. the formation of an activated acylphosphate derivative of UDP-MurNAc followed by a nucleophilic attack by the amino group of the amino acid or dipeptide, ultimately resulting in the formation of a peptide bond and release of inorganic phosphate (Anderson et al. 1996; Falk et al. 1996; Liger et al. 1996; Tanner et al. 1996; Vaganay et al. 1996; Bertrand et al. 1999) (Fig. 3E).
Figure 3.
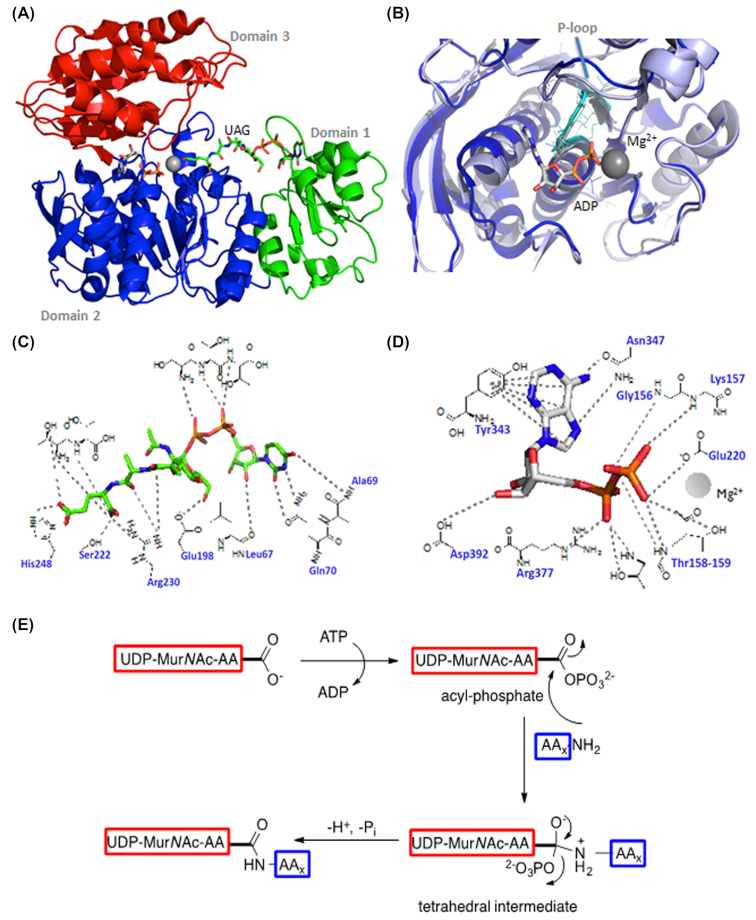
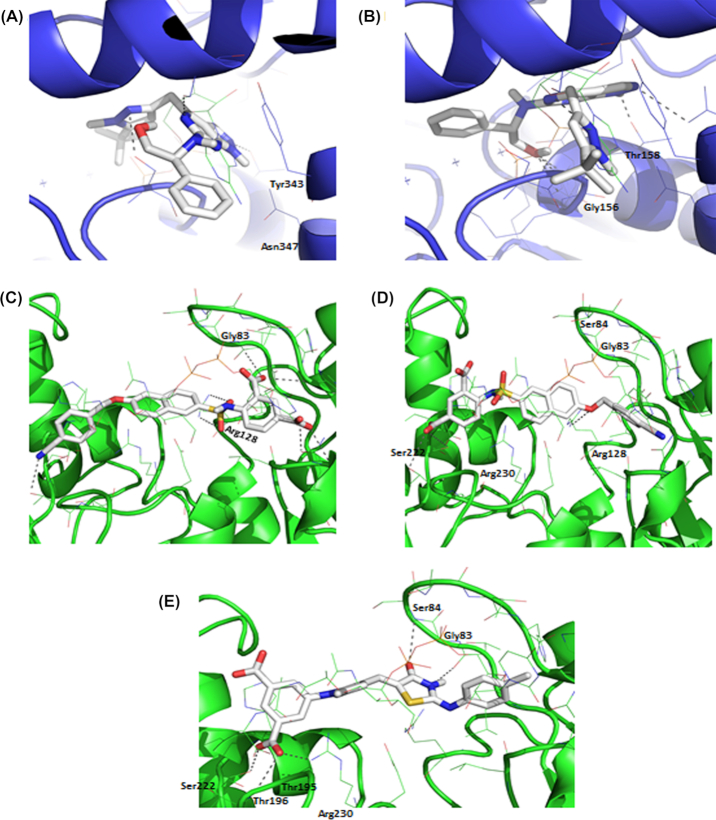
(A), Cartoon representation of the crystal structure of MurE from M. tuberculosis. The UDP substrate binding domain 1 is shown in green, the central ATPase domain 2 is shown in blue and the amino acid substrate binding domain 3 is shown in red. Image generated using PyMOL (PDB code: 2XJA). (B), Overlay of domain 2 from MurE from E. coli (PDB code: 1E8C, light blue) and M. tuberculosis (PDB code: 2XJA, dark blue). The P-loops are highlighted in cyan. (C), Interaction map of the UDP-MurNAc substrate with MurE from M. tuberculosis. (D), Interaction map of ADP with MurE from M. tuberculosis. (E), The reaction mechanism of Mur ligases. The ‘AA’ in the UDP-MurNAc-AA substrate represents the peptide stems of varying lengths whereas the AAx.NH2 represents the incoming amino acid.
All ATP dependent Mur ligases (Mur synthetases) contain common structural motifs despite their low amino acid sequence identities. ATP-dependent Mur ligases can be described as containing three distinct domains: (i) an N-terminal Rossmann-type α/β fold domain that binds to the UDP-substrate, (ii) a central ATPase domain (with a highly conserved WalkerA-like motif, TG[T/S]XGK[T/S(2)]) and (iii) a C-terminal domain that binds the appropriate amino acid (Smith 2006) (Fig. 3).
The first amino acid of the peptide stem (l-Ala) is ligated onto UDP-MurNAc by MurC. In M. leprae, l-Ala is replaced by Gly. Interestingly, M. tuberculosis MurC is able to incorporate both l-Ser and Gly into the growing peptide chain in vitro (Munshi et al. 2013), although it is questionable whether the UDP substrates containing these amino acids would be accepted by MurD in the subsequent reaction. This substrate flexibility has also been observed in some other organisms. Addition of d-Glu by MurD is followed by incorporation of mDAP onto the growing peptide chain by MurE. While over-expression of M. tuberculosis MurE in E. coli and Pseudomonas putida does not affect growth, a lytic effect was observed when the S. aureus MurE (which incorporates l-Lys instead of mDAP) was overexpressed in E. coli (Mengin-Lecreulx et al. 1999). In all mDAP-specific MurE ligases, an arginine residue at position 416 binds to the free end of mDAP, whereas this residue is replaced by an alanine or asparagine in all l-Lys-specific MurE ligases (Dementin 2001). MurE from M. tuberculosis contains three domains, similar to that reported for E. coli. However, unlike its E. coli homologue, the mycobacterial MurE coordinates a magnesium ion with the UDP-MurNAc-l-Ala-d-Glu substrate (Basavannacharya et al. 2010a; Basavannacharya et al. 2010b). The last two amino acids of the stem peptide are added by MurF as the dipeptide d-Ala-d-Ala Interestingly, MurF from E. coli accepts muropeptides with either mDAP or l-Lys with similar efficiencies and its N-terminus is distinct from the other Mur ligases (Yan et al. 2000).
The entire MurA-F pathway is unique to prokaryotes and all corresponding genes are single-copy and essential in M. tuberculosis (Sassetti, Boyd and Rubin 2003; Griffin et al. 2011; DeJesus et al. 2017) making them attractive therapeutic targets. MurC-F share conserved residues and mode of action, and are thus amenable to multi-target therapy. Development of high-throughput activity assays as well as a one-pot assay that screens for inhibitors of more than one enzyme in the Mur pathway could aid drug discovery (Basavannacharya et al. 2010a; Eniyan et al. 2016).
In addition to ATP-dependent Mur ligases, the enzymes catalysing the formation of substrates for Mur ligase reactions are also key components of PG biosynthetic machinery. MurI is a glutamate racemase, which provides the d-Glu required for PG synthesis. Unlike in Bacillus subtilis there is only one gene coding for MurI in M. tuberculosis. The available crystal structure of MurI (Poen et al. 2016) and new inhibitors of the enzyme (Prosser et al. 2016) strengthen its potential as druggable target in the pathogen. However, several issues question the value of MurI as a drug target. Homogeneous MurI purifies as a stable dimer that is enzymatically inactive in vitro. It is presumed that activator molecules such as UDP-MurNAc-l-Ala may dissociate the dimer to generate the active enzyme as observed in the case of E. coli MurI (Ho et al. 1995). This inability to extract and purify an enzymatically active form of the enzyme from heterologous expression systems poses a significant bottleneck for the development of high-throughput inhibitor screening assays. Additionally, there is some ambiguity over its essentiality (Sassetti, Boyd and Rubin 2003; Harth et al. 2005; Griffin et al. 2011; Morayya et al. 2015). Mortuza et al. identified a d‐amino acid transaminase in M. smegmatis that could complement the activity of MurI allowing murI mutants to grow in the absence of d-Glu (Mortuza et al. 2018). Whether its homologue in M. tuberculosis also retains d‐amino acid transaminase activity is not known. The presence of enzymes with overlapping functions indicates that resistance would develop easily against glutamate racemase inhibitors. However, as TB chemotherapy is always a combination, the rise of resistance may be controlled by administering a combination of drugs that work synergistically, such as glutamate racemase inhibitors and d-cycloserine (David 2001).
mDAP is synthesised from aspartate in eight biosynthetic steps (Usha et al. 2012). The enzymes in the pathway are essential and thus have the potential to become drug targets. Alpha-ketopimelic acid inhibits the activity of DapA (dihydrodipicolinate synthase) in the micromolar range indicating that it is a druggable target (Shrivastava et al. 2016). Mycobacterium tuberculosis DapE was found to be insensitive to the known DapE inhibitor L-captopril (Usha et al. 2016). Finally, DapF in Chlamydia trachomatis is bifunctional and involved in both mDAP synthesis and the racemisation of glutamate (Liechti et al. 2018). Whether the DapF from M. tuberculosis could also function as a d-Glu racemase is unknown. This essential enzyme converts ll-DAP into mDAP. Although little variation was observed in the two-domain α/β-fold of the apo structure of DapF from M. tuberculosis, when compared with DapF from Haemophilus and Bacillus species, it was noted that the M. tuberculosis DapF backbone is stabilised via a tyrosine residue specific to mycobacterial DAP epimerases. Any change of the interactions this residue is involved in would destabilize DapF (Usha et al. 2009) thereby providing a rationale for design of anti-tubercular specific DapF inhibitors.
The di-peptide substrate for MurF (d-Ala-d-Ala) is synthesised by the alanine racemase Alr, followed by the d-Ala-d-Ala ligase Ddl. Alr is a pyridoxal-5-phosphate-containing enzyme (van Heijenoort 2001b) and its crystal structure shows a homodimer with the domains adopting a similar fold to that observed in Alr structures from other species (LeMagueres et al. 2005). However, the observed hinge angle between the domains is different for Alr from M. tuberculosis. The interest in Alr as a drug target has been dampened by conflicting reports on its essentiality in M. smegmatis and M. tuberculosis. While Milligan et al. found deletion mutants require a steady supply ofd-Ala for growth, insertion mutants that did not show growth defects in the absence of d-Ala were selected for by Marshall et al. (Milligan et al. 2007; Awasthy et al. 2012, Marshall et al. 2017). Therefore, it is hypothesised that the function of Alr can be substituted by an unidentified transaminase that converts pyruvate and d-Glu to d-Ala so Alr-specific inhibitors would likely have no anti-mycobacterial effect.
Ddl catalyses the ATP-dependent ligation reaction to generate the dipeptide d-Ala-d-Ala. While only one copy of ddl was identified in mycobacteria, two ddl genes are present in E. coli (Zawadzke, Bugg and Walsh 1991). Ddl exists as a dimer, in which each monomer contains three domains with the ligand binding site formed at the intersection of these three domains (Bruning et al. 2011). The apo form of Ddl from M. tuberculosis is unique in that in the absence of substrate it adopts a closed conformation that has to undergo a significant structural rearrangement to bind to the substrates. The closed conformation of Ddl offers many unique binding pockets adjacent to the active site that may be useful for drug design. Ligand binding and catalytic activity, therefore, requires significantly greater structural rearrangement compared to orthologues from other species such as S. aureus. Biochemical analyses and classical enzyme kinetic studies of Ddl from M. tuberculosis revealed the mechanism of inhibition by d-cycloserine—knowledge that could be useful in the design of future drugs (Prosser and de Carvalho 2013). Currently, d-cycloserine is known to target Ddl and Alr in M. tuberculosis, with strong inhibition observed in the case of Ddl and weaker inhibition seen with Alr.
PG biosynthesis: the membrane-associated steps
A lipid carrier is essential for the transport of the hydrophilic PG precursors across the hydrophobic cell membrane. Mycobacteria differ from E. coli in terms of the molecule that serves as the lipid carrier (decaprenyl phosphate versus undecaprenyl-phosphate) and in the synthesis of this molecule. In fact, even within mycobacteria there are marked differences within this pathway. Based on sequence homology it is presumed that M. smegmatis genome codes for 4 to 10 prenyl diphosphate synthases, whereas M. tuberculosis could contain up to 4 synthases (Crick et al. 2000). Rv1086 and Rv2361 are two distinct prenyl diphosphate synthases, a ω,E,Z-farnesyl diphosphate synthase (Z-FPPS) and a decaprenyl diphosphate (DecaPP) synthase, respectively (Schulbach, Brennan and Crick 2000). These act sequentially to synthesise decaprenyl diphosphate from isopentenyl diphosphate and E-geranyl diphosphate (Schulbach, Brennan and Crick 2000; Schulbach et al. 2001; Kaur, Brennan and Crick 2004). Rv1086 adds one isoprene unit to geranyl diphosphate, generating the 15-carbon product (E,Z-farnesyl diphosphate). Rv2361c then processively adds a further seven isoprene units to E,Z-farnesyl diphosphate to generate the 50-carbon prenyl diphosphate. The crystal structures of the enzymes show that they are closely related, which partly explains how Rv2361 can compensate for the deletion of Rv1086 (Schulbach et al. 2001; Wang et al. 2008). The decaprenyl diphosphate, synthesised de novo at the cytoplasmic side of the cell membrane or released by the polymerisation of PG on the periplasmic face of the membrane, must be dephosphorylated for the transfer of the activated sugar molecule from lipid I. The dephosphorylation of decaprenyl diphosphate to decaprenyl phosphate is performed by membrane-bound pyrophosphatases. In spite of multiple undecaprenyl pyrophosphate phosphatases (Upp) identified in E. coli (BacA, PgpB, YbjG and LpxT) the majority of this activity (75%) is attributed to BacA (UppP); which also confers bacitracin resistance in E. coli (El Ghachi et al. 2005; Bickford and Nick 2013). PgpB and BacA interact with PBP1B (which polymerises PG) and stimulate its activity by dephosphorylation of the released undecaprenyl pyrophosphate (Hernández-Rocamora et al. 2018). The crystal structure of E. coli BacA shows a unique topology raising the possibility of its involvement as a lipid carrier flippase in addition to being a phosphatase that could potentially accept substrates from the cytoplasm as well as the periplasm (Workman, Worrall and Strynadka 2018). Despite the redundancy of the phosphatases, deletion mutants lacking bacA show marked phenotypic effects such as attenuation of virulence in Staphylococcus aureus and Streptococcus pneumoniae in mice models, and impaired biofilm formation in M. smegmatis (Chalker et al. 2000; Röse, Kaufmann and Daugelat 2004). However, the M. tuberculosis homologue of BacA was found to have no role in either acid resistance or in virulence in mice models (Darby et al. 2011). This suggests that there may be other enzymes in M. tuberculosis with decaprenyl pyrophosphate phosphatase activity. The synthesis and recycling of the lipid carrier is essential for the membrane-bound steps of PG synthesis as discussed below and is a verified target for antibiotics such as bacitracin (Siewert and Strominger 1967).
The first membrane step in the biosynthesis of PG involves the phospho-N-acetylmuramoyl-pentapeptide transferase or translocase MraY. MraY catalyses the transfer of the phospho-MurNAc-pentapeptide moiety of UDP-MurNAc-pentapeptide to the acceptor decaprenyl phosphate, yielding MurNAc-(pentapeptide)-pyrophosphorylundecaprenol or lipid I. This proceeds via a covalent intermediate, which is formed upon nucleophilic attack by an aspartate residue on the β-phosphate of the UDP-MurNAc-pentapeptide resulting in the release of UMP (Chung et al. 2013). While E. coli uses the undecaprenyl lipid, M. tuberculosis solely uses the decaprenyl lipid, however, both decaprenyl and heptaprenyl acceptors have been identified for M. smegmatis (Takayama, Schnoes and Semmler 1973). Comprehensive biochemical or structural studies on mycobacterial MraY have not been reported. However, active site mapping of MraY from E. coli and comparison with MraY from other Gram-negative and Gram-positive bacteria revealed five conserved hydrophilic sequences located on the cytoplasmic side of the membrane containing invariant amino acids (Al-Dabbagh et al. 2008). An X-ray crystal structure for MraY from Aquifex aeolicus and a method to quantify MraY activity has provided further insight into the active site residues (Chung et al. 2013). Improvements of the biochemical assay to quantify MraY activity (Siricilla et al. 2014) is a significant asset in the effort to develop antimycobacterial agents targeting this protein.
Following the synthesis of lipid I, the N-acetylglucosamine transferase MurG catalyses the transfer of GlcNAc from UDP-GlcNAc to lipid I, yielding GlcNAc-MurNAc-(pentapeptide)-pyrophosphoryl-decaprenol or lipid II.
MurT and GatD are responsible for the amidation of the d-Glu residue in the peptide stem of both, lipid I and lipid II in Staphylococcus aureus and Streptococcus pneumoniae (Figueiredo et al. 2012; Munch et al. 2012). The arrangement of the murT and gatD genes in an operon is conserved across various bacterial families (Figueiredo et al. 2012) and has also been confirmed in mycobacteria (Maitra et al., unpublished results). Crystal structures of the enzymes from S. aureus and S. pneumoniae reveal that the pair form a stable, functional heterodimer wherein GatD is responsible for the sequestration of the glutamine donor and its subsequent deamination and MurT is a Mur ligase-like protein that allows for the d-iGlu in the stem peptide to be amidated to d-iGln and utilises an ATP in the process (Leisico et al. 2018; Morlot et al. 2018; Noldeke et al. 2018). While GatD is inactive in the absence of MurT, the latter can functionally substitute the role of MurE (Munch et al. 2012; Leisico et al. 2018). Amidation of PG plays a crucial role in PG synthesis and maturation. It was hypothesised that amidation could facilitate the translocation of lipid II across the cytoplasmic membrane as a consequence of the reduction of polarity (Munch et al. 2012). It has also been reported that non-amidated lipid II is an inefficient substrate for transpeptidation reactions in S. pneumoniae (Zapun et al. 2013). Amidated muropeptides are recognized by PknB in M. tuberculosis and through the action of the kinase could regulate cell growth, division and PG turnover (Mir et al. 2011).
The homologous murT/gatD genes in M. tuberculosis are essential for in vitro growth of the pathogen (Sassetti, Boyd and Rubin 2003). The essentiality of MurT/GatD and the requirement for complex formation for activity may be exploited for designing drug scaffolds.
Mycobacterial lipid II also contains amidated mDAP residues (Mahapatra et al. 2005b). In Corynebacterium glutamicum and B. subtilis, ltsA and asnB encode for the amidotransferases responsible for the amidation of mDAP (Levefaudes et al. 2015; Dajkovic et al. 2017). LtsA and AsnB belong to the growing family of glutamine amidotransferases (such as GatD) whose members catalyse amide nitrogen transfer from glutamine to various specific acceptor substrates (mDAP in this case). Most of the investigations into the role of mDAP amidation have focussed on the host immune response modulation of such modification. Nucleotide-binding oligomerisation domain-containing protein 1 (NOD1) is a pattern recognition receptor that recognizes MurNAc-tripeptide fragments containing mDAP and activates the innate immune system (Girardin et al. 2003b). Amidation of mDAP enables evasion of recognition by NOD1 by interacting with residues away from the NOD1-leucine-rich repeat (LRR) recognition region (Roychowdhury, Wolfert and Boons 2005; Wolfert, Roychowdhury and Boons 2007; Vijayrajratnam et al. 2016; Wang et al. 2016). Unlike d-Glu amidation, the modification of mDAP residue does not influence the pattern of insertion of PG precursors into the cell wall or the overall cross-linking levels (Levefaudes et al. 2015; Dajkovic et al. 2017). However, the amidation of the mDAP residues in mycobacteria is essential for the formation of LD-cross-links (Ngadjeua et al. 2018). Additionally, the presence of zones of uncontrolled PG degradation in the ΔasnB mutant of B. subtilis suggests that mDAP amidation controls the activity of hydrolytic enzymes such as autolysins (Dajkovic et al. 2017). Once the modifications have been introduced, lipid II is flipped to the outer face of the cytoplasmic membrane.
The translocation of lipid II across the cytoplasmic membrane has been subject to some controversy, with the role of lipid II flippase having being contested between FtsW and MurJ. The evidence supporting FtsW as the lipid II flippase comes from an assay performed by Mohammadi et al. (Mohammadi et al. 2011) on an in vitro reconstitution system, in which it was observed that FtsW, and not MurJ, was able to translocate fluorescently labelled lipid II across the membrane of bacterial membrane vesicles. It was later suggested by the same research group that lipid II may be translocated via a pore-like structure in FtsW (Mohammadi et al. 2014). Conversely, it has been suggested based on genetic analyses and biochemical assays with RodA, that proteins from the SEDS family may be glycosyltranferases (GTases) rather than lipid II flippases (Meeske et al. 2016; Emami et al. 2017). However, FtsW was not active as GTase in another study in which it was also observed that FtsW, but not MurJ, binds lipid (Leclercq et al. 2017). However, FtsW does in fact have GTase activity, but only when in complex with its cognate penicillin-binding protein (Taguchi et al. 2019).
The initial hypothesis for MurJ as the lipid II flippase was based on its essentiality and the observation that PG synthesis stops and precursors accumulate upon depletion of the murJ gene in E. coli (Inoue et al. 2008; Ruiz 2008). A colicin M-based cellular assay suggested that MurJ might flip lipid II across the membrane (Sham et al. 2014). Native mass spectroscopy showed that MurJ binds lipid II and that this binding is affected by cardiolipin (Bolla et al. 2018). The crystal structures of MurJ from Thermosipho afticanus (Kuk, Mashalidis and Lee 2017) and E. coli (Zheng et al. 2018) confirmed similarity to MOP family of transporters and further rationalised possible mechanisms by which MurJ is likely to transport lipid II across the membrane. However, until there is direct evidence with the mycobacterial protein, the identity of the flippase in the pathogen remains uncertain.
Both PG precursor synthesis and their transport are regulated by PknB-mediated phosphorylation of CwlM (introduced earlier as regulating MurA activity). CwlM exists in two forms within the mycobacterial cell. The non-phosphorylated CwlM is membrane-associated, interacts with MurJ and presumably enables it to transport lipid II (Turapov et al. 2018). The phosphorylated CwlM is cytoplasmic and interacts with FhaA and MurA. The authors hypothesise that on binding to unpolymerised PG precursors in the periplasmic region, PknB undergoes autophosphorylation and in turn phophorylates its substrate CwlM, which dissociates from MurJ thereby reducing the flipping of PG precursors. Direct phosphorylation of MurJ by PknB was also proposed to produce the same outcome.
A number of compound classes bind to extracytoplasmic lipid II and prevent PG polymerisation. Glycopeptides such as vancomycin (MICMtb- 65 µg/mL) bind to d-Ala-d-Ala in lipid II and nascent PG. Antimicrobial peptides such as nisin (MICMtb- > 60 µg/mL), teixobactin (MICMtb- 0.125 µg/mL) and malacidin (MICMtb- not determined) bind primarily to the pyrophosphate moiety of lipid II and inhibit PG synthesis.
PG biosynthesis: linkage in mature PG
Once the precursors are translocated to the periplasmic space, lipid II is used as the substrate for the subsequent polymerisation reactions and the formation of mature PG. The final stages of PG maturation occurs in the periplasm, carried out by transglycosylases and transpeptidases, which are often bifunctional penicilloyl serine transferases or penicillin binding proteins (PBPs) (Crick, Mahapatra and Brennan 2001; Sauvage et al. 2008), resulting in the formation mature of PG chains and peptide with DD-cross-links (Goffin and Ghuysen 1998). The class A and B PBPs synthesize PG; whereas the class C PBPs are PG hydrolases with either DD-endopeptidase or DD-carboxypeptidase activity. Both reactions provide the tetrapeptide substrates for LD-transpeptidases. Class A PBPs (in mycobacteria PBP1 and PBP1A, encoded by ponA1 and ponA2, respectively) consist of bifunctional enzymes with an N-terminal glycosyltransferase and a C-terminal DD-transpeptidase (penicillin-binding) domain (Mahapatra et al. 2000; Bhakta and Basu 2002; Patru and Pavelka 2010). Class B PBPs (in mycobacteria PbpA and PbpB) are monofunctional DD-transpeptidases (Dasgupta et al. 2006; Plocinski et al. 2011). The glycosyltransferase domain catalyses the transfer of the terminal N‐glycolylmuramic acid residue from a nascent glycan chain to N‐acetylglucosamine of lipid II, releasing decaprenol pyrophosphate (Scheffers and Pinho 2005; van Heijenoort 2007). The C‐terminal DD-transpeptidase domain of PonA1 crosslinks the penultimate d‐Ala to the mDAP of another peptide (Filippova et al. 2016). The energy for the DD-transpeptidation reaction stems from the cleavage of the d-Ala-d-Ala peptide bond of the donor peptide resulting in the release of the terminal d-Ala residue (Ghuysen 1991). PonA localises to the poles and septum in mycobacteria and is essential in regulating cell length probably through its interaction with both PknB and the peptidoglycan hydrolase RipA (Hett, Chao and Rubin 2010; Kieser and Rubin 2014). The activities of PonA1 and PonA2 may be altered under different growth conditions as mutants lacking one of the corresponding genes show differential response to cell wall antibiotics (Kieser et al. 2015).
The ld-cross-link was first identified by Wietzerbin in 1974 (Wietzerbin et al. 1974). Its physiological significance, however, was revealed only in 2008 when Lavollay and colleagues discovered that the PG of stationary phase M. tuberculosis almost exclusively contained ld-cross-links (Lavollay et al. 2008). An LD-transpeptidase, LdtMt1, which catalysed the formation of ld-cross-links, was identified and covalent adduct formation with the β-lactamase drugs imipenem, meropenem and ertapenem was demonstrated through mass spectroscopy (Lavollay et al. 2008). Subsequently, a second LD-transpeptidase, LdtMt2 was identified by Gupta and colleagues, and loss of the ldtMt2 gene in M. tuberculosis was shown to result in altered colony morphology, loss of virulence and increased sensitivity to amoxicillin-clavulanate (Gupta et al. 2010). In addition, LdtMt2 is critical for growth in the absence of PonA1 or PonA2, suggesting PBPs and LdtMt2 together support new cell wall synthesis during growth (Kieser et al. 2015).
The influence of LD-cross-linking on mycobacterial physiology and drug susceptibility was further investigated by Baranowski and colleagues, making use of an M. smegmatis strain lacking Ldts as a model (Baranowski et al. 2018). It was observed that in the absence of LD-cross-links, the rigidity of aging cell wall was compromised leading to the formation of spherical blebs. The increased susceptibility of M. tuberculosis to DD-transpeptidase targeting β-lactams was also confirmed and it was demonstrated that inhibiting both DD- and LD-transpeptidases with amoxicillin and meropenem respectively, resulted in synergistic lowering of the MIC (Baranowski et al. 2018).
The two-step reaction of LD-transpeptidases begins with the acylation of the enzyme by the third residue (l-chiral centre) of the tetrapeptide donor, followed by deacylation of this acyl-enzyme intermediate by the third residue (d-chiral centre) of the adjacent acceptor stem resulting in the formation of a LD-peptide linkage. The crystal structure of the extra-membrane domain of LdtMt2 with a bound PG fragment (Erdemli et al. 2012) showed that each monomer consists of two globular domains: an N-terminal domain resembling an immunoglobulin domain, and a C-terminal catalytic domain, consisting of a β sandwich with two mixed β sheets characteristic of the YkuD fold (Bielnicki et al. 2006), with the two domains connected by a short linker molecule. Structural, kinetic and calorimetric data revealed a catalytic mechanism for LdtMt2 in which both acyl-acceptor and acyl-donor substrates enter from the same side to reach the catalytic site (Both et al. 2013; Triboulet et al. 2015; Bhattacharjee et al. 2019).
The PG layer was long regarded as a network with mainly DD-cross-links. However, the type of cross-links in PG can switch to mainly LD-cross-links in response to changes in the environment and growth phase in Mycobacteria (Gupta et al. 2010) and E. coli (Hugonnet et al. 2016). The shift to LD-cross-links allows cells to grow in the presence of most β-lactam antibiotics that target PBPs and not LD-transpeptidases, and the use of tetrapeptide donors by LD-transpeptidases allows for the generation of peptide cross-links in the absence of de novo synthesis. These features might facilitate the repair of damaged PG (Morè et al. 2019) or render PG resistant to hydrolytic enzymes (Lavollay et al. 2008), which could be vital during stationary phase or persistence.
PG transpeptidases are verified but difficult targets for antimicrobial drug discovery as (i) due to their redundancy, many of these enzymes can substitute the function of another and (ii) the drugs most commonly used to target them, the β-lactams, are prone to inactivation by β-lactamases.
Drug development efforts targeting the LD-transpeptidases have primarily focussed on further exploring the potential of carbapenems, particularly meropenem, as Ldt inhibitors. Shortly after the identification of LdtMt1, Hugonnet and colleagues demonstrated that a combination of meropenem and clavulanate was able to kill 13 different XDR strains of M. tuberculosis with MIC values > 1 µg/mL (with one exception at 1.25 µg/mL) (Hugonnet et al. 2009). This built upon previous work demonstrating that clavulanate can act as an irreversible inhibitor of the M. tuberculosis β-lactamase BlaC (Hugonnet and Blanchard 2007).
The direct targeting of LdtMt1 by carbapenems and cephalosporins was investigated by Dubee and colleagues and it was demonstrated that while both classes of β-lactamase are able to form adducts with LdtMt1, cephalosporins do so 7- to 1000-times slower than carbapenems (Dubee et al. 2012). Since then, Kim et al. showed that meropenem acts as a suicide inhibitor of LdtMt2 by inducing conformational changes in the protein; further informing drug design groups about inhibition mechanisms for the protein (Kim et al. 2013).
Recently, in a small retrospective observational case study, Paven and colleagues reported that of 18 patients with XDR-TB who were treated with meropenem-clavulanate, 15 had a ‘successful outcome’ (5 ‘cured’ and 10 ‘completed treatment’), while two patients failed to complete the treatment and one died (Payen et al. 2018). It is also notable that no adverse effects were observed as a result of the treatment, suggesting that with further study this may present a promising route to new β-lactamase-based TB treatment. However, treatment of Mycobacterium bovis BCG with meropenem was also reported to cause a significant increase in endogenous ATP levels and suppression of this ATP spike using drugs such as Bedaquiline attenuated the activity of the former (Shetty and Dick 2018). Therefore, the inclusion of new drugs into combination therapy poses complications of antagonistic activity.
PG biosynthesis: orientation of mature PG
The orientation of PG has been a subject of debate and speculation throughout the past decades. The classical depiction represents PG glycan strands as parallel to the bacterial membrane surface as this provides better flexibility to accommodate cell division (Koch 1998, Koch 2000; Vollmer and Holtje 2004; Scheffers and Pinho 2005; Vollmer 2008), however, others postulate that PG is arranged perpendicular/orthogonal to the membrane (Pink et al. 2000; Dmitriev et al. 2003; Dmitriev, Toukach and Ehlers 2005; Meroueh et al. 2006). Using atomic force microscopy the glycan strands of E. coli were seen running approximately along the short axis of the cell, and the PG was more ordered in rod shaped E. coli sacculi than spheroid shaped sacculi from mutant strains (Turner et al. 2018). Our current understanding of the order of PG glycan strands is largely derived from studies on rod-shaped E. coli and B. subtilis, where new glycan strands are inserted circumferentially forming new PG laterally along the side wall with the help of a protein complex called elongasome (Dominguez-Escobar et al. 2011; Garner et al. 2011; Hussain et al. 2018). Whether a similar order of PG glycan strands is present in mycobacteria is not known, as there are key differences between mycobacterial PG biosynthesis and other rod-shaped bacterial PG structure such as the incorporation of nascent PG at polar ends, the presence of a high proportion of LD-cross-linkages and the absence of MreB. The complexity of the mycobacterial cell wall structure, lack of pure samples and the presence of abundant secondary cell wall polymers make it challenging to determine its architecture.
PG biosynthesis: the dcw gene cluster
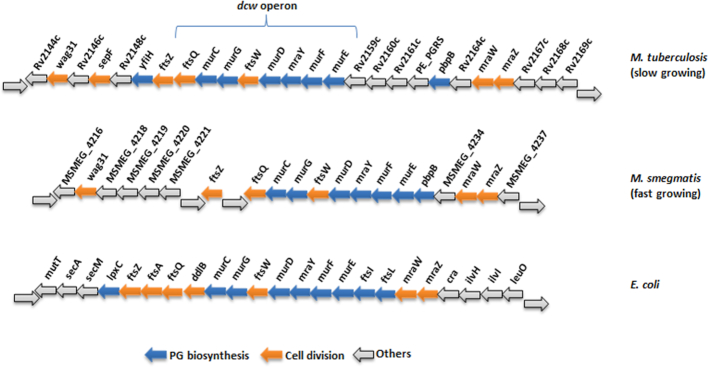
Although most of the genes involved in PG biosynthesis have been identified in E. coli and their mycobacterial orthologues are known, the differences in the structure of PG between the organisms, however, warrants further investigation into the products of these key genes. Since the sequencing the genome of M. tuberculosis (Cole et al. 1998), a plethora of information is available about the pathogen, aiding a more targeted approach towards drug discovery and development. The majority of PG biosynthetic genes in M. tuberculosis are located on the division-cell wall (dcw) cluster along with genes required for cell division (Munshi et al. 2013). This clustering of genes is also observed in E. coli (Eveland, Pompliano and Anderson 1997), with the exception of the Rv2161c-Rv2160c-Rv2159c region and the homologues present only in the slow growing mycobacterial species such as M. tuberculosis (Fig. 4). The clustering of cell division genes (ftsZ, ftsQ and ftsW) and PG biosynthesis genes in the dcw operon are indicative of a complex and spatio-temporal coordination between cell division proteins and cell wall precursor synthesis enzymes (Mingorance, Tamames and Vicente 2004; Vicente et al. 2006; Munshi et al. 2013; Carel et al. 2014).
Figure 4.
Conservation in genomic organization of dcw gene clusters. Here, we compare the differences in the gene content of the dcw clusters of fast and slow growing mycobacteria with that of E. coli.
Degradation and remodelling of PG
PG is a dynamic structure that is constantly synthesised and remodelled. Cell growth, division and septation require the co-ordination of the enzymes required for both PG synthesis and hydrolysis. PG hydrolases (PGH) form a vast group of enzymes including glycosidases, amidases, endopeptidases and carboxypeptidases, which together are capable of fully digesting the PG polymer into soluble fragments. They can act by cleaving bonds within polymeric or soluble fragments of PG (Loessner et al. 2002; Vollmer et al. 2008), and hence may have a periplasmic, membrane-associated or cytoplasmic location (Holtje 1998). Often called autolysins, they are capable of lysing whole cells when deregulated. Due to their selective ability to cleave PG, the hydrolases have been implicated in a plethora of cellular processes including cell growth, PG maturation, PG turnover, cell division, sporulation, resuscitation and pathogenicity (Goodell and Schwarz 1985; Heidrich et al. 2002; Morlot et al. 2010; Lee and Huang 2013). Due to their central role in the processes outlined above these enzymes offer a unique opportunity to alter the permeability of the mycobacterial cell wall and so make M. tuberculosis and related pathogens more susceptible to drug treatment. In support of their importance in regulation of bacterial physiology Machowski et al. recently identified multiple homologues of these PG remodelling genes in M. tuberculosis (Machowski et al. 2014).
Glycosidases such as N-acetyl-β-d-glucosaminidases and N-acetyl-β-d-muramidases cleave the polysaccharide backbone of PG, while N-acetylmuramyl-l-Ala amidases hydrolyse the amide bond between MurNAc and l-Ala of the stem peptide. Lytic transglycosylases are, by virtue of their enzymatic reaction, not ‘hydrolases’ but they are usually discussed amongst the PG hydrolases. Unlike other N-acetyl-β-d-muramidases like lysozyme the lytic transglycosylases catalyse the formation of a 1,6-anhydromuramyl ring by an intramolecular transglycosylation reaction in the MurNAc residue of the released PG fragment. Different forms of endopeptidases and carboxypeptidases are capable of hydrolyzing amide bonds either in the stem peptide or cross-linkages of PG (Scheurwater, Reid and Clarke 2008; Vollmer et al. 2008).
The presence of three major autolysins, N-MurGly-l-Ala amidase, an amino peptidase and an endopeptidase in M. smegmatis, was first reported in 1977 (Kilburn and Best 1977) followed later by partial characterisation of a β-glycosidase from M. phlei (Li et al. 1999).
DacB2 from M. tuberculosis is a DD-carboxypeptidase (Class C PBP) that removes d-Ala residues at position 5 of pentapeptides (Kumar et al. 2012). The overexpression of dacB2 results in altered morphology and defective biofilm formation, while its deletion leads to increased intracellular survival of the mutant in THP-1 cells (Bourai, Jacobs and Narayanan 2012). DacB2 was also reported to possess dd-endopeptidase activity (Baranowski et al. 2018). Thus, it can affect cell morphology by hydrolysing the DD-cross-links as well providing tetrapeptide substrates for LD-transpeptidases by removing the terminal d-Ala residue.
Early investigations by Votyakova (Votyakova et al. 1994) and Mukamolova (Mukamolova et al. 1998b; Mukamolova et al. 1999; Mukamolova et al. 2002) in Micrococcus luteus suggested the presence of a resuscitation promoting factor (Rpf) with lytic transglycosylase activity responsible for the release of PG fragments (Fig. 2). This ground-breaking discovery led to the identification of five Rpfs (rpfA-rpfE) in Mycobacterium spp. (Cole et al. 1998; Mukamolova et al. 1998a; Sutcliffe and Harrington 2004), which significantly enhanced our understanding not only of the remodelling of cell wall PG during daughter cell formation, but also the processes behind dormancy and reactivation of bacilli during active and latent TB. Although all five Rpfs are dispensable for mycobacterial growth, the lack of three of the five rpfs leads to avirulence in mice and an inability to resuscitate from dormant bacilli (Downing et al. 2005). Furthermore, the lack of spontaneous reactivation of the rpf deletion mutants reveals a synergy between them, suggesting that their function cannot be substituted by another enzyme (Downing et al. 2005, Kana et al. 2008). Interestingly, an Rpf-related motif identified in mycobacteriophages is involved in PG degradation to facilitate phage infection during the stationary phase (Piuri and Hatfull 2006). This suggested that the Rpf-related motif was important for the breakdown of PG with extensive ld-cross-links such as those encountered in dormant M. tuberculosis. The Rpfs have detectable sequence homology with lysozymes and lytic transglycosylases (Cohen-Gonsaud et al. 2004; Mukamolova et al. 2006). The C-terminal domain of RfpB from M. tuberculosis comprises a PG binding cleft with two parts. One forms a compact lysozyme-like fold resembling that of c-lysozyme, whereas the other resembles soluble lytic transglycosylase proteins such as E. coli Slt70 (van Asselt, Thunnissen and Dijkstra 1999; Cohen-Gonsaud et al. 2005). Although the catalytic domains in RpfE and RpfC are similar to those in RfpB (Chauviac et al. 2014; Mavrici, Prigozhin and Alber 2014), the substrate-binding site in RpfE is more positively charged than that in RpfB. This suggests that these Rpfs either function optimally at different pH values or cleave different micro-domains of PG.
Due to the network-like structure of PG, its enlargement during cell growth and division requires the synergistic activity of both biosynthetic and hydrolytic enzymes for the incorporation of new glycan strands into the growing PG sacculus. PG hydrolases can destroy the integrity of the sacculus; hence their activity must be controlled in the cell to avoid autolysis. There is mounting evidence that the biosynthetic and hydrolytic enzymes interact with each other to form complexes capable of synergistic breaking and making of bonds in PG and regulating activity. In E. coli, Slt70 (a soluble lytic transglycosylase) forms a complex with PBP 3 (a PG transpeptidase) and PBP 7/8 (a dd-endopeptidase) and both enzymes synergistically degrade PG (Romeis and Holtje 1994). Slt70 also interacts with PBP1B, PBP1C and PBP 2 (von Rechenberg, Ursinus and Holtje 1996). The lytic transglycosylase MltA interacts with PBP1B (a bifunctional synthase) and MipA (MltA-interacting structural protein). MtlB interacts with PBP1B, PBP1C and PBP 3 (von Rechenberg, Ursinus and Holtje 1996; Vollmer, von Rechenberg and Holtje 1999). In Mycobacterium spp. the resuscitation promoting factors RpfB and RpfE interact with the endopeptidase RipA at the septum of dividing bacteria (Hett et al. 2007; Hett and Rubin 2008). An interaction between SltB1 and PBP2 has been observed in P. aeruginosa (Legaree and Clarke 2008; Nikolaidis et al. 2012).
Mycobacterial RipA is a d-Glu-mDAP endopeptidase that cleaves between positions 2 and 3 of the stem peptide (Both, Schneider and Schnell 2011). RipA is essential in M. tuberculosis (Sassetti, Boyd and Rubin 2003) and its depletion in M. smegmatis causes severe growth inhibition (Hett and Rubin 2008). Unregulated, RipA behaves like an autolysin in M. tuberculosis and M. smegmatis, resulting in spherical cells that eventually lyse (Chao et al. 2013). RipA, belonging to NlpC/P60 family, has a Cys-His-Glu catalytic triad residing in the C-terminal catalytic domain, (Ruggiero et al. 2010) and is regulated by proteolytic cleavage by MarP (Botella et al. 2017a). RipA becomes a lethal autolysin in the absence of protein interactions that are necessary to control its proteolytic activation (Chao et al. 2013). RipA-mediated cleavage in PG and its activation is more robust in the fast-growing environmental species M. smegmatis compared to the slow growing M. tuberculosis or M. bovis BCG—presumably a consequence of the varying demands for PG hydrolysis required at differing growth rates. RipB has unique features in its N-terminal segment, which runs across the catalytic core domain and forms a helix instead of the β-hairpin loop seen in the homologous module of RipA (Both, Schneider and Schnell 2011). These enzymes are not essential and so are not attractive drug targets.
N-acetylmuramyl-l-Ala amidases hydrolyse the bond between MurNAc and the peptide portion of the PG and have been implicated in PG degradation and turnover, cell separation, antibiotic resistance and spore formation (Heidrich et al. 2001; Vollmer et al. 2008). PG amidases fall into two groups; one containing the amidase_2 domain and other the amidase_3 zinc-dependent domain (which includes the E. coli AmiA, AmiB and AmiC and B. subtilisCwlB and CwlC enzymes). The first autolysin identified in M. tuberculosis was an amidase known as CwlM (Rv3915), which has since been established to have a regulatory role in PG synthesis and maintenance as mentioned in preceding sections. Another homologue (Rv3717) of the E. coli PG amidases recently identified in M. tuberculosis was found to lack a PG-binding domain and is active on muramyl dipeptide, but not on polymerised PG (Kumar et al. 2013; Prigozhin et al. 2013). The catalytic function identified by the structural analysis of Rv3717 indicates that the catalytic pocket of this enzyme is shaped as a blind tunnel and accommodates only PG monomers, as peptide cross-links in polymerised PG cannot enter the tunnel. It uses an overall positive charge on its surface to bind to its substrate and regulates its activity through an auto-regulatory β-hairpin (Kumar et al. 2013). As this is a cytosolic protein the activity of this enzyme depends on the action of another PG hydrolase cleaving the cross-links of PG fragments released from the cell wall and transported back into the cytoplasm. This indicates a possible involvement for Rv3717 in PG recycling (Prigozhin et al. 2013).
Transport and recycling of degraded PG components
A study of turnover rates in E. coli led to the serendipitous discovery of PG recycling (Goodell and Schwarz 1985). The extent of PG fragments released from the cell wall of growing bacteria, is in the range of 25%–50% in Gram-positive organisms such as B. subtilis (Mauck, Chan and Glaser 1971) and E. coli (Chaloupka and Strnadova 1972; Goodell 1985; Goodell and Schwarz 1985). However, E. coli recycles about 90% of the turnover products, releasing only 0%–5% of the PG material into the culture supernatant. Until recently, it was believed that PG recycling is a characteristic of Gram-negative bacteria only (Herve et al. 2007). However, the identification of orthologues of recycling enzymes in B. subtilis and subsequent muropeptide recovery studies have uncovered evidence of PG recycling in Gram-positive organisms as well (Reith and Mayer 2011; Kluj et al. 2018). Apart from the obvious advantages associated with conservation of resources and energy, PG recycling also plays a significant role in monitoring the integrity of the cell wall, cell signalling and antibiotic resistance via β-lactamase induction (Jacobs et al. 1994). Additionally PG recycling plays a crucial role in the suppression of the innate immune response, by effectively recovering cell wall muropeptides that would otherwise stimulate immunity, via proteins that recognise PG (Boudreau, Fisher and Mobashery 2012; Johnson, Fisher and Mobashery 2013).
Although there is little knowledge about PG recycling in M. tuberculosis, some similarities have been observed with the well-studied system in E. coli. Recycling of PG precursors entails lateral wall and septal PG degradation to form GlcNAc-(1→4)-1,6-anhydro-MurNAc-peptide monomers, GlcNAc-(1→4)-1,6-anhydro-MurNAc dissacharides and free peptides (Goodell 1985). Pulse-chase experiments with tritiated mDAP provided evidence to suggest that many of the cell wall precursors were recycled (Goodell and Schwarz 1985). AmpG from E. coli, also required to induce AmpC (a β-lactamase), was identified as the permease transporting the primary products of the action of lytic transglycosylases on PG into the cytoplasm (Korfmann and Sanders 1989; Jacobs et al. 1994; Cheng and Park 2002). Although Gram-positive organisms lack an AmpG homologue, the presence of muropeptides with various length peptide side chains (tetrapeptides, dipeptides) (Mahapatra et al. 2008), as well as inducible β-lactamases (Flores, Parsons and Pavelka 2005), suggests that an equivalent pathway for the recycling pathway for PG exists in mycobacteria. Once the muropeptides have been transported back to the cytoplasm, various amidases and epimerases, which are non-essential for growth, further process them to products that can be utilised for de novo PG precursor synthesis (Park and Uehara 2008).
Homologues of the enzymes involved in the import, breakdown and recycling of muropeptide in the cytoplasm are yet to be identified in M. tuberculosis. In the preceding section we discussed the various enzymes involved in degradation and remodelling of the cell wall PG. Recent work by Moynihan et al. provided evidence for the uptake of muropeptide fragments in M. tuberculosis and M. bovis BCG; where they show cleavage of stem peptide followed by disaccharide cleavage by LpqI (NagZ) and finally lactyl-ether removal from the MurNAc moiety (Moynihan et al.2019). NagA is involved in PG recycling through the deacetylation of GlcNAc-6P in a number of other organisms, including E. coli, B. subtilis, S. aureus and Streptomyces coelicolor. A putative nagA gene in an operon distinct from other bacterial organisms, has been identified in M. tuberculosis (Ahangar et al. 2018). Mycobacterium tuberculosis nagA is essential and encoded along with genes involved in PG biosynthesis and carbohydrate uptake, indicating that it may play a role in the recycling of GlcNAc6P derived from the breakdown of PG.
Structural characterisation of NagA reveal it to be a dimer, with each unit organised in two domains with active site pockets occurring in domain I (Ahangar et al. 2018). The crystal structure reveals the binding of the GlcNAc-6P substrate to M. smegmatis NagA and indicates that the active site does not undergo any major structural change on binding the substrate. However, two loop regions demonstrate a capacity for conformational flexibility and an importance in facilitation of substrate binding, providing opportunity for designing inhibitor molecules (Ahangar et al. 2018). Biochemical characterization shows a clear preference of NagA for GlcNAc6P over other amino sugar analogues. The stringent recognition of this fragment is hypothesised to ensure the integrity of PG recycling (Ahangar et al. 2018).
The essentiality of nagA makes it a feasible drug target. Eukaryotic homologues of the enzyme have low sequence identity and accommodate larger, glycolyl substituted substrates. This property makes the two homologues distinct, however, small inhibitors acting on M. tuberculosis NagA may be accepted by mammalian homologues thereby giving rise to cytotoxicity (Bergfeld et al. 2012).
Recycling of stem peptides is facilitated by murein peptide ligase (Mpl) in Gram-negative organisms. Mpl ligates di-, tri- and tetrapeptide stems onto UDP-MurNAc in an ATP-dependent step thereby replacing the functions of MurC-F playing an important role in the transition of cell towards stationary phase, as observed in E. coli (Talukder et al. 1996). An mpl homologue has not been identified in Gram positive bacteria or in any mycobacterial species.
Role of PG in cell signalling
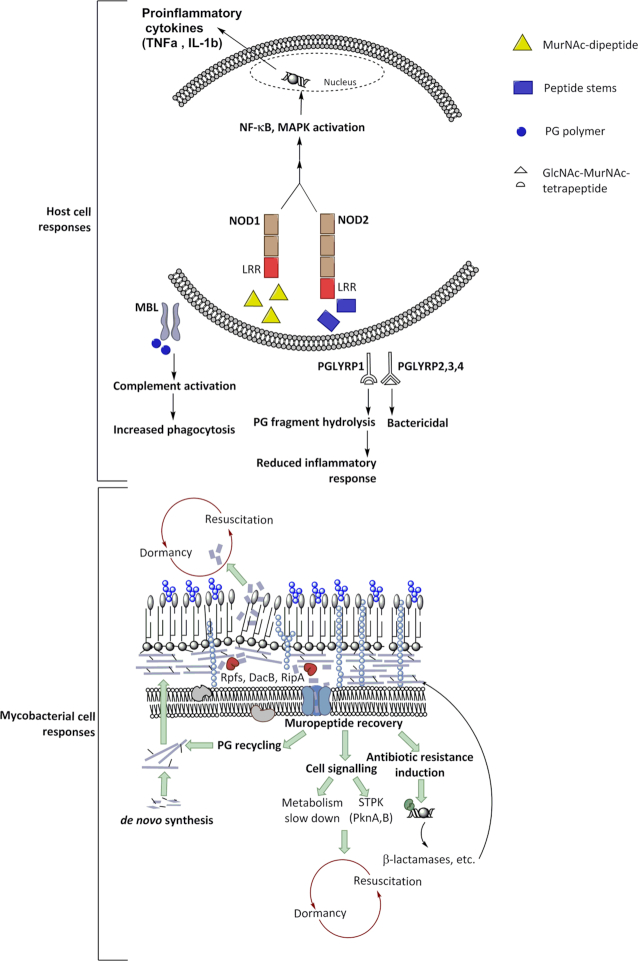
PG fragments released from the cell envelope during growth and cell division play important roles in cellular signalling and inter-bacterial communication (Humann and Lenz 2009) (Fig. 5). Rpfs produce PG fragments that lead to the resuscitation of dormant mycobacteria (Mukamolova et al. 1998a; Mukamolova et al. 1999; Nikitushkin et al. 2013). RpfC and RpfD are restricted to pathogenic and environmental mycobacteria and are secreted into the culture medium, unlike RpfA, RpfB and RpfE, which are membrane-bound (Machowski et al. 2014). This implies that specific PG products are likely to act as messengers, which can act as a signal to reawaken dormant cells (Mukamolova et al. 1998a; Zhang et al. 2001; Machowski et al. 2014). The release of mDAP-containing muropeptides during growth triggers sporulation in B. subtilis and by binding of the serine threonine protein kinase (STPK) PrkC to the PG fragment (Shah et al. 2008). It is possible that in mycobacteria the PG fragments released by Rpfs bind PknB, triggering a cascade of signals leading to resuscitation of the bacteria (Fig. 5). Furthermore, Russell-Golman et al. also showed that mycobacterial double mutants of RpfA and RpfE show a reactivation-deficient and attenuated phenotype (Russell-Goldman et al. 2008), suggesting that Rpfs serve as virulence factors (Romano et al. 2012). In addition, Rpfs induce elevated levels of cytokines in cultures from individuals with latent TB (Riano et al. 2012).
Figure 5.
The role of PG and its breakdown products in signalling. The digestion of PG by hydrolases generates products like muramyl dipeptide and muramyl tripeptide that are key signalling molecules to resuscitate bacterial cells and may induce β-lactamase. PG fragments are encountered by the host immune surveillance systems either extracellularly (e.g. in serum) or are phagocytosed triggering various pro-inflammatory host immune responses.
Although PG has been the subject of intense study over the last number of decades, the changes that occur when a cell enters and exits stationary phase are not well understood. In some bacteria, d-amino acids are incorporated into the PG polymer in the stationary phase, which affects the amount of PG per cell (Lam et al. 2009; Cava et al. 2011). d-Ala inhibits the germination of spores in B. subtilis (Chesnokova et al. 2009). d-Met and d-Leu were the most prominent non-canonical d-amino acids incorporated into the PG during the stationary phase in Vibrio cholerae (Lam et al. 2009).
The ability of a host to recognise bacterial components and trigger an innate immune response is the first line of defence against infection. The fact that eukaryotes do not produce PG has led to the evolution of several strategies to detect PG or PG fragments (Humann and Lenz 2009; Otten et al. 2018). The germline-encoded pattern-recognition receptors (PRRs) are expressed mainly on immune cells and recognise pathogen-associated molecular patterns (PAMPs) and mediate the production of immunoregulatory cytokines such as tumour necrosis factor (TNF) and type I interferons (IFNs) (Medzhitov and Janeway 1997). Toll-like receptors (TLR) are activated by immunomodulatory lipoproteins in mycobacteria such as lipomannan (LM), lipoarabinomannan (LAM), mannosyl capped LAM (ManLAM) or phosphoinositide capped LAM (PILAM) and PG components present on the cell surface or endosome/lysosome membranes (Gilleron, Quesniaux and Puzo 2003; Quesniaux et al. 2004; Singh et al. 2012; Esin et al. 2013; Xie et al. 2013). The cytoplasm of the cell is also under scrutiny for PAMPs that breach the cell membrane. These PAMPs are recognised by constitutively expressed Nod-like receptors (NLR) like NOD, NALP and NAIP. They consist of the NACHT or NOD oligomerisation domain and a motif similar to the microbial sensing leucine-rich repeat (LRR) receptors present in the TLRs. While NOD1 specifically recognises the γ-d-glutamyl-mDAP peptide of PG fragments, NOD2 and NALP3 are activated by intracellular muramyl dipeptides (Mukamolova et al. 1998a; Zhang et al. 2001; Chamaillard et al. 2003a; Girardin et al. 2003a; Chamaillard et al. 2003b; Uehara et al. 2006; Humann and Lenz 2009; Pandey et al. 2009; Killick et al. 2013; Machowski et al. 2014). Thus, any changes in the structure of PG could impair the innate immune responses of the host.
Targeting PG biosynthesis to tackle antibiotic resistance
(R)The ‘Golden Age’ of antibacterial drug discovery appears to be over: since the 1990s there has been a void in the development of new drugs for the treatment of bacterial infections (Silver 2011). More specifically, for TB the current frontline treatment regimen is a cocktail of four drugs (isoniazid, rifampicin, pyrazinamide and ethambutol) that was first introduced in the 1960’s (Zumla, Nahid and Cole 2013). The emergence of MDR- and XDR-TB strains, however, have reinvigorated the search for antimycobacterial agents with novel modes of action (Young et al. 2008). Recent progress in this area includes the approval of Bedaquiline (Sirturo) (Mahajan 2013; Koul et al. 2014) and Delamanid (Deltyba) (Ryan and Lo 2014).
Pathways involved in the biosynthesis of cell wall PG have historically been recognised as important drug targets. This is demonstrated by the fact that a significant number of the antibiotics in current clinical use, primarily β-lactams (e.g. penicillins and cephalosporins) and glycopeptides, act by inhibiting the later stages of PG biosynthesis (Fig. 2). Mycobaterium tuberculosis is considered resistant to many β-lactams, due to a combination of the following factors: the thick, complex and hydrophobic nature of its cell envelope, the presence of one or more active β-lactamases (Nampoothiri et al. 2008) and low-affinity PBPs (Chambers et al. 1995). A number of clinically useful TB drugs, however, do target cell wall processes. These include: (i) the first line anti-TB drug isoniazid which inhibits MA biosynthesis (Zhang et al. 1992), (ii) ethambutol, another frontline drug, which inhibits AG synthesis (Mikusova et al. 1995) and (iii) a second line drug d-cycloserine which is a broad spectrum antibiotic that inhibits PG biosynthesis via inhibition of Alr and Ddl (Zhang 2005). Delamanid, like isoniazid, also targets genes in MA biosynthesis (Ryan and Lo 2014); and Pretomanid (PA-824), an antitubercular drug under regulatory review by the FDA, has also been shown to affect not only respiratory genes, but also transcription of genes responsive to known cell wall inhibitors like isoniazid (Manjunatha, Boshoff and Barry 2009). The efficacy of these drugs highlights the essentiality of the mycobacterial cell wall PG and highlights the opportunity to develop novel chemical entities, which target enzymes involved in the synthesis of the mycobacterial cell envelope (Feng and Barletta 2003; Silver 2006; Prosser and de Carvalho 2013). Further research to identify the unique features of the mycobacterial PG enzymes with respect to their structure, function and regulation can advance the development of narrow-spectrum, targeted antibiotics.
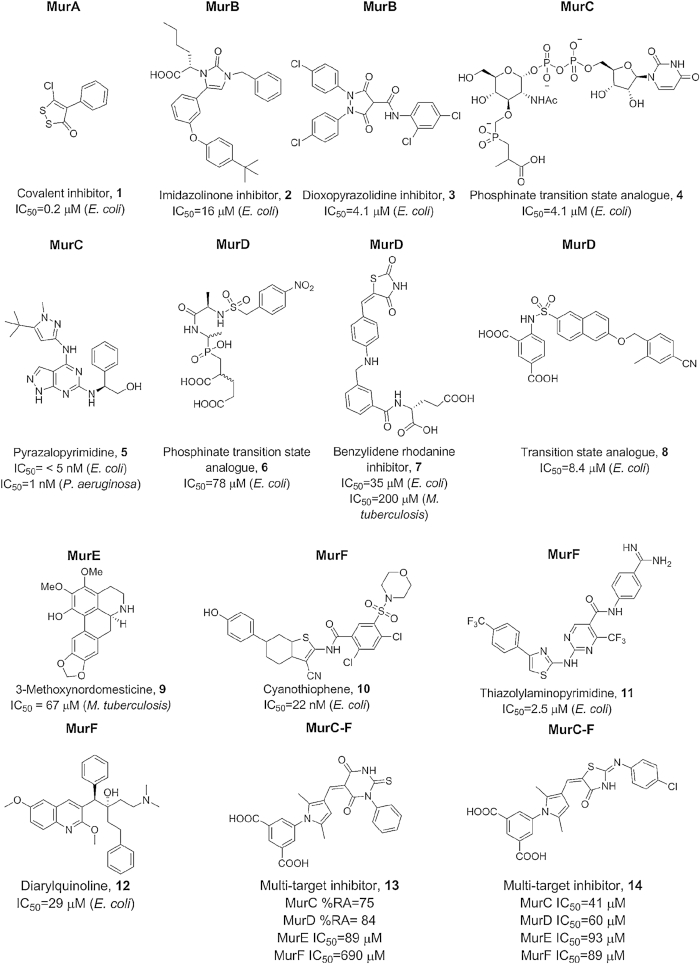
To date the early stages of PG biosynthesis have been poorly exploited as antibacterial targets, and so present a significant opportunity for the development of the next generation of antimycobacterial drugs. In particular, the enzymes involved in the cytoplasmic stage of the PG biosynthetic pathway are currently considered as new targets for drug discovery. This interest, however, has yet to translate to the development of a clinically useful molecule. Although inhibitors against MurA and MurB have been reported previously (Baum et al. 2001; Bronson et al. 2003; Yang et al.2006) (1, 2, 3, Fig. 6.) herein, we will review the progress made to date towards the development of inhibitors targeting the ATP-dependent Mur ligases MurC–F involved in the cytoplasmic phase of PG biosynthesis.
Figure 6.
Structures of representative inhibitors of MurA-F with inhibitory data.
The Mur ligases are found in all clinically significant bacterial pathogens, and so inhibitors would be expected to be broad spectrum. In addition, (i) there are conserved structural and sequence motifs and 3 binding sites in MurC-F against which antimicrobial agents can be designed (e.g. MurE Fig. 3) and (ii) partial inhibition at multiple steps of PG synthesis (various Mur enzymes) compared to inhibition of a single enzyme in the pathway may provide a more efficient route to obtaining bactericidal activity and decrease the likelihood of development of resistance. As a result, pan or multi-target inhibitors are conceivable and these may provide a better approach to obtaining robust antibacterial activity.
Adding to the appeal of the ATP-dependent Mur ligases as targets for drug design, many X-ray crystal structures have been solved for proteins from different bacterial species (Bertrand et al. 1999; Deva et al. 2006; Basavannacharya et al. 2010a). However, only the X-ray crystal structure of MurE from M. tuberculosis is currently available (Basavannacharya et al. 2010a; Basavannacharya et al. 2010b). Homology models of MurC and MurD from M. tuberculosis and MurC-F from M. leprae have been built based on sequence identification with the corresponding homologue from either E. coli or H. influenzae (Anuradha et al. 2010; Arvind et al. 2012; Shanmugam, Anbazhagan and Natarajan 2012); Shanmugam and Natarajan 2012). Docking of native substrates to these models aligned well with known X-ray crystal structures and good homology was observed between conserved binding site residues (Anuradha et al. 2010; Arvind et al. 2012; Shanmugam and Natarajan 2012). In silico docking of naphthalene sulphonamide inhibitors recapitulated the binding mode observed in X-ray crystal structures and highlighted the importance of the Glu moiety as a driver of affinity for MurD (Arvind et al. 2012). One may expect that inhibitors of Mur ligases from other species may also inhibit the mycobacterial enzymes. These models, therefore, provide a good starting point for the design of novel ligands, however, experimental validation of the predicted structures is currently lacking.
The progress made to date in the design of Mur ligase inhibitors has recently been reviewed, although it is noteworthy that activity data on M. tuberculosis are woefully missing (Silver 2011; Perdih et al. 2014; Sangshetti et al. 2017). The inhibitors discovered to date fall primarily into three classes: (i) transition state analogues, which contain for example phosphinate or sulphonamide functional groups which mimic the tetrahedral transition state formed during the enzyme-catalysed reaction (Fig. 3) (Marmor et al. 2001; Reck et al. 2001), (ii) phosphate/diphosphate mimics which typically contain a heterocyclic moiety (e.g. rhodanine, thiazolidindione) (Barreteau et al. 2012) and (iii) inhibitors that bind to the ATP binding site (Hameed P et al. 2014). A number of core functional groups are repeatedly found in inhibitors of MurA to F, particularly those which act as mimics of the tetrahedral transition state or are phosphate bioisosteres. The majority of inhibitors developed to date have arisen from HTS ( in vitro or in silico) followed by structure-guided design, however, natural products have also been the source of a number of inhibitors (Guzman et al. 2010; Guzman et al. 2013b).
The majority of published MurC inhibitors are either phosphinate transition state analogues (4, Fig. 6) (Marmor et al. 2001; Reck et al. 2001) or l-Ala analogues (El Zoeiby et al. 2003). To the best of our knowledge, however, antibacterial activity has not been demonstrated for either class, which is perhaps unsurprising as the above compounds are very polar and are unlikely to be membrane permeable. The most promising MurC inhibitors identified to date were reported in a study by Astra Zeneca in which a series of potent pyrazolopyrimidines were developed (Fig. 6, 5). Compound 5 was proposed to bind into the ATP binding site and encouragingly, also demonstrated bactericidal activity, albeit against efflux deficient mutants (MIC = 3.1 and 1.5 µM against E. coli and P. aeruginosa respectively). Importantly, 5 also demonstrated selectivity by not inhibiting a panel of human kinases. From mutation resistance maps 5 was suggested to be selective for MurC over the other Mur ligases, however, biochemical data to test for inhibition of MurD-F is lacking. Most significantly, the mode of action of the pyrazolopyrimidines was conclusively demonstrated to be as a result of inhibition of MurC. This is a particularly noteworthy advance, as it is the first example in which an antibacterial activity of a Mur ligase inhibitor has been demonstrated to be due to target engagement (Hameed P et al. 2014).
MurC phosphinate transition state analogues are also potent, sub-nanomolar inhibitors of MurD. Replacement of the uridine core with an aryl sulphonamide as a phosphate mimic afforded the simplified inhibitor 6 (Fig. 6), which retained an IC50 in the low micromolar range against MurD and was more amenable to optimisation by medicinal chemistry (Strancar et al.2006). The d-glutamate moiety or conformationally restrained analogues are present in the majority of published inhibitors, as they are key to potent inhibition of MurD (7, 8, Fig. 6.) (Barreteau et al. 2012). Another class of MurD inhibitors feature a benzylidene rhodanine moiety, which occupies the uridine diphosphate binding site (7, Fig. 6). Although 7 is a low micromolar inhibitor of the E. coli enzyme, it inhibits the mycobacterial enzyme with an IC50 of 200 µM, highlighting the challenges associated with the design of broad spectrum agents (Barreteau et al. 2012).
A number of the reported MurD inhibitors also inhibit MurE and, again, contain phosphinate groups (or sulphonamides) that mimic the tetrahedral intermediate formed during the enzyme-catalysed reaction (Tanner et al. 1996; Strancar et al. 2007). Alternative scaffolds have been identified from natural product sources and are of particular interest here due to their demonstrated antimycobacterial activity. For example N-methyl-2-alkenyl-4-quinolone derivatives isolated from Evodia rutaecarpaand aporphine alkaloids isolated from Ocotea macrophylla inhibit the activity of MurE from M. tuberculosis (9, Fig. 6,) and prevent cell growth at micromolar concentrations (Guzman et al. 2010; Guzman et al. 2011; Wube et al. 2011; Guzman et al. 2013b). These natural products also inhibit the growth of a number of other Gram-positive and Gram-negative bacteria, but at much higher concentrations than those required against mycobacteria. It has yet to be demonstrated that the observed antibacterial activity is due to inhibition of MurE only, however, given the relatively low affinity and lipophilicity of these compounds it is unlikely that they are highly selective for their target. The natural product S-leucoxine from Rhodostemonodaphne crenaticupula and related synthetic 1-tetrahydroquinolines have also been demonstrated to inhibit MurE and mycobacterial cell growth, further investigation, however suggested a pleiotropic mechanism of action (Pesnot et al. 2011; Guzman et al. 2015).
A significant number of potent nanomolar inhibitors have been developed for MurF. Abbott Laboratories, for example, developed a series of cyanothiophene ligands with micromolar to nanomolar affinity (10, Fig. 6) (Gu et al. 2004; Stamper et al. 2006) and Johnson & Johnson developed a series of potent thiazolylaminopyrimidine containing inhibitors (11, Fig. 6) (Baum et al. 2006). However, none of the compounds tested demonstrated any significant antibacterial activity. Finally, the identification of diarylquinolines as low micromolar inhibitors of MurF led to the development of 12 (Fig. 6), which exhibited an MIC of 8 µg/mL against a number of bacterial species (Enterococcus faecalis, E. faecium and S. aureus). Compound 12 binds MurF in the presence of ATP and so is likely to occupy the substrate binding site. In addition, treatment of LPS deficient E. coli with 12 resulted in the accumulation of the substrate of MurF, however, over expression of MurF did not significantly affect the MIC. Hence, although the diarlyquinolines represent a promising start point for the development of novel antibacterial agents, the mechanism of action warrants further examination.
Strategies for the development of multi-target inhibitors of the Mur ligases fall in to two categories: (i) ligands that mimic the shared tetrahedral intermediate of the Mur ligases and (ii) ligands designed to target the ATP-binding site, which has the greatest sequence and structural homology amongst the enzymes. Alternatively, for dual target inhibitors, the substrate binding sites of MurC/D and MurE/F are likely to have sufficient similarity to allow the design of dual inhibitors. The majority of efforts to date have focussed on the first strategy leading to benzene-1,3-dicarboxylic acid 2,5-dimethylpyrrole derivatives (Fig. 6, 13 & 14) (Perdih et al. 2014) and napthyl tetronic acids (Mansour et al. 2007) as first multi-target lead compounds. To evaluate the potential of these Mur ligase inhibitors as inhibitors of the mycobacterial enzymes, we performed a docking study (Autodock Vina) (Trott and Olson 2010), using a select number of inhibitors and MurE (pdb code: 2XJA) from M. tuberculosis as a model system. The pyrazalopyrimidine 5 was designed to inhibit MurC from E. coli (Deva et al. 2006). Docking of 5 to MurE from M. tuberculosis resulted in the prediction of two major binding modes. In the first binding mode there is good overlap between the pyrazolpyrimidine ring and the adenosine ring of ATP (as observed X-ray crystal structure) allowing a π-π stacking interaction with Tyr343 (an aromatic residue at this position is highly conserved amongst the Mur ligases); the pyrazole ring is predicted to occupy the phosphate binding site and forms a H-bonding interaction with Thr158 (part of the conserved diphosphate binding motif) (Fig. 7). In the second binding mode the pyrazole ring forms a π-π stacking interaction with Tyr343, the pyrazolopyrimidine moiety is predicted to form polar contacts with two key residues (Asn347 and Thr158) and the hydroxyl group interacts with residues in the highly conserved phosphate binding loop (Gly156, Lys157, Thr158) (Fig. 7B). Given the recapitulation of key interactions formed by ATP within the binding site and similarities with the binding mode predicted by Hameed et al. it is feasible that 5 and analogues thereof may provide a starting points for the development of inhibitors for the mycobacterial Mur ligases (Hameed P et al. 2014).
Figure 7.
Representative predictive binding pockets for selected compounds to MurE (PDB code 2XJA) from M. tuberculosis (overlaid with ATP or UAG as appropriate, shown in lines) (A), Compound 5, predicted binding affinity −9.2 kcal/mol (B), Compound 5, predicted binding affinity −8.9 kcal/mol (C), Compound 8, predicted binding affinity -7.8 kcal/mol (D) Compound 8, predicted binding affinity −7.2 kcal/mol (E) Compound 14, predicted binding affinity −6.8 kcal/mol.
The benzylidene rhodanine inhibitor 7 was designed as an inhibitor of MurD from E. coli (Zidar et al. 2010). In the X-ray crystal structure 7 was found to span the substrate binding site with the rhodanine heterocycle acting as a diphosphate mimic. This binding mode is recapitulated when docked into MurE from M. tuberculosis, however, because of the larger binding site the rhodanine heterocycle is predicted to bind in the hydrophobic pocket occupied by the uracil moiety of UAG and not to the diphosphate binding site as in E. coli MurD.
Transition state analogue, 8, another inhibitor of MurD, which contains a conformationally restrained Glu analogue and a sulphonamide moiety as a transition state analogue, was also docked to MurE. This inhibitor was predicted to span the substrate binding site and two alternate binding modes were predicted: (i) in which the Glu mimic occupied the uracil binding site and the sulphonamide the diphosphate binding site and (ii) in which the cyanobenzyl functional group occupied the uracil binding site and the sulphonamide and Glu mimic mapped well onto the observed conformation of UDP-MurNac-peptide in the X-ray crystal structure of MurE (Fig. 7C, D). However, it is unlikely that the sulphonamide moiety will be able to bind in a similar conformation to the tetrahedral transition state due to the larger and more open binding site of MurE in comparision to MurD. It is important to note that the N-terminal domain exhibits the greatest sequence and structural variation i.e. the N-terminal domain in MurC/D is significantly different to that in MurE/F. This domain is also more flexible and undergoes significant conformational changes on ligand binding; this flexibility is required to enable the Mur ligase to accommodate the growing peptide chain of the PG precursor (Basavannacharya et al. 2010b). As docking experiments are based solely on and restricted by the structures/conformations available, these need to be integrated with in vitro experiments such as enzyme inhibition kinetics to gain confidence in the targeting of inhibitors.
A related set of compounds designed as multi-target Mur ligase inhibitors are compounds 13(a dual inhibitor of MurD/E) and 14(an inhibitor of MurC-F). These were docked against MurE from M. tuberculosis (Tomasic et al. 2010; Basavannacharya et al. 2010a; Perdih et al. 2014). For 13, as expected, the dihydopyrimidine moiety was predicted to act as a diphosphate mimic and the phenyl ring to occupy the uracil binding site. The remainder of the inhibitor maps well onto the conformation of UAG observed in the X-ray crystal structure ( 13 is, however, truncated compared to UAG and so does not span the entire binding site). 14, which is a moderate inhibitor of MurC-F, is predicted to adopt a similar conformation within the substrate binding site with the rhodanine heterocycle again acting as a diphosphate mimic and the chlorophenyl moiety occupying the uracil binding site. The increased flexibility in 14, compared to its precursor 13, allows the heterocycle to form polar contacts with key residues in the diphosphate binding loop (Gly83, Ser84). Interactions with key residues in the substrate binding site are picked up by the isophthalic acid moiety (Arg230, Ser222, Thr195 and Thr196) (Fig. 7E). The recapitulation of key interactions between substrate and protein supports the conclusion that these ligands likely bind to and inhibit mycobacterial MurE, although the inherent flexibility of the Mur ligases is likely to complicate this picture. Experimental evidence is needed to confirm inhibition of mycobacterial Mur ligases by any of the inhibitors discussed here.
As can be seen from this survey of the literature, much progress towards the development of potent inhibitors of the Mur ligases has been made. However, the translation from effective enzyme inhibition to antibacterial action is a significant challenge. Membrane permeability and active efflux are likely to present the biggest hurdles and many inhibitors developed to date are very polar, particularly the phosphinate transition state analogues and amino acid derivatives, making them unlikely to easily traverse the membrane (Jarlier and Nikaido 1994). For other classes of inhibitors, the explanation is less clear.
It is also important to highlight that for those compounds with a demonstrated antibacterial activity there is often no evidence for the observed bactericidal effect being caused by inhibition of a Mur ligase (Silver 2011). The IC50/MIC data in conjunction with molecular modelling or X-ray crystallography, would significantly inform the design of next generation of inhibitors. The study by Barreteau et al. (Barreteau et al. 2012), which evaluated a selection of inhibitors designed to target MurD from E. coli against orthologues from different bacteria is a step in this direction.
Many of the inhibitors discussed contain what would typically be considered undesirable functional groups, for example rhodanine and thiazolidindione moieties are notorious pan assay interference compounds (PAINS) (Baell 2010; Baell and Holloway 2010). These compounds often prove to be not amenable to optimisation, preventing the progression of hit/lead molecules. The pitiful success of HTS as a source of novel antibacterial agents has been the subject of a comprehensive commentary in the literature (Tomašić and Mašič 2012). Hits derived from HTS campaigns are typically quite lipophilic, whereas antibiotics are typically larger and more polar than drugs designed for human targets (Payne et al. 2007). It is, therefore, questionable whether the libraries amassed by the pharmaceutical industry are optimal for antibacterial drug discovery.
Fragment-based drug discovery may offer a viable alternative (Hajduk and Greer 2007). This approach employs libraries of low molecular weight compounds or fragments, these could include commercial fragment libraries, repurposed kinase focussed libraries to target the ATP binding site and libraries of phosphate bioisosteres (Huang et al. 2009; Volkamer et al. 2015). The fragments are screened against the protein targets of interest using a range of biophysical assays which have a much lower false positive hit rate compared to the biochemical assays typically used in HTS screens (Schulz and Hubbard 2009). One of the major advantages of this approach, and which is particularly relevant to antimicrobial drug discovery, is that due to the nature of the hits identified, i.e. low molecular weight, low-affinity and high-ligand efficiency, one can exert greater control over the physiochemical properties of the inhibitor during the process of hit to lead development (Hajduk 2006). The process of fragment optimisation is typically design intensive and structure guided, however, given the current lack of structural information for the mycobacterial enzymes, in situ optimisation using kinetic target guided synthesis has great potential and would allow rapid exploration of the protein binding landscape and the inherent flexibility of the Mur ligases (Hajduk 2006).
It is clear from the progress to date that to obtain anti-tubercular agents that target the Mur ligases a number of hurdles need to be overcome. As a result, further exploration of the Mur ligases as drug targets and the mechanisms of the inhibitors developed to date are necessary if therapeutically useful molecules are to be developed.
CONCLUSIONS AND FUTURE OUTLOOK
Due to the protective role of PG and its absence in humans, its biosynthetic pathway remains an excellent therapeutic target (Strancar et al. 2007; Zawadzke et al. 2008; Tomasic et al. 2010). Encouragingly, technologies for high throughput screening against these targets have advanced (Kristan et al. 2009; Turk et al. 2009; Guzman et al. 2013a). We would hope that these developments and the new insights into the molecular mechanisms of cell wall synthesis could lead to the development of targeted anti-mycobacterials.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Professor Simon Gibbons and Professor Timothy D. McHugh for their helpful suggestions. SB would like to thank MRC, UK for a New Investigators Research Grant (GO801956). AM and LTM would like to thank the Wellcome Trust (108877/Z/15/Z) for their PhD studentship. WV received support from the BBSRC (BB/P001289/1).
The authors acknowledge that the work of many eminent researchers could not be cited in the review due to limited space. We are grateful for their collective research contributing to advances in technology and our understanding of the mycobacterial cell envelope.
Contributor Information
Arundhati Maitra, Mycobacteria Research Laboratory, Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, Malet Street, London WC1E 7HX, UK.
Tulika Munshi, Mycobacteria Research Laboratory, Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, Malet Street, London WC1E 7HX, UK.
Jess Healy, Department of Pharmaceutical and Biological Chemistry, UCL School of Pharmacy, 29–39 Brunswick Square, London WC1N 1AX, UK.
Liam T Martin, Mycobacteria Research Laboratory, Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, Malet Street, London WC1E 7HX, UK.
Waldemar Vollmer, The Centre for Bacterial Cell Biology, Institute for Cell and Molecular Biosciences, Newcastle University, Richardson Road, Newcastle upon Tyne, NE2 4AX, UK.
Nicholas H Keep, Mycobacteria Research Laboratory, Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, Malet Street, London WC1E 7HX, UK.
Sanjib Bhakta, Mycobacteria Research Laboratory, Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, Malet Street, London WC1E 7HX, UK.
Conflict of interest. None declared.
REFERENCES
- Ahangar MS, Furze CM, Guy CSet al.. Structural and functional determination of homologs of the Mycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA). J Biol Chem. 2018;293:9770–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dabbagh B, Henry X, Ghachi MEet al.. Active Site Mapping of MraY, a Member of the Polyprenyl-phosphate N-Acetylhexosamine 1-Phosphate Transferase Superfamily, Catalyzing the First Membrane Step of Peptidoglycan Biosynthesis†. Biochemistry. 2008;47:8919–28. [DOI] [PubMed] [Google Scholar]
- Aldridge BB, Fernandez-Suarez M, Heller Det al.. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335:100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Eveland SS, Onishi HRet al.. Kinetic mechanism of the Escherichia coli UDPMurNAc-tripeptide D-alanyl-D-alanine-adding enzyme: use of a glutathione S-transferase fusion. Biochemistry. 1996;35:16264–9. [DOI] [PubMed] [Google Scholar]
- Anuradha CM, Mulakayala C, Babajan Bet al.. Probing ligand binding modes of Mycobacterium tuberculosis MurC ligase by molecular modeling, dynamics simulation and docking. J Mol Model. 2010;16:77–85. [DOI] [PubMed] [Google Scholar]
- Arvind A, Kumar V, Saravanan Pet al.. Homology modeling, molecular dynamics and inhibitor binding study on MurD ligase of Mycobacterium tuberculosis. Interdiscip Sci, Comput Life Sci. 2012;4:223–38. [DOI] [PubMed] [Google Scholar]
- Awasthy D, Bharath S, Subbulakshmi Vet al.. Alanine racemase mutants of Mycobacterium tuberculosis require D-alanine for growth and are defective for survival in macrophages and mice. Microbiology. 2012;158:319–27. [DOI] [PubMed] [Google Scholar]
- Babajan B, Chaitanya M, Rajsekhar Cet al.. Comprehensive structural and functional characterization of Mycobacterium tuberculosis UDP-NAG enolpyruvyl transferase (Mtb-MurA) and prediction of its accurate binding affinities with inhibitors. Interdiscip Sci, Comput Life Sci. 2011;3:204–16. [DOI] [PubMed] [Google Scholar]
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–40. [DOI] [PubMed] [Google Scholar]
- Baell JB. Observations on screening-based research and some concerning trends in the literature. Future Med Chem. 2010;2:1529–46. [DOI] [PubMed] [Google Scholar]
- Baranowski C, Welsh MA, Sham L-Tet al.. Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife. 2018;7:e37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau H, Sosic I, Turk Set al.. MurD enzymes from different bacteria: evaluation of inhibitors. Biochem Pharmacol. 2012;84:625–32. [DOI] [PubMed] [Google Scholar]
- Basavannacharya C, Moody PR, Munshi Tet al.. Essential residues for the enzyme activity of ATP-dependent MurE ligase from Mycobacterium tuberculosis. Protein Cell. 2010b;1: 1011–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavannacharya C, Robertson G, Munshi Tet al.. ATP-dependent MurE ligase in Mycobacterium tuberculosis: biochemical and structural characterisation. Tuberculosis (Edinb). 2010a;90:16–24. [DOI] [PubMed] [Google Scholar]
- Baum EZ, Crespo-Carbone SM, Abbanat Det al.. Utility of muropeptide ligase for identification of inhibitors of the cell wall biosynthesis enzyme MurF. Antimicrob Agents Chemother. 2006;50:230–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum EZ, Montenegro DA, Licata Let al. Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme. Antimicrob. Agents Chemother. 2001; 45:3182–8.. 11600375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Sander P. Mycobacterium tuberculosis lipoproteins in virulence and immunity–fighting with a double‐edged sword. FEBS Lett. 2016;590:3800–19. [DOI] [PubMed] [Google Scholar]
- Benson TE, Marquardt JL, Marquardt ACet al.. Overexpression, purification, and mechanistic study of UDP-N-acetylenolpyruvylglucosamine reductase. Biochemistry. 1993;32:2024–30. [DOI] [PubMed] [Google Scholar]
- Bergfeld AK, Pearce OM, Diaz SLet al.. Metabolism of vertebrate amino sugars with N-glycolyl groups: elucidating the intracellular fate of the non-human sialic acid N-glycolylneuraminic acid. J Biol Chem. 2012;287:28865–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JA, Auger G, Martin Let al.. Determination of the MurD mechanism through crystallographic analysis of enzyme complexes. J Mol Biol. 1999;289:579–90. [DOI] [PubMed] [Google Scholar]
- Bhakta S, Basu J. Overexpression, purification and biochemical characterization of a class A high-molecular-mass penicillin-binding protein (PBP), PBP1* and its soluble derivative from Mycobacterium tuberculosis. Biochem J. 2002;361:635–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee N, Triboulet S, Dubée Vet al.. Negative impact of carbapenem methylation on the reactivity of β-lactams for cysteine acylation revealed by quantum calculations and kinetic analyses. Antimicrob Agents Chemother. 2019;AAC. e02039-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford JS, Nick HS. Conservation of the PTEN catalytic motif in the bacterial undecaprenyl pyrophosphate phosphatase, BacA/UppP. Microbiology. 2013;159:2444–55. [DOI] [PubMed] [Google Scholar]
- Bielnicki J, Devedjiev Y, Derewenda Uet al.. B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins. 2006;62:144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolla JR, Sauer JB, Wu Det al.. Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ. Nature Chem. 2018;10:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella H, Vaubourgeix J, Lee MHet al.. Mycobacterium tuberculosis protease MarP activates a peptidoglycan hydrolase during acid stress. EMBO J. 2017a;36:536–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella H, Yang G, Ouerfelli Oet al.. Distinct spatiotemporal dynamics of peptidoglycan synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. MBio. 2017b;8:e01183–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both D, Schneider G, Schnell R. Peptidoglycan remodeling in Mycobacterium tuberculosis: comparison of structures and catalytic activities of RipA and RipB. J Mol Biol. 2011;413:247–60. [DOI] [PubMed] [Google Scholar]
- Both D, Steiner EM, Stadler Det al.. Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis. Acta Crystallogr Sec D, Biol crystallogr. 2013;69: 432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau MA, Fisher JF, Mobashery S. Messenger functions of the bacterial cell wall-derived muropeptides. Biochemistry. 2012;51:2974–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourai N, Jacobs WR Jr., Narayanan S. Deletion and overexpression studies on DacB2, a putative low molecular mass penicillin binding protein from Mycobacterium tuberculosis H(37)Rv. Microb Pathog. 2012;52: 109–16. [DOI] [PubMed] [Google Scholar]
- Boutte CC, Baer CE, Papavinasasundaram Ket al.. A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. eLife. 2016;5:e14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. [DOI] [PubMed] [Google Scholar]
- Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb). 2003;83:91–7. [DOI] [PubMed] [Google Scholar]
- Bronson JJ, DenBleyker KL, Falk PJet al. Discovery of the first antibacterial small molecule inhibitors of MurB. Bioorg. Med. Chem. Lett. 2003;13::873–5.. 12617911 [DOI] [PubMed] [Google Scholar]
- Brown ED, Vivas EI, Walsh CTet al.. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol. 1995;177: 4194–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning JB, Murillo AC, Chacon Oet al.. Structure of the Mycobacterium tuberculosis D-alanine:D-alanine ligase, a target of the antituberculosis drug D-cycloserine. Antimicrob Agents Chemother. 2011;55:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carel C, Nukdee K, Cantaloube Set al.. Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum. PLoS One. 2014;9:e97148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam Het al.. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30:3442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker AF, Ingraham KA, Lunsford RDet al.. The bacA gene, which determines bacitracin susceptibility in Streptococcus pneumoniae and Staphylococcus aureus, is also required for virulence. Microbiology. 2000;146:1547–53. [DOI] [PubMed] [Google Scholar]
- Chaloupka J, Strnadova M. Turnover of murein in a diaminopimelic acid dependent mutant of Escherichia coli. Folia Microbiol (Praha). 1972;17:446–55. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Girardin SE, Viala Jet al.. Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell Microbiol. 2003a;5:581–92. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Yet al.. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003b;4:702–7. [DOI] [PubMed] [Google Scholar]
- Chambers HF, Moreau D, Yajko Det al.. Can penicillins and other beta-lactam antibiotics be used to treat tuberculosis? Antimicrob Agents Chemother. 1995;39:2620–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MC, Kieser KJ, Minami Set al.. Protein complexes and proteolytic activation of the cell wall hydrolase RipA regulate septal resolution in mycobacteria. PLoS Pathog. 2013;9:e1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauviac FX, Robertson G, Quay DHet al.. The RpfC (Rv1884) atomic structure shows high structural conservation within the resuscitation-promoting factor catalytic domain. Acta Crystallogr F Struct Biol Commun. 2014;70:1022–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Park JT. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J Bacteriol. 2002;184:6434–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova ON, McPherson SA, Steichen CTet al.. The spore-specific alanine racemase of Bacillus anthracis and its role in suppressing germination during spore development. J Bacteriol. 2009;191:1303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BC, Zhao J, Gillespie RAet al.. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Gonsaud M, Barthe P, Bagneris Cet al.. The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes. Nat Struct Mol Biol. 2005;12:270–3. [DOI] [PubMed] [Google Scholar]
- Cohen-Gonsaud M, Keep NH, Davies APet al.. Resuscitation-promoting factors possess a lysozyme-like domain. Trends Biochem Sci. 2004;29:7–10. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill Jet al.. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. [DOI] [PubMed] [Google Scholar]
- Craggs PD, Mouilleron S, Rejzek Met al.. The mechanism of acetyl transfer catalyzed by Mycobacterium tuberculosis GlmU. Biochemistry. 2018;57:3387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick DC, Mahapatra S, Brennan PJ. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology. 2001;11:107R–18R. [DOI] [PubMed] [Google Scholar]
- Crick DC, Schulbach MC, Zink EEet al.. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol. 2000;182:5771–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Tesson B, Chauhan Set al.. Hydrolysis of peptidoglycan is modulated by amidation of meso-diaminopimelic acid and Mg(2+) in Bacillus subtilis. Mol Microbiol. 2017;104:972–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–76. [DOI] [PubMed] [Google Scholar]
- Darby CM, Venugopal A, Ehrt Set al.. Mycobacterium tuberculosis gene Rv2136c is dispensable for acid resistance and virulence in mice. Tuberculosis. 2011;91:343–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Datta P, Kundu Met al.. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006;152:493–504. [DOI] [PubMed] [Google Scholar]
- David S. Synergic activity of d-cycloserine and β-chloro-d-alanine against Mycobacterium tuberculosis. J Antimicrob Chemother. 2001;47:203–6. [DOI] [PubMed] [Google Scholar]
- DeJesus MA, Gerrick ER, Xu Wet al.. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. MBio. 2017;8:e02133–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementin S. Etude du mécanisme réactionnel des Mur synthétases, enzymes impliquées dans la biosynthèse du peptidoglycane bactérien. 2001.
- den Blaauwen T, de Pedro MA, Nguyen-Disteche Met al.. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–44. [DOI] [PubMed] [Google Scholar]
- De Smet KA, Kempsell KE, Gallagher Aet al.. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology. 1999;145:3177–84. [DOI] [PubMed] [Google Scholar]
- Deva T, Baker EN, Squire CJet al.. Structure of Escherichia coli UDP-N-acetylmuramoyl:L-alanine ligase (MurC). Acta crystallographica Section D, Biological crystallography. 2006;62:1466–74. [DOI] [PubMed] [Google Scholar]
- Dmitriev B, Toukach F, Ehlers S. Towards a comprehensive view of the bacterial cell wall. Trends Microbiol. 2005;13:569–74. [DOI] [PubMed] [Google Scholar]
- Dmitriev BA, Toukach FV, Schaper KJet al.. Tertiary structure of bacterial murein: the scaffold model. J Bacteriol. 2003;185:3458–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Escobar J, Chastanet A, Crevenna AHet al.. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–8. [DOI] [PubMed] [Google Scholar]
- Domínguez-Escobar J, Chastanet A, Crevenna AHet al.. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–8. [DOI] [PubMed] [Google Scholar]
- Downing KJ, Mischenko VV, Shleeva MOet al.. Mutants of Mycobacterium tuberculosis lacking three of the five rpf-like genes are defective for growth in vivo and for resuscitation in vitro. Infect Immun. 2005;73:3038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubee V, Triboulet S, Mainardi JLet al.. Inactivation of Mycobacterium tuberculosis L,D-Transpeptidase Ldt(Mt1) by Carbapenems and Cephalosporins. Antimicrob Agents Chemother. 2012;56:4189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Brown JR, Sylvester DRet al.. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J Bacteriol. 2000;182:4146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziadek B, Brzostek A, Grzybowski Met al.. Mycobacterium tuberculosis AtsG (Rv0296c), GlmU (Rv1018c) and SahH (Rv3248c) Proteins Function as the Human IL-8-Binding Effectors and Contribute to Pathogen Entry into Human Neutrophils. PLoS One. 2016;11:e0148030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghachi M, Derbise A, Bouhss Aet al.. Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. J Biol Chem. 2005;280:18689–95. [DOI] [PubMed] [Google Scholar]
- El Zoeiby A, Sanschagrin F, Darveau Aet al.. Identification of novel inhibitors of Pseudomonas aeruginosa MurC enzyme derived from phage-displayed peptide libraries. J Antimicrob Chemother. 2003;51:531–43. [DOI] [PubMed] [Google Scholar]
- Emami K, Guyet A, Kawai Yet al.. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol. 2017;2:16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eniyan K, Dharavath S, Vijayan Ret al.. Crystal structure of UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) from Mycobacterium tuberculosis. Biochimica et biophysica acta Proteins and proteomics. 2018;1866:397–406. [DOI] [PubMed] [Google Scholar]
- Eniyan K, Kumar A, Rayasam GVet al.. Development of a one-pot assay for screening and identification of Mur pathway inhibitors in Mycobacterium tuberculosis. Sci Rep. 2016;6:35134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdemli SB, Gupta R, Bishai WRet al.. Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of L,D-transpeptidase 2. Structure. 2012;20:2103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. L-form bacteria, cell walls and the origins of life. Open biology. 2013;3:120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esin S, Counoupas C, Aulicino Aet al.. Interaction of Mycobacterium tuberculosis cell wall components with the human natural killer cell receptors NKp44 and Toll-like receptor 2. Scand J Immunol. 2013;77:460–9. [DOI] [PubMed] [Google Scholar]
- Eveland SS, Pompliano DL, Anderson MS. Conditionally lethal Escherichia coli murein mutants contain point defects that map to regions conserved among murein and folyl poly-gamma-glutamate ligases: identification of a ligase superfamily. Biochemistry. 1997;36:6223–9. [DOI] [PubMed] [Google Scholar]
- Falk PJ, Ervin KM, Volk KSet al.. Biochemical evidence for the formation of a covalent acyl-phosphate linkage between UDP-N-acetylmuramate and ATP in the Escherichia coli UDP-N-acetylmuramate:L-alanine ligase-catalyzed reaction. Biochemistry. 1996;35:1417–22. [DOI] [PubMed] [Google Scholar]
- Feng Z, Barletta RG. Roles of Mycobacterium smegmatis D-alanine:D-alanine ligase and D-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor D-cycloserine. Antimicrob Agents Chemother. 2003;47:283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo TA, Sobral RG, Ludovice AMet al.. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog. 2012;8:e1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova EV, Kieser KJ, Luan CHet al.. Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin‐binding protein PonA1 reveal potential mechanisms of antibiotic resistance. FEBS J. 2016;283: 2206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores AR, Parsons LM, Pavelka MS Jr. Characterization of novel Mycobacterium tuberculosis and Mycobacterium smegmatis mutants hypersusceptible to beta-lactam antibiotics. J Bacteriol. 2005;187:1892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrellad MA, Klepp LI, Gioffré Aet al.. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Heredia A, Pohane AA, Melzer ESet al.. Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. eLife. 2018;7:e37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang Wet al.. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen JM. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. [DOI] [PubMed] [Google Scholar]
- Gilleron M, Quesniaux VF, Puzo G. Acylation state of the phosphatidylinositol hexamannosides from Mycobacterium bovis bacillus Calmette Guerin and Mycobacterium tuberculosisH37Rv and its implication in Toll-like receptor response. J Biol Chem. 2003;278:29880–9. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala Jet al.. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003a;278:8869–72. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Travassos LH, Herve Met al.. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003b;278:41702–8. [DOI] [PubMed] [Google Scholar]
- Glauner B, Holtje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–95. [PubMed] [Google Scholar]
- Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell EW, Schwarz U. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J Bacteriol. 1985;162:391–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell EW. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Gawronski JD, DeJesus MAet al.. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz AE, De Sousa-d'Auria C, McNeil MRet al.. Assembling of the Mycobacterium tuberculosis cell wall core. J Biol Chem. 2016;291:18867–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Lavollay M, Mainardi JLet al.. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med. 2010;16:466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YG, Florjancic AS, Clark RFet al.. Structure-activity relationships of novel potent MurF inhibitors. Bioorganic & medicinal chemistry letters. 2004;14:267–70. [DOI] [PubMed] [Google Scholar]
- Guzman JD, Evangelopoulos D, Gupta Aet al.. Antimycobacterials from lovage root (Ligusticum officinale Koch). Phytotherapy research: PTR. 2013b;27:993–8. [DOI] [PubMed] [Google Scholar]
- Guzman JD, Evangelopoulos D, Gupta Aet al.. Antitubercular specific activity of ibuprofen and the other 2-arylpropanoic acids using the HT-SPOTi whole-cell phenotypic assay. BMJ open. 2013a;3:e002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman JD, Gupta A, Evangelopoulos Det al.. Anti-tubercular screening of natural products from Colombian plants: 3-methoxynordomesticine, an inhibitor of MurE ligase of Mycobacterium tuberculosis. J Antimicrob Chemother. 2010;65:2101–7. [DOI] [PubMed] [Google Scholar]
- Guzman JD, Pesnot T, Barrera DAet al.. Tetrahydroisoquinolines affect the whole-cell phenotype of Mycobacterium tuberculosis by inhibiting the ATP-dependent MurE ligase. J Antimicrob Chemother. 2015;70:1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman JD, Wube A, Evangelopoulos Det al.. Interaction of N-methyl-2-alkenyl-4-quinolones with ATP-dependent MurE ligase of Mycobacterium tuberculosis: antibacterial activity, molecular docking and inhibition kinetics. J Antimicrob Chemother. 2011;66:1766–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discovery. 2007;6:211. [DOI] [PubMed] [Google Scholar]
- Hajduk PJ. Fragment-based drug design: how big is too big?. J Med Chem. 2006;49:6972–6. [DOI] [PubMed] [Google Scholar]
- Hameed P S, Manjrekar P, Chinnapattu Met al.. Pyrazolopyrimidines establish MurC as a vulnerable target in Pseudomonas aeruginosa and Escherichia coli. ACS Chem Biol. 2014;9:2274–82. [DOI] [PubMed] [Google Scholar]
- Hansen JM, Golchin SA, Veyrier FJet al.. N-glycolylated peptidoglycan contributes to the immunogenicity but not pathogenicity of Mycobacterium tuberculosis. J Infect Dis. 2014;209:1045–54. [DOI] [PubMed] [Google Scholar]
- Harth G, Masleša‐Galić S, Tullius MVet al.. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol Microbiol. 2005;58: 1157–72. [DOI] [PubMed] [Google Scholar]
- Hayashi JM, Richardson K, Melzer ESet al.. Stress-induced reorganization of the mycobacterial membrane domain. MBio. 2018;9:e01823-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus Aet al.. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–78. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger Jet al.. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Rocamora VM, Otten CF, Radkov Aet al.. Coupling of polymerase and carrier lipid phosphatase prevents product inhibition in peptidoglycan synthesis. Cell Surface. 2018;2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve M, Boniface A, Gobec Set al.. Biochemical characterization and physiological properties of Escherichia coli UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase. J Bacteriol. 2007;189: 3987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hett EC, Chao MC, Rubin EJ. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog. 2010;6:e1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hett EC, Chao MC, Steyn AJet al.. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol. 2007;66:658–68. [DOI] [PubMed] [Google Scholar]
- Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72:126–56., table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H-T, Falk PJ, Ervin KMet al.. UDP-N-acetylmuramyl-L-alanine functions as an activator in the regulation of the Escherichia coli glutamate racemase activity. Biochemistry. 1995;34:2464–70. [DOI] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Zhou T, Lafleur Ket al.. Kinase selectivity potential for inhibitors targeting the ATP binding site: a network analysis. Bioinformatics. 2009;26:198–204. [DOI] [PubMed] [Google Scholar]
- Hugonnet J-E, Blanchard JS. Irreversible inhibition of the Mycobacterium tuberculosis β-lactamase by clavulanate. Biochemistry. 2007;46:11998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J-E, Mengin-Lecreulx D, Monton Aet al.. Factors essential for L, D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. eLife. 2016;5:e19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J-E, Tremblay LW, Boshoff HIet al.. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humann J, Lenz LL. Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection. J Innate Immun. 2009;1:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Wivagg CN, Szwedziak Pet al.. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife. 2018;7:e32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Murata Y, Takahashi Het al.. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J Bacteriol. 2008;190:7298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Huang LJ, Bartowsky Eet al.. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 1994;13:4684–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap PK, Soni V, Vithani Net al.. Substrate-bound crystal structures reveal features unique to Mycobacterium tuberculosis N-acetyl-glucosamine 1-phosphate uridyltransferase and a catalytic mechanism for acetyl transfer. J Biol Chem. 2012;287:39524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankute M, Cox JA, Harrison Jet al.. Assembly of the mycobacterial cell wall. Annu Rev Microbiol. 2015;69:405–23. [DOI] [PubMed] [Google Scholar]
- Jarlier V, Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett. 1994;123:11–8. [DOI] [PubMed] [Google Scholar]
- Johnson JW, Fisher JF, Mobashery S. Bacterial cell-wall recycling. Ann N Y Acad Sci. 2013;1277:54–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce G, Williams KJ, Robb Met al.. Cell division site placement and asymmetric growth in mycobacteria. PLoS One. 2012;7:e44582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana BD, Gordhan BG, Downing KJet al.. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrinsky DB, Barry CE. Synthesis of labeled meropenem for the analysis of M. tuberculosis transpeptidases. Tetrahedron Lett. 2010;51: 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur D, Brennan PJ, Crick DC. Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J Bacteriol. 2004;186:7564–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser KJ, Boutte CC, Kester JCet al.. Phosphorylation of the peptidoglycan synthase PonA1 governs the rate of polar elongation in mycobacteria. PLoS Pathog. 2015;11:e1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser KJ, Rubin EJ. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol. 2014;12:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn JO, Best GK. Characterization of autolysins from Mycobacterium smegmatis. J Bacteriol. 1977;129:750–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick KE, Ni Cheallaigh C, O'Farrelly Cet al.. Receptor-mediated recognition of mycobacterial pathogens. Cell Microbiol. 2013;15:1484–95. [DOI] [PubMed] [Google Scholar]
- Kim HS, Kim J, Im HNet al.. Structural basis for the inhibition of Mycobacterium tuberculosis L, D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains. Acta Crystallogr Sect D. 2013;69:420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluj RM, Ebner P, Adamek Met al.. Recovery of the Peptidoglycan Turnover Product released by the Autolysin Atl in Staphylococcus aureus involves the Phosphotransferase System Transporter MurP and the Novel 6-phospho-N-acetylmuramidase MupG. Front Microbiol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL. Orientation of the peptidoglycan chains in the sacculus of Escherichia coli. Res Microbiol. 1998;149:689–701. [DOI] [PubMed] [Google Scholar]
- Koch AL. Simulation of the conformation of the murein fabric: the oligoglycan, penta-muropeptide, and cross-linked nona-muropeptide. Arch Microbiol. 2000;174:429–39. [DOI] [PubMed] [Google Scholar]
- Korfmann G, Sanders CC. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother. 1989;33:1946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani S, Yanagida I, Kato Ket al.. Studies on peptides, glycopeptides and antigenic polysaccharide-glycopeptide complexes isolated from an L-11 enzyme lysate of the cell walls of Mycobacterium tuberculosis strain H37Rv. Biken J. 1970;13:249–75. [PubMed] [Google Scholar]
- Koul A, Vranckx L, Dhar Net al.. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nature communications. 2014;5:3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristan K, Kotnik M, Oblak Met al.. New high-throughput fluorimetric assay for discovering inhibitors of UDP-N-acetylmuramyl-L-alanine: D-glutamate (MurD) ligase. J Biomol Screen. 2009;14:412–8. [DOI] [PubMed] [Google Scholar]
- Kuk AC, Mashalidis EH, Lee SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol. 2017;24:171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kumar S, Kumar Det al.. The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis. Acta Crystallogr Sec D, Biol Crystallogr. 2013;69:2543–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Arora K, Lloyd JRet al.. Meropenem inhibits D, D‐carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol. 2012;86:367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Saravanan P, Arvind Aet al.. Identification of hotspot regions of MurB oxidoreductase enzyme using homology modeling, molecular dynamics and molecular docking techniques. J Mol Model. 2011;17:939–53. [DOI] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava Fet al.. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud Met al.. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Derouaux A, Olatunji Set al.. Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep. 2017;7:43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Huang KC. The role of hydrolases in bacterial cell-wall growth. Curr Opin Microbiol. 2013;16:760–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaree BA, Clarke AJ. Interaction of penicillin-binding protein 2 with soluble lytic transglycosylase B1 in Pseudomonas aeruginosa. J Bacteriol. 2008;190:6922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisico F, D VV, Figueiredo TAet al.. First insights of peptidoglycan amidation in Gram-positive bacteria - the high-resolution crystal structure of Staphylococcus aureus glutamine amidotransferase GatD. Sci Rep. 2018;8:5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMagueres P, Im H, Ebalunode Jet al.. The 1.9 A crystal structure of alanine racemase from Mycobacterium tuberculosis contains a conserved entryway into the active site. Biochemistry. 2005;44:1471–81. [DOI] [PubMed] [Google Scholar]
- Levefaudes M, Patin D, De Sousa-D'Auria Cet al.. Diaminopimelic acid amidation in Corynebacteriales: new insights into the role of LtsA in peptidoglycan modification. J Biol Chem. 2015:jbc.M115. 642843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti G, Singh R, Rossi PLet al.. Chlamydia trachomatis dapF encodes a bifunctional enzyme capable of both d-glutamate racemase and diaminopimelate epimerase activities. mBio. 2018;9:e00204–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liger D, Masson A, Blanot Det al.. Study of the overproduced uridine-diphosphate-N-acetylmuramate:L-alanine ligase from Escherichia coli. Microb Drug Resist. 1996;2: 25–7. [DOI] [PubMed] [Google Scholar]
- Li S, Kang J, Yu Wet al.. Identification of M. tuberculosis Rv3441c and M. smegmatis MSMEG_1556 and essentiality of M. smegmatis MSMEG_1556. PLoS One. 2012;7:e42769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZS, Beveridge TJ, Betts Jet al.. Partial characterization of a major autolysin from Mycobacterium phlei. Microbiology. 1999;145:169–76. [DOI] [PubMed] [Google Scholar]
- Loessner MJ, Kramer K, Ebel Fet al.. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol. 2002;44:335–49. [DOI] [PubMed] [Google Scholar]
- Machowski EE, Senzani S, Ealand Cet al.. Comparative genomics for mycobacterial peptidoglycan remodelling enzymes reveals extensive genetic multiplicity. BMC Microbiol. 2014;14:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R. Bedaquiline: first FDA-approved tuberculosis drug in 40 years. Int J Appl Basic Med Res. 2013;3:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Bhakta S, Ahamed Jet al.. Characterization of derivatives of the high-molecular-mass penicillin-binding protein (PBP) 1 of Mycobacterium leprae. Biochem J. 2000;350:Pt 1:75–80. [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Crick DC, McNeil MRet al.. Unique structural features of the peptidoglycan of Mycobacterium leprae. J Bacteriol. 2008;190:655–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Scherman H, Brennan PJet al.. N Glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. J Bacteriol. 2005a;187:2341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Yagi T, Belisle JTet al.. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J Bacteriol. 2005b;187:2747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunatha U, Boshoff HI, Barry CE. The mechanism of action of PA-824: novel insights from transcriptional profiling. Commun Integr Biol. 2009;2:215–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour TS, Caufield CE, Rasmussen Bet al.. Naphthyl tetronic acids as multi-target inhibitors of bacterial peptidoglycan biosynthesis. Chem Med Chem. 2007;2:1414–7. [DOI] [PubMed] [Google Scholar]
- Marmor S, Petersen CP, Reck Fet al.. Biochemical characterization of a phosphinate inhibitor of Escherichia coli MurC. Biochemistry. 2001;40:12207–14. [DOI] [PubMed] [Google Scholar]
- Marshall DD, Halouska S, Zinniel DKet al.. Assessment of metabolic changes in Mycobacterium smegmatis wild-type and alr mutant strains: evidence of a new pathway of d-alanine biosynthesis. J Proteome Res. 2017;16:1270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauck J, Chan L, Glaser L. Turnover of the cell wall of Gram-positive bacteria. J Biol Chem. 1971;246:1820–7. [PubMed] [Google Scholar]
- Mavrici D, Prigozhin DM, Alber T. Mycobacterium tuberculosis RpfE crystal structure reveals a positively charged catalytic cleft. Protein Sci. 2014;23:481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–8. [DOI] [PubMed] [Google Scholar]
- Meeske AJ, Riley EP, Robins WPet al.. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Falla T, Blanot Det al.. Expression of the Staphylococcus aureus UDP-N-acetylmuramoyl- L-alanyl-D-glutamate:L-lysine ligase in Escherichia coli and effects on peptidoglycan biosynthesis and cell growth. J Bacteriol. 1999;181:5909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meniche X, Otten R, Siegrist MSet al.. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci. 2014;111:E3243–E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroueh SO, Bencze KZ, Hesek Det al.. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci U S A. 2006;103:4404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikusova K, Slayden RA, Besra GSet al.. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother. 1995;39:2484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan DL, Tran SL, Strych Uet al.. The alanine racemase of Mycobacterium smegmatis is essential for growth in the absence of D-alanine. J Bacteriol. 2007;189:8381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingorance J, Tamames J, Vicente M. Genomic channeling in bacterial cell division. J Mol Recognit. 2004;17:481–7. [DOI] [PubMed] [Google Scholar]
- Mir M, Asong J, Li Xet al.. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizyed S, Oddone A, Byczynski Bet al.. UDP-N-acetylmuramic acid (UDP-MurNAc) is a potent inhibitor of MurA (enolpyruvyl-UDP-GlcNAc synthase). Biochemistry. 2005;44:4011–7. [DOI] [PubMed] [Google Scholar]
- Mohammadi T, Sijbrandi R, Lutters Met al.. Specificity of the transport of lipid II by FtsW in Escherichia coli. J Biol Chem. 2014;289:14707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi Ret al.. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30: 1425–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morayya S, Awasthy D, Yadav Ret al.. Revisiting the essentiality of glutamate racemase in Mycobacterium tuberculosis. Gene. 2015;555:269–76. [DOI] [PubMed] [Google Scholar]
- Morlot C, Straume D, Peters Ket al.. Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae. Nature Commun. 2018;9:3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KAet al.. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortuza R, Aung HL, Taiaroa Get al.. Overexpression of a newly identified d‐amino acid transaminase in Mycobacterium smegmatis complements glutamate racemase deletion. Mol Microbiol. 2018;107:198–213. [DOI] [PubMed] [Google Scholar]
- Morè N, Martorana AM, Biboy Jet al.. Peptidoglycan remodeling enables escherichia coli to survive severe outer membrane assembly defect. mBio. 2019;10:e02729–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Cadby IT, Veerapen Net al. The hydrolase LpqI primes mycobacterial peptidoglycan recycling. Nat Commun. 2019; 10:2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamolova GV, Kaprelyants AS, Young DIet al.. A bacterial cytokine. Proc Natl Acad Sci U S A. 1998a;95:8916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamolova GV, Kormer SS, Kell DBet al.. Stimulation of the multiplication of Micrococcus luteus by an autocrine growth factor. Arch Microbiol. 1999;172:9–14. [DOI] [PubMed] [Google Scholar]
- Mukamolova GV, Murzin AG, Salina EGet al.. Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol Microbiol. 2006;59:84–98. [DOI] [PubMed] [Google Scholar]
- Mukamolova GV, Turapov OA, Kazarian Ket al.. The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol Microbiol. 2002;46:611–21. [DOI] [PubMed] [Google Scholar]
- Mukamolova GV, Yanopolskaya ND, Kell DBet al.. On resuscitation from the dormant state of Micrococcus luteus. Antonie Van Leeuwenhoek. 1998b;73:237–43. [DOI] [PubMed] [Google Scholar]
- Munch D, Roemer T, Lee SHet al.. Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog. 2012;8:e1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi T, Gupta A, Evangelopoulos Det al.. Characterisation of ATP-Dependent Mur Ligases Involved in the Biogenesis of Cell Wall Peptidoglycan in Mycobacterium tuberculosis. PLoS One. 2013;8:e60143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakel M, Ghuysen JM, Kandler O. Wall peptidoglycan in Aerococcus viridans strains 201 Evans and ATCC 11563 and in Gaffkya homari strain ATCC 10400. Biochemistry. 1971;10:2170–5. [DOI] [PubMed] [Google Scholar]
- Nampoothiri KM, Rubex R, Patel AKet al.. Molecular cloning, overexpression and biochemical characterization of hypothetical beta-lactamases of Mycobacterium tuberculosis H37Rv. J Appl Microbiol. 2008;105:59–67. [DOI] [PubMed] [Google Scholar]
- Ngadjeua F, Braud E, Saidjalolov Set al.. Critical impact of peptidoglycan precursor amidation on the activity of L,D‐Transpeptidases from Enterococcus faecium and Mycobacterium tuberculosis. Chem Eur J. 2018;24:5743–7. [DOI] [PubMed] [Google Scholar]
- Nikitushkin VD, Demina GR, Shleeva MOet al.. Peptidoglycan fragments stimulate resuscitation of “non-culturable” mycobacteria. Antonie Van Leeuwenhoek. 2013;103: 37–46. [DOI] [PubMed] [Google Scholar]
- Nikolaidis I, Izoré T, Job Vet al.. Calcium-dependent complex formation between PBP2 and lytic transglycosylase SltB1 of Pseudomonas aeruginosa. Microb Drug Resist. 2012;18:298–305. [DOI] [PubMed] [Google Scholar]
- Noldeke ER, Muckenfuss LM, Niemann Vet al.. Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus. Sci Rep. 2018;8:12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten C, Brilli M, Vollmer Wet al.. Peptidoglycan in obligate intracellular bacteria. Mol Microbiol. 2018;107:142–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Yang Y, Jiang Zet al.. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh A, Verma SK, Khan Set al.. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J Mol Biol. 2009;386:451–64. [DOI] [PubMed] [Google Scholar]
- Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev. 2008;72: 211–27., table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JT. Uridine-5'-pyrophosphate derivatives. II. Isolation from Staphylococcus aureus. J Biol Chem. 1952;194: 877–84. [PubMed] [Google Scholar]
- Patru MM, Pavelka MS Jr. A role for the class A penicillin-binding protein PonA2 in the survival of Mycobacterium smegmatis under conditions of nonreplication. J Bacteriol. 2010;192:3043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen M, Muylle I, Vandenberg Oet al.. Meropenem-clavulanate for drug-resistant tuberculosis: a follow-up of relapse-free cases. Int J Tuberc Lung Dis. 2018;22:34–9. [DOI] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJet al.. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discovery. 2007;6:29–40. [DOI] [PubMed] [Google Scholar]
- Perdih A, Hrast M, Barreteau Het al.. Benzene-1,3-dicarboxylic acid 2,5-dimethylpyrrole derivatives as multiple inhibitors of bacterial Mur ligases (MurC-MurF). Bioorganic Med Chem. 2014;22:4124–34. [DOI] [PubMed] [Google Scholar]
- Pesnot T, Gershater MC, Ward JMet al.. Phosphate mediated biomimetic synthesis of tetrahydroisoquinoline alkaloids. Chem Commun. 2011;47:3242–4. [DOI] [PubMed] [Google Scholar]
- Petit JF, Adam A, Wietzerbin-Falszpan Jet al.. Chemical structure of the cell wall of Mycobacterium smegmatis. I. Isolation and partial characterization of the peptidoglycan. Biochem Biophys Res Commun. 1969;35:478–85. [DOI] [PubMed] [Google Scholar]
- Pink D, Moeller J, Quinn Bet al.. On the architecture of the gram-negative bacterial murein sacculus. J Bacteriol. 2000;182:5925–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisabarro AG, de Pedro MA, Vazquez D. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol. 1985;161:238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piuri M, Hatfull GF.. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiol. 2006;62:1569–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plocinski P, Ziolkiewicz M, Kiran Met al.. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J Bacteriol. 2011;193:3246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poen S, Nakatani Y, Opel-Reading HKet al.. Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery. Biochem J. 2016;473:1267–80. [DOI] [PubMed] [Google Scholar]
- Pompeo F, van Heijenoort J, Mengin-Lecreulx D. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J Bacteriol. 1998;180:4799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhin DM, Mavrici D, Huizar JPet al.. Structural and biochemical analyses of Mycobacterium tuberculosis N-acetylmuramyl-L-alanine amidase Rv3717 point to a role in peptidoglycan fragment recycling. J Biol Chem. 2013;288:31549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser GA, de Carvalho LP. Kinetic mechanism and inhibition of Mycobacterium tuberculosis D-alanine:D-alanine ligase by the antibiotic D-cycloserine. FEBS J. 2013;280:1150–66. [DOI] [PubMed] [Google Scholar]
- Prosser GA, Rodenburg A, Khoury Het al.. Glutamate racemase is the primary target of β-chloro-D-alanine in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2016: AAC. 01249-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesniaux V, Fremond C, Jacobs Met al.. Toll-like receptor pathways in the immune responses to mycobacteria. Microbes Infect. 2004;6:946–59. [DOI] [PubMed] [Google Scholar]
- Radkov AD, Hsu Y-P, Booher Get al.. Imaging bacterial cell wall biosynthesis. Annu Rev Biochem. 2018;87:991–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond JB, Mahapatra S, Crick DCet al.. Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J Biol Chem. 2005;280:326–33. [DOI] [PubMed] [Google Scholar]
- Reck F, Marmor S, Fisher Set al.. Inhibitors of the bacterial cell wall biosynthesis enzyme MurC. Bioorganic Med Chem Letters. 2001;11:1451–4. [DOI] [PubMed] [Google Scholar]
- Reith J, Mayer C. Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl Microbiol Biotechnol. 2011;92:1–11. [DOI] [PubMed] [Google Scholar]
- Riano F, Arroyo L, Paris Set al.. T cell responses to DosR and Rpf proteins in actively and latently infected individuals from Colombia. Tuberculosis (Edinb). 2012;92:148–59. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Rivera FP, Zhou X, Theriot JAet al.. Acute Modulation of Mycobacterial Cell Envelope Biogenesis by Front‐Line Tuberculosis Drugs. Angew Chem Int Ed. 2018;57:5267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M, Aryan E, Korf Het al.. Potential of Mycobacterium tuberculosis resuscitation-promoting factors as antigens in novel tuberculosis sub-unit vaccines. Microbes Infect. 2012;14:86–95. [DOI] [PubMed] [Google Scholar]
- Romeis T, Holtje JV. Penicillin-binding protein 7/8 of Escherichia coli is a DD-endopeptidase. Eur J Biochem. 1994;224:597–604. [DOI] [PubMed] [Google Scholar]
- Roychowdhury A, Wolfert MA, Boons GJ. Synthesis and proinflammatory properties of muramyl tripeptides containing lysine and diaminopimelic acid moieties. ChemBioChem. 2005;6:2088–97. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Marasco D, Squeglia Fet al.. Structure and functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation. Structure. 2010;18:1184–90. [DOI] [PubMed] [Google Scholar]
- Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci. 2008;105:15553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell-Goldman E, Xu J, Wang Xet al.. A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes. Infect Immun. 2008;76:4269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan NJ, Lo JH. Delamanid: first global approval. Drugs. 2014;74: 1041–5. [DOI] [PubMed] [Google Scholar]
- Röse L, Kaufmann SH, Daugelat S. Involvement of Mycobacterium smegmatis undecaprenyl phosphokinase in biofilm and smegma formation. Microbes Infect. 2004;6: 965–71. [DOI] [PubMed] [Google Scholar]
- Sangshetti JN, Joshi SS, PatilRHet al.. Mur Ligase Inhibitors as Anti-bacterials: A Comprehensive Review. Curr Pharm Des. 2017;23:3164–96. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. [DOI] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak Met al.. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–58. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol. 2008;40:586–91. [DOI] [PubMed] [Google Scholar]
- Schubert K, Sieger B, Meyer Fet al.. The antituberculosis drug ethambutol selectively blocks apical growth in CMN group bacteria. MBio. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulbach MC, Brennan PJ, Crick DC. Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J Biol Chem. 2000: 22876-81. [DOI] [PubMed] [Google Scholar]
- Schulbach MC, Mahapatra S, Macchia Met al.. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J Biol Chem. 2001;276: 11624–30. [DOI] [PubMed] [Google Scholar]
- Schulz MN, Hubbard RE. Recent progress in fragment-based lead discovery. Curr Opin Pharmacol. 2009;9:615–21. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki MH, Popham DLet al.. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham L-T, Butler EK, Lebar MDet al.. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam A, Anbazhagan V, Natarajan J. Virtual screening of phenylsulfonamido-3-morpholinopropan-2-yl dihydrogen phosphate derivatives as novel inhibitors of MurC–MurF ligases from Mycobacterium leprae. Med Chem Res. 2012;21:4341–51. [Google Scholar]
- Shanmugam A, Natarajan J. Comparative modeling of UDP-N-acetylmuramoyl-glycyl-D-glutamate-2, 6-diaminopimelate ligase from Mycobacterium leprae and analysis of its binding features through molecular docking studies. J Mol Model. 2012;18:115–25. [DOI] [PubMed] [Google Scholar]
- Shetty A, Dick T. Mycobacterial cell wall synthesis inhibitors cause lethal ATP burst. Front Microbiol. 2018;9:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava P, Navratna V, Silla Yet al.. Inhibition of Mycobacterium tuberculosis dihydrodipicolinate synthase by alpha-ketopimelic acid and its other structural analogues. Sci Rep. 2016;6:30827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Whiteside S, Jewett JCet al.. D-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8: 500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert G, Strominger JL. Bacitracin: an inhibitor of the dephosphorylation of lipid pyrophosphate, an intermediate in the biosynthesis of the peptidoglycan of bacterial cell walls. Proc Natl Acad Sci. 1967;57:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LL. Does the cell wall of bacteria remain a viable source of targets for novel antibiotics? Biochem Pharmacol. 2006;71:996–1005. [DOI] [PubMed] [Google Scholar]
- Singh PP, Smith VL, Karakousis PCet al.. Exosomes isolated from mycobacteria-infected mice or cultured macrophages can recruit and activate immune cells in vitro and in vivo. J Immunol. 2012;189:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siricilla S, Mitachi K, Skorupinska-Tudek Ket al.. Biosynthesis of a water-soluble lipid I analogue and a convenient assay for translocase I. Anal Biochem. 2014;461: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J Mol Biol. 2006;362:640–55. [DOI] [PubMed] [Google Scholar]
- Soni V, Upadhayay S, Suryadevara Pet al.. Depletion of M. tuberculosis GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen. PLoS Pathog. 2015;11:e1005235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper GF, Longenecker KL, Fry EHet al.. Structure-based optimization of MurF inhibitors. Chem Biol Drug Design. 2006;67:58–65. [DOI] [PubMed] [Google Scholar]
- Strancar K, Blanot D, Gobec Set al. Design, synthesis and structure-activity relationships of new phosphinate inhibitors of MurD. Bioorg. Med. Chem. Lett. 2006; 16:343–8.. 16271472 [DOI] [PubMed] [Google Scholar]
- Strancar K, Boniface A, Blanot Det al.. Phosphinate inhibitors of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate: L-lysine ligase (MurE). Arch Pharm (Weinheim). 2007;340:127–34. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G, Gal L, Peneff Cet al.. Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture. J Biol Chem. 2001;276:11844–51. [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev. 2004;28:645–59. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Welsh MA, Marmont LSet al.. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nature Microbiol. 2019:4:587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Schnoes HK, Semmler EJ. Characterization of the alkali-stable mannophospholipids of Mycobacterium smegmatis. Biochem Biophys Acta. 1973;316:212–21. [DOI] [PubMed] [Google Scholar]
- Talukder AA, Yanai S, Nitta Tet al.. RpoS-dependent regulation of genes expressed at late stationary phase in Escherichia coli. FEBS Lett. 1996;386:177–80. [DOI] [PubMed] [Google Scholar]
- Tanner ME, Vaganay S, van Heijenoort Jet al.. Phosphinate inhibitors of the D-glutamic acid-adding enzyme of peptidoglycan biosynthesis. J Org Chem. 1996;61:1756–60. [DOI] [PubMed] [Google Scholar]
- Templin MF, Ursinus A, Holtje JV. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 1999;18:4108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanky NR, Young DB, Robertson BD. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb). 2007;87:231–6. [DOI] [PubMed] [Google Scholar]
- Tomasic T, Zidar N, Kovac Aet al.. 5-Benzylidenethiazolidin-4-ones as multitarget inhibitors of bacterial Mur ligases. Chem Med Chem. 2010;5:286–95. [DOI] [PubMed] [Google Scholar]
- Tomašić T, Mašič LP. Rhodanine as a scaffold in drug discovery: a critical review of its biological activities and mechanisms of target modulation. Expert Opin Drug Discovery. 2012;7:549–60. [DOI] [PubMed] [Google Scholar]
- Triboulet S, Bougault CM, Laguri Cet al.. Acyl acceptor recognition by Enterococcus faecium L,D‐transpeptidase Ldtfm. Mol Microbiol. 2015;98:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turapov O, Forti F, Kadhim Bet al.. Two Faces of CwlM, an Essential PknB Substrate, in Mycobacterium tuberculosis. Cell reports. 2018;25:57–67. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk S, Kovac A, Boniface Aet al.. Discovery of new inhibitors of the bacterial peptidoglycan biosynthesis enzymes MurD and MurF by structure-based virtual screening. Bioorganic Med Chem. 2009;17:1884–9. [DOI] [PubMed] [Google Scholar]
- Turner RD, Mesnage S, Hobbs JKet al.. Molecular imaging of glycan chains couples cell-wall polysaccharide architecture to bacterial cell morphology. Nature Commun. 2018;9:1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udou T, Ogawa M, Mizuguchi Y. Spheroplast formation of Mycobacterium smegmatis and morphological aspects of their reversion to the bacillary form. J Bacteriol. 1982;151:1035–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara A, Fujimoto Y, Kawasaki Aet al.. Meso-diaminopimelic acid and meso-lanthionine, amino acids specific to bacterial peptidoglycans, activate human epithelial cells through NOD1. J Immunol. 2006;177:1796–804. [DOI] [PubMed] [Google Scholar]
- Usha V, Dover LG, Roper DIet al.. Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis. Acta Crystallogr Sec D. 2009;65:383–7. [DOI] [PubMed] [Google Scholar]
- Usha V, Lloyd AJ, Lovering ALet al.. Structure and function of Mycobacterium tuberculosis meso-diaminopimelic acid (DAP) biosynthetic enzymes. FEMS Microbiol Lett. 2012;330:10–6. [DOI] [PubMed] [Google Scholar]
- Usha V, Lloyd AJ, Roper DIet al.. Reconstruction of diaminopimelic acid biosynthesis allows characterisation of Mycobacterium tuberculosis N-succinyl-L, L-diaminopimelic acid desuccinylase. Sci Rep. 2016;6:23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaganay S, Tanner ME, van Heijenoort Jet al.. Study of the reaction mechanism of the D-glutamic acid-adding enzyme from Escherichia coli. Microb Drug Resist. 1996;2:51–4. [DOI] [PubMed] [Google Scholar]
- van Asselt EJ, Thunnissen AM, Dijkstra BW. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J Mol Biol. 1999;291:877–98. [DOI] [PubMed] [Google Scholar]
- van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001a;11:25R–36R. [DOI] [PubMed] [Google Scholar]
- van Heijenoort J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev. 2007;71:620–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001b;18:503–19. [DOI] [PubMed] [Google Scholar]
- Vicente M, Rico AI, Martinez-Arteaga Ret al.. Septum enlightenment: assembly of bacterial division proteins. J Bacteriol. 2006;188:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayrajratnam S, Pushkaran AC, Balakrishnan Aet al.. Bacterial peptidoglycan with amidated meso-diaminopimelic acid evades NOD1 recognition: an insight on NOD1 structure-recognition study. Biochem J. 2016:473:4573–92. [DOI] [PubMed] [Google Scholar]
- Vincent AT, Nyongesa S, Morneau Iet al.. The mycobacterial cell envelope: a relict from the past or the result of recent evolution?. Front Microbiol. 2018;9:2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkamer A, Eid S, Turk Set al.. Pocketome of human kinases: prioritizing the ATP binding sites of (yet) untapped protein kinases for drug discovery. J Chem Inf Model. 2015;55:538–49. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Holtje JV. The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)?. J Bacteriol. 2004;186:5978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier Pet al.. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–86. [DOI] [PubMed] [Google Scholar]
- Vollmer W, von Rechenberg M, Holtje JV. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem. 1999;274: 6726–34. [DOI] [PubMed] [Google Scholar]
- Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32:287–306. [DOI] [PubMed] [Google Scholar]
- von Rechenberg M, Ursinus A, Holtje J-V. Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb Drug Resist. 1996;2:155–7. [DOI] [PubMed] [Google Scholar]
- Votyakova TV, Kaprelyants AS, Kell DB. Influence of viable cells on the resuscitation of dormant cells in micrococcus luteus cultures held in an extended stationary phase: the population effect. Appl Environ Microbiol. 1994;60:3284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Matsuo Y, Pradipta ARet al.. Synthesis of characteristic Mycobacterium peptidoglycan (PGN) fragments utilizing with chemoenzymatic preparation of meso-diaminopimelic acid (DAP), and their modulation of innate immune responses. Organic Biomol Chem. 2016;14:1013–23. [DOI] [PubMed] [Google Scholar]
- Wang W, Dong C, McNeil Met al.. The structural basis of chain length control in Rv1086. J Mol Biol. 2008;381:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Global Tuberculosis Report Geneva. Geneva, Switzerland: World Health Organisation, 2018. [Google Scholar]
- Wietzerbin J, Das BC, Petit JFet al.. Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry. 1974;13:3471–6. [DOI] [PubMed] [Google Scholar]
- Wolfert MA, Roychowdhury A, Boons G-J. Modification of the structure of peptidoglycan is a strategy to avoid detection by nucleotide-binding oligomerization domain protein 1. Infect Immun. 2007;75:706–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman SD, Worrall LJ, Strynadka NC. Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling. Nature Commun. 2018;9:1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wube AA, Hüfner A, Thomaschitz Cet al.. Design, synthesis and antimycobacterial activities of 1-methyl-2-alkenyl-4 (1H)-quinolones. Bioorganic Med Chem. 2011;19:567–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Hodgkinson JW, Katzenback BAet al.. Characterization of three Nod-like receptors and their role in antimicrobial responses of goldfish (Carassius auratus L.) macrophages to Aeromonas salmonicida and Mycobacterium marinum. Dev Comp Immunol. 2013;39:180–7. [DOI] [PubMed] [Google Scholar]
- Xu L, Wu D, Liu Let al.. Characterization of mycobacterial UDP-N-acetylglucosamine enolpyruvyle transferase (MurA). Res Microbiol. 2014;165:91–101. [DOI] [PubMed] [Google Scholar]
- Yang Y, Severin A, Chopra Ret al. 3,5-dioxopyrazolidines, novel inhibitors of UDP-N- acetylenolpyruvylglucosamine reductase (MurB) with activity against gram-positive bacteria. Antimicrob. Agents Chemother. 2006;50:556–64.. 16436710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Munshi S, Leiting Bet al.. Crystal structure of Escherichia coli UDPMurNAc-tripeptide d-alanyl-d-alanine-adding enzyme (MurF) at 2.3 A resolution. J Mol Biol. 2000;304:435–45. [DOI] [PubMed] [Google Scholar]
- Young DB, Perkins MD, Duncan Ket al.. Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest. 2008;118:1255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Philippe J, Abrahams KAet al.. In vitro reconstitution of peptidoglycan assembly from the Gram-positive pathogen Streptococcus pneumoniae. ACS Chem Biol. 2013;8:2688–96. [DOI] [PubMed] [Google Scholar]
- Zawadzke LE, Bugg TD, Walsh CT. Existence of two D-alanine:D-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry. 1991;30:1673–82. [DOI] [PubMed] [Google Scholar]
- Zawadzke LE, Norcia M, Desbonnet CRet al.. Identification of an inhibitor of the MurC enzyme, which catalyzes an essential step in the peptidoglycan precursor synthesis pathway. Assay Drug Dev Technol. 2008;6: 95–103. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Heym B, Allen Bet al.. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Woods Aet al.. Resuscitation of dormant Mycobacterium tuberculosis by phospholipids or specific peptides. Biochem Biophys Res Commun. 2001;284:542–7. [DOI] [PubMed] [Google Scholar]
- Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol. 2005;45:529–64. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Bulloch EM, Bunker RDet al.. Structure and function of GlmU from Mycobacterium tuberculosis. Acta crystallogr Sec D, Biol Crystallogr. 2009;65:275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Sham LT, Rubino FAet al.. Structure and mutagenic analysis of the lipid II flippase MurJ from Escherichia coli. Proc Natl Acad Sci U S A. 2018;115:6709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidar N, Tomasic T, Sink Ret al.. Discovery of novel 5-benzylidenerhodanine and 5-benzylidenethiazolidine-2,4-dione inhibitors of MurD ligase. J Med Chem. 2010;53:6584–94. [DOI] [PubMed] [Google Scholar]
- Zumla A, Nahid P, Cole ST. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discovery. 2013;12:388–404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.