Abstract
Lipids are critical for neurodevelopment; therefore, disruption of lipid homeostasis by environmental chemicals is expected to have detrimental effects on this process. Previously, we demonstrated that the benzalkonium chlorides (BACs), a class of commonly used disinfectants, alter cholesterol biosynthesis and lipid homeostasis in neuronal cell cultures in a manner dependent on their alkyl chain length. However, the ability of BACs to reach the neonatal brain and alter sterol and lipid homeostasis during neurodevelopment in vivo has not been characterized. Therefore, the goal of this study was to use targeted and untargeted mass spectrometry and transcriptomics to investigate the effect of BACs on sterol and lipid homeostasis and to predict the mechanism of toxicity of BACs on neurodevelopmental processes. After maternal dietary exposure to 120 mg BAC/kg body weight/day, we quantified BAC levels in the mouse neonatal brain, demonstrating for the first time that BACs can cross the blood-placental barrier and enter the developing brain. Transcriptomic analysis of neonatal brains using RNA sequencing revealed alterations in canonical pathways related to cholesterol biosynthesis, liver X receptor-retinoid X receptor (LXR/RXR) signaling, and glutamate receptor signaling. Mass spectrometry analysis revealed decreases in total sterol levels and downregulation of triglycerides and diglycerides, which were consistent with the upregulation of genes involved in sterol biosynthesis and uptake as well as inhibition of LXR signaling. In conclusion, these findings demonstrate that BACs target sterol and lipid homeostasis and provide new insights for the possible mechanisms of action of BACs as developmental neurotoxicants.
Keywords: in utero exposure, benzalkonium chloride, sterols, lipids, neurodevelopment, multiomics
The brain is the most lipid-rich organ, and lipids play important roles in neurodevelopment. For example, docosahexaenoic acid, a ω-3 long-chain polyunsaturated fatty acid, is essential for early neurodevelopmental events such as neurogenesis (Coti Bertrand et al., 2006; Katakura et al., 2009). Cholesterol is another critical molecule—serving an essential role in hedgehog signaling (Porter et al., 1996), neurogenesis (Komada et al., 2008), synapse formation and function (Koudinov and Koudinova, 2001; Mauch et al., 2001), and myelination (Saher et al., 2005). Importantly, the brain synthesizes its cholesterol locally and independently (Bjorkhem and Meaney, 2004). The role of cholesterol in neurodevelopment is underscored by the cholesterol biosynthesis disorder, Smith–Lemli–Opitz syndrome (SLOS), which is caused by a defect in the last step of the postsqualene cholesterol biosynthetic pathway, 3β-hydroxysterol-Δ7-reductase (DHCR7) (Porter and Herman, 2011; Thurm et al., 2016). This defect results in increased levels of cholesterol precursors, such as 7-dehydrocholesterol (7-DHC), and reduced cholesterol levels in all tissues and fluids of SLOS patients (Tint et al., 1994, 1995). The SLOS phenotype includes multiple congenital malformations, developmental delay, cognitive impairment, and behavior problems (Porter and Herman, 2011; Thurm et al., 2016). A variety of other malformation disorders are also attributed to alterations in cholesterol homeostasis (Porter and Herman, 2011).
Some known developmental neurotoxicants, such as retinoic acid and ethanol, have been shown to modulate cholesterol homeostasis (Chen et al., 2011; Zhou et al., 2014). Thus, it is plausible that environmental chemicals capable of disrupting cholesterol and lipid homeostasis could be potential developmental neurotoxicants. The small molecule and known teratogen, AY9944, induces a pharmacological model of SLOS in rats by potently inhibiting DHCR7, which leads to the same biochemical defects as observed in SLOS (Kolf-Clauw et al., 1996; Roux et al., 1979, 1980). Previously, we screened for environmental molecules and drugs that could inhibit cholesterol biosynthesis in a manner similar to AY9944. We reported that benzalkonium chloride (BAC), a widely used quaternary ammonium compound (QAC) antimicrobial, displays high structural similarity to AY9944 (Herron et al., 2016). We further found that short-chain benzalkonium chlorides (BACs) (C10 and C12) potently inhibit DHCR7, while long-chain BACs (C14 and C16) suppress the formation of other cholesterol precursors, such as zymosterol, lathosterol, and desmosterol (Herron et al., 2016). Exposure to BACs also altered the lipidome of neuronal cells, in a manner dependent on the BAC alkyl chain length (Hines et al., 2017). Given the important role of lipids in neurodevelopment, our findings indicate that environmental exposure to BACs during developmental stages could adversely affect neurodevelopment.
QACs, including BACs, are disinfectants widely used in residential, commercial, and medical settings, as well as the food processing industry. They are found in disinfecting solutions and wipes, hand sanitizers, eye drops, and nasal spray, indicating a variety of exposure scenarios (Gilbert and Moore, 2005; Holah et al., 2002; McDonnell and Russell, 1999; Ratani et al., 2012). BACs are also directly applied to eating utensils, egg shells, milking equipment and udders, and medical instruments (EPA, Programs, AD, 2006). High levels of QACs have been found in grapefruit seed extracts (Takeoka et al., 2005), food additives (Kröckel et al., 2003), fruits, and processed food such as milk and other dairy products (Slimani et al., 2017). Therefore, exposure to QACs is prevalent given the diversity of applications and presence in food products.
Recently, QACs have been suggested to be bona fide teratogens, based on an increased incidence of neural tube defects (NTDs) observed in both mice and rats exposed in utero to an environmentally relevant mixture of QACs that contains BACs (Hrubec et al., 2017). Neural tube defects are closely associated with defects in neural stem/progenitor cell (NSPC) proliferation and neurogenesis (Hirata et al., 2001; Ishibashi et al., 1995; Zhong et al., 2000). Lipid metabolism has been shown to influence NSPC proliferation and neurogenesis, as a variety of lipids are used as building blocks for membranes, energy sources, and signaling entities (Bieberich, 2012; Dietschy and Turley, 2004; Edmond, 1992; Knobloch et al., 2013; Salem et al., 2001; Spitzer, 1973; Warshaw and Terry, 1976). Thus, disruption of lipid metabolism by BACs might contribute to the previously reported NTDs and other neurodevelopmental outcomes. However, the ability of BACs to enter the fetal brain and alter sterol and lipid homeostasis has not yet been investigated. Therefore, the objective of the present study was to test the hypothesis that in utero exposure to BACs alters sterol and lipid homeostasis in the developing brain.
MATERIALS AND METHODS
Chemicals
Optima LC/MS solvents (acetonitrile, chloroform, methanol, methylene chloride, and water), 2-methylbutane, ammonium acetate (Optima LC/MS), formic acid (Optima LC/MS), and sodium chloride were purchased from Thermo Fisher Scientific (Grand Island, New York). The deuterated (d7-) sterol standard d7-7-dehydrocholesterol was prepared as reported previously (Xu et al., 2011a,b). d7-cholesterol was purchased from Avanti Polar Lipids (Alabaster, Alabama). 13C3-desmosterol and 13C3-lanosterol were purchased from Kerafast (Boston, Massachusetts). d7-BAC C12 and d7-BAC C16 were prepared as described previously (Herron et al., 2016).
Animals
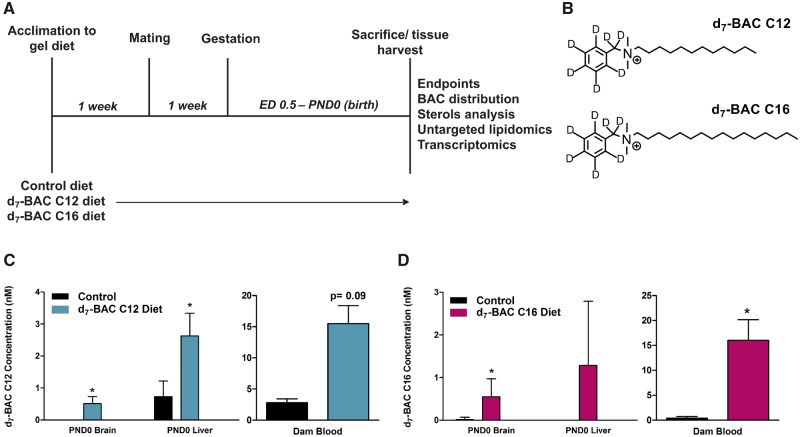
C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, Maine). The University of Washington Institutional Animal Care and Use Committee approved all animal protocols. Experiments conducted were also in accordance with the Guiding Principles in the Use of Animals in Toxicology. Adult male and female mice (4–6 months old) were acclimated to untreated Nutra-Gel diet (Product #F5769, Bio-Serv, Frenchtown, New Jersey) for 1 week prior to dietary BAC exposure (Figure 1A). Our previous pilot studies have shown that exogenous BAC contamination on the mass spectrometer source led to artifact levels of BAC in control samples, preventing accurate quantitation. Therefore, to accurately quantify the ability of BACs to enter the neonatal brain, deuterated BACs were used. Thus, females were randomly assigned to exposure groups (n = 3–4 per group) as described: control Nutra-Gel diet, 120 mg/kg body weight/day d7-BAC C12 in Nutra-Gel diet, or 120 mg/kg body weight/day d7-BAC C16 in Nutra-Gel diet. Excess gel diet was provided fresh each day so as not to restrict food intake. No difference in food consumption was observed between exposure groups.
Figure 1.
Experimental design, structure of deuterium (d7)-labeled BACs, and BAC tissue distribution analysis. A, Dams were exposed prior to mating and gestation. At birth, neonates and dams were sacrificed and tissues were harvested to be used for the described endpoints. B, Structures of d7-BAC C12 (top) and d7-BAC C16 (bottom). C, Concentration of d7-BAC C12 in control and exposed neonatal brain and liver and dam blood determined by ultra-high-performance liquid chromatography tandem mass spectrometry. D, Concentration of d7-BAC C16 in control and exposed neonatal brain and liver and dam blood determined by ultra-high-performance liquid chromatography tandem mass spectrometry. n = 5–7 biological replicates per condition for PND0 tissues and n = 2–3 biological replicates per condition for dam blood; all statistical analyses are relative to control using the Student’s t test assuming unequal variances; *p < .05; Abbreviation: BAC, benzalkonium chloride.
Following the 1-week acclimation period, females were exposed to control or BAC diet for 1 week prior to mating. Breeding pairs consisted of 1 male per 2 females (2 breeding cages per exposure group) and were housed together for 1 week. Sires were then removed from the cage and dams were fed the control or BAC diet throughout gestation until postnatal day 0 (PND0). At PND0, neonates were sacrificed, and the brain and liver were dissected, flash-frozen in 2-methylbutane on dry ice and stored at −80°C until subsequent analyses. Sex was not determined at this age. The weight of each frozen tissue was recorded. Dams were sacrificed at this time to collect blood samples for BAC measurements.
Lipid extraction
Prior to lipid extraction, isotopically labeled sterol internal standards (to be used for quantification in the sterols analysis) and unlabeled d0-BAC C12 and d0-BAC C16 internal standards (to be used for quantification of the fed d7-BAC in tissues and fluids) were added to each tissue (sagittal-cut half PND0 brain, PND0 liver, or dam blood). Note that the other half of the PND0 brain was used for RNA sequencing analysis as described below. For sterols analysis, 5 µg of d7-cholesterol and d7-7-dehydrocholesterol, and 1 µg of 13C3-lanosterol and 13C3-desmosterol were added to each sample. For BAC analysis, 0.03 nmol of d0-BAC C12 and d0-BAC C16 were added to each sample. Lipid extraction was carried out as previously described in detail (Herron et al., 2018). To extract the lipids, samples were homogenized in Folch solution (4 ml chloroform/methanol [2/1, v/v]), using a blade homogenizer. NaCl aqueous solution (0.9% [w/v], 1 ml) was added and the resulting mixture was briefly vortexed and then centrifuged for 5 min in a clinical tabletop centrifuge at 10°C. The lower (organic) phase was recovered, transferred to a separate glass tube, and the solvent was removed in vacuo using a SpeedVac® (Thermo Fisher Savant). Finally, the resulting dried tissue or blood extracts were redissolved in 0.3 ml methylene chloride prior to analysis.
UHPLC-MS/MS analysis of BAC and BAC-metabolite levels
Analysis of BAC levels was performed by ultra-high-performance liquid chromatography tandem mass spectrometry (UHPLC-MS/MS) using a triple-quadrupole mass spectrometer (API 6500TM; SCIEX, Vaughan, Ontario, Canada) equipped with electrospray ionization (ESI). For analysis, 40 µl of lipid extract was transferred to an LC vial, dried under a stream of argon, and reconstituted in 40 µl water/methanol (1/1, v/v). Reverse-phase chromatography was performed with the following conditions: C18 column (Hypersil GOLD®, 100 mm × 2.1 mm, 1.9 µm particle diameter; ThermoFisher Scientific, Grand Island, New York); flow rate, 0.4 ml/min; and gradient elution method with solvent A (0.1% formic acid, 2 mM ammonium formate in water) and solvent B (acetonitrile) was used: 0 min, 30% B; 9.5 min, 85% B; 10–12 min, 30% B. MS conditions: declustering potential, 50 V; entrance potential, 10 V; collision energy, 31 V; collision cell exit potential, 10 V. ESI parameters: spray voltage 3500 V; temperature, 400°C; curtain gas, 35 psi; ion source gas, 45 psi. For MS analysis, selective reaction monitoring (SRM) was employed to monitor the mass-to-charge ratio (m/z) of the parent ion (Q1) for each BAC (isotopically labeled analyte and corresponding internal standard) and the m/z of the characteristic fragment (Q3), formed from the loss of benzyl group (C7H8) (see Table 1 for mass transitions). d7-BAC concentrations were calculated based on the ratio of the analyte peak area to the internal standard peak area and internal standard concentration in each sample. The major BAC metabolites were also monitored, including the +1O- and +1O-2H-metabolites of d7-BAC C12 and the +1O-metabolite of d7-BAC C16 in dam blood and neonatal brain samples (see Table 1 for mass transitions). Presence of a metabolite peak was confirmed by comparison to a standard sample containing BAC metabolites that had been generated in NADPH-fortified human liver microsomes (further details on generation and characterization of BAC metabolites can be found in our manuscript in submission, by Seguin R.; Herron, J.; Dempsey, J.; Xu, L.). All data are presented as mean ± standard deviation. Statistical analyses relative to control were conducted using the Student’s t test assuming unequal variance (p value < .05).
Table 1.
The m/z of the Parent Ion (Q1) and Characteristic Fragment (Q3) for Each BAC Analyte of Interest Was Monitored, Including the Isotopically Labeled Analyte, Major Metabolite/s, and Corresponding Internal Standard
| Analyte | Q1 Parent Ion (m/z) | Q3 Fragment Ion (m/z) |
|---|---|---|
| d7-BAC C12 | 311.2 | 212.2 |
| d7-BAC C12 +1O | 327.2 | 228.2 |
| d7-BAC C12 +1O-2H | 325.2 | 226.2 |
| d0-BAC C12 | 304.2 | 212.2 |
| d7-BAC C16 | 367.2 | 268.2 |
| d7-BAC C16 +1O | 383.2 | 284.2 |
| d0-BAC C16 | 360.2 | 268.2 |
UHPLC-MS/MS analysis of sterol levels
Analysis of sterols was performed by UHPLC-MS/MS using a triple-quadrupole mass spectrometer (API 4000TM; SCIEX, Vaughan, Ontario, Canada) equipped with atmospheric pressure chemical ionization as described previously (Herron et al., 2018). For analysis, 10 µl of lipid extract was transferred to an LC vial, dried under a stream of argon, and reconstituted in 100 µl 90% MeOH with 0.1% formic acid. See previously published protocol for further details on UHPLC-MS/MS analysis, including MS conditions and quantification method (Herron et al., 2018). Data are presented as mean ± standard deviation. Statistical analyses relative to control were conducted using Student’s t test assuming unequal variance (p value < .05).
Untargeted lipidomics and data analysis
Chromatographic separation was performed using a Waters Acquity FTN ultraperformance liquid chromatography (Waters Corp, Milford, Massachusetts) with a hydrophilic interaction column (HILIC; Phenomenex Kinetex, 2.1 × 100 mm, 1.7 µm) maintained at 40°C. An injection volume of 5 μl was used. Ion mobility-mass spectrometry (IM-MS) analysis was performed on a Waters Synapt G2-Si HDMS (Waters Corp) equipped with an ESI source. Detailed chromatographic and IM-MS conditions and mass calibration parameters were as previously published (Hines et al., 2017). Data alignment, peak detection, and normalization were performed in Progenesis QI (Nonlinear Dynamics). The chromatographic region from 0.4 to 8.4 min was considered for peak detection. The reference sample for alignment was automatically selected by Progenesis QI, and data were normalized to tissue weight. The resulting data set was filtered by analysis of variance p value < .05. Principal components analysis was performed in EZInfo (Umetrics). Lipid identifications were made against the METLIN database within 20 ppm mass accuracy. Data are presented as mean ± standard deviation. Statistical analyses relative to control were conducted using Student’s t test assuming unequal variance with a Bonferroni correction for multiple comparisons (p value < .025).
Total RNA isolation
PND0 brain halves (sagittal-cut) from control, BAC C12, or BAC C16 exposure groups (n = 4 per exposure group) were homogenized in 1 ml of QIAzol Lysis Reagent (Qiagen) using a blade homogenizer. Total RNA was extracted from each sample using the RNeasy Lipid Tissue Mini Kit (Qiagen, Germantown, Maryland) according to the manufacturer’s protocol. RNA concentration was quantified using a microplate spectrophotometer (Bio-Tek, Winooski, Vermont) to quantify the absorbance at 260 nm. The RNA integrity was evaluated by formaldehyde agarose gel electrophoresis to visualize the 18S and 28S rRNA bands. Novogene (Chula Vista, California) performed the cDNA library construction and sequencing using the Illumina NovaSeq 6000 platform (150 base pairs paired-end, with sequencing depth above 20 million reads per sample). Novogene confirmed the RNA integrity and purity using the Agilent 2100 BioAnalyzer (Agilent Technologies Inc, Santa Clara, California). Samples with RNA Integrity Number (RIN) of 8.6 or above were submitted to RNA sequencing.
RNA sequencing and data analysis
Raw RNA sequencing reads in FASTQ format were mapped to the mouse genome using HISAT (https://ccb.jhu.edu/software/hisat/; Last accessed June 19, 2019), and format conversions were performed using Samtools. Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/; Last accessed June 19, 2019) was used to estimate relative abundances of transcripts from each RNA sample. Cuffdiff, a module of Cufflinks, was then used to determine differentially expressed genes (DEGs) between comparison control versus BAC C12 or control versus BAC C16. Differentially expressed genes met the following criteria: adjusted p value < .05 (corresponding to the allowed false discovery rate of 5%).
Differentially expressed genes were plotted in a Venn diagram to identify the common and uniquely expressed genes for each exposure (BAC C12 or BAC C16). Canonical Pathway analysis and Upstream Regulator analysis of DEGs was performed using the Core Analysis feature of Ingenuity Pathway Analysis (IPA, Ingenuity Systems). The Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/summary.jsp; Last accessed June 19, 2019) annotation enrichment analysis was used to identify enriched lists of DEGs related to sterol and lipid metabolism from each comparison pair (control vs BAC C12 or control vs BAC C16) (Huang et al., 2009). A two-way hierarchical clustering dendrogram was conducted on the enriched lists of DEGs related to sterol and lipid metabolism for the control versus BAC C12 comparison and the control versus BAC C16 comparison using ClustVis (https://biit.cs.ut.ee/clustvis/; Last accessed June 19, 2019) (Metsalu and Vilo, 2015). Correlation distance and average linkage was used for clustering of rows and columns.
Validation of genes using real-time quantitative polymerase chain reaction (RT-qPCR)
A subset of genes related to sterol and lipid metabolism were selected for validation using RT-qPCR. Total RNA isolated from the PND0 brains (n = 4 per exposure group) was reverse-transcribed into cDNA using the SuperScript IV First-Strand Synthesis System (Invitrogen, Carlsbad, California). The resulting cDNA was amplified by qPCR, using the TaqMan Gene Expression Mastermix (Thermo Fisher Scientific, Waltham, Massachusetts) in a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). TaqMan gene expression assays were used for all qPCR reactions, see Supplemental Table 1 for assay ID (Thermo Fisher Scientific). Data are expressed as mean percentage of the expression of the housekeeping gene β-actin ± standard error of the mean. Statistical analyses relative to control were conducted using the Student’s t test assuming unequal variance (p value < .05).
Joint pathway analysis of genes and metabolites
To integrate results obtained from sterols analysis, untargeted lipidomics, and transcriptomics analyses, the joint pathway analysis module of MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/MetaboAnalyst/faces/home.xhtml; Last accessed June 19, 2019) was used. Joint pathway analysis exploits the KEGG (Kyoto Encyclopedia of Genes and Genomes) metabolic pathway database to discover pathways involved in underlying biological processes using gene expression and metabolite data. Lists of DEGs and significantly altered sterols and lipids from either BAC C12 or BAC C16 exposed neonatal brains were uploaded into the module. Features were then mapped to KEGG metabolic pathways for over-representation analysis and pathway topology analysis. For over-representation analysis, pathway enrichment was determined by hypergeometric test and reported with an FDR-adjusted p value. For topology analysis, degree centrality was used to assess the position of a gene or compound within a pathway.
RESULTS
Distribution of BACs in PND0 Tissues and Maternal Circulation
To examine whether BACs can enter the embryonic brain, dams were exposed 1-week prior to mating and throughout gestation to control Nutra-Gel diet or Nutra-Gel diet with added d7-BAC C12 or d7-BAC C16 (120 mg/kg/day) (Figs. 1A and 1B). This dosing regimen was adapted from Hrubec et al., who had previously demonstrated that 120 mg/kg/day of an environmentally relevant mixture of QACs, including BACs, increased the incidence of NTDs in both mice and rats (Hrubec et al., 2017). In the current study, BACs were found at low nM levels in various PND0 tissues, including the brain and liver (Figs. 1C and 1D). d7-BAC C12 levels were higher in the brain and liver of neonates than in the control neonates (Figure 1C), whereas d7-BAC C16 levels were only significantly higher in the brain (Figure 1D). Levels in the neonatal tissues corresponded to approximately 20 nM BAC in the maternal circulation (Figs. 1C and 1D). As additional evidence of systemic BAC exposure, BAC metabolites that entailed cytochrome P450-mediated oxidations of the alkyl chain were observed in the neonatal brain exposed to d7-BAC C12 and in the circulation of dams exposed to either BAC (Supplemental Figure 1) but not in the control mice (identification of these metabolites can be found in our manuscript under review by Seguin R.; Herron, J.; Dempsey, J.; Xu, L.). Overall, these results indicate that BACs do indeed cross the blood-placenta barrier, enter the fetus, and reach the developing brain.
Assessment of Sterol Homeostasis in PND0 Brains
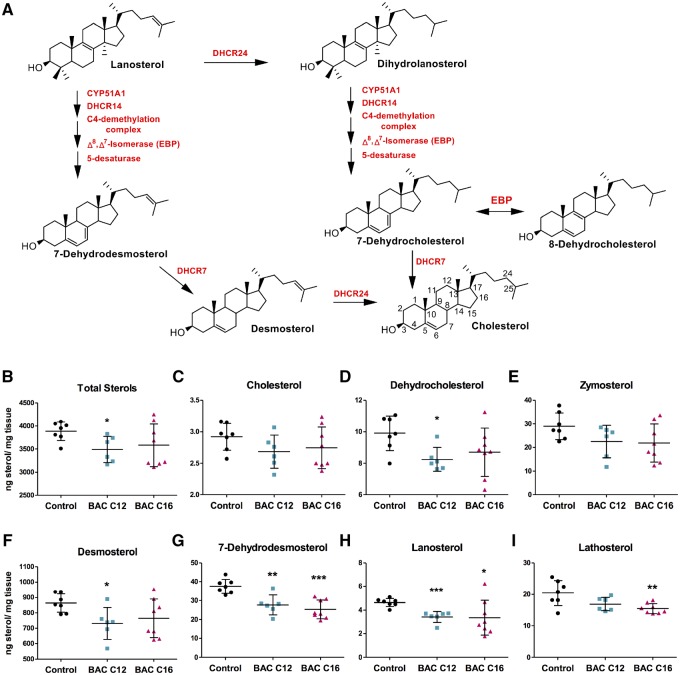
BAC compounds of different alkyl chain lengths have been shown to alter cholesterol biosynthesis (Figure 2A) in neuronal cell lines (Herron et al., 2016). However, the ability of BACs to alter sterol homeostasis during neurodevelopment has not been characterized. Therefore, levels of sterols in lipid extracts from control and exposed PND0 brains were assessed using UHPLC-MS/MS (Herron et al., 2018). Decreased sterol levels were observed in both BAC C12 and BAC C16 exposed brains (Figs. 2B–I). Exposure to BAC C12 had a more pronounced effect on sterol levels, significantly decreasing levels of total sterols as well as the precursors, 7- and 8-DHC, 7-DHD, desmosterol, and lanosterol, with a trend of decreased cholesterol and other cholesterol precursors. BAC C16 also lead to significantly decreased levels of 7-DHD, lanosterol, and lathosterol with a trend of decreased total sterols, cholesterol, and other cholesterol precursors. The decrease in lanosterol, as well as sterol metabolites downstream of lanosterol, indicate that BACs inhibit biosynthesis upstream of the postsqualene cholesterol biosynthetic pathway in neonatal brains.
Figure 2.
Lipid extracts from neonatal brains exposed in utero to BAC C12 or C16 were analyzed to assess changes to sterols involved in the post-squalene cholesterol biosynthetic pathway (see cholesterol biosynthesis scheme in panel A). B, Total sterol levels, as well as (C) cholesterol, (D) dehydrocholesterol, (E) zymosterol, (F) desmosterol, (G) 7-dehydrodesmosterol, (H) lanosterol, and (I) lathosterol were quantified in each neonatal brain. N = 5–7 biological replicates per condition; all statistical analyses are relative to control using Student’s t test assuming unequal variances; * p < .05; ** p < .005; *** p < .0005. Abbreviation: BAC, benzalkonium chloride.
Untargeted Analysis of the PND0 Brain Lipidome
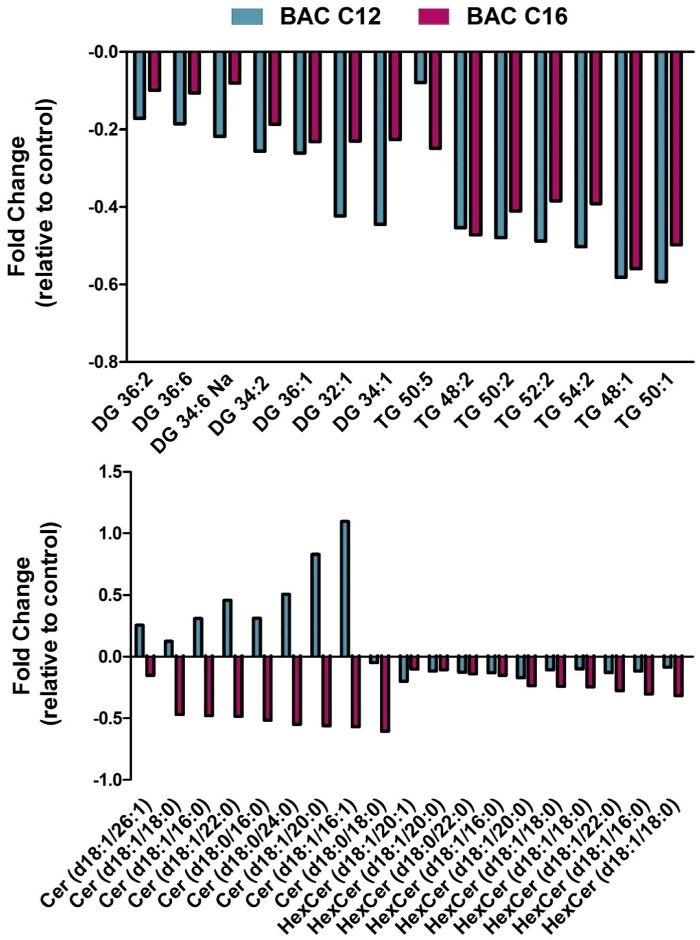
We have previously demonstrated that BAC compounds altered lipid homeostasis in treated neuronal cells, leading to decreased levels of triacylglycerols (TGs), diacylglycerols (DGs), and ceramides (Cers) by both BAC-C12 and BAC-C16 and varied changes to phospholipids and sphingomyelins in a manner dependent on the alkyl chain length (Hines et al., 2017). In the current study, significant alterations in the lipidome of BAC C12- and BAC C16-exposed neonatal brains were also observed, as determined by our HILIC-IM-MS lipidomics method (Figure 3). Principal component analysis (PCA) revealed group separation dependent on exposure, with BAC-exposed brains separating from control brains along PC-1 (accounting for 64.0% of the variance), although some overlap is observed (Supplemental Figure 2). BAC-C12 exposed brains further separated from BAC-C16 samples along PC-2 (accounting for 13.3% of the variance).
Figure 3.
Lipidomic changes in neonatal brains exposed to BAC C12 or BAC C16 relative to control group. Various lipid species including DGs, TGs, Cers, and HexCers are main contributors to the group separation in the principal component analysis (Supplemental Figure 1). Both BACs decreased DGs and TGs, although this trend did not reach statistical significance for BAC C16. BAC C16 significantly decreased Cers and HexCers, whereas BAC C12 had little effect on these lipids. n = 5–7 biological replicates per condition; data filtered by analysis of variance p value < .05. Abbreviations: BAC, benzalkonium chloride; DGs, diacylglycerols; TGs, triacylglycerols.
Significantly altered lipid species were identified by retention time and m/z (Supplemental Table 2). Changes in lipid classes, including DGs, TGs, hexosylceramides (HexCers), and Cers, contributed to the group separations observed in the PCA. BACs altered these lipid species in a manner dependent on the alkyl chain length. BAC C12 exposure led to an overall decrease in DGs and TGs in the neonatal brains, whereas BAC C16 exposure also decreased the levels of these lipids albeit not reaching statistical significance due to large error. Furthermore, BAC C16 significantly decreased the levels of Cers and HexCers (Figure 3). The reduction in the levels of TGs, DGs, and Cers is consistent with what was previously observed in neuronal cells exposed to BACs (Hines et al., 2017).
Global Changes in Gene Expression Profiles in BAC-Exposed PND0 Brains
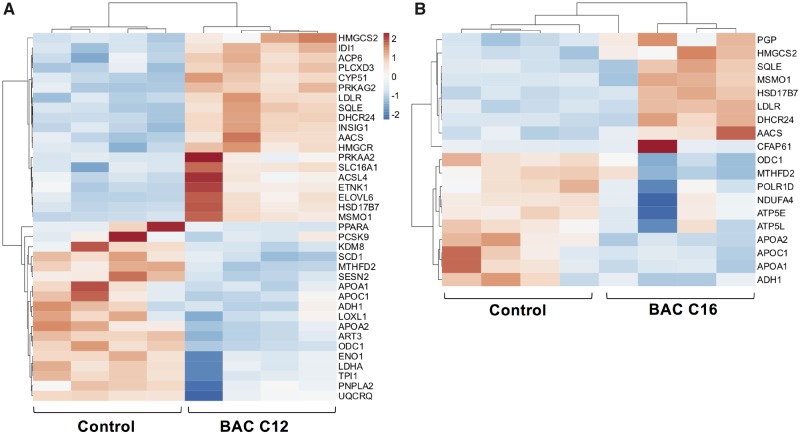
To elucidate mechanisms underlying the observed decreases in sterols and lipids, transcriptomic changes in control and BAC-exposed neonatal brains were investigated using RNA sequencing. Overall, BAC C12 showed more significant gene expression changes than BAC C16. Compared to controls, in utero exposure to BAC C12 altered the expression of 507 genes in PND0 brains, whereas in utero exposure to BAC C16 altered the expression of 139 genes (adjusted p < .05) (Figs. 4A and 4B). BAC C12 upregulated 308 genes and downregulated 199 genes, whereas BAC C16 upregulated 65 genes and downregulated 74 genes. Among the DEGs, 77 genes were coregulated by both BACs, whereas 430 genes were uniquely regulated by BAC C12 and 62 genes were uniquely regulated by BAC C16. Among the commonly regulated genes, 46 of the genes were upregulated and 31 were downregulated.
Figure 4.
Venn diagrams of genes commonly or uniquely expressed in neonatal brains exposed in utero to BAC C12 or BAC C16: (A) upregulated genes or (B) downregulated genes. BAC C12 exposure led to an upregulation of 308 genes, whereas BAC C16 upregulated 65 genes. Of the genes with increased expression, 46 were coregulated by the BACs. BAC C12 exposure also led to a downregulation of 199 genes, whereas BAC C16 upregulated 74 genes. Of the genes with decreased expression, 31 were coregulated by the BACs. C, Top 5 canonical signaling pathways significantly altered in neonatal brains exposed in utero to BAC C12 or BAC C16. The BACs altered many of the same pathways, however TR/RXR activation was unique to BAC C12 and EIF2 signaling was unique to BAC C16. Z-score activation values to right of bar for each pathway, positive number indicates activation whereas negative number indicates inhibition of pathway. n = 4 biological replicates per condition; adjusted p < .05. Abbreviations: BAC, benzalkonium chloride; LXR/RXR, liver X receptor-retinoid X receptor; TR/RXR, thyroid hormone receptor-retinoid X receptor.
Ingenuity Pathway Analysis of DEGs in BAC-Exposed PND0 Brains
Functional characterization of DEGs was conducted using IPA. Ingenuity Pathway Analysis revealed the most significantly altered canonical pathways of DEGs in the control versus BAC C12 comparison and the control versus BAC C16 comparison (Figure 4C;Supplemental Table 3). For the control versus BAC C12 comparison, the top 5 significantly altered canonical pathways included the “Superpathway of Cholesterol Biosynthesis” (8 genes), “liver X receptor-retinoid X receptor (LXR/RXR) activation” (14 genes), “thyroid hormone receptor-retinoid X receptor (TR/RXR) Activation” (11 genes), “Glutamate Receptor Signaling” (8 genes), and “Acute Phase Response Signaling” (13 genes) (Figure 4C;Supplemental Table 3). The “Superpathway of Cholesterol Biosynthesis” was predicted to be activated, whereas the “LXR/RXR activation” and “Acute Phase Response Signaling” pathways were predicted to be inhibited. Predictions of activation or inhibition is inferred from the activation z-score which is calculated based on the match between the gene expression pattern expected from the IPA Knowledge Base and the expression (“increase” or “decreased”) of DEGs within the dataset. Activation z-scores were not predicted for the “TR/RXR Activation” or “Glutamate Receptor Signaling” pathways, likely due to ambiguity in the direction of gene expression changes in these pathways. For the control versus BAC C16 comparison, the top 5 significantly altered canonical pathways included “EIF2 signaling”, “Superpathway of Cholesterol Biosynthesis” (5 genes), “LXR/RXR Activation” (7 genes), “Glutamate Receptor Signaling” (4 genes), and “Acute Phase Response Signaling” (6 genes) (Figure 4C;Supplemental Table 3). The “Superpathway of Cholesterol Biosynthesis” was predicted to be activated, whereas the “EIF2 signaling” and “LXR/RXR activation” pathways were predicted to be inhibited. Activation z-scores were not predicted for the “Acute Phase Response Signaling” or “Glutamate Receptor Signaling” pathways.
The upstream analysis module of IPA also predicted transcriptional regulators of gene expression and whether they were activated or inhibited (Supplemental Tables 4 and 5). Of particular interest was the identification of the upstream regulator of sterol and lipid homeostasis, SREBP cleavage-activating protein (SCAP), as one of the most significantly activated transcriptional regulators in both the control versus BAC C12 and control versus BAC C16 comparisons (Figure 5). This prediction was based on the overlap p value, which measures the significance of the overlap between dataset genes and genes regulated by the transcriptional regulator (control vs. BAC C12, p = 2.02E−12; control vs BAC C16, p = 4.37E−07). SCAP activation was predicted in both IPA comparisons based on the expression pattern of a variety of DEGs involved in sterol and lipid biosynthesis and transport (Table 2). SCAP is an escort protein required for cholesterol and lipid homeostasis. When sterol levels are low, SCAP, a sterol sensor, binds sterol regulatory element binding proteins (SREBPs) and mediates their transport from the endoplasmic reticulum (ER) to the Golgi (Camargo et al., 2009), where SREBPs are cleaved by proteases. Cleaved cytoplasmic portion of SREBPs subsequently translocates to the nucleus and activate genes involved in sterol and lipid biosynthesis, transport, and metabolism. SREBP1c governs the activation of genes involved in fatty acid and triacylglyceride biosynthesis, whereas SREBP2 is involved in cholesterol biosynthesis (Camargo et al., 2009).
Figure 5.
Ingenuity pathway analysis identifies the upstream regulator SREBP cleavage-activating protein as significantly activated in neonatal brains exposed in utero to BAC C12 (A) or BAC C16 (B). Red, increased gene expression; green, decreased gene expression; orange, predicted activation; yellow, findings inconsistent with state of downstream molecule; and gray, effect not predicted. n = 4 biological replicates per condition; adjusted p < .05. Abbreviation: BAC, benzalkonium chloride.
Table 2.
Differentially Expressed Genes Regulated by SREBP Cleavage-Activating Protein in Neonatal Brains Exposed In Utero to BAC C12 or BAC C16
| BAC C12 |
BAC C16 |
||||
|---|---|---|---|---|---|
| Gene ID | Description | Log2(FC) | Adjusted p Value | Log2(FC) | Adjusted p Value |
| AACS | Acetoacetyl-CoA synthetase | 0.42 | 9.81E−03 | 0.35 | 3.84E–02 |
| ACSL4 | Acyl-CoA synthetase long-chain family member 4 | 0.51 | 4.44E–03 | ||
| ALB | Albumin | –2.19 | 4.44E–03 | –3.69 | 9.98E–03 |
| CYP51 | Cytochrome P450, family 51 | 0.58 | 3.23E–02 | ||
| DLK1 | Delta like non–canonical Notch ligand 1 | –0.53 | 2.70E–02 | ||
| ELOVL6 | ELOVL family member 6, elongation of long-chain fatty acids | 0.58 | 4.44E–03 | ||
| HMGCR | 3-hydroxy-3-methylglutaryl-CoenzymeA reductase | 0.57 | 4.44E–03 | ||
| HMGCS2 | 3-hydroxy-3-methylglutaryl-CoenzymeA synthase 2 | 0.69 | 4.44E–03 | 0.61 | 9.98E–03 |
| IDI1 | Isopentenyl-diphosphate delta isomerase | 0.91 | 4.44E–03 | ||
| INSIG1 | Insulin induced gene 1 | 0.41 | 9.81E–03 | ||
| LDLR | Low-density lipoprotein receptor | 0.87 | 4.44E–03 | 0.73 | 9.98E–03 |
| MSMO1 | Methylsterol monooxygenase 1 | 0.66 | 4.44E–03 | 0.48 | 9.98E–03 |
| MT2 | Metallothionein 2 | –0.48 | 1.66E–02 | ||
| MTHFD2 | Methylenetetrahydrofolate cyclohydrolase | –0.66 | 4.44E–03 | –0.53 | 9.98E–03 |
| PCSK9 | Proprotein convertase subtilisin/kexin type 9 | –0.81 | 4.44E–03 | ||
| SCD | Stearoyl-Coenzyme A desaturase | –0.36 | 1.98E–02 | ||
| SQLE | Squalene epoxidase | 0.62 | 4.44E–03 | 0.45 | 9.98E–03 |
n = 4 biological replicates per condition; adjusted p < .05.
Hierarchical Clustering Analysis of DEGs Involved in Sterol and Lipid Metabolism
Given the identification of canonical signaling pathways, prediction of upstream regulators of sterol and lipid homeostasis, and the apparent sterol- and lipid-lowering effects of BACs on neonatal brains, expression patterns of genes involved in sterol and lipid homeostasis were examined in detail. DAVID functional annotation clustering analysis identified an enriched subset of genes functionally-related to sterol and lipid metabolism in both control versus BAC C12 and control versus BAC C16 comparisons (Supplemental Table 6). The gene cluster related to sterol and lipid homeostasis was found to be enriched in both BAC C12 (37 genes) and BAC C16 (19 genes) groups, with an enrichment score of 3.33 and 2.00, respectively (Supplemental Table 7). Two-way hierarchical clustering analysis revealed distinct gene expression patterns between the control and exposed samples for each comparison (Figure 6), except for 1 BAC C16-sample which clustered remotely with the controls. For the BAC C12 versus control comparison group, increased expression of genes known to play key roles in sterol biosynthesis and homeostasis was observed in the exposed group (Figure 6A;Supplemental Table 6), including Hmgcs2, Idi1, Cyp51, Ldlr, Sqle, Dhcr24, Insig1, Hmgcr, Hsd17b7 and Msmo1. Increases in Hmgcs2, Ldlr, Sqle, Dhcr24, Hsd17b7, and Msmo1 was also observed in the BAC C16 samples (Figure 6B;Supplemental Table 7). Both BACs also decreased the expression of genes encoding lipoproteins, including Apoa1, Apoa2, Apoc1, indicating decreased transport of lipids.
Figure 6.
Two-way hierarchical clustering dendrograms of differentially expressed genes involved in sterol and lipid homeostasis in neonatal brains exposed in utero to (A) BAC C12 (A) or (B) BAC C16; adjusted p < .05. n = 4 biological replicates were tested for each group (shown as columns). Each row represents a different gene. Dendrogram was generated for each comparison (A) control versus BAC C12 and (B) control versus BAC C16 using standardized means and correlation distance and average linkage was used for clustering both rows and columns. Adjusted p < .05. Abbreviation: BAC, benzalkonium chloride.
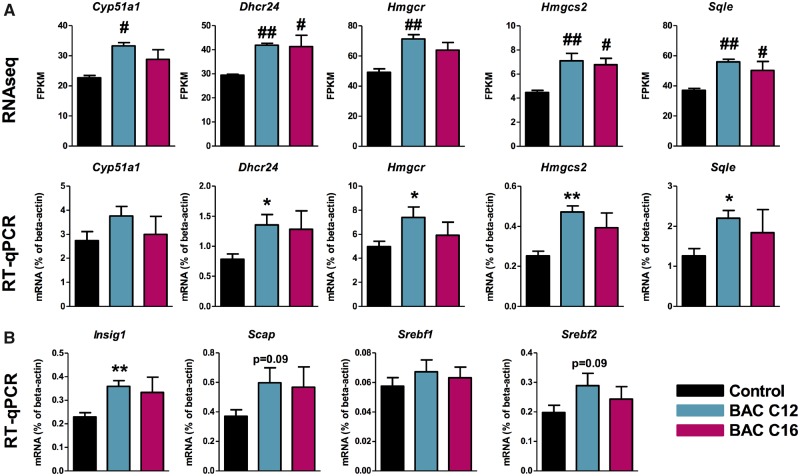
Targeted Validation of Sterol- and Lipid-Related Genes Identified by RNA Sequencing
Differentially expressed genes involved in sterol and lipid homeostasis were validated using RT-qPCR (Figure 7). Real-time quantitative polymerase chain reaction results were consistent with the RNA sequencing data, with BAC C12 showing a more significant effect than BAC C16 (Figs. 7A and B). Genes involved in cholesterol biosynthesis, including Dhcr24, Hmgcr, Hmgcs2, and Sqle were significantly increased by both BACs in the RNA sequencing analyses, but significant increases were only observed with BAC C12 exposure when validated by RT-qPCR although the trend was consistent. Changes in genes involved in the regulation of sterol and lipid biosynthesis and uptake were also assessed using RT-qPCR. Insig1, which was significantly altered by exposure to either BAC in the RNA sequencing analyses, also showed significant increases in expression with BAC C12 when measured by RT-qPCR. Although not significantly increased based on the RNA sequencing analysis, Scap, Srebf1, and Srebf2 were also included in the RT-qPCR analysis, as Scap had been predicted as an activated upstream regulator in the IPA analysis. Scap and Srebf2 showed a trend toward increased expression in the BAC C12 samples, but not the control or BAC C16 samples (Figure 7C).
Figure 7.
Validation of sterol- and lipid-related differentially expressed genes using RT-qPCR. A, BAC C12 and BAC C16 exposure increased the expression of cholesterol biosynthetic genes. (B) Genes involved in regulation of sterol biosynthesis trended toward increased expression in neonatal brains exposed in utero to BAC C12. For RNA sequencing, differentially expressed genes met the criteria of having an adjusted p value < .05, corresponding to the allowed false discovery rate of 5; #p < .05, ##p < .005; n = 4 biological replicates per condition. For RT-qPCR, all statistical analyses are relative to control using Student’s t test assuming unequal variances, * p < .05, ** p < .005; n = 4 biological replicates per condition. Abbreviations: BAC, benzalkonium chloride; RT-qPCR, real-time quantitative polymerase chain reaction.
Joint Pathway Analysis of Genes and Metabolites
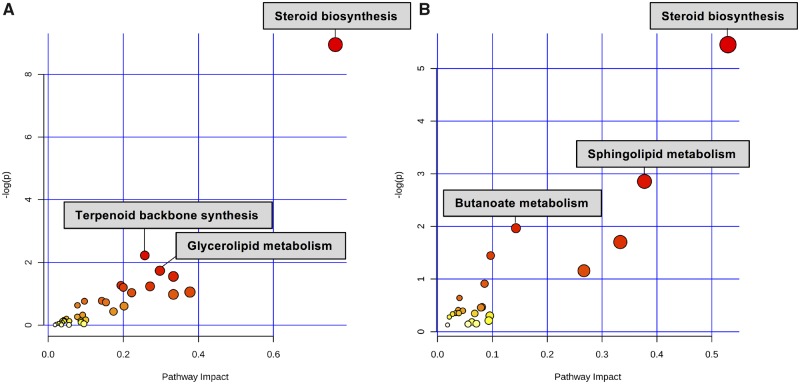
MetaboAnalyst 4.0 was used to conduct joint pathway analysis which integrates significant features generated from mass spectrometry and transcriptomic analyses. In both BAC C12 and BAC C16 exposed neonatal brains, joint pathway analysis of DEGs and significantly altered sterols and lipids confirmed the importance of steroid biosynthesis. This pathway was the top-most enriched pathway identified, based on the significantly altered genes, sterols, and lipids, in BAC C12 (adjusted p value = .01, Impact = .76) and BAC C16 (adjusted p value = .34, Impact = .53) exposed neonatal brains (Figure 8). This pathway was more significantly enriched in BAC C12 exposed brains, which corresponds with previous findings that BAC C12 is more effective at altering sterol homeostasis.
Figure 8.
Joint pathway analysis of significantly altered genes, sterols, and lipids from (A) BAC C12- and (B) BAC C16- exposed neonatal brains. Pathway enrichment determined by hypergeometric test and reported with –log (p value) on y-axis, and pathway impact determined by degree centrality measures and reported by pathway impact values on x-axis.
DISCUSSION
We previously demonstrated that BAC disinfectants can alter sterol and lipid homeostasis in neuronal cells (Herron et al., 2016; Hines et al., 2017). The current study aimed at determining whether BACs could alter sterol and lipid homeostasis in a similar manner in the developing mouse brain. This study demonstrated for the first time that BACs can accumulate in the neonatal brain and alter the transcriptome, sterol profiles, and the lipidome following in utero exposure. Specifically, decreases in levels of sterols in the post-squalene cholesterol biosynthetic pathway and in a variety of lipid species, particularly TGs and DGs, were observed in neonatal brains exposed in utero to BAC C12 or BAC C16. Given the important roles of sterols and lipids during neurodevelopment, these data provide further evidence that in utero exposure to BACs may have significant adverse effects on neurodevelopment.
BACs directly inhibit cholesterol biosynthesis in neuronal cell lines, as evidenced by altered sterol levels and increases in the sterol and lipid biosynthetic genes Hmgcr, Dhcr7, Ebp, Srebf2, and Fasn (Herron et al., 2016). In the current study, no 7-DHC was accumulated in BAC C12-treated neonatal brains, but both BACs inhibited cholesterol biosynthesis. The decreases in the levels of lanosterol (the first sterol in the pathway) and other cholesterol precursors suggest that the inhibition of cholesterol biosynthesis by BACs occurs before the formation of lanosterol. Further transcriptomic studies revealed the “Superpathway of Cholesterol Biosynthesis” as one of the most significantly altered canonical pathways in neonatal brains exposed to either BAC compound. Consistent with our previous results in neuronal cell lines, in utero BAC exposure upregulated genes involved in sterol biosynthesis in the neonatal brain, including Dhcr24, Hmgcr, Hmgcs2, and Sqle, supporting direct inhibition of cholesterol biosynthesis by BACs. BACs also significantly upregulated Insig, with a trend toward upregulation of Srebf2 and Scap, which are all involved in the regulation of sterol and lipid biosynthesis. When sterol levels are low, SCAP binds SREBPs and mediates the translocation of the protein complex from the ER to the Golgi. Cleaved SREBPs subsequently increase the expression of sterol biosynthetic genes upon translocation to the nucleus. Decreased sterol levels observed in both BAC C12 and BAC C16-exposed neonatal brains are consistent with the finding that BACs upregulate sterol biosynthesis via SCAP signaling.
The LXR/RXR signaling pathway was predicted to be inhibited by IPA in neonatal brains exposed to either BAC. Liver X receptor is responsive to intracellular cholesterol homeostasis as the cholesterol precursor, desmosterol, and cholesterol-derived side-chain oxysterols serve as ligands for LXR signaling (Janowski et al., 1999; Yang et al., 2006). Thus, the downregulation of desmosterol (Figure 2) could contribute to the inhibition of LXR signaling.
Of interest, is the fact that LXRs have been shown to regulate both cholesterol homeostasis and neurodevelopment. Liver X receptors form heterodimers with Retinoid X Receptors (RXRs) to bind sequence-specific elements which initiates the transcription of genes such as apolipoprotein E (ApoE) (Laffitte et al., 2001; Mak et al., 2002), a lipoprotein that mediates lipid transport between astrocytes and neurons, and the cholesterol efflux transporters, Abca1 and Abcg1 (Venkateswaran et al., 2000). Moreover, LXR regulates lipogenesis, upregulating fatty acid synthase (FAS) expression through direct interaction with the FAS promoter when the pathway is activated (Joseph et al., 2002; Schultz et al., 2000). Finally, LXR has been shown to play a role in a variety of neurodevelopmental processes, including formation of the cortical layers (Fan et al., 2008), neuronal migration (Xing et al., 2010), myelination (Makoukji et al., 2011; Meffre et al., 2015), and neurogenesis (Theofilopoulos et al., 2013).
When LXR signaling is inhibited, lipogenesis is expected to be downregulated, including the synthesis of TGs (Joseph et al., 2002; Schultz et al., 2000). Previously, we have shown that BACs alter the lipidome of neuronal cells, including significant changes to several lipid classes, such as glycerides, sphingolipids, and phospholipids (Hines et al., 2017). In this study, we found that TGs and DGs are downregulated by BACs, which further supports the notion that BAC exposure leads to the inhibition of LXR/RXR signaling.
Sphingolipids, such as HexCers and Cers, were also significantly affected in neonatal brains exposed to either BAC. The exact mechanism underlying these changes is not clear, but inhibition of LXR signaling and downregulation of lipid synthesis could be contributing factors. Regardless, sphingolipids, including HexCers and Cers, play fundamental roles in a variety of cellular processes, which could be another way that BAC exposure can affect neurodevelopment (Ishibashi et al., 2013).
Another novel finding of this study was the effect of BAC exposure on genes in the glutamate receptor signaling pathway, one of the top 5 significantly altered canonical pathways in BAC-exposed neonatal brains. Genes encoding various glutamate ionotropic and metabotropic receptors as well as proteins involved in glutamate signaling at the synapse were affected by both BACs (Supplemental Table 3). Glutamatergic signaling regulates various aspects of neurodevelopment, including neuronal migration, differentiation, neurite outgrowth, synaptogenesis and survival (Choudhury et al., 2012). Glutamate also plays essential roles in motor control, synaptic plasticity, learning and memory, and cognition (Choudhury et al., 2012). Of particular relevance to the current study is the fact that lipid rafts, membrane domains enriched in cholesterol, sphingolipids, and gangliosides, are critical for various aspects of synaptic signaling, including glutamate receptor signaling (Hering et al., 2003; Wang, 2014). It has been hypothesized that changes in cholesterol biosynthesis and subsequent alterations in membrane raft events could contribute to SLOS pathophysiology (Korade and Kenworthy, 2008). Brain raft fractions prepared from AY9944-treated rats show altered gel electrophoresis profiles, suggesting that altered sterol composition perturbs raft protein content (Keller et al., 2004). Smith–Lemli–Opitz syndrome pathology also includes impaired neuronal response to glutamate (Wassif et al., 2001). Therefore, the altered sterol and lipid homeostasis observed in BAC-exposed neonatal brains could very well contribute to the alterations in glutamate receptor signaling pathway, which may have significant consequences in neurodevelopment.
This study is not without limitations. First, although 2 different BACs were evaluated for their effects on the neonatal brain, the use of a single dose prohibited the evaluation of a dose-response relationship. However, the selected dose was derived from the 2017 study by Hrubec et al., which found an increase in NTDs and late gestation fetal deaths with exposure to a mixture of BAC-containing QACs (Hrubec et al., 2017). Second, the effects of BACs at a single time-point of development was examined in this study. The mouse embryo depends on maternal cholesterol until embryonic day 11 (E11) (Herrera and Ortega-Senovilla, 2010; Tint et al., 2006). Therefore, earlier time-points, such as the transition from maternal- to embryonic-derived cholesterol, might be more susceptible to the effects of BACs.
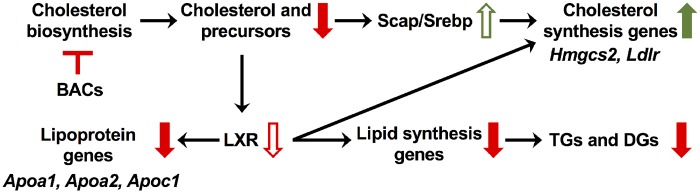
In conclusion, we have demonstrated that BAC disinfectants cross the blood-placental barrier and embryonic blood-brain barrier and alter sterol and lipid homeostasis (Figure 9). Alterations in signaling pathways important for neurodevelopment (ie, LXR/RXR and glutamatergic signaling) were also observed in this study, possibly arising from altered sterol and lipid homeostasis. Since the developmental neurotoxicants ethanol and retinoic acid are known to modulate cholesterol homeostasis, the potential for BACs to cause adverse neurodevelopmental outcomes should be seriously considered. Therefore, for future work, it is imperative to characterize the neurological phenotype resulting from in utero exposure to BAC disinfectants, including morphological and behavioral outcomes.
Figure 9.
Summary of observed alterations in sterol and lipid homeostasis resulting from in utero exposure to BAC disinfectants. Inhibition of cholesterol biosynthesis by BACs leads to decreased levels of sterols, which results in increased SREBP cleavage-activating protein-mediated signaling and expression of cholesterol synthesis genes. Downregulation of the cholesterol precursor desmosterol, which is an LXR ligand, could contribute to the predicted inhibition of LXR signaling. Inhibition of LXR signaling leads to downregulation of apolipoprotein genes and lipogenesis genes, the latter of which subsequently results in downregulation of TGs and DGs that were observed by lipidomics. Abbreviations: BAC, benzalkonium chloride; LXR/RXR, liver X receptor-retinoid X receptor; TR/RXR, thyroid hormone receptor-retinoid X receptor.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
University of Washington Environmental Pathology/Toxicology Training Program (National Institutes of Health (NIH) T32 ES007032-39), the NIH National Institute of Child Health and Human Development (R01HD092659), Interdisciplinary Center for Exposures, Diseases, Genomics and Environment (UW EDGE) (NIH P30ES007033), Sheldon D. Murphy Endowment, and the Department of Medicinal Chemistry.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs Theo Bammler and Lu Wang of the UW EDGE Center for assisting with the Ingenuity Pathway Analysis.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Bieberich E. (2012). It’s a lipid’s world: Bioactive lipid metabolism and signaling in neural stem cell differentiation. Neurochem. Res. 37, 1208–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I., Meaney S. (2004). Brain cholesterol: Long secret life behind a barrier. Arterioscler. Thromb. Vasc. Biol. 24, 806–815. [DOI] [PubMed] [Google Scholar]
- Camargo N., Smit A. B., Verheijen M. H. G. (2009). SREBPs: SREBP function in glia-neuron interactions. FEBS J. 276, 628–636. [DOI] [PubMed] [Google Scholar]
- Chen J., Costa L. G., Guizzetti M. (2011). Retinoic acid isomers up-regulate ATP binding cassette A1 and G1 and cholesterol efflux in rat astrocytes: Implications for their therapeutic and teratogenic effects. J. Pharmacol. Exp. Ther. 338, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury P. R., Lahiri S., Rajamma U. (2012). Glutamate mediated signaling in the pathophysiology of autism spectrum disorders. Pharmacol. Biochem. Behav. 100, 841–849. [DOI] [PubMed] [Google Scholar]
- Coti Bertrand P., O'Kusky J. R., Innis S. M. (2006). Maternal dietary (n-3) fatty acid deficiency alters neurogenesis in the embryonic rat brain. J. Nutr. 136, 1570–1575. [DOI] [PubMed] [Google Scholar]
- Dietschy J. M., Turley S. D. (2004). Thematic review series: Brain lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J. Lipid Res. 45, 1375–1397. [DOI] [PubMed] [Google Scholar]
- Edmond J. (1992). Energy metabolism in developing brain cells. Can. J. Physiol. Pharmacol. 70 Suppl, S118–S129. [DOI] [PubMed] [Google Scholar]
- EPA, U. S., Programs, O. O. P., AD. (2006). US EPA—Pesticides—Registration Eligibility Decision for Alkyl Dimethyl Benzyl Ammonium Chloride (ADBAC), 1–126.
- Fan X., Kim H.-J., Bouton D., Warner M., Gustafsson J. -A. (2008). Expression of liver X receptor beta is essential for formation of superficial cortical layers and migration of later-born neurons. Proc. Natl. Acad. Sci. U S A 105, 13445–13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P., Moore L. E. (2005). Cationic antiseptics: Diversity of action under a common epithet. J. Appl. Microbiol. 99, 703–715. [DOI] [PubMed] [Google Scholar]
- Hering H., Lin C.-C., Sheng M. (2003). Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J. Neurosci. 23, 3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, E., and Ortega-Senovilla, H. (2010) Maternal lipid metabolism during normal pregnancy and its implications in fetal development, Clin. Lipidol., 5:6, 899–911. [Google Scholar]
- Herron J., Hines K. M., Xu L. (2018). Assessment of altered cholesterol homeostasis by xenobiotics using ultra-high performance liquid chromatography-tandem mass spectrometry. Curr. Protoc. Toxicol. 16, e65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron J., Reese R. C., Tallman K. A., Narayanaswamy R., Porter N. A., Xu L. (2016). Identification of environmental quaternary ammonium compounds as direct inhibitors of cholesterol biosynthesis. Toxicol. Sci. 151, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines K. M., Herron J., Xu L. (2017). Assessment of altered lipid homeostasis by HILIC-ion mobility-mass spectrometry-based lipidomics. J. Lipid Res. 58, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Tomita K., Bessho Y., Kageyama R. (2001). Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. EMBO J. 20, 4454–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holah J. T., Taylor J. H., Dawson D. J., Hall K. E. (2002). Biocide use in the food industry and the disinfectant resistance of persistent strains of Listeria monocytogenes and Escherichia coli. J. Appl. Microbiol 92, 111S–120S. [PubMed] [Google Scholar]
- Hrubec T. C., Melin V. E., Shea C. S., Ferguson E. E., Garofola C., Repine C. M., et al. (2017). Ambient and dosed exposure to quaternary ammonium disinfectants causes neural tube defects in rodents. Birth Defects Res. 16, 81–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ishibashi M., Ang S. L., Shiota K., Nakanishi S., Kageyama R., Guillemot F. (1995). Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9, 3136–3148. [DOI] [PubMed] [Google Scholar]
- Ishibashi Y., Kohyama-Koganeya A., Hirabayashi Y. (2013). New insights on glucosylated lipids: Metabolism and functions. Biochim. Biophys. Acta 1831, 1475–1485. [DOI] [PubMed] [Google Scholar]
- Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. (1999). Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. U S A 96, 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. B., Laffitte B. A., Patel P. H., Watson M. A., Matsukuma K. E., Walczak R., Collins J. L., Osborne T. F., Tontonoz P. (2002). Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277, 11019–11025. [DOI] [PubMed] [Google Scholar]
- Katakura M., Hashimoto M., Shahdat H. M., Gamoh S., Okui T., Matsuzaki K., Shido O. (2009). Docosahexaenoic acid promotes neuronal differentiation by regulating basic helix-loop-helix transcription factors and cell cycle in neural stem cells. Neuroscience 160, 651–660. [DOI] [PubMed] [Google Scholar]
- Keller R. K., Arnold T. P., Fliesler S. J. (2004). Formation of 7-dehydrocholesterol-containing membrane rafts in vitro and in vivo, with relevance to the Smith-Lemli-Opitz syndrome. J. Lipid Res. 45, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch M., Braun S. M. G., Zurkirchen L., von Schoultz C., Zamboni N., Araúzo-Bravo M. J., Kovacs W. J., Karalay Ö., Suter U., Machado R. A. C., et al. (2013). Metabolic control of adult neural stem cell activity by FASN-dependent lipogenesis. Nature 493, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolf-Clauw M., Chevy F., Wolf C., Siliart B., Citadelle D., Roux C. (1996). Inhibition of 7-dehydrocholesterol reductase by the teratogen AY9944: A rat model for Smith-Lemli-Opitz syndrome. Teratology 54, 115–125. [DOI] [PubMed] [Google Scholar]
- Komada M., Saitsu H., Kinboshi M., Miura T., Shiota K., Ishibashi M. (2008). Hedgehog signaling is involved in development of the neocortex. Development 135, 2717–2727. [DOI] [PubMed] [Google Scholar]
- Korade Z., Kenworthy A. K. (2008). Lipid rafts, cholesterol, and the brain. Neuropharmacology 55, 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudinov A. R., Koudinova N. V. (2001). Essential role for cholesterol in synaptic plasticity and neuronal degeneration. FASEB J. 15, 1858–1860. [DOI] [PubMed] [Google Scholar]
- Kröckel L., Jira, W., and Wild, D. (2003) Identification of benzalkonium chloride in food additives and its efficacy against bacteria in minced meat and raw sausage batters. Eur. Food Res. Technol, 216:402–406. [Google Scholar]
- Laffitte B. A., Repa J. J., Joseph S. B., Wilpitz D. C., Kast H. R., Mangelsdorf D. J., Tontonoz P. (2001). LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. U S A 98, 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P. A., Laffitte B. A., Desrumaux C., Joseph S. B., Curtiss L. K., Mangelsdorf D. J., Tontonoz P., Edwards P. A. (2002). Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J. Biol. Chem. 277, 31900–31908. [DOI] [PubMed] [Google Scholar]
- Makoukji J., Shackleford G., Meffre D., Grenier J., Liere P., Lobaccaro J. -M. A., Schumacher M., Massaad C. (2011). Interplay between LXR and Wnt/β-catenin signaling in the negative regulation of peripheral myelin genes by oxysterols. J. Neurosci. 31, 9620–9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch D. H., Nägler K., Schumacher S., Göritz C., Müller E. C., Otto A., Pfrieger F. W. (2001). CNS synaptogenesis promoted by glia-derived cholesterol. Science 294, 1354–1357. [DOI] [PubMed] [Google Scholar]
- McDonnell G., Russell A. D. (1999). Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre D., Shackleford G., Hichor M., Gorgievski V., Tzavara E. T., Trousson A., Ghoumari A. M., Deboux C., Nait Oumesmar B., Liere P., et al. (2015). Liver X receptors alpha and beta promote myelination and remyelination in the cerebellum. Proc. Natl. Acad. Sci. U S A 112, 7587–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T., Vilo J. (2015). ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F. D., Herman G. E. (2011). Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 52, 6–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J. A., Young K. E., Beachy P. A. (1996). Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255–259. [DOI] [PubMed] [Google Scholar]
- Ratani S. S., Siletzky R. M., Dutta V., Yildirim S., Osborne J. A., Lin W., Hitchins A. D., Ward T. J., Kathariou S. (2012). Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl. Environ. Microbiol. 78, 6938–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C., Dupuis R., Horvath C., Talbot J. N. (1980). Teratogenic effect of an inhibitor of cholesterol synthesis (AY 9944) in rats: Correlation with maternal cholesterolemia. J. Nutr. 110, 2310–2312. [DOI] [PubMed] [Google Scholar]
- Roux C., Horvath C., Dupuis R. (1979). Teratogenic action and embryo lethality of AY 9944R. Prevention by a hypercholesterolemia-provoking diet. Teratology 19, 35–38. [DOI] [PubMed] [Google Scholar]
- Saher G., Brügger B., Lappe-Siefke C., Möbius W., Tozawa R-i., Wehr M. C., Wieland F., Ishibashi S., Nave K.-A. (2005). High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 8, 468–475. [DOI] [PubMed] [Google Scholar]
- Salem N., Litman B., Kim H. Y., Gawrisch K. (2001). Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids 36, 945–959. [DOI] [PubMed] [Google Scholar]
- Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., et al. (2000). Role of LXRs in control of lipogenesis. Genes Dev. 14, 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slimani K., Féret A., Pirotais Y., Maris P., Abjean J. -P., Hurtaud-Pessel D. (2017). Liquid chromatography-tandem mass spectrometry multiresidue method for the analysis of quaternary ammonium compounds in cheese and milk products: Development and validation using the total error approach. J. Chromatogr. A 1517, 86–96. [DOI] [PubMed] [Google Scholar]
- Spitzer J. J. (1973). CNS and fatty acid metabolism. Physiologist 16, 55–68. [PubMed] [Google Scholar]
- Takeoka G. R., Dao L. T., Wong R. Y., Harden L. A. (2005). Identification of benzalkonium chloride in commercial grapefruit seed extracts. J. Agric. Food Chem. 53, 7630–7636. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos S., Wang Y., Kitambi S. S., Sacchetti P., Sousa K. M., Bodin K., Kirk J., Saltó C., Gustafsson M., Toledo E. M., et al. (2013). Brain endogenous liver X receptor ligands selectively promote midbrain neurogenesis. Nat. Chem. Biol. 9, 126–133. [DOI] [PubMed] [Google Scholar]
- Thurm A., Tierney E., Farmer C., Albert P., Joseph L., Swedo S., et al. (2016). Development, behavior, and biomarker characterization of Smith-Lemli-Opitz syndrome: An update. J. Neurodev. Disord. 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tint G. S., Irons M., Elias E. R., Batta A. K., Frieden R., Chen T. S., Salen G. (1994). Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N. Engl. J. Med. 330, 107–113. [DOI] [PubMed] [Google Scholar]
- Tint G. S., Seller M., Hughes-Benzie R., Batta A. K., Shefer S., Genest D. (1995). Markedly increased tissue concentrations of 7-dehydrocholesterol combined with low levels of cholesterol are characteristic of the Smith-Lemli-Opitz syndrome. J. Lipid Res. 36, 89–95. [PubMed] [Google Scholar]
- Tint G. S., Yu H., Shang Q., Xu G., Patel S. B. (2006). The use of the Dhcr7 knockout mouse to accurately determine the origin of fetal sterols. J. Lipid Res. 47, 1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US EPA (2006). Reregistration Eligibility Decision for Alkyl Dimethyl Benzyl Ammonium Chloride (ADBAC). EPA-HQ-OPP-2006-0339, Office of Prevention, Pesticides and Toxic Substances, Antimicrobials Division, U.S. Environmental Protection Agency. Signed on August 3, 2006. [Google Scholar]
- Venkateswaran A., Laffitte B. A., Joseph S. B., Mak P. A., Wilpitz D. C., Edwards P. A., Tontonoz P. (2000). Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc. Natl. Acad. Sci. U S A 97, 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. (2014). Lipid rafts: A signaling platform linking cholesterol metabolism to synaptic deficits in autism spectrum disorders. Front. Behav. Neurosci. 8, 104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshaw J. B., Terry M. L. (1976). Cellular energy metabolism during fetal development. VI. Fatty acid oxidation by developing brain. Dev. Biol. 52, 161–166. [DOI] [PubMed] [Google Scholar]
- Wassif C. A., Zhu P., Kratz L., Krakowiak P. A., Battaile K. P., Weight F. F., et al. (2001). Biochemical, phenotypic and neurophysiological characterization of a genetic mouse model of RSH/Smith–Lemli–Opitz syndrome. Hum. Mol. Genet. 10, 555–564. [DOI] [PubMed] [Google Scholar]
- Xing Y., Fan X., Ying D. (2010). Liver X receptor agonist treatment promotes the migration of granule neurons during cerebellar development. J. Neurochem. 115, 1486–1494. [DOI] [PubMed] [Google Scholar]
- Xu L., Korade Z., Rosado D. A., Liu W., Lamberson C. R., Porter N. A. (2011a). An oxysterol biomarker for 7-dehydrocholesterol oxidation in cell/mouse models for Smith-Lemli-Opitz syndrome. J. Lipid Res. 52, 1222–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Liu W., Sheflin L. G., Fliesler S. J., Porter N. A. (2011b). Novel oxysterols observed in tissues and fluids of AY9944-treated rats: A model for Smith-Lemli-Opitz syndrome. J. Lipid Res. 52, 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., McDonald J. G., Patel A., Zhang Y., Umetani M., Xu F., Westover E. J., Covey D. F., Mangelsdorf D. J., Cohen J. C., et al. (2006). Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 281, 27816–27826. [DOI] [PubMed] [Google Scholar]
- Zhong W., Jiang M. M., Schonemann M. D., Meneses J. J., Pedersen R. A., Jan L. Y., Jan Y. N. (2000). Mouse numb is an essential gene involved in cortical neurogenesis. Proc. Natl. Acad. Sci. U S A 97, 6844–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Chen J., Zhang X., Costa L. G., Guizzetti M. (2014). Prenatal ethanol exposure up-regulates the cholesterol transporters ATP-binding cassette A1 and G1 and reduces cholesterol levels in the developing rat brain. Alcohol . 49, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.