Abstract
Meiotic recombination generates genetic diversity upon which selection can act. Recombination rates are highly variable between species, populations, individuals, sexes, chromosomes, and chromosomal regions. The underlying mechanisms are controlled at the genetic and epigenetic level and show plasticity toward the environment. Environmental plasticity may be divided into short- and long-term responses. We estimated recombination rates in natural populations of wild barley and domesticated landraces using a population genetics approach. We analyzed recombination landscapes in wild barley and domesticated landraces at high resolution. In wild barley, high recombination rates are found in more interstitial chromosome regions in contrast to distal chromosome regions in domesticated barley. Among subpopulations of wild barley, natural variation in effective recombination rate is correlated with temperature, isothermality, and solar radiation in a nonlinear manner. A positive linear correlation was found between effective recombination rate and annual precipitation. We discuss our findings with respect to how the environment might shape effective recombination rates in natural populations. Higher recombination rates in wild barley populations subjected to specific environmental conditions could be a means to maintain fitness in a strictly inbreeding species.
Keywords: recombination landscape, natural variation, climate conditions, domestication
Introduction
Meiotic recombination, the exchange of DNA between homologous chromosomes, and the random segregation of chromosomes into gametes are fundamental to eukaryotic reproduction. Novel allelic combinations are generated through recombination upon which selection can act, which facilitates adaptation. Recombination may create novel beneficial combinations of alleles as well as break apart favorable ones (Barton 1995; Charlesworth and Barton 1996; Otto 2009). In addition to recombination and random segregation, different breeding systems are thought to have an impact on maintaining genetic diversity in a population (Jain 1976). Among plants, there are outbreeding species such as Secale cereale or Hordeum bulbosum showing high levels of heterozygosity, and strictly inbreeding species such as Triticum aestivum and Hordeum vulgare showing high levels of homozygosity (Johnston et al. 2009; International Barley Genome Sequencing Consortium et al. 2012; Bauer et al. 2017; Mascher et al. 2017; International Wheat Genome Sequencing Consortium et al. 2018). In strictly inbreeding species such as barley (H. vulgare) and its wild relative H. vulgare spp. spontaneum (K. Koch) Thell (Brown et al. 1978), the loss of flexibility may provide short-term advantages coupled with long-term disadvantages (Jain 1976). As a main driver of genetic variation, recombination rates are highly variable at multiple scales, for example, between different species (Auton et al. 2012; Stapley et al. 2017), populations of the same species (Kong et al. 2010; Salomé et al. 2012; Spence JP, Song YS, unpublished data), individuals of the same population (Wang et al. 2012), and sexes (Kong et al. 2010; Kianian et al. 2018). Recombination rates vary even along chromosomes, with elevated recombination rates found near the distal ends of the chromosomes and low recombination rates surrounding the pericentromere in most organisms with large genomes (Stapley et al. 2017; Haenel et al. 2018). At fine scales, recombination tends to be focused in narrow hotspots determined by the zinc finger DNA binding protein PRDM9 in most mammals (Boulton et al. 1997; Oliver et al. 2009; Baudat et al. 2010; Myers et al. 2010). In plants, which lack PRDM9, recombination hotspots are determined by chromatin features and are often found within gene promoters (Choi and Henderson 2015). However, similar to rapidly evolving PRDM9 hotspots in mammals (Boulton et al. 1997; Oliver et al. 2009; Baudat et al. 2010; Myers et al. 2010), different Theobroma cacao and rice populations show largely divergent hotspot locations influenced by retrotransposon abundance and genetic divergence (Marand et al. 2019; Schwarzkopf EJ, Motamayor JC, Cornejo OE, unpublished data).
Variation in recombination rate is influenced by genetic, epigenetic, and environmental factors. Genetic divergence and copy number variation of recombination modifiers impact patterns of recombination in plants (Ziolkowski et al. 2015, 2017). Recombination rates are influenced by DNA methylation (Melamed-Bessudo and Levy 2012; Mirouze et al. 2012; Yelina et al. 2012; Habu et al. 2015), histone modifications (Choi et al. 2013; Underwood et al. 2018), and nucleosome occupancy (Choi et al. 2018). Environmental effects on patterns of recombination reveal no clear consensus owing to differences between species and experimental systems. For example, the relationship between temperature and recombination was found to resemble an U-shaped curve, with elevated recombination rates found at low and high temperatures (Plough 1917; Plough 1921; Lloyd et al. 2018; Modliszewski et al. 2018), or a reverse U-shaped curve (Yanney Wilson 1959). Additionally, positive or negative linear correlations were described (Bomblies et al. 2015; Jackson et al. 2015; Phillips et al. 2015). In Drosophila, desiccation, hypoxia, and hyperoxia affect recombination rates, indicating fitness-related plasticity (Aggarwal et al. 2015; Aggarwal DD, Rybnikov SR, Cohen I, Rashkovetsky E, Michalak P, Korol AB, unpublished data). Recombination rates are further influenced by nutritional status or pathogen attack (Law 1963; Kovalchuk et al. 2003; Andronic 2012; Fuchs et al. 2018; Rey et al. 2018). As mentioned by Bomblies et al. (2015), these various correlations bring to mind the question of whether environmental factors act independently or in concert. However, most studies focused on the effect of extreme environmental stress over a single generation, with exceptions in Drosophila, where short breeding cycles allowed observations over a few hundred generations (Law 1963; Kovalchuk et al. 2003; Aggarwal et al. 2015; Phillips et al. 2015; Lloyd et al. 2018).
In the present study, we used a population genetic approach to estimate effective recombination rates in a strictly inbreeding grass species, H. vulgare, and asked whether natural variation in recombination rates is associated with present (1970–2000) and past (6,000–22,000 years before present [BP]) environmental conditions. We find strong similarities between the recombination landscape of wild and domesticated barley based on population genetic data. However, fine-scale differences regarding the physical distribution of recombination events are detected. Finally, we observe natural variation in recombination rate among subpopulations of wild barley and find strong associations with temperature, isothermality, solar radiation, and precipitation.
Results and Discussion
Recombination Landscapes Are Highly Conserved between Wild and Domesticated Barley
The physical distribution and frequency of recombination events, that is, recombination landscape, play a role in plant adaptation as some genes are more likely to recombine than others. The barley genome is highly compartmentalized, with for example, disease resistance genes located in highly recombining distal regions of the chromosomes and genes involved in photosynthesis in low-recombining interstitial regions (Mascher et al. 2017). Previous characterizations of the recombination landscape of barley focused on domesticated barley (Künzel et al. 2000; Higgins et al. 2012; Phillips et al. 2012, 2015; Dreissig et al. 2015, 2017), except for cytological studies revealing a slightly higher number of chiasmata in domesticated barley than in wild barley (Ross-Ibarra 2004). Here, we asked whether the fine-scale physical distribution of recombination events differs between domesticated and wild barleys. We hypothesized different recombination landscapes might be a consequence of adaptation to different environments, for example, natural habitats versus post-Neolithic farming.
In order to estimate recombination rates in wild barley (H. vulgare spp. spontaneum (K. Koch) Thell), we applied coalescent theory to a single nucleotide polymorphism (SNP) data set comprising 26,417 positions in a natural population of 74 geo-referenced accessions (fig. 1) (Milner et al. 2019). The Interval program from the LDhat package was used to estimate the population-scaled recombination rate (ρ) along the seven chromosomes of barley. We validated ancestral recombination rates estimated from population genetic data by comparing them to experimental measurements obtained through pollen nuclei sequencing of an F1 hybrid between two modern barley cultivars (Dreissig et al. 2017). We observed a positive correlation of 0.81 between ancestral and experimental recombination landscape across relative chromosomal intervals of 0.01 (% of total chromosome length, P = 1.48 × 10−24). These correlations are comparable to previous work in Arabidopsis (Choi et al. 2013) and wheat (Darrier et al. 2017), showing that coalescent-based methods provide reliable estimates of recombination landscapes. Next, we attempted to compare wild barley and domesticated landraces in order to test if the domestication process had an impact on their respective recombination landscapes. We estimated the population-scaled recombination rate in barley landraces using a large SNP data set comprising 26,334 SNPs and 100 randomly selected geo-referenced barley landraces (fig. 1, Milner et al. 2019). For both wild barley and landraces, ρ was summed over relative chromosomal intervals and averaged across all seven chromosomes. Based on Spearman’s rank correlation, the two recombination landscapes are highly similar (r = 0.91, P = 2.08 × 10−39). Elevated recombination rates are strictly confined to distal chromosomal regions, leaving about 80% of the chromosomes nearly devoid of recombination (fig. 2). In both, the extent of the recombining region is smaller on the short arm and greater on the long arm of all chromosomes. At the fine-scale, however, differences became visible. On the long arm, elevated recombination rates are detected in more interstitial regions in wild barley (fig. 2, 80–90% relative chromosome length), which is most pronounced on chromosome 2H, 3H, 5H, 6H, and 7H (supplementary fig. 2, Supplementary Material online). In domesticated landraces, elevated recombination rates are more distally confined on the long arm (90–100% relative chromosome length), with no striking difference between chromosomes except for 7H (supplementary fig. 2, Supplementary Material online). On the short arm, however, this does not seem to be the case, as elevated recombination rates are strictly confined to the first 5% of the chromosome in both groups.
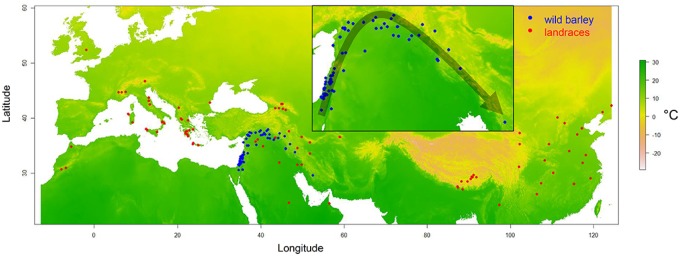
Fig. 1.
Origin of wild barley and landrace accessions. Collection sites of wild barley accessions (blue) and barley landraces (red) are shown. Colouring represents annual mean temperature under present conditions (°C). Within the inlet, which is zoomed in on the Fertile Crescent, the black arrow indicates the direction in which wild barley sub-populations were sampled.
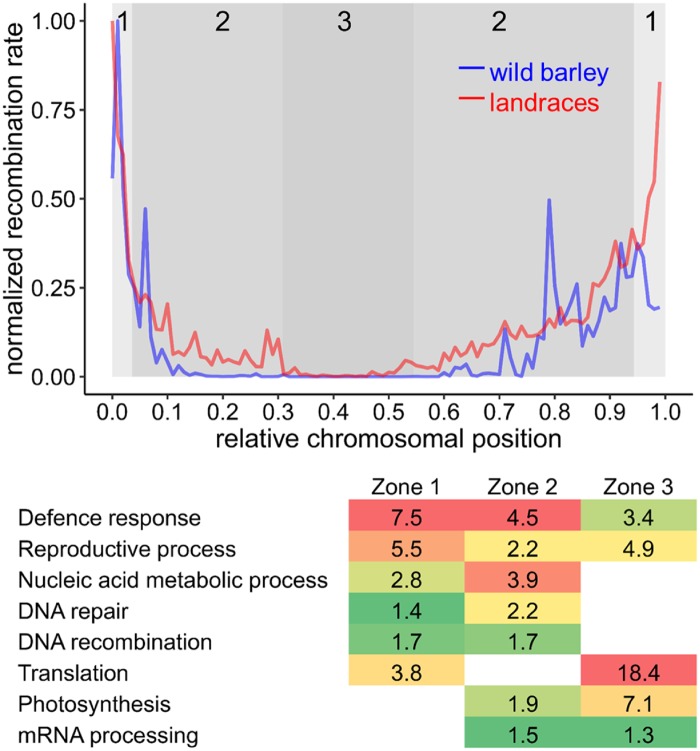
Fig. 2.
Comparison of recombination landscapes between wild barley and domesticated landraces. Normalized recombination rate (0 = lowest value, 1 = highest value within a population) of wild barley (blue) and domesticated landraces (red) along relative chromosomal positions (0 = distal end of the short arm, 1 = distal end of the long arm) derived from the average of all seven chromosomes. On the short arm, highest values in both wild and domesticated barley overlap within the distal tip (5%) of the chromosome. On the long arm, highest values of wild barley reside within 80–90% of chromosome length, whereas highest values of domesticated barley reside within the distal tip (90–100% chromosome length). Enrichment of Gene Ontology (GO) terms in genomic compartments are adopted from Mascher et al. (2017). Coloured rectangles indicate −log10-transformed P-values from 1.3 (green) to 18.4 (red).
Wild barley was estimated to have diverged from its most recent common ancestor approximately 4 Ma (Brassac and Blattner 2015), and domestication began approximately 10,000 years ago (Badr et al. 2000). By comparing the recombination landscape of wild barley with that of domesticated barley landraces, we show that recombination landscapes are highly conserved throughout domestication. Our data provide evidence for a strict separation between chromosomal regions permissive for recombination and chromosomal regions suppressive for recombination, even in long-term ancestral recombination data. Previous work demonstrated meiotic recombination is largely suppressed in heterochromatic regions enriched in CG, CHG, and CHH DNA methylation (Melamed-Bessudo and Levy 2012; Mirouze et al. 2012; Yelina et al. 2012), and histone modifications such as H3K27me3, H3K9me3, H3K27me1, and H3K9me2 (Aliyeva-Schnorr et al. 2015; Baker et al. 2015). A possible explanation for our observations could be that the chromatin environment suppressive for recombination is highly conserved across evolutionary time-scales. At the fine-scale, however, elevated recombination rates are shifted toward more distal regions on the long arm of the chromosomes in domesticated barley. Distal and interstitial regions show different gene contexts, with distal regions enriched for defense response genes and interstitial regions rather enriched for genes involved in basic cellular processes, such as nucleic acid metabolism, DNA repair, photosynthesis, and mRNA processing (fig. 2). As a consequence of this compartmentalization, differences in recombination rate might be caused by selection for elevated recombination rates in regions harboring defense response genes throughout barley’s domestication, as recombination hotspots tend to be found near disease resistance genes (Serra et al. 2018). Wild barley, which is not exposed to high pathogen pressure and does not show strong selection on resistance genes (Stukenbrock and McDonald 2008; Ma et al. 2019), may therefore show a different ancestral recombination landscape.
Natural Variation in Recombination Rate
Recombination rates are highly variable at multiple scales, such as along chromosomes, sexes, individuals, populations, and species (Stapley et al. 2017). In a strictly inbreeding species such as barley (Brown et al. 1978), recombination may be under selection to counterbalance inbreeding depressions and maintain fitness (Charlesworth et al. 1977). Previous studies have shown increased chiasmata frequency in inbreeding plants (Stebbins 1950; Rees and Ahmad 1963; Zarchi et al. 1972; Gibbs et al. 1975). In this study, we asked whether recombination rates differ among natural populations of wild barley.
To analyze variation in recombination rate within a wild barley population, subpopulations needed to be defined for which recombination rates could be estimated. We first performed a principal component analysis (PCA) to test for population structure. The first two principal components explained 8.28% of the observed variance and revealed a continuous gradient along the Fertile Crescent, resembling its shape in the PCA space (supplementary fig. 3A, Supplementary Material online). We analyzed population admixture using sNMF (Frichot et al. 2014) with the number of ancestral populations (K) ranging from 1 to 20. As K increased, the cross-entropy criterion decreased and no local minimum was reached (supplementary fig. 3B and C, Supplementary Material online). This suggested a continuous genetic gradient along the Fertile Crescent, which is supported by the absence of major geographic obstacles.
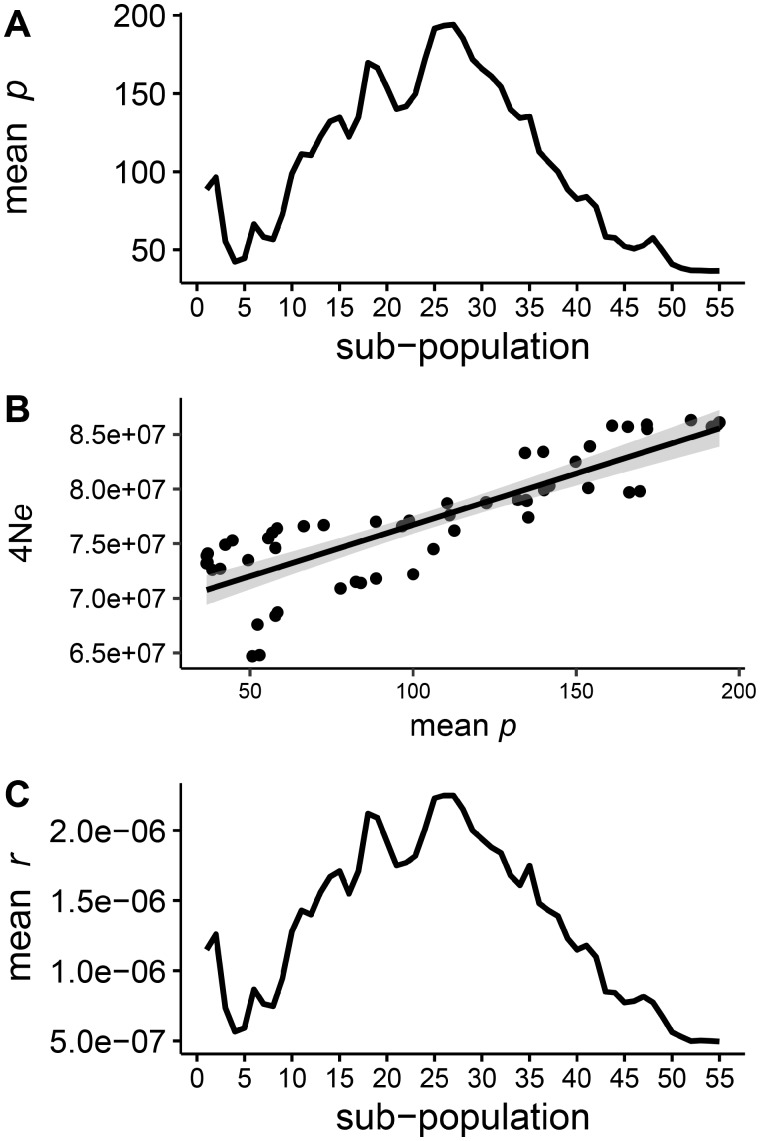
Since it was not feasible to define subpopulations based on ancestry coefficients, we instead defined overlapping subpopulations based on the geographical distribution of wild barley in agreement with their distribution in the PCA space. Subpopulations were defined following a sliding window approach comprising 20 accessions per window with a step size of 1 accession. Sliding windows were moved along the geographical distribution of wild barley in the Fertile Crescent (fig. 1, Russell et al. 2014, 2016). In total, population-scaled recombination rates (ρ) were estimated in 55 subpopulations and averaged across all 7 chromosomes. Our analysis revealed substantial variation among subpopulations (fig. 3A). Importantly, similar trends were observed between individual chromosomes (supplementary fig. 4, Supplementary Material online). For example, the lowest and highest genome-wide average ρ varied by a factor of 5.3. Since ρ is affected by effective population size (ρ = 4Ne × r), we estimated 4Ne based on nucleotide diversity (thetaW, Watterson’s theta) in each subpopulation and an assumed mutation rate (mu) of 3.5 × 10−9 per bp per year (Lin et al. 2002). Effective population size varied between subpopulations by a factor of 1.33 and was positively correlated with ρ (fig. 3B, r = 0.79, P = 4.85 × 10−13). We used the estimates of 4Ne to obtain the effective recombination rate per generation in each subpopulation (re = ρ/4Ne). After correcting for differences in effective population size, variation in effective recombination rate (re) remained largely unchanged, with the lowest and highest genome-wide average re varying by a factor of 4.54 (fig. 3C, Kruskal–Wallis test, P < 2.2 × 10−16). Therefore, variation in the population-scaled recombination rate seemed unlikely to be entirely caused by differences in effective population size. However, it cannot be excluded that different populations may experience different mutation rates. We also tested whether the observed pattern is explained by geographical distance within subpopulations, which may result in higher genetic diversity in subpopulations spanning wider geographical ranges (Owuor et al. 1997; Hübner et al. 2009; Russell et al. 2014) affecting estimates of the population-scaled recombination rate. For each subpopulation, we calculated the longitudinal, latitudinal, and altitudinal range as a measure of geographical distance. For example, almost the whole range of recombination rate values was found twice over distinct geographical ranges (e.g., 50–70 and 300–400 km, supplementary fig. 5, Supplementary Material online). A low number of haplotypes in narrow populations, and a lack of contact in extremely dispersed populations could cause low recombination rates in extremely narrow or dispersed populations. On the other hand, the absence of a linear association and our observation of the full range of recombination rate values over similar geographical ranges suggest that geographical distance does not primarily explain variation in the effective recombination rate. Finally, estimates of effective recombination rates may be influenced by selection. Based on previous work showing that wild barley is not subjected to strong selection and rather found in a state of random genetic drift (Russell et al. 2016; Milner et al. 2019), we conclude the observed differences are unlikely to be caused by patterns of selection. Taken together, effective recombination rates are potentially affected by a multitude of population genetic factors, as well as actual differences in meiotic recombination rate.
Fig. 3.
Subpopulation analysis of ρ, 4Ne, and r in geo-referenced wild barley accessions. Seventy-four geo-referenced wild barley accessions were divided into 55 sub-populations of 20 accessions per sub-population according to a sliding window approach with a step size of 1 accession. Sliding windows are moved along the geographical distribution of wild barley across the Fertile Crescent. (A) Estimation of genome-wide mean population-scaled recombination rate (ρ). (B) Correlation between effective population size (4Ne), based on estimates of Watterson’s theta (thetaW) and an assumed mutation rate (mu) of 3.5 × 10−9, and population-scaled recombination rate (ρ). (C) Genome-wide mean effective recombination rate (re) corrected for differences in effective population size.
Environmental Factors Shape Effective Recombination Rate in Natural Populations
There is a large body of experimental evidence showing correlations between recombination rates and environmental conditions. Particularly, the effect of temperature on meiotic recombination was studied in a number of experimental systems, such as Drosophila, Arabidopsis, barley, and other plants (Dowrick 1957; Mange 1968; Zhuchenko et al. 1985; Jackson et al. 2015; Phillips et al. 2015; Lloyd et al. 2018; Modliszewski et al. 2018). However, as mentioned by Lloyd et al. (2018), an interesting question is whether these observations are reflective of what occurs in natural populations. Therefore, we sought to explore the relationship between effective recombination rate and environmental conditions in natural populations.
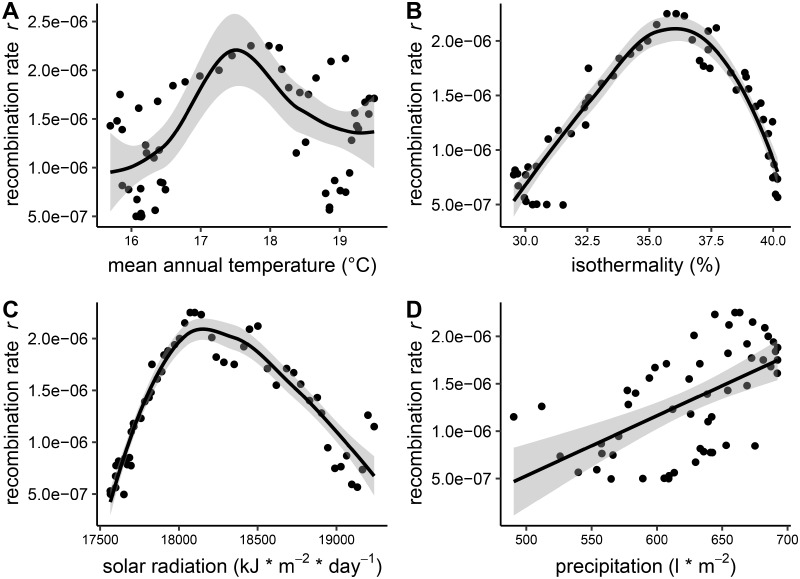
To address this question in natural populations of wild barley, we extracted annual mean temperature values for 74 geo-referenced wild barley accessions under present (1970–2000), Mid Holocene (MH, about 6,000 years BP), and Last Glacial Maximum (LGM, about 22,000 years BP) conditions. After the LGM and throughout the MH, wild barley showed a range expansion across the Fertile Crescent reflecting its current geographical distribution (Russell et al. 2014). We therefore focused on environmental conditions during the MH. Plotting recombination rate against annual mean temperature revealed a nonlinear relationship between temperature and recombination rate (fig. 4A). Across an annual mean temperature range from 15.6 to 19.5 °C, recombination rate was lower in subpopulations at either end of the scale and higher over the intermediate range, showing a reverse U-shaped curve. The same trend was observed by correlating recombination with different temperature conditions, that is, present conditions, MH conditions, and LGM conditions (supplementary figs. 6 and 7, Supplementary Material online). Considering the contribution of meiotic recombination to the effective recombination rate, it is important to consider the timing of meiosis. Meiosis usually takes place in spring under temperatures, which may be different to the annual mean. We therefore used present climate data to test if the trend observed for annual temperature is similar to that observed for approximate spring temperatures, which we considered the mean of March and April. A significant positive correlation indicated that annual mean temperature values reveal a similar trend as spring temperatures ranging from 12 to 16 °C (Pearson’s r = 0.978, P = 1.28 × 10−37).
Fig. 4.
Correlation between recombination rate and environmental variables. Recombination rate estimated in 55 subpopulations of wild barley is plotted against environmental variables. The black line represents a smoothed curve over the data and the gray area represents the 95% confidence interval of the smoothed curve. (A) Relationship between recombination rate and annual mean temperature under Mid Holocene conditions. (B) Reverse U-shaped relationship between recombination rate and isothermality under Mid Holocene conditions. (C) Reverse U-shaped relationship between recombination rate and annual mean solar radiation under present conditions. (D) Correlation between recombination rate and annual precipitation under Mid Holocene conditions.
Interestingly, variation in recombination rate was best explained by isothermality (fig. 4B), which describes temperature variability based on the day-to-night temperature range relative to the summer-to-winter temperature range (i.e., higher values indicating larger temperature variation and vice versa). We observed a reverse U-shaped curve, showing higher recombination rates across an intermediate isothermality range and lower recombination rates at either end of the scale. To test for a systematic bias in our sliding window approach, we performed the same analysis on a set of 55 randomized subpopulations, that is, randomly grouped accessions contrary to grouped according to geographical distribution, focusing on one representative chromosome. When subpopulations were chosen randomly, no correlation with temperature or isothermality was found (MH annual mean temperature: P = 0.11; MH isothermality: P = 0.11; supplementary fig. 8, Supplementary Material online).
In addition to temperature, we also observed a nonlinear relationship between recombination rate and annual solar radiation (fig. 4C). A positive linear relationship was observed with annual precipitation across the three different climate conditions (fig. 4D, present: r = 0.298, P = 0.027; MH: r = 0.572, P = 5.1 × 10−6; LGM: r = 0.765, P = 1.1 × 10−11). Across time, from past (LGM) to present conditions, annual precipitation generally decreased in the Fertile Crescent. Interestingly, higher precipitation under LGM conditions appears to be better suited to explain differences in recombination rate. This is in agreement with a positive correlation between outcrossing rate and annual precipitation (Abdel-Ghani et al. 2004), which also results in higher effective recombination rates (Nordborg 2000). In barley, self-fertilization occurs while the spike is still enclosed in the flag leaf sheath (Alqudah and Schnurbusch 2017), which results in a high inbreeding coefficient in our data (F = 0.978, CV = 0.02%). Therefore, although outcrossing does play a role, we conclude it is likely to have a minor effect in strictly inbreeding barley.
The differences in effective recombination rate between subpopulations were strictly confined to distal regions of the chromosome and no difference was observed in interstitial regions (fig. 5). Considering the contribution of the meiotic recombination rate to the effective recombination rate, it is tempting to speculate on the molecular mechanisms leading to an increase in physically confined regions of the chromosomes. In plants, Drosophila, and yeast, temperature was shown to affect the frequency and distribution of class I crossover as well as meiotic chromosome axis and synaptonemal complex length (Börner et al. 2004; Higgins et al. 2012; Aggarwal et al. 2015; Phillips et al. 2015; Lloyd et al. 2018; Modliszewski et al. 2018). This implies a mechanistic role of some proteins involved in meiotic chromosome axis, synaptonemal complex, and/or CO formation in mediating temperature-dependent plasticity of the recombination landscape which might in part be of biophysical origin as various proteins involved in these processes and their activity are temperature sensitive (Morgan et al. 2017). In addition, UV-radiation and the nutritional state of the plant affect the formation of DNA double strand breaks and meiotic recombination (Grant 1952; Griffing and Langridge 1963; Ries et al. 2000; Knoll et al. 2014; Aggarwal et al. 2015; Mercier et al. 2015; Si et al. 2015; Martín et al. 2017; Rey et al. 2018; Aggarwal DD, Rybnikov SR, Cohen I, Rashkovetsky E, Michalak P, Korol AB, unpublished data).
Fig. 5.
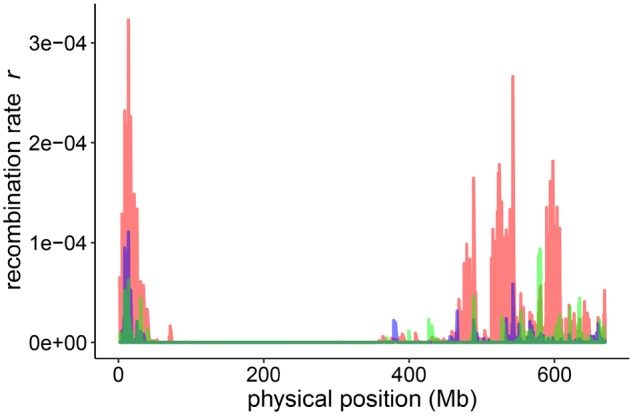
Recombination landscapes of different sub-populations. Recombination rates of different sub-populations (red = #27, blue = #4, green = #55) along the physical length of chromosome 5H. These three subpopulation were selected to represent the observed minima and maxima among all subpopulations. Variation in recombination rate is strictly confined to distal regions of the chromosome and no variation is observed in the low-recombining pericentromeric region.
Our observations suggest that effective recombination rates are higher in populations subjected to intermediate annual temperature, isothermality, and solar radiation rather than extremes. This seems to be contradictory of what was observed in experimental studies in Arabidopsis and barley (Phillips et al. 2015; Lloyd et al. 2018; Modliszewski et al. 2018), where temperature extremes were associated with increased recombination rate. However, key differences between our observations and experimental studies are the number of generations analyzed, the multitude of environmental conditions considered, and population genetic factors. Effective recombination rates estimated by patterns of linkage disequilibrium (LD) are the result of thousands of rounds of recombination and patterns of selection, outcrossing, and ancestral admixture. On the other hand, experimental studies are often limited to one generation owing to the time it takes to cultivate plants. We therefore think of our observations as long-term effective recombination rates, which may differ from short-term responses of the recombination machinery to environmental stress. These differences between long- and short-term effects might be influenced by different alleles of meiotic regulators, leading to differences in recombination rates (Sidhu et al. 2017; Ziolkowski et al. 2017). However, since key meiotic regulators generally show high sequence conservation (Villeneuve and Hillers 2001; Sidhu et al. 2017), our observations may be explained by meiotic plasticity toward the environment. Our data do not enable us to dissect the precise environmental conditions every population/individual faced during each meiotic cycle over time within the Fertile Crescent. However, they allow us to describe broad environmental effects on the recombination landscape of natural populations. For a strictly inbreeding species such as barley, the observed pattern can be interpreted as a measure to balance the loss of fitness caused by inbreeding. Interestingly, Morrell et al. (2005) observed surprisingly low levels of LD in wild barley, comparable to that of Zea mays, an outbreeding species. Our data provide evidence for high effective recombination rates under specific environmental conditions, which could explain the rapid LD decay observed by Morrell et al. (2005).
Taken together, our observations imply differences in how effective recombination rates are correlated with environmental conditions over long periods of time versus short-term responses of the recombination machinery to environmental stress. Under natural conditions, plant populations are subjected to varying environmental conditions, which are tightly linked, such as temperature, light, and precipitation. In controlled experiments, however, often only a single parameter of interest is changed in order to study its effects. Our observations suggest an interplay between temperature, light, and precipitation shaping variation in effective recombination rate in wild barley. A maximum effective recombination rate under intermediate temperature and light, as well as high precipitation may be interpreted as a means of generating genetic diversity upon which selection can act (Presgraves 2005). In a strictly inbreeding species, this might be a mechanism to counteract the negative effects of inbreeding and maintain fitness.
Materials and Methods
Estimation of Population-Scaled Recombination Rate Using LDhat
In the present study, we used genotyping-by-sequencing (GBS) data with imputed SNP calls of 74 geo-referenced wild barley accessions (H. vulgare spp. spontaneum (K. Koch) Thell) and 100 domesticated barley landraces (supplementary file 1, Supplementary Material online) (Milner et al. 2019). Imputation of missing genotypes was performed with FILLIN (Swarts et al. 2014). SNPs with (1) less than 95% missing data, (2) less than 2% heterozygous calls, and (3) a minor allele count ≧10 in a set of 1,140 wild barleys were used as input for imputation (data stored on e!DAL [Arend et al. 2014] http://dx.doi.org/10.5447/IPK/2019/3). The 74 wild barley accessions were chosen out of a total of 1,140 accessions based on the availability of exact geographical coordinates of their collection sites. Accessions with identical collection coordinates were removed to avoid analyzing potential duplications. Barley landraces were randomly chosen out of a total of 19,778, considering only those with exact geographical coordinates available. We selected physically mapped positions in the barley reference genome (Mascher et al. 2017) with <10% missing data, >10% minor allele frequency. Since barley is a strictly inbreeding species, heterozygous SNPs were removed and the data were treated as haploid and phased (Brown et al. 1978). We used the Interval program from the LDhat package (Hudson 2001; McVean et al. 2004; Auton and McVean 2007; Choi et al. 2013) to estimate the population-scaled recombination rate (ρ/kb, ρ = 4Ne × re, where Ne is the effective population size and re is the effective recombination rate per generation in a population) between pairs of SNPs. In total, 26,417 SNPs out of 689,178 imputed SNPs were used for wild barley with an average resolution of 180.85 kb (supplementary fig. 1, Supplementary Material online). For barley landraces, a total of 26,334 SNPs out of 306,049 imputed SNPs was used with an average resolution of 174.01 kb (supplementary fig. 1, Supplementary Material online). The Interval program was run in 60,000,000 iterations, sampling every 5,000 updates, with a block penalty of 5, and using a population-scaled mutation rate per site of theta (theta = 4Ne × mu) = 0.01. To obtain a reproducible output from the Interval program, different iteration numbers (10,000,000 vs. 60,000,000), block penalties (5, 10, 15, 20), and theta values (0.1, 0.01, 0.001) were tested and selected based on visual comparison of multiple runs. The Stat program from the LDhat package was used to summarize ρ/kb values and the first 25,000 iterations were discarded as burn in.
In order to compare recombination rate estimates inferred from population genetic data to experimental measurements based on pollen nuclei sequencing (Dreissig et al. 2017), ρ/kb was converted to ρ by multiplying by interval width (kb). All chromosomes were partitioned into relative intervals of 0.01% and ρ as well as experimental measurements (cM/Mb) were summed over the same relative intervals. The average of all chromosomes was calculated across the relative intervals to obtain a genome-wide overview. In order to compare recombination landscapes regarding the distribution of maxima and minima, ρ values were normalized within each group by dividing by the maximum value to obtain the normalized recombination rate. Spearman’s rank correlation between population-scaled recombination rate and experimental recombination rate was calculated to test if the observed distributions differ and Student’s t-tests were performed to infer statistical significance.
Analysis of Population Structure
PCA was performed using all SNPs employing the snpgdsPCA() function of the SNPrelate package in R (R Core Team, https://www.r-project.org/contributors.html; last accessed June 18, 2019) which implements the FastPCA algorithm of Galinsky et al. (2016). Partitioning of wild barley accessions into K ancestral groups and estimation of individual ancestry coefficients were performed with sNMF (Frichot et al. 2014) using all SNPs. The sNMF algorithm was run for K values between 1 and 20 to identify the optimal K value using the cross-entropy criterion. We used K = 20 to estimate individual ancestry coefficients. Ancestry coefficients were averaged across 15 replications using CLUMPP (Jakobsson and Rosenberg 2007) applying the LargeKGreedy algorithm with 500 permutations.
Subpopulation Analysis in Sliding Windows
To analyze natural variation in the population-scaled recombination rate within the 74 geo-referenced wild barley accessions, we divided the entire wild barley population into 55 subpopulations of 20 accessions per group according to a sliding window approach with a difference of 1 accession between subpopulations. Sliding windows were selected according to the geographical distribution of wild barley across the Fertile Crescent, starting from the south of Israel, moving further north across the Lebanon to the north-western part of Syria and southern Turkey, further east to the north-eastern part of Syria and south-eastern Turkey, and further south-east across the northern part of Iraq and western part of Iran (fig. 1, Russell et al. 2014). ρ/kb was estimated for each subpopulation using the Interval program with identical parameters as mentioned above.
To account for potential differences in 4Ne between subpopulations, we estimated 4Ne by estimating nucleotide diversity (thetaW, Watterson’s theta) in each subpopulation and assuming a mutation rate per bp (mu) of 3.5 × 10−9 which was previously described for wild barley (Lin et al. 2002). We estimated thetaW using the Convert program of the LDhat package and used the genome-wide average thetaW value of each subpopulation to estimate the effective size of each subpopulation. Nucleotide diversity (thetaW) was divided by mutation rate per bp (mu) to estimate 4Ne (4Ne = thetaW/mu). We then used the estimates of 4Ne to calculate the mean effective recombination rate per generation (re) for each subpopulation (re = ρ/4Ne). Accessions were also randomly assigned to subpopulations and analyzed in the same way to subsequently test for a systematic bias.
The inbreeding coefficient (F) was calculated in each subpopulation using the –het function of vcftools including all SNPs (Danecek et al. 2011).
Climate Data and Correlation with Recombination Rate
Climate data were obtained from the global climate database (worldclim.org). Bioclimatic variables, including annual mean temperature (°C), isothermality (%), and annual precipitation (mm) were extracted for present conditions (1970–2000), MH conditions (about 6000 years BP), and LGM conditions (about 22,000 years BP) from the CMIP5 MultiModel Ensemble data set (MPI-ESM-P) from the 30 arc-second and 2.5 arc-minute rasters. Solar radiation values (kJ × m−2 × day−1) were extracted for present conditions from 30 arc-second (0.86 km2 at the equator) and 2.5 arc-minute (21.6 km2 at the equator) rasters. We focused on the above mentioned bioclimatic variables because of the extensively studied effects of temperature, precipitation, and solar radiation on meiosis (see Results and Discussion). To estimate spring temperature, we calculated the average of March and April under present conditions, since no monthly climate data are available under MH or LGM conditions. Additionally, elevation data from the NASA Shuttle Radar Topographic Mission (SRTM) was obtained from the CGIAR Consortium for Spatial Information (http://srtm.csi.cgiar.org/) at 90 m resolution.
We used the “extract” function of the “raster” package of the R statistical environment (R Core Team, https://www.r-project.org/contributors.html) to extract the respective bioclimatic values at the locations of the geo-referenced barley accessions. All values were extracted in a 5 km radius using the “buffer” function and averaged using the “fun=mean” function of the “raster” package.
For each subpopulation, we calculated mean temperature, mean isothermality, mean precipitation, mean solar radiation, and mean elevation. For linear relationships (e.g., precipitation), we calculated Pearson’s correlation coefficient between effective recombination rate (re) and bioclimatic variables in each subpopulation. Student’s t-tests were performed to infer statistical significance. In case of nonlinear relationships (e.g., annual mean temperature, isothermality, solar radiation), a Kruskal–Wallis test was performed to compare between subpopulations.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to acknowledge all of those who actually collected all the data in previous studies and made their data available. We would also like to thank Danuta Schueler for providing us with the exact collection sites of all wild barleys and landraces, Albert Wilhelm Schulthess for valuable feedback, all members of the Meiosis team at the IPK for fruitful discussions, Andreas Houben, Joerg Fuchs, and Ingo Schubert for critical reading of the manuscript, as well as the German Federal Ministry of Education and Research (BMBF—FKZ 031B0188) and the IPK Gatersleben for financial support. M.M. acknowledges support from the BMBF (Grant FKZ 031B0190) and Pakt für Forschung und Innovation: SAW-2015-IPK-1 ‘BRIDGE’.
References
- Abdel-Ghani AH, Parzies HK, Omary A, Geiger HH.. 2004. Estimating the outcrossing rate of barley landraces and wild barley populations collected from ecologically different regions of Jordan. Theor Appl Genet. 1093:588–595. [DOI] [PubMed] [Google Scholar]
- Aggarwal DD, Rashkovetsky E, Michalak P, Cohen I, Ronin Y, Zhou D, Haddad GG, Korol AB.. 2015. Experimental evolution of recombination and crossover interference in Drosophila caused by directional selection for stress-related traits. BMC Biol. 13:101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliyeva-Schnorr L, Beier S, Karafiátová M, Schmutzer T, Scholz U, Doležel J, Stein N, Houben A.. 2015. Cytogenetic mapping with centromeric bacterial artificial chromosomes contigs shows that this recombination-poor region comprises more than half of barley chromosome 3H. Plant J. 842:385–394. [DOI] [PubMed] [Google Scholar]
- Alqudah AM, Schnurbusch T.. 2017. Heading date is not flowering time in spring barley. Front Plant Sci. 8:896.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronic L. 2012. Viruses as triggers of DNA rearrangements in host plants. Can J Plant Sci. 926:1083–1091. [Google Scholar]
- Arend D, Lange M, Chen J, Colmsee C, Flemming S, Hecht D, Scholz U.. 2014. e!DAL—a framework to store, share and publish research data. BMC Bioinformatics 15:214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Ségurel L, Street T, Leffler EM, Bowden R, Aneas I, Broxholme J, et al. 2012. A fine-scale chimpanzee genetic map from population sequencing. Science 3366078:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, McVean G.. 2007. Recombination rate estimation in the presence of hotspots. Genome Res. 178:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr A, Müller K, Sch R, Rabey HE, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F.. 2000. On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol. 174:499–510. [DOI] [PubMed] [Google Scholar]
- Baker K, Dhillon T, Colas I, Cook N, Milne I, Milne L, Bayer M, Flavell AJ.. 2015. Chromatin state analysis of the barley epigenome reveals a higher-order structure defined by H3K27me1 and H3K27me3 abundance. Plant J. 841:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. 1995. A general model for the evolution of recombination. Genet Res. 652:123–145. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B.. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 3275967:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E, Schmutzer T, Barilar I, Mascher M, Gundlach H, Martis MM, Twardziok SO, Hackauf B, Gordillo A, Wilde P, et al. 2017. Towards a whole-genome sequence for rye (Secale cereale L.). Plant J. 895:853–869. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Higgins JD, Yant L.. 2015. Meiosis evolves: adaptation to external and internal environments. New Phytol. 2082:306–323. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N.. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 1171:29–45. [DOI] [PubMed] [Google Scholar]
- Boulton A, Myers RS, Redfield RJ.. 1997. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci U S A. 9415:8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassac J, Blattner FR.. 2015. Species-level phylogeny and polyploid relationships in Hordeum (Poaceae) inferred by next-generation sequencing and in silico cloning of multiple nuclear loci. Syst Biol. 645:792–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AHD, Zohary D, Nevo E.. 1978. Outcrossing rates and heterozygosity in natural populations of Hordeum spontaneum Koch in Israel. Heredity 411:49–62. [Google Scholar]
- Charlesworth B, Barton NH.. 1996. Recombination load associated with selection for increased recombination. Genet Res. 671:27–41. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Strobeck C.. 1977. Effects of selfing on selection for recombination. Genetics 861:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Henderson IR.. 2015. Meiotic recombination hotspots—a comparative view. Plant J. 831:52–61. [DOI] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FCH, et al. 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet. 4511:1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Tock AJ, Lambing C, Underwood CJ, Hardcastle TJ, Serra H, Kim J, Cho HS, Kim J, et al. 2018. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 284:532–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 2715:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrier B, Rimbert H, Balfourier F, Pingault L, Josselin A-A, Servin B, Navarro J, Choulet F, Paux E, Sourdille P.. 2017. High-resolution mapping of crossover events in the hexaploid wheat genome suggests a universal recombination mechanism. Genetics 2063:1373–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowrick GJ. 1957. The influence of temperature on meiosis. Heredity 111:37. [Google Scholar]
- Dreissig S, Fuchs J, Cápal P, Kettles N, Byrne E, Houben A.. 2015. Measuring meiotic crossovers via multi-locus genotyping of single pollen grains in barley. PLoS One 109:e0137677.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreissig S, Fuchs J, Himmelbach A, Mascher M, Houben A.. 2017. Sequencing of single pollen nuclei reveals meiotic recombination events at megabase resolution and circumvents segregation distortion caused by postmeiotic processes. Front Plant Sci. 8:1620.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frichot E, Mathieu F, Trouillon T, Bouchard G, François O.. 2014. Fast and efficient estimation of individual ancestry coefficients. Genetics 1964:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs L, Jenkins G, Phillips D.. 2018. Anthropogenic impacts on meiosis in plants. Front Plant Sci. 9:1429.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinsky KJ, Bhatia G, Loh P-R, Georgiev S, Mukherjee S, Patterson NJ, Price AL.. 2016. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 983:456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PE, Milne C, Carrillo MV.. 1975. Correlation between the breeding system and recombination index in five species of Senecio. New Phytol. 753:619–626. [Google Scholar]
- Grant V. 1952. Cytogenetics of the hybrid Gilia millefoliata × achilleaefolia. Chromosoma 54:372–390. [PubMed] [Google Scholar]
- Griffing B, Langridge J.. 1963. Factors affecting crossing over in the tomato. Aust J Biol Sci. 164:826–837. [Google Scholar]
- Habu Y, Ando T, Ito S, Nagaki K, Kishimoto N, Taguchi-Shiobara F, Numa H, Yamaguchi K, Shigenobu S, Murata M, et al. 2015. Epigenomic modification in rice controls meiotic recombination and segregation distortion. Mol Breed. 35:103. [Google Scholar]
- Haenel Q, Laurentino TG, Roesti M, Berner D.. 2018. Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics. Mol Ecol. 2711:2477–2497. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin F.. 2012. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 2410:4096–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner S, Höffken M, Oren E, Haseneyer G, Stein N, Graner A, Schmid K, Fridman E.. 2009. Strong correlation of wild barley (Hordeum spontaneum) population structure with temperature and precipitation variation. Mol Ecol. 187:1523–1536. [DOI] [PubMed] [Google Scholar]
- Hudson RR. 2001. Two-locus sampling distributions and their application. Genetics 1594:1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium, Mayer KFX, Waugh R, Brown JWS, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, et al. 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium, IWGSC RefSeq principal investigators, Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N IWGSC whole-genome assembly principal investigators Pozniak CJ, et al. 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:6403.. [DOI] [PubMed] [Google Scholar]
- Jackson S, Nielsen DM, Singh ND.. 2015. Increased exposure to acute thermal stress is associated with a non-linear increase in recombination frequency and an independent linear decrease in fitness in Drosophila. BMC Evol Biol. 15:175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain SK. 1976. The evolution of inbreeding in plants. Annu Rev Ecol Syst. 71:469. [Google Scholar]
- Jakobsson M, Rosenberg NA.. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2314:1801–1806. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Timmerman-Vaughan GM, Farnden KJF, Pickering R.. 2009. Marker development and characterisation of Hordeum bulbosum introgression lines: a resource for barley improvement. Theor Appl Genet. 1188:1429–1437. [DOI] [PubMed] [Google Scholar]
- Kianian PMA, Wang M, Simons K, Ghavami F, He Y, Dukowic-Schulze S, Sundararajan A, Sun Q, Pillardy J, Mudge J, et al. 2018. High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat Commun. 91:2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A, Fauser F, Puchta H.. 2014. DNA recombination in somatic plant cells: mechanisms and evolutionary consequences. Chromosome Res. 222:191–201. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 4677319:1099–1103. [DOI] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B.. 2003. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 4236941:760–762. [DOI] [PubMed] [Google Scholar]
- Künzel G, Korzun L, Meister A.. 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 1541:397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CN. 1963. An effect of potassium on chiasma frequency and recombination. Genetica 331:313–329. [Google Scholar]
- Lin J-Z, Morrell PL, Clegg MT.. 2002. The influence of linkage and inbreeding on patterns of nucleotide sequence diversity at duplicate alcohol dehydrogenase loci in wild barley (Hordeum vulgare ssp. spontaneum). Genetics 1624:2007–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Morgan C, Franklin FCH, Bomblies K.. 2018. Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 2084:1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Liu M, Stiller J, Liu C.. 2019. A pan-transcriptome analysis shows that disease resistance genes have undergone more selection pressure during barley domestication. BMC Genomics 201:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mange EJ. 1968. Temperature sensitivity of segregation-distortion in Drosophila melanogaster. Genetics 583:399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Zhao H, Zhang W, Zeng Z, Fang C, Jiang J.. 2019. Historical meiotic crossover hotspots fueled patterns of evolutionary divergence in rice. Plant Cell 31(3):645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, Rey M-D, Shaw P, Moore G.. 2017. Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 126:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 5447651:427–433. [DOI] [PubMed] [Google Scholar]
- McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P.. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 3045670:581–584. [DOI] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Levy AA.. 2012. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci U S A. 10916:E981–E988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M.. 2015. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 66:297–327. [DOI] [PubMed] [Google Scholar]
- Milner SG, Jost M, Taketa S, Mazón ER, Himmelbach A, Oppermann M, Weise S, Knüpffer H, Basterrechea M, König P, et al. 2019. Genebank genomics highlights the diversity of a global barley collection. Nat Genet. 512:319–326. [DOI] [PubMed] [Google Scholar]
- Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J.. 2012. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A. 10915:5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modliszewski JL, Wang H, Albright AR, Lewis SM, Bennett AR, Huang J, Ma H, Wang Y, Copenhaver GP.. 2018. Elevated temperature increases meiotic crossover frequency via the interfering (type I) pathway in Arabidopsis thaliana. PLoS Genet. 145:e1007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CH, Zhang H, Bomblies K.. 2017. Are the effects of elevated temperature on meiotic recombination and thermotolerance linked via the axis and synaptonemal complex? Philos Trans R Soc Lond B Biol Sci. 3721736:20160470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell PL, Toleno DM, Lundy KE, Clegg MT.. 2005. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc Natl Acad Sci U S A. 1027:2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P.. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 3275967:876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M. 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 1542:923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP.. 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 512:e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. 2009. The evolutionary enigma of sex. Am Nat. 174(Suppl 1):S1–S14. [DOI] [PubMed] [Google Scholar]
- Owuor ED, Fahima T, Beiles A, Korol A, Nevo E.. 1997. Population genetic response to microsite ecological stress in wild barley, Hordeum spontaneum. Mol Ecol. 612:1177–1187. [Google Scholar]
- Phillips D, Jenkins G, Macaulay M, Nibau C, Wnetrzak J, Fallding D, Colas I, Oakey H, Waugh R, Ramsay L.. 2015. The effect of temperature on the male and female recombination landscape of barley. New Phytol. 2082:421–429. [DOI] [PubMed] [Google Scholar]
- Phillips D, Nibau C, Wnetrzak J, Jenkins G.. 2012. High resolution analysis of meiotic chromosome structure and behaviour in barley (Hordeum vulgare L.). PLoS One 76:e39539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plough HH. 1917. The effect of temperature on crossing over in Drosophila. J Exp Zool. 242:147–209. [Google Scholar]
- Plough HH. 1921. Further studies on the effect of temperature on crossing over. J Exp Zool. 322:187–202. [Google Scholar]
- Presgraves DC. 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Curr Biol. 1518:1651–1656. [DOI] [PubMed] [Google Scholar]
- Rees H, Ahmad K.. 1963. Chiasma frequencies in Lolium populations. Evolution 174:575–579. [Google Scholar]
- Rey M-D, Martín AC, Smedley M, Hayta S, Harwood W, Shaw P, Moore G.. 2018. Magnesium increases homoeologous crossover frequency during meiosis in ZIP4 (Ph1 gene) mutant wheat-wild relative hybrids. Front Plant Sci. 9:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries G, Heller W, Puchta H, Sandermann H, Seidlitz HK, Hohn B.. 2000. Elevated UV-B radiation reduces genome stability in plants. Nature 4066791:98–101. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra J. 2004. The evolution of recombination under domestication: a test of two hypotheses. Am Nat. 1631:105–112. [DOI] [PubMed] [Google Scholar]
- Russell J, Mascher M, Dawson IK, Kyriakidis S, Calixto C, Freund F, Bayer M, Milne I, Marshall-Griffiths T, Heinen S, et al. 2016. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat Genet. 489:1024–1030. [DOI] [PubMed] [Google Scholar]
- Russell J, van Zonneveld M, Dawson IK, Booth A, Waugh R, Steffenson B.. 2014. Genetic diversity and ecological niche modelling of wild barley: refugia, large-scale post-LGM range expansion and limited mid-future climate threats? PLoS One 92:e86021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Fitz J, Laitinen RAE, Warthmann N, Yant L, Weigel D.. 2012. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity 1084:447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra H, Choi K, Zhao X, Blackwell AR, Kim J, Henderson IR.. 2018. Interhomolog polymorphism shapes meiotic crossover within the Arabidopsis RAC1 and RPP13 disease resistance genes. PLoS Genet. 1412:e1007843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si W, Yuan Y, Huang J, Zhang X, Zhang Y, Zhang Y, Tian D, Wang C, Yang Y, Yang S.. 2015. Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F2 plants. New Phytol. 2064:1491–1502. [DOI] [PubMed] [Google Scholar]
- Sidhu GK, Warzecha T, Pawlowski WP.. 2017. Evolution of meiotic recombination genes in maize and teosinte. BMC Genomics 181:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM.. 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos Trans R Soc Lond B Biol Sci. 3721736:20160455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. London: Geoffrey Cumberlege. [Google Scholar]
- Stukenbrock EH, McDonald BA.. 2008. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol. 46:75–100. [DOI] [PubMed] [Google Scholar]
- Swarts K, Li H, Romero Navarro JA, An D, Romay MC, Hearne S, Acharya C, Glaubitz JC, Mitchell S, Elshire RJ, et al. 2014. Novel methods to optimize genotypic imputation for low-coverage, next-generation sequence data in crop plants. Plant Genome 73. [Google Scholar]
- Underwood CJ, Choi K, Lambing C, Zhao X, Serra H, Borges F, Simorowski J, Ernst E, Jacob Y, Henderson IR, et al. 2018. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 284:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM, Hillers KJ.. 2001. Whence meiosis? Cell 1066:647–650. [DOI] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR.. 2012. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 1502:402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanney Wilson J. 1959. Chiasma frequency in relation to temperature. Genetica 291:290–303. [Google Scholar]
- Yelina NE, Choi K, Chelysheva L, Macaulay M, de Snoo B, Wijnker E, Miller N, Drouaud J, Grelon M, Copenhaver GP, et al. 2012. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 88:e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarchi Y, Simchen G, Hillel J, Schaap T.. 1972. Chiasmata and the breeding system in wild populations of diploid wheats. Chromosoma 381:77–94. [Google Scholar]
- Zhuchenko AA, Korol AB, Kovtyukh LP.. 1985. Change of the crossing-over frequency in Drosophila during selection for resistance to temperature fluctuations. Genetica 671:73–78. [Google Scholar]
- Ziolkowski PA, Berchowitz LE, Lambing C, Yelina NE, Zhao X, Kelly KA, Choi K, Ziolkowska L, June V, Sanchez-Moran E, et al. 2015. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife 4:e03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Underwood CJ, Lambing C, Martinez-Garcia M, Lawrence EJ, Ziolkowska L, Griffin C, Choi K, Franklin FCH, Martienssen RA, et al. 2017. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 313:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.