Abstract
Background
Despite tremendous interest in modulating the microbiome to improve health, the association between diet and the colonic mucosa–associated gut microbiome in healthy individuals has not been examined.
Objective
To investigate the associations between Healthy Eating Index (HEI)–2005 and the colonic mucosa–associated microbiota.
Methods
In this cross-sectional observational study, we analyzed bacterial community composition and structure using 16S rRNA gene (V4 region) sequencing of 97 colonic mucosal biopsies obtained endoscopically from different colon segments of 34 polyp-free participants. Dietary consumption was ascertained using an FFQ. Differences in α- and β-diversity and taxonomic relative abundances between the higher and lower score of total HEI and its components were compared, followed by multivariable analyses.
Results
The structure of the microbiota significantly differed by the scores for total HEI, total and whole fruits (HEI 1 and HEI 2), whole grains (HEI 6), milk products and soy beverages (HEI 7), and solid fat, alcohol, and added sugar (HEI 12). A lower score for total HEI and HEIs 2, 7, and 12 was associated with significantly lower richness. A lower score for total HEI was associated with significantly reduced relative abundance of Parabacteroides, Roseburia, and Subdoligranulum but higher Fusobacterium. A lower score for HEI 2 was associated with lower Roseburia but higher Bacteroides. A lower score for HEI 7 was associated with lower Faecalibacterium and Fusobacterium but higher Bacteroides. A lower score for HEI 12 was associated with lower Subdoligranulum but higher Escherichia and Fusobacterium (false discovery rate–adjusted P values <0.05). The findings were confirmed by multivariate analysis. Less abundant bacteria such as Alistipes, Odoribacter, Bilophila, and Tyzzerella were also associated with dietary quality.
Conclusions
A lower score for total HEI–2005 was significantly associated with reduced relative abundance of potentially beneficial bacteria but increased potentially harmful bacteria in the colonic mucosa of endoscopically normal individuals.
Keywords: diet, dietary pattern, healthy eating index, microbiota, colon, fruit, dairy products, fat
Introduction
Diet is a potentially modifiable risk factor of multiple diseases. The Western-style, low-fiber diet rich in processed meat, fat, sugar, and sodium has been associated with increased risk of metabolic diseases including inflammatory bowel diseases (IBDs) and colorectal cancer (1), whereas a plant-based, high-fiber diet rich in fruit, vegetables, and whole grains has been associated with reduced risk of these diseases (2–4). The Healthy Eating Index (HEI)–2005 has been inversely associated with risk of incident cancers, including pancreatic cancer, in our prior research (5).
Ample evidence suggests that diet is a principal factor modulating gut microbial composition (6, 7). In turn, the human gut microbiota has a major impact on colonization resistance against intestinal pathogens, nutrient uptake, vitamin synthesis, energy harvest, carcinogen metabolism, chronic inflammation, and host immune response (8, 9). Previous studies examining the fecal microbiota of human volunteers revealed that a high-fat, low-fiber diet was associated with increased inflammation-associated Bacteroides, Bilophila, and Escherichia coli and decreased Roseburia, which metabolizes dietary plant-derived polysaccharides (9, 10). The diet-driven shift in microbial composition leads to variations in producing SCFAs, which affect host metabolism, epithelial barrier function, and mucosal inflammation and proliferation (11).
There has been tremendous interest in modulating the microbiota to improve health. However, the association between dietary quality and the colonic mucosa–associated gut microbiota in healthy individuals has not been rigorously investigated. By far, most human-gut-microbiota studies have used fecal samples. Fecal microbiota are different from colonic-adherent microbiota that interact more directly with the host immune system. Therefore, mucosa-associated and fecal microbiota may fulfill distinct roles within the colon ecosystem (12, 13) and, thus, their associations with diet may also differ.
In this study, our objective was to examine the association between the total HEI–2005 score and its 12 food-based components and the community composition and structure of the colonic mucosa–associated microbiota.
Methods
Study population and design
In our cross-sectional case-control study designed to examine the association between the gut microbiome and risk of colorectal tumor, we prospectively and consecutively enrolled participants aged 50–75 y who underwent a clinically ordered colonoscopy at the Michael E DeBakey Veterans Affairs Medical Center, Houston, between 2013 and 2017. We did not recruit patients with a history of: 1) familial or hereditary colon diseases or IBD; 2) invasive cancer, except for nonmelanoma skin cancer; 3) colorectal polyps in the past 3 y; 4) end-stage renal disease requiring dialysis; 5) severe mental disabilities; 6) surgery or hospitalization within the past year; 7) oral or systemic use of antibiotics in the past 3 mo; 8) hepatitis B virus, hepatitis C virus, and HIV, or methicillin-resistant Staphylococcus aureus-positive infection; or 9) bleeding disorders and anticoagulant use. Participants who had changed dietary habits in the past 3 mo were also not included. Participants were advised to stop taking routinely used medications 7 d before the procedure and to stop antidiabetic medications 1 d before.
All participants provided written informed consent. The procedures followed were in accordance with the ethical standards of the institution. The protocol was approved by the Institutional Review Boards at both Baylor College of Medicine and the Michael E DeBakey Veterans Affairs Medical Center.
Data collection
Each participant attended an education session 1–2 wk before the colonoscopy procedure. The research coordinator administered a questionnaire to collect information on lifestyle and medical history and obtained anthropometric measurements using a calibrated scale. We assessed dietary consumption in the past year using the validated 110-item semiquantitative 2005 Block FFQ (14). Participants completed this survey at home and mailed it back before the colonoscopy, including a reminder call if necessary, with a response rate of 87%. We called participants to complete sporadic missing responses on the FFQ.
Dietary quality was defined by the HEI–2005, based on the key recommendations from the Dietary Guidelines for Americans. It comprises 12 food-based components, including 9 adequacy components and 3 moderation components (15).
Collection of colonic mucosal biopsies
We enrolled 612 participants during the study period, 562 completed the colonoscopy procedure, and 172 had normal colons and therefore were eligible to be included in this study. Among them, 133 had colonic biopsies (1–6 pieces) collected from the 6 colon segments when possible: cecum, ascending colon, transverse colon, descending colon, sigmoid colon, and rectum. All biopsies were immediately placed in a sterile tube on dry ice and transferred to a –80°C freezer within 15 min.
Bacterial DNA isolation, library preparation, 16S rRNA gene sequencing, and bioinformatics
DNA extraction and bacterial 16S rRNA gene sequencing were conducted at the Center for Metagenomics and Microbiome Research at Baylor College of Medicine (16). DNA was extracted from the colonic biopsy using the Powerlyzer UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories) and immediately stored at –20°C before the amplification step. We included negative control (buffer blank) samples. These were blinded and processed alongside mucosal biopsies during data generation and processing.
We targeted the fouth hypervariable (V4) region of the 16S rRNA gene because of its high domain specificity and broad coverage of gastrointestinal bacteria compared with other variable regions (17, 18). The V4 region was amplified by PCR using primers 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT). Each resulting amplicon set was barcoded with a unique 12mer tag (19). Successful amplicons were pooled at a similar equal molar DNA concentration, purified, and sequenced in the MiSeq platform (Illumina), using the 2 × 250-bp paired-end protocol yielding pair-end reads that overlap almost completely. This protocol targets ≥10,000 reads per sample.
We used a pipeline developed at the Center for Metagenomics and Microbiome Research for the bioinformatics analysis. Briefly, the read pairs were demultiplexed based on the unique molecular barcodes added via PCR during library generation, then merged using the Ultrafast Sequence Analysis (20). Sequences were assigned into operational taxonomic units (OTUs) at a similarity cut-off value of 97% using the UPARSE algorithm (21). The OTUs were subsequently mapped to an optimized version of the SILVA database to determine taxonomies (22). We defined major taxa as having a relative abundance of >1.5% and rare taxa as 0.05–1.5%. To increase reproducibility, our bioinformatic processing includes raw sequence quality control, mate pair stitching, and removal of spurious sequence and chimeras (23).
The mucosal samples from 69 participants were sent for 16S rRNA gene sequencing. The sequencing run was performed in 4 batches. The mucosal samples from the same participant were processed in the same batch. Among these 69 participants, 40 returned the FFQ, and 5 were excluded because they had a self-reported energy intake <800 or >5000 kcal per day. These 35 participants contributed 99 pieces of mucosal biopsies to the study. After the negative control (reads <100), low-quality (reads <1654), spurious, and singleton sequences were removed, the dataset was rarefied to 1654 reads per sample. Two mucosal samples (included 1 single biopsy from 1 participant with lower dietary quality) with poor sequencing results were excluded further. Therefore, we included 97 mucosal biopsies from 34 participants in the final analysis (Supplemental Figure 1). Of the 34 participants, 27 did not have a history of polyps and 7 had polyps >3 y earlier.
Statistical analysis
We categorized higher compared with lower dietary quality score based on the median scores of the total HEI and each of its 12 individual components in 34 participants. The gut microbiota profile was the single primary endpoint of the present study. The general characteristics of the participants based on dietary quality were compared using the Student's t test or Fisher's exact test.
The α-diversity was measured by the Shannon diversity index that measures both community richness and evenness. The Shannon index and taxonomic relative abundances (at phylum and genus levels) were compared based on the HEI scores using the Mann–Whitney test. β-Diversity (microbial structures) was compared using the Weighted UniFrac as the distance matrix (24). The distances were visualized by a Principal Coordinate Analysis plot and the Monte-Carlo permutation test was performed to estimate P values.
When we observed an association between major taxa and dietary quality in the univariate analysis, we further calculated the coefficient of fold change (FC) of the relative counts based on higher compared with lower dietary quality score using the empirical Bayes shrinkage method based on negative binomial distribution (DESeq2) (25). The relative sequencing counts were normalized by dividing the raw counts by the DESeq2 size factor for each sample. The multivariable model was adjusted for age, race, BMI, smoking status, alcohol use, type 2 diabetes, colon segment, and other major OTUs. In the DESeq2 package, we used the segment variable to adjust for within-sample variation. To account for the dependent microbiome sequence and covariates from the same participants, we created a cluster identification (ID) variable from the original study ID to distinguish participants who contributed multiple samples to the analysis. The cluster ID was created within each level of the main confounding variable (e.g., smoking, obesity).
In addition, we conducted a sensitivity analysis to examine the association between major bacterial genera and the total HEI using only the sigmoid specimen because 23 of 34 participants had a sigmoid colon biopsy. To alleviate concern regarding the reproducibility of the sequencing assay by different batches, we included the samples from the single sequencing batch (56 mucosal biopsies from 13 participants) in the sensitivity analysis. Lastly, we conducted the exploratory univariate linear regression on the correlation between the bacterial relative abundance and each HEI component on a continuous scale.
All statistical analyses were performed using SAS 9.4 (SAS Inc.) and R statistical software (version 3.4.4, R Foundation). A P value <0.05 denoted statistical significance for general analyses. All P values were adjusted for multiple comparisons using the false discovery rate (FDR) algorithm in the microbiome analysis (26). An FDR-adjusted 2-sided P value <0.05 denoted statistical significance.
Results
General characteristics of participants based on dietary quality and their associations with the gut microbiome
Demographic characteristics of 34 participants (1 woman) who underwent colonic mucosal biopsies are summarized in Table 1. A low-quality diet was defined as one with a total HEI score <60 and a high-quality diet with a total HEI score ≥60. There was no significant difference in the distribution of the demographic, medical history, and exposure variables between 2 quality groups, or the number and segment of biopsies used. In addition, our previous study showed that the bacterial composition did not differ by colon segments (Supplemental Figure 2). Because the relative abundance of major bacterial genera differed significantly by smoking, alcohol use, hypertension, BMI, and obesity (Supplemental Figure 3), these factors were adjusted in the multivariable analysis.
TABLE 1.
Basic characteristics of study participants between high and low total HEI groups1
| HEI <602 | HEI ≥60 | ||
|---|---|---|---|
| Characteristics | (n = 17) 46 mucosal samples | (n = 17) 51 mucosal samples | P3 |
| Age, y | |||
| Mean (SD) | 61.2 (5.3) | 63.0 (5.7) | 0.36 |
| Sex, % | |||
| Male | 97.8 | 100 | 0.47 |
| Race, n (%) | |||
| Non-Hispanic white | 13 (76.4) | 11 (64.7) | 0.69 |
| African American | 2 (11.8) | 4 (23.5) | |
| Hispanic | 2 (11.8) | 2 (11.8) | |
| BMI (kg/m2), n (%) | |||
| <30 | 3 (17.6) | 6 (35.3) | 0.44 |
| ≥30 (obese) | 14 (82.4) | 11 (64.7) | |
| Hypertension, n (%) | |||
| Yes | 14 (82.4) | 11 (64.7) | 0.44 |
| No | 3 (17.6) | 6 (35.3) | |
| Diabetes, n (%) | |||
| Yes | 11 (64.7) | 6 (35.3) | 0.17 |
| No | 6 (35.3) | 11 (64.7) | |
| Smoking status, n (%) | |||
| Never | 5 (29.4) | 8 (47.1) | 0.48 |
| Ever | 12 (70.6) | 9 (52.9) | |
| Alcohol drinking, n (%) | |||
| Never | 3 (17.7) | 2 (11.8) | 0.80 |
| Former | 4 (23.5) | 6 (35.3) | |
| Current | 10 (58.8) | 9 (52.9) | |
| Segment sites, n (%) | |||
| Cecum | 7 (15.2) | 10 (19.6) | 0.97 |
| Ascending | 10 (21.7) | 8 (15.7) | |
| Transverse | 5 (10.9) | 7 (13.7) | |
| Descending | 5 (10.9) | 6 (11.8) | |
| Sigmoid | 11 (23.9) | 12 (23.5) | |
| Rectum | 8 (17.4) | 8 (15.7) | |
| No. of mucosal biopsies from the same individual | |||
| 1 | 6 | 10 | 0.44 |
| 2 | 1 | 0 | |
| 3 | 4 | 2 | |
| 4 | 1 | 0 | |
| 5 | 4 | 5 | |
1HEI, Healthy Eating Index.
The median cut-off point for total HEI was 60 in this study population.
P values were for the Student's t test or Fisher's exact test
Dietary quality and α- and β-diversity of the colonic gut microbiome
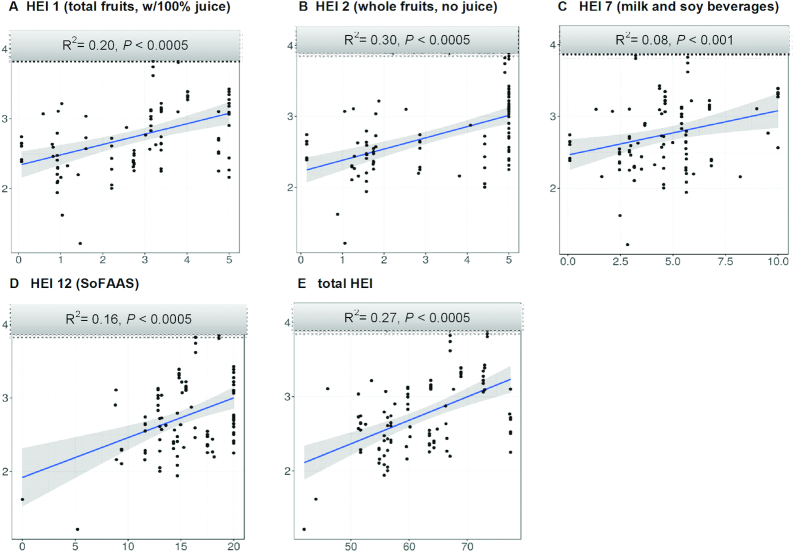
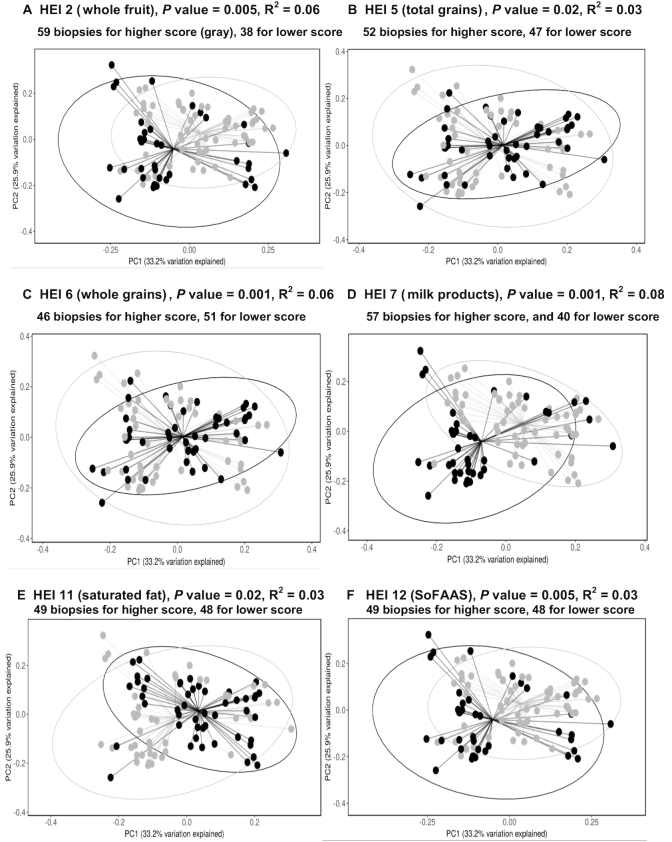
The sequencing data were classified into 1141 OTUs, and 120 OTUs had a relative abundance >0.05%. Participants with a lower score for total HEI, total fruits (HEI 1), whole fruits (HEI 2), milk products and soy beverages (HEI 7), or calories from solid fats, alcohol, and added sugar (SoFAAS, HEI 12) had significantly lower microbial α-diversity than participants with a higher score (FDR-adjusted P value <0.01) (Figure 1). The β-diversity also differed by higher compared with lower scores for HEIs 2, 5, 6, 7, 11, and 12 (FDR-adjusted P values <0.05) (Figure 2).
FIGURE 1.
α-Diversity of the OTUs of the colonic mucosa-associated gut bacteria based on dietary quality (panels A–E). Bacterial Shannon index (y-axis) was regressed against the score of each component and the HEI (x-axis) in a linear regression model. Each symbol represents a sample (97 biopsies from 34 participants). R2 indicates the coefficient of determination. P value was for the significance of the correlation. Only the dietary components with false discovery rate-adjusted P values <0.05 are presented. HEI, Healthy Eating Index; OTU, operational taxonomic unit; SoFAAS, calories from solid fats, alcoholic beverages, and added sugars.
FIGURE 2.
β-Diversity of the OTUs of the colonic mucosa–associated bacterial composition based on dietary quality (panels A–F). Principal coordinate plot used the weighted UniFrac as the distance matrix. The Monte-Carlo permutation test was used to estimate P values. The lower quality group (black) was separated from the higher quality group (gray). The proportion of variance explained by the first 2 principal coordinates is denoted in the corresponding axis label. For all the panels, x-axis: PC1 (33.2% variation explained); y-axis: PC2 (25.9% variation explained). HEI, Healthy Eating Index; OTU, operational taxonomic unit; PC, principal coordinate; SoFAAS, calories from solid fats, alcoholic beverages, and added sugars.
Dietary quality and relative abundance of colonic bacterial phylum and genus
At the phylum level, a lower score for total HEI, HEIs 2, 5, 7, and 12 was associated with altered relative abundance of 6 major phyla (Tables 2 and 3).
TABLE 2.
The relative abundance of major bacterial phyla based on higher or lower quality score of total HEI and HEI components1
| Relative abundance (mean %) in higher/lower score of HEI groups | |||||||
|---|---|---|---|---|---|---|---|
| Diet components (median, range) | No. of mucosa (H/L) | Firmicutes | Bacteroidetes | Proteobacteria | Verrucomicrobia | Fusobacteria | Actinobacteria |
| Total HEI (60, 0–100) | 51/46 | 50/45 | 37/37 | 9.6/10.1 | 2.3/3.8* | 0.2/3.9** | 0.67/0.34* |
| Adequacy components | |||||||
| HEI 1: total fruit with juice (2.7, 0–5) | 58/39 | 48/47 | 36/39 | 8.9/11.3 | 4.2/1.3** | 2.3/1.4 | 0.61/0.37 |
| HEI 2: whole fruit, no juice (2.5, 0–5) | 59/39 | 50/44 | 34/42* | 9.3/10.6 | 3.9/1.6*** | 2.3/1.4 | 0.64/0.32* |
| HEI 3: total vegetables (2.4, 0–5) | 47/50 | 50/46 | 37/36 | 8.5/11.1 | 3.7/2.4 | 0.4/3.4 | 0.37/0.66 |
| HEI 4: dark green and orange vegetables & legumes (2.3, 0–5) | 62/35 | 50/44 | 35/41 | 8.9/11.5 | 2.9/3.2 | 2.7/0.6* | 0.61/0.35 |
| HEI 5: total grains (4.5, 0–5) | 52/47 | 44/52* | 38/36 | 12.2/7.1** | 4.1/1.8 | 1.2/2.7 | 0.32/0.74** |
| HEI 6: whole grains (1.4, 0–5) | 46/51 | 40/54*** | 41/33* | 12.8/7.1*** | 4.6/1.6 | 0.5/3.3 | 0.62/0.42 |
| HEI 7: milk & soy beverages (4.4, 0–10) | 57/40 | 50/44 | 33/43* | 9.8/9.9 | 3.9/1.7* | 2.7/0.9* | 0.68/0.29* |
| HEI 8: meat & beans (8.01, 0–10) | 48/49 | 45/49 | 38/36 | 10.4/9.2 | 4.3/1.7 | 0.7/3.1 | 0.66/0.38 |
| HEI 9: oils (8.3, 0–10) | 52/47 | 49/45 | 40/34 | 8.3/11.6 | 1.2/5.2 | 1.0/3.0 | 0.45/0.59 |
| Moderation components | |||||||
| HEI 10: sodium (3.5, 0–10) | 46/51 | 47/48 | 36/37 | 10/9.6 | 2/4 | 3.3/0.7 | 0.71/0.34 |
| HEI 11: saturated fat (5.8, 0–10) | 49/48 | 46/49 | 40/34 | 9.6/10.1 | 2.6/3.4 | 0.9/2.9* | 0.35/0.69 |
| HEI 12: SoFAAS (15, 0–20) | 49/48 | 47/48 | 42/32* | 7.6/12.1 | 2.3/3.8* | 0.4/3.5*** | 0.63/0.41 |
The false discovery rate–adjusted P values are reported using asterisks, with *P < 0.05, **P < 0.005, ***P < 0.0005. The Mann–Whitney test was used to compare the mean relative abundance of the taxa based on dietary quality. HEI, Healthy Eating Index; H/L, higher/lower score of the HEI groups; SoFAAS, calories from solid fats, alcoholic beverages, and added sugars.
TABLE 3.
Fold changes of the relative counts of major bacterial phyla and genera and dietary quality1
| Total HEI (n = 58 for H, 39 for L) | HEI 2 (whole fruits) (n = 59 for H, 38 for L) | HEI 5 (total grains) (n = 52 for H, 47 for L) | HEI 7 (milk & soy beverages) (n = 57 for H, 40 for H) | HEI 12 (SoFAAS) (n = 49 for H, 48 for L) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Taxa | Median count2 H/L | FC (95% CI)3 | Median count2 H/L | FC (95% CI)3 | Median count2 H/L | FC (95% CI)3 | Median count2 H/L | FC (95% CI)3 | Median count2 H/L | FC (95% CI)3 |
| Phylum | ||||||||||
| Bacteroidetes | — | — | 551/551 | 0.95 (0.72, 1.23) | — | — | 551/551 | 1.51 (1.28, 1.80) | 551/551 | 0.41 (0.34, 0.51) |
| Firmicutes | — | — | 752/752 | 1.11 (0.89, 1.39) | 752/799 | 1.09 (0.92, 1.29) | — | — | — | — |
| Fusobacteria | 0/8.2 | 9.55 (4.01, 22.7) | — | — | 0.66/1.71 | 5.05 (1.92, 13.2) | 4.5/0 | 0.43 (0.24, 0.76) | 0/7.50 | 21.3 (9.11, 50.0) |
| Proteobacteria | — | — | — | — | 130/113 | 0.87 (0.61, 1.23) | 113/113 | 0.54 (0.40, 0.73) | — | — |
| Verrucomicrobia | — | — | 19.4/0 | 0.43 (0.26, 0.71) | — | — | 15.4/0.4 | 0.49 (0.27, 0.87) | 2.9/0 | 1.12 (0.38, 3.30) |
| Genus | ||||||||||
| Bacteroides | — | — | 392/680 | 1.73 (1.20, 2.50) | — | — | 369/804 | 2.59 (1.98, 3.40) | — | — |
| Parabacteroides | 17/5.1 | 0.21 (0.10, 0.42) | — | — | — | — | — | — | — | — |
| Faecalibacterium | — | — | — | — | — | — | 140/110 | 0.25 (0.17, 0.38) | — | — |
| Subdoligranulum | 31/11 | 0.10 (0.05, 0.19) | — | — | — | — | — | — | 29.2/12 | 0.31 (0.21, 0.46) |
| Roseburia | — | — | 28/15 | 0.29 (0.15, 0.56) | — | — | — | — | — | — |
| Akkermansia | 6.5/0 | 0.07 (0.03, 0.18) | — | — | — | — | — | — | — | — |
| Escherichia | — | — | — | — | — | — | — | — | 10/33.2 | 5.99 (3.37, 10.64) |
| Fusobacterium | 0/5.5 | 3.94 (1.39, 11.1) | — | — | — | — | 2.9/0 | 0.29 (0.14, 0.60) | 0/6.4 | 23.0 (5.81, 91.4) |
1FC, fold change; H, higher score; HEI, Healthy Eating Index; L, lower score; SoFAAS, calories from solid fats, alcoholic beverages, and added sugars.
Normalized median count calculated by dividing the raw count by a size factor in the DESeq2 function.
Coefficients of FC were estimated using the empirical Bayes shrinkage method based on negative binomial distribution. Model was adjusted for age, BMI (<25, 25–<30, and ≥30 kg/m2), race (white and nonwhite), smoking (never, former, and current), alcohol consumption (never, former, and current), Hispanic (yes or no), type 2 diabetes (yes compared with no), colon segment (cecum, ascending, transverse, descending, sigmoid, and rectum), and cluster ID.
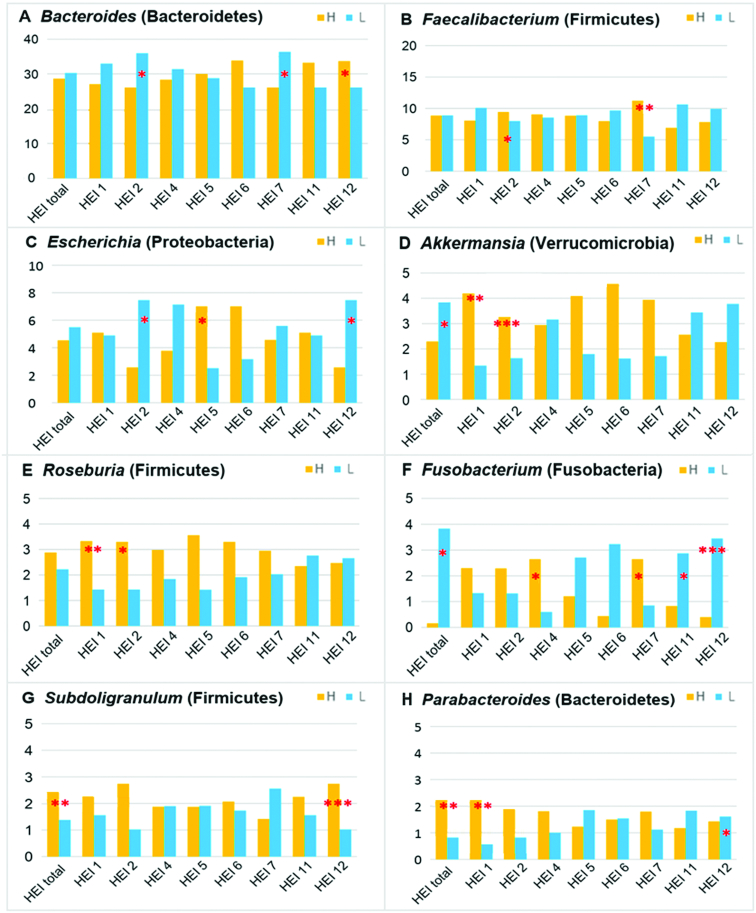
There were 99 genera with a relative abundance >0.05%. We found that the relative abundance of 27 bacterial genera, mostly in Lachnospiraceae and Ruminococcaceae families, differed by total HEI (FDR-adjusted P <0.05) (Supplemental Figure 4). We summarized the relative abundance of major genera based on dietary quality in Figure 3. A lower score for total HEI was associated with more Fusobacterium and Akkermansia, but less Subdoligranulum and Parabacteroides. A lower score for HEI 2 was associated with more Bacteroides and Escherichia but less Faecalibacterium, Roseburia, and Akkermansia. A lower score for HEI 7 was associated with more Bacteroides but less Faecalibacterium and Fusobacterium. A lower score for HEI 12 was associated with more Escherichia and Fusobacterium but less Subdoligranulum (FDR-adjusted P values <0.05). Most of the findings were confirmed in multivariate analyses. However, the relative count of Akkermansia was significantly lower with a lower total HEI (Table 3).
FIGURE 3.
Relative abundance of the major bacterial genera based on dietary quality [panels A–H, by bacterial genus (phylum)]. Bacterial mean relative abundances (y-axis, %) were compared between high (H, yellow) and low (L, blue) HEI components using the Mann–Whitney test. The asterisks indicate significantly different taxa, with false discovery rate-adjusted P value *<0.05, **<0.005, ***<0.0005. The analysis was based on 97 biopsies from 34 participants. The number of biopsies for total HEI and each component by H/L status: total HEI (51/46), HEI 1 (58/39), HEI 2 (59/38), HEI 4 (62/35), HEI 5 (52/47), HEI 6 (46/51), HEI 7 (57/40), HEI 11 (49/48), and HEI 12 (49/48). H, higher score; HEI, Healthy Eating Index; L, lower score.
The univariate linear regression showed a significant correlation between Roseburia, Parabacteroides, Subdoligranulum, and Fusobacterium and total HEI; Roseburia and Bacteroides and HEI 2; Bacteroides, Faecalibacterium, and Fusobacterium and HEI 7; Fusobacterium and Escherichia and HEI 12; and 6 bacterial genera and HEI 6. However, the linear regression analysis did not show a significant correlation between Akkermansia and total HEI. In addition, the unclassified (Unc) OTUs Prevotellaceae(Unc00yx7) and Lachnospiraceae (Unc8782) were also significantly influenced by multiple dietary factors. The strength of the correlation is indicated by the R2 value in Table 4.
TABLE 4.
Linear regression analysis on the correlation between relative abundance of the bacterial phyla and genera and dietary quality based on 97 mucosal samples from 34 participants1
| Total HEI | HEI 1 | HEI 2 | HEI 3 | HEI 4 | HEI 5 | HEI 6 | HEI 7 | HEI 9 | HEI 11 | HEI 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | |||||||||||
| Actinobacteria | — | — | — | — | — | — | — | — | ↓ 0.083 | — | — |
| Bacteroidetes | — | — | ↓ 0.091 | — | ↓ 0.12 | — | — | ↓ 0.083 | — | — | ↑ 0.10 |
| Firmicutes | — | — | ↑ 0.072 | ↑ 0.10 | ↑ 0.18 | ↓ 0.13 | ↓ 0.16 | — | — | — | — |
| Fusobacteria | ↓ 0.14 | — | — | — | — | — | ↓ 0.083 | — | — | ↓ 0.095 | ↓ 0.12 |
| Proteobacteria | — | — | — | — | — | ↑ 0.066 | ↑ 0.10 | — | — | — | ↓ 0.13 |
| Verrucomicrobia | — | — | — | — | — | — | ↑ 0.10 | — | ↓ 0.17 | — | — |
| Genus | |||||||||||
| Increased with good dietary quality | |||||||||||
| Roseburia | ↑ 0.16 | ↑ 0.16 | ↑ 0.15 | — | — | ↑ 0.092 | ↑ 0.25 | — | — | — | — |
| Alistipes | ↑ 0.059 | — | ↑ 0.059 | — | — | ↑ 0.13 | — | ↑ 0.13 | — | — | — |
| Parabacteroides | ↑ 0.13 | ↑ 0.11 | ↑ 0.13 | — | — | — | — | ↑ 0.24 | — | — | — |
| Subdoligranulum | ↑ 0.14 | — | — | — | — | — | — | — | ↑ 0.073 | ↑ 0.11 | ↑ 0.34 |
| Sutterella | ↑ 0.079 | — | — | — | — | — | ↑ 0.067 | — | ↑ 0.074 | — | ↑ 0.076 |
| Odoribacter | — | ↑ 0.091 | ↑ 0.082 | — | — | — | — | — | — | — | ↑ 0.11 |
| Fusicatenibacter | — | — | ↑ 0.10 | — | — | — | — | ↑ 0.29 | — | — | — |
| Reduced with good dietary quality | |||||||||||
| Prevotellaceae (Unc00yx7) | — | ↓ 0.16 | ↓ 0.12 | ↓ 0.079 | ↓ 0.13 | — | — | ↓ 0.082 | — | — | — |
| Fusobacterium | ↓ 0.14 | — | — | — | — | — | ↓ 0.094 | — | — | ↓ 0.094 | ↓ 0.12 |
| Bilophila | — | ↓ 0.072 | — | — | — | — | — | — | — | ↓ 0.12 | ↓ 0.12 |
| Bacteroides | — | — | ↓ 0.14 | — | — | — | — | ↓ 0.15 | — | — | — |
| Lachnoclostridium | — | — | ↓ 0.081 | — | — | — | — | — | — | — | — |
| Tyzzerella | ↓ 0.086 | — | — | — | — | — | — | — | — | — | ↓ 0.14 |
| Escherichia | — | — | — | — | — | — | — | — | — | — | ↓ 0.15 |
| Mixed change with dietary quality | |||||||||||
| Lachnospiraceae (UncO8782) | ↑ 0.064 | ↑0.078 | — | ↑0.12 | — | ↓ 0.078 | — | — | — | — | ↑ 0.15 |
| Barnesiella | ↑ 0.091 | ↑ 0.063 | ↑ 0.13 | — | — | ↓ 0.19 | — | ↑ 0.090 | — | — | — |
| Desulfovibrio | — | — | ↑ 0.064 | — | — | — | ↑ 0.087 | — | ↓ 0.13 | — | — |
| Akkermansia | — | — | — | — | — | — | ↑ 0.10 | — | ↓ 0.17 | — | — |
| Erysipelatoclostridium | — | — | — | — | ↑ 0.097 | — | — | — | — | — | ↓ 0.077 |
| Faecalibacterium | — | — | — | — | — | — | — | ↑ 0.16 | — | ↓ 0.081 | — |
Only the significant [false discovery rate-adjusted P values <0.05 (6 phyla and 25 genera)] linear correlation between the relative abundance of the bacteria and dietary quality (as the dependent variable) is presented. HEIs 8 and 10 are not presented because no significant association was observed. The symbol ↓ denotes the significantly reduced abundance with the higher score; ↑ denotes the significantly increased abundance with the higher score. The values shown are R2, indicating the coefficient of determination. HEI, Healthy Eating Index; HEI 1, total fruits (with juice); HEI 2, whole fruits (without juice); HEI 3, total vegetables; HEI 4, dark-green vegetables; HEI 5, total grains; HEI 6, whole grains; HEI 7, milk products and soy beverages; HEI 9, oils; HEI 11, saturated fat; HEI 12, calories from solid fats, alcoholic beverages, and added sugars; Unc, unclassified.
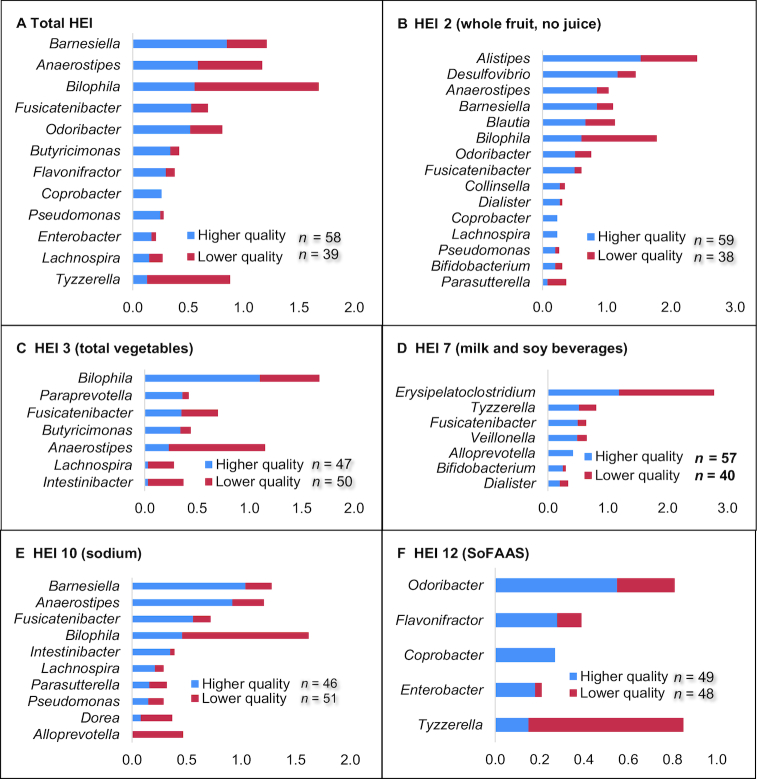
For uncommon bacteria, a lower abundance of Barnesiella, Blautia, Enterobacter, Fusicatenibacter, and Odoribacter and a higher abundance of Tyzzerella or Bilophila were related to a lower score of total HEI, HEI 2, and HEI 12; a lower abundance of Bifidobacterium and Dialister was related to a lower score of HEIs 2 and 7 (FDR-adjusted P values <0.05) (Figure 4).
FIGURE 4.
Stacked bar chart of relative abundance of the less abundant genera (<1.5%) differed statistically significantly based on dietary quality (panels A–F) (false discovery rate-adjusted P values <0.05). The x-axis represents the relative abundance (%). HEI, Healthy Eating Index; SoFAAS, calories from solid fats, alcoholic beverages, and added sugars.
The sensitivity analysis based on 23 sigmoid samples showed consistent alteration in the gut microbiota by the total HEI score, albeit the differences were not statistically significant (Supplemental Figure 4). The sensitivity analysis based on a single sequencing batch also confirmed the significant associations. This analysis also revealed the positive association between total HEI and Faecalibacterium, as well as between whole grains intake and Akkermansia (Supplemental Figure 5).
Discussion
In this cross-sectional study, we examined the association between dietary quality and the gut bacterial community composition and structure in colonic mucosal biopsies from participants with an endoscopically normal colon. We found that major phyla and genera members mostly in the Lachnospiraceae and Ruminococcaceae families of the Clostridiales order were associated with dietary quality. A lower dietary quality was significantly associated with a lower bacterial α-diversity and less abundant Roseburia, Subdoligranulum, and Parabacteroides but more abundant Fusobacterium and Escherichia. In addition, we found that uncommon bacterial genera Alistipes, Barnesiella, Bifidobacterium, Fusicatenibacter, and Odoribacter were related to higher dietary quality and Bilophila and Tyzzerella were related to lower dietary quality. Our study may offer a potential biological explanation of the observational associations between dietary quality and risk of diseases.
First, we found that lower dietary quality was associated with a reduction in Roseburia, Subdoligranulum, and Parabacteroides. A lower score of whole fruits was also associated with less Roseburia. Subdoligranulum and Roseburia both belong to the Clostridiales order of the Firmicutes phylum, and are highly efficient at producing SCFAs (27) via the fermentation of dietary fibers (11). An experimental study has shown that dietary fiber increases anti-inflammatory and anticarcinogenic SCFA-producing microbiota via induction of T-regulatory cells in the colonic tissues and may explain its potential protective effect against cancer (11, 28). Low-fiber consumption has also been associated with a shift from Parabacteroides to Bacteroides (29). We found that a lower score for fruits (whole or total) was associated with increased Bacteroides but decreased Parabacteroides. In addition, the relative abundance of Bifidobacterium was higher in those with higher whole fruit consumption. Bifidobacterium is a common probiotic bacterium used in the treatment of patients with IBD (30). These results indicate that a higher score for total HEI and fruit was associated with more SCFA producers in the human gut.
On the other hand, lower dietary quality was associated with more Fusobacteria and Fusobacterium. Fusobacterium nucleatum (F. nucleatum) induces a proinflammatory immune response, promotes carcinogenesis, and has been positively associated with IBD and colorectal cancer (31–33). A previous dietary intervention study reported that fecal F. nucleatum was markedly increased after participants switched from a plant-based diet to a Western-style diet (34). Our study found that a lower score of SoFAAS was associated with more abundant Fusobacteria and Escherichia. Increased prevalence of Escherichia has been suggested as a marker for an unstable microbial community and intestinal inflammation (35). Overall, a lower score for total HEI and SoFAAS was associated with more potentially pathogenic bacteria in our study.
We observed that a lower score of milk products was also associated with less Fusobacteria, as well as less Faecalibacterium, Bifidobacterium, and Parabacteroides. This finding indicates that milk products can promote the growth of both potentially harmful and beneficial bacteria. Faecalibacterium is a major genus in the Ruminococcaceae family of the Firmicutes phylum and a major butyrate producer in the colon. Its anti-inflammatory effect on the host has been reported (36). Certain fermented dairy products (such as yogurt) are associated with probiotic effects, whereas other high-fat dairy products (such as butter or cheese) stimulate bile acid synthesis in the intestines and may increase cancer risk (37, 38). Nevertheless, we did not observe a positive association between Fusobacteria and milk products in the linear regression analysis. Further studies need to evaluate whether different dairy products differentially affect the gut microbiome.
Although not shown in the dichotomized analysis, linear regression analysis showed that a lower quality score for whole grain (HEI 6) was associated with more abundant Fusobacteria and less abundant Akkermansia and Roseburia. Akkermansia has been inversely associated with the onset of inflammation and metabolic disorders in obese mice (39). A lower abundance of Akkermansia muciniphila results in a thinner intestinal mucus layer and increased gut permeability (40). Therefore, having a higher score of whole-grain consumption may promote the growth of potentially beneficial bacteria and inhibit Fusobacteria. In our study, although Akkermansia was significantly more abundant in those with lower total HEI, the relative count of Akkermansia was significantly higher in participants with higher total HEI after adjusting for confounding factors. Overall, our study would support the beneficial effect of Akkermansia related to higher dietary quality.
We have provided novel evidence on the association between diet and several less abundant bacteria. Alistipes, Anaerostipes, Barnesiella, Bifidobacterium, Bilophila, Dialister, Enterobacter, Fusicatenibacter, and Odoribacter were in general related to a better dietary quality, whereas Bilophila and Tyzzerella were related to a lower dietary quality. Future studies need to elucidate the functional significance of the less abundant bacteria in the human host.
We did not have compelling evidence to show the influence of the dietary quality of meat and beans, vegetables, salt, and oil on the gut microbiota; more in-depth analysis is needed to examine the association between these food items and the gut microbiota that may have been missed by HEI–2005. The HEI–2005 was later updated to HEI–2010 and HEI–2015, mostly with updated scores for vegetables, grains, oil, and fatty acids (41). A recent report showed that the HEI–2010 explains most variance in human fecal microbiota attributable to habitual diet compared with 2 other indices (42). It will be of interest to incorporate microbiome information in refining dietary guidelines.
Our study had multiple strengths. We provided novel and fundamental data on the association between dietary quality and the colonic-adherent microbiota in endoscopically normal individuals. Our study had high internal validity because the study was conducted in a homogeneous study population recruited from the same colonoscopy clinic, and we used the same study protocol for all the samples. The response rate to the FFQ was 87%, and there was no missing information on the survey. We minimized the potential influence of medications and antibiotics on the gut microbiota. Several limitations should be noted. First, the generalizability of our findings may be limited because our participants were all veterans whose dietary habits may be different from those of the general population (43). Our study findings may not be generalized to other studies that use feces samples or use different protocols for DNA extraction, sequencing (such as targeting the V3–V4 region), or bioinformatics pipelines (44, 45). Second, whole-genome shotgun sequencing is needed to identify Unc genera or species and define their functional significance related to dietary quality. Third, despite the fact we implemented robust quality control measures, the reproducibility of the 16s rRNA sequencing may not be optimal (23). The findings on the less abundant genera were preliminary. Lastly, self-reported dietary intake may be subject to reporting bias and, thus, measurement error.
In conclusion, our study showed that total dietary quality, fruit, milk products and soy beverages, added sugar, alcohol, and saturated fat had the greatest influences on the mucosa-associated gut microbiota. Dietary components can either serve as prebiotics or disturb the symbiotic relation between the gut microbiota and the host. Whether the gut microbiota can be used as a biomarker for dietary intake should be further investigated. As diet only partially explains the variability in the gut microbiome composition and structure, other exogenous and endogenous factors such as host genetics (46, 47) should also be considered in future research. The molecular pathologic epidemiology that incorporates diet and lifestyle, genomics, metagenomics, metabolites, immune- and mutagenesis-related biomarkers, and other molecular features of the disease will aid in elucidating the etiopathogenesis of various diseases and promoting precision medicine (48, 49).
Supplementary Material
Acknowledgments
We thank David Ramsey at Baylor College of Medicine for data management; Ashely Johnson, Preksha Shah, Kathryn Royse, Ava Smith, Mahmoud Al-Saadi, and Jocelyn Uriostegui for collecting and processing samples; the nurses at the endoscopy unit of the Michael E DeBakey VA Medical Center for sample collection; Ms Sonora Hudson for editing the manuscript.
The authors’ contributions were as follows—LJ, JFP, and NJA: designed the research; YL, NJY, HBE, CH, DYG, DLW, LC, SP, JK, RC, RH, JH, NH, MJ, FK, GK, YN, RS, MV, JFP, and LJ: conducted the research; ZW, LC, and LJ: analyzed the data; YL, LJ, and HBE: wrote the manuscript; LJ: had primary responsibility for the final content; NJA, HBE, DYG, DLW, RH, GK, NM, and JFP: reviewed the manuscript; and all authors: read and approved the final manuscript. None of the authors report a conflict of interest related to research presented in this article.
Notes
This study was supported by the Cancer Prevention and Research Institute of Texas (RP140767, principal investigators: LJ, JFP), Gillson Longenbaugh Foundation, Golfers Against Cancer organization (principal investigator: LJ), and the NIH grant K07CA181480 (principal investigator: YL) and partly supported with the use of facilities and resources of the Houston VA HSR&D Center for Innovations in Quality, Effectiveness, and Safety (CIN13-413). Research Training Grant from the Cancer Prevention and Research Institute of Texas (RP160097, principal investigator: MS). The opinions expressed reflect those of the authors and not necessarily those of the Department of Veterans Affairs, the US government, or Baylor College of Medicine.
Supplemental Figures 1–5 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: FC, fold change; FDR, false discovery rate; HEI, Healthy Eating Index; IBD, inflammatory bowel disease; ID, identification; OTU, operational taxonomic unit; SoFAAS, calories from solid fat, alcoholic beverages, and added sugar; Unc, unclassified.
References
- 1. Zinocker MK, Lindseth IA. The Western diet-microbiome–host interaction and its role in metabolic disease. Nutrients. 2018;10(3):E365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li WQ, Park Y, Wu JW, Ren JS, Goldstein AM, Taylor PR, Hollenbeck AR, Freedman ND, Abnet CC. Index-based dietary patterns and risk of esophageal and gastric cancer in a large cohort study. Clin Gastroenterol Hepatol. 2013;11(9):1130–6. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller PE, Lazarus P, Lesko SM, Muscat JE, Harper G, Cross AJ, Sinha R, Ryczak K, Escobar G, Mauger DT et al.. Diet index-based and empirically derived dietary patterns are associated with colorectal cancer risk. J Nutr. 2010;140(7):1267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Solbak NM, Xu JY, Vena JE, Csizmadi I, Whelan HK, Robson PJ. Diet quality is associated with reduced incidence of cancer and self-reported chronic disease: observations from Alberta's Tomorrow Project. Prev Med. 2017;101:178–87. [DOI] [PubMed] [Google Scholar]
- 5. Arem H, Reedy J, Sampson J, Jiao L, Hollenbeck AR, Risch H, Mayne ST, Stolzenberg-Solomon RZ. The Healthy Eating Index 2005 and risk for pancreatic cancer in the NIH-AARP study. J Natl Cancer Inst. 2013;105(17):1298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sonnenburg JL, Backhed F. Diet–microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R et al.. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM et al.. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. [DOI] [PubMed] [Google Scholar]
- 10. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al.. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones RB, Zhu X, Moan E, Murff HJ, Ness RM, Seidner DL, Sun S, Yu C, Dai Q, Fodor AA et al.. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep. 2018;8(1):4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Block G, Hartman AM, Dresser CM, Carroll MD, Gannon J, Gardner L. A data-based approach to diet questionnaire design and testing. Am J Epidemiol. 1986;124(3):453–69. [DOI] [PubMed] [Google Scholar]
- 15. Guenther PM, Reedy J, Krebs-Smith SM. Development of the Healthy Eating Index-2005. J Am Diet Assoc. 2008;108(11):1896–901. [DOI] [PubMed] [Google Scholar]
- 16. Mejia-Leon ME, Petrosino JF, Ajami NJ, Dominguez-Bello MG, de la Barca AM. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014;4:3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, Schwager E, Crabtree J, Ma S, Microbiome Quality Control Project C et al.. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol. 2017;35(11):1077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N et al.. Moving pictures of the human microbiome. Genome Biol. 2011;12(5):R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. [DOI] [PubMed] [Google Scholar]
- 21. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. [DOI] [PubMed] [Google Scholar]
- 22. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wen C, Wu L, Qin Y, Van Nostrand JD, Ning D, Sun B, Xue K, Liu F, Deng Y, Liang Y et al.. Evaluation of the reproducibility of amplicon sequencing with Illumina MiSeq platform. PLoS One. 2017;12(4):e0176716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benjamini Y, Yekutieli D. Quantitative trait loci analysis using the false discovery rate. Genetics. 2005;171(2):783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, Bonnaure-Mallet M, Jolivet-Gougeon A. Roseburia spp.: a marker of health?. Future Microbiol. 2017;12:157–70. [DOI] [PubMed] [Google Scholar]
- 28. Heerdt BG, Houston MA, Augenlicht LH. Short-chain fatty acid-initiated cell cycle arrest and apoptosis of colonic epithelial cells is linked to mitochondrial function. Cell Growth Differ. 1997;8(5):523–32. [PubMed] [Google Scholar]
- 29. Graf D, Di Cagno R, Fak F, Flint HJ, Nyman M, Saarela M, Watzl B. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. 2015;26:26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt EG, Claesson MH, Jensen SS, Ravn P, Kristensen NN. Antigen-presenting cells exposed to Lactobacillus acidophilus NCFM, Bifidobacterium bifidum BI-98, and BI-504 reduce regulatory T cell activity. Inflamm Bowel Dis. 2010;16(3):390–400. [DOI] [PubMed] [Google Scholar]
- 31. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA et al.. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17(9):1971–8. [DOI] [PubMed] [Google Scholar]
- 34. O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E et al.. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Selvanantham T, Lin Q, Guo CX, Surendra A, Fieve S, Escalante NK, Guttman DS, Streutker CJ, Robertson SJ, Philpott DJ et al.. NKT cell-deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J Immunol. 2016;197(11):4464–72. [DOI] [PubMed] [Google Scholar]
- 36. Ferreira-Halder CV, Faria AVS, Andrade SS. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol. 2017;31(6):643–8. [DOI] [PubMed] [Google Scholar]
- 37. Thirabunyanon M, Boonprasom P, Niamsup P. Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol Lett. 2009;31(4):571–6. [DOI] [PubMed] [Google Scholar]
- 38. Lai LR, Hsieh SC, Huang HY, Chou CC. Effect of lactic fermentation on the total phenolic, saponin and phytic acid contents as well as anti-colon cancer cell proliferation activity of soymilk. J Biosci Bioeng. 2013;115(5):552–6. [DOI] [PubMed] [Google Scholar]
- 39. Schneeberger M, Everard A, Gomez-Valades AG, Matamoros S, Ramirez S, Delzenne NM, Gomis R, Claret M, Cani PD. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, de Vos WM, Satokari R. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol. 2015;81(11):3655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guenther PM, Casavale KO, Reedy J, Kirkpatrick SI, Hiza HA, Kuczynski KJ, Kahle LL, Krebs-Smith SM. Update of the Healthy Eating Index: HEI-2010. J Acad Nutr Diet. 2013;113(4):569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bowyer RCE, Jackson MA, Pallister T, Skinner J, Spector TD, Welch AA, Steves CJ. Use of dietary indices to control for diet in human gut microbiota studies. Microbiome. 2018;6(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoerster KD, Lehavot K, Simpson T, McFall M, Reiber G, Nelson KM. Health and health behavior differences: U.S. military, veteran, and civilian men. Am J Prev Med. 2012;43(5):483–9. [DOI] [PubMed] [Google Scholar]
- 44. Song EJ, Lee ES, Nam YD. Progress of analytical tools and techniques for human gut microbiome research. J Microbiol. 2018;56(10):693–705. [DOI] [PubMed] [Google Scholar]
- 45. Poussin C, Sierro N, Boue S, Battey J, Scotti E, Belcastro V, Peitsch MC, Ivanov NV, Hoeng J. Interrogating the microbiome: experimental and computational considerations in support of study reproducibility. Drug Discov Today. 2018;23(9):1644–57. [DOI] [PubMed] [Google Scholar]
- 46. Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19(5):731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Awany D, Allali I, Dalvie S, Hemmings S, Mwaikono KS, Thomford NE, Gomez A, Mulder N, Chimusa ER. Host and microbiome genome-wide association studies: current state and challenges. Front Genet. 2018;9:637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hamada T, Nowak JA, Milner DA Jr., Song M, Ogino S. Integration of microbiology, molecular pathology, and epidemiology: a new paradigm to explore the pathogenesis of microbiome-driven neoplasms. J Pathol. 2019;247(5):615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.