Abstract
Polycomb repressive complex 2 (PRC2) is a histone methyltransferase that maintains cell identity during development in multicellular organisms by marking repressed genes and chromatin domains. In addition to four core subunits, PRC2 comprises multiple accessory subunits that vary in their composition during cellular differentiation and define two major holo-PRC2 complexes: PRC2.1 and PRC2.2. PRC2 binds to RNA, which inhibits its enzymatic activity, but the mechanism of RNA-mediated inhibition of holo-PRC2 is poorly understood. Here we present in vivo and in vitro protein–RNA interaction maps and identify an RNA-binding patch within the allosteric regulatory site of human and mouse PRC2, adjacent to the methyltransferase centre. RNA-mediated inhibition of holo-PRC2 is relieved by allosteric activation of PRC2 by H3K27me3 and JARID2-K116me3 peptides. Both holo-PRC2.1 and -PRC2.2 bind RNA, providing a unified model to explain how RNA and allosteric stimuli antagonistically regulate the enzymatic activity of PRC2.
Introduction
PRC2 is a histone methyltransferase (HMTase) that methylates H3 histones at lysine 27 to form the H3K27me3 mark of facultative heterochromatin (reviewed in1–4). The H3K27me3 mark is essential for the epigenetic maintenance of transcriptional repression at developmentally-expressed genes. The core PRC2 complex includes a histone methyltransferase subunit—EZH2 or EZH1; the regulatory subunit EED; one histone-binding subunit—RBBP4 or RBBP7; and SUZ12, which serves as a scaffold5–7. The recruitment of PRC2 to chromatin and its HMTase activity are tightly regulated. For instance, after EZH2 introduces the H3K27me3 histone mark, the methylated histone peptide binds to a regulatory site within EED to stimulate the methyltransferase activity of PRC28 through allosteric activation9–12.
The function of PRC2 is also regulated by its accessory subunits: sub-stoichiometric subunits of PRC2 that are differentially expressed during development1,2. For instance, JARID2 is methylated by PRC2 at lysine 116 (JARID2-K116me3) and then binds to the regulatory centre in EED to allosterically activate PRC2 during de novo methylation at target genes11. AEBP2 and the three polycomb-like (PCL) proteins—PHF1, PHF19 and MTF2—facilitate DNA binding by PRC2 through direct interactions13–17. EPOP (previously termed C17ORF96 or esPRC2p48) is another accessory subunit of PRC2 that facilitates gene repression18,19. Unbiased proteomic studies identified these factors as the most abundant accessory subunits of PRC218–22 and determined that they form two types of holo-PRC2 complexes, including the core subunits and different accessory subunits21: PRC2.1 includes one of the PCL accessory subunits (PHF1, PHF19 or MTF2) and EPOP, while PRC2.2 includes AEBP2 and JARID221.
Direct interactions with RNA have been proposed to recruit PRC2 to target genes for epigenetic repression, to evict it from active genes, and to retain it in a poised state at lowly expressed genes (reviewed in23–27). More recently, RNA was shown to inhibit the HMTase activity of PRC228–30. Experiments using isolated subunits, partial complexes and the core PRC2 complex attributed RNA binding to the core subunits EZH2, EED and SUZ1230–35. Yet, the question of how RNA inhibits different types of holo-PRC2 complexes remains unanswered. RNA competes for nucleosome36 and DNA binding by PRC2 and the automethylation activity of EZH2 is not affected by RNA15, suggesting that the competition with DNA might explain the inhibitory effect of RNA on PRC215. This however leaves an unresolved conundrum: how does PRC2 overcome RNA inhibition at target genes while within the RNA-rich environment of the nucleus? Moreover, an earlier study had demonstrated that RNA inhibits the HMTase activity of PRC2 also toward biotinylated histone tail peptides29, which is inconsistent with a model whereby RNA inhibits PRC2 activity exclusively by competing with DNA binding. This suggests the possibility that RNA may inhibit the methyltransferase activity of PRC2 via multiple mechanisms. Testing this possibility is important not only to understand how RNA regulates the HMTase activity of PRC2 at the molecular level, but also because PRC2 methylates non-histone substrates, including transcriptional regulators (37 and references therein).
Here, we show that RNA binds and inhibits both types of holo-PRC2 complexes—PRC2.1 and PRC2.2. Using in vivo UV crosslinking and mass spectrometry in mouse embryonic stem cells (mESCs), we mapped RNA-binding regions on the core complex to both regulatory and catalytic centres. In vitro studies on active holo-PRC2 complexes confirmed binding of RNA to the regulatory site of PRC2, at the interface between EZH2 and EED, near the catalytic centre. In agreement with this observation, RNA-mediated inhibition of PRC2 is relieved by peptides that bind to the allosteric regulatory centre and RNA inhibited the methyltransferase activity of PRC2 also toward DNA-free substrates. Based on these findings, we provide a mechanistic framework to explain how RNA-mediated inhibition of the two major types of holo-PRC2 complexes takes place and further generalise it for RNA-mediated inhibition of methyltransferase activity toward non-histone substrates.
Results
RNA binds both PRC2.1 and PRC2.2 in vivo
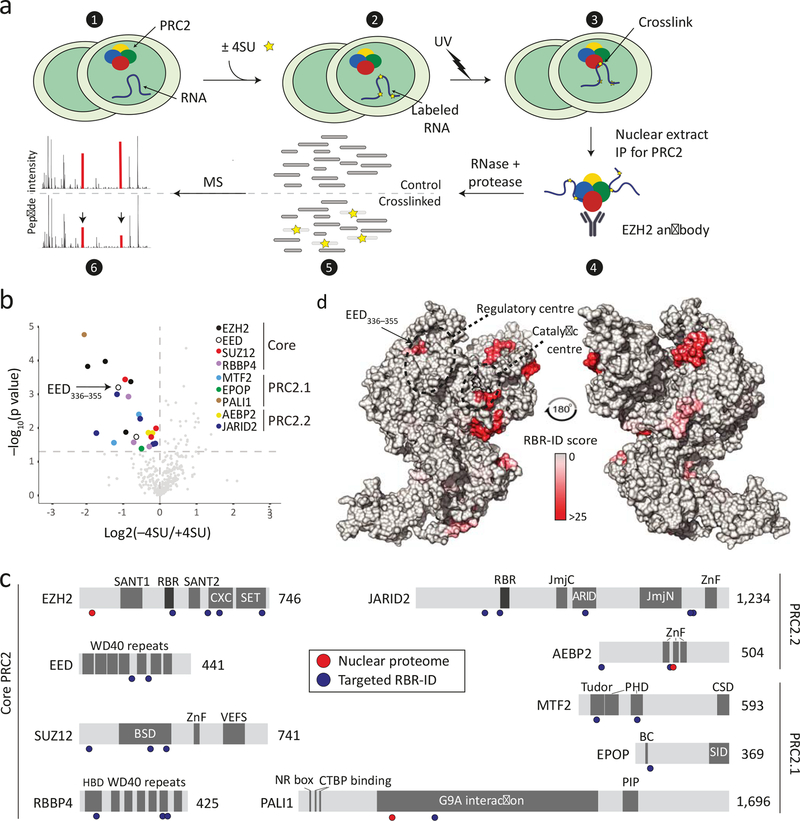
We previously used protein–RNA cross-linking with mass spectrometry for RNA-binding region identification (RBR-ID) in the nuclear proteome of mESCs38. Through reanalysing these data, we detected significant peptide hits within core PRC2 subunits EZH2 and SUZ12, consistent with previous observations30–35. We also identified the accessory PRC2.2 subunit AEBP2 (Supplementary Fig. 1a), consistent with our own previous work34,39. However, we did not detect peptides within JARID2, which is known to crosslink to RNA in vivo40. We reasoned that the low coverage of this and other PRC2 subunits in the whole nuclear proteome (Supplementary Fig. 1a) might have limited our ability to detect protein–RNA interaction sites in this complex.
To overcome this obstacle, we developed a “targeted” variant of RBR-ID and utilised an immunoprecipitation step to focus the mass spectrometry analysis on PRC2 (Fig. 1a, Supplementary Fig 1b and Supplementary Data Set 1 and 2). The portion of mass spectrometry signal that could be attributed to PRC2 increased 100-fold, from 0.4% in the proteome-wide data to 40% in the targeted RBR-ID experiments (Supplementary Fig. 1c), which allowed us to identify several additional PRC2 peptides that crosslinked to RNA (Fig. 1b and Supplementary Fig. 1d). We recovered multiple significant hits in all core subunits, in the PRC2.1 accessory subunits MTF2 and PALI, and in both accessory subunits of PRC2.2—AEBP2 and JARID2 (Fig. 1b and Supplementary Fig. 1a). These hits accumulated on the catalytic lobe of PRC2 (Fig. 1c–d and Supplementary Fig. 1e) and several mapped to domains previously proposed to bind to RNA, such as the EZH2 RBR (residues 342–36832), the JARID2 RBR (residues 332–35840), the EZH2 CXC and SET domains35, and the RRM-like beta-sheet domain of SUZ125. In addition to these, we noticed a highly significant crosslinked peptide on EED, very close to the regulatory centre, near the stimulatory recognition motif9 (SRM) of EZH2 (Fig. 1b, d; EED 336–355 is highlighted).
Fig. 1: Targeted RBR-ID of PRC2.
a, Targeted RBR-ID experimental design. Mouse ESCs treated with or without 4SU (1 and 2) were crosslinked with UV to generate RNA–protein crosslinks (3). After preparing nuclear extracts, we ‘targeted’ the RBR-ID technique by performing immunoprecipitations for endogenous PRC2 using an EZH2 antibody (4). Following IP, we treated eluted proteins with RNase and protease to remove crosslinked RNA and generate peptides (5), which were analysed via high resolution LC-MS/MS to identify decreases in apparent peptide abundance caused by the crosslink with RNA (6). b, Volcano plot of peptide intensities comparing material from 4SU-pulsed and control (−4SU) cells. Dashed horizontal line indicates P value of 0.05. Peptides on core and accessory PRC2 subunits are highlighted. P values were calculated using paired two-sided Student’s t-tests from three independent experiments and 10 total replicates. c, Mapping to PRC2 subunits of RNA-interacting peptides detected by targeted RBR-ID (blue circles, this study) or proteome-wide RBR-ID38 (red circles). Known protein domains, including previously identified RNA-binding regions (RBRs) on EZH232 and JARID240 are shown. d, RBR-ID structural mapping. Residue-level RBR-ID scores were calculated according to the level of 4SU-depletion and statistical significance and the resulting heatmap was used to colour the surface of a composite PRC2 model using two published PRC2 structures (PDB: 5WAI and 6C23, see Methods section). The substrate peptide in the catalytic centre is shown in black.
Thus, our targeted RBR-ID approach not only identified interactions between RNA and the PRC2.2 subunits AEBP2 and JARID215,30,34,38–40, but also MTF2 (also known as PCL2) and PALI—accessory subunits of the PRC2.1 complex that were previously not known to bind RNA. This strongly suggests that PRC2.1 also binds RNA in vivo.
RNA binds and inhibits both PRC2.1 and PRC2.2 in vitro
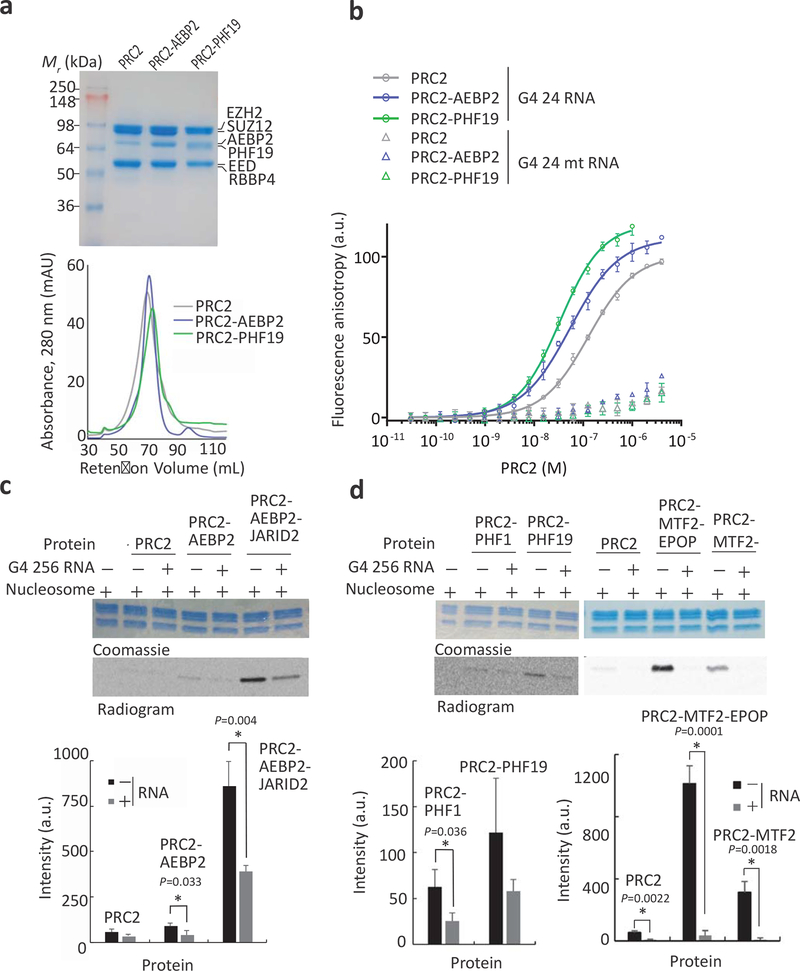
To further investigate the molecular nature and biochemical function of PRC2–RNA interactions in the context of holo-PRC2 complexes, we continued our studies in vitro, by reconstituting the human core PRC2 complex and a PRC2 complex with either human AEBP2 (PRC2-AEBP2) or the PCL protein PHF19 (PRC2-PHF19) (Fig. 2a). Given that PRC2 preferably binds to RNA that contains short repeats of consecutive guanines34, and since fluorescence anisotropy requires a small labelled ligand, we quantified affinity of PRC2 for an RNA composed of four UUAGGG repeats (G4 24 RNA, Fig. 2b and Table 1). These four repeats originate from TERRA RNA, fold into a G-quadruplex RNA structure and bind PRC2 in cells34,41. As a negative control, we used a size-matched mutant RNA without G-tracts, composed of four UGAGUG repeats (G4 mt 24 RNA, Fig. 2b and Table 1). In good agreement with earlier observations39, the affinity of the core PRC2 complex to RNA (Kd = 129 ± 6 nM) increased by approximately 2-fold when AEBP2 is in the complex (Kd = 54.6 ± 3.8 nM). Strikingly, the addition of PHF19 increased the affinity of PRC2 to RNA by nearly 4-fold compared to that of the core PRC2 complex alone. The affinity of PRC2-PFH19 (Kd = 34.0 ± 1.8 nM) to RNA was even greater than that of the PRC2-AEBP2 complex. The PRC2.1 accessory subunits MTF2 and EPOP also increased the affinity of PRC2 for RNA, with the PRC2-MTF2-EPOP complex having an approximately 2-fold higher affinity for RNA (Kd = 40.9 ± 3.9 nM) compared to PRC2-MTF2 (Kd = 82.4 ± 9.8 nM) and approximately 3-fold higher compared to core PRC2 (Kd = 129 ± 6 nM; Fig 2b, Table 1 and Supplementary Fig. S2g–i). These results indicate that both PRC2.1 and PRC2.2 holo-complexes bind to RNA. None of the PRC2 complexes bound to the mutant RNA (triangles in Fig. 2b and Table 1), indicating that the RNA-binding specificity of PRC2 toward this G-tract motif34 is preserved in the presence of a PCL subunit.
Fig. 2: RNA binds to and inhibits both PRC2.1 and PRC2.2.
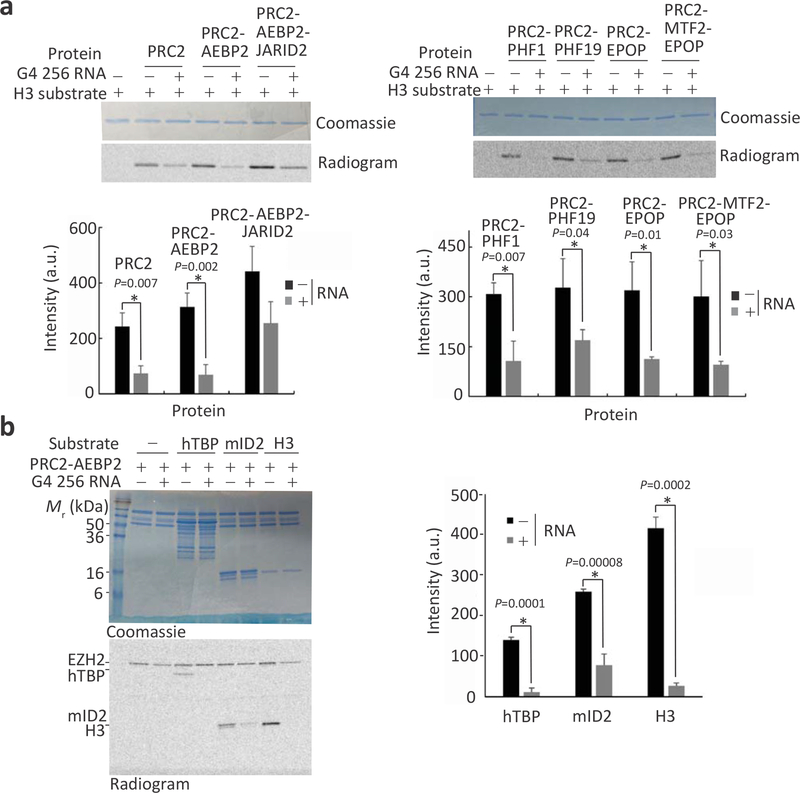
a, Coomassie blue-stained SDS-PAGE (top) and gel filtration chromatography (bottom, HiPrep 16/600 Sephacryl S-400 HR) of the PRC2 complexes that were used for binding assays. b Fluorescence anisotropy used to quantify the affinity of PRC2 complexes to G4 24 and G4 24 mutant (mt) RNAs. Data represent the mean of three independent experiments that were carried out on different days, error bars represent standard deviation. See Table 1 for dissociation constants (Kd) and Hill coefficients. c,d, HMTase assays of PRC2.2 (c) and PRC2.1 (d) complexes toward nucleosome substrates were carried out in the presence or absence of 8 μM G4 256 RNA. Histone proteins were visualised using Coomassie (upper gel) and methylation levels of H3 were determined by14C-autoradiography (bottom). Bar plots represent the mean of quantification using densitometry and error bars represent standard deviation based on three independent experiments. P values were determined using unpaired two-tailed Student’s t-test; *, P < 0.05. See Supplementary Fig. 2 for complete gel scans, SDS-PAGE and gel filtration chromatography of holo-PRC2 complexes and evidence for nucleosome reconstitution. Source data are available in Supplementary Data Set 5.
Table 1: Affinities of PRC2 complexes to G4 24 and G4 24 mt RNAs.
Dissociation constants (Kd) and Hill coefficients were derived from the binding curves in Fig. 2b. Standard errors were calculated from three independent replicates that were performed on different days.
| Protein | RNA | Kd (nM) | Hill |
|---|---|---|---|
| PRC2 | G4 24 | 129 ± 6 | 0.92 ± 0.03 |
| PRC2-AEBP2 | G4 24 | 54.6 ± 3.8 | 0.88 ± 0.04 |
| PRC2-PHF19 | G4 24 | 34.0 ± 1.8 | 1.01 ± 0.04 |
| PRC2 | G4 24 mt | n.d. | n.d. |
| PRC2-AEBP2 | G4 24 mt | n.d. | n.d. |
| PRC2-PHF19 | G4 24 mt | n.d. | n.d. |
Since the PCL protein MTF2 interacts with RNA in vivo and in vitro and the PRC2-PHF19 complex interacts with RNA in vitro, we wished to determine if PCL proteins allow for RNA-mediated inhibition of PRC2. We performed in vitro histone methyltransferase assays using recombinant nucleosome substrates in the presence or absence of a 256-base-long RNA that includes ten UUAGGG repeats flanked by sequences devoid of G-tracts (G4 256 RNA, see Methods section), which also bound to PRC2 with nanomolar affinity (Supplementary Fig. 3k–l). In agreement with previous studies, the RNA inhibited the HMTase activity of the core PRC2 complex28–30, the PRC2-AEBP2 complex15 and the PRC2-AEBP2-JARID2 complex15,28 toward nucleosome substrates (Fig. 2c and Supplementary Fig. 2a). We then performed the same experiments with three reconstituted human PRC2.1 complexes: PRC2-PHF1, PRC2-PHF19 and PRC2-MTF2-EPOP (Fig. 2a and Supplementary Fig. 2c,d). Despite variations in the baseline activity, RNA exerted an inhibitory effect regardless of which PCL protein (PHF1, PHF19 or MTF2) was present or whether EPOP was included (Fig. 2d and Supplementary Fig. 2b,j,k). This indicates that RNA inhibits the HMTase activity of PRC2 even when assembled with its accessory subunits to form either the PRC2.1 or PRC2.2 complexes.
RNA binds to the allosteric regulatory centre of PRC2
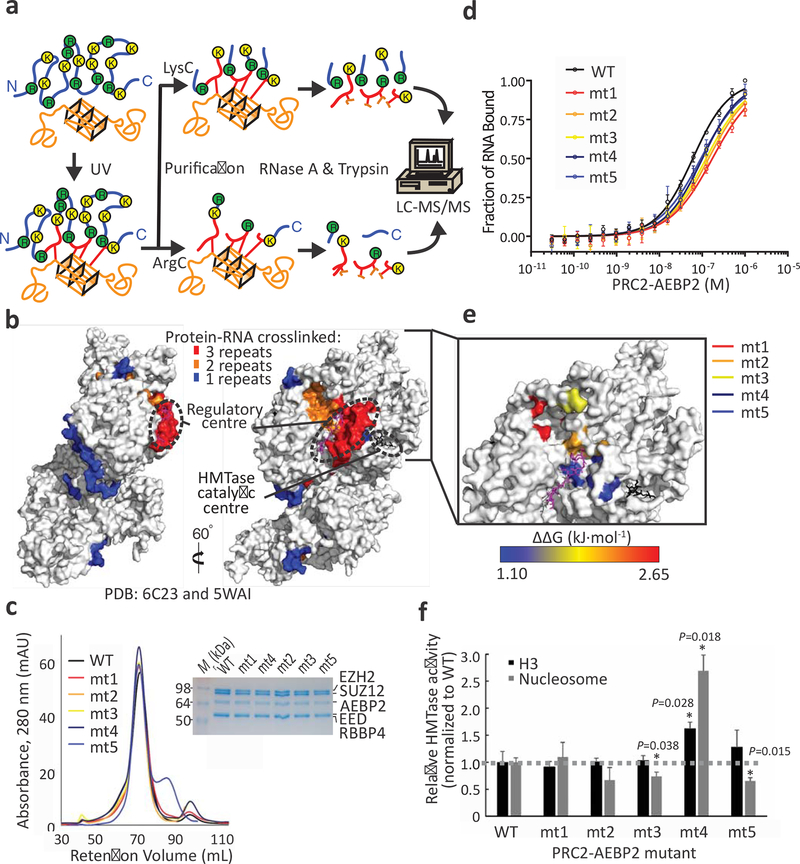
Under the notion that RNA binds to the PRC2 core complex and to the two types of holo-PRC2 complexes with similar affinity (up to 4-fold Kd, Fig. 2b and Table 1), specificity and inhibitory activity, we reasoned that RNA inhibits PRC2 through interactions with the core PRC2 subunits. To determine the site of PRC2 that binds to the inhibitory G-tract-containing RNA, we combined an in vitro UV cross-linking method34 with the RBDmap approach42 for mapping protein-RNA interactions using mass spectrometry (Fig. 3a and Methods). We used this approach to detect protein-RNA interactions between the reconstituted PRC2-AEBP2 complex and the G4 256 RNA (Fig. 3b and Supplementary Fig. 3a) that, similar to the G4 24 RNA, binds to PRC2 with high affinity (Supplementary Fig. 3k,l). Remarkably, most of the RNA-linked peptides that were identified in independent in vitro RBDmap replicates clustered within the same site at the interface between EED and EZH2 (Fig. 3b and Supplementary Data Set 3), overlapping the mouse EED peptide 336–355 identified in vivo by RBR-ID (Fig. 1d). These results are also in agreement with earlier UV crosslinking experiments that were carried out with SDS-PAGE, without high-resolution mapping, and identified EED and EZH2 as the two subunits that crosslinked to RNA within the context of an assembled PRC2-AEBP2 complex34. Intriguingly, this RNA-binding site overlaps with the regulatory site that was previously shown to regulate the HMTase activity of PRC2 through allosteric stimulation9–12 (Fig. 3b).
Fig. 3: Mapping of protein–RNA interactions within PRC2-AEBP2 in vitro.
a, Schematic representation of the in vitro RBDmap workflow (see Methods section): in vitro reconstituted protein–RNA complexes are crosslinked, followed by tandem proteolytic digestion and LC-MS/MS to reveal peptides adjacent (blue) to the protein–RNA crosslink (red). RNA is shown in orange. b, RBDmap results: amino acids within the PRC2-AEBP2 structure were coloured in blue, orange or red if they resided within peptides that were crosslinked to RNA in 1, 2 or 3 independent RBDmap experiments, respectively. A methylated peptide in the regulatory centre is coloured magenta and the substrate peptide in the catalytic centre is coloured black (PDB accession: 6C23 and 5WAI). c-d, Validation using point mutations. The purity and integrity of the mutant complexes were assessed using SDS-PAGE and gel filtration chromatography (HiPrep 16/600 Sephacryl S-400 HR) (c). Fluorescence anisotropy was used to quantify the affinity of the mutants to G4 24 RNA (d). The resulting dissociation constant (Kd), Hill coefficients and the derived ΔΔG are indicated together with details of the mutated amino acids in EZH2 and EED in Table 2. Error bars in (d) represent standard deviation based on three independent experiments that were done on different days. e, The impaired capacity of the mutants to bind RNA is represented in a ΔΔG heat map using the PRC2-AEBP2 structure. Mutated amino acids are mapped to the structure and ΔΔG colour code is indicated (bottom). f, Mean HMTase activity of indicated PRC2 mutants toward H3 histones (black bars), or nucleosomes (grey bars) normalised to the activity of wild-type PRC2-AEBP2 (dashed line). Error bars represent standard deviation based on three independent experiments. P values were determined using paired two-tailed Student’s t-test; *, P < 0.05. See Supplementary Fig. 3 for HMTase radiograms and gel scans, SDS-PAGE analyses and mass spectrometry intensities resulting from the RBDmap process and additional mutants that were assayed. Source data are available in Supplementary Data Set 6.
To validate this finding, we introduced point mutations in EZH2 and EED within the identified RNA-binding site or its vicinity (mt1–8). We reconstituted mutant PRC2-AEBP2 complexes (Fig. 3c and Supplementary Fig. 3b) and measured their affinity to G4 24 RNA (Fig. 3d–e, Table 2 and Supplementary Fig. 3c–d). The mutations reduced the affinity of PRC2 to RNA (Fig. 3e, Table 2 and Supplementary Fig. 3d–g), with ΔΔG in the range of 1.1–2.7 kJ/mol. The mutations mt1–3 and mt6–7 caused the largest reduction in affinity (ΔΔG 1.8–2.7 kJ/mol, Fig 3d,e, Table 2 and Supplementary Fig. 3c–g), which supports a direct function of the mutated amino acids in RNA binding. Importantly, these mutations did not lead to adverse effects on complex assembly (Fig. 3c and Supplementary Fig. 3b) or on PRC2 activity toward histone substrates, and only mt3, mt5 and mt8 displayed a negative impact on the activity of PRC2 toward nucleosome substrates (Fig. 3f and Supplementary Fig. 3h–j). The mutant mt4 displayed a positive effect on HMTase activity, possibly by stabilising PRC2 in a conformation resembling its allosterically stimulated state, given its location near the regulatory centre. The mutants led to only a modest reduction in the affinity of PRC2 to RNA and even in case of the complex bearing the most effective set of mutations, as determined by its in vitro affinity for RNA (mt1, ~3-fold reduction of affinity), we could not detect any change in the extent of RNA-mediated inhibition (Supplementary Fig. 4a–d). This is consistent with our photocrosslink mapping (Fig. 1d and 3b), and previous hydrogen deuterium exchange analyses35, which pointed to multiple relatively large RNA-binding surfaces within PRC2 that might be involved in RNA-mediated inhibition of the complex (see below).
Table 2: Affinities of PRC2-AEBP2 mutant complexes to G4 24 RNA.
Dissociation constants (Kd), Hill coefficients and the derived ΔΔG values were calculated from the binding curves in Fig. 3d. Details of the mutated amino acids in EZH2 and EED are indicated. Standard errors were calculated from three independent replicates.
| PRC2-AEBP2 mutant | Mutation sites | Kd (nM) | Hill | ΔΔG (kJ·mol−1) |
|---|---|---|---|---|
| WT | n/a | 60.5 ± 2.2 | 1.03 ± 0.04 | 0 |
| mt1 | EZH2 R27A/R31A/F165A,EED R355A | 173 ± 7 | 0.87 ± 0.03 | 2.65 |
| mt2 | EZH2 H129A/K156A/H158A/G159A/R161A, EED R306A | 141 ± 8 | 0.91 ± 0.04 | 2.13 |
| mt3 | EZH2 R16A/K17A/R18A/K20A | 124 ± 7 | 0.91 ± 0.04 | 1.81 |
| mt4 | EZH2 K661A | 102 ± 4 | 0.99 ± 0.04 | 1.32 |
| mt5 | EZH2 Y133A/T144A/F145A/Y153A, EED Y308A | 93.7 ± 6.3 | 0.89 ± 0.05 | 1.10 |
In addition to mutations guided by our RBDmap results (Fig. 3b), we assayed two RNA-binding-deficient mutants, mt4 (Fig. 3c–e) and mt8 (Supplementary Fig. 3c–d,g), that reside externally to the regulatory site and were previously analysed35. In the original study, these mutants were tested in the context of the minimal PRC2 core complex (EZH2, EED and the VEFS domain of SUZ12) and found to reduce the affinity of PRC2 to RNA by up to 12-fold Kd35. Within the PRC2-AEBP2 complex, mt4 and mt8 exhibited an affinity change of approximately 2-fold Kd (ΔΔG of 1.3 and 1.1 kJ/mol, respectively). These results are in good agreement with our and others’ observations that AEBP2 and regions of SUZ12—beyond the VEFS domain5—interact with RNA (Fig. 1) and that AEBP2 increases the affinity of PRC2 to RNA (Fig. 2b and Table 1).
Our results thus indicate that amino acids within the allosteric site of PRC2 and in its immediate vicinity are directly involved in RNA binding. These findings reinforce previous observations of dispersed RNA-binding sites in EZH235, but also show additional significant protein–RNA contacts within the regulatory subunit EED, and point to the regulatory site of PRC2 as an important determinant for RNA binding.
Stimulatory peptides relieve RNA-mediated inhibition of PRC2
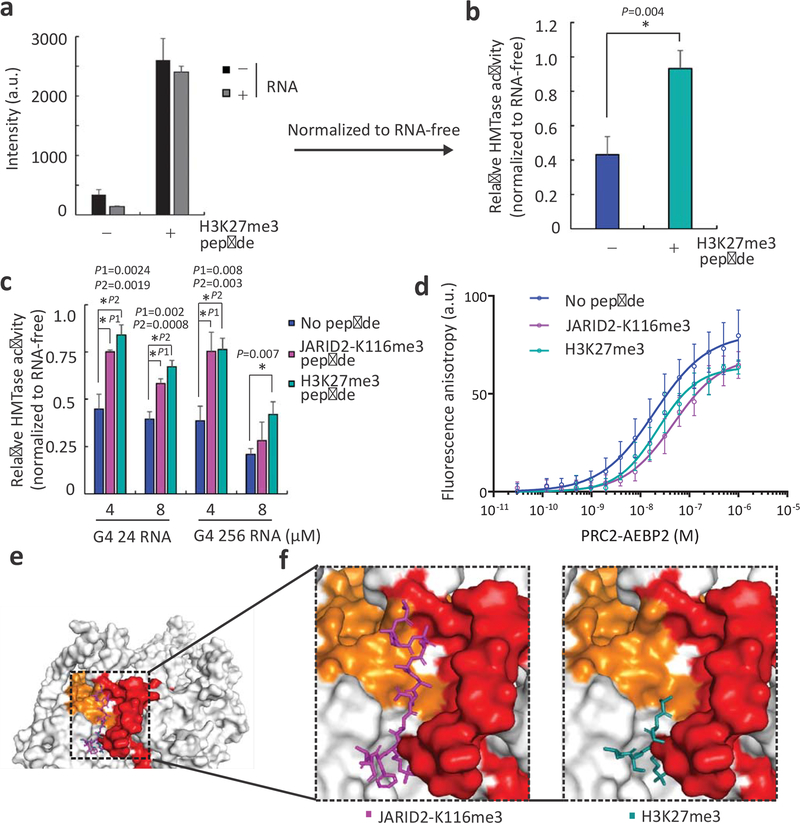
Given the location of a prominent RNA-binding site within the regulatory centre of PRC2, we wished to determine the regulatory interplay between peptide ligands that bind to this site and stimulate PRC2 (H3K27me3 and JARID2-K116me3) and inhibitory RNAs (e.g. G4 256 RNA and G4 24 RNA). Both stimulatory peptides H3K27me3 and JARID2-K116me3 peptides could significantly overcome RNA-mediated inhibition by G4 24 and G4 256 RNAs (Fig. 4a–c, Supplementary Fig. 4a–d). Quantitative binding assays indicated that the JARID2-K116me3 peptide competed with RNA for binding to PRC2, decreasing the ΔG by 2.24 kJ/mol, but the H3K27me3 peptide did not (Fig. 4d and Table 3). This is consistent with the observation that the longer JARID2-K116me3 peptide extends into the pocket formed at the EED-EZH2 interface (Fig. 4e,f, in purple), which was identified as a primary site of PRC2–RNA interactions by both RBR-ID (Fig. 1) and RBDmap (Fig. 2), whereas the short H3K27me3 peptide used in these assays did not (Fig 4f, in green). Similarly, the PRC2 allosteric inhibitor A39543 does not bind to the RNA-binding surface (Supplementary Fig. 4i) and it did not compete with RNA binding (Supplementary Fig. 4g,h).
Fig. 4: Stimulatory peptides relieve the RNA-mediated inhibition of PRC2.
a, HMTase assays of PRC2 in the presence (+) or absence (−) of 80 μM H3K27me3 peptide and in the presence (+) or absence (−) of 4.0 μM G4 256 RNA. b, HMTase activities of PRC2 in its basal and stimulated states, relative to the HMTase activity of an RNA-free PRC2 within the same state: bar plot based on the same data as in panel a, after normalising each RNA-containing sample (grey bars in panel a) to the corresponding RNA-free sample (black bars in panel a) to yield the relative HMTase activity of PRC2 in either its stimulated (H3K27me3 peptide, in green) or basal (no peptide, in blue) state. c, HMTase activities, relative to an RNA-free sample, of PRC2-AEBP2 in its stimulated (10 μM JARID2-K116me2 peptide, in magenta, or 80 μM H3K27me3 peptide, in green) or its basal (no peptide, in blue) state and in the presence of RNA as indicated (relative activities were calculated as in panel b). Error bars in panels a-c represent standard deviations based on 3 independent experiments. All bar plots are represented means. P values were determined using unpaired two-tailed Student’s t-test; *, P < 0.05. d, The affinity of the PRC2-AEBP2 complex to G4 24 RNA was quantified using fluorescence anisotropy in the presence or absence of 100 μM H3K27me3 or 10 μM JARID2-K116me3 peptides. The KCl concentration in the binding buffer was reduced to 100 mM (rather than 200 mM KCl that was used in Fig 2b–c) in order to mimic the conditions used in the HMTase assays presented in this figure. Error bars represent standard deviations in 11, 6 and 4 independent replicates for the binding curves plotted in blue, purple and green, respectively. Values are represented means. See Table 3 for dissociation constants and Hill coefficients. e, The stimulatory peptides’ binding sites in PRC2 (coordinates: PDB 6C23): Orange and red represent RNA-linked polypeptides (colour code as in Fig. 3b), after superimposing the JARID2-K116me3 peptide (magenta, from PDB: 6C23) and the H3K27me3 peptide (dark green, from PDB: 3IIW). f, Close-up of the two peptides’ binding sites. Source data are available in Supplementary Data Set 7.
Table 3: Affinities of PRC2-AEBP2 complexes to G4 24 RNA in the presence or absence of stimulatory peptides.
Binding assays were performed in the presence of 100 μM H3K27me3 or 10 μM JARID2-K116me3 peptides as indicated, and binding curves are presented in Fig. 4d. Resulting dissociation constants (Kd), Hill coefficients, the derived ΔΔG values and the number of independent replicates (n) are indicated. Peptide sequences are indicated, with the trimethylated lysines in red. Highlighted in grey are amino acids within the peptide sequence that were previously traced in the regulatory centre using high-resolution cryo-EM (PDB: 6C23) or x-ray crystallography (PDB: 3IIW). Standard errors were calculated from three independent replicates and are indicated in the table.
| Competitive peptide | Sequence | Kd (nM) | Hill | ΔΔG (kJ·mol−1) | n |
|---|---|---|---|---|---|
| No peptide | n/a | 19.2 ± 2.7 | 0.79 ± 0.06 | 0 | 11 |
| JARID2-K116me3 | KRPRLQAQRK (me3) FAQSQ | 46.7 ± 6.9 | 0.88 ± 0.08 | 2.24 | 6 |
| H3K27me3 | TKAARK (me3) SAPAT | 22.5 ± 3.1 | 1.05 ± 0.12 | 0.4 | 4 |
These data indicate that stimulatory peptides of PRC2 relieve the inhibitory activity of RNA, possibly through allosteric modulation and, at least for JARID2-K116me3, in part through competition with RNA at the regulatory centre.
RNA inhibits PRC2 in a DNA-independent manner
The RNA-binding sites that we identified in PRC2 using RBDmap and targeted RBR-ID are adjacent to and, in some cases, overlap the substrate-binding site in the methyltransferase centre (Supplementary Fig. 5a). We therefore wished to determine if RNA can inhibit the methyltransferase activity of PRC2 through a mechanism other than competition for nucleosomes36 or DNA binding15. We repeated the HMTase assays using DNA-free H3 histones as substrates (Fig. 5a), rather than nucleosomes. In agreement with the hypothesis of a DNA-independent RNA-mediated inhibition of PRC2, G4 256 RNA inhibited the activity of both PRC2.2 (Fig. 5a, left) and PRC2.1 (Fig. 5a, right) toward H3 substrate. Unlike the longer G4 256 RNA, the G4 24 RNA did not inhibit PRC2 toward H3 histones (Supplementary Fig. 5d,e and Supplementary Data Set 4), suggesting that its observed inhibitory effect in the context of nucleosomal substrates (Fig. 4c) relies on competition with nucleosomal DNA15. This indicates that although a 24-base-long G-quadruplex RNA is sufficient to bind PRC2 (Fig. 2 and34) and inhibit HMTase toward nucleosome substrates (Fig. 4c), longer RNAs might be required for methyltransferase inhibition toward non-nucleosome substrates. Contributing factors would likely be an increased affinity of the long RNA for PRC2 (G4 256 RNA, Kd = 1.09 ± 0.13 nM; Supplementary Fig. 3k,l) over the short RNA (G4 24 RNA, Kd = 19.2 ± 2.7 nM; Fig. 4d, and Table 3) and additional steric hindrances that are likely offered by the longer RNA. Although PRC2 activity toward H3 histones outside nucleosomes has limited biological significance, these data indicate that RNA-mediated HMTase inhibition of PRC2 can take place independent of competition for DNA or nucleosome binding. Indeed, the JARID2-K116 unmethylated peptide, as well as the H3K27M oncogenic peptide that binds to the catalytic site but not the regulatory centre of PRC210 (Supplementary Fig. 5a), reduced the affinity of PRC2 for RNA (Supplementary Fig 5b,c). This suggests that these peptides can directly compete with RNA binding at the catalytic site of the complex in addition to the regulatory centre, further supporting our mapping of protein-RNA interactions to this region (Fig. 1, Supplementary Fig. 6e).
Fig. 5: DNA-independent RNA-mediated inhibition of PRC2.
a, HMTase assays carried out in the presence of 0.5 μM PRC2 complexes as indicated, 4 μM H3 histone substrate and in the presence or absence of 1 μM G4 256 RNA. The bar plot (bottom) represents the activity, as recorded by densitometry after SDS-PAGE (top). b, HMTase assays were carried out in the presence of 0.5 μM PRC2-AEBP2, 1 μM H3 or non-histone substrates human TBP (hTBP, 20 μM) or mouse ID2 (mID2, 15 μM), and in the presence or absence of 8 μM G4 256 RNA. The bar plot (right) represents the activity, as recorded by densitometry after SDS-PAGE (left). In all plots within the figure, bars represent means and error bars represent standard deviation based on three independent experiments and P values were determined using unpaired two-tailed Student’s t-test; *, P < 0.05. Complete gel scans are in Supplementary Fig. 5. Source data are available in Supplementary Data Set 8.
We next assayed the methyltransferase activity of PRC2 toward two non-histone substrates of PRC2—human TBP (hTBP) and mouse ID2 (mID2)37—and confirmed inhibition of PRC2 activity by RNA also for these substrates (Fig. 5b). Collectively, these results demonstrate that RNA can inhibit the methyltransferase activity of PRC2 toward a DNA-free substrate, including non-histone substrates.
The regulatory centre and the RNA-binding site are exposed in PRC2.1
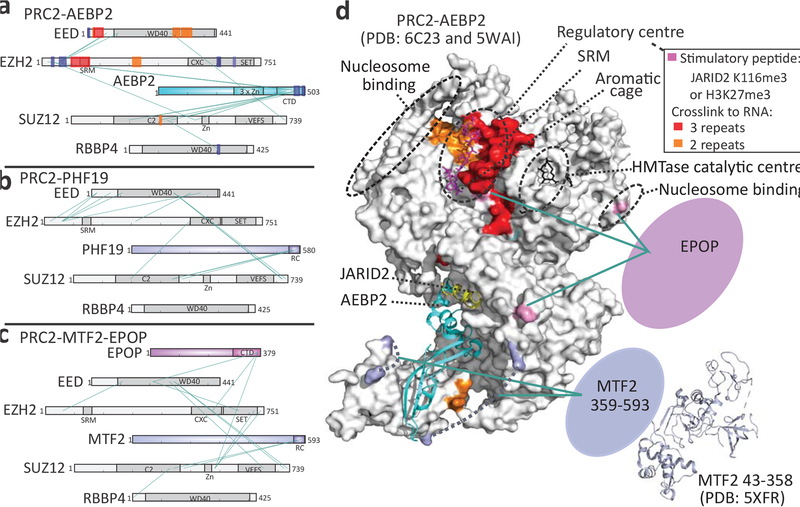
Since RNA binds and inhibits both PRC2.1 and PRC2.2 (Fig. 2), we hypothesised that the PRC2.1 complex adopts a similar architecture to PRC2-AEBP2, leaving the regulatory site exposed. We mapped protein-protein interactions within PRC2-PHF19 and PRC2-MTF2-EPOP using bis(sulfosuccinimidyl)suberate (BS3) crosslinking with mass spectrometry (BS3 XL-MS, Fig. 6b,c). For a direct comparison, we also mapped interactions within the PRC2-AEBP2 complex (Fig. 6a), including the 216 amino acids that complete the N-terminal domain of the canonical AEBP2 isoform but were not included in previous structural investigations into PRC2-AEBP25,7. Distances between crosslinked lysine pairs within PRC2 core subunits were measured using the high-resolution structure of the PRC2-AEBP2-JARID2 complex5 and resulted in similar distance distributions for the three complexes (Supplementary Fig. 6a,b), supporting a similar structural organisation of the core subunits in the PRC2.1 and PRC2.2.
Fig. 6: The RNA-binding site in the regulatory centre of PRC2 is exposed within both PRC2.1 and PRC2.2.
a-c BS3 crosslinking with mass spectrometry (BS3 XL-MS) results for PRC2-AEBP2 (a), PRC2-PHF19 (b) and PRC2-MTF2-EPOP (c). Core subunits are coloured grey, accessory subunits are indicated in assorted colours and selected domains are shown in dark colours (see Supplementary Fig. 6d, middle structure, for the same view with the core subunits in assorted colours). Green lines represent inter-molecular protein-protein BS3 crosslinks. Blue, orange and red boxes on the protein representation in panel a represent RNA–protein crosslinks that were identified in 1, 2, or 3 independent RBDmap experiments, respectively (same data as in the 3D representation in Fig. 3b). d, Accessory proteins and RNA-binding sites within the holo-PRC2 complex: surface view of PRC2 was generated as in Fig. 3b. AEBP2 (cyan) and JARID2 (yellow) fragments are shown as a ribbon representation and the N-terminus of MTF2 that was determined crystallographically (PDB: 5XFR) is in light blue, to approximate scale. EPOP (pink) and the C-terminal region of MTF2 (light blue) are indicated as blobs, to approximate scale. Green lines indicate crosslinks between PRC2 core subunits to PCL proteins and EPOP. Residues within core PRC2 subunits that were crosslinked to EPOP are indicated in pink. Residues within core PRC2 subunits that reside at the termini of unstructured loops that were crosslinked to PCL proteins are indicated in light blue and linked with dashed arcs. Protein–RNA contacts that were determined in 2 or 3 independent RBDmap replicates (see Fig. 3 for complete data) are indicated in orange and red, respectively. Other key functional centres or structural features are highlighted using dashed black circles. See Supplementary Fig. 6 for distances histograms of BS3 XL-MS and different views of the structure presented in (d).
We next mapped crosslinking sites of the accessory subunits PHF19, MTF2 and EPOP from the PRC2-PHF19 and the PRC2-MTF2-EPOP complexes to the core subunits (Fig. 6d, see Methods for full description). We identified interactions between the C-terminal ‘reversed chromodomain’ (RC domain)44 of the PCL proteins PHF19 and MTF2 to the C2 domain of SUZ12 (Fig. 6b,c), in good agreement with binding assays from previous studies6,44. Importantly, no interactions were detected between PCL proteins and domains of core subunits within the catalytic lobe of PRC2. Although the C-terminal of EPOP crosslinked to residues within the catalytic lobe of PRC2, under the SRM of EZH2, amino acids in the regulatory centre or the RNA-binding site were not crosslinked (Fig. 6c,d and Supplementary Fig. 6c).
While the structure of the N-terminal portion of EED was never determined, our BS3 XL-MS results indicate that it resides within the vicinity of the N-terminal of EZH2 in all the examined PRC2 complexes (Fig. 6a–c). Similar crosslinking was previously observed in the PRC2-AEBP2-JARID2 complex5. In addition to mutual protein–protein crosslinks (BS3 XL-MS: green lines in Fig. 6a–c), the N-terminal portions of EED and EZH2 cluster multiple RNA–protein crosslinked peptides (RBDmap: blue and red spots in Fig. 6a). The simplest explanation for these observations is that the N-terminal regions of EZH2 and EED reside in close proximity and form a single RNA-binding site that is likely exposed in both the PRC2.1 and PRC2.2 complexes. This is in good agreement with the ability of an RNA containing short repeats of consecutive guanines to bind the two types of holo-PRC2 complexes with similar affinity and specificity, and to inhibit the methyltransferase activity of both of them (Fig. 2), possibly through an identical mechanism.
Discussion
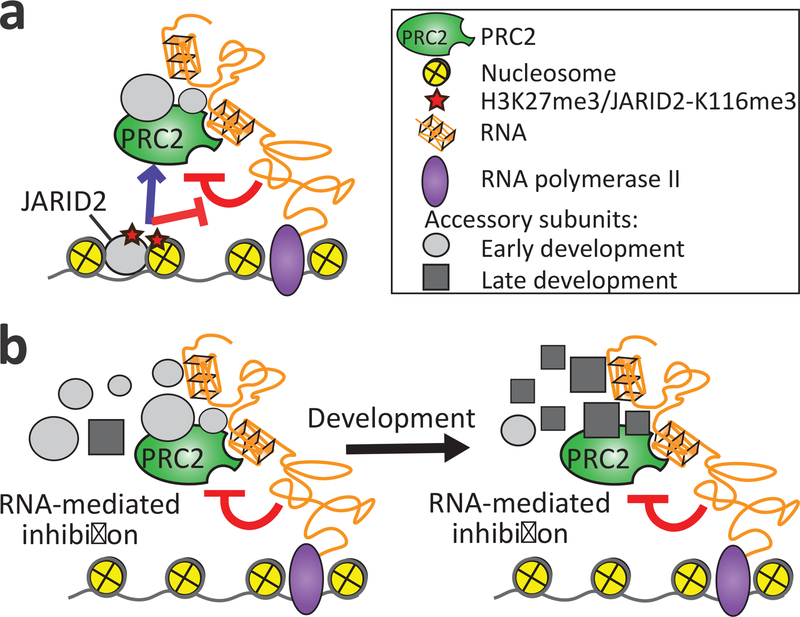
The recruitment of PRC2 to chromatin and its regulation at target genes are determined by interactions with multiple factors, including accessory proteins, specific DNA sequences, and RNA2. Among the protein factors, unbiased proteomic approaches identified JARID2, AEBP2, EPOP, PALI and PCL proteins as key accessory subunits that were reproducibly identified across studies and experimental systems20–22,45, albeit in relative abundances that change across developmental stages20. Our study indicates that regardless of subunit composition, RNA binds to and inhibits various subtypes of PRC2 complexes in vivo and in vitro (Fig. 1, 2). Mechanistically, this is achieved through exploiting multiple surfaces on the catalytic lobe of the core complex, including the regulatory centre formed at the interface of EED and EZH2 (Fig. 3), which is exposed both within the PRC2.1 and PRC2.2 (PRC2-AEBP2-JARID2) complexes (Fig. 6). It implies that PRC2 can bind RNA throughout various stages of development, even when the composition of its accessory subunits varies significantly (Fig. 7b).
Fig. 7: A model for RNA-mediated inhibition of PRC2.
a, Stimulatory effectors of PRC2 — JARID2-K116me3 and H3K27me3 — relieve the inhibitory activity of RNA simultaneously with HMTase stimulation. This process provides a molecular mechanism to overcome RNA-mediated inhibition during the nucleation, spreading and propagation of the H3K27me3 mark at polycomb-target genes8,11,46. b, A model for RNA-mediated inhibition of PRC2 during development: RNA binds to the allosteric regulatory centre of PRC2 and inhibits methyltransferase activity towards histone and non-histone substrates, either when PRC2 is in complex with AEBP2 and JARID2 (PRC2.2) or PHF1, PHF19, MTF2, PALI and EPOP (PRC2.1). RNA-mediated inhibition of PRC2 provides a fail-safe mechanism to prevent substrate methylation by RNA-bound PRC2 at non-target genes, even if the stoichiometry of its common accessory subunits changes during development.
One face of PRC2 clusters binding sites for multiple regulatory factors
Our analysis of PRC2.1 complexes, together with data from previous structural investigations into the architecture of the PRC2-AEBP2-JARID2 complex5,6, indicates that binding sites for multiple ligands and factors cluster on one face of PRC2 (Fig. 6d and Supplementary Fig. 6c, d). This face also contains binding sites for the PRC2.1 and PRC2.2 accessory subunits that regulate the recruitment of PRC2 to chromatin1–3. The same face also includes the catalytic site and the allosteric regulatory centre that stimulates HMTase activity upon binding of methylated H3 or JARID2 peptides8,11,12. All these stimulatory, regulatory, and catalytic modules are concentrated on the same face, along with the most prominent RNA-binding region identified by both RBR-ID and RBDmap (Supplementary Fig. 6). This structural organisation might provide RNA with simple means to block methyltransferase activity and to simultaneously interact with other RNA-binding regions of EZH2 such as the CXC and SET domains35, as well as with the accessory subunits JARID240, AEBP238, MTF2 (Fig. 1) and possibly other PCL proteins (Fig. 2).
Interplay of RNA and stimulatory peptides at the allosteric regulatory centre
Amino acids of EED and EZH2 involved in allosteric activation of PRC28–11, such as for example EZH2 F14512, reside within the main RNA-binding site identified by RBR-ID and RBDmap or in its immediate vicinity (Fig. 6). The RNA-binding site is also in the immediate vicinity of the SRM (Fig. 6d), which stabilises the methyltransferase centre within EZH29 during allosteric activation8. Here, we report that allosteric stimulation of PRC2 though either H3K27me3 or JARID2-K116me3 peptides relieve RNA-mediated inhibition (Fig. 4), providing a direct link between the RNA-binding site that we identified in the regulatory centre of PRC2 and the process of effector-induced stimulation.
The JARID2-K116me3 stimulatory peptide was assigned a significant role in nucleating the H3K27me3 mark11, which in turn stimulates PRC2 selectively at repressed polycomb-target genes8 to allow for the nucleation and, eventually, spreading and propagation of the H3K27me3 mark8,11,46. It is plausible that RNA-mediated inhibition of PRC2 is suspended at new PRC2 target sites through stimulatory interactions with the JARID2-K116me3 moiety (Fig. 7a and Supplementary Discussion in the Supplementary Note 1).
Multi-modal inhibition of PRC2 by RNA
The mechanistic details of RNA-mediated PRC2 inhibition are of great importance to understand the biological meaning of the still poorly understood PRC2-RNA interactions. The fact that RNA does not inhibit the prominent automethylation activity of EZH2 (Supplementary Fig. 2a,b, 5 and15) previously suggested a model whereby RNA inhibits PRC2 by competing with nucleosomal DNA for binding15. Indeed, some of the amino acids that were altered within our mt1 and mt3 mutants, which showed decreased RNA affinity, were previously identified as part of a nucleosome-binding site47. Yet, most of the mutated amino acids that affect RNA binding in our experiments (Fig. 3d and Table 2) do not interact with nucleosomes47, but rather reside within the regulatory centre or its immediate vicinity (Fig. 3e). Moreover, recent investigations into the automethylation of EZH248,49 suggest that it occurs intramolecularly (in cis). Thus, this activity benefits from a high local substrate concentration that could overcome RNA-mediated inhibition. It is thus possible that additional bases, beyond the mere G-quadruplex motif, sterically block the substrate-binding site of PRC2. Indeed, a direct—and possibly inhibitory—interaction between RNA and the SET domain in vivo is supported by our targeted RBR-ID data (Fig. 1c–d), in vitro by the competition of JARID2 and H3 substrate peptides for RNA binding—presumably at the catalytic site (Supplementary Fig. 5b,c)—and indirectly by hydrogen deuterium exchange (HDX) results from an independent study35. Therefore, RNA can inhibit PRC2 through different mechanisms, including competition for nucleosome binding36, competition for DNA binding15 and through blocking methyltransferase activity directly.
Future studies may reveal if the inhibitory activity of RNA binding to PRC2 could be relieved by specific transcripts, during specific stages of development, in disease states or at specific loci, and possibly through the involvement of cell-type-specific factors beyond the most abundant accessory subunits of PRC220–22. The location of an exposed RNA-binding site within the allosteric centre of the two common forms of holo-PRC2 complexes provides means for RNA-mediated regulation of PRC2 in most cell lineages and during most stages of normal development and suggest a mechanism by which stimulatory peptides relieve RNA-mediated inhibition to allow de novo methylation at PRC2 target loci.
Methods
Targeted RBR-ID
For label-free experiments, mESCs were cultured on gelatin-coated dishes in KnockOut DMEM (Gibco #10829018) supplemented with 15% FBS (Gibco #10437028), 100 mM nonessential amino acids (Sigma #M7145), 0.1 mM 2-mercaptoethanol (Gibco #21985023), 1 mM L-glutamine (Sigma #G7513), 100 U/mL leukemia inhibitory factor (LIF) (Millipore #ESG1107), 3 μM CHIR99021 (Millipore #361559), 1 μM PD0325901 (Millipore #444966), 50 U/mL penicillin and 50 μg/mL streptomycin.
For SILAC-assisted quantification, cells were cultured in medium containing 15% dialyzed FBS (Thermo Fisher #88440), 2 mM Proline and 0.47 mM conventional or heavy isotope-labelled amino acids (Arginine-10 + Lysine-8). Following several passages in heavy media, cell extracts were analysed via mass spectrometry and only used if >98% labelling could be confirmed. Cells were crosslinked as previously described38,50. Briefly, cells treated for 2 hrs with 500 μM 4-thiouridine (4SU) were crosslinked with 1 J/cm2 UVB light. SILAC-labelled cell extracts were prepared as follows: cells were lysed in 50 mM Tris (pH 8 at 25 °C), 150 mM NaCl, 1% IGEPAL CA-630, 0.2 mM EDTA, 2 mM MgCl2, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin and 0.2 mM PMSF, then treated with 2,500 U/mL Pierce Universal nuclease (Thermo #88700) for 30 min at 25 °C. Extracts were sonicated briefly, then NaCl was added to bring the final concentration to 300 mM and extracts were rotated at 4 °C for 30 min to complete lysis. Extracts were centrifuged at 18,000 RCF for 10 min at 4 °C and supernatants were collected. We quantified cell extracts via Bradford protein assay and prepared 1:1 mixtures of ± 4SU-treated heavy and light labelled extracts. For label-free quantification replicates, nuclear extracts were prepared from cells as previously described38,50. Polyclonal anti-EZH2 antibody was generated by immunising rabbits with a fragment of mouse EZH2 spanning amino acids 1 to 370 (Uniprot Q61188) and affinity purifying using the same antigen. Immunoprecipitations were performed by adding EZH2 antibody to extracts and incubating overnight at 4 °C, then recovering protein-antibody complexes with protein G Dynabeads (Thermo #10003D) pre-blocked with 1 mg/mL BSA. Beads were washed 3x with IP wash buffer: (20 mM Tris [pH 7.9 at 4 °C], 0.2 mM EDTA, 200 mM KCl and 0.05% IGEPAL CA-630). To elute bound proteins, 0.1 M glycine (pH 2.4) was added to beads and incubated at 25 °C with gentle shaking for 10 min, then eluate was transferred to a fresh tube containing 0.1 M Tris (pH 8 at 25 °C) to neutralise. Fractions of eluted proteins were taken for western blot, and the remainder diluted in trypsin digestion buffer (final: 50 mM NH4HCO3 [pH 8], 1 mM MgCl2 and 1 mM CaCl2) or chymotrypsin digestion buffer (final: 100 mM Tris [pH 8 at 25 °C], 1 mM MgCl2 and 10 mM CaCl2,). To remove crosslinked RNA, 12.5 μg/mL RNase A was added to samples and incubated for 30 min at 37 °C. To reduce proteins, samples were treated with 5 mM dithiothreitol for 60 min at 25 °C, then cysteines were alkylated with 14 mM iodoacetamide for 30 min in the dark, before additional reduction with 5 mM dithiothreitol for additional 15 min in the dark. Proteins were digested with trypsin or chymotrypsin at a protease:sample ratio of 1:20 and incubated overnight at 37 °C or 25 °C, respectively. Peptides were quenched with formic acid and desalted on C18 stage tips, then dried and resuspended in 0.1% formic acid prior to mass spectrometry analysis.
For mass spectrometry, a reverse phase gradient with a nano-LC was performed on a C18 column with a 2%–60% binary gradient (mobile phase A: 0.1% formic acid in aqueous; mobile phase B: 80% acetonitrile, 0.1% formic acid) for 90 minutes. The gradient continued to 95% over 2 minutes and held for 13 minutes at 95% mobile phase B. MS was performed using a Thermo Orbitrap Fusion instrument and MS/MS data was collected in centroid mode using the Orbitrap mass analyser. Fragmentation of peptides for MS/MS was performed using HCD and only charge states of 2–5 were included for fragmentation. MS/MS spectra were processed through MaxQuant51 using a FASTA file comprising PRC2 complex proteins. SILAC and unlabelled samples were processed with the same parameters. To generate the protein-level analyses shown in Supplemental Data Set 2, MS/MS spectra were processed using a mouse proteome FASTA file.
RBR-ID analysis
After removal of suspected contaminants, MaxQuant peptide abundances were normalised by the mean of all peptide intensities in each MS run, or in the case of SILAC data by the mean heavy or light labelled peptide intensity in each run. For each peptide, a log2-converted ratio was calculated between samples treated with or without 4SU to assess depletion mediated by RNA-crosslinking. P values for peptides observed only in SILAC samples were analysed via a paired, two-sided Student’s t-test, while an unpaired two-sided Student’s t-test was performed for peptides common between SILAC and label-free quantification samples, to account for missing values in the data matrix. RBR-ID scores, which reflect both the degree and consistency of 4SU-mediated depletion of a peptide38, were calculated as follows:
To visualise targeted RBR-ID scores on the high-resolution structure of the human PRC2-AEBP2, RBR-ID scores at each residue were calculated as the sum of the RBR-ID scores of all overlapping identified peptides, and then mouse peptide sequences were aligned to corresponding human protein sequence using ClustalOmega. Since a large number of amino acids were not resolved in the high-resolution cryo-EM structure of the PRC2-AEBP2-JARID2 complex (PDB: 6C23)5, and in order to allow for maximal coverage of this complex, the crystal structure of SUZ12-RBBP4-JARID2-AEBP2 (PDB: 5WAI)6 was superimposed on this structure and the non-catalytic lobe of 6C23 was omitted, with the exception of SUZ12 amino acids 497–518, which is absent in 5WAI6.
Protein expression and purification
See Supplementary Note 2.
Fluorescence anisotropy assay
The 3′ fluorescein labelled G4 24 RNA (UUAGGG)4 and G4 mt 24 RNA (UGAGUG)4 were synthesised by IDT. RNA was incubated for 2 min at 95 °C in 10 mM Tris-HCl pH 7.5 (at 25 °C) and was then immediately snap-cooled on ice for 2 min. Next, RNA was allowed to fold for 30 min at 37 °C in binding buffer (50 mM Tris-HCl pH 7.5 at 25 °C, 200 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2, 2 mM 2-mercaptoethanol, 0.1 mg/mL bovine serum albumin (NEB #B9000S), 0.05% Nonidet P40 (Roche #11754599001) and 0.1 mg/mL fragmented yeast tRNA (Sigma #R5636)). For assaying the RNA-binding affinity in the presence or absence of stimulatory or substrate peptides, binding buffer was adjusted to mimic conditions used for HMTase assays with these peptides and consisted of 50 mM Tris-HCl pH 7.5 (at 25 °C), 100 mM KCl, 2 mM 2-mercaptoethanol, 0.05% Nonidet P40 (Roche #11754599001) and 0.1 mg/mL bovine serum albumin (NEB #B9000S). Serial dilutions of the protein were made separately, in the same buffer, and combined with the RNA solution for a final reaction volume of 40 μL containing 5 nM fluorescently-labelled RNA and the desired final protein concentration. Samples were equilibrated at 30 °C for 30 min before measurement. Fluorescence anisotropy data were collected using a PHERAstar plate reader (BMG Labtech) at 30 °C (λex = 485 nm, λem = 520 nm). The background was subtracted from protein-free samples. Kd, Hill and standard error values were calculated with GraphPad Prism 7 software using non-linear regression for specific binding with Hill slope function. For comparison between different complexes, non-linear regression was carried out using the background-subtracted anisotropy values. For comparison between wild-type and mutants of the same complex, RNA fraction bound was calculated using the background-subtracted anisotropy of the fully-bound wild-type protein sample, which were run on the same plate. Independent replicates of all fluorescent anisotropy experiments were performed on different days.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using 32P end-labelled RNA as previously described34,52.
Mapping protein-RNA interactions of an in vitro reconstituted complex using RBDmap
The RBDmap workflow was adopted from42 and was modified to allow for mapping of protein-RNA interactions in an in vitro reconstituted complex. DNA templates for in vitro transcription of G4 256 RNA were generated through PCR amplification using a synthetic gene (GenScript, see Supplementary Note 3 for the sequence) and forward and reverse primers that were designed to form a 5′ T7 promoter and a 3′ 25-mer polyA region respectively (see Supplementary Note 3 for primer sequences). 30 μL of 5 μM RNA in 20 mM Tris-HCl (pH 7.5 at 25 °C) was incubated for 2 min at 95 °C and was then snap-cooled on ice for 2 min. 90 μL of ice-cold Milli-Q ultrapure water and 30 μL of ice-cold 5X RNA binding buffer (250 mM Tris-HCl pH 7.5 at 25 °C], 500 mM KCl, 12.5 mM MgCl2, 0.5 mM ZnCl2 and 10 mM 2-mercaptoethanol) was added to a final volume of 150 μL. The RNA was then allowed to fold at 37 °C for 30 min. Separately, PRC2-AEBP2 was prepared at 4 °C by adding purified protein stock to 1X binding buffer, as above, to a final volume of 150 μL. The entire RNA solution was then combined with the entire protein solution in a single well of a 24-well plate to final concentrations of 0.5 μM G4 256 RNA and 1.0 μM PRC2-AEBP2. The mixture was then incubated at room temperature for 30 min to allow for the formation of ribonucleoprotein complexes. The 24-well plate was then placed onto an aluminium block within a container of ice and was irradiated by UV (254 nm, 6 rounds of 0.83 J/cm2 each) in a UVP CL-1000 Ultraviolet Crosslinker (Scientifix #CL-1000) with the top of the 24-well plate at a distance of ~10 cm below five 8-watt tube lamps. Digestion of the crosslinked ribonucleoprotein complexes by ArgC (Promega #V1881) and LysC (NEB #P8109S) was then performed by adding 5 μL of 100 ng/μL ArgC or LysC stock solution to the appropriate samples, after which the samples were incubated at room temperature overnight. As input sample, 30 μL was taken and stored at 4 °C until RNase digestion of all samples was performed (see below). The remaining sample was applied to 1 mL of oligo d(T)25 magnetic beads (NEB #S1419S) that were pre-washed with- and pre-incubated in 1X RNA-binding buffer for 10 min at 25 °C with gentle agitation. After applying the sample, the beads were incubated with gentle agitation for 10 minutes at room temperature, and then for 1 hour at 4 °C before capture with a magnet. The supernatant was kept for SDS-PAGE analysis (‘flow-through’). All steps from this point until the elution were done at 4 °C. After removal of the flow-through, the beads were transferred into a 50 mL conical tube and resuspended with 17 mL of buffer 1 (20 mM Tris-HCl pH 7.5 at 25 °C, 500 mM LiCl, 0.5% Lithium Dodecyl Sulfate (w/v) (Sigma-Aldrich #L9781), 1 mM EDTA and 5 mM DTT) with gentle agitation for 5 min before capture by a magnet. The beads were then washed twice as above with bead buffer 2 (20 mM Tris-HCl pH 7.5 at 25 °C, 500 mM LiCl, 0.1% Lithium Dodecyl Sulfate, 1 mM EDTA and 5 mM DTT) and twice with bead buffer 3 (20 mM Tris-HCl pH 7.5 at 25 °C, 500 mM LiCl, 1 mM EDTA and 5 mM DTT). Then the beads were washed twice with bead buffer 4 (20 mM Tris-HCl pH 7.5 at 25 °C, 200 mM LiCl, 1mM EDTA and 5 mM DTT) as above, and residual bead buffer 4 was used to transfer beads to a 1.7 mL microcentrifuge tube before capture by a magnet, and the remaining bead buffer 4 was removed. RNA-linked polypeptides were then eluted by heating at 55 °C for 2 min in 300 μL of bead elution buffer (20 mM Tris-HCl pH 7.5 at 25 °C and 1 mM EDTA) before capturing the beads using a magnet, and transferring the supernatant (‘eluate’) to another tube. Beads were discarded and never reused in order to avoid cross-contamination.
The ‘input’ and ‘eluate’ samples were then processed for mass spectrometric analysis. The minimal amount of RNase A that was required in order to completely digest the RNA was determined empirically. The appropriate amount of RNase A (Thermo Fisher Scientific #EN0531) was then added to the samples, followed by a 1 h incubation at 37 °C. The pH was then adjusted to 8.0 by adding 1 M Tris-HCl (pH 8.0 at 25 °C). Disulphide bonds were reduced by adding TCEP to a final concentration of 10 mM and the samples were incubated for 30 min at 65 °C. Free thiol groups were alkylated by adding 2-chloroacetamide (Sigma #C0267) to a final concentration of 40 mM and samples were incubated for 20 min at room temperature in the dark. The pH of each sample was adjusted again to 8.0 using 1 M Tris-HCl (pH 8.0 at 25 °C). Trypsin was added to each sample at a trypsin:protein mass ratio of 1:4, where protein mass before crosslinking was considered. The samples were then incubated for 14–18 h at 37 °C, in an orbital shaker at 200 rpm. Next, additional trypsin was added, using the same amount as above, and the samples were incubated for additional 2 h at 37 °C in an orbital shaker at 200 rpm. The digestion was stopped by the addition of formic acid to pH 3.0. Tryptic peptides were purified using OMIX C18 pipette tips (Agilent Technologies) according to the manufacturer’s instructions. The samples were then dried using a centrifugal vacuum concentrator (Labconco Acid-Resistant CentriVap Concentrator #7810041; CentriVap −105 Cold Trap, #7385037; Javac Vector LT-5 High Vacuum Pump, #VectorLT), and stored at −80 °C until mass spectrometric analysis.
Prior to mass spectrometric analysis, the peptides were resuspended in 20 μL of 0.1% formic acid, sonicated in a sonicator water bath for 10 min (Grant Instruments XUBA3 Analogue Ultrasonic Bath), and centrifuged for 5 min at 21,000 RCF. The samples were then transferred to mass spectrometric vials and analysed by liquid chromatography tandem-mass spectrometry (LC-MS/MS) within 48 hours.
Tandem mass spectrometry for RBDmap
See Supplementary Note 2.
Bis[sulfosuccinimidyl]suberate crosslinking mass spectrometry (BS3 XL-MS)
0.5 to 1.0 μM PRC2 complex with its accessory subunits was cross-linked with 5–20 μg bis[sulfosuccinimidyl] suberate (BS3) (ThermoFisher #21585) in 20 mM HEPES pH 7.5 (at 25 °C), 150 mM NaCl, 1 mM TCEP, in a total reaction volume of 85 μL. Reactions were allowed to proceed for 30 min at 25 °C before Tris-HCl pH 8.0 (at 25 °C) was added to a final concentration of 30 mM, to quench the reaction, for 15 min at 25 °C. Crosslinking efficiency was assessed by subjecting a portion of the product to 10% SDS-PAGE. The crosslinked product solutions were adjusted to pH 8.0 by adding 20 μL of 1 M Tris-HCl pH 8.0 (at 25 °C), reduced in 10 mM TCEP for 30 min at 60 °C and then alkylated in 40 mM 2-chloroacetamide for 20 min at 25 °C in the dark. Mixtures were then digested with 1:100 trypsin:protein mass ratio overnight at 37 °C with shaking on a Thermomixer (Eppendorf). Protein digestions were stopped by acidification through adding formic acid to 1% (v/v). Digested peptides were purified using OMIX C18 Mini-Bed pipette tips according to the manufacturer’s instructions, dried in a vacuum concentrator and reconstituted into 20 μL of 0.1% formic acid prior to mass spectrometric analysis.
The peptides were analysed by LC-MS/MS on an Orbitrap Fusion Tribrid Instrument connected to an UltiMate 3000 UHPLC liquid chromatography system (Thermo-Fisher Scientific) as described above for RBDmap.
pLink and pLink253 were used to identify BS3-crosslinked peptides with a false discovery rate of 0.05. In addition, the crosslinked peptides were kept for downstream structural analysis only if they had been identified in at least two independent replicates or if they have been identified with a P value of less than 10−4. 2D representations of crosslinked peptides and proteins were generated using xiNET54 . R scripts used for data retention and analysis downstream of pLink can be downloaded from GitHub: https://github.com/egmg726/crisscrosslinker.
Nucleosome reconstitution
Mononucleosomes were reconstituted as previously described55. In brief, recombinant histones were purified from inclusion bodies and reconstituted into histone octamers. Histone octamers and 182 bp ‘601’ DNA were combined in a buffer containing 20 mM Tris-HCl pH 7.5 (at 25 °C), 2 M KCl, 1 mM EDTA, 1 mM DTT and were dialysed for 55 h at 4–8 °C against a buffer consisting of 20 mM Tris-HCl pH 7.5 (at 25 °C), 25 mM KCl, 1 mM EDTA and 1 mM DTT with a salt concentration that was gradually reduced during the process. The quality of reconstituted nucleosomes was assessed by a 4% TBE gel and negative stain EM.
Negative stain EM
Negative-stain EM experiments were carried out for assessing mononucleosome samples. 2 μL of 0.03 mg/mL mononucleosome solution was applied to a glow-discharged continuous carbon grid (EMgrid Australia) and incubated for 30 sec before blotting with filter paper and immediately staining by successive two times of 15 sec and one time of 30 sec incubations in drops of 2% (w/v) uranyl acetate. The stain was removed by blotting with filter paper and the grid was air-dried before imaging. The grid was imaged at a magnification of 52,000x using an FEI Tecnai 12 transmission electron microscope operating at 120 keV.
HMTase activity assays
Unless indicated otherwise, the HMTase activity assays were performed as previously described15 with minor modifications. Briefly, RNAs were folded as described for binding assays above and were then allowed to bind PRC2 in RNA binding buffer (50 mM Tris-HCl pH 7.5 at 25 °C, 100 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2 and 2 mM 2-mercaptoethanol) at 25 °C for 30 min. Each 10 μL HMTase reaction contained 500 nM PRC2 complex, RNA as indicated throughout the text, 4 μM H3.1 histone protein (NEB M2503S) or 2 μM mononucleosomes (see above for in vitro nucleosome reconstitution), and 5.0 μM S-[methyl-14C]-adenosyl-L-methionine (PerkinElmer #NEC363050UC). The reactions were incubated for 1 h at 30 °C in HMTase buffer (77.5 mM Tris-HCl pH 8.0 at 30 °C, 155 mM KCl, 3.88 mM MgCl2, 0.2 mM ZnCl2, 3.1 mM 2-mercaptoethanol, 0.1 mg/mL BSA (NEB B9000) and 5% v/v glycerol). For HMTase assays in the presence or absence of stimulatory peptides, reaction buffer consisted of 50 mM Tris-HCl (pH 8.0 at 30 °C), 100 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2, 2 mM 2-mercaptoethanol, 0.1 mg/mL bovine serum albumin and 5% v/v glycerol. The reactions were then stopped by adding 4× LDS sample buffer (ThermoFisher #NP0007) to a final concentration of 1×. Samples were then incubated at 95 °C for 5 min and the samples were subjected to 16.5% SDS-PAGE. Gel were first stained with InstantBlue Coomassie protein stain (Expedeon #ISB1L) and then vacuum-dried for 60 min at 80 °C with the aid of a VE-11 electric aspirator pump (Jeio Tech). Dried gels were exposed to a storage phosphor screen (GE healthcare) for 1–8 days and the signal was acquired using Typhoon Trio imager (GE healthcare). Densitometry was carried out using ImageJ56. Relative HMTase activities of mutant PRC2 complexes were obtained through a normalisation to the signal obtained from the corresponding wild-type PRC2 complex that was loaded in a different lane on the same gel. All experiments were performed in triplicates.
For assaying RNA inhibition of non-histone substrates, experiments were performed as above, with the exception that the final reaction buffer was 25 mM HEPES pH 8.0 (at 25 °C), 40 mM KCl, 50 mM NaCl, 3 mM MgCl2, 2 mM 2-mercaptoethanol and 10% v/v glycerol.
Structures used for visualisation
The high-resolution cryo-EM structure of the PRC2-AEBP2-JARID2 complex (PDB accession 6C23)5 and the crystal structure of SUZ12-RBBP4-JARID2-AEBP2 (PDB accession 5WAI)6 were used for PRC2 data representation as described above. The coordinates of the EED-bound H3K27me3, H3K27M and A395 are from PDB accessions 3IIW8, 5HYN10, and 5K0M43 respectively. The coordinates of MTF2 were obtained from PDB accession 5XFR16.
Cell lines
RBR-ID experiments were carried out in E14Tg2A.4 (E14) mESCs that were obtained from the Reinberg lab, as previously used by Kaneko et al.57. When cultured in the described conditions, these E14 cells display the typical morphology of mESCs. In addition, we have accumulated extensive genomic and functional data that confirm their pluripotent state and identity. All cell lines used in the Bonasio Lab are routinely tested for mycoplasma. The E14 cells used in these experiments tested negative as recently as October 2017.
Code availability
R scripts used for XL-MS and RBDmap data analysis downstream of pLink and Andromeda, respectively, can be downloaded from GitHub: https://github.com/egmg726/crisscrosslinker.
Sequences
For construct and primer sequences, see the Supplementary Note 3.
Statistics and Reproducibility
All P values were calculated using paired or unpaired two-tailed Student’s t-test, as explicitly indicated in the respective figure legends. P values were given when P < 0.05. All binding curves and bar plots represent means averaged across the number of samples indicated in the respective figure legend.
Data Availability
LC-MS raw data for targeted RBR-ID experiments has been deposited at the Chorus project with ID 1560 (https://chorusproject.org). Mass spectrometry data for RBDmap and BS3 XL-MS experiments were deposited at FigShare, with DOIs https://doi.org/10.26180/5c3d9751c64ae and https://doi.org/10.26180/5c3d8dd45651b, respectively. Source data for Fig. 2–5 and Supplementary Fig. 2–5 are available within Supplementary Data Set 5–8, respectively.
Supplementary Material
Acknowledgments
We would like to thank the Monash Biomedical Proteomics Facility for providing instrumentation and technical support. Q.Z. holds an Australian Research Council (ARC) Discovery Early Career Researcher Award (DE180100219). N.J.M. is the Isabella and Marcus Foundation Charlee Ferrar Scholar and is also supported through an Australian Government Research Training Program (RTP) Scholarship. R.W-T. was supported by NIH training grant T32GM008216. E.H.G. holds a Biomedicine Discovery Scholarship and is an EMBL-Australia PhD student. B.M.O. supported through an Australian Government RTP Scholarship and also by the Monash Graduate Excellence Scholarship. R.B. acknowledges support from the NIH (R01GM127408) and the March of Dimes Foundation (1-FY-15–344). C.D. is an EMBL-Australia Group Leader and acknowledges support from the ARC (DP190103407) and the NHMRC (APP1162921).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Schuettengruber B, Bourbon HM, Di Croce L & Cavalli G Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34–57 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Margueron R & Reinberg D The Polycomb complex PRC2 and its mark in life. Nature 469, 343–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon JA & Kingston RE Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49, 808–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comet I, Riising EM, Leblanc B & Helin K Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 16, 803–810 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kasinath V et al. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359, 940–944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Jiao L, Shubbar M, Yang X & Liu X Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding. Mol Cell 69, 840–852 e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciferri C et al. Molecular architecture of human polycomb repressive complex 2. Elife 1, e00005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margueron R et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao L & Liu X Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justin N et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 7, 11316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanulli S et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol Cell 57, 769–783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CH et al. Allosteric Activation Dictates PRC2 Activity Independent of Its Recruitment to Chromatin. Mol Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perino M et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet (2018). [DOI] [PubMed] [Google Scholar]
- 14.Lee CH et al. Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol 24, 1028–1038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287–291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi J et al. DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat Struct Mol Biol 24, 1039–1047 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Beringer M et al. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol Cell 64, 645–658 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29, 229–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloet SL et al. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat Struct Mol Biol 23, 682–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauri S et al. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep 17, 583–595 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Smits AH, Jansen PW, Poser I, Hyman AA & Vermeulen M Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res 41, e28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brockdorff N Noncoding RNA and Polycomb recruitment. RNA 19, 429–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidovich C & Cech TR The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21, 2007–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hekimoglu B & Ringrose L Non-coding RNAs in polycomb/trithorax regulation. RNA Biol 6, 129–37 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Ringrose L Noncoding RNAs in Polycomb and Trithorax Regulation: A Quantitative Perspective. Annu Rev Genet 51, 385–411 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Bonasio R & Shiekhattar R Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48, 433–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko S, Son J, Bonasio R, Shen SS & Reinberg D Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev 28, 1983–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herzog VA et al. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet 46, 973–981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cifuentes-Rojas C, Hernandez AJ, Sarma K & Lee JT Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55, 171–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Sun BK, Erwin JA, Song JJ & Lee JT Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko S et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24, 2615–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanhere A et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38, 675–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell 65, 1056–1067 e5 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Long Y et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beltran M et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res 26, 896–907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ardehali MB et al. Polycomb Repressive Complex 2 Methylates Elongin A to Regulate Transcription. Mol Cell 68, 872–884 e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He C et al. High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Mol Cell 64, 416–430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidovich C, Zheng L, Goodrich KJ & Cech TR Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20, 1250–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneko S et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 53, 290–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montero JJ et al. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat Commun 9, 1548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castello A et al. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol Cell 63, 696–710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He Y et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat Chem Biol 13, 389–395 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Ballare C et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19, 1257–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conway E et al. A Family of Vertebrate-Specific Polycombs Encoded by the LCOR/LCORL Genes Balance PRC2 Subtype Activities. Mol Cell 70, 408–421 e8 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Oksuz O et al. Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol Cell 70, 1149–1162 e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poepsel S, Kasinath V & Nogales E Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol 25, 154–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee G, Yu LeRoy, Stafford Reinberg. Automethylation of PRC2 fine-tunes its catalytic activity on chromatin. bioRxiv 10.1101/349449(2018). [DOI] [Google Scholar]
- 49.Wang L, Paucek Gooding, Lee Cech. Regulation of histone methylation by automethylation of PRC2. bioRxiv 10.1101/343020(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warneford-Thomson R, He C, Sidoli S, Garcia BA & Bonasio R Sample Preparation for Mass Spectrometry-based Identification of RNA-binding Regions. J Vis Exp (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–72 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Davidovich C, Goodrich KJ, Gooding AR & Cech TR A dimeric state for PRC2. Nucleic Acids Res 42, 9236–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang B et al. Identification of cross-linked peptides from complex samples. Nat Methods 9, 904–6 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Combe CW, Fischer L & Rappsilber J xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics 14, 1137–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luger K, Rechsteiner TJ & Richmond TJ Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaneko S, Son J, Shen SS, Reinberg D & Bonasio R PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20, 1258–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC-MS raw data for targeted RBR-ID experiments has been deposited at the Chorus project with ID 1560 (https://chorusproject.org). Mass spectrometry data for RBDmap and BS3 XL-MS experiments were deposited at FigShare, with DOIs https://doi.org/10.26180/5c3d9751c64ae and https://doi.org/10.26180/5c3d8dd45651b, respectively. Source data for Fig. 2–5 and Supplementary Fig. 2–5 are available within Supplementary Data Set 5–8, respectively.