Abstract
The fibrillary aggregation of the protein alpha synuclein (Asyn) is a hallmark of Parkinson’s disease, and the identification of small molecule binding sites on fibrils is essential to the development of diagnostic imaging probes. A series of molecular modelling, photoaffinity labeling, mass spectrometry, and radioligand binding studies were conducted on Asyn fibrils. The results of these studies revealed the presence of three different binding sites within fibrillar Asyn capable of binding small molecules with moderate to high affinity. A knowledge of the amino acid residues in these binding sites will be important in the design of high affinity probes capable of imaging fibrillary species of Asyn.
Keywords: Alpha synuclein, Lewy Bodies, Lewy Neurites, Parkinson’s disease
Graphical Abstract
INTRODUCTION
Alpha synuclein (Asyn) is a small (140 amino acid), intrinsically disordered protein that is localized in presynaptic nerve terminals in the central nervous system (CNS).1–2 Although its biological function is poorly understood, Asyn is currently believed to play a key role in docking of neurotransmitter vesicles to active zones of nerve terminals to facilitate the release of neurotransmitters into the synaptic cleft.3 In Parkinson’s disease (PD), Asyn forms fibrillar protein aggregates containing a beta pleated sheet structure.4 These fibrillar protein aggregates accumulate to the level where they form insoluble intraneuronal inclusions termed Lewy bodies (LBs) and Lewy neurites (LNs). The accumulation of LBs and LNs in the substantia nigra and other brain regions represents a hallmark feature of PD.5 A second form of fibrillary Asyn, termed glia cell inclusions (GCIs), is found in the oligodendrocytes of white matter tracts in patients diagnosed with Multiple System Atrophy (MSA).6 The identification of dominant mutations in SNCA, the gene encoding Asyn, in familial PD, including point mutations that promote fibril formation and triplication of the SNCA gene provides a strong link between fibrillary Asyn and the pathogenesis of PD.7–8
The diagnosis of PD has traditionally relied on the clinical presentation of a number of motoric deficits including bradykinesia, rigidity, rest tremor and postural instability. Although accuracy of the clinical diagnosis of PD is very good in later stages of the disease, the early diagnosis of PD is problematic since not all features are present in early PD. Furthermore, the same clinical features can be present in other Parkinsonian syndromes such as vascular parkinsonism, MSA, corticobasal degeneration (CBD), and progressive supranuclear palsy (PSP).9 With the exception of MSA, these other Parkinsonian syndrones do not include the pathologic accumulation of Asyn.
Positron Emission Tomography (PET) is an in vivo imaging technique that is capable of quantifying a change in protein levels, rates of cellular metabolism, or protein-protein interactions that serve as the foundation of the molecular basis of disease. Perhaps the most significant advance in the past decade in PET imaging studies is the development of radiotracers capable of imaging the presence of amyloid β (Aβ) plaques10–13 and tau-based neurofibrillary tangles (NFTs)14–16 in patients diagnosed with Alzheimer’s disease (AD). Because of the availability of Aβ- and tau-specific probes, it is now clear that Aβ plaques represent an “antecedent” marker for identifying patients at risk for developing AD, whereas NFTs occur relatively late in the disease process when the neuronal loss leading to cognitive impairment has already occurred.
The success of Aβ plaque and tau imaging in AD has led to a similar effort towards developing a PET radiotracer for imaging insoluble Asyn that occurs in LBs and LNs in PD, and GCIs in MSA.9, 17 Since Aβ and tau are found in both AD and PD postmortem brain samples,18 a suitable PET probe for imaging insoluble Asyn aggregates must have a high affinity for Asyn and low affinity for fibrillary Aβ and tau. Unfortunately, most ligands developed to date have not shown this desired selectivity for Asyn versus Aβ and tau for PET imaging studies in PD.
Our group has recently reported a number of structurally-diverse compounds that are capable of binding to Asyn fibrils.19–22 Although structure-activity relationship studies have identified compounds displaying a higher affinity for Asyn versus Aβ and tau, the properties of the ligand that possess this property are not ideal for functioning as a PET radiotracer for imaging Asyn.21, 23 In this regard, a greater understanding of the location of the binding sites for small molecules on Asyn, and the amino acid residues that comprise the binding sites, would be useful in guiding the design of a PET radiotracer having the affinity, selectivity, and physicochemical properties required for imaging LBs and LNs. The publication of the solid-state NMR structure of Asyn fibrils by Reinstra and colleagues24 served as the starting point for the series of studies described in this communication.
RESULTS AND DISCUSSION
The structure of Asyn was imported from the Protein Data Bank25 (PDB ID: 2N0A) and visualized with the 3-dimension visualization program PyMol (pymol.org). Docking studies were conducted using the AutoDock 4.226 plugin on PyMol. The test set for docking studies included four different classes of compounds shown in Figure 1. A grid box that covered the major beta-sheet structure of residue K34 to K97 was applied to the Asyn fibril structure for molecular blind docking to search the potential binding sites. The blind docking studies allow for predicting potential sites of interaction with the protein by calculating a % probability of interaction and binding energy for each putative site. The values for each of these measurements for the eight test compounds are shown in Table S1 and S2.
Figure 1.
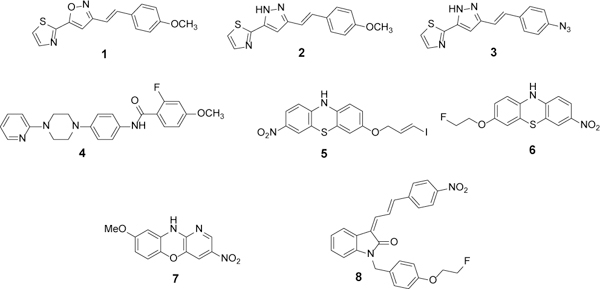
Structures of the compounds used in the docking and in vitro binding assays.
The results of the docking studies revealed thirteen putative binding sites in Asyn for small molecules (Figure S1 and Table S1). However, it was possible to exclude a number of these sites based on their low % probability of interaction. This reduced the number of putative binding sites to six. Furthermore, site 1 was excluded as a credible binding site since it is located in the N-terminal region that is highly disordered in the NMR structure. We note that some evidence indicates that these regions are more ordered in fibrils formed under different conditions, so sites in terminal regions could be investigated in subsequent studies.27 Because the side chains of site 6 were reported disordered in the fibril structure24, and the inner core of the Asyn fibril was also reported as a relatively inaccessible region for small molecule binding, site 6 was also excluded.24, 28 The remaining binding sites included: 1) site 2, which contains Y39-S42-T44; 2) site 9, which contains G86-F94-K96; 3) site 3, which contains L45-V48-H50; and 4) site 13, which contains L43-L45-V48-H50. Since site 3 is contained within site 13, these two sites can collectively be termed site 3/13 (Figure 2).
Figure 2.
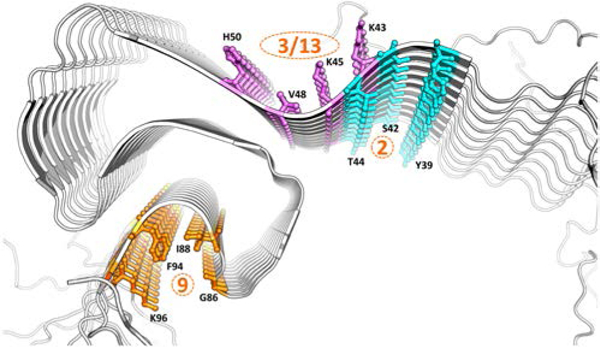
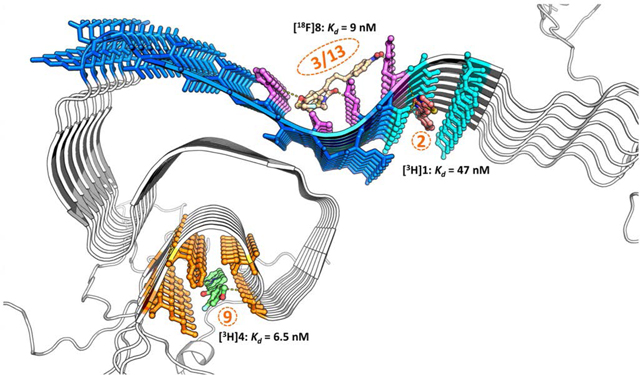
Solid-state NMR structure of Asyn showing the three putative binding sites for small molecules. Color labeled cyan, site 2; purple, Site 3/13; orange, site 9.
The docking studies also revealed that the different classes of compounds displayed selectivity among the binding sites. For example, the stryrene-based analogs (1–3) and the piperazine analog (4) showed a preference for sites 2 and 9, whereas the tricyclic compounds (5–7) and the indolinone-diene analog (8) showed a near exclusive preference for site 3/13 (Table S2). The identification of sites 2, 9 and 3/13 as potential binding sites is also supported by the high degree of hydration and stability of these sites using the molecular dynamics program NAMD29 with the VMD30 visualization and analysis module. The results of these studies can be found in Figure S2 and Table S3.
In order to confirm the location of these binding sites, photoaffinity labeling of Asyn fibrils was conducted using the azide-containing styrene analog, 3. In addition to sites 2 and 9, this compound also has a modest probability of interacting with site 3/13. Disaggregation of the photoaffinity-labeled Asyn fibrils into their corresponding monomers followed by positive ion and negative ion matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) revealed the labeling of peptide fragments corresponding to residues T44-K58 or E46-K60 (Figure 3 and S3, and Table S4). While we favor assignment of the site of crosslinking to T44-K58 due to the absence of the E46-K58 photo-adduct, we were unable to definitively assign this by MS-MS sequencing. These results indicate that azido analog 3 binds to either site 2 or 3/13, and site 9 can be excluded as a potential binding site for this compound. Given their structural similarity, we surmised that 1 and 2, also bind to site 2 or 3/13.
Figure 3.
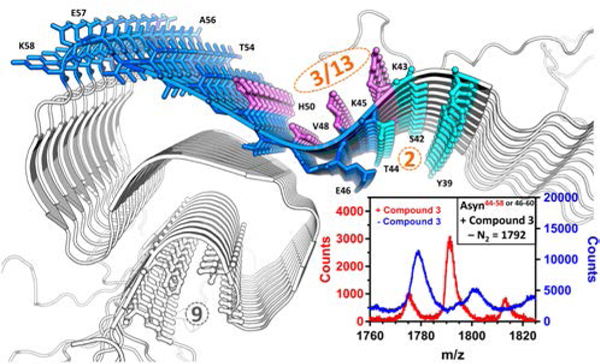
Site of the peptide fragments T44-K58 labeled (blue) by the photoaffinity probe, 3. This peptide fragment contains sites 2 and 3/13.
As a means of further characterizing the binding of small molecules to the three putative binding sites, a series of in vitro binding studies in Asyn fibrils was conducted using two different tritiated ligands, [3H]1 and [3H]4. These radioligands were synthesized for in vitro binding studies aimed at screening potential ligands for PET imaging studies, and in vitro autoradiography studies for imaging LBs and LNs in postmortem samples of PD brain. The in vitro characterization of these radioligands will be published separately. However, homologous competition studies revealed that [3H]1 has a Kd value of 47 nM and [3H]4 has a Kd value of 6.5 nM for recombinant Asyn fibrils, respectively. In vitro competition curves measuring the displacement of [3H]1 and [3H]4 from Asyn fibrils revealed different Ki values for compound 2. That is, 2 had a Ki value of 71 nM for displacing [3H]1, and a Ki value of 488 nM for displacing [3H]4 from Asyn fibrils (Figure 4). These data are consistent with the in silico prediction that the styrene analogs (1-3) bind at with higher affinity to site 2 than site 9, while 4 binds with highest affinity to site 9.
Figure 4.
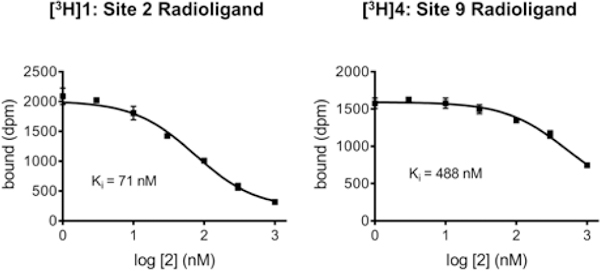
In vitro binding studies of 2 using either [3H]1 or [3H]4 as the radioligand. Since 2 is predicted to bind to site 2, its affinity for Asyn fibrils was higher when a site 2 preferring ligand (i.e., [3H]1) was used in the binding assay.
The next set of experiments was designed to confirm that the styrene-based analogs 1–3 bind to site 2 and not site 3/13. Since analogs 6, 7 and 8 were predicted to bind near exclusively to site 3/13, in vitro competition studies were conducted measuring the potency of these compounds to displace [3H]1 from Asyn fibrils. Previous studies have shown 6 and 7 bind to Asyn fibrils with an affinity of 16 nM (for 6) and ~70 nM (for 7) in in vitro binding studies using [125I]5 as the radioligand.19 However, neither compound was able to displace the binding of [3H]1 from Asyn fibrils (Ki > 1,000 nM). Furthermore, [18F]8 was shown to bind to Asyn fibrils with a Kd value of 9 nM,21 but it also did not displace [3H]1 from Asyn fibrils (Figure S4). These data support the binding of the styrene-based analogs (1-3) to site 2 and not site 3/13.
To confirm that compounds predicted to bind to site 3/13 do not bind to site 9, in vitro binding assays were conducted measuring the ability of compounds 7 and 8 to displace [3H]4 from Asyn fibrils. The Ki values under both assays conditions were >1,000 nM showing that they indeed bind to distinct sites (Figure S4). Taken together, our competition and photo-crosslinking studies allow us to generate a self-consistent model for binding site selectivity that is in agreement with the predictions from our docking studies. The binding preferences are as follows: stryrene-based analogs (1–3) bind to site 2 (with weaker affinity for site 9), the piperazine analog (4) binds to site 9 (with weaker affinity for site 2), and the tricyclic compounds (5–7) and indolinone-diene analog (8) bind to site 3/13. The interactions of representative compounds with each of the binding sites is shown in Figure 5.
Figure 5.
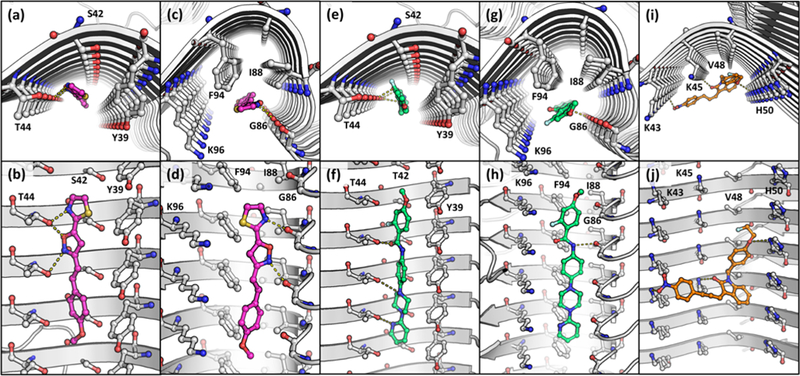
Docking studies of 1 to site 2 (a and b) and 9 (c and d), 4 to sites 2 (e and f) and 9 (g and h), and 8 to site 3/13 (i and j). (a, c, e, g, and i) are viewed from the fibril axis, and (b, d, f, h, and j) are viewed from the fibril side view.
The importance in understanding where small molecules are predicted to bind to Asyn in designing an in vitro binding assay to measure their affinity to Asyn fibrils is evident from the data shown in Figure 4 and Figure S4. The Ki value of 2 for displacing [3H]1 from Asyn fibrils was 71 nM, and would be considered to be a good lead compound for radiotracer development (Figure 4). In contrast, the Ki value for displacing [3H]4 from Asyn fibrils was 488 nM, an affinity that would exclude its consideration from further radiotracer development. Similarly, one would conclude that the indolinone-diene analog 8 has no affinity for Asyn fibrils based on the competition data using [3H]1 and [3H]4 as the radioligands (Figure S4), whereas direct binding assays of the 18F-labeled compound reveals that it has a Kd value of 9 nM for Asyn fibrils. For protein structures such as fibrils, with many shallow surfaces, having a set of compounds with varied site selectivity is essential to profiling how changes to candidate molecules affects not only affinity, but binding site preference.
In summary, we have utilized a combination on in silico docking, photoaffinity labeling, and radioligand binding studies to identify three putative binding sites for small molecules in Asyn fibrils. The location of these binding sites is critical in designing radioligand binding studies aimed at determining the affinity of small molecules for Asyn fibrils. The identification of high-affinity compounds for Asyn fibrils represents the first step in developing a radiotracer for PET imaging studies of LBs and LNs in PD. We are currently using the strategy described in this study in the screening of chemical libraries as a means of identifying suitable lead compounds for the development of a radiotracer for imaging insoluble Asyn aggregates with PET.
METHODS
Chemistry.
Reagents were purchased from Sigma-Aldrich and Fisher Scientific. Silica gel chromatography was carried out on a Biotage Isolera™ Spektra One chromatograph system. All synthesized compounds were analyzed and confirmed to have purity over 95% with a Waters Alliance LC-MS system. NMR spectra were measured on a Brucker 500 or 360 MHz spectrometer as indicated. Chemical shifts (δ values) were reported in ppm relative to TMS. For multiplicity, s = singlet, d = doublet, t = triplet, m = multiplet. 1H NMR spectra data were presented as follows: Chemical shifts (multiplicity, coupling constants, integration).
Synthesis of (E)-3-(4-methoxystyryl)-5-(thiazol-2-yl)isoxazole (1)
To a solution of (2Z,4E)-3-hydroxy-5-(4-methoxyphenyl)-1-(thiazol-2-yl)penta-2,4-dien-1-one22 (250 mg, 0.87 mmol) in EtOH (5.00 mL) was added NH2OH-HCl (121 mg, 1.74 mmol) and stirred at 80 °C for 3 h. The reaction mixture was cooled down to the ambient temperature and removed EtOH in vacuo. The crude compound was purified by Biotage purification system (Hex: EtOAc = 3:1) which gave a 1:1 mixture of two isomers. Further purification with dichloromethane yield compound 1 (40 mg, 16.0%) as a colorless solid. 1H NMR (500 MHz, CDCl3) δ ppm δ 3.84 (s, 3H), 6.93 (d, J = 8.7 Hz, 2H), 7.03 (d, J = 16.5 Hz, 1H), 7.07 (s, 1H), 7.20 (d, J = 16.5, 1H), 7.49 (d, J = 8.7 Hz, 2H), 7.53 (d, J = 3.1 Hz, 1H), 7.99 (d, J = 3.1 Hz, 1H). 13C NMR (126 MHz, CDCl3) δ ppm 55.33, 98.88, 113.02, 114.30, 121.28, 128.32, 128.51, 136.35, 144.40, 154.39, 160.45. 162.73, 163.68. MS (ESI) m/z 285 (M+H)+.
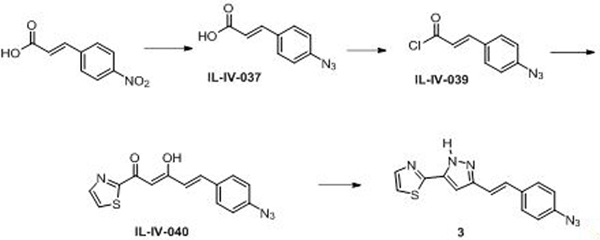
(E)-3-(4-Azidophenyl)acrylic acid (IL-IV-037)
Metal zinc (1.95 g, 29.9 mmol) was added to AcOH (45.0 mL) and water (5.0 mL) followed by the addition of (E)-3-(4-nitrophenyl)acrylic acid (2.89 g, 14.9 mmol). The reaction mixture was stirred at 25 °C for 0.5 h and filtered. The filtrate was cooled to 0 °C for 10 min, and a solution of sodium nitrite (1.24 g, 17.9 mmol) in water (10 ml) was added dropwise at 0 °C. The reaction mixture was stirred at 0 °C for 10 min. Then sodium azide (1.16 g, 17.9 mmol) in water (10.00 ml) was added dropwise to the reaction mixture. The mixture was kept stirred at 0 °C for another 15 min. After the reaction, the solvent was filtered, and washed with water. The solid was dried over via high vacuum, recrystallized with ethyl acetate/hexane, washed with hexane, and dried over to yield (E)-3-(4-azidophenyl) acrylic acid (738.6 mg, 3.90 mmol, 26.1 %) as a light yellow solid. 1H NMR (500 MHz, DMSO-d6) δ ppm 6.48 (d, J = 16.1 Hz, 1H), 7.14 (d, J = 8.5 Hz, 2H), 7.56 (d, J = 16.0 Hz, 1H), 7.73 (d, J = 8.5 Hz, 1H), 12.35 (s, 1H).
(E)-3-(4-azidophenyl)acryloyl chloride (IL-IV-039)
Sulfurous dichloride (0.30 ml, 4.19 mmol) was added to a solution of (E)-3-(4-azidophenyl)acrylic acid (720 mg, 3.81 mmol) in dichloromethane (10 ml). The reaction mixture was stirred at 80 °C for 2.5 h. The solvent was then evaporated and the solid formed was dried via high vacuum. 1H NMR (500 MHz, CDCl3) δ ppm 6.60 (d, J = 15.5 Hz, 1H), 7.09 (d, J = 8.5 Hz, 2H), 7.58 (d, J = 8.5 Hz, 2H), 7.79 (d, J = 15.5 Hz, 1H).
(2Z,4E)-5-(4-Azidophenyl)-3-hydroxy-1-(thiazol-2-yl)penta-2,4-dien-1-one (IL-IV-040)
LiHMDS (2.65 mL, 2.65 mmol) was added dropwise to a solution of 1-(thiazol-2-yl)ethan-1-one (306 mg, 2.408 mmol) at −78 °C, and the mixture was stirred for 1 h at the same temperature. (E)-3-(4-Azidophenyl)acryloyl chloride (IL-IV-039, 600 mg, 2.89 mmol) in THF (2.145 mL) was added to the reaction mixture dropwise at −78 °C, and then the mixture was warmed up to 25 °C and stirred for 3 h at room temperature. After the reaction, saturated NH4Cl aqueous (100 mL) was added to a reaction mixture, which was then extracted with ethyl acetate (50 mL) twice. The combined organic layer was dried over Na2SO4 and concentrated. The residue was recrystallized with MeOH giving the product as an orange solid. 1H NMR (500 MHz, CDCl3) δ ppm 6.59 (d, J = 15.8 Hz, 1H), 6.76 (s, 1H), 7.06 (d, J = 8.5 Hz, 2H), 7.55 (d, J = 8.5 Hz, 1H), 7.65 (d, J = 15.8 Hz, 1H), 7.67 (d, J = 3.0 Hz, 1H), 8.02 (d, J = 3.0 Hz, 1H).
(E)-2-(3-(4-Azidostyryl)-1H-pyrazol-5-yl)thiazole (3)
Hydrazine hydrate (171mg, 3.42 mmol) was added to a solution of (2Z,4E)-5-(4-azidophenyl)-3-hydroxy-1-(thiazol-2-yl)penta-2,4-dien-1-one (500 mg, 1.676 mmol) in Ethanol (4 ml), stirred at 80 °C for 3 h. After the reaction, EtOH was removed in vacuo, purufied by Biotage (IL-IV-42). The purification gave (E)-2-(5-(4-azidostyryl)-1H-pyrazol-3-yl)thiazole (150 mg, 0.510 mmol, 30.4 % yield) as a light yellow solid. 1H NMR (360 MHz, DMSO-d6) δ ppm 6.95 (s, 1H), 7.08–1.16 (m, 3H), 7.29 (d, J = 7.2 Hz, 1H), 7.61 (d, J = 8.2 Hz, 2H). 7.67 (d, J = 3.4 Hz, 1H), 7.86 (d, J = 3.4 Hz, 1H).
Molecular Docking.
All the structures of compound were drew on ChemDraw Profession 15.1 (PerkinElmer Informatics, Inc.), then imported to Chem3D Ultra 15.1 (PerkinElmer Informatics, Inc.) to minimize individual structures by MMFF94 force field for preparation of molecular docking. Molecular blind docking studies were performed via AutoDock 4.226 plugin on PyMOL (pymol.org). The solid-state NMR structure of full-length Asyn fibril (PDB ID: 2N0A) was obtained from RCSB protein data bank (https://www.rcsb.org/) as a target protein for blind docking. Non-polar hydrogens were removed from both compounds and protein structures. A grid box with a dimension of 95 × 50 × 95 Å3 was applied to Asyn fibril structure, majorly including beta-sheet section (residue K34-K97) with parts of N- and C-terminus disorder residues that close to the beta-sheet area. The Lamarckian Genetic Algorithm with a maximum of 2,500,000 energy evaluations was used to calculate 1,000 protein-ligand binding poses for each compound. The % probability, predicted best binding energy, and average binding energy of each binding site that determined from blind docking were reported.
Molecular Dynamics Simulation.
In order to investigate the stability and accessibility of the Asyn fibril structure in solution, molecular dynamics simulation was performed via NAMD 2.1229 with CHARMM22 force field, and analyzed by using visualization program VMD30. The protein was solvated in a TIP3P water box with a minimized volume of 160 × 168 × 120 Å3, and then 90 sodium ions were randomly added to the solvated system for charge neutralization. Periodic boundary conditions were used for the molecular dynamics simulation. A cut-off of 12 Å was set to non-bonded interaction, and long-range electrostatics interaction was calculated by using Particle Mesh Ewald (PME) method. SHAKE algorithm was used to constrain bonds involving hydrogen atoms. A 50,000 steps of energy minimization was implemented. Then, the minimized system was heated in NVT ensemble with constant volume at 310 K for 100 ps with a time step of 1 fs. The system was then equilibrated in NPT ensemble at 310 K and 1 atm for 200 ps with 1 fs time step. The Langevin dynamics and Langevin piston method were used to maintain the pressure at 1 atm and temperature at 310 K. A final molecular dynamics simulation was run in a NPT ensemble for 10 ns with a time step of 2 fs, and saved the trajectory every 1 ps.
Photocrosslinking.
Asyn was expressed recombinantly in E. Coli from a pT-T7 plasmid and was purified by size exclusion chromatography followed by ion exchange chromatography as previously described.31 Following purification purified protein was dialyzed against 20 mM tris, 100 mM NaCl pH7.5 at 4 °C overnight. Subsequently, samples were prepared containing 100 μM Asyn which were shaken for 48 hours at 1500 rpm and 37 °C producing fibrils. To remove residual monomers post-aggregation, fibrils were pelleted at 13,500 rpm for 1.5 hours in a bench top centrifuge and resuspended in the same volume and buffer that was removed. Compound 3 was dissolved in DMSO producing a 136 mM stock. Samples containing ~100 μM Asyn fibrils with and without 900 μM Compound 3 were prepared in glass vials which were irradiated with a standard TLC lamp for 5 hours. Following irradiation, samples were disaggregated by adding sodium dodecyl sulfate (SDS) to a final concentration of 15 mM and boiling for 1 hour. After disaggregating the Asyn fibrils, SDS was removed via protein precipitation using choloform and methanol.32 Trypsin was added to monomeric protein samples at a final concentration of 0.05 mg/mL and samples were incubated overnight at 37 °C overnight. Mass spectra of each digested sample were acquired via matrix assisted laser desorption ionization (MALDI-MS) on a Bruker μLtraflex III (Billerica, MA, USA). Samples were shot using two methods, one measuring positive ions with a reflection (RP) and the other measuring negative ions via a linear method (LN).
Binding Assay.
The radioligand competition binding assays used Asyn fibrils as binding substrate. Asyn fibrils were prepared from recombinant human Asyn protein as previously described.33 Binding mixtures contained Asyn fibrils at a concentration of 25 nM for assays with [3H]4 and 150 nM for assays with [3H]1. At radioligand concentrations of 2 nM in the binding assays, these fibril concentrations resulted in 5% - 10% of the radioligand bound to fibrils in the absence of competitor. Different concentrations of competitor compounds, ranging from 1 nM to 1000 nM were combined with Asyn fibrils and 2 nM radioligand in 30 mM Tris-HCL pH 7.4, 0.1% BSA, in a total volume of 150 μl. The binding mixtures were incubated at 37 °C for 2 hrs in 96 well plates. Bound and free radioligand were then separated by vacuum filtration through 1.0 μm Glass Fiber filters in 96-well filter plates (Millipore), followed by three 200 μl/well washes with cold assay buffer. Filters containing the bound ligand were mixed with 150 μL of Optiphase Supermix scintillation cocktail (PerkinElmer) and counted after overnight incubation. All data points were performed in triplicate. Data were analyzed using Graphpad Prism 7 software to obtain EC50 values by fitting the data to the equation Y=bottom+(top-bottom)/(1+10(x-logEC50)). Ki values were calculated from EC50 values using the equation Ki=EC50/(1+[radioligand]/Kd).
Supplementary Material
Acknowledgments
Funding
This research was supported by the Michael J. Fox Foundation and the National Institutes of Health (NIH NS081033 to E.J.P.). J.J.F. thanks NSF(DGE-1321851) and the Parkinson’s Disease Foundation (PF-RVSA-SFW-1754) for fellowship support. The other members of the Michael J. Fox Alpha Synuclein Imaging Consortium include Jamie Eberling, Eugene Johnson, and Kalpana M. Merchant from the Michael J. Fox Foundation; Chester A. Mathis, William E. Klunk, and N. Scott Mason from University of Pittsburgh Medical Center; Dale Mitchell, Wolfgang Schmidt, and David Hardick from BioFocus-Charles River; Edilio Borroni, Luca Golbi, Michael Horner, and Kevin Nash from Roche; and Joel Mercier from UCB.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Details for the docking studies, photocrosslinking, and representative in vitro binding data. (PDF)
The authors declare no competing financial interests.
REFERENCES
- 1.Clayton DF; George JM, Synucleins in synaptic plasticity and neurodegenerative disorders. J Neurosci.Res. 1999, 58 (1), 120–129. [PubMed] [Google Scholar]
- 2.Ueda K; Fukushima H; Masliah E; Xia Y; Iwai A; Yoshimoto M; Otero DA; Kondo J; Ihara Y; Saitoh T, Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc.Natl.Acad.Sci U.S.A 1993, 90 (23), 11282–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lautenschlager J; Stephens AD; Fusco G; Strohl F; Curry N; Zacharopoulou M; Michel CH; Laine R; Nespovitaya N; Fantham M; Pinotsi D; Zago W; Fraser P; Tandon A; St George-Hyslop P; Rees E; Phillips JJ; De Simone A; Kaminski CF; Schierle GSK, C-terminal calcium binding of alpha-synuclein modulates synaptic vesicle interaction. Nature communications 2018, 9 (1), 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vilar M; Chou HT; Luhrs T; Maji SK; Riek-Loher D; Verel R; Manning G; Stahlberg H; Riek R, The fold of alpha-synuclein fibrils. Proc.Natl.Acad.Sci U.S.A 2008, 105 (25), 8637–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spillantini MG; Crowther RA; Jakes R; Hasegawa M; Goedert M, alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc.Natl.Acad.Sci.U.S.A 1998, 95 (11), 6469–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormack A; Chegeni N; Chegini F; Colella A; Power J; Keating D; Chataway T, Purification of alpha-synuclein containing inclusions from human post mortem brain tissue. Journal of neuroscience methods 2016, 266, 141–50. [DOI] [PubMed] [Google Scholar]
- 7.Chartier-Harlin MC; Kachergus J; Roumier C; Mouroux V; Douay X; Lincoln S; Levecque C; Larvor L; Andrieux J; Hulihan M; Waucquier N; Defebvre L; Amouyel P; Farrer M; Destee A, Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 2004, 364 (9440), 1167–1169. [DOI] [PubMed] [Google Scholar]
- 8.Singleton AB; Farrer M; Johnson J; Singleton A; Hague S; Kachergus J; Hulihan M; Peuralinna T; Dutra A; Nussbaum R; Lincoln S; Crawley A; Hanson M; Maraganore D; Adler C; Cookson MR; Muenter M; Baptista M; Miller D; Blancato J; Hardy J; Gwinn-Hardy K, alpha-Synuclein locus triplication causes Parkinson’s disease. Science 2003, 302 (5646), 841. [DOI] [PubMed] [Google Scholar]
- 9.Kotzbauer PT; Tu Z; Mach RH, Current status of the development of PET radiotracers for imaging alpha synuclein aggregates in Lewy bodies and Lewy neurites. Clinical and Translational Imaging 2017, 5 (1), 3–14. [Google Scholar]
- 10.Klunk WE; Engler H; Nordberg A; Wang Y; Blomqvist G; Holt DP; Bergström M; Savitcheva I; Huang GF; Estrada S, Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound‐B. Annals of neurology 2004, 55 (3), 306–319. [DOI] [PubMed] [Google Scholar]
- 11.Wong DF; Rosenberg PB; Zhou Y; Kumar A; Raymont V; Ravert HT; Dannals RF; Nandi A; Brašić JR; Ye W, In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (flobetapir F 18). Journal of Nuclear Medicine 2010, 51 (6), 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowe CC; Ackerman U; Browne W; Mulligan R; Pike KL; O’Keefe G; Tochon-Danguy H; Chan G; Berlangieri SU; Jones G, Imaging of amyloid β in Alzheimer’s disease with 18F-BAY94–9172, a novel PET tracer: proof of mechanism. The Lancet Neurology 2008, 7 (2), 129–135. [DOI] [PubMed] [Google Scholar]
- 13.Kudo Y; Okamura N; Furumoto S; Tashiro M; Furukawa K; Maruyama M; Itoh M; Iwata R; Yanai K; Arai H, 2-(2-[2-Dimethylaminothiazol-5-yl] ethenyl)-6-(2-[fluoro] ethoxy) benzoxazole: a novel PET agent for in vivo detection of dense amyloid plaques in Alzheimer’s disease patients. Journal of Nuclear Medicine 2007, 48 (4), 553–561. [DOI] [PubMed] [Google Scholar]
- 14.Chien DT; Bahri S; Szardenings AK; Walsh JC; Mu F; Su M-Y; Shankle WR; Elizarov A; Kolb HC, Early clinical PET imaging results with the novel PHF-tau radioligand [F-18]-T807. Journal of Alzheimer’s Disease 2013, 34 (2), 457–468. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart SN; Baker SL; Okamura N; Furukawa K; Ishiki A; Furumoto S; Tashiro M; Yanai K; Arai H; Kudo Y, Dynamic PET measures of tau accumulation in cognitively normal older adults and Alzheimer’s disease patients measured using [18F] THK-5351. PLoS One 2016, 11 (6), e0158460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama M; Shimada H; Suhara T; Shinotoh H; Ji B; Maeda J; Zhang M-R; Trojanowski JQ; Lee VM-Y; Ono M, Imaging of tau pathology in a tauopathy mouse model and in Alzheimer patients compared to normal controls. Neuron 2013, 79 (6), 1094–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotzbauer PT; Cairns NJ; Campbell MC; Willis AW; Racette BA; Tabbal SD; Perlmutter JS, Pathologic accumulation of alpha-synuclein and Abeta in Parkinson disease patients with dementia. Archives of neurology 2012, 69 (10), 1326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muntane G; Dalfo E; Martinez A; Ferrer I, Phosphorylation of tau and α-synuclein in synaptic-enriched fractions of the frontal cortex in Alzheimer’s disease, and in Parkinson’s disease and related α-synucleinopathies. Neuroscience 2008, 152 (4), 913–923. [DOI] [PubMed] [Google Scholar]
- 19.Bagchi DP; Yu L; Perlmutter JS; Xu J; Mach RH; Tu Z; Kotzbauer PT, Binding of the radioligand SIL23 to alpha-synuclein fibrils in Parkinson disease brain tissue establishes feasibility and screening approaches for developing a Parkinson disease imaging agent. PLoS One 2013, 8 (2), e55031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue X; Dhavale DD; Li J; Luo Z; Liu J; Yang H; Mach RH; Kotzbauer PT; Tu Z, Design, synthesis, and in vitro evaluation of quinolinyl analogues for alpha-synuclein aggregation. Bioorganic & medicinal chemistry letters 2018, 28 (6), 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu W; Zhou D; Gaba V; Liu J; Li S; Peng X; Xu J; Dhavale D; Bagchi DP; d’Avignon A, Design, synthesis, and characterization of 3-(benzylidene) indolin-2-one derivatives as ligands for α-synuclein fibrils. Journal of medicinal chemistry 2015, 58 (15), 6002–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh C-J; Xu K; Lee I; Graham TJA; Tu Z; Dhavale D; Kotzbauer P; Mach RH, Chalcones and 5-Membered Heterocyclic Isosteres Bind to Alpha Synuclein Fibrils In Vitro. ACS Omega 2018. Acceppted, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X; Jin H; Padakanti PK; Li J; Yang H; Fan J; Mach RH; Kotzbauer P; Tu Z, Radiosynthesis and in vivo evaluation of two PET radioligands for imaging α-synuclein. Applied Sciences 2014, 4 (1), 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuttle MD; Comellas G; Nieuwkoop AJ; Covell DJ; Berthold DA; Kloepper KD; Courtney JM; Kim JK; Barclay AM; Kendall A; Wan W; Stubbs G; Schwieters CD; Lee VM; George JM; Rienstra CM, Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nature structural & molecular biology 2016, 23 (5), 409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abola EE; Bernstein FC; Koetzle TF, The protein data bank In Neutrons in Biology, Springer: 1984; pp 441–441. [Google Scholar]
- 26.Morris GM; Huey R; Lindstrom W; Sanner MF; Belew RK; Goodsell DS; Olson AJ, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry 2009, 30 (16), 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haney CM; Petersson EJ, Fluorescence spectroscopy reveals N-terminal order in fibrillar forms of α-synuclein. Chemical Communications 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peelaerts W; Baekelandt V, α-synuclein folds: the cards are on the table. Nature Structural and Molecular Biology 2016, 23 (5), 359. [DOI] [PubMed] [Google Scholar]
- 29.Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K, Scalable molecular dynamics with NAMD. Journal of computational chemistry 2005, 26 (16), 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphrey W; Dalke A; Schulten K, VMD: visual molecular dynamics. Journal of molecular graphics 1996, 14 (1), 33–38. [DOI] [PubMed] [Google Scholar]
- 31.Haney CM; Wissner RF; Warner JB; Wang YJ; Ferrie JJ; Covell DJ; Karpowicz RJ; Lee VM-Y; Petersson EJ, Comparison of strategies for non-perturbing labeling of α-synuclein to study amyloidogenesis. Organic & biomolecular chemistry 2016, 14 (5), 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puchades M; Westman A; Blennow K; Davidsson P, Removal of sodium dodecyl sulfate from protein samples prior to matrix‐assisted laser desorption/ionization mass spectrometry. Rapid Communications in Mass Spectrometry 1999, 13 (5), 344–349. [DOI] [PubMed] [Google Scholar]
- 33.Bagchi DP; Yu L; Perlmutter JS; Xu J; Mach RH; Tu Z; Kotzbauer PT, Binding of the radioligand SIL23 to α-synuclein fibrils in Parkinson disease brain tissue establishes feasibility and screening approaches for developing a Parkinson disease imaging agent. PloS one 2013, 8 (2), e55031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


