Abstract
African trypanosomes infect a broad range of mammals, but humans and some higher primates are protected by serum trypanosome lytic factors that contain Apo-L1. In the human infective subspecies of Trypanosoma brucei, T. b. rhodesiense, a gene product derived from the variant surface glycoprotein gene family member, serum-resistance associated protein (SRA protein) protects against Apol-L1-mediated lysis. Protection against trypanosome lytic factor requires the direct interaction between SRA protein and Apo-LI within the endocytic apparatus of the trypanosome, but some uncertainty remains as to the precise mechanism and location of this interaction. In order to provide more insight into the mechanism of SRA-mediated resistance to trypanosome lytic factor, we assessed the localisation of SRA in T.b. rhodesiense EATRO3, using a novel monoclonal antibody raised against SRA together with a set of well-characterized endosomal markers. By 3D-IF single cell analysis, combined with double-labeling immunoelectron microscopy, we found that ≈50% of SRA protein localized to the lysosome, with the remaining population being distributed through the endocytic pathway, but apparently absent from the flagellar pocket membrane. These data suggest that the SRA/TLF interaction is intracellular, with the concentration within the endosomes potentially crucial for ensuring a high efficiency.
Keywords: Trypanosoma brucei rhodesiense, Serum Associated Protein, Monoclonal Antibody
Introduction
Immune evasion for African trypanosomes has multiple components. Antigenic variation is the primary mechanism, based on the mono-allelic expression of the variant surface glycoproteins (VSG) (Cross, 1975). Additional mechanisms assisting in defence against the host immune system include rapid capping, endocytosis and degradation of surface-bound immune complexes(Barry, 1979, Pal et al., 2003, Engstler et al., 2007). A third component is responsible for resistance to innate immune factors and dictates host range. In particular this governs the ability to infect humans and a number of higher primates (Pays et al., 2014).
The immune effectors responsible for host restriction are trypanolytic factor (TLF) 1 and 2, serum macromolecular complexes which contain two primate-specific proteins: haptoglobin-related protein (Hpr) and apolipoprotein L-1 (ApoL1). TLF1 enters the trypanosome through binding of Hpr to the haptoglobin-haemoglobin receptor (HpHbR) and clathrin-mediated endocytosis, while ApoL1 causes trypanolysis via the transmembrane flux of chloride and the osmotic swelling of the lysosome (Vanhamme et al., 2003, Perez-Morga et al., 2005, Thomson et al., 2015). Trypanosomes that infect humans have evolved mechanisms that negate TLF-mediated killing. In the West Africa subspecies Trypanosoma brucei gambiense, the resistance mechanism appears to involve multiple factors, including alterations to TLF endocytosis by a polymorphism in the HpHbR lowering the affinity for ligand(Vanhollebeke et al., 2008, Higgins et al., 2013), expression of the T. b. gambiense specific glycoprotein TgsGP and reduction of sensitivity to ApoL1 through altered cysteine protease activity (Uzureau et al., 2013). In the East African form, T. b. rhodesiense, a single serum resistance-associated (SRA) gene, is both necessary for resistance to TLFs and sufficient to confer resistance on otherwise sensitive T. b. brucei isolates (De Greef et al., 1994, Xong et al., 1998). The mechanism of resistance is a stoichiometric binding of SRA to ApoL1 that prevents ApoL1 trypanocidal action (Pays et al., 2014).
To gain access to the lysosome via endocytosis, ApoL1 is trafficked through the endosomal system. Endocytosis is exclusively clathrin-mediated in trypanosomes, and extremely rapid in the mammalian stages. Following the initial endocytosis event at the flagellar pocket membrane, cargo progresses through a series of endosomal sub compartments that have been defined to some degree, and which correspond broadly to early, middle and late endosomes of mammalian cells plus recycling sorting/endosomes; these are defined by the presence of the small GTPases Rab 5, 21, 28 and 7 respectively for the endocytic arm, and Rab11 for the recycling pathways (Manna et al., 2014).
Despite a considerable body of literature, it remains unknown where the initial interaction between SRA and ApoL1 occurs within the trypanosome endosomal system, while descriptions of the localisation of SRA within the cell are not always consistent. Initial reports localized endogenous SRA to solely the lysosome by immunofluorescence, using p67 as the lysosomal marker (Vanhamme et al., 2003). Subsequently, a wider distribution within the endosomal system, extending from the flagellar pocket to the lysosome and including several intermediate endosomes was reported in a cell line over-expressing a Ty epitope-tagged SRA transgene. In this study, visualization of SRA in the lysosome required the use of the lysosomal protease inhibitor FMK-024, presumably to delay degradation of the SRA protein or Ty epitope (Stephens et al., 2011). These conflicting reports have significant impact in understanding the function of SRA, and in particular the likely site(s) of interaction; does SRA bind and inactivate TLF early in the endocytic pathway or is the encounter later, at a point close to the lysosome? These points are important as they also influence the mechanism by which TLF is prevented from accessing the lysosome; specifically the intracellular routes that are required to simply recycle TLF from early endosomes are well established, but pathways to bring material from late or terminal endosomes back to the surface are poorly documented in trypanosomes. Here, with high resolution 3D deconvolved immunofluorescence (3D-IF), the subcellular localisation of SRA protein has been investigated in cell lines over a range of expression levels that spans the endogenous expression level in T. b. rhodesiense and two T. b. brucei cell lines containing SRA transgenes.
Results and discussion
Characterisation of an SRA monoclonal antibody and transgenic cell lines
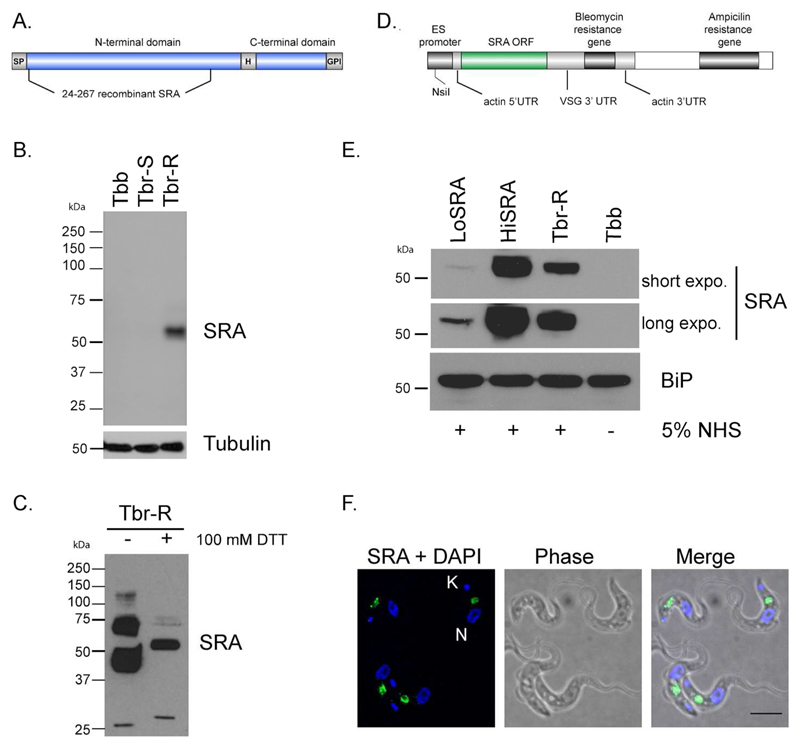
The mature SRA polypeptide has ~360 residues, three potential N-linked glycosylation sites and a glycosylphosphatidylinositol (GPI) anchor at the C-terminus (Vanhamme et al., 2003, Oli et al., 2006). SRA has structural similarity to VSG, and in particular the division into a large N-terminal and smaller C-terminal domain. The N-terminal domain, corresponding to residues 24 to 267, has been modelled using a VSG template (Campillo et al., 2003). The region included in that model was expressed as a recombinant protein in E. coli (Thomson et al., 2009) and used to generate monoclonal antibodies in mice (Figure 1A). A panel of hybridomas were screened against recombinant SRA by ELISA (data not shown) and positive clones rescreened against recombinant SRA by Western blot and finally whole cell lysates from T. b. rhodesiense and T. b. brucei (Figure 1B). Two monoclonal antibodies remained at this point and the monoclonal anti-SRA (5D2) that gave the strongest reactivity by Western blot was selected. MAb5D2 recognized a polypeptide of ~50 kDa in lysates of T. b. rhodesiense grown in the presence of human serum, but did not recognize any antigens in lysates from T. b. brucei or T. b. rhodesiense cultured for an extensive period in the absence of human serum, a treatment frequently resulting in silencing of the SRA gene (De Greef et al., 1994, Xong et al., 1998). MAb5D2 therefore represents a defined anti-SRA reagent with high specificity.
Figure 1. Characterization of an SRA monoclonal antibody and transgenic cell lines expressing.
A. Schematic representation of the recombinant SRA protein used to generate the monoclonal antibody. SP: Signal Peptide, HD: Hinge Domain, GPI: glycosylphosphatidylinositol anchor. B. Western blot analysis with a mouse monoclonal antibody generated against SRA (mAb 5D2). Tubulin was used as loading control. Tbb, T. b. brucei, Tbr-S, T. b. rhodesiense-Human Serum Sensitive, Tbr-R, T. b. rhodesiense-Human Serum Resistant. C. WB analysis with anti-SRA of T. b. rhodesiense protein extracts in presence or absence of 100mM DTT. D. Schematic representation of the plasmid used to ectopically express SRA in T. b. brucei. E. WB analysis of T. b. brucei cell lines ectopically expressing SRA (LoSRA and HiSRA). BiP was used as loading control. Two exposure times are shown (30 s as short exposure and 3 min as long exposure). F. Immunofluorescence (IF) analysis of the endogenous SRA (green) expression pattern in T. b. rhodesiense. DNA is stained with DAPI. Scale bar, 5 μm.
As a member of the VSG family, SRA is predicted to share similar structures with both VSGs and the trypanosome transferrin receptor (De Greef et al., 1994, Carrington et al., 1996). Evidence from in silico modelling of the SRA protein suggests there are two disulphide bonds in the N-terminal domain of SRA. The small C-terminal domain of SRA shares conserved cysteine resides with the VSG C-terminal domains (Campillo et al., 2003), but, in SRA, there is an additional cysteine present in the linker between the N-terminal and C-terminal domain. Significantly, a minority of VSGs also possess a cysteine in an equivalent position which forms an inter-chain disulphide bond in the VSG dimer. To test if SRA is a disulphide linked dimer, protein extracts of Tbr-R were prepared in the absence or presence of 100mM of DTT. After WB analysis, DTT-treated SRA protein migrated as the expected 50kDa single band, while two major species, at ~40kDa and ~70kDa and a minor one at 130kDa were detected for the untreated extract (Figure 1C). The most likely interpretation is that the 40 kDa form is the SRA monomer with disulphide bonds intact and the 70 kDa form is an SRA dimer, suggesting that SRA is present as both disulphide bonded dimers and non-disulphidebonded forms within the cell. The higher molecular weight form may represent an additional oligomeric state.
To create cell lines expressing different levels of SRA protein, a transgene encoding the complete SRA open reading frame, flanked by an actin 5’UTR and VSG 3’UTR, was introduced ~400 bp downstream of the VSG promoter in an expression site (Penate et al., 2009) (Figure 1D). Cell lines containing the transgene were screened by Western blotting with the monoclonal anti-SRA and two cell lines were selected for experiments, with one expressing significantly less SRA (LoSRA) than T. b. rhodesiense and one expressing more SRA (HiSRA) (Figure 1E).
To confirm the correct folding of SRA and its ability to confer resistance to normal human serum, both clones were grown in 5% fresh and inactivated NHS, together with the T. b. brucei S16 parental cell line. While S16 cells died within 24 hours in such conditions, the two transgenic SRA clones proliferated indistinguishably from T. b. rhodesiense (data not shown) validating that the transgenic SRA was both folding and being targeted correctly.
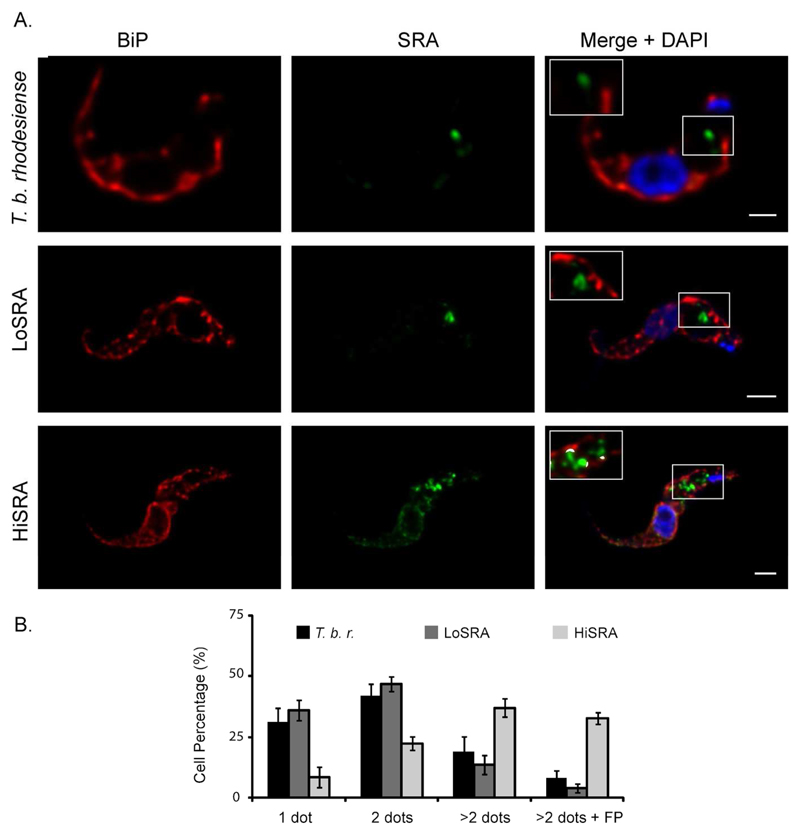
To localize SRA in T. b. rhodesiense cells, we carried out 3D-deconvolved immuno-fluorescence microscopy on cells grown in the presence of 5% human serum. Statistical analysis of the SRA distribution was performed using interphase cells (i.e. 1K1N), and specifically excluding cells undergoing mitosis or cytokinesis where SRA localisation is significantly more complex, most likely due to the replication and segregation of cellular organelles. Consistent with earlier studies, SRA demonstrated significant distribution to regions of the cell between the nucleus and kinetoplast, and which is consistent with a location throughout the endocytic pathway (Figure 1F). Significantly we were unable to detect SRA at the surface of the parasite or in a region close to the kinetoplast, where the flagellar pocket resides, suggesting that the majority of the SRA we were able to visualize was present within internal cellular structures. For 70.5% of the cells (n=156), SRA concentrates in one or two areas more proximal to the nucleus than kinetoplast (see quantitation in Figure 4B). Under our fixation and permeabilization conditions (2% PFA w/v and 1% NP40 v/v), SRA immunoreactivity was found close to the flagellar pocket only in a very small proportion of the cells (3.2%).
Figure 4. Overexpression of SRA affects its subcellular localisation.
A. Double indirect 3D-immunofluorescence (3D-IF) was performed in bloodstream T. brucei (T. b. rhodesiense, T. b. brucei LoSRA and T. b. brucei HiSRA) with anti-BiP (red), anti-SRA mAb (green) and DAPI staining (blue). Higher magnification is shown in inset, showing anti-SRA and anti-BiP fluorescence signals colocalisation mask (white). Scale bars, 1 μm. B. Statistical analysis from the SRA pattern observed by 3D-IF depending on the presence of 1, 2, more than 2 dots and its localisation in the flagellar pocket area (done from 3 independent experiments, n=572).
Endogenous SRA is partially associated with the lysosome in T. b. rhodesiense
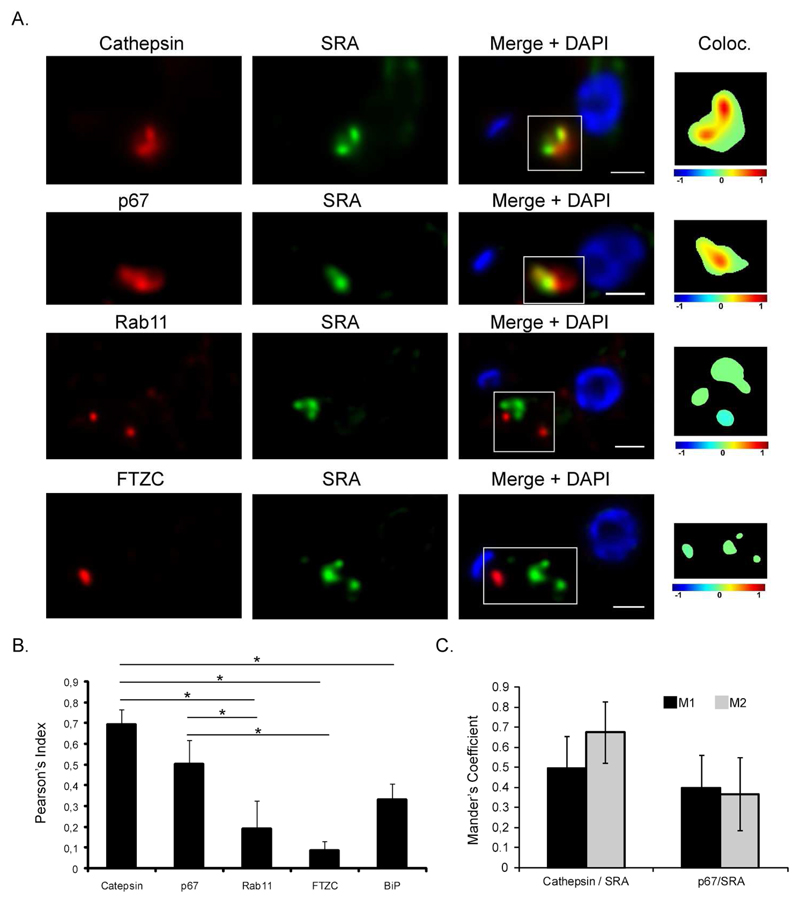
Since the subcellular localisation of SRA has not been definitively described in the literature (Vanhamme et al., 2003, Stephens et al., 2011) we re-examined SRA localisation taking advantage of MAb5D2, and combined this with high resolution 3D-deconvolution imaging (Landeira et al., 2009), performed in T. b. rhodesiense. To determine the proportion of colocalisation between SRA and the lysosome, two independent lysosomal markers were used: p67 and TbCathepsin (TbCatL), a lysosomal protease, also known as trypanopain (Caffrey et al., 2001) (Figure2A). Single-cell quantification was performed from the 3D-deconvolved images using JACoP, a component of ImageJ. Under permeabilizing conditions, SRA partially localized with both lysosomal markers; Pearson’s coefficients were statistically significant when comparing SRA/TbCAtL (rp=0.69±0.07) and SRA/p67 (rp=0.50±0.11) with the Pearson’s coefficient calculated to examine the colocalisation between SRA and BiP, and ER marker (rp=0.33±0.08) (Figure 2B). As a marker of recycling endosomes we used antibody to TbRab11 and double IF was done with anti-SRA MAb5D2 (Figure 2A, lower panel). No colocalisation was found between both proteins (rp=0.19±0.13), discarding the possibility of a significant presence for SRA in the exocytic or recycling pathways. Finally, to assess a possible location of SRA in the flagellar pocket area as previously suggested (Stephens et al., 2011), we combined anti-SRA with a rabbit polyclonal anti-FTZC antibody that was described as a component of the flagellum transition zone, located at the base of the flagellum (Bringaud et al., 2000). Double IF followed by single cell analysis failed to detect any significant colocalisation between SRA and FTZC (rp=0.08±0.04) (Figure 2A and 2B), suggesting that in endogenous conditions, SRA does not traffic through flagellar pocket area.
Figure 2. SRA partially associates with lysosome.
A. Representative pictures of double 3D-IF combining rabbit polyclonal anti-Cathepsin, anti-p67, anti-Rab11 or anti-FTZC antibodies (red) with the mouse monoclonal anti-SRA (5D2) antibody (green). The colocalisation color map (inset) shows the intensity of colocalisation between SRA and the red-labeled endocytic markers as indicated by the color bar. In these color maps, the −1 to +1 heat map depicts the measured icorr values. Negatively correlated relationships (icorr values between −1 and 0) are shown in blue-green colors, whereas positive correlations (icorr values between 0 and 1) are represented by warmer yellow-red colors. Scale bar 1 μm. B and C. Colocalisation analysis. Pearson’s coefficient was calculated for each individual 3D deconvolved image (n=70). * p-value <0.05. Mander’s coefficients were calculated (when Pearson’s coefficient≥0.5) to define the M1 and M2 as the percentages of pixels in one channel (M1=red) that intersect with some signal in other channel (M2=green). Both coefficients were assessed using “JACoP”, a colocalisation plugin available in Image J.
For both lysosomal markers that showed significant colocalisation, we decided to apply Manders' M1 and M2 coefficients that quantitate the portion of the intensity in each channel that coincides with some intensity in the other channel. These data represent the ratio of intersecting volumes, but considering each channel separately. For cells stained with TbCatL and SRA, 49.5% (±16.1%) of the signal detected for TbCatL (M1) coincided with SRA signal, while 67.6% (±16.3%) of SRA (M2) coincided with TbCatL (Figure 2C). Congruently with the Pearson’s coefficient data calculated for p67 and SRA, Mander’s coefficients decreased in comparison to the TbCatL/SRA relation with 39.9% (±15.2%) of p67 signal (M1) that coincided with SRA, and 36.8% (±18.1%) of SRA signal (M2) coinciding with p67. These results strongly support that SRA traffics to the lysosome, as approximately half of the SRA is detected in this compartment.
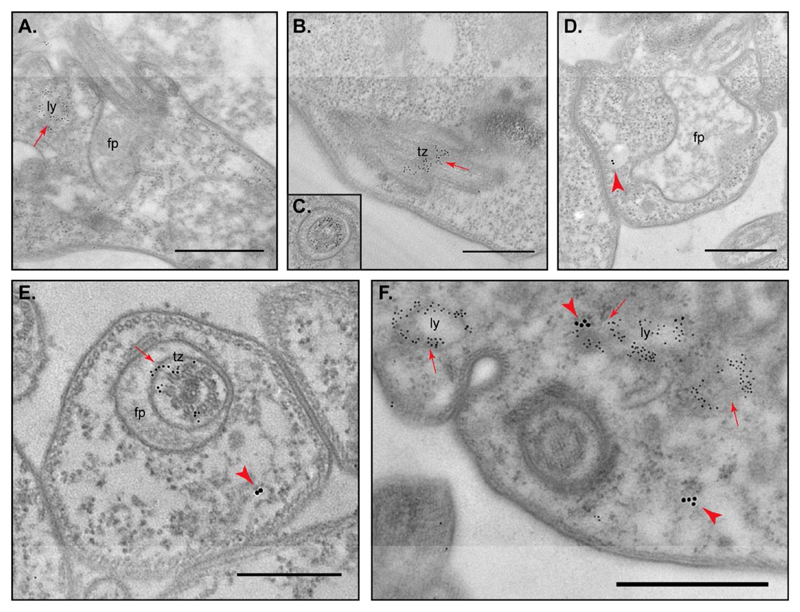
To confirm the SRA subcellular distribution analyzed by 3D-IF, we decided to conduct immunoelectron microscopy (IEM) combining anti-p67 (lysosome) and anti-FTZC with the monoclonal anti-SRA antibody. To guarantee relevant IEM labeling conditions, antibodies were first studied separately. As expected, both rabbit polyclonal antibodies gave a characteristic pattern shown Figure 3A for anti-p67 and Figure 3B and 3C for anti-FTZC. Despite the low sensitity of the antibody, a specific anti-SRA MAb5D2 signal was observed; 15 nm gold particles were associated with vesicles as revealed in Figure 3D. We thus decided to perform double IEM, combining anti-SRA either with anti-p67 or anti-FTZC. We first ensured the lack of cross-reaction between the first and the second antibodies (data not shown) and then performed double IEM. Congruently with the 3D-IF observations, SRA was never found associated with FTZC or the flagellar pocket area (Figure 3E) while partial colocalization SRA/p67 was confirmed (Figure 3F). From all these data, we can also hypothesize that the remaining SRA signal that does not associate with the lysosome vesicles could be connected with the endosomal structures.
Figure 3. Localisation of SRA to the vicinity of lysosomes in T. b. rhodesiense NHS resistant cell line by double-labeling immunoelectron microscopy (IEM).
Cells were first labeled either with rabbit anti-p67 (A), or rabbit anti-FTZC (B and C), or mouse anti-SRA (D) antibodies. Double-labeling was then performed with anti-SRA/anti-FTZC (E) and anti-SRA/anti-p67 (F). Arrowheads indicate SRA labeling (15nm gold particles conjugated) and arrows indicate p67 or FTZC labeling (5nm gold particles conjugated). fp, flagellar pocket; ly, lysosome; tz, transition zone. Scale bar 1 μm.
Overexpression of SRA affects subcellular localisation
To determine if the location of SRA is influenced by copy number, we conducted SRA immunofluorescence on our transgenic T. b. brucei cells that are under- or over-expressing SRA (Figure 4). As for endogenous expression in T. b. rhodesiense, statistical analysis was performed of the SRA distribution in 1K1N cells (Figure 4A for IF and Figure 4B for statistical data). For the loSRA line, the localisation of SRA was comparable with that obtained from T. b. rhodesiense, with ~70 to 80% of the cells exhibiting SRA as one or two punctae suggesting restriction to the lysosome and endosomes. No association was observed with the endoplasmatic reticulum (ER), as revealed by a rabbit polyclonal anti-BiP antibody (Figure 4A). For the hiSRA cell line, we found that SRA localisation was significantly more extensive, with 36.9% (±3.8%) of the cells displaying a localisation that was distinct to T. b. rhodesiense, while 32.6% (±2.4%) of cells exhibited SRA in the flagellar pocket area. Additionally, for these cells, colocalisation with BiP was observed, suggesting that when over-expressed SRA is not efficiently folded or exported and remains within the ER and possibly Golgi apparatus (Figure 4A).
Inhibition of endocytosis does not affect the endogenous SRA location in T. b. rhodesiense
In order to better assess the possibility that SRA trafficks into the flagellar pocket, despite having an undetectable presence at steady state, we used two approaches to disrupt endocytosis. We reasoned that if even small levels of SRA were present within the flagellar pocket, the blockade to endocytic trafficking would result in accumulation of SRA at that site, and facilitate the detection of SRA that was essentially en route via the surface.
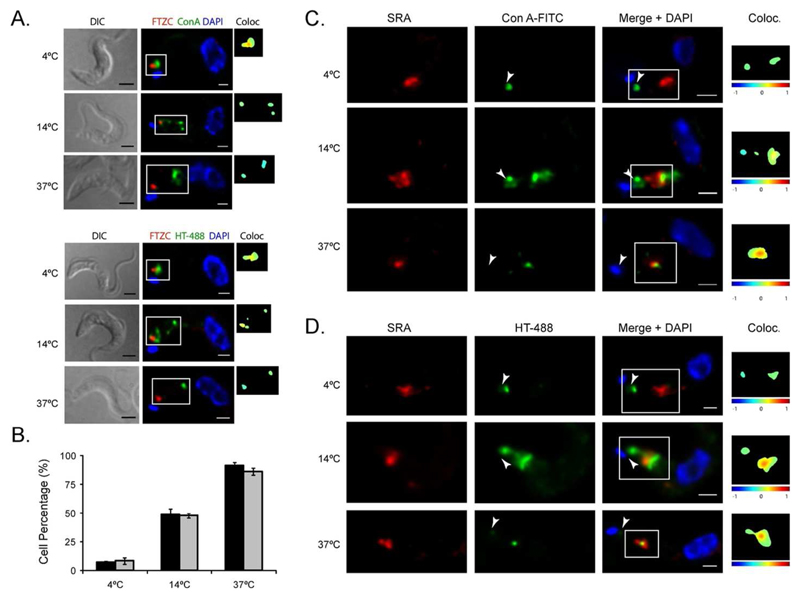
Firstly, we monitored the location of fluorescein isothiocyanate (FITC)-labeled lectin Concanavalin A (ConA) and 488-labeled human transferrin (HT-488) in mid-log phase BSF T. b. rhodesiense at 4°C, 14°C and 37°C. Both reagents report on endocytosis of surface protein; ConA by binding oligomannose N-glycans (essentially VSG) and transferrin by binding the ESAG6/7 transferrin receptor. ConA and HT-488 labelling was followed by immunostaining with anti-FTZC (Bringaud et al., 2000) after fixation and permeabilization (Figure 5A). For both reagents, low temperature clearly led to a decrease in endocytosis as expected. At 4°C, ConA and HT-488 were retained at the flagellar pocket, as colocalization with FTZC proved. This association was progressively lost when temperature increased. At 14°C, Con A and HT-488 are internalized to the endocytic system and at 37°C, both compounds reached the lysosome (Figure 5A). We thus decided to use such conditions to assess the subcellular location of SRA upon endocytic perturbation (Figure 5C and 5D). Significantly, the localisation of SRA was unaltered by these low temperature treatments, retaining indistinguishable distributions to our previous physiological conditions. While no evidence of SRA enrichment in the flagellar pocket area was detected, colocalisation of SRA with ConA and HT was significant when these fluorophores were endocytosed and trafficked through endosomes to the lysosome (see statistics in Figure 5B).
Figure 5. Inhibition of endocytosis pathway induced by low temperature does not affect endogenous SRA localisation in T. b. rhodesiense.
A. To set up the temperature-mediated endocytic inhibition, Con A-FITC and HT-488 (green) uptake was combined with 3D-IF with anti-FTZC antibody (red) upon three different temperatures (4°C, 14°C and 37°C). At 4°C, both reagents are retained at the flagellar pocket and associate with FTZC. At 14°C, Con A and HT-488 are internalized to the endocytic system. At 37°C, both compounds are delivered to the terminal lysosome and do not associate anymore with FTZC.B. Con A-FITC (green) uptake was combined with 3D-IF with anti-SRA antibody (red). After fixation, cells were subjected to IF with the mouse monoclonal anti-SRA antibody (red). Arrowheads indicate flagellar pocket area. C. Human Transferrin (HT)-488 (green) uptake combined with 3D-IF with anti-SRA antibody (red). The same protocol was applied as in B. Scale bars, 2μm for DIC and 1 μm for IF. D. Statistical analysis of the colocalisation of SRA with ConA-FITC or HT-488 depending on the temperatures (4°C, 14°C or 37°C), calculated from two independent experiments for each reagent (n=500).
Low temperature is not a specific endocytic pathway inhibition, but also perturbs exocytosis and many other cellular processes (Yeaman et al., 2001). Exocytosis to the cell surface is via the flagellar pocket (Grunfelder et al., 2003, Sutterwala et al., 2007) and alterations to exocytosis could modulate the inhibition of endocytosis and mask a putative enrichment of SRA in the flagellar pocket. Therefore, to inhibit endocytosis in a very distinct manner, we suppressed expression of the clathrin heavy chain (TbCLH) by RNAi. Knockdown of clathrin results in an enlargement of the flagellar pocket, suggesting that membrane delivery is unaffected but removal is blocked (Allen et al., 2003). As T. b. rhodesiense EATRO3 does not contain the relevant transgenic elements for RNAi, we decided to take advantage of the T. b. brucei cell line expressing SRA (LoSRA).
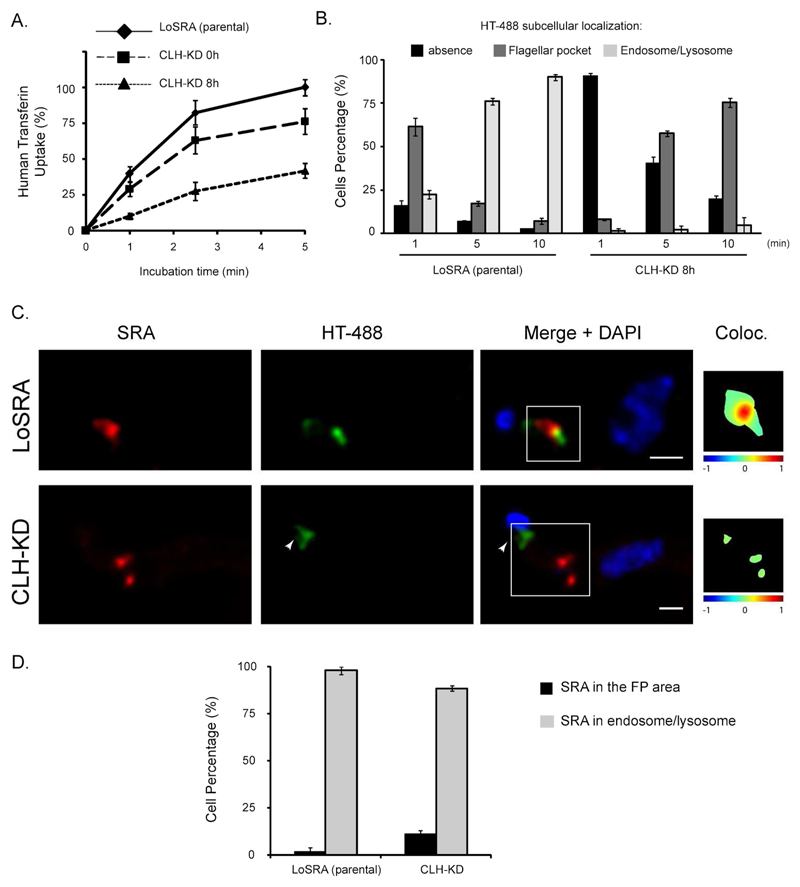
We first monitored TbCLH depletion by measuring uptake of Alexa-488 (HT-488) human transferrin by flow cytometry. After only 8 hours of RNAi induction, HT-488 uptake was reduced by 58.2% (±5.1%) (Figure 6A). To confirm this fluorescence microscopy was also used. Statistical analysis performed on a total of 372 cells, analyzed in two independent experiments, showed that under normal conditions, for 90.1% (±1.8%) of the cells, human transferrin was localized in the endosomes/lysosome area after 10min of uptake (Figure 6B). Consistent with the flow cytometry, under TbCLH-KD, 75.4% (±2.7%) of the cells retain HT in the flagellar pocket area, while the remaining cells did not show any labeling (Figure 6B).
Figure 6. Inhibition of endocytosis pathway by TbClathrin KD does not affect SRA localisation.
A. HT-488 uptake analysis by flow cytometry, upon KD of TbClathrin. B. Statistical analysis of the HT-488 subcellular localisation, upon KD of TbCLH, after 1 min, 5 min or 10 min of HT incubation. C. Representative IF pictures combining HT-488 (green) 5 min uptake with SRA (red) immunostaining with the 5D2 monoclonal antibody. Upper panel: parental cell line. Lower panel: TbCLH KD. The colocalisation color map (inset) shows the intensity of colocalisation between SRA and HT-488 as indicated by the color bar. Arrowheads indicate flagellar pocket area. Scale bars, 1μm. D. Statistical analysis of the presence/absence of SRA in the flagellar pocket area upon TbCLH KD (n=88). No statistical significance (p-value=0.111) was observed.
Single cell analysis (n=88) of 3D deconvolved pictures was performed using a colocalisation color map to ascertain the impact on SRA. Rapid transferrin uptake in the parental cell line leads to partial colocalisation between the HT-488 and SRA in the endosomes/lysosome area (98% of the cells, Figure 6C). For the TbCLH induced RNAi line (LoSRA-TbCLH-KD), SRA was not significantly associated with human transferrin. A colocalisation mask indicated 11.5% of the cells with overlapping between HT-488 and SRA (with a non-significant p-value=0.111 with Fischer exact test), suggesting than upon such condition of strong endocytic inhibition, SRA did not significantly enrich in the flagellar pocket area (Figures 6C and 6D), suggesting that SRA does not commonly traffic there.
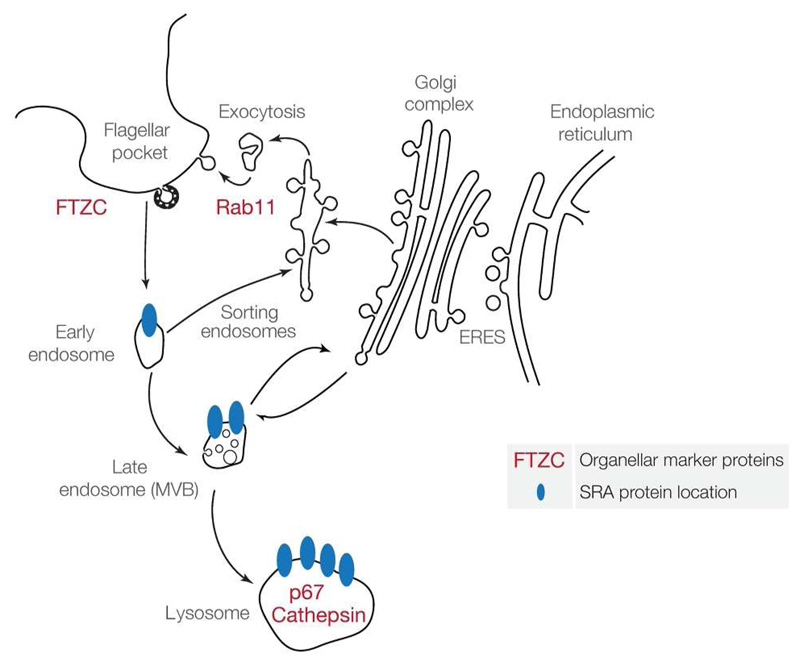
SRA represents a key protein that confers full human serum resistance when expressed in T. brucei and its intracellular itinerary is crucial to understanding the function, as the location of SRA determines where it interacts with TLF. Previous work on SRA subcelular localisation lead to contradictory data (Vanhamme et al., 2003, Stephens et al., 2011). Here we attempt, using new reagent combined with modulation of expression and trafficking, to determine specifically if SRA is either at the surface, in the flagellar pocket area, or in the endocytic pathway. In summary, our data suggest SRA is present at internal membranes, but that even with an endocytosis block, its detection at the cell surface is not significant, suggesting that the flagellar pocket does not represent a major site for interaction between SRA and TLF (Figure 7). The concentration of SRA within endosomes may act as a mechanism to increase the local concentration, and therefore ensure efficient sequestration of TLF.
Figure 7. Schematic model of SRA subcellular localisation in T. b. rhodesiense, inferred from data obtained with the mouse monoclonal anti-SRA antibody.
Double IFs and IEMs, combining anti-SRA with various endocytic pathway markers, through endogenous conditions and/or compromising physically or molecularly the endocytosis, indicate that SRA traffics from lysosomes to endosomes, avoiding the flagellar pocket area. Our data suggest that the SRA interacts with the trypanolytic factor in an intracellular manner, with the concentration within the endosomes potentially crucial for increasing efficiency.
Experimental procedures
Ethics statement
Human blood samples were taken from healthy donors, who provided written informed consent for the collection of samples. These samples were specifically obtained for this study. The procedure was approved by ethical committee of Institute of Parasitology and Biomedicine Lopez-Neyra (Spanish National Research Council).
Trypanosomes
T. b. rhodesiense East African Trypanosomiasis Research Organization (EATRO3 ETat1.2 TREU164 (Turner et al., 2004)) and T. b. brucei single marker (derived from T. brucei bloodstream, Molteno Institute Trypanozoon antigenic type (MITat) 1.2; clone 221a) cell lines (Wirtz et al., 1999), grown in HMI-9 medium supplemented of 10% inactivated Fetal Calf Serum (Hirumi et al., 1989) or 5% inactivated Normal Human Serum, were used for all the experimental procedures.
Production of the monoclonal antibody raised against SRA
His-tagged recombinant SRA (residues 24 to 267) was expressed in E. coli from plasmid p3303, a pET15b derivative (Thomson et al., 2009). The polypeptide was purified using nickel ion affinity chromatography under standard conditions. A BALB/c mouse was immunized by three intraperitoneal injections of 50 μg of purified recombinant SRA. Three days before fusion, 40 μg of the protein was injected intravenously. Normal antibody-producing spleen cells were fused with immortal myeloma cells. The hybridoma that produced the desired antibody was selected by ELISA and subcloned to obtain a single epitope antibody, which was named 5D2. This monoclonal antibody was tested by WB and IF against the NHS-resistant T.b. rhodesiense (Tbr-R), NHS-sensitive T.b. rhodesiense (Tbr-S) and T.b. brucei cell lines.
SRA expressing Trypanosome
The SRA open reading frame was cloned by PCR using oligonucleotides 5’GGATCCATGCCCCGAAATTCGGGCCG-3’and 5’-GGATCCTTAAAACAGAAAGGCCACAA-3’, digested with BamHI (underlined nucleotides) and cloned into an expression vector (pMig104) designed to integrate 405 bp downstream of the active ES promoter (Penate et al., 2009). Correct orientation was confirmed by sequencing. The 221-VSG 3’UTR was cloned by PCR using oligonucleotides 5’-ATGGATCCACCCCTCTTTGGCTTGCAGTT-3’and 5’-CTCGGGCCCG TCCAGGATAGAGGTGTGAGTTAAA-3’, digested with BamHI and SmaI respectively, and inserted downstream of the SRA ORF stop codon. T. b. brucei bloodstream form single marker trypanosomes were transfected and selected with 5 μg.ml-1 Bleomycin using procedures previously described (Wirtz et al., 1999). Transformants were called LoSRA and HiSRA.
RNAi procedure
To generate clathrin depleted cell lines, the plasmid p2T7TiCLH was transfected into the LoSRA (Allen et al., 2003). Transformants were selected with 2.5 μg.ml-1 G418, 5 μg.ml-1Hygromycin and 5 μg.ml-1Bleomycin. Clones were induced with tetracycline and selected for growth defect and transferrin uptake decrease (see Uptake assays).
Protein electophoresis and western blotting
For protein extraction, mid-log phase cells were harvested, washed in TDB-glucose, and resuspended in SDS–polyacrylamide sample buffer at a concentration of 1.106 cell equivalents per microlitre. Samples were electrophoresed on 10% SDS–PAGE gels at 5.106 cell equivalents per lane and then transferred to Hybond-ECL nitrocellulose membrane (Amersham Pharmacia Biotech). Transfer and equivalence of loading were checked by staining the membrane with Ponceau S solution (Sigma) prior to blocking for 20 min at room temperature in blocking solution (PBS supplemented by 5% dried milk). Membranes were incubated 1 h at room temperature with primary antibody at the appropriate dilution in 0.5% blocking solution (mouse monoclonal anti-TbSRA antibody was diluted 1/10, the mouse monoclonal anti-Tubulin, 1/5000 and the rabbit anti-Bip 1/20000), washed 3 ×10 min in PBS/0.05% Tween-20, then incubated for one hour with horseradish peroxidase goat anti-mouse conjugate (GE). After washing, bound antibodies were detected by reaction with luminol and visualized by exposure to X-ray film.
Uptake assays
For colocalisation studies of SRA with Fluorescein isothiocyanate (FITC)-labeled Concanavalin A (Con A) (Vector Laboratories) and human holo-Tf Alexa Fluor 488-conjugated (HT-488) (Invitrogen), uptake assays were performed as followed. 2.106 parasites, previously washed with TDB-glucose plus 1% BSA and cooled down 10 min at 4°C, were incubated with 10 μg FITC-Con A or 5 μg HT-488 at the appropriate temperatures (4°C, 14°C and 37°C) for 30 min in a volume of 250 μl. After incubation, cells were fixed for one hour at 4 °C in 1% PFA diluted in cold TDB. Cells were finally washed twice with TDB to remove unlabelled Con A or HT-488 and PFA, and keep at 4°C before immunofluorescence.
For analysis of human transferrin uptake by flow cytometry, 2 x 106 trypanosomes were harvested and washed with TDB-glucose plus 1% BSA, resuspended in 250 μl in the same buffer and incubated for 10 minutes at 37 °C. Then, 5 μg of HT-488 were added. Incubations were carried out for 0 (no Tf), 1, 2.5, 5 and 10 minutes at 37 °C. Cells were then fixed at least for one hour at 4 °C in 1% PFA diluted in cold PBS. Parasites were finally washed twice with PBS and labelled cells were analysed with a Becton Dickinson FACS Calibur TM flow cytometer (BD Biosciences) using BD CellQuest™ Pro version 4.0.2 software (Cordon-Obras et al., 2013).
3D Immunofluorescence
Cellular localisation analysis was performed by 3D immunofluorescence as described previously (Landeira et al., 2009). Images displayed in the figures are maximum intensity projections from digitally deconvolved multichannel 3D image data sets using Huygens Essential software v. 2.9 (Scientific Volume Imaging). Cells were fixed with 2% PFA for one hour and permeabilized with 1% NP40 for 30 min. Immunofluorescence analysis was carried out in 0.5% blocking reagent (Roche) in PBS (Sigma) using the mouse monoclonal anti-TbSRA antibody (5D2) (1:50), the rabbit polyclonal anti-BiP (1/20000), anti-p67 (1/4000), anti-TbCat (1/4000) and anti-Rab11 (1/250) and anti-FTZC (1/2000) antibodies. Pseudocoloring and maximum intensity projections were performed using ImageJ software v. 1.43 (National Institutes of Health). Deconvolution of 3D images was performed using Huygens Essential software (version 2.9; Scientific Volume Imaging) using an experimentally calculated point-spread function with 0.2-μm TetraSpeck microspheres (Invitrogen). For the colocalisation analysis, JACoP and Colocalization Colormap, available under Image J, were used. Live-cell imaging was performed at 37 °C, and cells were visualized under differential interference contrast (DIC) optics.
Immunoelectron microscopy
Parasites were washed in TDB-glucose and fixed in 4% paraformaldehyde and 0.25% glutaraldehyde in PBS, pH 7.4, 1 hour at 4°C. The cell pellet was embedded in LRWhite resin (Electron Microscopy Science) and polymerized at -20°C with UV light. Thin sections (50-70nm) were cut and probed with anti-SRA (1/2), anti-p67 (1/4000) and/or anti-FTZC (1/2000) antibodies diluted PBS with 1% BSA, 0.5% Tween 20 (Sigma), 2 hours at RT. Primary antibody incubation was followed by 1:30 dilution goat anti-mouse (15nm) and/or anti-rabbit (5nm) gold conjugated antibodies (British Biocell International) incubation for 1 hour. Control preparations processed without primary antibody and irrelevant primary antibodies were all negative.
Statistical analysis
We used one-way ANOVA to compare the Pearson’s coefficient value generated by the JACoP analysis. Confidence intervals (CI) were set at 95%. IBM SPSS Statistics software (version 20) was used to perform the statistical analysis.
Acknowledgments
We thank James Bangs (Buffalo, USA) for providing anti-TbCat and anti-p67 antibodies and Frédéric Bringaud (Bordeaux, France) for anti-FTZC antibody.
Funding
MN is funded by grants from the Spanish Ministerio de Ciencia e Innovación, (SAF2012-40029), Junta de Andalucia (CTS-5841) and VI PN de I+D+I 2008-2011, ISCIII -Subdirección General de Redes y Centros de Investigación Cooperativa (RICET) RD12/0018/0001 and RD12/0018/0015. JMB is supported by a Miguel Servet Fellowship (CP09/00300) and funded by “Fondo de Investigación Sanitaria” (FIS) PI10/01128. JR and MC were funded by a Wellcome Trust Project Grant 093008/Z/10/Z. Work in the Dundee laboratory was funded by the Wellcome Trust (program grant 093008/Z/10/Z) and the Medical Research Council.
References
- Allen CL, Goulding D, Field MC. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. The EMBO journal. 2003;22:4991–5002. doi: 10.1093/emboj/cdg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JD. Capping of variable antigen on Trypanosoma brucei, and its immunological and biological significance. Journal of cell science. 1979;37:287–302. doi: 10.1242/jcs.37.1.287. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Robinson DR, Barradeau S, Biteau N, Baltz D, Baltz T. Characterization and disruption of a new Trypanosoma brucei repetitive flagellum protein, using double-stranded RNA inhibition. Molecular and biochemical parasitology. 2000;111:283–297. doi: 10.1016/s0166-6851(00)00319-4. [DOI] [PubMed] [Google Scholar]
- Caffrey CR, Hansell E, Lucas KD, Brinen LS, Alvarez Hernandez A, Cheng J, et al. Active site mapping, biochemical properties and subcellular localization of rhodesain, the major cysteine protease of Trypanosoma brucei rhodesiense. Molecular and biochemical parasitology. 2001;118:61–73. doi: 10.1016/s0166-6851(01)00368-1. [DOI] [PubMed] [Google Scholar]
- Campillo N, Carrington M. The origin of the serum resistance associated (SRA) gene and a model of the structure of the SRA polypeptide from Trypanosoma brucei rhodesiense. Molecular and biochemical parasitology. 2003;127:79–84. doi: 10.1016/s0166-6851(02)00306-7. [DOI] [PubMed] [Google Scholar]
- Carrington M, Boothroyd J. Implications of conserved structural motifs in disparate trypanosome surface proteins. Molecular and biochemical parasitology. 1996;81:119–126. doi: 10.1016/0166-6851(96)02706-5. [DOI] [PubMed] [Google Scholar]
- Cordon-Obras C, Cano J, Gonzalez-Pacanowska D, Benito A, Navarro M, Bart JM. Trypanosoma brucei gambiense Adaptation to Different Mammalian Sera Is Associated with VSG Expression Site Plasticity. PLoS ONE. 2013;8:e85072. doi: 10.1371/journal.pone.0085072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GA. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- De Greef C, Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Molecular and biochemical parasitology. 1994;68:277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Engstler M, Pfohl T, Herminghaus S, Boshart M, Wiegertjes G, Heddergott N, Overath P. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131:505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- Grunfelder CG, Engstler M, Weise F, Schwarz H, Stierhof YD, Morgan GW, et al. Endocytosis of a glycosylphosphatidylinositol-anchored protein via clathrin-coated vesicles, sorting by default in endosomes, and exocytosis via RAB11-positive carriers. Mol Biol Cell. 2003;14:2029–2040. doi: 10.1091/mbc.E02-10-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins MK, Tkachenko O, Brown A, Reed J, Raper J, Carrington M. Structure of the trypanosome haptoglobin-hemoglobin receptor and implications for nutrient uptake and innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1905–1910. doi: 10.1073/pnas.1214943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. The Journal of parasitology. 1989;75:985–989. [PubMed] [Google Scholar]
- Landeira D, Bart JM, Van Tyne D, Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. The Journal of cell biology. 2009;186:243–254. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna PT, Boehm C, Leung KF, Natesan SK, Field MC. Life and times: synthesis, trafficking, and evolution of VSG. Trends in parasitology. 2014 doi: 10.1016/j.pt.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oli MW, Cotlin LF, Shiflett AM, Hajduk SL. Serum resistance-associated protein blocks lysosomal targeting of trypanosome lytic factor in Trypanosoma brucei. Eukaryotic cell. 2006;5:132–139. doi: 10.1128/EC.5.1.132-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Hall BS, Jeffries TR, Field MC. Rab5 and Rab11 mediate transferrin and anti-variant surface glycoprotein antibody recycling in Trypanosoma brucei. The Biochemical journal. 2003;374:443–451. doi: 10.1042/BJ20030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Perez-Morga D. The molecular arms race between African trypanosomes and humans. Nature reviews. 2014;12:575–584. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- Penate X, Lopez-Farfan D, Landeira D, Wentland A, Vidal I, Navarro M. RNA pol II subunit RPB7 is required for RNA pol I-mediated transcription in Trypanosoma brucei. EMBO reports. 2009;10:252–257. doi: 10.1038/embor.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homble F, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science (New York, N.Y. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- Stephens NA, Hajduk SL. Endosomal localization of the serum resistance-associated protein in African trypanosomes confers human infectivity. Eukaryotic cell. 2011;10:1023–1033. doi: 10.1128/EC.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterwala SS, Creswell CH, Sanyal S, Menon AK, Bangs JD. De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol-anchored proteins, in African trypanosomes. Eukaryotic cell. 2007;6:454–464. doi: 10.1128/EC.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: Relevance to trypanosome lysis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2894–2899. doi: 10.1073/pnas.1421953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Molina-Portela P, Mott H, Carrington M, Raper J. Hydrodynamic gene delivery of baboon trypanosome lytic factor eliminates both animal and human-infective African trypanosomes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19509–19514. doi: 10.1073/pnas.0905669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CM, McLellan S, Lindergard LA, Bisoni L, Tait A, MacLeod A. Human infectivity trait in Trypanosoma brucei: stability, heritability and relationship to sra expression. Parasitology. 2004;129:445–454. doi: 10.1017/s0031182004005906. [DOI] [PubMed] [Google Scholar]
- Uzureau P, Uzureau S, Lecordier L, Fontaine F, Tebabi P, Homble F, et al. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature. 2013;501:430–434. doi: 10.1038/nature12516. [DOI] [PubMed] [Google Scholar]
- Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- Vanhollebeke B, De Muylder G, Nielsen MJ, Pays A, Tebabi P, Dieu M, et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science (New York, N.Y. 2008;320:677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Molecular and biochemical parasitology. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, Pays A, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- Yeaman C, Grindstaff KK, Wright JR, Nelson WJ. Sec6/8 complexes on trans-Golgi network and plasma membrane regulate late stages of exocytosis in mammalian cells. The Journal of cell biology. 2001;155:593–604. doi: 10.1083/jcb.200107088. [DOI] [PMC free article] [PubMed] [Google Scholar]