Abstract
Translation initiation in eukaryotes requires eIF4E, the cap binding protein, which mediates its function through an interaction with the scaffolding protein eIF4G, as part of the eIF4F complex. In trypanosomatids, four eIF4E homologues have been described but the specific function of each is not well characterized. Here, we report a study of these proteins in Trypanosoma brucei (TbEIF4E1 through 4). At the sequence level, they can be assigned to two groups; TbEIF4E1 and 2, similar in size to metazoan eIF4E1; and TbEIF4E3 and 4, with long N-terminal extensions. All are constitutively expressed, but whilst TbEIF4E1 and 2 localise to both the nucleus and cytoplasm, TbEIF4E3 and 4 are strictly cytoplasmic and are also more abundant. After knockdown through RNAi, TbEIF4E3 was the only homologue confirmed to be essential for viability of the insect procyclic form. In contrast, TbEIF4E1, 3 and 4 were all essential for the mammalian bloodstream form. Simultaneous RNAi knockdown of TbEIF4E1 and 2 caused cessation of growth and death in procyclics, but with a delayed impact on translation, whilst knockdown of TbEIF4E3 alone or a combined TbEIF4E1 and 4 knockdown led to substantial translation inhibition which preceded cellular death by several days, at least. Only TbEIF4E3 and 4 were found to interact with T. brucei eIF4G homologues; TbEIF4E3 bound both TbEIF4G3 and 4 whilst TbEIF4E4 bound only to TbEIF4G3. These results are consistent with TbEIF4E3 and 4 having distinct but relevant roles in initiation of protein synthesis.
Keywords: translation, eIF4E, eIF4G, Trypanosomatid, Leishmania
1. Introduction
The trypanosomatids, including the various Leishmania and Trypanosoma species, are pathogenic protozoans well known not only for the diseases they cause, but also for their complex biology and the unusual molecular mechanisms required for their gene expression. Transcription of protein coding genes occurs polycistronically [1, 2] and processing to monocistronic mRNAs occurs through coupled trans-splicing and polyadenylation (reviewed in [3]). The result is that mRNAs have a common 39nt long spliced-leader (or mini-exon) sequence at the distal end of the 5’ UTR, which is identical for all mRNAs of a given species. At the 5’ end of the spliced-leader sequence lies the 7-methyl-GTP cap nucleotide (m7G), followed by four methylated nucleotides in the spliced-leader sequence, the cap4 structure [4]. Regulation of gene expression in trypanosomatids is accomplished mainly through post-transcriptional mechanisms such as control of mRNA stability and possibly translation (for reviews see [5–7]). However, in contrast to the considerable characterization of their mechanisms of mRNA synthesis and processing, relatively little is understood regarding protein synthesis and how it can be regulated.
Translation initiation is the most complex stage of protein synthesis and the one which can vary more significantly between different taxonomic groups [8]. A major player in this process in eukaryotes, and also a major target for translation control, is eIF4E (eukaryotic initiation factor 4E), the m7G cap binding protein. eIF4E is a small polypeptide (24-25 kDa in mammals) which, apart from protein synthesis, has been implicated in a number of processes involved in mRNA metabolism such as transport and control of its stability (reviewed in [9–12]). In translation it is part of the heterotrimeric complex eIF4F, which also includes the RNA helicase eIF4A and the large scaffolding protein eIF4G [13]. eIF4F facilitates the recruitment of the small ribosomal subunit to the mRNA, which is accomplished with the help of another translation initiation complex, eIF3. eIF4E mediates the binding of the complex to the mRNA and, as part of eIF4F, can bind simultaneously to both the cap and eIF4G, which can then mediate interactions with other translation factors [14–16]. Despite its well described function in translation in the cytoplasm, substantial levels of eIF4E localize to the nucleus (reviewed in [17]), where it promotes the nuclear export of selected mRNAs [10, 18, 19].
The structure of eIF4E bound to 7-methyl-GDP [20, 21] is shaped like a cupped hand, with the cap analogue located in a narrow cap-binding slot on the concave side of the protein. eIF4E is characterized by eight tryptophan residues located at conserved positions along the protein. Cap recognition is mediated by, among other interactions, base sandwich-stacking between W56 and W102 (mammalian eIF4E-1 numbering) while W73 is involved in the interaction with eIF4G [9, 13, 20]. The activity of eIF4E can be regulated in a number of ways such as through the action of eIF4E interacting proteins, eIF4E-BPs, or directly through phosphorylation (reviewed in [22–25]).
In the yeast Saccharomyces cerevisae only one eIF4E homologue, essential for viability, is found [26]. More usually, multiple forms of this protein are present in any specific organism (reviewed by [27]). In mammals, three different classes of eIF4E have been described: eIF4E-1, the prototypical cap-binding protein; eIF4E-2 (also known as 4EHP), which binds the cap less well, does not interact with eIF4G and has a regulatory role [28, 29]; and eIF4E-3, which binds both cap and eIF4G but has a restricted, tissue specific, expression pattern [30]. An extensive analysis of hundreds of eIF4E-related sequences has indicated that different events of duplication of the eIF4E gene may have occurred in distinct eukaryotic groups after divergence, resulting in new forms of the protein with novel or modified functions [31].
In trypanosomatids, the initial description of a single putative eIF4E homologue [32] was followed by the identification of four homologues in genome sequences from L. major, all of which are also conserved in Trypanosoma species [33]. The four Leishmania eIF4E homologues (LmEIF4E1 through 4, LeishIF4E1 through 4 or simply EIF4E1 through 4) were shown to diverge in binding affinity for different synthetic cap analogues, in expression levels and in polysomal association in sucrose sedimentation gradients. None could rescue the growth of a yeast strain lacking a functional eIF4E and none could confidently be implicated as having a predominant role in translation or its control [33, 34].
Here we describe work focused on the four T. brucei proteins (here named TbEIF4E1 through 4). First, sequence analysis identified features which distinguish TbEIF4E1 and 2 from TbEIF4E3 and 4. For all four homologues, the cap-binding affinities, subcellular localization, and the expression in two stages of the T. brucei life cycle were characterised. The effect of depletion, through RNAi, on cellular growth and translation was determined and the interaction between putative eIF4E/eIF4G homologues was also analysed. The results presented allow the separation of the four proteins into two distinct groups. The first comprises TbEIF4E1 and 2, which localise both to the nucleus and the cytoplasm, do not seem to be directly involved in translation but perform functions essential for cellular viability. The second group is formed by the TbEIF4E3 and 4 homologues, more abundant, strictly cytoplasmic proteins which seem to be required for translation and take part in the formation of distinct eIF4F-like complexes.
2. Materials and methods
2.1. Sequence analysis
BLAST searches were carried out with the T. brucei genome sequences available at the Gene DB website of the Sanger Institute Pathogen Sequencing Unit (www.genedb.org). Further sequence searches and Clustal W alignments were done as previously described [33]. Relevant trypanosomatid GeneDB accessions: TbEIF4E1 – Tb11.18.0004; TbEIF4E2 – Tb927.10.16070; TbEIF4E3 – Tb11.01.3630; TbEIF4E4 –Tb927.6.1870; Lm (L. major) EIF4E1 – LmjF27.1620; LmEIF4E2 – LmjF19.1500; LmEIF4E3 – LmjF28.2500; LmEIF4E4 – LmjF30.0450 (the annotated sequence for LmEIF4E4 is missing 139 residues from what we believe is the N-terminus of the protein – they are encoded upstream of the annotated open reading frames and are homologous to the equivalent segment in the T. brucei orthologue).
2.2. PCR and cloning methods
All T. brucei eIF4E and eIF4G coding sequences were amplified from Lister 427 total genomic DNA and first cloned into the pGEM-T Easy vector (Promega). All amplified fragments were first sequenced, and the resulting sequences compared with those from the T. brucei genome sequencing project, prior to their use in the subcloning reactions. In order to express N-terminal His-tagged fusion proteins the TbEIF4E1 through 4 full length sequences were subcloned into the modified pET15b vector as described previously for the T. brucei eIF4A homologues [35]. Alternatively, to generate the eYFP and HA-tagged fusions and for the RNAi experiments, the same fragments were cloned respectively into the transfection vectors p2216 [35,36], p2477 [36] or p2T7-177 [37]. To generate the plasmids used in the transcription and translation reactions they were also subcloned into the pGEM3zf+ plasmid (Promega). To express the full length proteins as N-terminal GST tagged fusions, the TbEIF4G3 and 4 sequences were subcloned into the pGEX4T3 plasmid (GE Healthcare). His-tagged TbEIF4G3 and 4 were also expressed, with a C-terminal tag, as fragments consisting of the proteins’ N-terminus plus the central MIF4G/HEAT domain (TbEIF4G31-228 and TbEIF4G41-319 – numbers indicating the residues from the wild type protein remaining in the recombinant product) using the pET21a vector (Novagen). Supplementary TABLE I lists all oligonucleotides used in the amplifications reactions as well as the strategies and restrictions enzymes used in the subsequent subcloning events cited above. For the double TbEIF4E1/2 construct, for the RNAi experiments, the TbEIF4E2 gene was reamplified flanked by sites for Bgl II and BamH I and cloned into the dephosphorylated BamH I site of the p2T7-177-TbEIF4E1 construct. For the double TbEIF4E1/4 construct, first the p2T7-177-TbEIF4E1 was digested with BamH I, which cuts internally to the TbEIFE1 gene and at its 3’ end at the site introduced after the original PCR reaction, releasing a ~360bp fragment consisting of the second half of the target gene. This fragment was then cloned into the linearised BamH I site of the p2T7-177-TbEIF4E4 construct in a similar procedure as described for TbEIF4E1/2. Recombinant protein expression was performed according to standard procedures using the pET15, pET21 or pGEX4T3 derived constructs after transformation into Escherichia coli.
2.3. Cap binding assay
The four [35S]- methionine labeled TbEIF4Es were synthesized after in vitro transcription of their respective genes cloned in the pGEM3zf+ vector, following linearization with Xba I (TbEIF4E1 and 3) or BamH I (TbEIF4E2 and 4), with T7 RNA polymerase followed by translation with the nuclease treated Rabbit Reticulocyte Lysate System (Promega). For the LmEIF4E4 homologue, its gene was amplified from L. major (MHOM/IL/81/Friedlin) genomic DNA flanked by sites for Afl III and Not I (5’ primer - CT GAC ATG TCT ACC CCT CTC GAT GTG; 3’ primer - TA TGC GGC CGC GTA GCG ACG ACG GTT CTT TTT C) and cloned into the Nco I/Not I sites of pET21D (Novagen). The resulting plasmid was sequenced and linearized with Not I prior to transcription and translation, as described above. Assays were performed essentially as described previously [33], with non-specific binding removed by washes with 0.1 mM GTP and specific elution achieved with 50 μM cap analogue. As positive control, the labeled Xenopus laevis eIF4E cDNA was used.
2.4. Parasite growth, transfection and RNA interference
Cultivation of procyclic and bloodstream forms of T. brucei, Lister 427, as well as the procyclic cell line used for the ectopic expression and RNAi experiments, Lister 427 29-13 [38], were performed as previously described [35]. Cultures grown to mid-log phase (106 – 107 cells/ml for procyclic and 105 – 106 for bloodstream forms) were harvested for the production of total protein extract. For the RNAi of the bloodstream forms, Lister 427 90-13 was used [38], grown in the presence of 5 μg/ml of G418 and hygromycin. Transfection procedures were performed using standard conditions with selection of stable DNA integration being carried out using phleomycin (2.5 μg/ml for procyclic and 0.5 μg/ml for bloodstream cells). For the RNAi experiments, and induction of eYFP and HA-tagged fusion proteins, 1 μg/ml of tetracycline was added to mid-log phase cultures of transfected cells.
For metabolic labeling, methionine-free SDM-79 medium was used. Cultures were washed once in the same medium prior to being resuspended to a concentration of 1x107 cells/ml in the medium supplemented with 50 μCi/ml [35S]- methionine and incubated at 28°C. Aliquots were then taken to measure the trichloroacetic acid precipitable incorporation into protein and for analysis through SDS-PAGE and autoradiography.
2.5. RNA and protein analysis
RNA extraction and Northern blots were performed using standard methods [35, 39, 40]. DNA fragments containing complete open reading frames were used as probes for TbEIF4E1 through 4. Polyclonal serum against TbEIF4E1 through 4 and TbEIF4G3 and 4 were produced in rabbits using the corresponding recombinant His-tagged proteins. The different sets of antibodies were affinity purified and used in western blotting assays with the respective recombinant proteins and different samples of total protein extract from both procyclic and bloodstream forms of T. brucei. The western-blots were generally developed by ECL.
2.6. Fluorescence microscopy
For the indirect immunofluorescence assay, wild type procyclic cells grown to mid-log phase were harvested and washed with SDM-79 minus serum. The cells were fixed at a density of 5x106/ml with 3% paraformaldehyde, washed once in PBS and adhered to poly-L-lysine coated slides. Permeabilization was carried out with 0.1% Triton X-100 followed by blocking with 1% BSA. Antibody detection of the eIF4E homologues followed standard procedures using the various rabbit anti-sera and goat anti-rabbit IgG Alexa Fluor 488 (Invitrogen – Molecular Probes, USA). For the eYFP fusions, after induction with tetracycline, over-expressing transfected cells were directly observed in 35 mm culture plates. All the samples were visualized using a Leica SPII-AOBS confocal microscope.
2.7. Pull-down assays
Pull-down assays were essentially performed as described previously [33] using Glutathione-Sepharose beads (GE Healthcare). Beads alone, GST or GST-tagged TbEIF4G3 and 4 were immobilized on the beads and assayed for their ability to bind to [35S]-labelled TbEIF4E1 through 4, obtained after transcription and translation of the full-length genes cloned in the pGEM3zf+ plasmid.
2.8. Immunoprecipitation assays
Immunoprecipitations (IPs) were carried out with T. brucei cytoplasmic extracts produced from exponentially grown procyclic cells lysed in lysis buffer (20 mM Hepes-KOH, pH7.4, 75 mM potassium acetate, 4 mM magnesium acetate, 2 mM DTT) at a concentration of 2x108 cells/ml. IPs were carried out at 4°C using standard procedures. For the assays with the anti-TbEIF4E3 and 4 sera, protein-A Sepharose beads (GE Healthcare) were used with the affinity purified antibodies, or the corresponding total pre-immune sera as controls and cytoplasmic extracts from wild type cells. For the IPs of the HA-tagged proteins, cytoplasmic extracts from transfected cells expressing each recombinant protein were directly incubated with the Monoclonal Anti-HA Agarose Conjugate (Sigma). In both instances, proteins were eluted in SDS-PAGE samples buffer and assayed in western blots with affinity purified antibodies directed against the different T. brucei eIF4E, eIF4G or eIF4A homologues, as described in the text. For the Western blots of the IPs using the anti-TbEIF4E3 and 4 sera, proteins transferred to the PVDF membranes were stained with Rouge Ponceau and the band corresponding to the heavy weight chain of IgG from the immunoprecipitated complexes was cut out prior to blocking and incubation with the primary and secondary antibodies.
3. Results
3.1. The Trypanosoma eIF4E homologues fall in two distinct sequence groups
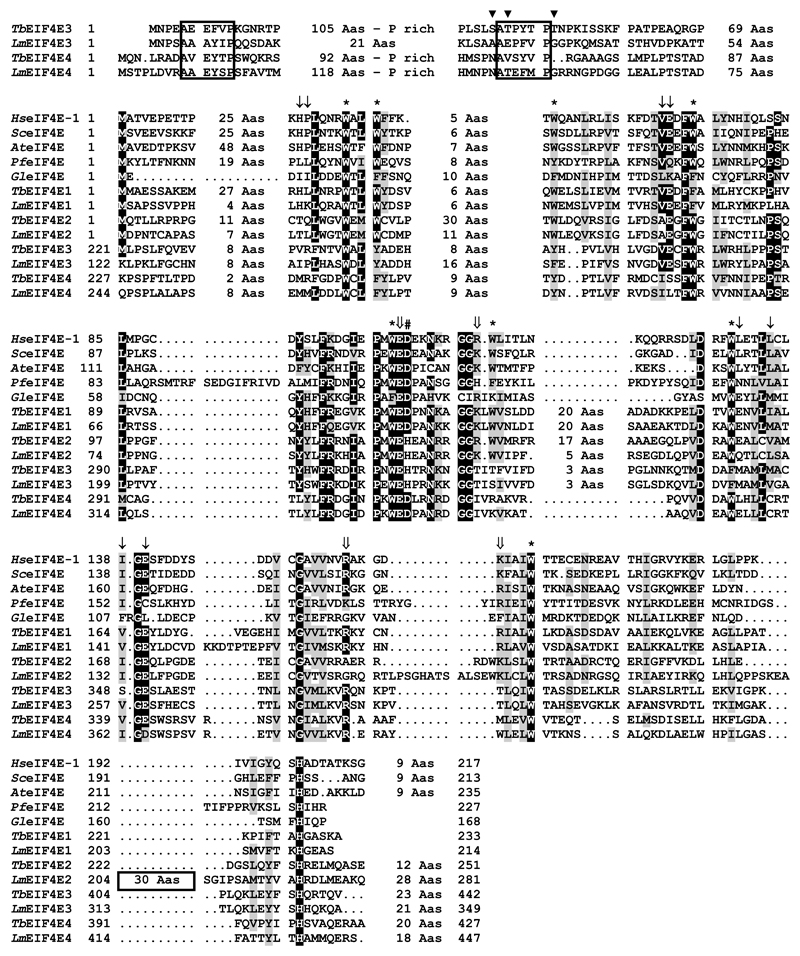
Based on the alignment shown in Figure 1, which compares the T. brucei and L. major eIF4E sequences with representative homologues from different eukaryotic lineages, including metazoans, plants, fungi and unrelated protozoans, the four trypanosomatid sequences can be broadly classified into two different groups. Group 1 comprises the EIF4E1 and 2 sequences, which are more similar in size to the human and yeast sequences, whilst group 2 includes EIF4E3 and 4, which share a few common features absent from the remaining proteins. These include a long N-terminus of more than 150 amino acids which share extensive homology between different orthologues of say, the EIF4E3 sequences, and also contain short segments of limited homology which seem to be conserved between the EIF4E3 and 4 sequences. Of the eight conserved tryptophan residues typical of eIF4E sequences, most are either conserved in the various trypanosomatid homologues or are replaced by other aromatic residues. The only exception is W112 (human eIF4E numbering), present in the EIF4E1 and 2 sequences but which is replaced by non-aromatic hydrophilic residues in EIF4E3 and 4. Additional noteworthy substitutions in the trypanosomatid sequences are D104, next to the near universally conserved W102E103, involved in cap binding [20] and conserved in all sequences shown but which is replaced by a histidine in EIF4E2 and 3; and V69E70, implicated in binding to eIF4G [41] but which is missing in the EIF4E4 homologues.
Fig. 1.
Sequence analysis of the four T. brucei eIF4E sequences. Clustal W alignment of the T. brucei and L. major eIF4E homologues with sequences from representative eukaryotic lineages. Amino acids identical in more than 60% of the sequences are highlighted in dark gray, while amino acids defined as similar, based on the BLOSUM 62 Matrix, on more than 60% of the sequences, are shown in pale gray. When necessary, spaces were inserted within the various sequences (dots) to allow better alignment. The first two boxes highlight regions which share conserved elements in the N-terminus of the EIF4E3 and 4 homologues from Leishmania and Trypanosoma species, with phosphorylated residues identified in TbEIF4E3 [49] indicated by the symbol ▾. The third box highlight the unique insertion observed in LmEIF4E2. * indicates the conserved tryptophan residues. Large arrows indicate non-tryptophan residues required for the interaction with the cap structure (based on [20]). Thin arrows indicate conserved non-tryptophan residues shown to be involved in eIF4G binding [41]. The symbol # marks the conserved D residue which is replaced by H solely in the trypanosomatid EIF4E2 and EIF4E3 sequences. GenBank Accessions for the non-trypanosomatid sequences: Hs (human) eIF4E-1 – NP_001959; Sc (Saccharomyces cerevisaei) eIF4E – NP_014502; At (Arabidopsis thaliana) eIF4E – NP_193538; Pf (Plasmodium falciparum) eIF4E1 – XP_001351220; Gl (Giardia lamblia) – XP_001710318.
3.2. TbEIF4E1, 2 and 4, but not TbEIF4E3, can efficiently bind the m7G cap
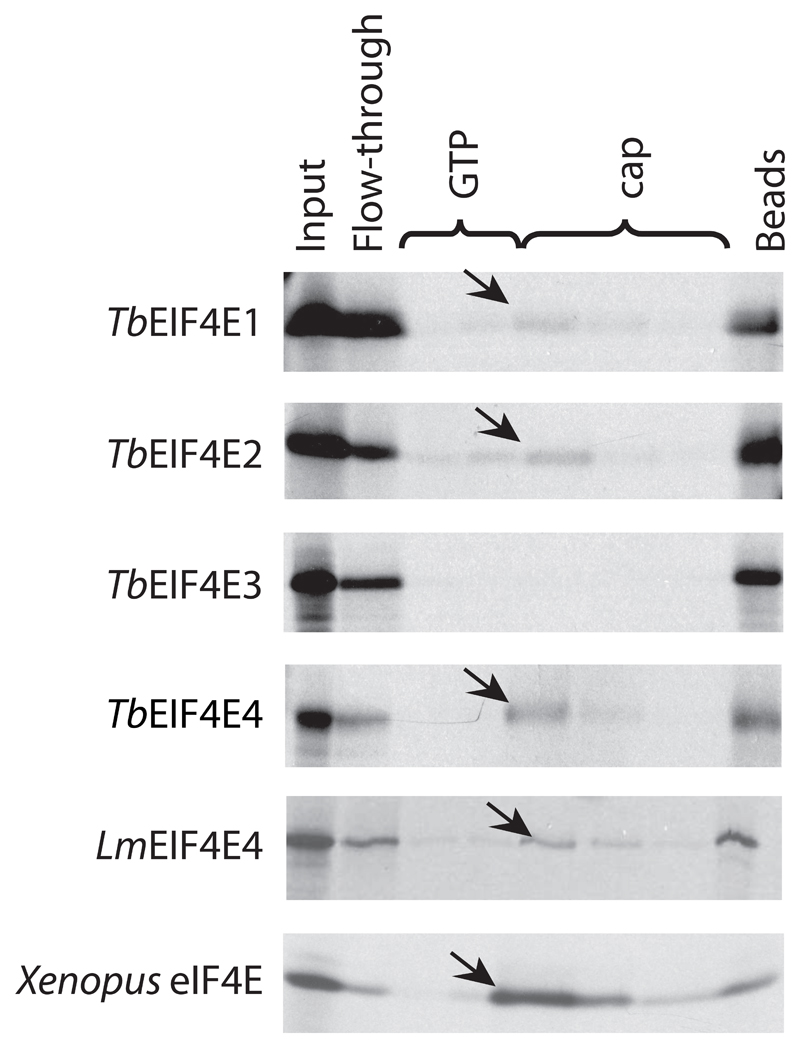
To start the study of the TbEIF4E proteins, their genes were first cloned and the respective [35S]-labeled proteins, synthesized in vitro, assayed as to their ability to bind to the commercially available m7G cap bound to Sepharose beads. Despite the significant differences between the more complex cap4 from trypanosomatids and the m7G cap from their metazoan hosts, this assay is useful to compare the substrate binding properties of the various homologues. TbEIF4E1, 2 and 4 were all found to be able to bind to the beads, and were specifically eluted with the soluble cap analogue, whilst TbEIF4E3 did not show any binding (Figure 2). A similar approach has been previously carried out for the first three L. major eIF4E homologues [33], where neither LmEIF4E2 nor LmEIF4E3 bound to the cap-beads, while LmEIF4E1 bound efficiently. Here, L. major LmEIF4E4 was also assayed and bound efficiently to the resin, as its T. brucei orthologue. The sole difference between the two organisms then lies with the EIF4E2 orthologues whose sequences diverge by the presence of a unique insertion in the Leishmania protein, found near its C-terminus, which is absent from its Trypanosoma counterpart (Figure 1).
Fig. 2.
Cap binding assays. Autoradiography of the cap binding chromatography performed with the four 35S-labelled T. brucei eIF4E homologues, the Leishmania LmEIF4E4 and the Xenopus leavis eIF4E homologue as positive control. Aliquots of the various washes were ran on SDS-PAGE and compared with samples from the original translation reaction (Input) as well as the non-bound fraction (Flow-through) and any protein remaining bound to the beads (Beads). The arrows indicate the proteins eluted by the cap analogue. The assay shows the affinity of TbEIF4E1, 2 and 4, as well as LmEIF4E4, for the 7-methyl-GTP Sepharose resin.
3.3. All four eIF4E homologues are expressed in both procyclic and bloodstream forms
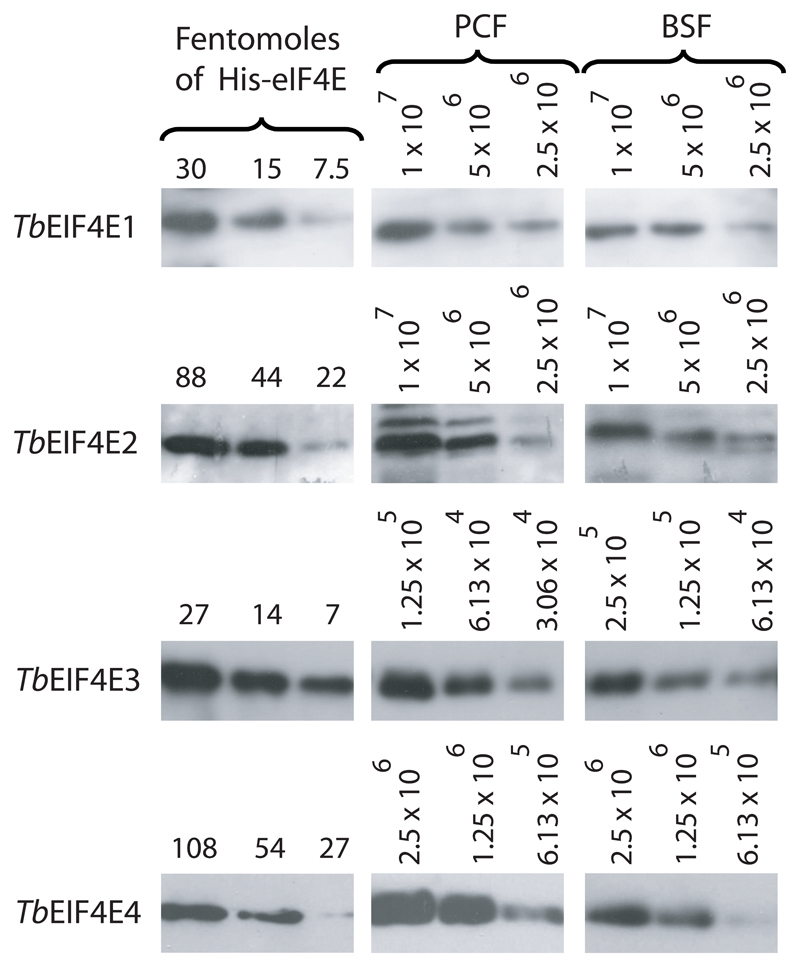
The four T. brucei eIF4Es were expressed as His-tagged recombinant proteins in Escherichia coli and used to raise specific polyclonal antisera in rabbits (Supplementary Figure 1). Cloned DNAs and sera were then used in Northern and Western blots to assay for expression of each gene in exponentially growing procyclic and bloodstream cells. All four mRNAs were expressed with little variation in levels between the two stages (Supplementary Figure 2) and the same applied for the corresponding proteins (Figure 3). The Western blot results were then quantitated and used to produce an estimate of the copy number of each protein (summarized in Table 1). TbEIF4E3 is by far the most abundant protein, especially in procyclic cells (>5x104 molecules per cell), followed by TbEIF4E4, which also is present at levels above 1x104 molecules per cell. In contrast, both TbEIF4E1 and 2 are present at much lower levels (compatible with the Northern results for TbEIF4E2 where its mRNA required a longer exposure than the remaining homologues in order to be detected). For most proteins the levels observed are slightly reduced in bloodstream forms, when compared with procyclics, which is consistent with the smaller cell volume. With the exception of EIF4E4, which was not previously quantified, the values observed for the T. brucei proteins are equivalent to those obtained for the L. major orthologues (Table 1 and reference [33]), presumably a reflection of conserved function.
Fig. 3.
Expression analysis of TbEIF4E1 through 4 in procyclic and bloodstream procyclic cells. Quantitation and expression analysis of TbEIF4E1, 3 and 4 proteins in procyclic/bloodstreams forms. Recombinant His tagged TbEIF4E1, 2, 3 and 4 were quantitated, diluted to defined concentrations (in fentomoles) and run on SDS-PAGE gels with whole parasite extract obtained from known number of cells from both procyclic and bloodstream forms.
Table 1. Estimate of the intracellular levels of the various T. brucei and L. major eIF4E homologues. ND – Not determined.
| Homologue | Protein levels (molecules/cell) |
||
|---|---|---|---|
|
T. brucei procyclics |
T. brucei bloodstream |
1
L. major promastigotes |
|
| EIF4E1 | 3-8 x 103 | 1.5-5 x 103 | 2-4 x 103 |
| EIF4E2 | 1-5 x 103 | 1-5 x 103 | ~1 x 103 |
| EIF4E3 | 5-10 x 104 | 2-4 x 104 | 4-10 x 104 |
| EIF4E4 | 2-4 x 104 | 1-2 x 104 | ND |
Based on reference [33].
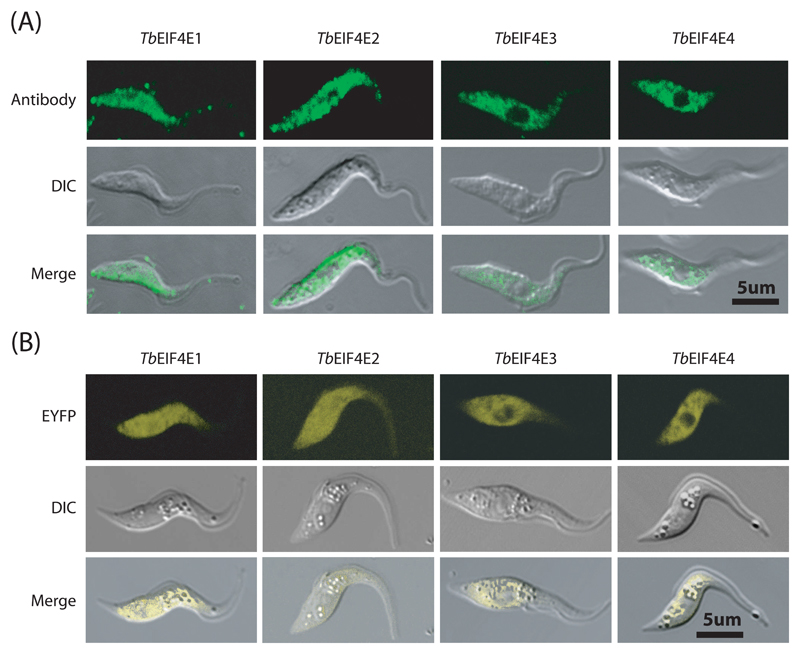
3.4. TbEIF4E1 and 2 are found in both cytoplasm and nuclei, whereas TbEIF4E3 and 4 are confined to the cytoplasm
Immunofluorescence experiments were carried out for TbEIF4E proteins in procyclic cells. TbEIF4E3 and 4 have a very similar localization and both are found throughout the cytoplasm with very little or no localization to the nucleus (Figure 4A and Supplementary Figure 3). In contrast, TbEIF4E1 and 2 can be clearly detected both in the cytoplasm as well as in the nucleus in the majority of cells. These results were confirmed in over-expression experiments using EIF4E-enhanced yellow fluorescent protein (eYFP) fusions in transgenic procyclic T. brucei cells (Figure 4B). TbEIF4E3 and 4 again were found confined to the cytoplasm, whilst TbEIF4E1 and 2 were found in the cytoplasm and in the nucleus, in good agreement with previously reported results [42].
Fig. 4.
Subcellular localisation of TbEIF4E1 through 4 in T. brucei procyclic forms. (A) Subcellular localisation of the four eIF4E homologues in wild type T. brucei procyclic cells (WT 427) by indirect immunofluorescence. This was performed using the antibodies directed to the various homologues followed by incubation with the Alexa Fluor 488 conjugated secondary antibody. (B) The localisation of TbEIF4E1 through 4 was also confirmed through the expression of eYFP fusion proteins in transfected T. brucei procyclic cells examined under the confocal microscope.
3.5. RNAi of TbEIF4E3, but not the remaining eIF4E homologues, in procyclic forms prevents cell viability whilst RNAi of TbEIF4E1, 3 and 4 in bloodstream forms impacts on cell proliferation and survival
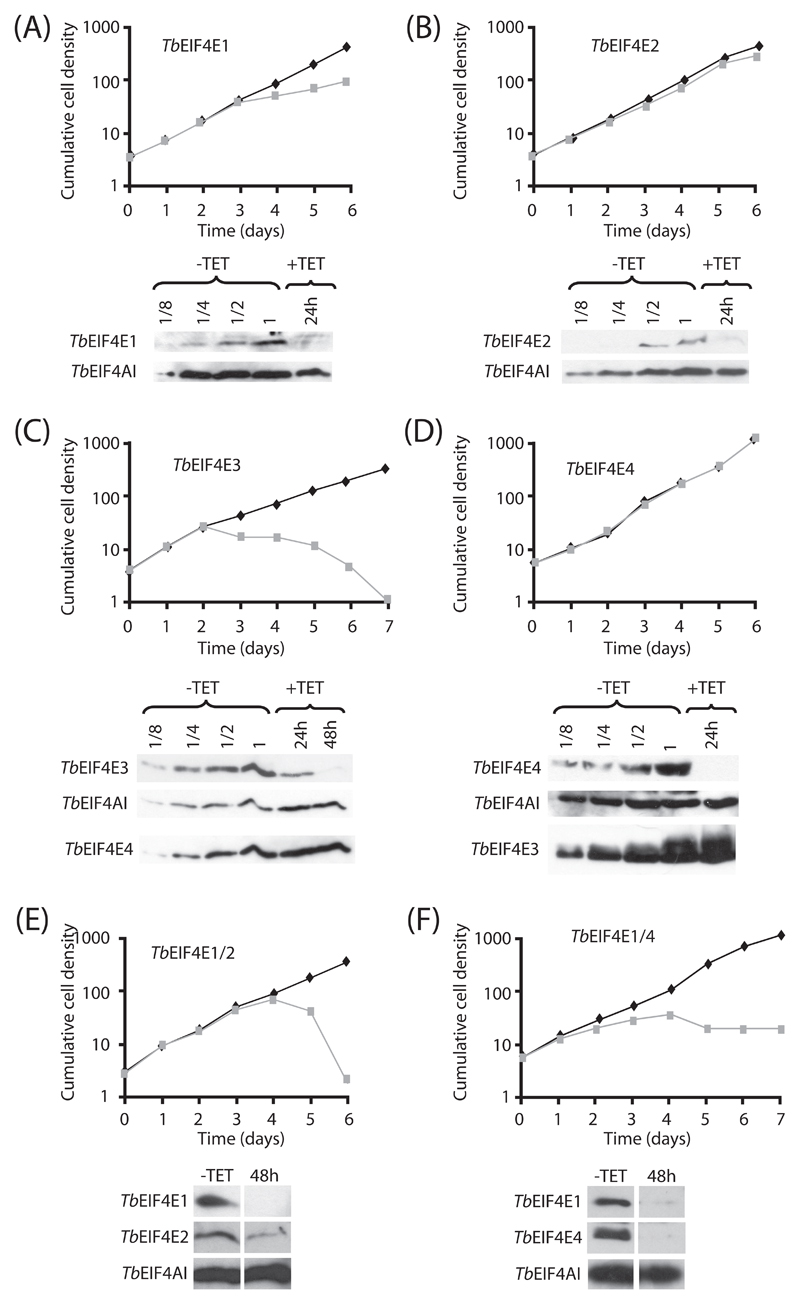
The requirement for the various eIF4E homologues for cellular viability was then investigated in T. brucei procyclic and bloodstream forms by knockdown of their expression through RNA interference. Representative growth curves of the various knockdowns for procyclic cells are shown in Figure 5. The phenotype of cells after RNAi ablation of TbEIF4E1, 2 and 4 (Figures 5A, 5B and 5D) was similar and no significant impact on cell survival was observed after knockdown of any of the three proteins. For TbEIF4E1, a reduction in the rate of cellular growth was observed four days after addition of tetracycline, but the cells remained viable and cell numbers kept increasing. Western blotting, shown below each curve, confirmed major depletions of the respective proteins (over 90% depletion for TbEIF4E1 and 4; 70 to 80% depletion for TbEIF4E2) within two to three days of RNAi induction, with no effect observed on the levels of the endogenous loading control, the T. brucei homologue of eIF4AI, TbEIF4AI. Knockdown of TbEIF4E3 produced a very distinct phenotype, since proliferation stopped two days after RNAi induction, followed by subsequent cell death (Figure 5C). The western-blotting confirmed substantial reduction (over 90%) in levels of TbEIF4E3 within 48 hours of tetracycline addition whilst the control, TbEIF4AI, remained unaffected. For both TbEIF4E3 and 4 knockdowns, western-blots were also carried out to see if depletion of TbEIF4E3 had any impact on TbEIF4E4 and vice-versa. Whilst no effect was observed on the levels of TbEIF4E4 after depletion of TbEIF4E3, knockdown of TbEIF4E4 seemed to provoke a slight increase on the levels of TbEIF4E3 (Figures 5C and 5D). Two double RNAi experiments were also performed to test for redundancy: TbEIF4E1 and 2, as these two are resident in the nucleus, and TbEIF4E1 and 4, as their L. major orthologues are both reported to interact with an eIF4G homologue [43]. When the simultaneous knockdown of TbEIF4E1 and 2 was performed, cells ceased proliferation and died (Figure 5E), with cellular proliferation stopping within four days of tetracycline induction. A distinct phenotype was observed by the simultaneous knockdown of TbEIF4E1 and 4 (Figure 5F). Although cellular proliferation stopped after RNAi induction, at about the same timeframe as the TbEIF4E1/2 RNAi, cell death did not occur for more than 15 days onwards (data after day 7 not shown).
Fig. 5.
RNA interference of TbEIF4E1 through 4 in procyclic cells. Procyclic T. brucei cells were transfected with the p2T7-177 derived plasmid containing the individual eIF4E genes (TbEIF4E1 – (A), TbEIF4E2 – (B), TbEIF4E3 – (C), TbEIF4E4 – (D)) or the double constructs TbEIF4E1/2 (E) and TbEIF4E1/4 (F). Transfected cells were selected after growth in the presence of phleomycin and RNA interference induced after tetracycline addition. At regular intervals, cellular growth was monitored by counting the number of viable cells of cultures with and without tetracycline and the resulting values used to plot the curves shown (plus tetracycline – gray; minus tetracylcline – black). The values shown are indicative of the cumulative cell density in 105 cells per ml. Below each graph are Western-blot analysis of the proteins being targeted, using the affinity-purified antisera specific for each protein. These were performed with a single sample derived from the curves minus tetracycline (-TET) as well as selected samples from the curves produced after the RNAi induction (in hours after tetracycline addition). For the single protein knock-downs, note the various dilutions of the -TET sample (1 to 1/8 cell dilution – 1 equals to the amount of cells used in the +TET lane) which allows for a better estimate of the RNAi efficiency. In general the same blot was reprobed with affinity purified antibodies against the endogenous control TbEIF4AI. The blot from the TbEIF4E3 RNAi was further incubated with the anti-TbEIF4E4 antibodies and likewise the blot from the TbEIF4E4 experiment was assayed with the anti-TbEIF4E3 antibodies. All RNAi results shown are representative of at least three different experiments performed with at least two distinct transfection events. In F only the first 7 days of the curve are shown but +TET RNAi curves, from different transfection events, were kept for 15 days or more without any increase in cell numbers.
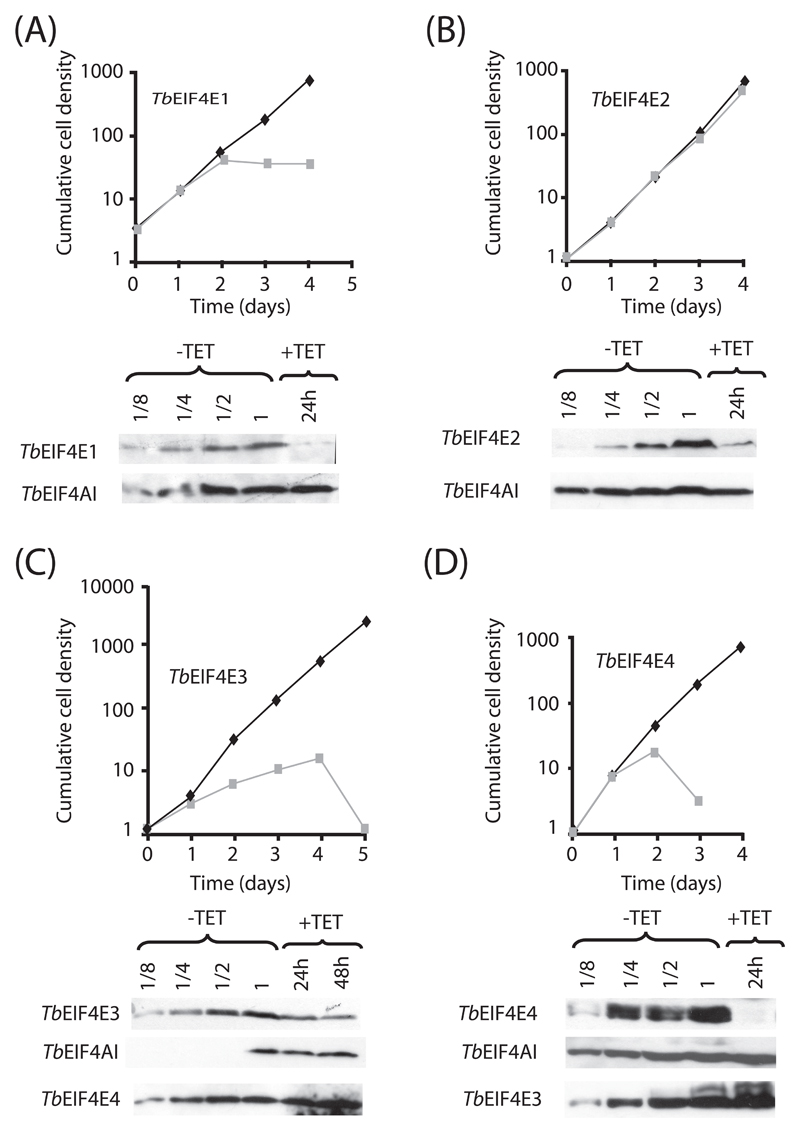
The RNAi procedure was then repeated with transfected bloodstream cells. Knockdown of TbEIF4E3 led to a cessation of proliferation around 24 h after induction followed by cell death (Figure 6C). Knockdown of TbEIF4E1 or 4 also caused the cells to stop growing between 24 and 48 hours after induction (Figures 6A and 6D). Cell death ensued for the TbEIF4E4 RNAi cells, whilst the cells depleted of TbEIF4E1 stopped growing but did not subsequently die, a phenotype which resembles what was observed for the double knockdown of TbEIF4E1/4 in procyclic cells. As for TbEIF4E2, no effect was observed in cellular growth after induction of the RNAi procedure (Figure 6B), as seen for procyclic cells. Western-blotting of selected samples confirmed the efficient depletion of the four proteins within their corresponding knockdown experiments (over 90% depletion for TbEIF4E1 and 4; 70 to 80% depletion for TbEIF4E2 and 3). As observed for procyclic cells, knock-down of TbEIF4E4 also seemed to provoke a slight increase on the levels of TbEIF4E3.
Fig. 6.
RNA interference of TbEIF4E1-4 in bloodstream cells. Bloodstream T. brucei cells were transfected with the p2T7-177 derived plasmids containing the eIF4E genes and monitored for cellular growth in the presence (gray curve) or absence (black curve) of tetracycline (TbEIF4E1 – (A), TbEIF4E2 – (B), TbEIF4E3 – (C), TbEIF4E4 – (D)). For all four RNAi procedures, expression of the targeted genes as well as the endogenous TbEIF4AI control was assayed in a sample from the control curve (-TET, diluted from 1 to 1/8 – 1 equals to the amount of cells used in the +TET lane) as well as in selected samples from the curve supplemented with tetracycline (+TET). The TbEIF4E3 and 4 RNAi blots were further assayed with the anti-TbEIF4E4 and anti-TbEIF4E3 antibodies, respectively. All procedures were done as described for Fig. 5.
3.6. TbEIF4E3 and TbEIF4E1/4 RNAi directly affects protein synthesis
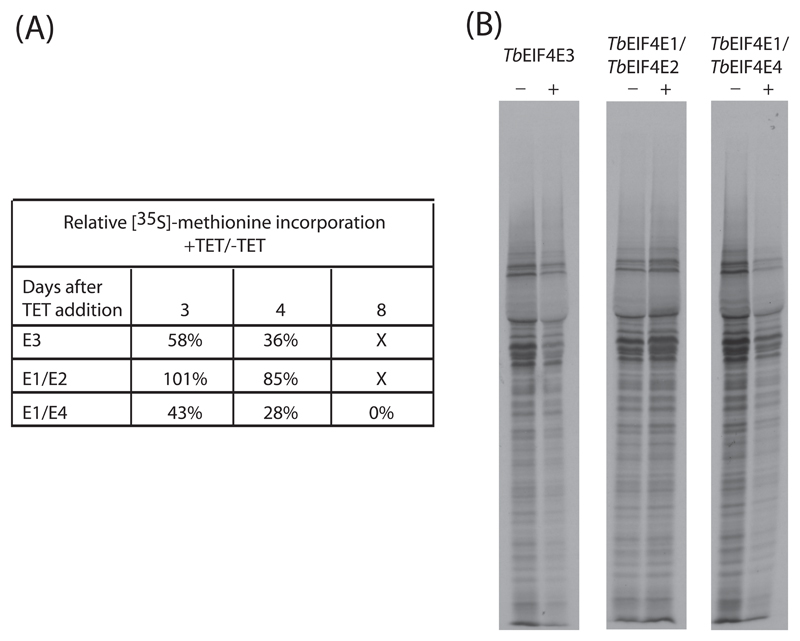
To investigate if the no-growth or cellular death phenotypes observed after the knock-down of TbEIF4E3 or the double constructs TbEIF4E1/2 and TbEIF4E1/4 was due to an inhibition of protein synthesis or to some unrelated process, we performed metabolic labeling with 35S-methionine at selected time points after tetracycline addition for the respective RNAi curves. Incorporation of labeled methionine was monitored and the results obtained were compared with those derived from the control curves without tetracycline. These values were used to calculate the total rate of protein synthesis between the two curves (plus and minus tetracycline) and estimate the effect on protein synthesis of the knockdown of the various proteins. As shown in Figure 7A at three days after RNAi induction, no effect on protein synthesis is observed for the TbEIF4E1/2 knockdown whilst depletion of TbEIF4E3 and TbEIF4E1/4 had significantly impaired translation. These results are consistent with both TbEIF4E3 and TbEIF4E1/4 knockdowns directly affecting protein synthesis since substantial translation inhibition preceded cell death for a minimum of three days for the TbEIF4E3 RNAi or for much longer in the case of the TbEIF4E1/4 knockdown. Nevertheless, in both cases no substantial changes in the profile of proteins synthesized were detected even in conditions of severe protein inhibition (Figure 7B). In contrast, for the TbEIF4E1/2 knockdown any inhibition of translation is only observed late in the RNAi curve, very near to the time of cell death. Although this does not rule out a direct role in protein synthesis as well, the inhibition observed could more likely be an indirect consequence of the various metabolic processes associated with cell death induced by the RNAi.
Fig. 7.
Metabolic labelling of procyclic cells from select RNAi curves. (A) Total protein synthesis was estimated after RNAi for TbEIF4E3 (E3) and the double constructs TbEIF4E1/2 (E1/E2) and TbEIF4E1/4 (E1/E4). At selected time points (3 and 4 days for the first two samples and 3, 4 and 8 days for TbEIF4E1/4), aliquots of the cells were incubated in the presence of 35S methionine followed by TCA precipitation and quantitation of the incorporated radioactivity. To estimate the efficiency of translation in the RNAi treated cells in comparison with the minus tetracycline controls, the normalized incorporation values after two hours of incubation were used to calculate the ratios shown in the figure. (B) For a qualitative analysis of the proteins being synthesized, aliquots of 2x106 cells derived from an independent labeling experiment (4 days after tetracycline addition) were ran on an SDS-PAGE gel followed by autoradiography.
3.7. TbEIF4E3 interacts with TbEIF4G3 and 4, while TbEIF4E4 binds TbEIF4G3
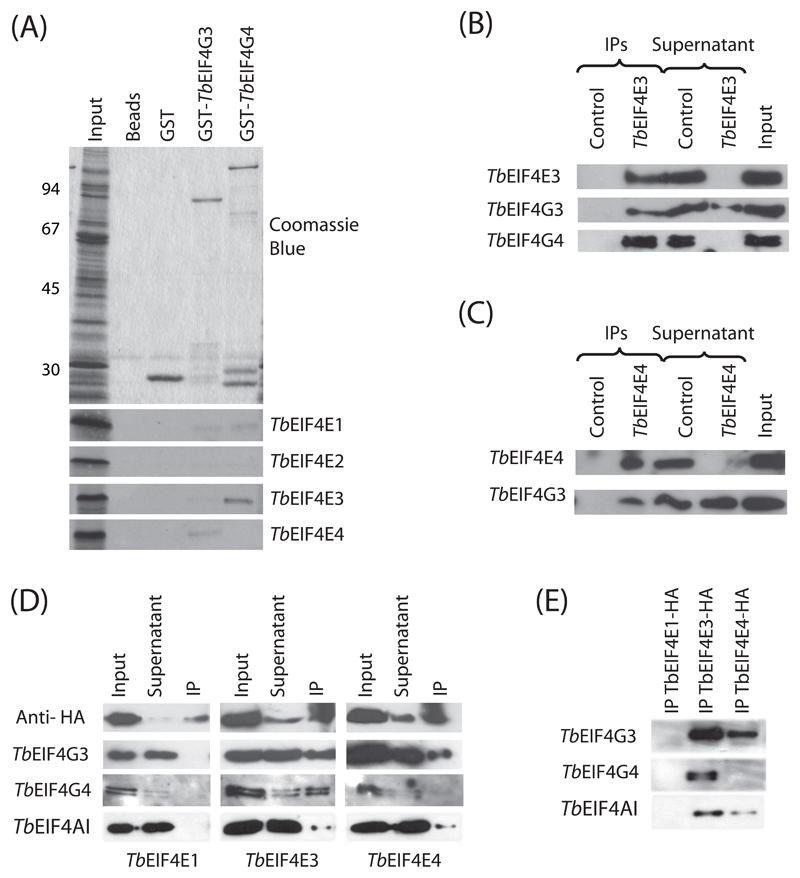
Since eIF4E participates in translation within the context of the eIF4F complex, bound to the eIF4G subunit, to investigate more directly the role of the four T. brucei eIF4Es in translation we opted to study their interaction with selected trypanosome eIF4G homologues. Of the five eIF4G homologues conserved in trypanosomatid species, only two (EIF4G3 and 4) share conserved features outside the HEAT domain, such as a short N-terminus, containing limited segments of similarity, and regions of homology on the C-terminal half. Furthermore, the L. major EIF4G3 orthologue has been shown to be able to bind to EIF4AI [33] as well as to orthologues of TbEIF4E1 and 4 [43], therefore confirming a potential role in translation. Here, to investigate possible interactions between the eIF4G and eIF4E homologues in vitro, the genes encoding TbEIF4G3 and 4 were cloned and expressed as N-terminal fusions with glutathione-S sepharose (GST). The resulting proteins, and GST alone used as negative control, were used in pull-down assays to analyze binding to the four [35S]-labeled T. brucei eIF4E homologues. GST-TbEIF4G3 specifically and efficiently bound to labeled TbEIF4E4, whilst GST-TbEIF4G4 bound to TbEIF4E3 (Figure 8A). Minor bindings were also observed for both TbEIF4E1 and 2 with the two eIF4G homologues and between TbEIF4E3 and GST-TbEIF4G3.
Fig. 8.
Analysis of the interactions between TbEIF4E3/TbEIF4E4 and TbEIF4G3/TbEIF4G4. (A) Pull-down assay using GST-tagged TbEIF4G3 and 4 and 35S-labelled TbEIF4E1 through TbEIF4E4. As controls GST on its own or only the glutathione sepharose beads were tested for their ability to bind the labeled protein. Upper panel: Coomassie Blue stained gel showing total translation extract (Input) as well as recombinant GST or GST- TbEIF4G3 and 4. Bottom panels: autoradiographies showing strong and specific binding between GST-TbEIF4G3 and labeled TbEIF4E4 and between GST-TbEIF4G4 and labeled TbEIF4E3. (B) Immunoprecipitation (IP) assay confirming interactions between TbEIF4E3 and TbEIF4G4 and between TbEIF4E3 and TbEIF4G3 in vivo. T. brucei cytoplasmic extracts were immunoprecipitated with the anti-TbEIF4E3 affinity purified antibodies (2 μg) or 5 μl of the pre-immune total serum as control (Control). Precipitated immunocomplexes were then used in Western blot assays with the TbEIF4E3, TbEIF4G3 or TbEIF4G4 antisera. In the same blots, an aliquot of the non-bound supernatant (Supernatant) was compared with an equivalent aliquot of the cytoplasmic extract used in the IP reaction and blot (Input). Equivalent aliquot means that if 20% of an IP was blotted (the real % values varied according to the protein assayed), then 20% of the same volume of the original cytoplasmic extract used for the IP, or its supernatant, was blotted as well. (C) Assay confirming the interaction between TbEIF4E4 and TbEIF4G3 in vivo. Immunoprecipitation was carried out as described in B with anti-TbEIF4E4 antibodies followed by Western-blots with the TbEIF4E4 and TbEIF4G3 antisera. (D) IP of HA-tagged eIF4E homologues. Cytoplasmic extracts from transfected T. brucei cells expressing HA-tagged TbEIF4E1, 3 and 4 (left, middle and right panels, respectively) were incubated with anti-HA beads and the precipitated samples, as well as equivalent aliquots of the non-bound supernatant and the input extract, were assayed for the presence of the HA-tagged proteins, TbEIF4G3 and 4 and TbEIF4AI. (E) To highlight the results from the IP lanes shown in D, these were ran on another gel and blotted again with the TbEIF4G3 and 4 and TbEIF4AI antisera, confirming the potential for both TbEIF4E3 and 4 to reconstitute functional eIF4F complexes.
To investigate if the same interactions occurred in vivo we used TbEIF4E3 and 4 antibodies in immunoprecipitation reactions with T. brucei cytoplasmic extracts. Immunoprecipitation of TbEIF4E3 was first carried out, with its presence in the immunoprecipitate as well as its depletion from the non-bound supernatant assessed by western- blotting with the anti-TbEIF4E3 antibody (Figure 8B). The same blot was then re-probed with antibodies raised against TbEIF4G3 and 4. All TbEIF4G4, and to a lesser extent TbEIF4G3, co-precipitated with TbEIF4E3. Immunoprecipitation of TbEIF4E4 was performed likewise followed by western-blotting with its own antibody as well as the one directed against TbEIF4G3. All TbEIF4E4 was brought down by the procedure and some, but not all, TbEIF4G3 co-precipitated as well (Figure 8C). To compare the binding of TbEIF4E1, 3 and 4 with the same eIF4G homologues in vivo under equivalent conditions, we expressed HA-tagged versions of these proteins in T. brucei cells followed by immunoprecipitation of the resulting cellular extracts with anti-HA antibody and western blotting (Figure 8D and E). In agreement with the pull down data above, TbEIF4G3 co-precipitated with both HA-tagged TbEIF4E3 and 4, but no co-precipitation with TbEIF4E1 was observed. In contrast, TbEIF4G4 only co-precipitated with TbEIF4E3. To confirm that functional eIF4F complexes were being formed, the blots were also incubated with anti-TbEIF4AI anti-sera. Both TbEIF4E3 and 4 were able to bring down the eIF4AI homologue, but no interaction was observed with TbEIF4E1 (Figure 8D and E). These binding results then confirmed specific interactions of TbEIF4E3 with both TbEIF4G3 and 4 and of TbEIF4E4 with TbEIF4G3, and highlight their potential to reconstitute three functional trypanosomatid eIF4F complexes.
4. Discussion
In cells with multiple eIF4E homologues, it has been proposed that only one acts as a general initiation factor functioning during the translation of the majority of the mRNAs, whilst the remainder perform more specialized functions, related or not, to protein synthesis [27, 44]. Most of the evidence for this proposal comes from multicellular organisms whereas the need for distinctions in the translation apparatus between different tissues or during development may be the driving force behind the evolution of various eIF4E homologues. In unicellular eukaryotes this may not apply although one initial aspect investigated here was the possibility that different eIF4E homologues would be differentially expressed during distinct life stages of the trypanosomatid life cycle, as indeed has been proposed for Leishmania EIF4E1 [34]. Overall the data presented rule out large differences in expression for the four eIF4E homologues between the two stages of the T. brucei cycle, consistent with the different homologues acting on different aspects of mRNA metabolism or on different mRNA populations, whilst reinforcing differences in abundance previously observed for the L. major proteins, with EIF4E3, and also EIF4E4, being present at significantly higher levels than EIF4E1 and 2. The distinct RNAi phenotypes in procyclic and bloodstream cells might suggest differences in the translation machinery, but the differential sensibilities of the two stages to the knockdowns could also be a consequence of their different metabolic rates, with bloodstream cells, which grow much faster, being most sensitive to changes in translation rate.
The localization data suggests that TbEIF4E3 and 4 do not function in the nucleus though TbEIF4E1 and 2 may have nuclear roles. In mammals, at least, nuclear eIF4E localize to speckles which are also the sites where splicing factors are found [45]. This localization pattern differs from what was seen for the endogenous or the over-expressed TbEIF4E1 and 2 which are spread throughout the nucleoplasm. The RNAi data does implicate both TbEIF4E1 and 2 as having important and possibly redundant roles for cellular survival, which could be associated with their nuclear localization, although the possibility remains that the two proteins participate in independent processes which, upon simultaneous inhibition by the double RNAi procedures, result in sufficient overall damage to produce the phenotype observed (the same applies to the TbEIF4E1/4 knockdown discussed below). So far EIF4E2 seems to be the least conserved of the four trypanosomatid eIF4E homologues studied. Although TbEIF4E2 binds to the m7G Sepharose in a similar manner to TbEIF4E1 and 4, Leishmania LmEIF4E2 does not bind this cap [33], but rather, preferentially binds the methylated cap 4 [34]. This difference, plus the existence of insertions in the Leishmania EIF4E2 which are missing from its T. brucei or T. cruzi orthologues, or even the human and yeast sequences, imply a divergence in function unique to the Leishmania protein.
In L. major, both EIF4E1 and 4 bind the cap4 structure, physically interact with EIF4G3 and all co-migrate with polysomes in sucrose gradients, strongly implicating these proteins in translation [34, 43]. Leishmania EIF4G3 also binds efficiently to the eIF4A homologue involved in translation [33, 35] and T. brucei EIF4G3 acts in protein synthesis, since its depletion rapidly blocks translation (D. Moura, unpublished data). Both TbEIF4E1 and 4 are required for growth of the bloodstream cells although the lack of a phenotype after knockdown in procyclics does not rule out a minor requirement for them in levels below the threshold produced after RNAi (considering the efficiency of the RNAi procedure, any requirements are nevertheless well below their original intracellular levels). Here we have confirmed, by both in vitro and in vivo approaches, the interaction between the EIF4G3 and EIF4E4 in T. brucei which clearly implicates the latter in protein synthesis. However, contrary to a previously reported interaction [43], neither in vitro (with L. major and T. brucei) nor in vivo (T. brucei) assays detected any interaction between EIF4G3 and EIF4E1 (this work and unpublished data). This discrepancy remains unresolved but it is nevertheless possible that in Figure 8D minor amounts of TbEIF4G3 co-precipitated with HA-tagged TbEIF4E1 but was not detected through the western-blotting procedure.
The similarities observed between TbEIF4E3 and 4 at the sequence level, their similar subcellular localization, abundance and their ability to bind to eIF4G partners are consistent with both performing related roles as part of distinct eIF4F complexes. So far, the existence of multiple eIF4F complexes, with distinct eIF4E and eIF4G subunits and within the same organism, has only been reported from plants where they were named eIF4F and eIF(iso)4F [46]. These two complexes can co-exist and differ in their ability to translate different classes of mRNAs [47, 48], raising the possibility that something similar may be happening with the trypanosomatid proteins. A potential role for phosphorylation in regulating their activity is also possible since TbEIF4E3 has been identified as a phosphoprotein [49] and TbEIF4E4 migrates in gels with a pattern indicative of post-translational modifications. Indeed the phosphorylated residues in TbEIF4E3 are located next to one of the two blocs of conserved residues unique to the N-terminus of both EIF4E3 and 4 sequences (see Figure 1).
EIF4E3, the most abundant Trypanosoma and Leishmania eIF4E, is the only confirmed essential homologue in procyclic and bloodstream T. brucei. Nevertheless, when compared with EIF4E4 in L. major, it binds much less efficiently to the trypanosomatid cap4 [34]. Moreover, L. major EIF4E3 does not seem to associate with polysomes [34], although this does not necessarily exclude a role in translation since the majority of mammalian eIF4E is not bound to ribosomes either [50]. To date, eIF4F-like complexes participate only in translation, so it seems plausible to suggest then that both EIF4E3 and 4 participate in protein synthesis; however their distinct cap binding affinities may indicate distinct modes of mRNA recognition leading to binding to different target mRNAs. It is possible that EIF4E3 may only bind to mRNAs in the context of an eIF4F complex or when associated with some other RNA binding protein. In metazoans the eIF4E-2 homologue (4EHP), involved in the selective translation repression of target messages, is recruited to selected mRNAs by binding proteins associated with their 3’UTRs [28, 29, 51]. This protein binds very weakly to the cap [52], so it cannot compete with eIF4E-1 in the absence of its co-factors. It is plausible to suggest then that, despite its abundance, for the parasite EIF4E3 to bind to the message RNAs it needs some other protein partner which can mediate selectivity in mRNA binding with a direct consequence on translation.
Supplementary Material
Acknowledgements
This work was partially funded with grants provided from the Brazilian funding agencies FACEPE (006/2003-CNPq/PPP) and CNPq (475471/2008-3 and 476255/2004-0). ERF, RD, DMNM, TDCL, RPL and CRSR received studentships from CAPES and FACEPE. Work in MC's lab is funded by the Wellcome Trust. Work in NS's lab is funded by the Wellcome Trust and the BBSRC. Technical assistance to grow the Leishmania and Trypanosoma cells was provided by B. S. Lima. We would like to thank C. Docena and the PDTIS-FIOCRUZ Program for Technological Development in Tools for Health for access and help with the confocal microscopy and sequencing.
References
- [1].Johnson PJ, Kooter JM, Borst P. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–81. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- [2].Martinez-Calvillo S, Yan S, Nguyen D, et al. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11:1291–9. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- [3].Liang XH, Haritan A, Uliel S, et al. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–40. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bangs JD, Crain PF, Hashizume T, et al. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem. 1992;267:9805–15. [PubMed] [Google Scholar]
- [5].Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–8. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- [7].Haile S, Papadopoulou B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr Opin Microbiol. 2007;10:569–77. doi: 10.1016/j.mib.2007.10.001. [DOI] [PubMed] [Google Scholar]
- [8].Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- [9].von der Haar T, Gross JD, Wagner G, et al. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol. 2004;11:503–11. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- [10].Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle. 2007;6:65–9. doi: 10.4161/cc.6.1.3688. [DOI] [PubMed] [Google Scholar]
- [11].Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86:178–83. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- [12].Goodfellow IG, Roberts LO. Eukaryotic initiation factor 4E. Int J Biochem Cell Biol. 2008;40:2675–80. doi: 10.1016/j.biocel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- [14].Hershey JWB, Merrick WC. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- [15].Preiss T, Hentze MW. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–11. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- [16].Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Strudwick S, Borden KL. The emerging roles of translation factor eIF4E in the nucleus. Differentiation. 2002;70:10–22. doi: 10.1046/j.1432-0436.2002.700102.x. [DOI] [PubMed] [Google Scholar]
- [18].Culjkovic B, Topisirovic I, Skrabanek L, et al. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–26. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Topisirovic I, Siddiqui N, Lapointe VL, et al. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009;28:1087–98. doi: 10.1038/emboj.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Marcotrigiano J, Gingras AC, Sonenberg N, et al. Cocrystal structure of the messenger RNA 5' cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–61. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- [21].Matsuo H, Li H, McGuire AM, et al. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–24. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- [22].Sonenberg N, Gingras AC. The mRNA 5' cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–75. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- [23].Raught B, Gingras AC. eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol. 1999;31:43–57. doi: 10.1016/s1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- [24].Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269:5350–9. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lasko P, Sonenberg N. Coordinated transcriptional and translational control in metabolic homeostasis in flies. Genes Dev. 2007;21:235–7. doi: 10.1101/gad.1524707. [DOI] [PubMed] [Google Scholar]
- [26].Altmann M, Handschin C, Trachsel H. mRNA cap-binding protein: cloning of the gene encoding protein synthesis initiation factor eIF-4E from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:998–1003. doi: 10.1128/mcb.7.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rhoads RE. eIF4E: new family members, new binding partners, new roles. J Biol Chem. 2009;284:16711–5. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lasko P, Cho P, Poulin F, et al. Contrasting mechanisms of regulating translation of specific Drosophila germline mRNAs at the level of 5'-cap structure binding. Biochem Soc Trans. 2005;33:1544–6. doi: 10.1042/BST0331544. [DOI] [PubMed] [Google Scholar]
- [29].Cho PF, Gamberi C, Cho-Park YA, et al. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16:2035–41. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Joshi B, Cameron A, Jagus R. Characterization of mammalian eIF4E-family members. Eur J Biochem. 2004;271:2189–203. doi: 10.1111/j.1432-1033.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- [31].Joshi B, Lee K, Maeder DL, et al. Phylogenetic analysis of eIF4E-family members. BMC Evol Biol. 2005;5:48. doi: 10.1186/1471-2148-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yoffe Y, Zuberek J, Lewdorowicz M, et al. Cap-binding activity of an eIF4E homolog from Leishmania. RNA. 2004;10:1764–75. doi: 10.1261/rna.7520404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dhalia R, Reis CR, Freire ER, et al. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol Biochem Parasitol. 2005;140:23–41. doi: 10.1016/j.molbiopara.2004.12.001. [DOI] [PubMed] [Google Scholar]
- [34].Yoffe Y, Zuberek J, Lerer A, et al. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot Cell. 2006;5:1969–79. doi: 10.1128/EC.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dhalia R, Marinsek N, Reis CR, et al. The two eIF4A helicases in Trypanosoma brucei are functionally distinct. Nucleic Acids Res. 2006;34:2495–507. doi: 10.1093/nar/gkl290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kelly S, Reed J, Kramer S, et al. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol. 2007;154:103–9. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125:211–6. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- [38].Wirtz E, Leal S, Ochatt C, et al. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- [39].Carrington M, Roditi I, Williams RO. The structure and transcription of an element interspersed between tandem arrays of mini-exon donor RNA genes in Trypanosoma brucei. Nucleic Acids Res. 1987;15:10179–98. doi: 10.1093/nar/15.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Webb H, Burns R, Ellis L, et al. Developmentally regulated instability of the GPI-PLC mRNA is dependent on a short-lived protein factor. Nucleic Acids Res. 2005;33:1503–12. doi: 10.1093/nar/gki298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Marcotrigiano J, Gingras AC, Sonenberg N, et al. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–16. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- [42].Kramer S, Queiroz R, Ellis L, et al. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci. 2008;121:3002–14. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yoffe Y, Leger M, Zinoviev A, et al. Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions. Nucleic Acids Res. 2009;37:3243–53. doi: 10.1093/nar/gkp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hernandez G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech Dev. 2005;122:865–76. doi: 10.1016/j.mod.2005.04.002. [DOI] [PubMed] [Google Scholar]
- [45].Dostie J, Lejbkowicz F, Sonenberg N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J Cell Biol. 2000;148:239–47. doi: 10.1083/jcb.148.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Browning KS, Webster C, Roberts JK, et al. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J Biol Chem. 1992;267:10096–100. [PubMed] [Google Scholar]
- [47].Gallie DR, Browning KS. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem. 2001;276:36951–60. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- [48].Mayberry LK, Allen ML, Dennis MD, et al. Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs. Plant Physiol. 2009;150:1844–54. doi: 10.1104/pp.109.138438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nett IR, Martin DM, Miranda-Saavedra D, et al. The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol Cell Proteomics. 2009;8:1527–38. doi: 10.1074/mcp.M800556-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rau M, Ohlmann T, Morley SJ, et al. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996;271:8983–90. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- [51].Cho PF, Poulin F, Cho-Park YA, et al. A new paradigm for translational control: inhibition via 5'-3' mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–23. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- [52].Zuberek J, Kubacka D, Jablonowska A, et al. Weak binding affinity of human 4EHP for mRNA cap analogs. RNA. 2007;13:691–7. doi: 10.1261/rna.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.