Abstract
Objective:
We study the performance of machine learning (ML) methods, including neural networks (NNs), to extract mutational test results from pathology reports collected by cancer registries. Given the lack of hand-labeled datasets for mutational test result extraction, we focus on the particular use-case of extracting Epidermal Growth Factor Receptor mutation results in non-small cell lung cancers. We explore the generalization of NNs across different registries where our goals are two-fold: (1) to assess how well models trained on a registry’s data port to test data from a different registry and (2) to assess whether and to what extent such models can be improved using state-of-the-art neural domain adaptation techniques under different assumptions about what is available (labeled vs unlabeled data) at the target registry site.
Materials and Methods:
We collected data from two registries: the Kentucky Cancer Registry (KCR) and the Fred Hutchinson Cancer Research Center (FH) Cancer Surveillance System. We combine NNs with adversarial domain adaptation to improve cross-registry performance. We compare to other classifiers in the standard supervised classification, unsupervised domain adaptation, and supervised domain adaptation scenarios.
Results:
The performance of ML methods varied between registries. To extract positive results, the basic convolutional neural network (CNN) had an F1 of 71.5% on the KCR dataset and 95.7% on the FH dataset. For the KCR dataset, the CNN F1 results were low when trained on FH data (Positive F1: 23%). Using our proposed adversarial CNN, we match the F1 of the models trained directly on each registry’s data. The adversarial CNN F1 improved when trained on FH and applied to KCR dataset (Positive F1: 70.8%). We found similar performance improvements when we trained on KCR and tested on FH reports (Positive F1: 45% to 96%).
Conclusion:
Adversarial domain adaptation improves the performance of NNs applied to pathology reports. In the unsupervised domain adaptation setting, we match the performance of models that are trained directly on target registry's data by using source registry's labeled data and unlabeled examples from the target registry.
Keywords: Natural Language Processing, Cancer Registry, Domain Adaptation, Neural Networks, Text Classification, Text Mining
Graphical Abstract
Convolutional neural networks are used to extract EGFR results from pathology reports Classifiers trained on local data do not perform well on reports from different registries Adversarial domain adaptation improves cross-registry generalization Using data from multiple registries with adversarial learning improves prediction
1. Introduction
Population-based cancer registries are the most valid source for measuring the incidence of cancer in a population. Registry data are essential to guide and evaluate evidence-based cancer prevention and control activities, including playing an increasingly important role in rapidly identifying patient cohorts and biospecimen cohorts across the spectrum of basic, clinical and population-based translational science. The Surveillance, Epidemiology, and End Results (SEER) program, sponsored by the National Cancer Institute (NCI) and the National Program of Cancer Registries, reports population-based cancer statistics for the United States. For example, SEER registries manually assign International Classification of Disease for Oncology Version 3 (ICD-O-3) codes based on pathology reports to designate the site (topography) and histology and behavior code (morphology) of neoplasms [24]. Although considerable amounts of information regarding cancer diagnoses is documented in pathology reports [1], much of it is in the form of unstructured text. Manually extracting all of the relevant information is costly due to the growing number of cancer cases and the increasing number of tumor characteristics that cancer registries are required to report.
Molecular testing is an important tool to identify personalized treatments for patients with actionable mutations, a crucial enabler of precision medicine. While mutational test results may be discussed in pathology reports, they are not coded in structured data sources collected by cancer registries. Therefore, SEER registries do not currently report results of mutation tests. Thus, automatically extracting mutational test results as disclosed in pathology reports offers an excellent opportunity to enable personalized treatments and clinical trial recruitment. To this end, the SEER program funded rapid response studies at the Kentucky Cancer Registry (KCR) and the Fred Hutchinson Cancer Research Center (FH) to develop automated methods to extract mutational test results for Epidermal Growth Factor Receptor (EGFR) and Anaplastic lymphoma kinase (ALK) in non-small cell lung cancers (NSCLCs). NSCLC patients with EGFR mutations are often candidates for targeted therapy directed at their mutation and have longer survival than with chemotherapy [17, 5]. While the pathology reports collected by different cancer registries may follow annotation standards established by the College of American Pathologists, they may differ with respect to writing styles, jargon and document formatting such that simple machine learning tools, or rule-based systems, are ineffective at extracting the relevant information with high recall and accuracy.
In this paper, we present generalization techniques to extract EGFR test results from unstructured text in pathology reports of NSCLC patients using hand-labeled datasets created by the SEER sponsored studies1 at KCR and FH. This information will provide cancer registries with an efficient tool to rapidly report genetic biomarkers that carry a relevant clinical implication in non-squamous NSCLC for the selection of candidates for effective oral targeted therapies. SEER registries currently lack the capacity of reporting genetic testing results. The use of validated tools to ascertain EGFR tests will potentially assist SEER registries to provide modern, updated clinical, and genomic information in nationally representative population samples of cancer patients. The tools will enable population health researchers to conduct epidemiological and outcomes research in clinically relevant, molecularly defined subgroups of NSCLC patients.
1.1. Biomedical Text Classification
Extracting information from biomedical documents has been studied for a wide variety of problems. Methods have been developed to extract diagnosis and procedure codes (ICD-9-CM) from electronic medical records [21, 13, 28]. ICD-9-CM codes are used by all healthcare facilities to standardize diagnosis reporting for billing purposes. Similarly, there are machine learning methods that extract ICD-O-3 codes from pathology reports [26].
Biomedical text classification methods generally fall into one of two groups: linear or neural network models. Tsoumakas et al. (2013) trained nearly 27 thousand Linear SVM models, one for each MeSH term [31]. For medical coding, Perotte et al. (2013) developed a hierarchical SVM to extract diagnosis and procedure codes from electronic medical records [25]. Goulart et al. [8] show that simple rules can be combined with an SVM model to extract EGFR and ALK results from pathology reports. Compared to their effort, our work differs in two major ways. First, we explore neural network-based methods to extract EGFR results – not only linear models. As we show in the discussion section of this paper, simple rules do not work well on pathology reports collected at all registries. Second, our work focuses on cross-registry performance. We test whether and to what extent machine learning methods generalize across different cancer registries.
Deep neural networks have been advancing state-of-the-art results across a wide range of biomedical tasks including bioinformatics [14, 16] and healthcare [19]. For text classification, Mullenbach et al. (2018) introduced a label-wise attention mechanism for medical coding [21]. In Rios and Kavuluru (2018), we introduced a matching network-based method – originally developed for few-shot learning – that further improved medical coding results [28]. Similar to this paper, Qiu et al. (2018) apply convolutional neural networks (CNNs) to pathology reports [26]. However, they focus on extracting topography and histology ICD-O-3 codes from the reports, not genetic testing results.
1.2. Generalization in Deep Learning
Generalization in deep learning has been studied theoretically [11] and empirically. At a high level, generalization of machine learning methods is important for many biomedical applications. Small and/or rural healthcare institutions and cancer registries may not have access to the data required to train neural networks or may not have the resources to annotate large amounts of data. If large institutions are able to share models and/or data with smaller institutions, then the smaller institution can dedicate resources to other tasks. With respect to cancer registries, if our models do not perform well on pathology reports collected by different registries, then our models have poor generalization. Developing methods that generalize across varying data distributions is known as domain adaptation. Both domain adaptation, and similar methods such as transfer learning, have been applied to medical documents [9, 32, 29]. There are two main domain adaptation settings studied by researchers: supervised and unsupervised. For both adaptation settings, we have access to two datasets — a source dataset and a target dataset. To illustrate, assume we have data from two cancer registries, C1 and C2. C2 (source) shares their data with C1 (target). C1 wants to use a classifier based on data from C2, however, their objective is to maximize the performance of the classifier on their own data. In the supervised adaptation setting, data from both registries (C1 and C2) have ground truth annotations. Multi-task learning [6] is a methodology in machine learning where multiple problems (tasks) are solved simultaneously. Similar to multi-task learning methodologies, models can be trained on both datasets simultaneously. For the unsupervised setting, only the source dataset (data from C2) is annotated. Therefore, the C1 registry must incorporate their unlabeled target data – similar to semi-supervised learning. Contrary to multi-task and semi-supervised learning, domain adaptation tries to make better use of the auxiliary data by matching the data distributions. For instance, if the C2 registry’s dataset differs substantially from C1’s data in terms of topic or style, then simply combining the two datasets could reduce the overall performance of the model [30].
There are several methods proposed for domain adaptation. Jiang and Zhai (2017) used an instance weighting-based approach for text [10]. They simply remove source instances that are significantly different from the target data. Daumé (2007) used a feature augmentation method where they have special features for the source data, the target data, and also shared feature representations for both the source and target data [4]. Ganin and Lempitsky (2015) developed an adversarial domain adaptation technique for neural networks [7]. Intuitively, they train a domain classifier that takes a mid-level CNN representation as input for each example, and predicts if the example comes from the source or target datasets. Using the domain classifier, the CNN parameters are modified to make the performance of domain classifier worse, thereby matching the data distributions. In Rios et al. (2018), for relation classification, we use a two stage approach for adversarial learning [30]. First, we train on the source data, then we fine-tune the model to match the source and target data distributions.
1.3. Domain Adaptation Scenarios in Cross-Registry Settings
In this paper, we present a neural network-based adversarial domain adaptation method to extract EGFR results from pathology reports. There are a few scenarios in which both supervised and unsupervised domain adaptation may arise for cancer registries. Therefore, our experiments investigate the following research questions (RQs):
RQ1. Can machine learning methods, including neural networks, accurately extract EGFR information from pathology reports? Before studying the various domain adaptation scenarios, we first examine the performance of machine learning methods when applied to the EGFR classification task. For this scenario, we assume each registry has access only to its datasets.
RQ2. Are EGFR test results described differently in pathology reports collected at different SEER registries? Will EGFR models trained on pathology reports from a single cancer registry generalize to other registries? In this setting, we analyze the cross-registry performance without any adaptation. For example, if a registry shares a pre-trained model, but did not give access to their data, then we want to examine how it will perform.
RQ3. Can unsupervised adversarial domain adaptation techniques improve the cross-registry performance of neural networks? This setting assumes a registry has access to another registry’s annotated data; however, we assume the registry’s own data is not annotated. If the registry does not have the resources to create a large high-quality annotated dataset, then this scenario can arise. We test if our unsupervised domain adaptation method can match the results produced when training on the source data directly.
RQ4. Do supervised domain adaptation methods improve the performance of models trained jointly on two registries’ labeled datasets? To answer this question, we assume that a registry has access to high quality annotated data from their registry and labeled data from another registry. We hypothesize that if we apply our supervised adversarial domain adaptation method, then we can improve on the performance of models simply trained on the combination (union) of both datasets.
Overall, to the best of our knowledge, this is the first effort to explore generalization of text classifiers across different registries. Furthermore, we evaluate the use of state-of-the-art techniques for domain adaptation on our novel task.
2. MATERIALS AND METHODS
RQ1 and RQ2 are based on conventional ML configurations, whose methodological aspects are well-known and hence are briefly mentioned in the Results section. In this section, we specifically discuss neural adversarial adaptation techniques to answer RQ3 and RQ4, the main contributions in this paper.
2.1. Datasets
This study is based on data from patients diagnosed with histologically confirmed, stage IV non-squamous NSCLC between 2011 and 2013, and reported to two SEER registries: KCR and the Cancer Surveillance System of FH. For each NSCLC case, staff at each registry retrieved electronic pathology reports, and following manual review labeled each as one of three categories: Unknown/Technical Difficulties, Positive, and Negative. The Unknown class is assigned to reports if it is not clear if the EGFR test was done or if the result cannot be ascertained properly; this class also represents instances where the test was done, but the result was not reported. If the test results are reported, then the positive and negative classes are used, respectively. Both datasets are annotated for EGFR mutation results and Anaplastic Lymphoma Kinase (ALK) fusion results. However, the KCR dataset only has 3 pathology reports that mention a positive ALK result. Therefore, for the purposes of this study, we focus on extracting EGFR test results because we have more positive examples in both datasets. The relative counts of different classes for both datasets are shown in Table 1.
Table 1:
Number of pathology reports for the three classes in the FH and KCR datasets.
| Cancer Registry | # Unknown | # Positive | # Negative |
|---|---|---|---|
| Fred Hutch. (FH) | 2921 | 232 | 1126 |
| KCR | 599 | 47 | 354 |
2.2. Method overview.
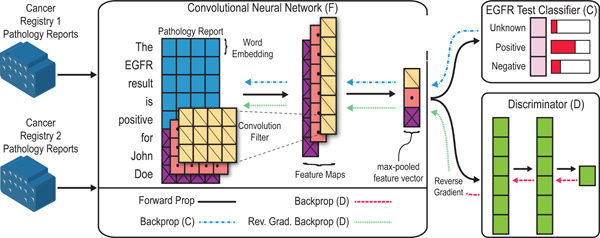
In Figure 1, we provide an overview of our method. Our model has three main components: the CNN (F), the classifier (C), and the discriminator (D). The discriminator is a multi-layer neural network where the final layer is a single sigmoid unit. During training, the discriminator is trained to learn if a given pathology report comes from the KCR or FH datasets. The CNN, F, is the adversary of the discriminator, D. Intuitively, the CNN parameters are updated to minimize the classification loss, while maximizing the error of the discriminator. The CNN and the discriminator compete during training, with the CNN eventually producing representations that are indistinguishable by the discriminator.
Figure 1:
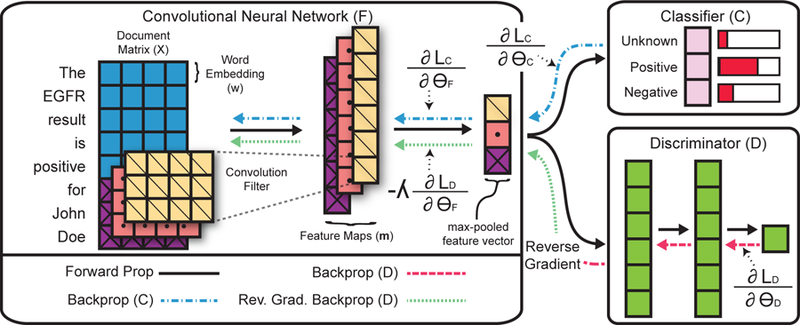
High-level overview of method for determining EGFR status of NSCLC cases from pathology reports. The architecture has three main components: a CNN (F), classifier (C), and discriminator (D). F returns a fixed size feature representation of a pathology report. C is a standard softmax output layer, and D is an MLP that predicts which registry a report originates. The reverse gradient changes the sign of the gradient, such that C is optimized to maximize the loss involving D, while D is optimized to minimize the loss (i.e., correctly predict the original registry).
2.3. Convolutional neural networks for text classification.
For the CNN component, we use a standard model from Kim (2014) [12]. Word vectors form the base element of the model. Given a pathology report, let represent the j-th word’s vector in the i-th document. As shown in Figure 1, each pathology report is represented as a matrix by concatenating the word vectors, where Ni is the number of words in the i-th document, and d is the size of the word vectors. Given the document matrix Xi, the CNN produces a fixed size feature representation. To produce a fixed size vector, the CNN uses max-over-time pooling. Each convolution filter q produces a feature map where c is the span of the convolution filter (the filters in Figure 1 span 3 words). To produce a fixed-sized vector with max-over-time pooling we take the max value for each feature map to represent a pathology report. We define the final output of the neural network as
where , s is the number of convolution filter sizes, and f is the number of filters per size.
2.4. Classification loss.
Extracting EGFR test results from pathology reports is a multi-class classification problem. Therefore, we pass the feature vector returned by F(Xi) to a fully-connected softmax layer
where and r is the number of classes. Next, we can train the classifier C(), and the CNN F(), using a multi-class cross-entropy loss
| (1) |
where is a binary indicator for the i-th pathology report’s j-th class and C()j is the prediction for the j-th class. S and T represent the index set of source and target instances. In the unsupervised domain adaptation setting, we only include S. Likewise, T is only used when training on the target dataset – the supervised classification setting. The combination of F() and C() form the CNN model introduced by Kim (2014) and has been shown to work well on a wide variety of biomedical text classification tasks [2, 27].
2.5. Adversarial Loss.
We are interested in training a model to extract EGFR test results from pathology reports. We want the model to generalize across datasets collected by different cancer registries. However, the pathology reports collected by separate cancer registries may differ in terms of writing style, format, and jargon. Some registries may collect reports that are thorough and document everything, while others may only have high-level summaries. Furthermore, EGFR testing may be outsourced by the reporting lab with results incorporated as addenda in a variety of formats across different labs. To overcome these issues we combine the CNN with an adversarial domain adaptation method.
First, we define the discriminator for report Xi as
where MLP(F(Xi)) is a multi-layer feed-forward neural network with a single sigmoid unit for the final layer. The discriminator is trained using a binary cross-entropy loss
| (2) |
where θF represents the parameters of the CNN, θD represents the parameters of the discriminator, and represents the expected value of the loss over different input instances. For example, represents the expectation over source instances. Intuitively, the loss is minimized (gradient descent) with respect to θD such that the discriminator is trained to predict which cancer registry each pathology report is from. The CNN weights, θF, are updated to confuse the discriminator by maximizing the loss (gradient ascent), making it hard for the discriminator to distinguish the CNN feature representations with respect to different cancer registries.
For gradient-based training, we use a gradient reversal layer (GRL) which takes a gradient as input and reverses the gradient’s sign [7]. As shown in Figure 1, we apply the GRL between the CNN and discriminator. Formally, this GRL is defined as
where is the gradient of the adversarial loss with respect to the CNN parameters. λ weights the intensity of the GRL. A large λ will encourage larger changes of the CNN, making the CNN a stronger adversary for the discriminator. Following Ganin and Lempitsky (2015), instead of using a static λ value, we modify its value over the course of training as
where p ∈ [0, 1] measures the training progress and is a hyperparameter. Following prior work, we set γ to 10 [7]. Likewise, after each epoch, we linearly increase p from 0 to 1 by increments of size 1/#epochs. By starting with a λ of 0, the CNN parameters are not initially affected by the discriminator. At the early stages of training, the CNN is mostly affected by the classification loss. Therefore, in the beginning, we control the noisy signal of the discriminator.
2.6. Training
For both the supervised and unsupervised domain adaptation scenarios, both the classification loss from Equation 1 and the adversarial loss from Equation 2 are combined as
such that the CNN, classifier, and discriminator parameters are learned jointly. In the base scenario, where we only have access to the labeled target dataset, is ignored. However, depending on the training scenario, the training process will slightly differ. Both loss functions are used in the unsupervised domain adaptation setting, with the exception that only the annotated source examples are used to train θF with . Finally, for the supervised domain adaptation scenario, the annotated source and target examples are used by .
3. RESULTS
3.1. Evaluation Method and Baselines
For this study, we use nested 5-fold cross-validation, where the inner-loop is used to pick the best hyperparameters [3]. Here by inner-loop we refer to the second level of cross-validation that lets us select potentially different sets of hyperparameters for each test fold in the main cross-validation setup. It should be noted that the neural network-based methods can vary run-to-run – especially for the positive class which has a small number of positive examples. Therefore, for each neural network-based model, and for each cross-validation fold, we train 5 models using a different random seed and we report the average of the 5 runs. When two datasets are used in a supervised and unsupervised domain adaptation scenario, for each fold of the target dataset, we append the entire source dataset to the target training split. For example, in the “FH + KCR” section of Table 2, we perform cross validation on the KCR dataset and we append the entire FH dataset to the training KCR fold. The testing fold will only contain KCR examples.
Table 2:
Model performances on the KCR dataset. Precision (P), Recall (R), and F1-Score (F1) are reported for the four major classes: Unknown, Positive, Negative, and Known. Note that the models were not trained on the Known class. The Known class metrics were calculated by merging the metrics for the Positive and Negative classes.
| Unknown |
Positive |
Negative |
Known |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| Rule-based | 0.620 | 0.881 | 0.728 | 0.200 | 0.064 | 0.097 | 0.524 | 0.203 | 0.292 | 0.524 | 0.196 | 0.284 | |
| KCR Only | SVM | 0.945 | 0.951 | 0.948 | 0.612 | 0.476 | 0.530 | 0.881 | 0.896 | 0.888 | 0.927 | 0.918 | 0.922 |
| BSVM | 0.934 | 0.958 | 0.946 | 0.767 | 0.329 | 0.446 | 0.893 | 0.867 | 0.879 | 0.937 | 0.898 | 0.916 | |
| CNN | 0.973 | 0.977 | 0.975 | 0.858 | 0.626 | 0.715 | 0.923 | 0.948 | 0.935 | 0.966 | 0.960 | 0.963 | |
| BioBERT | 0.937 | 0.933 | 0.935 | 0.826 | 0.791 | 0.804 | 0.884 | 0.895 | 0.889 | 0.901 | 0.907 | 0.904 | |
| FH + KCR | SVM | 0.956 | 0.946 | 0.951 | 0.761 | 0.540 | 0.612 | 0.901 | 0.913 | 0.906 | 0.935 | 0.918 | 0.926 |
| BSVM | 0.966 | 0.972 | 0.968 | 0.732 | 0.518 | 0.595 | 0.906 | 0.924 | 0.914 | 0.959 | 0.948 | 0.953 | |
| CNN | 0.980 | 0.969 | 0.974 | 0.885 | 0.667 | 0.746 | 0.925 | 0.968 | 0.945 | 0.956 | 0.970 | 0.962 | |
| CNN + Adv. | 0.973 | 0.980 | 0.976 | 0.899 | 0.729 | 0.801 | 0.941 | 0.952 | 0.946 | 0.970 | 0.959 | 0.964 | |
| FH ⇒ KCR | SVM | 0.848 | 0.820 | 0.832 | 0.100 | 0.147 | 0.118 | 0.742 | 0.749 | 0.742 | 0.746 | 0.779 | 0.760 |
| BSVM | 0.804 | 0.855 | 0.828 | 0.259 | 0.249 | 0.253 | 0.691 | 0.647 | 0.668 | 0.760 | 0.689 | 0.723 | |
| CNN | 0.867 | 0.888 | 0.875 | 0.273 | 0.230 | 0.244 | 0.741 | 0.737 | 0.732 | 0.832 | 0.791 | 0.805 | |
| CNN + Adv. | 0.965 | 0.979 | 0.972 | 0.900 | 0.708 | 0.780 | 0.938 | 0.938 | 0.938 | 0.969 | 0.948 | 0.958 | |
We report the precision (P), recall (R), and F1-Score (F1) for the unknown, positive, and negative classes, respectively. We also combine the Positive and Negative classes into a single class at test time. This combined class measures how well our models can predict if the results are stated in the report, even though we may predict the wrong result. We refer to this as the “known” class. Meaning, if we predict either Positive or Negative, we change the prediction to “known”.
For evaluation purposes, we experiment with 4 models: 2 linear models and 2 neural network-based models. We briefly describe the models below:
Rule-based – We experiment with a simple rule-based method that uses the following regular expressions: “positive (\w+\s){1,7}egfr”, “egfr[ :](\w+\s){1,7}positive”, and 2 similar regular expressions where the word “positive” is replaced with “negative”. If one of the regular expressions is matched, then we predict the class it represents, otherwise we return “Unknown”. For example, the string “the patient tested positive for an egfr mutation” matches the regular expressions we created. At a high level, this regular expression matches all strings that contain the word “positive” followed by, or preceded by, “egfr”; and the string “egfr” must be no more than seven words away from “positive”.
SVM – This model uses TF-IDF weighted ngrams as features to train a linear SVM. The model is trained using Scikit-Learn’s Linear SVC method [23]. We grid-search over the C regularization parameters [1e-4, 1e-3, 1e-2, 1e-1, 1., 10], the set of class weight options (“None” and “Balanced” in scikit-learn), and the combination of unigrams, bigrams, trigrams, and 4-grams.
BSVM – Similar to the SVM method, this model uses ngrams as features and we grid-search over the same parameters. However, instead of using a TF-IDF weighting scheme, we use a binary representation, 1 if a feature is present, and 0 otherwise.
BioBERT [15] – BioBERT is a method of pre-training neural networks. Specifically, BioBERT trains a general-purpose “language understanding” model on a total of 29 million PubMed citations. We fine-tune the parameters of BioBERT on our task.
CNN – A standard CNN model for text classification [12].
CNN + Adv. – The method proposed in this paper. As stated in the previous section, this method will vary slightly depending on the domain adaptation scenario, unsupervised or supervised.
For each CNN-based model, we use convolution filters that span 3, 4, and 5 words. We learn 300 filters for each size. We use dropout with a value of 0.5 and apply it after the CNN and before the classifier and discriminator. We also use L2 regularization over all CNN and classifier parameters with weight of 1e-3. For the discriminator, we use a 3-layer MLP, where the first two layers have 1024 hidden units with ReLU activation functions [22]. The final layer of the discriminator is a single output unit using a sigmoid activation function. Dropout is added between each layer of the discriminator with a value of 0.5. Furthermore, each CNN-based model is trained using the Adam optimizer with the learning rate 1e-3. Finally, we initialize the word embeddings with word2vec vectors trained on PubMed articles and abstracts. The word embedding size is 300.
With the exception of CNN Adv., each model is tested in three different scenarios:
“KCR/FH Only”, where we assume that each cancer registry only has access to its own data. Therefore, the “CNN + Adv.” method is not considered for this scenario.
“KCR + FH”, with which we test the supervised domain adaptation setting, where we assume we have access to ground truth annotations from both cancer registries regardless of which site is the target.
“KCR ⇒ FH”/“FH ⇒ KCR”, with which we assess the unsupervised domain adaptation setting, where we assume the target registry’s pathology reports have not been manually annotated. However, the registry has access to source registry’s annotated data. For this scenario, grid-search is performed on a subset of the source dataset’s training split because we assume that we do not have access to annotated target data.
3.2. Experiments
In this section, we address each of the research questions stated in Section 1.3.
RQ1. How difficult is it to extract EGFR test results for an unstructured pathology report? To answer this question, we analyze the results in the “KCR Only” and “FH Only” sections of Tables 2 and 3, respectively. For the KCR data, in Table 2, we find that the CNN outperforms both linear methods substantially across all four classes. For the Unknown class, the CNN outperforms the best linear method (SVM) by nearly 3%, from 0.948 to 0.975 F1. For the Positive class, which has only 47 examples in the dataset, the CNN outperforms the SVM method by 18%. Furthermore, we find that the SVM method, which uses TF-IDF weighted features, outperforms BSVM across all four classes. We find that the Unknown, Negative, and binary Known class F1 scores slightly vary across folds for the CNN in “KCR Only” with standard deviations 0.017, 0.034, and 0.026 (not shown in the table), respectively. For the linear models, and for all classes except the Positive class, the standard deviations are all around 0.015. Because of the limited number of Positive examples, the F1 standard deviation is much higher with a value around 0.11 for the SVM, BSVM, and CNN methods. We also find that BioBERT does not perform well overall on our task. For the Positive class on the KCR dataset, BioBERT achieves the best F1 of 0.804, but the performance on the Unknown and Negative classes is much worse. Because the Unknown and Negative classes occur more often, the Known F1 is also lower than the SVM.
Table 3:
Model performances on the FH dataset. Precision (P), Recall (R), and F1-Score (F1) are reported for the four major classes: Unknown, Positive, Negative, and Binary (Known). Note that the models were not trained on the Binary class. The Binary class metrics were calculated by merging the Positive and Negative classes.
| Unknown |
Positive |
Negative |
Known |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| Rule-based | 0.941 | 0.998 | 0.969 | 0.971 | 0.879 | 0.923 | 0.995 | 0.859 | 0.922 | 0.995 | 0.866 | 0.926 | |
| FH Only | SVM | 0.983 | 0.994 | 0.988 | 0.987 | 0.936 | 0.960 | 0.977 | 0.959 | 0.968 | 0.986 | 0.963 | 0.975 |
| BSVM | 0.983 | 0.996 | 0.989 | 0.979 | 0.952 | 0.963 | 0.985 | 0.959 | 0.972 | 0.990 | 0.964 | 0.977 | |
| CNN | 0.991 | 0.994 | 0.993 | 0.979 | 0.937 | 0.957 | 0.978 | 0.979 | 0.978 | 0.988 | 0.981 | 0.984 | |
| BioBERT | 0.982 | 0.979 | 0.981 | 0.935 | 0.840 | 0.885 | 0.930 | 0.955 | 0.943 | 0.956 | 0.961 | 0.958 | |
| KCR + FH | SVM | 0.983 | 0.995 | 0.988 | 0.996 | 0.940 | 0.967 | 0.979 | 0.960 | 0.969 | 0.988 | 0.962 | 0.975 |
| BSVM | 0.989 | 0.996 | 0.993 | 0.979 | 0.961 | 0.969 | 0.987 | 0.972 | 0.979 | 0.992 | 0.976 | 0.984 | |
| CNN | 0.991 | 0.994 | 0.993 | 0.972 | 0.948 | 0.959 | 0.982 | 0.978 | 0.980 | 0.988 | 0.981 | 0.984 | |
| CNN + Adv. | 0.989 | 0.993 | 0.991 | 0.981 | 0.940 | 0.960 | 0.976 | 0.974 | 0.975 | 0.985 | 0.977 | 0.981 | |
| KCR ⇒ FH | SVM | 0.792 | 0.981 | 0.876 | 0.234 | 0.116 | 0.155 | 0.780 | 0.375 | 0.504 | 0.916 | 0.445 | 0.597 |
| BSVM | 0.853 | 0.984 | 0.914 | 0.435 | 0.378 | 0.387 | 0.903 | 0.555 | 0.685 | 0.953 | 0.634 | 0.758 | |
| CNN | 0.956 | 0.994 | 0.975 | 0.545 | 0.389 | 0.452 | 0.882 | 0.843 | 0.862 | 0.986 | 0.902 | 0.942 | |
| CNN + Adv. | 0.988 | 0.993 | 0.990 | 0.984 | 0.941 | 0.962 | 0.976 | 0.973 | 0.974 | 0.984 | 0.974 | 0.979 | |
For the FH dataset, all 3 methods achieve an F1 score of 0.95 or higher across all four classes. Similar to what happened with the KCR dataset, the CNN method generally outperforms the two linear models, with an exception for the positive class. However, unlike the KCR dataset, the CNN model does not outperform the linear models by much. All the improvements are less than 1%. Given the high level of performance across all models, substantial improvements may not be possible in the FH dataset. We also find that BSVM slightly outperforms SVM. We find that machine learning may not be as important to extract EGFR test results for the FH dataset because the rule-based method achieves an F1 greater than 90% for every class. However, machine learned models still improve the results with a ≥ 4% improvement in F-score for the positive class. We do not report the standard deviations in Table 3, for the FH dataset. For all FH results, except for KCR ⇒ FH, the standard deviation is less than 0.01.
Overall, when the EGFR test results are recorded in the pathology report, we find that it is possible to extract the information using machine learning-based methods. However, we find that the performance can vary substantially at different registries. For example, the simple rule-based method achieves an F1 of 0.923 on the FH dataset in Table 3 for the Positive class. The rule-based F1 on the KCR dataset is only 0.097. This result suggests that the language used to describe EGFR results is more varied in the KCR dataset. We examine this more in the Discussion section.
RQ2. For the second research question, we test the cross-registry generalization of models trained on a single registry’s data. The results related to this question can be found in the “FH ⇒ KCR” and “KCR ⇒ FH” sections of Tables 2 and 3. For the KCR dataset in Table 2, generalization suffers across all classes when applying a model trained only on the FH dataset. For the Unknown class, the CNN’s F1 drops from 0.975 to 0.875 – a 9% drop in performance. The best method for the Positive class is the BSVM with an F1 of 0.253. However, the BSVM method still has a drop of 24% if we compare it to training on the KCR (target) dataset. For the frequent classes, TF-IDF weighting seems to help. Overall, we find that the CNN generalizes better than both linear models on three of the classes: Unknown, Negative, and Binary.
For the FH dataset, we find a large drop in performance in F1 for both the Positive and Negative classes. For the positive class, the SVM performance drops from 0.960 to 0.155. Likewise, the negative class results drop from 0.968 to 0.504. The BSVM linear model outperforms the SVM across all 4 classes. Nonetheless, while the linear models perform similar to the CNN when trained in the “FH only” setting, in the cross-registry scenario the CNN provides the best performance for all 4 classes.
RQ3. If a cancer registry has access to another registry’s annotated data, but does not have the time or resources to annotate their own data, do unsupervised domain adaptation methods help? The results for this question are in the “FH ⇒ KCR” and “KCR ⇒ FH” sections of Tables 2 and 3 (“CNN + Adv” rows). Overall, on both the KCR and FH datasets, we achieve substantial improvements using adversarial learning in the unsupervised domain adaptation setting. For the KCR dataset in Table 2, “CNN + Adv” performs similar to the CNN that is trained on both the FH and KCR datasets (KCR + FH). Also, compared to the CNN trained only on FH data and applied to the KCR dataset, the “CNN + Adv” improves the performance on the Unknown class by 10% and the Positive class improves by more than 50%, from 0.244 to 0.780 in F1. On the FH dataset, when we train on the KCR dataset and test on the FH dataset, we match the performance of the CNN that is trained directly on the FH dataset for all classes. For the Positive class, “CNN + Adv” models slightly outperform the corresponding models trained on the target datasets (“FH Only” and “KCR Only” in Tables 2 and 3) in this unsupervised domain adaptation setting (“KCR ⇒ FH”/“FH ⇒ KCR”).
RQ4. When annotated datasets are available for both registries, do adversarial learning techniques improve on methods that train on both datasets? For this research question, we focus on the “FH + KCR” sections in both results tables. First, for the KCR dataset, every method shows improvements when compared to training only on the KCR dataset. For example, for the positive class, SVM improves from 0.530 to 0.612, an 8% absolute improvement in F1. The CNN also improves by 3%. Furthermore, without adversarial learning, the CNN outperforms both SVM and BSVM when we use both datasets. When we train the CNN on both datasets and use adversarial learning, we outperform all other methods on the KCR dataset in the “FH + KCR” scenario. Finally, we achieve the largest improvement for the positive class where the F1 improves from 0.746 to 0.801.
For the FH dataset, unlike the KCR dataset, we achieve little to no improvement when we train on both datasets. Likewise, our proposed CNN adversarial method does not perform as well as the CNN which only trains on the FH dataset. Because the overall performance is very high when we only train on the FH dataset, there is not much to be gained by training on both datasets.
4. DISCUSSION
Based on our results in Tables 2 and 3, we find that there are multiple factors to consider if one is deciding to use adversarial domain adaptation techniques – at least for pathology reports from cancer registries. Does the cancer registry have access to annotations for their own data? If so, how well do the models perform when trained only on their data? As seen on the FH dataset, if the model performs very well (> 0.95 F1), then domain adaptation techniques may not help. However, as seen in the supervised domain adaptation setting on the KCR dataset, adversarial learning can help if the performance is not particularly good with locally annotated data. If the cancer registry does not have the resources to annotate their own data, but they have access to another registry’s annotated data, then adversarial domain adaptation techniques achieve similar results as if they were training on an annotated dataset from their registry.
Overall, the performance across all four classes is higher on the FH dataset when compared to the KCR results. There are two possible reasons for the differences in F1. First, the FH dataset is three times as large as the KCR dataset. The largest difference in performance between the two datasets is found with the Positive class, which has the smallest number of labeled examples. Are the differences in F1 on the Positive class between the two datasets caused by different dataset sizes? In Figure 2, we plot the learning curve of the BSVM method on the FH dataset. The best result on the KCR dataset is marked with an X (i.e., the CNN). With 1000 training examples, we find that the Positive class performance is still higher on the FH dataset. Via manual examination, we find that the reports in the FH dataset are much more consistent. For examples, the simple regular expression “positive (\w+\s){1,7}egfr” correctly predicts 188 out of 232 pathology reports for the Positive class in the FH dataset. Furthermore, the rule only results in 3 false positives. For the KCR dataset, using the same regular expression, we match 13 pathology reports. However, only 3 of the reports are correctly matched with the Positive class. This explains why the “KCR ⇒ FH” F1 scores were higher than the “FH ⇒ KCR” setting, even though the FH dataset was larger. Interestingly, we find that adversarial learning provides large improvements in the “FH ⇒ KCR” setting, even though the FH dataset is relatively simple. This suggests that if we can create training datasets for many genomic tests using simple rules (i.e., distant supervision [18]), then we may be able to use unsupervised adversarial domain adaptation to match the results we would obtain if we had a hand-labeled gold standard dataset.
Figure 2:
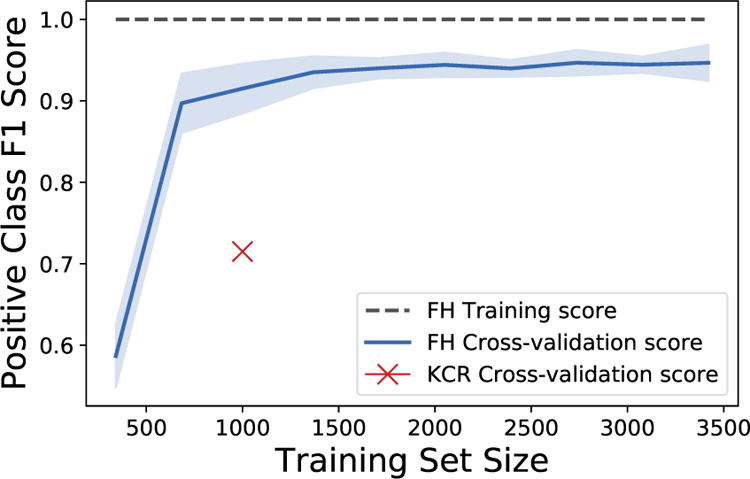
Learning curve measuring the F1 score of the Positive EGFR test result on the FH dataset. We also mark the CNN method’s KCR cross-validation results from Table 2, section “KCR Only”, with an X
5. CONCLUSION
In this paper, we study the generalization of machine learning methods across different cancer registries to understand many questions: Will models shared between registries perform well? Can we combine two registries datasets to improve performance? Do pathology reports collected by different registries substantially differ? Many machine learning methods, including neural networks, perform well when trained with carefully annotated data. However, if these models are shared with other registries, the performance may suffer. Therefore, we introduced an adversarial domain adaptation method for neural networks. Using adversarial learning, we improve the cross-registry generalization substantially, sometimes outperforming methods that were trained on datasets from both registries. There are two avenues we plan to explore in the future:
We performed adversarial learning via backpropagation using a GRL. The GRL method is known to have vanishing gradient issues [30]. If the discriminator becomes very accurate, then the gradients backpropagated to the CNN will be small. Therefore, the CNN will not overcome the discriminator to produce vector representations of the pathology reports that are indistinguishable between registries. Hence, we plan to explore other adversarial methods in the future.
There are many genomic tests of interest to cancer registries, including, but not limited to, EGFR, KRAS, and ALK. In this paper, we focused on extracting EGFR test result information. A natural next step would be to apply our methods to other genomic tests. However, curating datasets for each test is costly. In future work, we plan to use distance supervision in combination with domain adaptation to overcome curation issues.
Our method only extracts generic EGFR mutations in the context of NSCLC. Ideally, we should develop techniques to identify specific EGFR mutations. For example, EGFR exon 20 insertions predict tyrosine kinase inhibitors (TKI) resistance which occurs in approximately 10% of EGFR positive patients [20] at diagnosis. In addition, specific EGFR mutations have prognostic implications: exon 19 deletions are associated with longer overall survival compared with exon 21 L858R mutations, irrespective of treatment with EGFR TKIs. Future development will also focus on refinements to the current algorithms to extract specific EGFR mutations. Because of the high cost of annotating data for different mutation types, we expect distance supervision-based methods and our adversarial domain adaptation approach can be combined to reduce study costs.
9. ACKNOWLEDGEMENTS
We thank Tiffany Janes, one of the project managers at the Cancer Surveillance System (SEER FH registry) for her assistance with data transfer from FH to KCR; and Dr. Christina S Baik, for contribution as an annotator of FH and KCR data.
6. FUNDING
We are grateful for the support of the U.S. National Cancer Institute (NCI) through grant P30CA177558 and Surveillance, Epidemiology, and End Results Program (SEER) contracts HHSN261201300013I and HHSN261201800013I for enabling this effort. SMS and BG are supported through the NCI SEER contract HHSN26100007 and grant P30CA015704. RK’s efforts are also partially supported by the U.S. National Center for Advancing Translational Sciences via grant UL1TR001998.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS
None
Due to extremely small number of positive instances for the ALK mutations, this study is limited to EGFR test results
References
- [1].American College of Surgeons, 2012. Cancer program standards 2012: Ensuring patient-centered care https://www.facs.org/~/media/files/quality%20programs/cancer/coc/programstandards2012.ashx. Accessed: July 2, 2019.
- [2].Baumel T, Nassour-Kassis J, Cohen R, Elhadad M, Elhadad N, 2017. Multi-label classification of patient notes a case study on ICD code assignment. arXiv preprint arXiv:1709.09587
- [3].Cawley GC, Talbot NL, 2010. On over-fitting in model selection and subsequent selection bias in performance evaluation. Journal of Machine Learning Research 11, 2079–2107. [Google Scholar]
- [4].Daume H III, 2007. Frustratingly easy domain adaptation, in: Proceedings of the 45th Annual Meeting of the Association of Computational Linguistics, pp. 256–263. [Google Scholar]
- [5].Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D’Amico TA, Dilling TJ, et al. , 2016. NCCN guidelines insights: non–small cell lung cancer, version 4.2016. Journal of the National Comprehensive Cancer Network 14, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Evgeniou T, Pontil M, 2004. Regularized multi–task learning, in: Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining, ACM; pp. 109–117. [Google Scholar]
- [7].Ganin Y, Lempitsky V, 2015. Unsupervised domain adaptation by backpropagation, in: International Conference on Machine Learning, pp. 1180–1189. [Google Scholar]
- [8].Goulart BHL, Silgard ET, Baik CS, Bansal A, Sun Q, Durbin EB, Hands I, Shah D, Arnold SM, Ramsey SD, et al. , 2019. Validity of natural language processing for ascertainment of EGFR and ALK test results in SEER cases of stage iv non–small-cell lung cancer. JCO clinical cancer informatics 3, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hassanzadeh H, Nguyen A, Karimi S, Chu K, 2018. Transferability of artificial neural networks for clinical document classification across hospitals: A case study on abnormality detection from radiology reports. Journal of biomedical informatics 85, 68–79. [DOI] [PubMed] [Google Scholar]
- [10].Jiang J, Zhai C, 2007. Instance weighting for domain adaptation in NLP, in: Proceedings of the 45th annual meeting of the association of computational linguistics, pp. 264–271. [Google Scholar]
- [11].Kawaguchi K, Kaelbling LP, Bengio Y, 2017. Generalization in deep learning. arXiv preprint arXiv:1710.05468
- [12].Kim Y, 2014. Convolutional neural networks for sentence classification, in: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Association for Computational Linguistics, Doha, Qatar pp. 1746–1751. [Google Scholar]
- [13].Kocbek S, Cavedon L, Martinez D, Bain C, Mac Manus C, Haffari G, Zukerman I, Verspoor K, 2016. Text mining electronic hospital records to automatically classify admissions against disease: Measuring the impact of linking data sources. Journal of Biomedical Informatics 64, 158–167. [DOI] [PubMed] [Google Scholar]
- [14].Lan K, Wang D.t., Fong S, Liu L.s., Wong KK, Dey N, 2018. A survey of data mining and deep learning in bioinformatics. Journal of medical systems 42, 139. [DOI] [PubMed] [Google Scholar]
- [15].Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J, 2019. BioBERT: pre-trained biomedical language representation model for biomedical text mining. arXiv preprint arXiv:1901.08746 [DOI] [PMC free article] [PubMed]
- [16].Li Y, Huang C, Ding L, Li Z, Pan Y, Gao X, 2019. Deep learning in bioinformatics: introduction, application, and perspective in big data era. arXiv preprint arXiv:1903.00342 [DOI] [PubMed]
- [17].Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS, Squire J, et al. , 2013. Molecular testing guideline for selection of lung cancer patients for egfr and alk tyrosine kinase inhibitors: guideline from the college of american pathologists, international association for the study of lung cancer, and association for molecular pathology. Journal of Thoracic Oncology 8, 823–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mintz M, Bills S, Snow R, Jurafsky D, 2009. Distant supervision for relation extraction without labeled data, in: Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP: Volume 2-Volume 2, Association for Computational Linguistics; pp. 1003–1011. [Google Scholar]
- [19].Miotto R, Wang F, Wang S, Jiang X, Dudley JT, 2017. Deep learning for healthcare: review, opportunities and challenges. Briefings in bioinformatics 19, 1236–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morgillo F, Della Corte CM, Fasano M, Ciardiello F, 2016. Mechanisms of resistance to egfr-targeted drugs: lung cancer. ESMO open 1, e000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mullenbach J, Wiegreffe S, Duke J, Sun J, Eisenstein J, 2018. Explainable prediction of medical codes from clinical text, in: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), pp. 1101–1111. [Google Scholar]
- [22].Nair V, Hinton GE, 2010. Rectified linear units improve restricted Boltzmann machines, in: Proceedings of the 27th International Conference on Machine Learning (ICML-10), pp. 807–814. [Google Scholar]
- [23].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. , 2011. Scikit-learn: Machine learning in python. Journal of machine learning research 12, 2825–2830. [Google Scholar]
- [24].Percy C, Holten V.v., Muir CS, Organization WH, et al. , 1990. International classification of diseases for oncology
- [25].Perotte A, Pivovarov R, Natarajan K, Weiskopf N, Wood F, Elhadad N, 2013. Diagnosis code assignment: models and evaluation metrics. Journal of the American Medical Informatics Association 21, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Qiu JX, Yoon HJ, Fearn PA, Tourassi GD, 2018. Deep learning for automated extraction of primary sites from cancer pathology reports. IEEE journal of biomedical and health informatics 22, 244–251. [DOI] [PubMed] [Google Scholar]
- [27].Rios A, Kavuluru R, 2015. Convolutional neural networks for biomedical text classification: application in indexing biomedical articles, in: Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics, ACM; pp. 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rios A, Kavuluru R, 2018. EMR coding with semi-parametric multi-head matching networks, in: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), pp. 2081–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rios A, Kavuluru R, 2019. Neural transfer learning for assigning diagnosis codes to EMRs. Artificial intelligence in medicine 96, 116–122. [DOI] [PubMed] [Google Scholar]
- [30].Rios A, Kavuluru R, Lu Z, 2018. Generalizing biomedical relation classification with neural adversarial domain adaptation. Bioinformatics 34, 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tsoumakas G, Laliotis M, Markantonatos N, Vlahavas I, 2013. Large-scale semantic indexing of biomedical publications at bioasq, in: Proceedings of the First Workshop on Bio-Medical Semantic Indexing Question Answering, a Post-Conference Workshop of Conference Labs of the Evaluation Forum (CLEF). [Google Scholar]
- [32].Wiens J, Guttag J, Horvitz E, 2014. A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions. Journal of the American Medical Informatics Association 21, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]


