INTRODUCTION
TRK fusions are found in a variety of cancer types, lead to oncogenic addiction, and predict for tumor-agnostic efficacy to TRK inhibition1–8. With the recent approval of the first selective TRK inhibitor, larotrectinib, for patients with any TRK-fusion-positive adult or pediatric solid tumor, identifying mechanisms of treatment failure after initial response has become of immediate therapeutic relevance. To date, the only known resistance mechanism is the acquisition of on-target TRK kinase domain mutations, which interfere with drug binding and may be addressable through second-generation TRK inhibitors9–11. Here, we report the identification of off-target resistance in a series of TRK inhibitor-treated patients and patient-derived models mediated by genomic alterations that converge to activate the mitogen-activated protein kinase (MAPK) pathway. MAPK pathway-directed targeted therapy, administered alone or in combination with TRK inhibition, re-established disease control. Experimental modeling further suggests that upfront dual inhibition of TRK and MEK may delay time to progression in cancer types prone to the genomic acquisition of MAPK activating alterations. Collectively, these data suggest that a subset of patients will develop off-target mechanisms of resistance to TRK inhibition with potential implications for clinical management and future clinical trial design.
MAIN ARTICLE
To identify mechanisms of resistance to TRK inhibition in patients with TRK fusion-positive cancers, tumor biopsies and circulating cell-free DNA (cfDNA) were prospectively collected from patients treated with a variety of TRK inhibitors as part of prospective clinical trials and compassionate use programs. Paired sequencing was conducted (see Methods) to identify patients in which TRK kinase domain mutations were not detected or did not entirely explain resistance to the TRK inhibitor utilized. Acquired alterations involving upstream receptor tyrosine kinase or downstream MAPK pathway nodes were identified in six patients prompting further analysis of these cases.
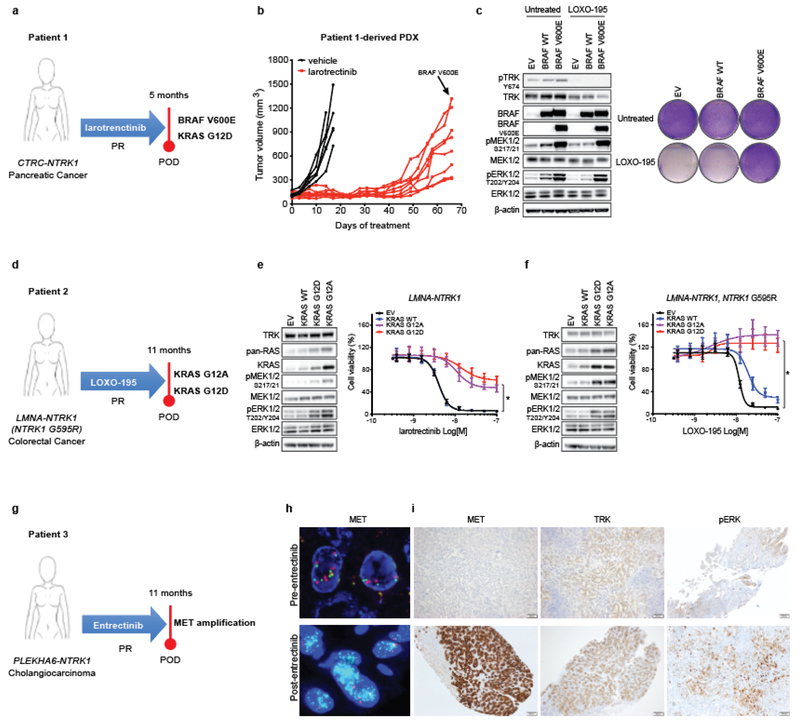
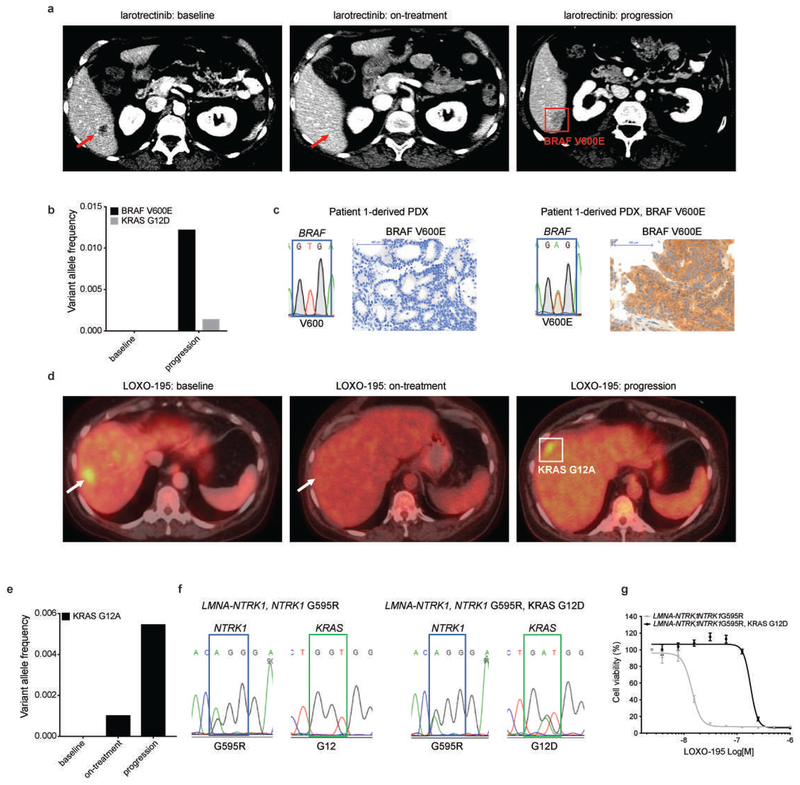
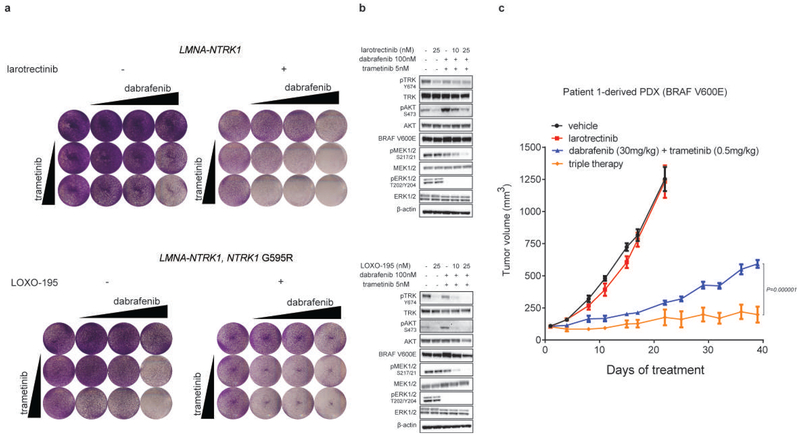
In the first patient (Patient 1), with a CTRC-NTRK1 fusion-positive pancreatic cancer that developed resistance to larotrectinib, targeted sequencing of paired pre-treatment and post-progression tumor biopsies revealed an acquired BRAF V600E mutation (Fig. 1a and Extended Data Fig. 1a). Sequencing of serial cfDNA samples orthogonally confirmed the acquisition of BRAF V600E along with a subclonal KRAS G12D mutation (Extended Data Fig. 1b). Patient-derived xenografts (PDXs) established from this patient’s tumor and treated with larotrectinib over time similarly demonstrated outgrowth of a BRAF V600E-positive subclone at the time of acquired resistance (Fig. 1b and Extended Data Fig. 1c). Consistent with the hypothesis that downstream MAPK pathway activation was responsible for TRK-independent bypass resistance, this patient rapidly progressed on subsequent treatment with LOXO-195, a 2nd-generation TRK inhibitor designed to maintain potency in the setting of TRK kinase domain mutations9. Further supporting the causative role of this alteration in mediating resistance, the ectopic expression of BRAF V600E in a NTRK1 fusion-positive pancreatic cancer cell line (TPR-NTRK1, NTRK1 G595R) conferred resistance to LOXO-195 (Fig. 1c).
Fig. 1: Alterations in the MAPK pathway or an upstream receptor tyrosine kinase confer resistance to TRK inhibitors in patients and preclinical models.
a, Schematic showing acquired BRAF V600E and KRAS G12D mutations in a CTRC-NTRK1-positive pancreatic adenocarcinoma patient with acquired resistance to the first-generation TRK inhibitor, larotrectinib. b, Tumor growth of larotrectinib-sensitive patient-derived xenografts established from this patient’s tumor treated with larotrectinib (200mg/Kg, 5 days/week). Genotyping at the time of acquired resistance identified outgrowth of a BRAF V600E-positive clone. c, Western blot and cell viability assay of a TPR-NTRK1, NTRK1 G595R pancreatic cancer cell line with ectopic expression of BRAF V600E and treated with 50nM of LOXO-195 for 24 (WB) or 72 (cell viability) hours. Total and phosphorylated proteins detected are indicated. Two biological replicates were performed for each experiment. d, Schematic showing presence of KRAS G12A and G12D mutations in a LMNA-NTRK1 fusion-positive colorectal cancer patient with acquired resistance to LOXO-195. Note that KRAS G12D emerged in cfDNA upon further disease progression (17 months on LOXO-195 therapy). e, f, Western blot for MAPK effectors and cell proliferation curves of a LMNA-NTRK1 (e) and a LMNA-NTRK1, NTRK1 G595R (f) colorectal cancer cell lines with ectopic expression of KRAS G12A and G12D, treated as indicated. Data are presented as mean ± SD. Two-tailed unpaired t-test was used to evaluate significant differences in % of viable cells. * indicates differences that were considered statistically significant (P<0.05). Exact P values are P=0.000000000000001 for the LMNA-NTRK1 cell line and P=0.000000001115265 for the LMNA-NTRK1, NTRK1 G595R cell line. Two biological replicates were performed for each experiment. g, Schematic showing acquired MET amplification in a PLEKHA6-NTRK1 fusion-positive cholangiocarcinoma patient with acquired resistance to entrectinib. h, Representative fluorescence in situ hybridization (FISH) and i, Immunohistochemistry (IHC) images of the pre- and post-entrectinib tumor biopsies from this patient. Lower panels show confirmed acquisition of MET amplification in the post-entrectinib sample (h) and increased MET and pERK staining by IHC (i). FISH and IHC were performed two independent times and similar results were obtained.
In the second patient (Patient 2), with a LMNA-NTRK1 fusion-positive colorectal cancer (CRC) that developed acquired resistance to LOXO-195, sequencing of tumor and serial cfDNA samples revealed emergence of multiple KRAS mutations consistent with polyclonal resistance mediated by a convergent mechanism (Fig 1d and Extended Data Fig. 1d, e). This patient previously had a prolonged response to larotrectinib followed by resistance driven by acquisition of an NTRK1 G595R solvent front mutation (the resultant substitution prevents drug binding11). Consistent with on-target NTRK-dependent resistance, subsequent treatment with LOXO-195 achieved a second response, eventually followed again by solitary site progression in the liver. Genomic analysis of the liver metastasis biopsy and serial cfDNA revealed the emergence of a KRAS G12A substitution (Fig. 1d and Extended Data Fig. 1d, e). This mutation disappeared in cfDNA after ablation of the liver metastasis and LOXO-195 continuation; however, a new KRAS G12D mutation emerged upon further disease progression (Fig. 1d). Consistent with this clinical observation, chronic treatment of a LMNA-NTRK1-positive, NTRK1 G595R-mutant CRC cell line (LMNA-NTRK1, NTRK1 G595R) with LOXO-195 likewise demonstrated KRAS G12D acquisition (Extended Data Fig. 1f, g). Further supporting the causative nature of these alterations in mediating resistance, ectopic expression of both KRAS G12A and G12D in TRK fusion-positive CRC cell lines was sufficient to increase MAPK pathway activation and confer resistance to both larotrectinib and LOXO-195 (Fig. 1e, f).
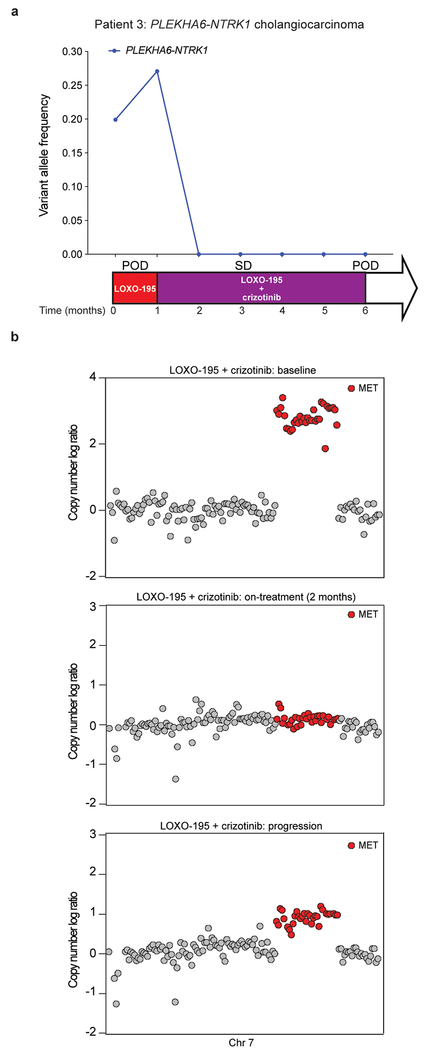
In the third patient (Patient 3), with a PLEKHA6-NTRK1 fusion-positive cholangiocarcinoma that developed acquired resistance to the 1st generation TRK inhibitor entrectinib, sequencing of both tissue and cfDNA identified an acquired high-level focal amplification of MET (Fig. 1g). Acquisition of MET high-level amplification and protein overexpression was orthogonally confirmed by MET FISH and immunohistochemistry, respectively (Fig. 1h, i), and sequencing of NTRK1 did not identify a kinase domain point mutation. Of note, MET amplification has been observed as a mechanism of off-target resistance in other oncogene-addicted cancers12–16. Consistent with the hypothesis that MET amplification drove TRK-independent resistance, the patient immediately progressed despite subsequent treatment with LOXO-195.
As all three index cases involved tumors of gastrointestinal origin, we next proceeded to broaden our analysis to all TRK fusion-positive gastrointestinal cancer patients for whom we had serial cfDNA samples (excluding gastrointestinal stromal tumors). Five additional patients were identified, 3 of whom developed emergent MAPK alterations while on TRK inhibitors (Supplementary Table 1). One patient with an ETV6-NTRK3 fusion-positive pancreatic cancer had a prolonged response to the multikinase TRK inhibitor PLX7486. No mechanism of resistance was identified at progression and he was subsequently treated with LOXO-195 with no response. At the time of progression on LOXO-195, cfDNA demonstrated acquisition of a hotspot MEK1 (MAP2K1) P124S mutation. While testing within the context of a TRK fusion suggests that this mutation has weak oncogenic potential, this alteration has previously been proposed to confer resistance to targeted therapy in BRAF V600E-mutant melanoma patients17. A second patient with TPR-NTRK1 fusion-positive pancreatic cancer had a prolonged response to entrectinib followed by resistance driven by acquisition of NTRK1 G595R. This patient was subsequently treated with LOXO-195 with a transient decline in tumor markers and resolution of tumor fevers, followed quickly by clinical deterioration and radiologic progression. Serial cfDNA sequencing on LOXO-195 revealed loss of the NTRK1 G595R mutation but emergence of the known activating ERBB2 S310F mutation18. Lastly, a TPM3-NTRK1 fusion-positive CRC patient developed polyclonal resistance to larotrectinib six months into therapy, with cfDNA demonstrating emergence of KRAS G12D, NTRK1 G595R, and NTRK1 F589L at the time of clinical progression (Supplementary Table 1).
Collectively, our analysis identified putative bypass-mediated resistance to first and next-generation TRK inhibitors in 75% (6/8) of TRK-fusion positive gastrointestinal cancers analyzed. These alterations are all predicted to restore MAPK signaling through parallel upstream receptor tyrosine kinase or downstream MAPK signaling nodes activation. We therefore reasoned that these alterations represent a recurrent and convergent mechanism of treatment failure to TRK inhibitors and that a subset of these alterations may be themselves actionable. To evaluate this hypothesis in the clinic, we treated patients 1 and 3 with targeted therapy directed at their respective acquired resistance mechanisms.
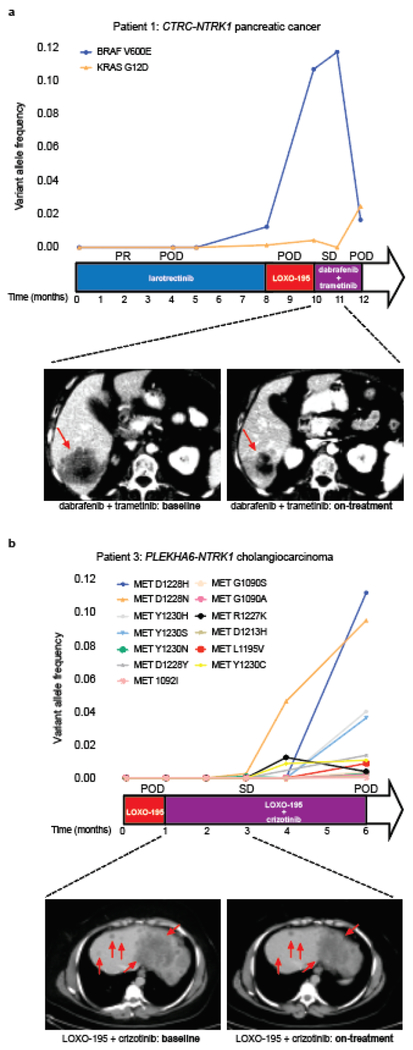
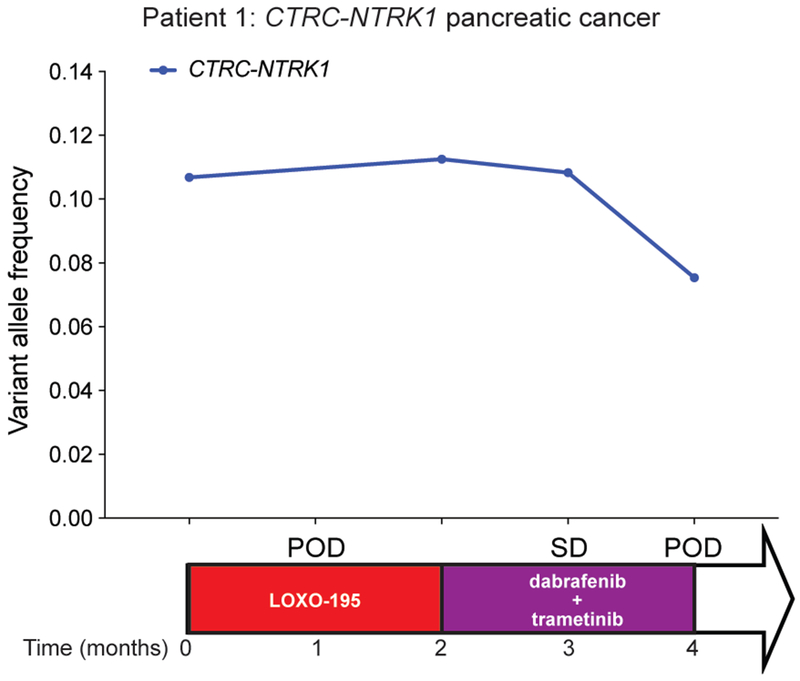
Patient 1 with pancreatic cancer and the emergent BRAF V600E mutation was treated with a combination of RAF and MEK inhibitors (dabrafenib and trametinib), resulting in early tumor regression, accompanied by a slight decrease in the allele frequency of the NTRK fusion and a ten-fold decrease in the mutant allele frequency of BRAF V600E detected in cfDNA (Fig. 2a and Extended Data Fig. 2). Simultaneously, a subclonal but preexisting KRAS G12D mutation rose in cfDNA and the patient developed radiographic progression shortly thereafter, suggesting that outgrowth of this alteration may have been responsible for the acquired resistance to RAF/MEK inhibition (Fig. 2a). It is likely, however, that this tumor was still partially driven by the TRK fusion. While a combination including a TRK inhibitor as a third agent was favored initially, it could not be secured in time. We thus tested whether the addition of a TRK inhibitor to the combination of dabrafenib and trametinib would enhance anti-tumor activity in TRK fusion-positive preclinical models transduced with BRAF V600E. Triple therapy (larotrectinib, dabrafenib, and trametinib) was significantly more effective than dabrafenib and trametinib at suppressing tumor growth and deeply inhibiting TRK-mediated signaling (AKT, ERK, MEK; Extended Data Fig. 3a, b). Triple therapy also enhanced tumor growth inhibition compared to dabrafenib and trametinib in PDXs derived from Patient 1 that harbor the BRAF V600E mutation (Extended Data Fig. 3c).
Fig. 2: Tailored combinatorial therapies are effective against tumors that developed bypass resistance to TRK inhibitors.
a,b, upper panel, Graph depicting the dynamics of select mutations detected in the cfDNA of the CTRC-NTRK1 positive pancreatic adenocarcinoma (Patient 1, a) and PLEKHA6-NTRK1 positive cholangiocarcinoma (Patient 3, b) patients while treated with a series of targeted therapies. middle panel, The time on treatment, best clinical response achieved (PR: partial response; SD: stable disease based on RECIST v1.1 criteria) and the time of progression (POD) for each of the indicated therapeutic regimens are displayed. Lower panel, Representative scans from the patients at baseline and on treatment with the combination of dabrafenib + trametinib (a) and LOXO-195 + crizotinib (b), respectively.
Patient 3 with cholangiocarcinoma and the emergent MET amplification was treated with the combination of LOXO-195, an agent on which her disease had just progressed, and the multikinase MET inhibitor, crizotinib. Marked tumor shrinkage was achieved at 8 weeks and disease control was maintained for 4.5 months, accompanied by the disappearance of detectable NTRK fusion and MET amplification in cfDNA (Fig. 2b and Extended Data Fig. 4a, b). Interestingly, post-progression cfDNA demonstrated reappearance of focal MET amplification in addition to 13 emergent missense mutations in MET (Fig. 2b and Extended Data Fig. 4b), several of which are known to impair crizotinib binding19,20. While multiple resistance mechanisms were observed, presumably secondary to inter/intratumoral heterogeneity, these alterations were remarkably convergent on MET reactivation. This on-target resistance to crizotinib further supports the mechanistic role for MET as an acquired driver of resistance to prior TRK inhibitor therapy.
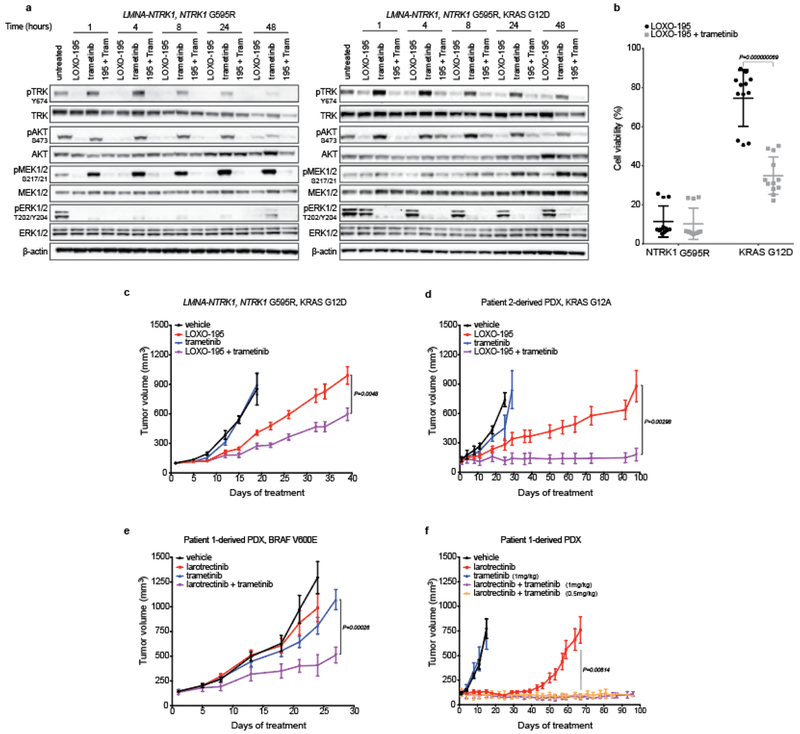
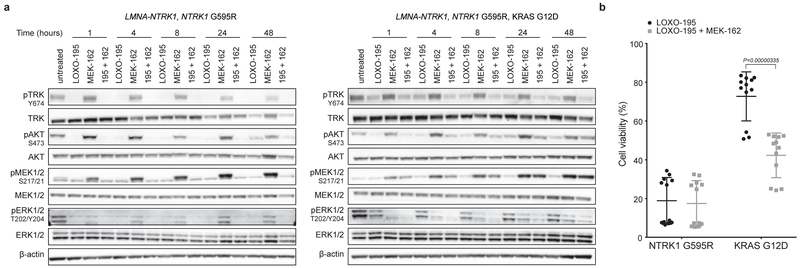
While the tumor regressions observed in patients with acquired BRAF V600E and MET amplifications were ultimately transient, they provide further clinical validation that the putative bypass mechanisms identified in these patients were biologically relevant. Given the convergence of these alterations on MAPK pathway activation, we explored the utility of combination TRK and MEK inhibition preclinically and clinically. Combinatorial treatment with LOXO-195 and a MEK inhibitor (trametinib or MEK-162) was more effective than either single agent alone in suppressing TRK and ERK activation and cell viability in the LMNA-NTRK1, NTRK1 G595R, KRAS G12D LOXO-195-resistant model (Fig 3a, b and Extended Data Fig. 5a, b). Furthermore, xenografts derived from this cell line were more sensitive to combinatorial therapy when compared to each of the single agent (Fig. 3c) and similar results were obtained from PDXs established from the LOXO-195-resistant tumor collected from patient 2 (KRAS G12A-mutant, LMNA-NTRK1-positive CRC; Fig. 3d). Despite these observations, Patient 2 was treated with the combination of LOXO-195 and trametinib and experienced rapid disease progression (Extended Data Fig. 6). While the KRAS mutations in the patient and the PDX had different G12 substitutions (a factor that can affect GTPase activity and consequent response to MEK inhibition21), this outcome was also consistent with prior clinical experience showing that KRAS mutations are insensitive to MEK inhibition at exposures achievable in people22,23.
Fig. 3: Dual TRK and MEK blockade is required to inhibit tumor growth in TRK fusion-positive models that acquired MAPK alterations.
a, Western blots from the two colorectal cancer cell lines LMNA-NTRK1, NTRK1 G595R and LMNA-NTRK1, NTRK1 G595R, KRAS G12D, treated as indicated. LOXO-195 (50nM), trametinib (10nM) or the combination of both drugs (195 + tram) were administered at the indicated time and protein lysates were probed with the indicated antibodies. While LOXO-195 was sufficient to inhibit both phospo-TRK and phospho-ERK in the KRAS wild type cell line, the combination of LOXO-195 and trametinib was required for this dual inhibition in the KRAS G12D mutated cell line. Three biological replicates were performed. b, Proliferation assays on the same cell lines (labeled NTRK1 G595R and KRAS G12D, respectively) treated for 72 hours with LOXO-195 (125nM), trametinib (2nM) or their combination. Data are presented as mean ± SD of four biological replicates. Two-tailed unpaired t-test was used to evaluate significant differences in % of viable cells. P values <0.05 were considered statistically significant. c, In vivo efficacy of LOXO-195 (100mg/Kg BID 5 days a week), trametinib (1mg/kg 4 days a week) or their combination on xenofgrafts established from the LMNA-NTRK1, NTRK1 G595R, KRAS G12D cell line (vehicle n=5, LOXO-195 n=6, trametinib n=5, LOXO-195 + trametinib n=6). d, In vivo efficacy of LOXO-195 (100mg/Kg BID 5 days a week), trametinib (3mg/kg 4 days a week) or their combination on PDXs established from the KRAS G12A positive liver biopsy collected at the time of LOXO-195 progression from patient 2 (Figure 1d; vehicle n=4, LOXO-195 n=5, trametinib n=5, LOXO-195 + trametinib n=5). e, In vivo efficacy of larotrectinib (200mg/kg daily 5 days a week), trametinib (1mg/kg daily 4 days per week) and the combination of both drugs in larotrectinib-resistant PDXs established from Patient 1 (Patient 1-derived PDX, BRAF V600E, Figure 1a; vehicle n=8, larotrectinib n=8, trametinib n=8, larotrectinib + trametinib n=7). f, In vivo efficacy of larotrectinib (200mg/kg daily 5 days a week), trametinib (1mg/kg 4 days a week) and the combination of both drugs in larotrectinib-sensitive PDXs established from Patient 1 (Patient 1-derived PDX, Figure 1a; note that trametinib was also tested in combination with larotrectinib at half of the dose: 0.5mg/kg 4 days a week, orange line; vehicle n=6, larotrectinib n=8, trametinib n=7, larotrectinib + trametinib (1mg/kg) n=7, larotrectinib + trametinib (0.5mg/kg) n=7). Combination therapy prevents the development of primary or acquired resistance (ongoing at 3 months). Two-tailed unpaired t-test was used to evaluate significant differences in the tumor volumes. Data are presented as mean±SEM. P values <0.05 were considered statistically significant.
In multiple oncogene-addicted cancers, however, the management of acquired resistance with next-generation inhibitors or combinatorial therapy has generally been less efficacious than upfront use of these agents24–26. We therefore reasoned that the upfront dual targeting of TRK and MEK might delay the emergence of off-target resistance that converges on downstream MAPK pathway activation in TRK fusion-positive models. To test this hypothesis, we treated the larotrectinib-resistant and sensitive PDXs established from Patient 1 with larotrectinib, trametinib or a combination of both. In larotrectinib-resistant PDXs, the combination of larotrectinib and trametinib delayed but did not prevent tumor growth compared to single agent treatments (Fig 3e). In larotrectinib-sensitive PDXs, while single-agent larotrectinib effectively controlled tumor growth for approximately a month, the combination resulted in complete and durable tumor regression (ongoing response at three months, Fig. 3f). Droplet digital PCR on residual responding tumors collected at the end of the experiment from mice treated with the combination found that the BRAF V600E mutation was indeed present, albeit at low variant allele frequency (Supplementary Table 2), suggesting that concomitant TRK and MEK inhibition limited the emergence of this resistant cell population. If recapitulated in additional models, these data suggest that the upfront combination may further delay the emergence of MEK-sensitive resistance mechanisms such as BRAF V600 mutations, when compared to sequential TRK inhibitor monotherapy followed by the combination upon development of clinical resistance27.
Together, our data suggest that a subset of TRK fusion-positive cancers will develop off-target resistance to TRK inhibition that will not be adequately addressed by next-generation TRK inhibitors alone. Intriguingly, while TRK fusions appear to predict for initial response to TRK inhibition in a tumor-agnostic manner, early clinical evidence suggests that the durability of response may be more limited in gastrointestinal cancer2,11,28. Our findings provide potential mechanistic insight into why this may be the case and have similarities to prior experience with targeted therapy in BRAF V600E-mutant or EGFR amplified colorectal cancers29–32. The bypass mechanisms we identified demonstrate remarkable convergence on the ERK signaling. A portion of these resistance mechanisms may be successfully managed with simultaneous TRK and MEK inhibition, drugs which have largely non-overlapping toxicity in patients, although upfront treatment with the combination may confer more durable responses.
METHODS
Ethical Compliance
We declare compliance with all relevant ethical regulation.
Patients
Patients were treated with TRK inhibitors as part of prospective IRB-approved research protocols or expanded access protocols. All patients provided written informed consent for genomic sequencing of tumor and cfDNA, and review of medical records for detailed demographic, pathologic, and clinical data and for publication of these information as part of an institutional IRB-approved research protocol (MSKCC; ). Research protocols for tumor collection and analysis were approved by the ethical committees of MSKCC.
Compounds
larotrectinib and LOXO-195 were obtained from Loxo Oncology. trametinib, MEK-162 were purchased from Selleckchem. All drugs were dissolved in DMSO to yield 10mM stocks and stored at −20 °C.
Targeted tumor sequencing
DNA from formalin-fixed paraffin-embedded tissue and matched germline DNA underwent targeted next-generation sequencing assay using (MSK-IMPACT)33. In brief, this assay uses a hybridization-based exon capture designed to capture all protein-coding exons and select introns of oncogenes, tumor-suppressor genes and key members of pathways that may be actionable by targeted therapies. In this study, either 410 or 468 key cancer-associated genes were analyzed. Sequencing data were analyzed as previously described to identify somatic single-nucleotide variants, small insertions and deletions, copy number alterations and structural rearrangements34. In addition, hotspot alterations were identified using an adaptation of a previously described method35 applied to a cohort of 24,592 sequenced human cancers36.
Targeted plasma sequencing
Cell-free DNA was extracted from all plasma samples and sequenced using a custom, ultra-deep coverage next-generation sequencing panel (MSK-ACCESS). The custom assay includes key exons and domains of 129 and introns of 10 genes harboring recurrent breakpoints, and uses duplex unique molecular identifiers (UMIs) and dual index barcodes to minimize background sequencing errors and sample-to-sample contamination. Sequencing data were analyzed using a custom bioinformatics pipeline that trims the UMIs, aligns the processed reads to the human genome, collapses PCR replicates into consensus sequences, and re-aligns the error-suppressed consensus reads. Consensus reads with representation from both strands of the original cfDNA duplex were used for de novo variant calling using VarDict (v1.5.1). Mutation calling required at least 1 collapsed read at a known cancer hotspot site or at least 3 collapsed reads at non-hotspot sites. All samples were sequenced to an average depth of approximately 20,000X coverage. Somatic mutations were identified and quantified as variant allele frequencies. Copy number alterations were identified across all samples using a previously described method34. NTRK fusions were identified and quantified using Manta (v1.5.0). All samples were manually reviewed to identify NTRK fusions, and cfDNA from Patient 3 was manually reviewed to identify copy number alterations, including MET. Variants were called against an unmatched healthy plasma donor to identify any specimen type-related artifacts. Mutations called at silent, intronic, and intergenic loci were removed.
Patient-derived primary cell lines
The LMNA-NTRK1 and the derived entrectinib-resistant LMNA-NTRK1, NTRK1 G595R CRC cell lines were obtained from Dr. Alberto Bardelli (Candiolo Cancer Institute, FPO, IRCCS, Turin, Italy). The LMNA-NTRK1, NTRK1 G595R, KRAS G12D cell line was established following chronic exposure of the LMNA-NTRK1, NTRK1 G595R to increasing concentrations of LOXO-195 (ranging from 1 to 200nM) for four months. The TPR-NTRK1, NTRK1 G595R pancreatic cancer cell line was established from a PDX engrafted with a biopsy of a patient at the time of progression on entrectinib. All cell lines were plated on collagen-coated petri dishes and cultured in DMEM/F12 + 10% FBS and 1% antibiotics.
Antibodies
Western blots: total protein lysates following the indicated treatment were extracted and separated using SDS-PAGE gels according to standard methods. Membranes were probed using the following antibodies: pan Trk clone A7H6R (92991S Cell Signaling Technology), phospho TrkA (Y674/675) clone C50F3 (4621S Cell Signaling Technology), phospho PLCƔ (Y783; 2821L Cell Signaling Technology), phospho MEK1/2 (S217/221) clone 41G9 (9154S Cell Signaling Technology), total MEK1/2 (9122L Cell Signaling Technology), BRAF clone D9T6S (14814S Cell Signaling Technology), BRAF V600E (ab228461 Abcam), phospho p44/42 MAPK (Erk1/2; T202/Y204) clone D13.14.4E (4370S Cell Signaling Technology), total ERK1/2 (9102S Cell Signaling Technology), phospho AKT (S473) clone D9E (4060L Cell Signaling Technology), total AKT (9272S Cell Signaling Technology), pan RAS (BK008, part AESA02 Cytoskeleton), KRAS (F234 Santa Cruz) and β-actin clone 13E5 (4970S Cell Signaling Technology).
Immunohistochemistry
C-Met immunohistochemistry was performed with clone SP44 (Ventana Medical Systems) at a concentration of 9.75 μg/ml and ready to use dilution. Trk A immunohistochemistry was performed with clone EP105BY (Abcam) at a concentration of 0.643 mg/ml and 1:750 dilution. Both C-Met and Trk A immunohistochemistry are clinical validated and were performed in a CLIA accredited laboratory. pERK immunohistochemistry was performed with clone D13.14.4E (Cell Signaling Technology) at a concentration of 1 μg/ml by the Molecular Cytology Core Facility at Memorial Sloan Kettering Cancer Center using Discovery XT processor (Ventana Medical Systems).
Fluorescence in situ hybridization (FISH)
MET FISH was performed using the ZytoLight SPEC dual-color MET/ CEN17 probe cocktail (Zytovision). MET FISH is a clinical validated assay that was performed in a CLIA accredited laboratory.
Plasmids and viral-particles production
pDONR223_KRAS_p.G12A (Cat. 81673) was purchased from Addgene. The pDONR223_KRAS_wt and p.G12D were generated by site directed mutagenesis using the following kit (Q5® Site-Directed Mutagenesis Kit; E0552S New England Biolabs) and primers: pDNRmutG12Atowt F GTTGGAGCTGGTGGCGTAGGC, pDNRmutG12AtoD F GTTGGAGCTGATGGCGTAGGC and the common reverse pDNRmut R TACCACAAGTTTATATTCAGTCATGGTGC. The pDONR plasmids were then subcloned in the pLX302 destination lentiviral vector (Addgene, Cat. 25896). Lentiviral particles were produced in HEK 293T cells as previously described37 and used to Infect the LMNA-NTRK1 and the LMNA-NTRK1, NTRK1 G595R cell lines. pLENTI6 lentiviral plasmids encoding for wild type and V600E BRAF were obtained from Dr. Neal Rosen’s laboratory and used to infect the TPR-NTRK1, NTRK1 G595R cell line. Transduced cell lines were used for Western blot analyses and proliferation assays.
Proliferation assays
Cell-titer glo-based assay: LMNA-NTRK1 and LMNA-NTRK1, NTRK1 G595R cell lines transduced with KRAS wild type or mutant-encoding plasmids were seeded in 96 well-plates (6,000 per well). The following day larotrectinib or LOXO-195 (1:2 dilutions starting with a maximum concentration of 100nM) was added. Cell-titer glo reagent was added 72 hours later and absorbance was read at 490nm according to the manufacturer protocol. Data are presented as % cell viability (mean±SD) normalized to the DMSO treated cells considered 100% viable. Cell-titer glo was also used to test the viability of LMNA-NTRK1, NTRK1 G595R and LMNA-NTRK1, NTRK1 G595R, KRAS G12D cell lines following treatment with LOXO-195 (125nM) or the combination of LOXO-195 (125nM) and trametinib (2nM). Colony formation assay: a TPR-NTRK1, NTRK1 G595R pancreatic cancer cell line transduced with wild type or V600E mutant BRAF was seeded in 6 well-plates (300,000 cells per well). The day after 50nM of LOXO-195 were added. After 72 hours of incubation cells were fixed in 4% glutaraldehyde and stained with crystal violet.
In vivo studies
Xenografts derived from the LMNA-NTRK1, NTRK1 G595R, KRAS G12D colorectal cancer primary cell line were generated by injecting 5 million of cells into the flank of six-weeks-old NSG female mice. Two weeks later tumors were collected and expanded in additional mice. PDXs were generated as follows: six-week-old NSG female mice were implanted subcutaneously with specimens freshly collected from patients at Memorial Sloan Kettering Hospital under an MSK approved IRB biospecimen protocol. Tumors developed within 2 to 4 months and were expanded into additional mice by serial transplantation. The generated PDXs were subjected to high coverage next generation sequencing with the MSK-IMPACT assay. For efficacy studies, treatment started when tumors reached approximately 100 mm3. Xenografts were randomized and dosed orally with LOXO-195 (100mg/kg BID 5 days per week), trametinib (1mg/kg 4 days per week) or a combination of the two agents. PDXs derived from Patient 1 were randomized and dosed orally with vehicle, larotrectinib (200mg/kg daily 5 days a week), trametinib (1mg/kg), or the combination. PDXs derived from Patient 2 were randomized and dosed with LOXO-195 (100mg/kg BID 5 days per week), trametinib (3mg/kg 4 days per week) or their combination. Mice were observed daily throughout the treatment period for signs of toxicity. Tumors were measured twice weekly using calipers, and tumor volume was calculated using the formula length × width2 × 0.52. Body weight was also assessed twice weekly. At the end of each treatment, animals were sacrificed and tumors were collected for biochemistry and histology analysis. Mice were cared for in accordance with guidelines approved by the MSK Institutional Animal Care and Use Committee and Research Animal Resource Center. Four to eight mice per group were included in each experiment.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 8 (GraphPad Software Inc. San Diego, CA). Two-tailed unpaired t-test was used to evaluate significant differences in % of viable cells in cell proliferation assays. Data are presented as mean ± SD. Exact P values are indicated. Two-tailed unpaired t-test was also used to evaluate significant differences in the tumor volumes in in vivo efficacy studies. Error bars represent SEM. Exact P values are indicated.
DATA AVAILABILITY STATEMENT
All genomic results and associated clinical data for all patients in this study are publically available in the cBioPortal for Cancer Genomics at the following URL: http://cbioportal.org/msk-impact. All relevant cell-free DNA sequencing data are included in the paper and/or supplementary files.
Extended Data
Extended Data Fig. 1: Hotspots mutations in KRAS and BRAF confer resistance to TRK inhibitors in patients and preclinical models.
a, Representative scans of Patient 1 at baseline, 4 weeks on larotrectinib treatment (responding) and at progression. Targeted sequencing of the tumor at progression identified a BRAF V600E mutation (red square). b, cfDNA analysis confirmed the emergence of BRAF V600E and identified a subclonal KRAS G12D mutation. c, Emergence of a BRAF V600E mutation in the larotrectinib-resistant PDXs presented in Fig. 1b demonstrated by Sanger sequencing and IHC staining using a BRAF V600E specific antibody to detect the mutant protein. d, Representative scans of Patient 2 at baseline, 4 weeks on LOXO-195 treatment (responding) and at progression. Targeted sequencing of the tumor at progression identified a KRAS G12A mutation (white square). e, cfDNA analysis confirmed the emergence of KRAS G12A. f, Sanger sequencing demonstrating the emergence of a KRAS G12D mutation in a LMNA-NTRK1, NTRK1 G595R positive primary CRC cell line treated with increasing concentrations of LOXO-195 for 4 months until the development of resistance. g, Cell proliferation on the LMNA-NTRK1, NTRK1 G595R and the LMNA-NTRK1, NTRK1 G595R, KRAS G12D primary cell lines treated for 72 hours with increasing concentrations (ranging from 0 to 1,000nM) of LOXO-195. Data are presented as mean±SD of two biological replicates.
Extended Data Fig. 2: Radiologic response to combined RAF/MEK inhibition in Patient 1 correlates with decreased allele frequency of the TRK fusion in cfDNA.
Graph depicting the allele frequencies of truncal NTRK fusion in the cfDNA of the CTRC-NTRK1 positive pancreatic adenocarcinoma) patient (Patient 1) while treated with LOXO-195 and the combination of dabrafenib and trametinib. The time on treatment, best clinical response (SD: stable disease based on RECIST v1.1 criteria) and the time of progression (POD) for each of the indicated therapeutic regimens are displayed.
Extended Data Fig. 3: TRK inhibition enhances the anti-tumor effect of the combination of RAF and MEK blockade in TRK fusion-positive preclinical models harboring a BRAF V600E mutation.
a, Activity of dual RAF/MEK inhibition (dabrafenib ranging from 50 to 500 nM and trametinib 1 and 5 nM) in the absence (left panel) or presence (right panel) of the TRK inhibitor [larotrectinib or LOXO-195 (25 nM)] on the proliferation of LMNA-NTRK1 and LMNA-NTRK1, NTRK1 G595R CRC cell lines transduced with the BRAF V600E mutation. Two biological replicates were performed. b, Western blot analysis on the same cell lines treated for 4 hours as indicated (larotrectinib/LOXO-195= 25nM, trametinib= 5nM, dabrafenib= 100 nM, the combination of dabrafenib= 100 nM and trametinib= 5 nM or the triple therapy at two different concentrations of larotrectinib/LOXO-195= 10 and 25 nM, respectively). The triple therapy is more potent than the combination of anti RAF/MEK alone in inhibiting MEK, ERK and AKT. Two biological replicates were performed. c, Efficacy of the triple therapy (larotrectinib + debrafenib + trametinib) against the Patient 1-derived PDX that harbors a V600E mutation. The triple therapy is significantly more efficacious than the combination of dabrafenib and trametinib alone in inhibiting tumor growth (P=0.000001). A minimum of six animals per group [vehicle (n=7), larotrectinib (n=6), dabrafenib + trametinib (n=7) and larotrectinib + dabrafenib + trametinib (n=6)] were used. Two-tailed unpaired t-test was used to evaluate significant differences in the tumor volumes. Data are presented as mean±SEM.
Extended Data Fig. 4: Radiologic response to combined TRK/MET inhibition in Patient 3 correlates with decreased allele frequency of the targeted alterations in cfDNA.
a, Graph depicting the allele frequencies of the truncal NTRK fusion in the cfDNA of the PLEKHA6-NTRK1 positive cholangiocarcinoma patient (Patient 3) while treated with LOXO-195 and the combination of LOXO-195 and crizotinib. The time on treatment, best clinical response (SD: stable disease based on RECIST v1.1 criteria) and the time of progression (POD) for each of the indicated therapeutic regimens are displayed. b, Copy number plots from this patient demonstrating disappearance of the MET amplification on treatment and reemergence at the time of disease progression.
Extended Data Fig. 5: Dual TRK and MEK blockade inhibits growth of the LOXO-195 resistant LMNA-NTRK1, NTRK1 G595R, KRAS G12D cancer cell line.
a, Western blot from the two colorectal cancer cell lines LMNA-NTRK1, NTRK1 G595R and LMNA-NTRK1, NTRK1 G595R, KRAS G12D, treated as indicated. LOXO-195 (50nM), MEK-162 (50nM) or the combination of both drugs (195 + 162) were administered at the indicated time and protein lysates were probed with the indicated antibodies. While LOXO-195 was sufficient to inhibit both phospo-TRK and phospho-ERK in the KRAS wild type cell line, the combination of LOXO-195 and MEK-162 was required for this dual inhibition in the KRAS G12D mutated cell line. Three biological replicates were performed. b, Proliferation assays on the same cell lines (labeled NTRK1 G595R and KRAS G12D, respectively) treated for 72 hours with LOXO-195 (125nM), MEK-162 (25nM) or their combination. Data are presented as mean ± SD of four biological replicates. Two-tailed unpaired t-test was used to evaluate significant differences in % of viable cells. P values <0.05 were considered statistically significant.
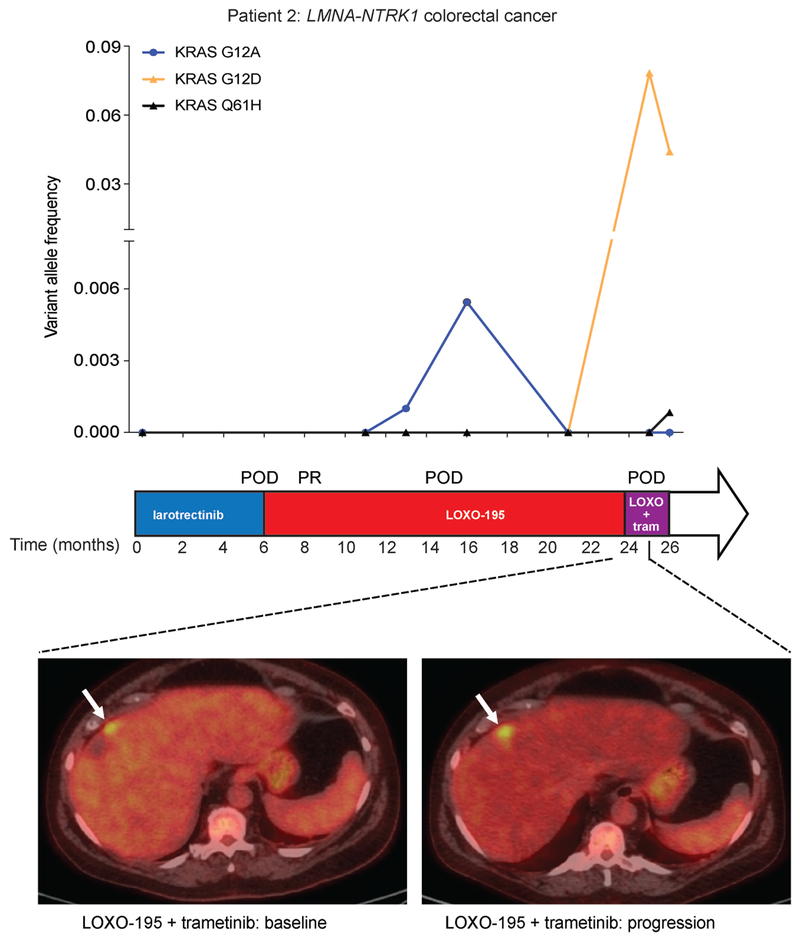
Extended Data Fig. 6: Radiologic and cfDNA correlates in a LOXO-195 resistant CRC patient treated with the combination of LOXO-195 and trametinib.
Graph depicting the dynamics of select mutations detected in the cfDNA of the LMNA-NTRK1, G595R mutated colorectal cancer patient while treated on targeted therapy (LOXO + tram: LOXO-195 + trametinib). The time on treatment, best clinical response (PR: partial response based on RECIST v1.1 criteria) and the time of progression (POD) for each of the indicated therapeutic regimens are displayed. Representative scans of Patient 2 are presented at baseline and at progression (4 weeks) with the combination of LOXO-195 and trametinib.
Supplementary Material
Supplementary Table 1: Characteristics of the TRK inhibitor-treated patients analyzed and genomic sequencing results obtained from serially collected cfDNA samples. Each column represents a cfDNA time point, with the timing of sample acquisition relative to targeted therapy and the sequencing platform used denoted on top (A: MSK-ACCESS; G: Guardant Rows; laro: larotrectinib; POD: Progression of disease). Single nucleotide variants and their variant allele frequencies (out of 1) are listed in rows. Known TRK mutations and MAPK alterations responsible for the resistance to TRK inhibitors are listed.
Supplementary Table 2: Frequency of the BRAF V600E mutation detected by droplet digital PCR sequencing on the Patient 1-derived PDX treated with vehicle, larotrectinib or the combination of larotrectinib and trametinib. Droplet counts for the wild type and the mutant allele and their ratio (Ratio [Mu]/[WT]) are reported.
Acknowledgements
This study was funded by the National Cancer Institute (NCI) under the MSK Cancer Center Support Grant/Core Grant (P30 CA008748) and the R01CA226864 (to MS and AD). This work was also partially funded by the Cycle for Survival (to AD) and LOXO Oncology. E.C. is a recipient of a MSK society scholar prize.
Competing interests
M.S. is in the Advisory Board of Bioscience Institute and Menarini Ricerche, received research funds from Puma Biotechnology, Daiichi-Sankio, Targimmune, Immunomedics and Menarini Ricerche, is a co-founder of Medendi Medical Travel and in the past two years he received honoraria from Menarini Ricerche and ADC Pharma.
A.D. has honoraria from Medscape, OncLive, PeerVoice, Physician Education Resources, Tyra Biosciences, Targeted Oncology, MORE Health,Research to Practice, Foundation Medicine, PeerView, AstraZeneca, Genentech/Roche, Bayer; consulting roles at Ignyta, Loxo Oncology, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Genentech/Roche, Takeda, Helsinn Therapeutics, BeiGene, Hengrui Therapeutics, Exelixis, and Bayer.
D.M.H. reports, regardless of potential relevance, personal fees from Atara Biotherapeutics, Chugai Pharma, CytomX Therapeutics, Boehringer Ingelheim, and AstraZeneca and research funding from Puma Biotechnology, AstraZeneca, and Loxo Oncology.
R.Y. has received research support from GlaxoSmithKline, Novartis and Array and consulting fees from GlaxoSmithKline.
J.F.H. has received honoraria from Medscape, the European Society of Medical Oncology, and Axiom Biotechnologies as well as research funding from Bayer.
R.S. has received research funding from Helsinn Therapeutics.
M.F.B. has received honoraria for advisory board participation from Roche and research support from Illumina.
M.L. has received honoraria for ad hoc advisory board participation from Astra-Zeneca, Bristol Myers Squibb, Takeda, and Bayer, and research support from LOXO Oncology (for expanded Archer targeted RNAseq testing) and Helsinn Therapeutics.
P.R. has received consulting fees from Novartis.
References
- 1.Cocco E, Scaltriti M & Drilon A NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 15, 731–747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demetri GD, et al. LBA17Efficacy and safety of entrectinib in patients with NTRK fusion-positive (NTRK-fp) Tumors: Pooled analysis of STARTRK-2, STARTRK-1 and ALKA-372-001. Annals of Oncology 29, mdy424.017–mdy424.017 (2018). [Google Scholar]
- 3.Drilon A, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 378, 731–739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drilon A, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1). Cancer Discov 7, 400–409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laetsch TW, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol 19, 705–714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Zanca D, Hughes SH & Barbacid M A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature 319, 743–748 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Schram AM, Chang MT, Jonsson P & Drilon A Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 14, 735–748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaishnavi A, Le AT & Doebele RC TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov 5, 25–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drilon A, et al. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer Discov 7, 963–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drilon A, et al. Repotrectinib (TPX-0005) Is a Next-Generation ROS1/TRK/ALK Inhibitor That Potently Inhibits ROS1/TRK/ALK Solvent- Front Mutations. Cancer Discov 8, 1227–1236 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Russo M, et al. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer Discov 6, 36–44 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Bardelli A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 3, 658–673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le X, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 24, 6195–6203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietrantonio F, et al. MET-Driven Resistance to Dual EGFR and BRAF Blockade May Be Overcome by Switching from EGFR to MET Inhibition in BRAF-Mutated Colorectal Cancer. Cancer Discov 6, 963–971 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Vega F, et al. EGFR and MET Amplifications Determine Response to HER2 Inhibition in ERBB2-Amplified Esophagogastric Cancer. Cancer Discov 9, 199–209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17, 77–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlino MS, et al. Preexisting MEK1P124 mutations diminish response to BRAF inhibitors in metastatic melanoma patients. Clin Cancer Res 21, 98–105 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Hyman DM, et al. Author Correction: HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 566, E11–E12 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Heist RS, et al. Acquired Resistance to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol 11, 1242–1245 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Qi J, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res 71, 1081–1091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter JC, et al. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Caunt CJ, Sale MJ, Smith PD & Cook SJ MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer 15, 577–592 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Falchook GS, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol 13, 782–789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long GV, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 371, 1877–1888 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Peters S, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 377, 829–838 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Soria JC, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 378, 113–125 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Misale S, et al. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nat Commun 6, 8305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathenson M, et al. O-020Activity of larotrectinib in patients with TRK fusion GI malignancies. Annals of Oncology 29(2018). [Google Scholar]
- 29.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo M, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov 6, 147–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siravegna G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21, 827 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Yaeger R, et al. Mechanisms of Acquired Resistance to BRAF V600E Inhibition in Colon Cancers Converge on RAF Dimerization and Are Sensitive to Its Inhibition. Cancer Res 77, 6513–6523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zehir A, et al. Erratum: Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 1004 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang MT, et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov 8, 174–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tkac J, et al. HELB Is a Feedback Inhibitor of DNA End Resection. Mol Cell 61, 405–418 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Characteristics of the TRK inhibitor-treated patients analyzed and genomic sequencing results obtained from serially collected cfDNA samples. Each column represents a cfDNA time point, with the timing of sample acquisition relative to targeted therapy and the sequencing platform used denoted on top (A: MSK-ACCESS; G: Guardant Rows; laro: larotrectinib; POD: Progression of disease). Single nucleotide variants and their variant allele frequencies (out of 1) are listed in rows. Known TRK mutations and MAPK alterations responsible for the resistance to TRK inhibitors are listed.
Supplementary Table 2: Frequency of the BRAF V600E mutation detected by droplet digital PCR sequencing on the Patient 1-derived PDX treated with vehicle, larotrectinib or the combination of larotrectinib and trametinib. Droplet counts for the wild type and the mutant allele and their ratio (Ratio [Mu]/[WT]) are reported.
Data Availability Statement
All genomic results and associated clinical data for all patients in this study are publically available in the cBioPortal for Cancer Genomics at the following URL: http://cbioportal.org/msk-impact. All relevant cell-free DNA sequencing data are included in the paper and/or supplementary files.