Abstract
Acetylation is a posttranslational modification conserved in all domains of life that is carried out by N-acetyltransferases. While acetylation can occur on Nα-amino groups, this review will focus on Nε-acetylation of lysyl residues and how the posttranslational modification changes the cellular physiology of bacteria. Up until the late 1990s, acetylation was studied in eukaryotes in the context of chromatin maintenance and gene expression. At present, bacterial protein acetylation plays a prominent role in central and secondary metabolism, virulence, transcription, and translation. Given the diversity of niches in the microbial world, it is not surprising that the targets of bacterial protein acetyltransferases are very diverse, making their biochemical characterization challenging. The paradigm for acetylation in bacteria involves the acetylation of acetyl-CoA synthetase, whose activity must be tightly regulated to maintain energy charge homeostasis. While this paradigm has provided much mechanistic detail for acetylation and deacetylation, in this review we discuss advances in the field that are changing our understanding of the physiological role of protein acetylation in bacteria.
Keywords: lysine acetylation, acetyltransferases, deacetylases, enzymatic, abiotic lysine acetylation
INTRODUCTION
The survival of bacteria in their environments is intimately linked to their ability to rapidly respond to stimuli that compromise their survival. In many cases, their survival depends on mechanisms that have evolved to quickly neutralize deleterious effects on diverse cellular processes. Posttranslational modification (PTM) is an efficient way to modulate protein function. Some modifications are reversible, but others cannot be reversed. Reversible modifications are especially useful to the cell because the function of a protein can be up- or downregulated in response to internal or external stimuli bypassing the energy-intensive processes of protein degradation, gene expression, and protein synthesis. PTMs include methylation (63), phosphorylation (11, 39), ADP-ribosylation (86), serine/threonine O-acetylation (41), succinylation (94), ubiquitinylation(62), adenylylation (51), S-nitrosylation (19), lipidation (108), glycosylation (35), phosphocholination (51), and many others (93). Here, we focus on protein acetylation and its effects on bacteria cell physiology.
Discovery of Protein Acetylation and Focus of This Review
The presence of acetyl groups on proteins was first detected in calf thymus histones (57). Acetylation of these lysyl residues abolished their positive charge, blocking the ability of the histone tails to interact with negatively charged phosphate ions of the DNA backbone. The net effect is a relaxation of DNA strands, which allows the transcriptional machinery to access and transcribe genes (75). Since the discovery of lysine acetylation, the scope of studies involving protein acetylation has greatly expanded. A recent review on this topic (25) discussed the advances in the field from the late 1990s to 2015. This review focuses on what has been learned since 2015.
Bacterial Protein Acetyltransferases
In prokaryotes, acetylation is carried out by homologs of the yeast Gcn5 histone N-acetyltransferase (GNATs, PF00583). GNATs transfer the acetyl moiety of acetyl-CoA to Nα-amino groups of proteins and small molecules or Nε-amino groups of lysyl residues of proteins (Figure 1). For clarity, hereafter we refer to bacterial GNATs as bGNATs.
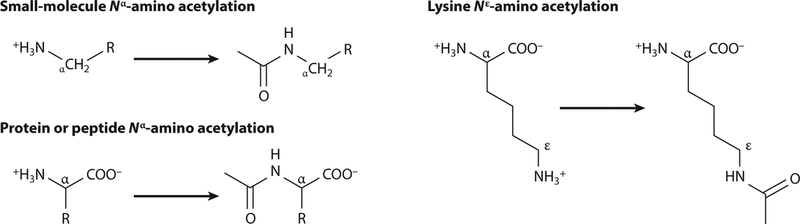
Figure 1.
Reaction schematic of Nα- and Nε-acetylation. Acetylation can occur by two primary mechanisms. (Left) Acetylation of small molecules or N termini of proteins is characterized by addition of an acetyl group to Nα-amino group. (Left, bottom) R represents an amino acid side chain of a protein or peptide. (Right) Generally, Nα-amino acetylation is irreversible. Acetylation of lysyl residues of proteins occurs posttranslationally at the Nε-amino group and can be reversed by deacetylases.
Protein acetylation can occur on Nα-amino groups (i.e., N termini of proteins) or on Nε-amino groups of lysyl residues. Nα-Acetylation is used to signal for protein degradation (24) and is irreversible, while Nε-acetylation of lysyl residues usually alters the biological activity of the protein. The observed reversibility of Nε-Lys acetylation by protein deacetylases (discussed below) allows for rapid modulation of a protein’s activity in vivo.
A bacterial genome can encode ~1–70 acetyltransferases, each predicted to have different targets. For example, the Streptomyces genome codes for 72 acetyltransferases, only a few of which have been characterized (25, 47, 83, 88). Salmonella enterica and Escherichia coli are model organisms for acetylation, and about half of their 26 acetyltransferases have experimentally validated enzymatic activities (8, 15, 27, 31, 32, 40, 48, 65, 71, 76, 78, 87, 90, 100).
While structures of GNATs have a conserved core domain consisting of β sheets that bind acetyl-CoA through interactions with the pyrophosphate and pantothenate moieties (92), it is currently not possible to determine acetyltransferase target preference based on sequence or structure alone. Therefore, it is difficult to establish whether a putative acetyltransferase will target a protein or a small molecule, especially since deletion or overexpression of acetyltransferases do not usually lead to growth phenotypes under standard laboratory conditions.
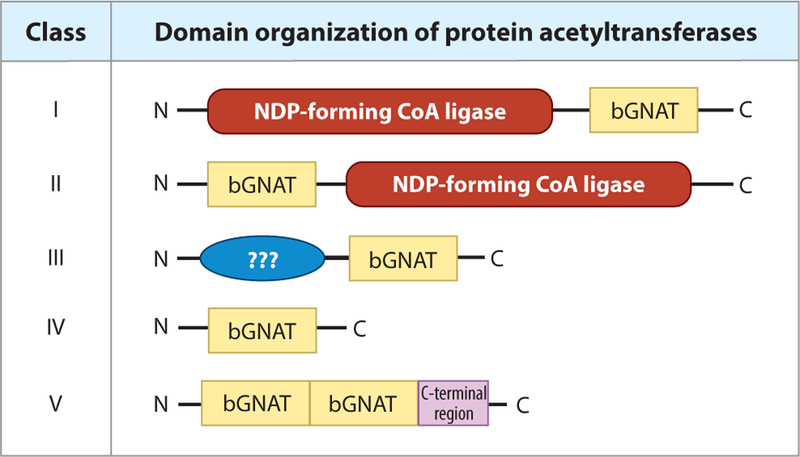
Regardless of substrate specificity, acetyltransferases are categorized based on their domain organization. To date, four domain organizations of acetyltransferases have been found (17, 71, 83, 95) (Figure 2). The S. enterica and E. coli Pat (protein acetyltransferase) was the first type I acetyltransferase to be discovered and consists of a large N-terminal domain (~700 residues) of unknown function and a GNAT-catalytic C-terminal domain (~200 residues) (71). While the physiological function of the large N-terminal domain is yet to be determined, studies showed that each monomer of SePat binds two molecules of acetyl-CoA, one on the N-terminal domain and one on the C-terminal domain (80). It is possible that binding of acetyl-CoA to the large N-terminal domain is a regulatory mechanism to cue acetylation, although no experimental evidence in support of this idea has been reported.
Figure 2.
Domain organization of bacterial protein acetyltransferases. Acetyltransferases belonging to classes I–IV have been identified within prokaryotic genomes. Type I acetyltransferases contain a large regulatory N-terminal domain of unknown function. These domains are homologous to NDP-forming CoA ligases, although they lack a critical active-site histidine residue and do not possess this activity. Within the C terminus of these proteins, a bGNAT catalytic domain carries out the acetylation event. Type II bGNATs are similar to type I, but their domain organization is reversed. The catalytic GNAT domain is within the N terminus, with a large regulatory domain on the C terminus. Type III bGNATs are also similar to type I, in that their regulatory domain lies on the N terminus. However, the regulatory domains of type III acetyltransferases are different in size from that of type I (~300–400 residues versus 800–900 residues, respectively). Type IV bGNATs are unique in that they only contain a catalytic bGNAT domain. It is thought that type I–III acetyltransferases target proteins while type IV are able to target both proteins and small molecules. Lastly, type V contains two sequential bGNAT domains before the C-terminal region. Abbreviations: bGNAT, bacterial GNAT; GNAT, Gcn5 histone N-acetyltransferase; NDP, nucleoside diphosphate.
Type II protein acetyltransferases were discovered in Streptomyces lividans (PatA) (83). This type of acetyltransferase is unique in domain orientation. Unlike type I bGNATs, the N-terminal domain of type II enzymes is the catalytic domain (~200 residues), and the C-terminal regulatory domain (~900 residues) is of unknown function. In addition to the reversed domain organization of type I acetyltransferases, the type II acetyltransferase of S. lividans contains a proline-rich linker that includes a collagen-like G-P-S motif. While the precise role of the regulatory C-terminal domain of SlPatA is unknown, removal of this domain renders the catalytic N terminus inactive (84). It appears that other actinomycetes have Pat enzymes that have similar domain organizations, suggesting that acetylases/deacetylases may have evolved in direct response to pressures unique to environmental niches. In both type I and type II acetyltransferases, the large regulatory domain has sequence homology to ADP-forming CoA synthetases (not to be confused with AMP-forming CoA synthetases that are regulated by acetylation, discussed below). These regulatory domains do not show catalytic activity as ADP-forming CoA synthetases, as the catalytic histidine residue has been substituted (71). It is not known why these enzymes have evolved these regulatory domains, and their function has not been shown. However, work by de Diego Puente and coworkers(14) with E. coli PatZ showed that binding of acetyl-CoA and subsequent autoacetylation of N-terminus residues altered the oligomeric state, triggering the formation of octamers from the stable tetrameric form of the enzyme. Work related to the function of the large domain of S. enterica Pat was performed by random mutagenesis. This approach identified five residues within the N-terminal domain of S. enterica Pat that were critical to function (80). Additional work is needed to determine why single amino acid changes have such a profound effect on the function of the protein. Notably, the type II GNAT from S. lividans contains a proline-rich region with a degenerate G-P-S motif (a signature of collagen) within the large domain (83). It is possible that octamer formation in S. lividans Pat is driven by the affinity among G-P-S regions and not by acetylation, as in E. coli PatZ. We speculate that the octameric state is required to trigger acetylation in vivo, although this has yet to be confirmed.
Type III protein acetyltransferases are similar to type I, in that they contain an N-terminal regulatory domain and a C-terminal GNAT catalytic domain (Figure 2). They differ, in that their regulatory N terminus is smaller (~300–400 residues) and their specific regulatory functions are known. For example, in Mycobacterium smegmatis, the N terminus of PatA (MsPatA) binds cyclic AMP (cAMP), which increases the activity of the enzyme (55), which acetylates a universal stress protein (54). In Micromonospora aurantiaca and Streptomyces lividans, a type III acetyltransferase (PatB) has an ACT-regulatory domain (ACT represents the enzymes initially found with this domain, aspartate kinase, chorismate mutase, and TyrA) that binds amino acids to increase its acetyltransferase activity (88, 97). In M. aurantiaca, binding of l-Cys or l-Arg increased the ability of PatB to acetylate acetyl-CoA synthetase (Acs) of this organism. These ACT-domain acetyltransferases are only found in actinomycetes, raising interesting questions about links in amino acid metabolism and acetylation in these bacteria.
Type IV acetyltransferases have no regulatory domain and consist only of a catalytic GNAT domain (Figure 2). Acetyltransferases with this domain organization are the most abundant within a genome. For example, S. enterica has a single type I acetyltransferase and 25 type IV acetyltransferases. It is generally found that within any given organism, its genome will code for one to two acetyltransferases that fall into the type I–III categories, with the rest of the acetyltransferases being type IV. There are few known exceptions to this rule, as seen in Bacillus subtilis, which has a genome coding for exclusively type IV acetyltransferases. It is generally assumed that type I–III acetyltransferases target protein substrates, with the most commonly known substrate being acyl-CoA synthetases, while type IV acetyltransferases target protein or small-molecule substrates. To date, no type I–III acetyltransferases that acetylate small molecules have been described. Lastly, type V acetyltransferases contain one N-terminal and one central GNAT domain prior to the C-terminal region. The Eis protein from Mycobacterium tuberculosis belongs to this type of acetyltransferases, but at present it is unclear whether the central GNAT domain is catalytically active (7).
Bacterial Protein Deacetylases
Reversibility of lysine protein acetylation is performed by deacetylases. Due to their discovery in yeast where they were shown to demodify acetylated lysyl residues of histone tails (33), deacetylase enzymes are categorized as HDACs (histone deacetylases, PF08295). There are four classes of HDACs, with classes I, II, and IV catalyzing lysine deacetylation without cofactors (28, 67). In these classes of HDACs, Zn(II) is required for deacetylation and binds to a water molecule that then triggers a nucleophilic attack on the carbonyl of the acetyl group. This reaction mechanism releases acetate as a by-product.
Class III HDACs, also known as sirtuins (PF02416, named after the yeast SIR2 protein), require NAD+ as a cofactor to remove the acetyl group from acetylated lysyl residues (29, 105). The acetyl moiety forms an intermediate with NAD+, which causes the release of nicotinamide (an inhibitor of sirtuin activity). This leads to deacetylation of the acetyl-lysyl residue and formation of O-acetyl-ADP-ribose (O-AADPR). The fates and physiological roles of O-AADPR in prokaryotes are not understood. Prokaryotic genomes tend to code for one to two sirtuin homologs, while some genomes also code for HDACs. For example, S. enterica has a single sirtuin NAD+-dependent deacetylase (CobB), while B. subtilis has one sirtuin deacetylase (SrtN) and one HDAC (AcuC) (16, 69). In human cells there are seven isoforms within the class III group of sirtuins (SIRT1–7), and the most commonly studied prokaryotic sirtuins are SIRT5 isoforms (69, 82). However, recent work in S. lividans showed that its genome coded for one SIRT5 deacetylase (CobB), one SIRT4 deacetylase (SrtA), and one HDAC (AcuC) (88). It is predicted these Streptomyces enzymes target different acetylated substrates, with CobB targeting acetoacetyl-CoA synthetase (83) and SrtA targeting Acs (88). How these deacetylases have evolved their substrate specificity is of interest.
Important to note, recent work has identified an additional class of sirtuins that do not catalyze a deacetylase reaction but rather carry out their own PTM. Known as SirTMs, these enzymes in Staphylococcus aureus lack the classic histidine active site of deacetylase sirtuins and instead possess ADP-ribosyltransferase activity. Lipoylation of Lys56 of GcvH (a lipoyl-carrier protein involved in oxidative stress) is required before SirTM can ADP-ribosylate an aspartate residue (60) of the lipoylated GcvH protein. It was predicted that ADP-ribosylation of lipoylated GcvH inhibited its interaction with oxidoreductase, allowing the cells to keep its response off under nonstress conditions (60).
The Paradigm of Reversible Lysine Acetylation
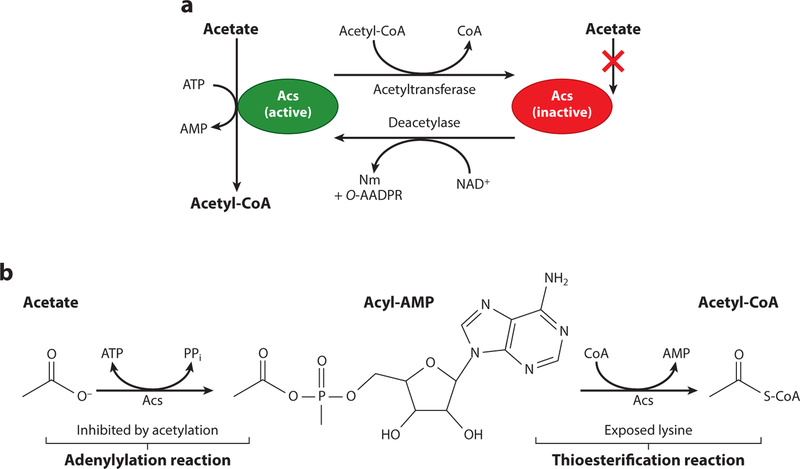
The paradigm for protein acetylation in bacteria is the modification of Acs (69–74) (Figure 3a). Acs is an AMP-forming CoA synthetase that has multiple conformations (apo, adenylylation, and thioesterification). The adenylylation conformation (the first half-reaction) allows acetate and ATP to bind to the enzyme. This conformation catalyzes the formation of an acyl-AMP intermediate (36, 69) (Figure 3b). The thioesterification conformation produces acetyl-CoA and releases AMP (Figure 3b). This same mechanism applies to many other bacterial CoA synthetases that utilize a wide range of fatty acids (20). High formation of AMP following this esterification can lead to energy disruptions in the cell, through inhibition of ADP synthesis (i.e., lower levels of ATP). In S. enterica, K609 of Acs coordinates the negatively charged carboxylate of acetate into the active site, where the protein forms the acyl-AMP intermediate (72). Crystal structures show that in the thioesterification conformation of CoA synthetases, the active-site lysyl residue is surface exposed, making it readily available for acetylation (12, 21). Under conditions where AMP levels are too high due to uncontrolled activity of Acs, an acetyltransferase acetylates residue K609, blocking acyl-AMP synthesis. When activity of Acs is needed, a deacetylase removes the acetyl group on this lysyl residue, returning the protein to its active form (69). This is a prime example of the importance and rapid nature of acetylation in prokaryotes and has been the basis for studying acetylation of other enzymes in bacteria.
Figure 3.
Reversible lysine acetylation of Acs. (a) Pictured is a model of Acs reversible lysine acetylation as studied in Salmonella enterica. Acs is a central metabolic enzyme that under low-acetate concentrations (10 mM) activates acetate to acetyl-CoA (green protein) via an acetyladenylate intermediate with the release of pyrophosphate. A catalytic lysyl residue (K609) coordinates the carboxylic acid of acetate into the active site, and acetylation of K609 halts Acs activity (red protein). The acetyl moiety on K609 can be removed by a protein deacetylase, returning the enzyme to its active form. In S. enterica, the type I bGNAT, Pat, acetylates Acs and the NAD+-dependent sirtuin deacetylase, CobB, removes this modification. (b) Activation of acetate to acetyl-CoA occurs in a two-step reaction mechanism. In the adenylation reaction, K609 of Acs binds ATP and acetate, forming an acyl-AMP intermediate with the release of pyrophosphate. In an esterification reaction, AMP is replaced by CoA, releasing free AMP. It is in this conformation that the active-site lysyl residue is exposed and likely targeted by acetylation. Acetylation of K609 blocks the adenylation reaction and stops the synthesis of acyl-AMP intermediates. Increased levels of AMP in the cell indirectly lead to lower levels of ATP synthesis, and this is the reason this enzyme is tightly regulated by reversible lysine acetylation. Abbreviations: Acs, acetyl-CoA synthetase; bGNAT, bacterial Gcn5 histone N-acetyltransferase; Nm, nicotinamide; O-AADPR, O-acetyl-ADP-ribose; PPi, pyrophosphate.
Sources and Fates of Acetyl-CoA
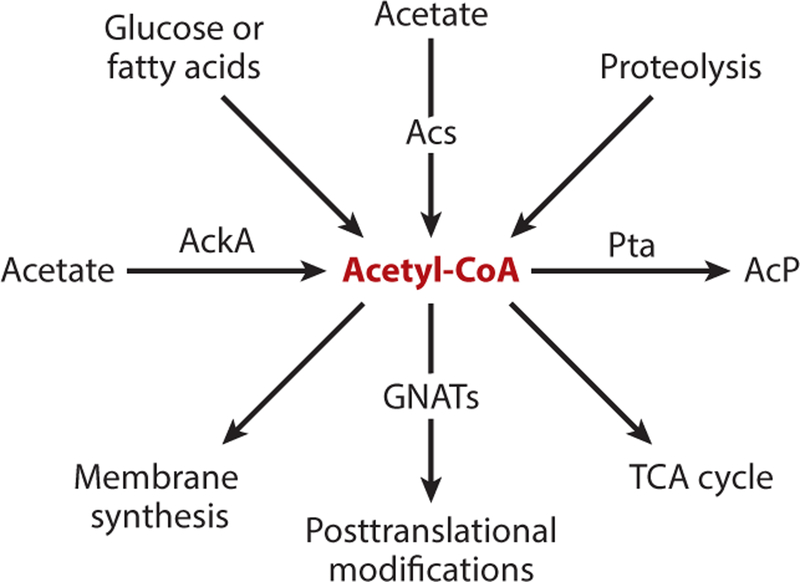
Acetyl donors for acetylation can come from two known sources, acetyl-CoA and acetyl-phosphate (AcP). Metabolically, AcP is generated under high-acetate conditions (50 mM) where acetate kinase activates acetate to acetyl-CoA. The latter is then converted to AcP by phosphotransacetylase (4, 9). The high-energy AcP molecule (ΔG°′ of hydrolysis = 44.8 kJ/mol) (81) can be used as an acetyl donor for chemical acetylation of lysyl residues of proteins (see below). Alternatively, under low-acetate conditions (10 mM), Acs activates acetate to acetyl-CoA. Independent of acetate metabolism, acetyl-CoA is derived from multiple sources. For example, oxidation of amino acids can lead to increased levels of acetyl-CoA (Figure 4). Once acetyl-CoA is produced, it has a variety of fates within the cell. It can be funneled into the TCA cycle for production of reducing equivalents, for the production of fatty acids for membrane synthesis, and, particularly relevant to this review, for use in posttranslational acetylation of proteins (Figure 4). It is clear that acetyl-CoA is the donor of acetyl moieties during enzyme-driven acetylation. However, the question of what triggers the acetylation of specific proteins is a bigger question that needs to be addressed within distinct physiological frameworks.
Figure 4.
Sources and fates of acetyl-CoA. Under low-acetate conditions (10 mM), acetate can be converted to acetyl-CoA via Acs (as described in Figure 3). Under high-acetate conditions (50 mM), AckA activates acetate to acetyl-CoA, while the Pta converts acetyl-CoA to AcP. Metabolism of glucose and other short-chain fatty acids also leads to the production of acetyl-CoA. Proteolysis leads to an increased accumulation of free amino acids, which are eventually deaminated and oxidized to acetyl-CoA. All of these sources of acetyl-CoA can funnel this high-energy molecule into synthesis of fatty acids for membrane production, lead into the TCA cycle for production of reducing equivalents, or be utilized for acetylation reactions. Abbreviations: AckA, acetate kinase; AcP, acetyl-phosphate; Acs, acetyl-CoA synthetase; GNAT, Gcn5 histone N-acetyltransferase; Pta, phosphotransacetylase.
PROTEIN ACETYLATION: ENYZMATIC VERSUS ABIOTIC
Enzymatic Acetylation Mechanism and How That Relates to In Vitro Assays
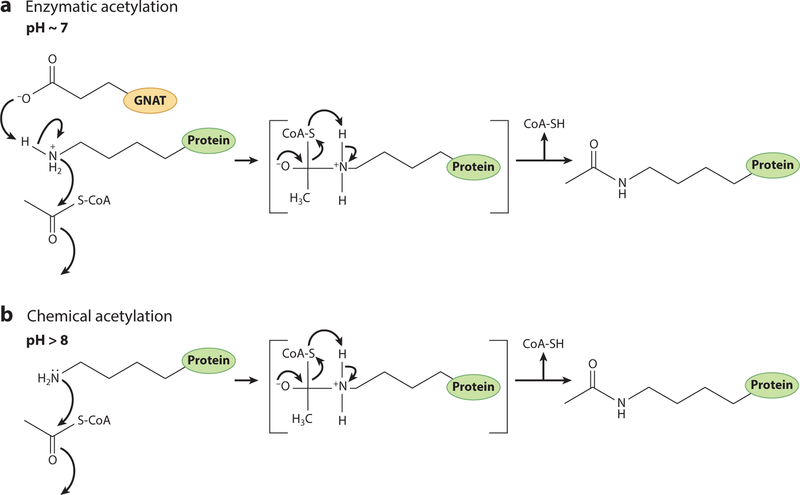
There are multiple examples in the literature that support the conclusion that, in bacteria, the enzymatic acetylation of proteins is of high physiological relevance. While it is not known what triggers the activity of acetyltransferases, we know that acetylation is necessary to maintain cellular homeostasis, modulate transcription of essential genes, and alter cellular metabolism based on environmental stressors or niches (42, 71, 79, 90, 107). This has been shown to be of importance because deletion of acetyltransferases under specific conditions changes the metabolism or physiology of a cell. It has been proposed that the activity of acetyltransferases proceeds via the extraction of a proton from the Nε group of the lysyl side chain by an active-site glutamate (92) (Figure 5). At cellular pH, lysyl residues are protonated and positively charged due to the pKa(10.53) of this side chain. The negatively charged catalytic glutamate residue acts as a base that facilitates a water-mediated proton extraction from the lysyl residue. The deprotonated, nucleophilic lysyl residue attacks the electrophilic carbonyl group of acetyl-CoA, generating acetyl-lysyl residues.
Figure 5.
Reaction mechanism of protein acetylation. (a) Bacterial Gcn5 histone N-acetyltransferase (bGNAT)-mediated protein acetylation involves a catalytic glutamate residue within the bGNAT core domain. The pKa of glutamate is 4.25, and at cellular pH (~7), the side chain is deprotonated. This allows the glutamate residue to act as a base that facilitates a water-mediated proton abstraction from the lysyl residue (pKa = 10.53) of the target protein. The epsilon amino group of the lysyl residue then undergoes a nucleophilic attack on the carbonyl carbon of the acetyl moiety of CoA, leading to the formation of an acetylated lysyl side chain. (b) Under high pH conditions, a larger portion of lysyl residues are deprotonated, bypassing the need for a catalytic proton abstraction from the side chain. Adapted with permission from Reference 25.
While the catalysis is specific and the core domain containing this glutamate is highly conserved, the residues important for recognition of acetyltransferase substrates is poorly understood. For this reason, the characterization of putative acetyltransferases is of utmost importance. Many papers discussed in this review report the identification of lysyl residues of many proteins that are acetylated, but not the acetyltransferases that modify them. We would argue that in order to truly understand the physiological relevance of acetylation, we must identify the enzymes that have evolved to control protein activity.
It is important to point out the relevance of performing in vitro acetylation assays at near neutral pH (~7). Alkaline pH should be avoided due to the lability of the thioester bond of acetyl-CoA to hydroxyl anions. In addition, when the pH of a reaction mixture is >8, a higher percentage of lysyl residues are deprotonated nonenzymatically. The combination of these two factors favors the nonenzymatic transfer of acetyl moiety to the epsilon amino group of lysine (Figure 5). Nonenzymatic acetylation is the biggest challenge to investigators in the field of protein acetylation who are interested in identifying lysyl residues that, if acetylated, can alter protein function. Analysis of published acetylomes reveals proteins that are modified at multiple locations, making it difficult to determine which residue may be responsible for alterations in protein function. It is therefore not only important to assess protein activity after an in vitro acetylation experiment but also equally important to identify the acetyltransferase that catalyzes the modification. Therefore, this review does not focus on global protein acetylome studies, as these experiments generally lead to large levels of chemical acetylation and the targets are, in the majority of cases, not verified with pure proteins in vitro.
Identification of Protein Targets of Putative Bacterial GNATs
Recent work published by Christensen et al. (10) reported the identification of protein targets of uncharacterized bGNATs in E. coli. The interesting part of this work was the use of a strain that reduced background levels of (de)acetylation. Generally, nonenzymatic acetylation is caused by AcP, which is produced from the abovementioned ackA pta pathway. For this study, the authors constructed a strain with pta, cobB, pat, and acs deleted in which the background noise of protein acetylation was substantially reduced, allowing them to identify new protein targets using a combination of Western blot analysis and mass spectrometry of the proteome when specific bGNATs were overproduced. Importantly, the authors substituted residues suspected to be required for bGNAT activity. Results from these initial studies allowed the authors to focus on four uncharacterized E. coli GNATs: RimI, YiaC, YjaB, and PhnO. Important to note, other bGNATs (AatA, ElaA, YiiD, and YafP) were not studied due to overexpression growth phenotypes. While these four GNATs were not analyzed, their growth phenotypes could be of use in the future for identifying their protein or small-molecule targets.
As mentioned above, the authors used mass spectrometry to analyze the acetylome, an approach that yielded a wealth of information. For example, the authors identified large numbers of proteins acetylated by YjaB (128 proteins) and YiaC (251 proteins). In contrast, the numbers of proteins acetylated by RimI and PhnO were substantially less (11 and 10 proteins, respectively). While it remains to be shown that each bGNAT directly acetylates all targets found, it is interesting to see the authors group the putative targets on a metabolic map and compare the specific lysine targets to results found from previous nonenzymatic AcP acetylation studies. Christensen et al. found that a majority of the bGNATs studied targeted proteins involved in early or late stage of glycolysis (prior to the formation of glyceraldehyde 3-phosphate), perhaps indicating a pivotal role for lysine acetylation in central metabolism. Also of interest is their finding that YiaC and YfiQ appeared to acetylate the transcription factor GntR, which controls the expression of genes whose protein products are part of the Entner-Doudoroff pathway.
While the identified targets from mass spectrometry and acetyl-lysine Western blots were not confirmed with pure proteins in vitro, Christensen et al. (10) found some interesting phenotypes associated with the overexpression of yiaC and yfiQ (a.k.a. patZ, pka). When either yiaC or yfiQ were overexpressed in E. coli, cells had a reduced migration on soft-agar motility assays. Notably, none of the proteins identified by mass spectrometry is known to be involved in motility, and the potential acetylation targets causing this phenotype still need to be identified. It was noted that many if not all bGNATs in E. coli are found to be upregulated in biofilms or during stationary phase, and this may be a hint of where to look for acetylation-dependent phenotypes.
In summary, the work presented by Christensen et al. (10) is an excellent example for beginning to identify acetyltransferase protein targets. While no targets were directly identified, this work opens up a lot of doors to continue to search for bGNAT protein targets.
CELLULAR PROCESSES UNDER REVERSIBLE LYSINE ACETYLATION CONTROL
Carbon-Nitrogen Metabolism
Every cell must undergo carbon and nitrogen metabolism for survival. In prokaryotes, the paradigm for protein acetylation is the AMP-forming Acs (69, 71) (Figure 3). Acetylation of Acs is necessary for maintaining energy-charge homeostasis (6), because excess Acs activity leads to the accumulation of AMP and depletion of ATP, leading to growth arrest (6). Since the discovery of Acs acetylation, the control mechanisms of this enzyme have been studied in depth.
While we know acetylation of central metabolic enzymes is necessary for adaptation, the cascade of events that trigger acetylation remain to be elucidated. Acs, like other members of the ANL superfamily of proteins (20) (https://pfam.xfam.org/clan/ANL), has AMP/ATP binding pockets that when occupied stimulate an open conformation of the protein (13). This open conformation exposes the active-site lysine of Acs, rendering it available for acetylation (13). It has recently been shown that acetylation is increased when cAMP, binds to the AMP pocket of Acs(23). While cAMP was bound to acetylated Acs, its deacetylation levels also decreased (23). This could imply that cAMP is a competitor for AMP, which serves as a cellular signal for protein acetylation and control. This was seen not only with Acs but also with other CoA synthetases (FadD and PrpE) of the cell.
An additional mechanism to control acetylation of Acs is through regulation of the posttranslational machinery that targets this enzyme. Multiple studies have shown that central regulatory enzymes control transcription of the genes coding for acetyltransferases and deacetylases (26, 101, 103). The nitrogen response regulator, GlnR, has been shown to upregulate expression of the Saccharopolyspora erythraea acetyltransferase (acuA), and deacetylase (srtN), under nitrogen-limiting conditions (101). In the absence of GlnR, S. erythraea displayed growth phenotypes with acetate that were related to lower levels of the acuA acetyltransferase and therefore dysregulation of Acs acetylation. This work linked nitrogen metabolism to the regulation of the central metabolism acetylation machinery. Other studies have shown that acetylation of GlnR alters its in vitro DNA-binding activity, although these studies require further analysis to assess physiological relevance (1).
Even though acetylation of Acs is thought to be conserved in all domains of life, how this acetylation event differs among bacterial species has recently been investigated. In S. enterica and E. coli, Acs is acetylated by the type I Pat (71) and its homolog PatZ (5), respectively (Figures 2, 3). This Pat protein contains a regulatory domain of unknown function (~700 amino acids) attached to the catalytic GNAT domain (~200 amino acids). Recent work also highlights the importance of Acs acetylation and deacetylation during bacterial infections (44). Using Vibrio cholerae and Drosophila melanogaster as an infection model, it was found that dysregulation of Acs acetylation reduced V. cholera virulence. This work emphasizes the impact of protein acetylation on the physiology of pathogenic bacteria that affect human health across the world (44). While many studies characterizing Acs acetylation by Pat have similarities, there are also differences worth discussing here. For example, work in M. aurantiaca identified an ACT-domain acetyltransferase that bound cysteine and arginine, which increased its activity for Acs acetylation (97). The homologous ACT-Pat protein of S. lividans was recently identified (88), and it also modified Acs in this bacterium. However, there were differences from the acetylation paradigm in S. enterica worth noting. The most important findings of these studies were: (a) Type I Pat enzymes show specificity for their own Acs substrate; that is, S. enterica Pat does not recognize S. lividans Acs, and conversely, S. lividans ACTPat does not recognize S. enterica Acs; and (b) unlike S. enterica Acs, the S. lividans Acs is acetylated on a serine residue within the acetylation motif of the protein. The alluded serine residue (S608) was located two positions upstream of the canonical acetylation site, i.e., K610. Notably, acetylation of S608 was required for ACT-Pat-mediated K610 acetylation (88). From a mechanistic standpoint, it is unclear why S608 needs to be acetylated for K610 to be modified. At present, the identity of the O-acetyltransferase enzyme that modifies S608 is unknown, but it is clear that Acs control in Streptomyces may be more complicated than in other bacteria. At the moment, one can only speculate about the existence of selective pressures unique to the environment occupied byS. lividans and M. aurantiaca (i.e., soil) that drove the evolution of a more elaborate control of Acs than the one observed in intestinal bacteria such as S. enterica and E. coli.
While acetylation of Acs has been and continues to be analyzed, studies are expanding to assess posttranslational modifications of additional central metabolic enzymes. A good place to start investigating lysine acetylation is to identify central metabolic enzymes that have active-site lysines that can be accessed by bGNATs (53). As mentioned previously, the positive charge of lysyl residues interacts with a substrate in the active site of an enzyme. Lysine acetylation abolishes this charge, blocking catalysis. Enolase, a central metabolic enzyme that converts 2-phosphoglycerate to phosphoenolpyruvate, has been found to be acetylated in many acetylome studies (104, 106). Work has been done to show in vitro that chemical acetylation of enolase decreases its activity and that acetylation mimics (e.g., Gln) of active-site enolase lysyl residues reduce its activity (53). Other work inM. tuberculosis identified two acetylation sites of isocitrate lyase (3). Acetylation of one lysyl residue increased the activity of the enzyme, while acetylation of another lysyl residue decreased enzyme activity. Interestingly, growth of M. tuberculosis on acetate and propionate increased acetylation levels of isocitrate lyase. It is possible the enzyme was chemically modified as a consequence of increased acetyl-CoA or AcP pools. This is a possibility that must be taken into consideration since acetyltransferases responsible for the modifications were not found. Enzyme-driven protein acetylation offers opportunities to better understand the impact of acetylation on the physiology of an organism. The powerful acetylating activity of AcP makes it more challenging to reach this goal.
Acetylome studies have revealed that many proteins are acetylated on multiple lysines. Therefore, verification of these acetylation events is pivotal for the advancement of the field. One such study characterized the acetylation of phosphoenolpyruvate carboxylase (PEPC), an enzyme that converts phosphoenolpyruvate to oxaloacetate (50, 52). Metabolic flux after production of oxaloacetate leads to increased synthesis of aspartate and ensures carbon flow through the citric acid cycle for production of glutamate. Corynebacterium glutamicum is a bacterium used for industrial production of glutamate, and under glutamate-producing conditions, the authors found that PEPC was acetylated at K653 (52). Although cells with a PEPCK653Q variant (an acetylation mimic) did not display growth phenotypes, glutamate production was completely abolished, and in vitro activity was greatly reduced compared to PEPCWT. A PEPCK643R (a deacetylation mimic variant) produced high levels of glutamate and maintained moderate activity in vitro. These results showed not only that the charge of residue K653 is necessary for enzyme activity, but also that by abolishing enzymatic activity in the PEPCK653Q variant, metabolic flux was greatly impacted. While the deacetylase of C. glutamicum was able to deacetylate artificially acetylated PEPC, no acetyltransferase was identified for this modification. Although AcP or acetyl-CoA could be a substrate for chemical modification of PEPC, we would argue PEPC acetylation would have to be tightly regulated and is likely enzymatic.
Other work has begun to investigate posttranslational modifications of enzymes involved in overproduction of antimicrobial-producing enzymes (96) and highlight that such modifications can greatly alter the derived products. Lysine malonylation was recently profiled in E. coli, where proteins involved in protein synthesis, energy metabolism, and fatty acid biosynthesis were enriched for this type of modification (59). Since the focus of this review is on acetylation, protein malonylation is not discussed in detail. However, it is important to note that CobB sirtuins can demalonylate modified proteins (56). Both of the abovementioned studies (52, 96) highlighted the importance of understanding the effects of protein acylation on metabolic flux. For industrial production of small molecules, we can be thinking of not only the present enzymes but also how those enzymes may be posttranslationally modified and how we can use that information to better engineer microbes to produce optimal yields of valuable chemicals.
Transcription
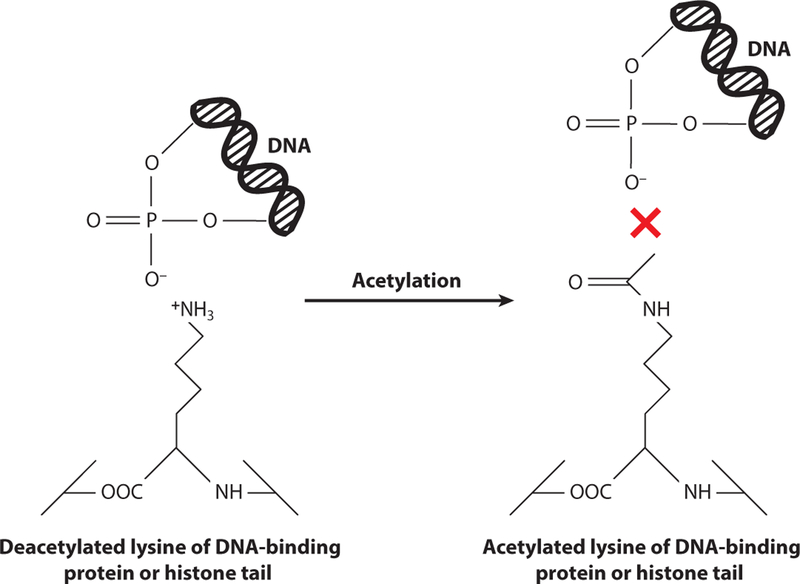
The ways in which acetylation alters gene regulation have been studied extensively in eukaryotes through the process of histone acetylation (57). Histones are nucleoid-associated proteins that have lysine-rich DNA-binding domains that interact with the negatively charged DNA backbone (Figure 6). The yeast Gcn5-acetyltransferase targets lysines within the DNA-binding domain of histones H3 and H2B (77), and yeast EsaI acetylates histone H4 and H2A (77). While histone acetylation has been given much attention, the prokaryotic mechanisms from which it evolved have recently been investigated. Although acetylation of transcription factors had been reported in acetylome studies (104, 106), it was not until recently that an acetyltransferase was found to acetylate and alter DNA binding of a prokaryotic transcription factor (79). Since then, an array of studies have been conducted to further elucidate these mechanisms.
Figure 6.
Acetylation schematic of lysyl residues of histone tails or prokaryotic DNA-binding proteins. Deacetylated lysyl residues (left) maintain a positive charge at cellular pH and interact with the negatively charged phosphate moieties on the DNA backbone. Upon acetylation of these lysyl residues by acetyltransferases (right), the charge on the side chain is abolished and can no longer interact with the DNA, causing a disruption in the interactions.
The prokaryotic DNA-binding protein HU is a small (10 kDa), basic, and thermostable protein that most closely resembles eukaryotic histones. HU is similar to histones, in that it causes DNA supercoiling with the assistance of topoisomerase (64). HU has been found to be acetylated in multiple acetylome studies; however, only in M. tuberculosis has an acetyltransferase been identified to directly acetylate HU (18). This acetyltransferase, Eis, modified MtHU and altered its DNA-binding capabilities in vitro (18). Mass spectrometry revealed 29 possible acetylation sites in the C-terminal domain, but currently it is unknown whether the Eis acetyltransferase targets one or multiple lysyl residues of MtHU. Interestingly, overexpression of eis in M. tuberculosis caused enhanced cell survival in macrophages, raising the possibility that acetylation plays an important role in pathogenesis of this organism. Further work identified a deacetylase, Rv1151c, that reversed the acetylation of HU by Eis (2). Deacetylation of MtHU restored its DNA-binding capabilities, and together this work encompassed a deacetylase/acetyltransferase system that targets a DNA-binding protein.
In addition to HU, another DNA-binding protein in M. tuberculosis, DosR, was found to be acetylated (98). A M. tuberculosis cAMP-domain acetyltransferase, RV0998, acetylated DosR in vitro. Residue K182 of DosR is known to interact with DNA, and under hypoxia it was found this lysyl residue had lower levels of acetylation. To replicate deacetylation of DosR under hypoxia, variants were constructed to mimic deacetylated DosR (DosRK182R). Presence of DosRK182R (a deacetylation mimic variant) led to a significant change in transcription of many genes (acr, dosR, fdxA, and others). The DosRK182R variant bound all DNA in electrophoretic mobility shift assays while the acetylation mimic variant, DosRK182Q, did not. Interestingly, cells where the gene encoding the RV0998 acetyltransferase was inactivated had reduced growth in macrophages, while DosRK182R-producing cells (a deacetylation mimic) inhibited intracellular survival of M. tuberculosis. Overall, it was hypothesized that hypoxia might induce deacetylation of DosR, which increases its DNA-binding ability to promote transcription of target genes needed for M. tuberculosis to adapt to hypoxia during virulence. While more work needs to be done to understand what triggers acetylation of DosR, this work highlighted an additional reversible lysine acetylation system of M. tuberculosis that is necessary for pathogenesis.
Much as in Mycobacterium, pathogenesis is affected via acetylation of transcriptional regulators in S. enterica. In this bacterium, the involvement of the Pat/CobB enzymes has been established by different research groups, but the conclusions reached by each group cannot yet be reconciled, suggesting that there may be additional information that needs to be obtained to bridge the differences in understanding how things work. Notably, both groups agree that the master regulator HilD, which controls the expression of the Salmonella pathogenicity island 1 (SPI1) is a substrate of the Pat enzyme (30, 66). Both groups agree that Pat acetylates HilD, but Hung et al. conclude that acetylated HilD (HilDAc) becomes unstable, while Sang et al. (66) conclude that Pat function is needed for HilD stability. Deacetylation of HilDAc by CobB was inferred by Hung et al. (30) from experiments that included known inhibitors of CobB sirtuins (e.g., splitomicin, nicotinamide). In the presence of such inhibitors a sevenfold reduction in the expression of a HilD-controlled reporter was measured, and the half-life of HilD was reduced fourfold. Additionally, HilDAc had a 60% reduced invasion rate in S. enterica compared to deacetylation mimics. Lastly, HilDAc reduced S. enterica colonization in ceca. Acetylation is used to fine-tune HilD activity during S. enterica infections, since it is hypothesized that too much HilD can be detrimental (66).
It seems that acetylation of prokaryotic regulators is often linked to a cell switching its metabolism or physiology. A recent proteomic study in B. subtilis identified a myriad of acetylated proteins, and a few of those targets were analyzed in depth (61). These authors showed that acetylation played a critical role in B. subtilis biofilm formation and swarming motility. The ability to modulate swarming or biofilm formation was achieved through acetylation of the YmcA biofilm regulator and the DegUS two-component system that modulates genes required for swarming motility (61). This study not only linked transcriptional regulation to acetylation but also linked multicellularity to acetylation mechanisms.
Work in Porphyromonas gingivalis highlighted a repressor involved in the oxidative stress response, RprY, whose coding region shared a transcription start site with pat (43). In vitro and in vivo studies showed that Pat enhanced acetylation of RprY and that acetylation changed the in vitro DNA-binding activity of RprY. Many of the abovementioned studies are a beginning to what is sure to be a fruitful field of transcription factor control via acetylation. Studying these processes in prokaryotes might further our understanding of how acetylation might have evolved in eukaryotes. Not only are regulators directly acetylated, but some of their small molecule ligands are acetylated (87, 89); however, that is outside of the scope of this review.
Translation
Acetylation has been considered to be a part of the mechanisms that control protein synthesis since the discovery of Rim acetyltransferases. Three acetyltransferases, RimI, RimJ, and RimL acetylate Nα-amine groups of ribosomal proteins S18, S5, and L12, respectively (78, 100). Since then, many indirect mechanisms of acetylation have been shown to alter mRNA translation efficiency in prokaryotes.
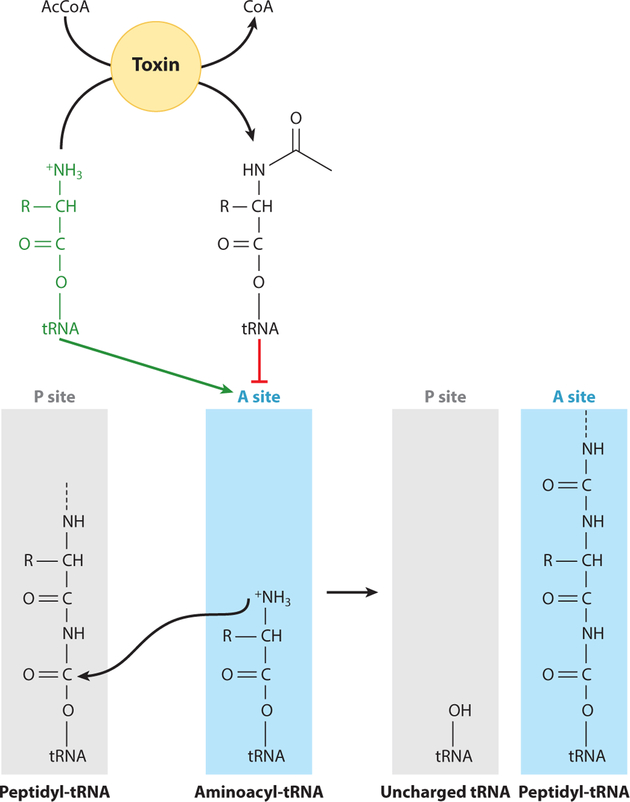
Exciting and noteworthy work in this research area has focused on toxin-antitoxin systems in bacteria where the toxin codes for an acetyltransferase. Work in S. enterica and E. coli has shown multiple toxin acetyltransferases that acetylate aminoacyl-tRNAs (8, 37, 65). It was found that three toxin-antitoxin acetyltransferase systems in S. enterica acetylated specific aminoacyl-tRNAs to varying degrees (8, 65). In E. coli, the acetyltransferase toxin acetylated only formyl-methionine tRNA; however, it is unclear where the site of acetylation is located, since there is no free amino group on fmet-tRNA (37). It is possible that the formyl group is lost immediately before the free amino group of methionine can be modified. Acetylation of free amino groups on aminoacyltRNAs blocks the ability of the ribosome to form peptide bonds (Figure 7), hence arresting protein synthesis, and in the case of some pathogenic bacteria, triggers a persister state that substantially reduces the activity of essential cellular machinery to evade host defenses. Once the cell has avoided the threat, it can reverse aminoacyl-tRNA acetylation and return to active growth.
Figure 7.
Acetylation of aminoacyl-tRNAs blocks peptide bond formation and inhibits protein synthesis. Under Salmonella enterica and Escherichia coli noninfectious states of growth, aminoacyl-tRNAs in the P site of the ribosome contain a free amino group that can attack the carbonyl group of the peptide chain in the P site. Many toxin acetyltransferases in these organisms have been found to acetylate this free amino group, effectively blocking protein synthesis. Abbreviation: AcCoA, acetyl-CoA.
It should be noted that it is unknown what triggers the acetylation of aminoacyl-tRNAs in these organisms, but hypotheses involving acetylation of the antitoxin have been studied (90). In this work, it was shown that acetylation of the antitoxin by its cognate toxin increased the toxin’s activity, increasing the level of acetylated aminoacyl-tRNAs, and reducing protein synthesis (90). In the alluded case, the CobB sirtuin deacetylated the antitoxin, reducing the activity of the toxin to its original level. This work was intriguing because it was the first instance of an acetyltransferase targeting a protein (antitoxin) and nonprotein (aminoacyl-tRNA) substrate. Additionally, this work highlighted a toxin that modulated its own activity through acetylation of its antitoxin.
In addition to the work in S. enterica and E. coli, other groups have identified toxin-antitoxin systems coding for acetyltransferase toxins that are predicted or shown to be involved in inhibiting protein synthesis (34, 46, 49, 58). It is important to study other toxin-antitoxin acetyltransferase systems to determine whether the toxins maintain their activity while in complex with their antitoxins and further determine the mechanisms in which cells enter and leave persister states. The current hypothesis in the field involves recycling of acetylated aminoacyl-tRNAs by peptidyltRNA hydrolase (Pth) (8) or through acetylation/deacetylation of the antitoxin to control the toxin’s aminoacyl-tRNA acetyltransferase activity (90). It is possible that a combination of both processes is at play, and this warrants further investigation. The reader is directed to many excellent reviews on toxin-antitoxin systems for further information about this fascinating topic (22, 38, 45, 85, 99).
Acetylation has been shown to affect the activity of tyrosyl-tRNA synthetase (TyrRS) in E. coli (91). Genetically encoded incorporation of acetyl-lysyl into specific locations of TyrRS resulted in variants with lower catalytic efficiencies. Importantly, the CobB sirtuin deacetylase removed these chemical modifications, returning TyrRS to its wild-type activity. In a noteworthy experiment of this work, the authors purified 25 E. coli acetyltransferases and tested them for in vitro acetylation of TyrRS. The only potential acetyltransferase candidate was Pat, but the activity of TyrRS was not tested after enzymatic acetylation. The authors suggested that perhaps AcP was the source of acetylation in vivo, although it would be interesting to further study the effects of Pat on this system.
CHALLENGES FACING CURRENT RESEARCH ON PROTEIN ACETYLATION
Below, we address some challenges that investigators interested in this research area face while trying to identify proteins whose function is modulated by acetylation.
Nonenzymatic acetylation. The fact that AcP is such a potent acetylating agent raises the background noise, making the task of identifying lysyl residues that are enzymatically modified laborious, hence slowing down the pace of progress. To circumvent this problem, investigators have introduced genetically encoded acetyl-lysyl residues at suspected acetylation sites. In other instances, AcP is used to acetylate all available sites and measurement of protein function is then correlated to degree of acetylation. In some cases, deacetylation by sirtuins or other deacetylases is used to infer that there is probably an acetyltransferase responsible for the modification. However, in the vast majority of cases, the identity of the acetyltransferase remains an enigma. These approaches leave unanswered, important questions regarding how the modifications are introduced and, most importantly, what physiological conditions caused by endogenous or exogenous stimuli lead to the introduction of such PTMs.
From our perspective, to accelerate progress in the field, more attention should be paid to the identification of the targets of bGNATs. Unfortunately, bioinformatics tools currently available to distinguish bGNATs that modify proteins versus those that modify small molecules are not effective. One way to circumvent this problem would be to couple the overexpression of genes encoding bGNATs with mass spectrometric analyses of the proteome followed by in vitro validation of the findings. The big question here is, under what conditions should cells be grown for these analyses? The answer should be guided by what is known about the lifestyles of the bacteria. Ideally, one should be able to find conditions that generate phenotypes that can be dissected genetically and biochemically.
Some researchers use anti-acetyl-lysine antibodies to probe for acetylation of proteins from in vivo overexpression experiments. This approach is frequently uninformative, due to the fact that in stationary phase, lysyl residues of proteins act as sinks for excess acetyl-CoA and AcP (68). If Western blots are used, we think it may be more informative to harvest in early or mid-log of growth, and to verify single acetylation sites via site-directed mutagenesis of targeted lysyl residues. It is generally thought that a physiologically relevant acetylation event occurs on a single lysyl residue, and therefore acetylation of a protein should be abolished (via in vitro assays, i.e., radiolabeled acetyl-CoA assays or in vivo assays, i.e., Western blots) when the single lysyl residue is altered. To the best of our knowledge, multiple enzyme-catalyzed acetylation events that have an effect on protein function have only been shown in the case of the S. erythraea Acs enzyme, which is acetylated four times by the S. erythraea AcuA acetyltransferase (102).
Why do proteomes contain so many acetylated lysyl residues? We speculate that the acetylation of lysyl residues plays an additional role that is connected to the metabolic value of acetyl moieties. When deacetylation is performed by enzymes that yield acetate, cells can readily activate it to acetyl-CoA, arguably the most important building block in metabolism. By acetylating lysyl residues, cells retain acetate rather than extruding into the environment where acetate can be consumed by other bacteria. In other words, it is possible that the acetylation of lysyl residues that do not affect protein function reflects on an acetate storage strategy that avoids loss of carbon.
OUTLOOK AND CONCLUDING REMARKS
There are many outstanding questions and opportunities in the emerging field of protein acylation. For example, it is of interest to know what controls and triggers protein acetylation and deacetylation. Are there AcP acetylation mechanisms that are physiologically relevant? Are there acetyltransferase enzymes that use AcP as substrate in lieu of acetyl-CoA? Or as mentioned above, is nonenzymatic AcP acetylation utilized as a carbon storage strategy? Regardless of mechanisms, the lack of validation of acetylation targets is mandatory for the assessment of the impact of protein acetylation on bacterial cell physiology. Many studies have shown that the function of a protein is not the only aspect affected by lysine acetylation, and perhaps we need to pay more attention to effects of acetylation on structure, oligomerization, protein-protein interaction, cellular localization, etc.
The information on protein acetylation is rapidly growing and is building a solid foundation for further characterization of bGNATs. As the field advances, we will find that many bGNATs, perhaps the majority, target small molecules and that this family of enzymes use not only acetate but an assortment of organic acids. Acetylation, paired with deacetylation, is a rapid way for a cell to adjust cellular metabolism or flux and is involved in a variety of important cellular processes ranging from virulence to central metabolism to DNA binding to protein synthesis. Acetylation therefore warrants much attention with regard to bacterial physiology and will provide valuable insights into how cells respond and adapt to endogenous and exogenous stimuli.
ACKNOWLEDGMENTS
This work was supported by NIH grant R35 GM130399 to J.C.E.-S.
Glossary
- PTM
posttranslational modification
- GNAT
Gcn5-related N-acetyltransferase
- bGNAT
bacterial GNAT
- Pat (Pat, PatA, PatB)
protein acetyltransferase
- ACT
aspartate kinase-chorismate mutase-TyrA domain
- Acs
acetyl-CoA synthetase
- CobB
a protein deacetylase
- AcP
acetyl-phosphate
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Amin R, Franz-Wachtel M, Tiffert Y, Heberer M, Meky M, et al. 2016. Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145. Front. Mol. Biosci 3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand C, Garg R, Ghosh S, Nagaraja V. 2017. A Sir2 family protein Rv1151c deacetylates HU to alter its DNA binding mode in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun 493:1204–9 [DOI] [PubMed] [Google Scholar]
- 3.Bi J, Wang Y, Yu H, Qian X, Wang H, et al. 2017. Modulation of central carbon metabolism by acetylation of isocitrate lyase in Mycobacterium tuberculosis. Sci. Rep 7:44826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinsmade SR, Escalante-Semerena JC. 2007. In vivo and in vitro analyses of single-amino acid variants of the Salmonella enterica phosphotransacetylase enzyme provide insights into the function of its N-terminal domain. J. Biol. Chem 282:12629–40 [DOI] [PubMed] [Google Scholar]
- 5.Castano-Cerezo S, Bernal V, Blanco-Catala J, Iborra JL, Canovas M. 2011. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol 82:1110–28 [DOI] [PubMed] [Google Scholar]
- 6.Chan CH, Garrity J, Crosby HA, Escalante-Semerena JC. 2011. In Salmonella enterica, the sirtuindependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Mol. Microbiol 80:168–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Biswas T, Porter VR, Tsodikov OV, Garneau-Tsodikova S. 2011. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. PNAS 108:9804–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheverton AM, Gollan B, Przydacz M, Wong CT, Mylona A, et al. 2016. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell 63:86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chittori S, Savithri HS, Murthy MR. 2012. Structural and mechanistic investigations on Salmonella typhimurium acetate kinase (AckA): identification of a putative ligand binding pocket at the dimeric interface. BMC Struct. Biol 12:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen DG, Meyer JG, Baumgartner JT, D’Souza AK, Nelson WC, et al. 2018. Identification of novel protein lysine acetyltransferases in Escherichia coli. mBio 9:e01905–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cousin C, Derouiche A, Shi L, Pagot Y, Poncet S, Mijakovic I. 2013. Protein-serine/threonine/tyrosine kinases in bacterial signaling and regulation. FEMS Microbiol. Lett 346:11–19 [DOI] [PubMed] [Google Scholar]
- 12.Crosby HA, Rank KC, Rayment I, Escalante-Semerena JC. 2012. Structural insights into the substrate specificity of the protein acetyltransferase RpPat: identification of a loop critical for recognition by RpPat. J. Biol. Chem 287:41392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crosby HA, Rank KC, Rayment I, Escalante-Semerena JC. 2012. Structure-guided expansion of the substrate range of methylmalonyl-CoA synthetase (MatB) of Rhodopseudomonas palustris. Appl. Environ. Microbiol 78:6619–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Diego Puente T, Gallego-Jara J, Castano-Cerezo S, Bernal Sanchez V, Fernandez Espin V, et al. 2015. The protein acetyltransferase PatZ from Escherichia coli is regulated by autoacetylation-induced oligomerization. J. Biol. Chem 290:23077–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuchi J, Kashiwagi K, Takio K, Igarashi K. 1994. Properties and structure of spermidine acetyltransferase in Escherichia coli. J. Biol. Chem 269:22581–85 [PubMed] [Google Scholar]
- 16.Gardner JG, Escalante-Semerena JC. 2009. In Bacillus subtilis, the sirtuin protein deacetylase encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl-CoA synthetase. J. Bacteriol 191:1749–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner JG, Grundy FJ, Henkin TM, Escalante-Semerena JC. 2006. Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD+ involvement in Bacillus subtilis. J. Bacteriol 188:5460–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, Padmanabhan B, Anand C, Nagaraja V. 2016. Lysine acetylation of the Mycobacterium tuberculosis HU protein modulates its DNA binding and genome organization. Mol. Microbiol 100:577–88 [DOI] [PubMed] [Google Scholar]
- 19.Gould N, Doulias PT, Tenopoulou M, Raju K, Ischiropoulos H. 2013. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J. Biol. Chem 288:26473–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulick AM. 2009. Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol 4:811–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. 2003. The 1.75 Å crystal structure of acetyl-CoA synthetase bound to adenosine-5 -propylphosphate and coenzyme A. Biochemistry 42:2866–73 [DOI] [PubMed] [Google Scholar]
- 22.Hall AM, Gollan B, Helaine S. 2017. Toxin-antitoxin systems: reversible toxicity. Curr. Opin. Microbiol 36:102–10 [DOI] [PubMed] [Google Scholar]
- 23.Han X, Shen L, Wang Q, Cen X, Wang J, et al. 2017. Cyclic AMP inhibits the activity and promotes the acetylation of acetyl-CoA synthetase through competitive binding to the ATP/AMP pocket. J. Biol. Chem 292:1374–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helbig AO, Gauci S, Raijmakers R, van Breukelen B, Slijper M, et al. 2010. Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol. Cell Proteom 9:928–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentchel KL, Escalante-Semerena JC. 2015. Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev 79:321–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentchel KL, Escalante-Semerena JC. 2015. Complex regulation of the sirtuin-dependent reversible lysine acetylation system of Salmonella enterica. Microbial. Cell 2:451–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hentchel KL, Escalante-Semerena JC. 2015. In Salmonella enterica, the Gcn5-related acetyltransferase MddA (formerly YncA) acetylates methionine sulfoximine and methionine sulfone, blocking their toxic effects. J. Bacteriol 197:314–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodawadekar SC, Marmorstein R. 2007. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene 26:5528–40 [DOI] [PubMed] [Google Scholar]
- 29.Hoff KG, Avalos JL, Sens K, Wolberger C. 2006. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure 14:1231–40 [DOI] [PubMed] [Google Scholar]
- 30.Hung CC, Eade CR, Altier C. 2016. The protein acyltransferase Pat post-transcriptionally controls HilD to repress Salmonella invasion. Mol. Microbiol 102:121–36 [DOI] [PubMed] [Google Scholar]
- 31.Hung MN, Rangarajan E, Munger C, Nadeau G, Sulea T, Matte A. 2006. Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis. J. Bacteriol 188:5606–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeuchi Y, Kitahara K, Suzuki T. 2008. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J 27:2194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai S, Johnson FB, Marciniak RA, McVey M, Park PU, Guarente L. 2000. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb. Symp. Quant. Biol 65:297–302 [DOI] [PubMed] [Google Scholar]
- 34.Iqbal N, Guerout AM, Krin E, Le Roux F, Mazel D. 2015. Comprehensive functional analysis of the 18 Vibrio cholerae N16961 toxin-antitoxin systems substantiates their role in stabilizing the superintegron.J. Bacteriol 197:2150–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarrell KF, Ding Y, Meyer BH, Albers SV, Kaminski L, Eichler J. 2014. N-Linked glycosylation in archaea: a structural, functional, and genetic analysis. Microbiol. Mol. Biol. Rev 78:304–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jogl G, Tong L. 2004. Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry 43:1425–31 [DOI] [PubMed] [Google Scholar]
- 37.Jurenas D, Chatterjee S, Konijnenberg A, Sobott F, Droogmans L, et al. 2017. AtaT blocks translation initiation by N-acetylation of the initiator tRNA(fMet). Nat. Chem. Biol 13:640–46 [DOI] [PubMed] [Google Scholar]
- 38.Jurenas D, Garcia-Pino A, Van Melderen L. 2017. Novel toxins from type II toxin-antitoxin systems with acetyltransferase activity. Plasmid 93:30–35 [DOI] [PubMed] [Google Scholar]
- 39.Kennelly PJ. 2014. Protein Ser/Thr/Tyr phosphorylation in the Archaea. J. Biol. Chem 289:9480–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leibowitz MJ, Soffer RL. 1970. Enzymatic modification of proteins. 3. Purification and properties of a leucyl, phenylalanyl transfer ribonucleic acid protein transferase from Escherichia coli. J. Biol. Chem 245:2066–73 [PubMed] [Google Scholar]
- 41.Li C, Wang D, Lv X, Jing R, Bi B, et al. 2017. Yersinia pestis acetyltransferase-mediated dual acetylation at the serine and lysine residues enhances the auto-ubiquitination of ubiquitin ligase MARCH8 in human cells. Cell Cycle 16:649–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R, Gu J, Chen YY, Xiao CL, Wang LW, et al. 2010. CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol 76:1162–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Krishnan K, Duncan MJ. 2018. Post-translational regulation of a Porphyromonas gingivalis regulator.J. Oral. Microbiol 10:1487743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liimatta K, Flaherty E, Ro G, Nguyen DK, Prado C, Purdy AE. 2018. A putative acetylation system in Vibrio cholerae modulates virulence in arthropod hosts. Appl. Environ. Microbiol 84:e01113–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobato-Marquez D, Diaz-Orejas R, Garcia-Del Portillo F. 2016. Toxin-antitoxins and bacterial virulence. FEMS Microbiol. Rev 40:592–609 [DOI] [PubMed] [Google Scholar]
- 46.Lobato-Marquez D, Moreno-Cordoba I, Figueroa V, Diaz-Orejas R, Garcia-del Portillo F. 2015. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep 5:9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu YX, Liu XX, Liu WB, Ye BC. 2017. Identification and characterization of two types of amino acid-regulated acetyltransferases in actinobacteria. Biosci. Rep 37:BSR20170157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marvil DK, Leisinger T. 1977. N-Acetylglutamate synthase of Escherichia coli: purification, characterization, and molecular properties. J. Biol. Chem 252:3295–303 [PubMed] [Google Scholar]
- 49.McVicker G, Tang CM. 2016. Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nat. Microbiol 2:16204. [DOI] [PubMed] [Google Scholar]
- 50.Mizuno Y, Nagano-Shoji M, Kubo S, Kawamura Y, Yoshida A, et al. 2016. Altered acetylation and succinylation profiles in Corynebacterium glutamicum in response to conditions inducing glutamate overproduction. MicrobiologyOpen 5:152–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller MP, Albers MF, Itzen A, Hedberg C. 2014. Exploring adenylylation and phosphocholination as post-translational modifications. ChemBioChem 15:19–26 [DOI] [PubMed] [Google Scholar]
- 52.Nagano-Shoji M, Hamamoto Y, Mizuno Y, Yamada A, Kikuchi M, et al. 2017. Characterization of lysine acetylation of a phosphoenolpyruvate carboxylase involved in glutamate overproduction in Corynebacterium glutamicum. Mol. Microbiol 104:677–89 [DOI] [PubMed] [Google Scholar]
- 53.Nakayasu ES, Burnet MC, Walukiewicz HE, Wilkins CS, Shukla AK, et al. 2017. Ancient regulatory role of lysine acetylation in central metabolism. mBio 8:e01894–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nambi S, Basu N, Visweswariah SS. 2010. Cyclic AMP-regulated protein lysine acetylases in mycobacteria. J. Biol. Chem 285:24313–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noy T, Xu H, Blanchard JS. 2014. Acetylation of acetyl-CoA synthetase from Mycobacterium tuberculosis leads to specific inactivation of the adenylation reaction. Arch. Biochem. Biophys 550–51:42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, et al. 2011. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteom 10:M111.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phillips DM. 1963. The presence of acetyl groups of histones. Biochem. J 87:258–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian H, Yao Q, Tai C, Deng Z, Gan J, Ou HY. 2018. Identification and characterization of acetyltransferase-type toxin-antitoxin locus in Klebsiella pneumoniae. Mol. Microbiol 108:336–49 [DOI] [PubMed] [Google Scholar]
- 59.Qian L, Nie L, Chen M, Liu P, Zhu J, et al. 2016. Global profiling of protein lysine malonylation in Escherichia coli reveals its role in energy metabolism. J. Proteome Res 15:2060–71 [DOI] [PubMed] [Google Scholar]
- 60.Rack JG, Morra R, Barkauskaite E, Kraehenbuehl R, Ariza A, et al. 2015. Identification of a class of protein ADP-ribosylating sirtuins in microbial pathogens. Mol. Cell 59:309–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reverdy A, Chen Y, Hunter E, Gozzi K, Chai Y. 2018. Protein lysine acetylation plays a regulatory role in Bacillus subtilis multicellularity. PLOS ONE 13:e0204687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rieser E, Cordier SM, Walczak H. 2013. Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem. Sci 38:94–102 [DOI] [PubMed] [Google Scholar]
- 63.Rivera C, Gurard-Levin ZA, Almouzni G, Loyola A. 2014. Histone lysine methylation and chromatin replication. Biochim. Biophys. Acta 1839:1433–39 [DOI] [PubMed] [Google Scholar]
- 64.Rouviere-Yaniv J, Yaniv M, Germond JE. 1979. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell 17:265–74 [DOI] [PubMed] [Google Scholar]
- 65.Rycroft JA, Gollan B, Grabe GJ, Hall A, Cheverton AM, et al. 2018. Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat. Commun 9:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sang Y, Ren J, Ni JJ, Tao J, Lu J, Yao YF. 2016. Protein acetylation is involved in Salmonella enterica serovar Typhimurium virulence. J. Infect. Dis 213:1836–45 [DOI] [PubMed] [Google Scholar]
- 67.Sauve AA, Wolberger C, Schramm VL, Boeke JD. 2006. The biochemistry of sirtuins. Annu. Rev. Biochem 75:435–65 [DOI] [PubMed] [Google Scholar]
- 68.Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, et al. 2015. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol 98:847–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390–92 [DOI] [PubMed] [Google Scholar]
- 70.Starai VJ, Escalante-Semerena JC. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci 61:2020–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Starai VJ, Escalante-Semerena JC. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol 340:1005–12 [DOI] [PubMed] [Google Scholar]
- 72.Starai VJ, Gardner JG, Escalante-Semerena JC. 2005. Residue Leu-641 of acetyl-CoA synthetase is critical for the acetylation of residue Lys-609 by the protein acetyltransferase enzyme of Salmonella enterica. J. Biol. Chem 280:26200–5 [DOI] [PubMed] [Google Scholar]
- 73.Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. 2003. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. 2004. A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. Curr. Opin. Microbiol 7:115–19 [DOI] [PubMed] [Google Scholar]
- 75.Sterner DE, Berger SL. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev 64:435–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuecker TN, Hodge KM, Escalante-Semerena JC. 2012. The missing link in coenzyme A biosynthesis: PanM (formerly YhhK), a yeast GCN5 acetyltransferase homologue triggers aspartate decarboxylase (PanD) maturation in Salmonella enterica. Mol. Microbiol 84:608–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473–79 [DOI] [PubMed] [Google Scholar]
- 78.Tanaka S, Matsushita Y, Yoshikawa A, Isono K. 1989. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet 217:289–93 [DOI] [PubMed] [Google Scholar]
- 79.Thao S, Chen CS, Zhu H, Escalante-Semerena JC. 2010. Nε-Lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLOS ONE 5:e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thao S, Escalante-Semerena JC. 2012. A positive selection approach identifies residues important for folding of Salmonella enterica Pat, an Nε-lysine acetyltransferase that regulates central metabolism enzymes. Res. Microbiol 163:427–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev 41:100–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsang AW, Escalante-Semerena JC. 1998. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem 273:31788–94 [DOI] [PubMed] [Google Scholar]
- 83.Tucker AC, Escalante-Semerena JC. 2013. Acetoacetyl-CoA synthetase activity is controlled by a protein acetyltransferase with unique domain organization in Streptomyces lividans. Mol. Microbiol 87:152–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tucker AC, Taylor KC, Rank KC, Rayment I, Escalante-Semerena JC. 2014. Insights into the specificity of lysine acetyltransferases. J. Biol. Chem 289:36249–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Melderen L, Jurenas D, Garcia-Pino A. 2017. Messing up translation from the start: how AtaT inhibits translation initiation in E. coli . RNA Biol 15:303–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Meter M, Mao Z, Gorbunova V, Seluanov A. 2011. Repairing split ends: SIRT6, mono-ADP ribosylation and DNA repair. Aging 3:829–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.VanDrisse CM, Escalante-Semerena JC. 2018. In Salmonella enterica, OatA (formerly YjgM) uses O-acetyl-serine and acetyl-CoA to synthesize N,O-diacetylserine, which upregulates cysteine biosynthesis. Front. Microbiol 9:2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.VanDrisse CM, Escalante-Semerena JC. 2018. In Streptomyces lividans, acetyl-CoA synthetase activity is controlled by O-serine and Nε-lysine acetylation. Mol. Microbiol 107:577–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.VanDrisse CM, Escalante-Semerena JC. 2018. Small-molecule acetylation controls the degradation of benzoate and photosynthesis in Rhodopseudomonas palustris. mBio 9:e01895–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.VanDrisse CM, Parks AR, Escalante-Semerena JC. 2017. A toxin involved in Salmonella persistence regulates its activity by acetylating its cognate antitoxin, a modification reversed by CobB sirtuin deacetylase. mBio 8:e00708–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Venkat S, Gregory C, Gan Q, Fan C. 2017. Biochemical characterization of the lysine acetylation of tyrosyl-tRNA synthetase in Escherichia coli. ChemBioChem 18:1928–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vetting MW, Carvalho LPS, Yu M, Hegde SS, Magnet S, et al. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys 433:212–26 [DOI] [PubMed] [Google Scholar]
- 93.Walsh CT. 2006. Posttranslational Modification of Proteins: Expanding Nature’s Inventory Greenwood Village, CO: Roberts and Co. [Google Scholar]
- 94.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, et al. 2013. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 4:842–51 [DOI] [PubMed] [Google Scholar]
- 95.Xu H, Hegde SS, Blanchard JS. 2011. Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 50:5883–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu JY, Xu Z, Liu X, Tan M, Ye BC. 2018. Protein acetylation and butyrylation regulate the phenotype and metabolic shifts of the endospore-forming Clostridium acetobutylicum. Mol. Cell Proteom 17:1156–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu JY, You D, Leng PQ, Ye BC. 2014. Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine. J. Biol. Chem 289:27034–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang H, Sha W, Liu Z, Tang T, Liu H, et al. 2018. Lysine acetylation of DosR regulates the hypoxia response of Mycobacterium tuberculosis. Emerg. Microbes Infect 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yeo CC. 2018. GNAT toxins of bacterial toxin-antitoxin systems: acetylation of charged tRNAs to inhibit translation. Mol. Microbiol 108:331–35 [DOI] [PubMed] [Google Scholar]
- 100.Yoshikawa A, Isono S, Sheback A, Isono K. 1987. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet 209:481–88 [DOI] [PubMed] [Google Scholar]
- 101.You D, Wang MM, Ye BC. 2017. Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol 103:845–59 [DOI] [PubMed] [Google Scholar]
- 102.You D, Yao LL, Huang D, Escalante-Semerena JC, Ye BC. 2014. Acetyl-CoA synthetase is acetylated on multiple lysine residues by a protein acetyltransferase with single GNAT domain in Saccharopolyspora erythraea. J. Bacteriol 196:3169–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.You D, Yin BC, Li ZH, Zhou Y, Yu WB, et al. 2016. Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. PNAS 113:6653–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. 2008. The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol 18:1529–36 [PubMed] [Google Scholar]
- 105.Yuan H, Marmorstein R. 2012. Structural basis for sirtuin activity and inhibition. J. Biol. Chem 287:42428–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang J, Sprung R, Pei J, Tan X, Kim S, et al. 2009. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell Proteom 8:215–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou Q, Zhou YN, Jin DJ, Tse-Dinh YC. 2017. Deacetylation of topoisomerase I is an important physiological function of E. coli CobB. Nucleic Acids Res 45:53349–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zverina EA, Lamphear CL, Wright EN, Fierke CA. 2012. Recent advances in protein prenyltransferases: substrate identification, regulation, and disease interventions. Curr. Opin. Chem. Biol 16:544–52 [DOI] [PMC free article] [PubMed] [Google Scholar]