Summary
Transmembrane chemoreceptors are widely present in Bacteria and Archaea. They play a critical role in sensing various signals outside and transmitting to the cell interior. Here, we report the structure of the periplasmic ligand-binding domain (LBD) of the transmembrane chemoreceptor MCP2201, which governs chemotaxis to citrate and other organic compounds in Comamonas testosteroni. The apo-form LBD crystal revealed a typical four-helix bundle homodimer, similar to previously well-studied chemoreceptors such as Tar and Tsr of Escherichia coli. However, the citrate-bound LBD revealed a four-helix bundle homotrimer that had not been observed in bacterial chemoreceptor LBDs. This homotrimer was further confirmed with size-exclusion chromatography, analytical ultracentrifugation and cross-linking experiments. The physiological importance of the homotrimer for chemotaxis was demonstrated with site-directed mutations of key amino acid residues in C. testosteroni mutants.
Keywords: Comamonas testosteroni chemoreceptor, methyl-accepting chemotaxis protein, ligand binding domain, crystal structure, homotrimer
Abbreviated Summary
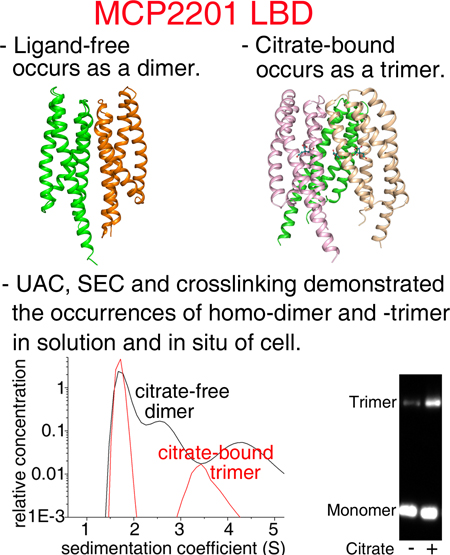
The structure of chemoreceptor MCP2201 LBD was solved. The apo-form (ligand-free) of MCP2201 LBD is a 4HB homodimer. In the presence of ligand citric acid, a previously unknown trimeric structure of MCP2201 LBD was observed. These results raised the possibility that ligand-binding induced oligomer-organization dynamics and subsequently signal generation within chemoreceptors.
Introduction
Many chemoreceptors, also known as methyl-accepting chemotaxis proteins (MCPs), sense environmental chemicals and other signals, trigger downstream signaling and drive chemotactic responses in Bacteria and Archaea (Hazelbauer et al., 2008, Wuichet & Zhulin, 2010, Parkinson et al., 2015). The chemoreceptors Tar and Tsr of E. coli represent the most ubiquitous type of bacterial chemoreceptors (Wuichet et al., 2007). Those chemoreceptors contain two transmembrane helices flanking a periplasmic ligand-binding domain (LBD) and cytoplasmic domains that modulate the activity of a histidine kinase CheA (Wuichet et al., 2007, Parkinson et al., 2015, Ortega et al., 2017). The cytoplasmic domains consist of a membrane-proximal HAMP domain and a signaling domain. The signaling domain includes distinct regions of sensory adaptation and kinase regulation and adopts the trimer-of-dimers (TOD) architecture (Kim et al., 1999). Furthermore, TODs are arranged into an extended hexagonal networked array, which is universally conserved in bacteria (Briegel et al., 2009). Two TODs, two molecules of CheW adaptor proteins and one CheA homodimer comprise the core signaling systems (Li & Hazelbauer, 2011).
Genomic studies revealed a wide repertoire of MCP LBDs (Ortega et al., 2017). The LBDs of Tar and Tsr from E. coli and Salmonella enterica, are the firstly identified ones, which fold in a four-helix bundle (4HB) (Milburn et al., 1991, Biemann & Koshland, 1994, Kim et al., 1999). Other LBD structures were also observed, for examples, the helical bimodular (HBM) in McpQ from Pseudomonas putida (Martin-Mora et al., 2016b), the sCACHE domain in TlpB from Helicobacter pylori (Goers Sweeney et al., 2012), the PAS (found in Per-Arnt-Sim proteins) in Aer2 and the dCACHE domain in PctA from P. aeruginosa (Rico-Jimenez et al., 2013) (Airola et al., 2013). So far, more than 80 types of LBDs have been described, but the 4HB, dCACHE, sCACHE and PAS are the most ubiquitous ones (Ortega et al., 2017).
In the well-studied E. coli Tar chemoreceptor, the periplasmic LBD is composed of 4HB homodimer in ligand-bound state (Milligan & Koshland, 1993, Biemann & Koshland, 1994). The ligand-binding site is located at the interface of two protomers (Milburn et al., 1991, Yeh et al., 1993), and ligands bind with a stoichiometry of one molecule per dimer (Milligan & Koshland, 1993). Ligand-binding initiates a piston-like downward sliding of the α4 helix within the integral chemoreceptors and the signal is further transmitted via piston and rotation movements of transmembrane helices generating conformational changes of the HAMP domains, and ultimately altering the CheA kinase activity (Chervitz & Falke, 1996, Ottemann et al., 1999, Hulko et al., 2006, Yu et al., 2015, Gushchin & Gordeliy, 2018).
Interestingly, MCP LBDs appear to behave differently upon ligand-binding. The individual LBD of Tar exists in a dynamic equilibrium between monomers and dimers and ligand binding induces dimerization since the ligand interacts with both monomers to stabilize the dimer (Milligan & Koshland, 1993, Biemann & Koshland, 1994, Yeh et al., 1993, Yeh et al., 1996). Similar to Tar, some other helical LBDs also alter the oligomeric state upon ligand binding, such as 4HB domains of PcaY_PP (Fernandez et al., 2017) and CtpH (Rico-Jimenez et al., 2016), HBM domains of McpS (Lacal et al., 2010) and McpQ (Martin-Mora et al., 2016b), and PilJ domain of McpN (Martin-Mora et al., 2019). On the other hand, the oligomerization of some dCACHE and sCACHE LBDs is not altered by ligand binding and their ligand binding sites are not located at the dimeric interface (Rico-Jimenez et al., 2013, Martin-Mora et al., 2018, Gavira et al., 2018). For example, the dCACHE of the P. aeruginosa PctA chemoreceptor was found as a monomer in both ligand-free and ligand-bound states (Rico-Jimenez et al., 2013), and the sCACHE domain of P. aeruginosa PA2652 chemoreceptor exists as dimer in both states (Martin-Mora et al., 2018).
The MCP2201 chemoreceptor in Comamonas testosteroni strain CNB-1 triggers chemotaxis towards tricarboxylic acid cycle intermediates such as citric acid and aromatic compounds (Ni et al., 2013). In this study, we obtained crystal structures of the MCP2201 LBD in both ligand-free and ligand-bound states. While the ligand-free LBD had a homodimer structure typical of the E. coli Tar and Tsr chemoreceptors, the citrate-bound LBD had a homotrimeric structure, which had not been previously seen in any bacterial chemoreceptor. Site-specific mutagenesis coupled with biochemical, physiological, and bioinformatics analyses provided further support for the physiological relevance of the homotrimeric structure.
RESULTS
The ligand-free MCP2201 LBD is a typical four-helix bundle homodimer
The ligand-free MCP2201 LBD was crystalized and its structure was determined at 2.8 Å resolution (Table S1). The monomer consists of four α-helices, α1 (Q59-K87), α2 (A91-L118), α3 (P122-A150) and α4 (A154-E195), and it adopts a 4HB fold (Fig. 1A). The 4HB was relatively relaxed and a large opening between helices α2 and α3 was observed (Fig. 1B), which allows a ligand to access the ligand-binding pocket.
Fig. 1. The structure of ligand-free MCP2201LBD.
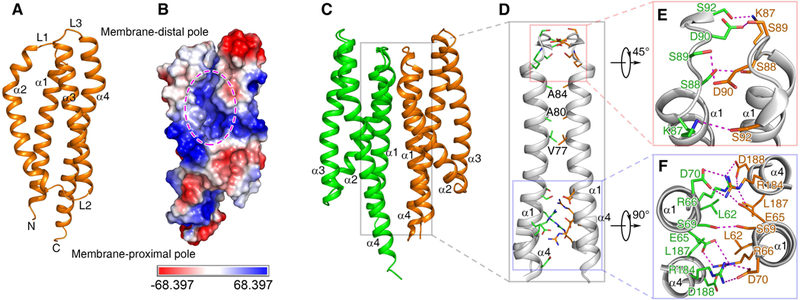
Cartoon (A) and electrostatic potential surface (B) representation of a monomer, the dimeric structure (C) and its interface (D) are shown. The ligand binding pocket in (B) is highlighted with ellipse. Two major parts of dimer interface, the membrane-distal and membrane-proximal ones, are enlarged in panels (E) and (F).
Two monomers related by a non-crystallography two-fold symmetric axis compact as a dimer (Fig. 1C) in the crystal structure. The dimeric interface is mainly composed of helices α1 and α4 (Fig. 1D, 1E, 1F), and is maintained via Van der Waals interactions and hydrogen bonds. The amino acid residues L62, V77, A80, A84, and L187 of helices α1 and α4 from one monomer interact with their counterparts of the other monomer to generate Van der Waals interactions. Similarly, amino acid residues E65, R66, K87, S88, S89, D90, S92, R184, and D188 interact with their counterparts of the other monomer to form 17 hydrogen bonds. In general, the monomeric structure and the dimer organization of the ligand-free MCP2201LBD are highly similar to that of the E. coli Tar and Tsr chemoreceptors (Tajima et al., 2011).
Identification of the citrate-binding pocket
Two structures of MCP2201 LBD in complex with ligand, the citric acid, were obtained at resolution of 2.5 Å with different unit cell parameters (Table S1). The two structures were identical to each other as evidenced by Cα r.m.s.d of 0.4 Å and only subtle conformational differences are found for those residues in N- and C-termini. We thus did not distinguish between the two structures in the following description. The citrate-binding site was located at an internal pocket of one monomer and was surrounded by all four helices (Fig. 2A, 2C). Seven amino acid residues (R81, T104, T108, Y138, R142, R135 and Y172) were involved in binding to citrate (Fig. 2B). The residues R135 and Y172 form hydrogen bonds with 1’-carboxyl group, the residues R81, T104, Y138 and R142 form hydrogen bonds with 2’-carboxyl group, and the residue T108 forms hydrogen bond with 2’-hydroxyl group, of the citric acid molecule. The 3’- carboxyl group does not form a hydrogen bond with protein and wedges into the space between helices α1 and α2 (Fig. 2A, 2B).
Fig. 2. The citrate-binding pocket and the effect of mutations in the binding pocket on chemotaxis to citrate.
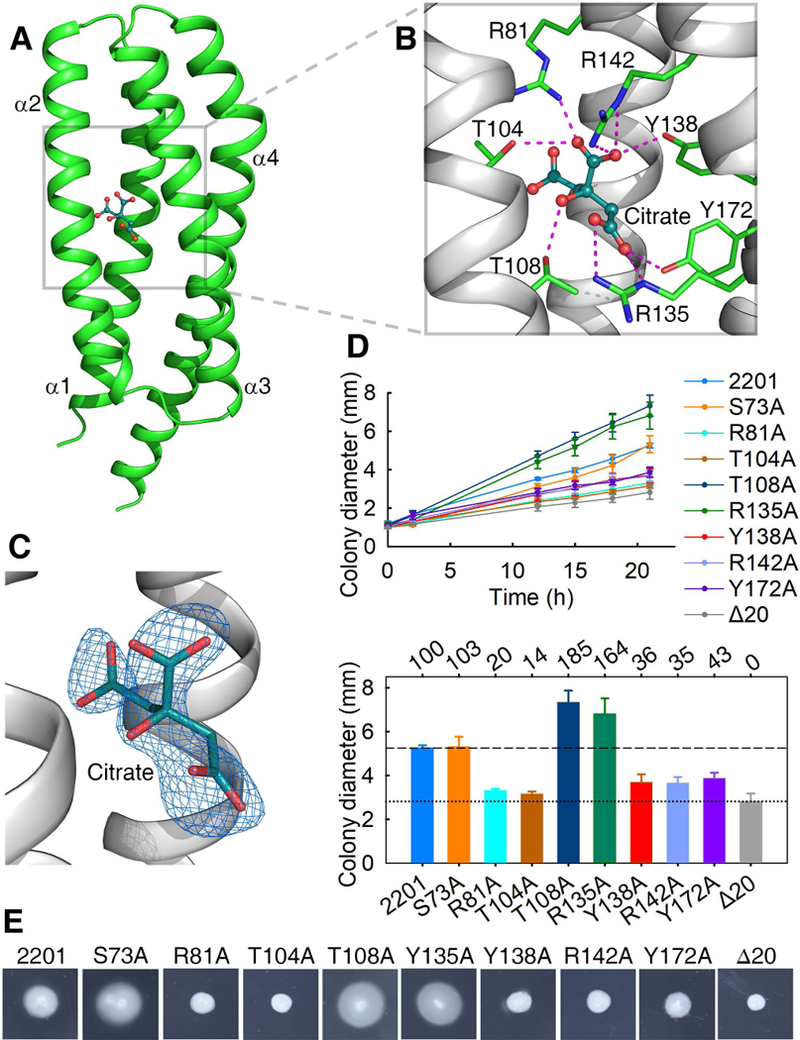
(A) The structure of a citrate-bound monomer. (B) The ligand-binding pocket. Residues involved in ligand-binding and the ligand (citric acid) are shown in stick and ball-and-stick models, respectively. (C) Citrate is surrounded by ligand-omit 1FoFc electron density map contoured at 3σ. (D) The plots of swimming ring diameter vs time for strains harboring MCP2201 mutants with substitutions in amino acid residues at the ligand binding pocket are shown in the upper panel. Averages of colony diameters of site-directed mutants were plotted, and standard deviations are shown, n=3. The relative chemotaxis abilities of mutants are shown in the bottom panel, with percentage labelled above histogram of panel. The levels of chemotactic response of chemotaxis-null mutant CNB-1Δ20 harboring or not wild-type MCP2201 towards citrate are normalized as 100% and 0%. Mutant S73A was neutral to chemotaxis and was chosen as a control. (E) One set of typical motility plate assay results for MCP2201 mutants.
To test the structural interpretation of the critical amino acid residues of the binding pocket, seven mutants of MCP2201 were constructed. The chemotactic responses of the resulted MCP2201 mutants were phenotypically characterized with the CNB-1Δ20 strain (in which all chemoreceptor genes have been disrupted) (Ni et al., 2013). The chemotaxis ability of mutants R81A, T104A, Y138A, R142A and Y172A decreased compared to the wild type, whereas mutants T108A and R135A increased chemotaxis (Fig. 2D, 2E).
Observation of a homotrimer for the citrate-bound LBD
Surprisingly, the citrate-bound MCP2201 LBD crystal structure revealed a homotrimer. This type of oligomerization had never been observed in bacterial chemoreceptor LBDs. The trimeric structure is composed of three citrate-bound monomers associated with a crystallographic three-fold symmetric axis, with helices α1 and α2 buried at the trimer interface (Fig. 3A, 3B and 3D). Key amino acid residues that maintain the stability of the trimeric structure were identified. Three hydrophobic residues L93, F96, and L97 were located at the trimer interface, and they packed against their counterparts from the other two monomers. These amino acid residues contribute to the inter-molecule hydrophobic contact and stabilize the trimeric structure. We also observed that residues S88 and residue S89 of one monomer interact with D90 of the other monomer via inter-molecule hydrogen bonds. The hydrogen bond of residue S89 with D90 of the other monomer is bridged by a water molecule (Fig. 3D). Residues V77, A80 and A84 were buried at the inter-molecule interface of the trimeric structure (Fig. 3C). The total area buried at the interface is approximately 425 Å2 per subunit, and the free energy of trimer formation calculated by PISA, ~ 6.7 kcal/mol, corresponds to a theoretical disassociation constant (Kd) of ~ 12 μM.
Fig. 3. The citrate-bound LBD trimer-of-monomers structure.
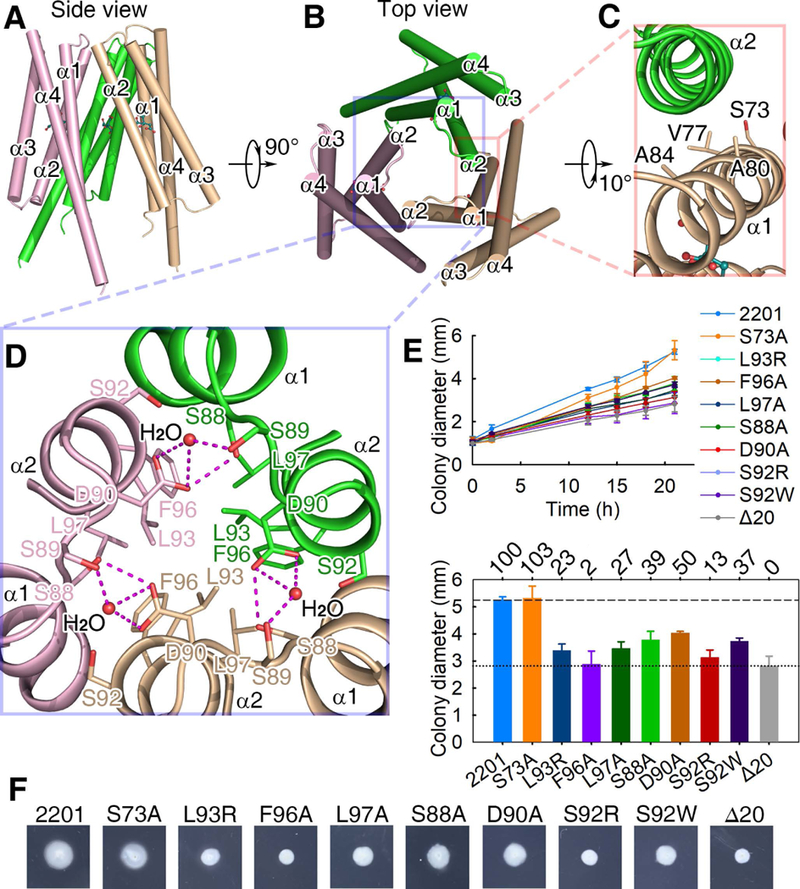
(A, B) The side (A) and top (B) views. (C) The interface between two monomers of the trimeric structure. (D) The three-fold symmetric interface of the trimeric structure. Residues L93, F96 and L97 form a hydrophobic patch and then interact with their counterparts in the other protomers. The distance between L97 of one protomer and F96 of another protomer is 4.2 Å. (E) The plots of swimming ring diameter vs time for strains harboring MCP2201 mutants. Substitutions in amino acid residues at the trimeric interface are shown in the upper panel, whereas the relative chemotaxis abilities of mutants are shown in the bottom panel with percentage labelled above histogram of panel. The standard deviations are shown, n=3. (f) One set of typical motility plate assay results for MCP2201 mutants.
The homotrimer of MCP2201 LBDs occurs in the presence of citric acid in solution
In order to confirm the citrate-bound homotrimer also occurs in solution, we further determined the oligomeric states of the ligand-free and citrate-bound MCP2201LBD using size-exclusion chromatography and analytical ultracentrifugation assays. In the size-exclusion chromatography assay, MCP2201 LBD was eluted with an apparent molecular mass of 35 kDa in the absence of citrate, suggesting the ligand-free MCP2201 LBD occurred as a homodimer (the calculated theoretic mass of a monomer is 19 kDa). In contrast, a fraction of MCP2201 LBD molecules were eluted with apparent molecular mass of 46 kDa in the presence of citrate (Fig. 4A), a value between the expected homotrimer and dimer (the calculated molecular mass of a homotrimer is 57 kDa). This result suggested the presence of virtual intermediate oligomeric species characterized by the rapid interchange of different oligomeric forms (such as trimer and dimer/monomer), similar to the cases of previously reported McpK and McpQ LBDs (Martin-Mora et al., 2016a, Martin-Mora et al., 2016b). The results of the analytical ultracentrifugation assay indicated the presence of homotrimers (apparent molecular mass of 57 kDa) and monomers (apparent molecular mass of 19 kDa) in the presence of citrate (Fig. 4B). Taken together, the size-exclusion chromatography and analytical ultracentrifugation results demonstrated that MCP2201 LBD homotrimer also occurred in solution, and the homotrimer formation is enhanced by citrate binding in solution.
Fig. 4. Size-exclusion chromatography, analytical ultracentrifugation and TMEA crosslinking assays support a trimeric structure of citrate-bound MCP2201LBD.
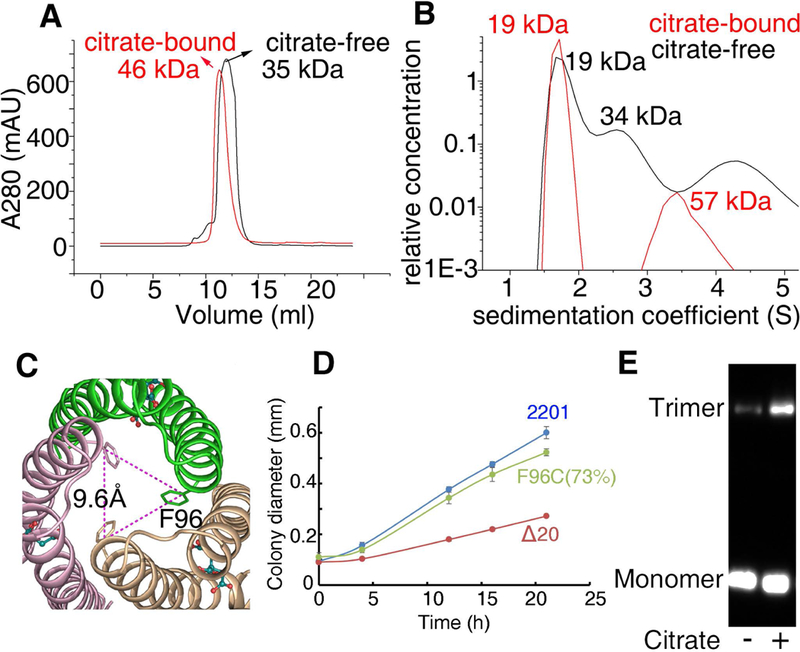
(A) Size-exclusion chromatography assay of MCP2201 LBD. (B) Analytical ultracentrifugation assay of MCP2201 LBD. For (A) and (B), the plots of proteins in the absence and presence of citrate are colored as black and red, respectively. (C) Top view of citrate-bound trimer-of-monomers structure of MCP2201 LBD showing F96 location. (D) The plots of swimming ring diameter vs time for strain harboring MCP2201 F96C mutant. (E) TMEA crosslinking of integral MCP2201 variant F96C. The bands of monomer and trimer are indicated.
TMEA crosslinking supports the LBD homotrimer in the full-length MCP2201 chemoreceptor
When analyzing the crystal structure, we observed that the residue F96 in each monomer of the trimeric structure was located at a distance of 9.6 Å (Fig. 4C), which is ideal for TMEA crosslinking. Thus, we used this method to gain additional support for the occurrence of the trimeric structure in the integral MCP2201. We found that F96C mutants remained 73% of the chemotaxis ability compared to the wildtype (Fig. 4D). As seen in Fig. 4E, a clear band of MCP2201-F96C trimer was detected after incubation with citrate. These results further suggest that the MCP2201 LBD homotrimer could be present as a physiological state of the MCP2201 chemoreceptor.
Trimer interface residues are important for triggering chemotaxis
The MCP2201 LBD-citrate complex structure (Fig. 3D) revealed amino acid residues critical for the stabilization of the homotrimer: The residues S88/D90 formed inter-molecular hydrogen bonds, and the residues L93/F96/L97 contributed to inter-molecule hydrophobic interactions. Although those amino acid residues were also located at the interface of homodimer, their contribution to stabilizing the dimeric structure was limited: residues S88/D90 are involved in only 3 of total 17 hydrogen bonds in the homodimer interface (Fig. 1E, 1F), and residues L93/F96/L97 do not pack against any hydrophobic residue of the other protomer in the homodimer (Fig. S1A). Thus, it was expected that replacements of these amino acid residues with amino acid residues of different physicochemical properties would disturb the trimeric structure, which in turn should negatively affect chemotaxis triggered by MCP2201. We thus assessed the importance of the trimeric structure by mutating the residues that affect the homotrimer but not the homodimer. Five mutants, namely S88A, D90A, L93R, F96A and L97A, were constructed and their chemotactic phenotypes were determined with CNB-1Δ20 harboring the mutated proteins. Our results showed that the chemotactic response of mutants S88A, D90A, L93R, F96A and L97A was significantly impaired in comparison to the wild type (Fig. 3E, 3F).
We further demonstrated the importance of the trimeric structure by mutating S92, a residue that is fully buried at the trimer interface, but exposed to solvent in the ligand-free LBD homodimer (Fig. S1B and S1C). Substitution of S92 with an aromatic amino acid residue such as tryptophan, or a charged residue such as arginine, should collapse the trimeric interface but not the dimeric interface. As shown in Fig. 3E, the chemotactic response in mutants S92W and S92R is severely impaired. These results suggest that the citrate-bound MCP2201 LBD homotrimer is of physiological importance.
The structural variation caused by citrate binding
Based on the crystal structures of ligand-free and citrate-bound MCP2201LBD, we explored the structural variation induced by citrate-binding (Fig. 5). In the citrate-bound trimeric structure, the distance between helices α1 and α2 are farther away than that in the ligand-free dimeric structure (Fig. 5A and 5B), because the 3’-carboxyl group of citric acid wedges into the space between the helices. Citric acid also interacts with residues from helices α2 and α3, thus the four-helix bundle of the citrate-bound LBD becomes more compact in comparison with the ligand-free dimeric structure. The citrate binding triggered a series of conformation changes, including the unbending of helix α2 (Fig. 5C) and a swing movement of helix α4 (Fig. 5D). These conformational changes would disrupt the dimeric interface observed in the ligand-free structure, thus preventing citrate-bound LBD from forming the dimeric structure (Fig. S2B). On the other hand, helix α2 in the ligand-free structure would prevent the formation of trimeric interface observed in the citrate-bound structure (Fig. S2A). Therefore, the ligand-free LBD could not form the trimeric structure.
Fig. 5.
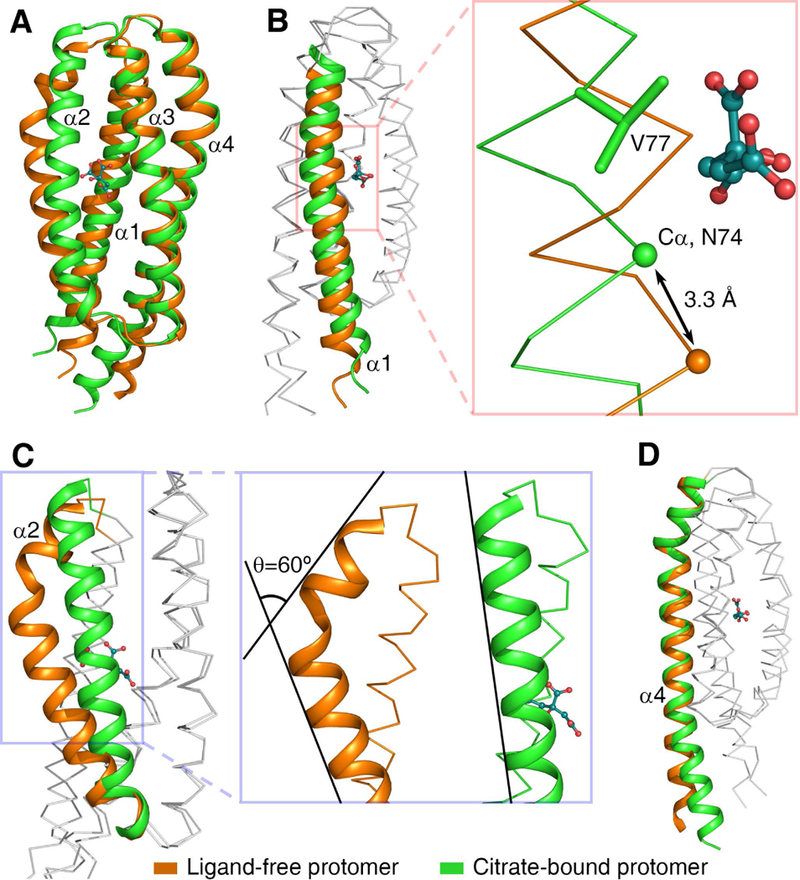
Structural comparison of apo- and citrate-bound MCP2201LBD. (A) The ligand-free (orange) and citrate-bound (green) structures were superimposed. The binding of citrate induced a series of conformation changes, including the movement of helix α1 (B), the unbending of helix α2 (C), and the swing of the C-terminus of helix α4 (D).
Residues involved in ligand-binding and in homodimer and homotrimer interfaces are conserved
In order to identify conserved residues in MCP2201 LBD, we have identified orthologous domains using reciprocal sequence similarity searches and constructed their multiple sequence alignment (Fig. S3). The final dataset contained sequences of chemoreceptors from 201 β- and γ-proteobacterial genomes representing 46 genera (Dataset S1). All enterobacterial orthologs from this dataset have been previously assigned to a specific Cluster of Orthologous Groups of chemoreceptors (COG2) (Ortega & Zhulin, 2016), therefore we conclude that all sequences in this data set including MCP2201 belong to COG2. The well-studied E. coli chemoreceptors Tar and Tsr belong to COG1 (Ortega & Zhulin, 2016) and the only experimentally studied representative of COG2 is the McpC chemoreceptor from Salmonella enterica, which was proposed to mediate chemotaxis away from a repellent L-cystine (Lazova et al., 2012). Multiple sequence alignment revealed 16 highly conserved (>95% sequences) amino acid residues within the MCP2201 LBD that are grouped in three categories: the ligand-binding site, the dimer interface and the trimer interface. Satisfactorily, R81, Y138, R135, R142 and Y172 were identified as the citrate-binding site and S88, D90 L93, and L97 were identified as trimer stabilizing residues during the structure analysis. Three conserved residues, E65, R66 and D70, contribute to the stabilization of the dimeric structure of apo-form (Fig. S4A). In detail, the positively charged E65 of one chain interacts with the negatively charged R66 of the other chain and their interaction is further enhanced via hydrogen bond interaction. D70 interacts with R184 in a similar fashion. The residue G151 contributes to a critical turn at the end of the α3 helix. K149 forms a hydrogen bond with the carboxyl oxygen atom of the loop L1 main chain (connecting helices α1 and α2) thus is critical to maintain the overall four-helix fold (Fig. S4B). E124 forms hydrogen bonds with main chain amino groups of T120 and Q121 along with the sidechain of K64, thus stabilizing the loop between helix α2 and α3 and the overall folding (Fig. S4C). Finally, W71 forms a hydrogen bond with Y172 to stabilize the binding pocket (Fig. S4D).
DISCUSSION
The sensory domains of bacterial chemoreceptor are diverse, for examples 4HB, HBM, dCACHE, sCACHE, and PAS domains. In this study we found that MCP2201 is a new member of 4HB family. The structure of MCP2201LBD could be superposed to that of E. coli Tar with Ca r.m.s.d of 2.7 Å (Fig. 6), revealing that the ligand binding site is closer to the membrane than that in Tar. Interestingly, MCP2201 binds to its ligand at the interior of a monomer. This binding mode only involves in amino acid residues of one MCP2201LBD molecule and suggests that the dimer is not a prerequisite for the ligand binding. Thus citrate-bound MCP2201LBD could exists as a oligomeric state (trimer) different from those helical sensory domains. In contrast, Tar binds to aspartate at its dimeric interface (Fig. 6A) and both monomers interact with aspartate (Yeh et al., 1993, Yeh et al., 1996). This binding mode enhances the dimer formation via the ligand linking both monomers and then alters the dynamic equilibrium between monomers and dimers (Yeh et al., 1996). The other helical sensory domains, i.e. 4HB family member Tsr (Kim et al., 1999), HBM family member McpS (Pineda-Molina et al., 2012), and PilJ family member McpN (Martin-Mora et al., 2019), also bind to the ligands at their dimeric interfaces in a way that amino acid residues from both monomers interacting with the ligands, resulting in that the dimer is a prerequisite for their ligand binding. Therefore, our work indicated that MCP2201 had a novel ligand binding mode different from other known bacterial chemoreceptors of the helical sensory domain families.
Fig. 6.
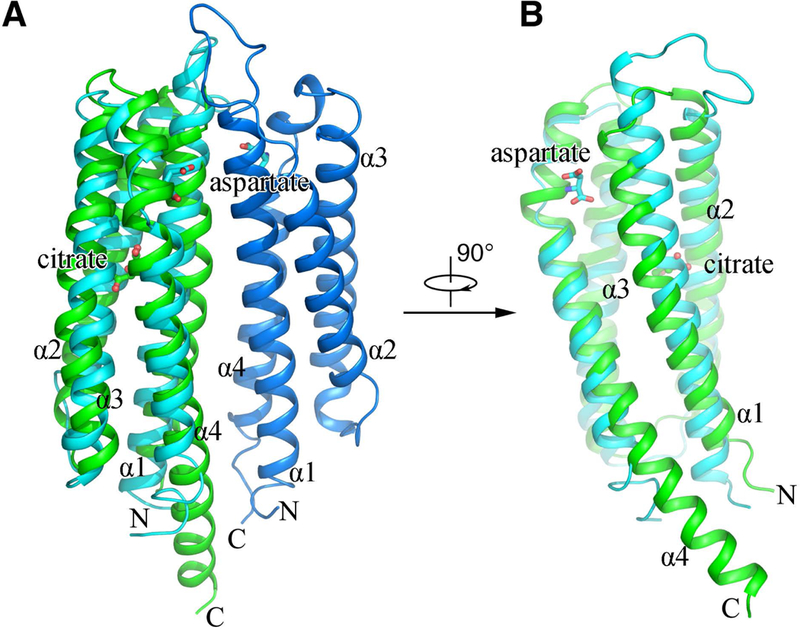
Structural comparison of the LBDs from MCP2201 and Tar. A citrate-bound MCP2201LBD molecule (green) is superposed with a aspartate-bound protomer (cyan) of Tar dimer (A, side view; B, view from the Tar dimeric interface).
The documentation of a previously unknown homotrimeric structure of MCP2201 LBD, when it bound with citric acid, is surprising. The physiological importance of the homotrimer for chemotaxis was demonstrated with site-directed mutations of amino acid residues that were structurally critical to the MCP2201 LBD homotrimer, i.e., S88, D90, L93, F96 and L97. Those residues are also involved in dimeric formation or buried nearby the dimeric interface, suggesting a possibility that the mutations of those residues would also affect the dimer. However, we further noticed that residues L93, L96and L97 were conserved as hydrophobic residues among MCP2201 orthologs, and these three residues did not pack against any hydrophobic residues of the other protomer in the dimer (Fig. S1A), but did in the homotrimer. We observed that the mutants F96A and F96C lost 98% and 27% of the chemotaxis ability, compared to that of wildtype MCP2201, indicating the substitution of F96 by amino acid residues with less hydrophobic sidechains lost more chemotaxis ability. This result revealed the importance of the hydrophobic packing by at least the F96 residue, and also suggested that the homotrimeric structure was critical to effect the functionality of MCP2201 in triggering chemotaxis.
Size-exclusion chromatography, analytical ultracentrifugation and cross-linking experiments demonstrated that MCP2201 LBD existed in three oligomer-states: monomer, homodimer (ligand free) and homotrimer (citrate-bound). Oligomerization is an intrinsic property of chemoreceptors. The current chemoreceptor model, which is based on a large body of experimental and theoretical evidences, describes that six chemoreceptors form a trimer-of-dimers that is arranged in networked hexagonal chemoreceptor arrays (Briegel et al., 2012, Liu et al., 2012). It is generally accepted that the LBDs of Tar, the central chemoreceptor model protein, exist as homodimers for signal transduction (Ottemann et al., 1999) and there is no any other type of interactions among chemoreceptor molecules outside the cytoplasmic kinase-regulating subdomain. On the other hand, direct experimental evidence for chemoreceptor organization in the membrane and periplasm is missing. Several studies involving recombinant LBDs from various chemoreceptors showed that they existed in both monomer and homodimer states and that ligand-binding shifted the dynamic equilibrium from one state to another (Rico-Jimenez et al., 2013, Liu et al., 2015, Martin-Mora et al., 2016b, Ortega et al., 2017). We have observed three oligo-organizations for MCP2201 LBD, i.e. monomer, homodimer and homotrimer, which is different from Tar and Tsr that have monomers and homodimers. We also demonstrated that the homotrimer state triggered chemotaxis towards citric acid, but the function of the homodimers remains unknown.
In conclusion, we observed that MCP2201 LBD has a previously unknown homotrimeric structure and it existed as monomer, dimer and trimer in solution. A recent survey focusing on sensory domains in bacterial chemoreceptors presented a wide diversity of ways and mechanisms for chemoreceptor sensing and signaling (Ortega et al., 2017). Based on those observations and results, it is rational to propose that the transition among the oligomerization states of the MCP22201 LBD contributes to the signal sensing and transducing into the cell interior.
Experimental Procedures
Strains, cultural conditions and chemotaxis assays
Comamonas testosteroni CNB-1 derivative strain CNB-1Δ20, in which all chemoreceptor genes have been were disrupted (Ni et al., 2013), were cultivated in LB broth at 30 °C, and a temperature of 37 °C was used for E. coli strains. Strains and plasmids used in this study are shown in Table S2. The MCP2201 gene, as well as its upstream 150 bp DNA that contains a native promoter, has been inserted into the plasmid pBBR1MCS-2 for chemotaxis assays. This plasmid has been used in the study of another C. testosteroni chemoreceptor MCP2901 (Huang et al., 2016) and its copy number is 6 per cell in C. testosteroni (Xiong & Maser, 2001). Swimming plate assays were performed as previously described (Huang et al., 2016). Briefly, soft agar plate containing 0.3% agarose in minimal medium were used. Carbon sources were added to a final concentration of 0.15 g/L. The genome of C. testosteroni does not encode a functional lac repressor and no ITPG was used. Plates were incubated at 30 oC for 21 h, and colony diameters were measured. The colony diameters were measured at various incubation times and were used to evaluate the chemotaxis ability of strains CNB-1Δ20 complemented with MCP2201 wildtype and mutant genes. All tests were conducted in triplicates and data were normalized as 100% with strain CNB-1Δ20 harboring MCP2201 and 0% with strains CNB-1Δ20 (not carrying MCP2201).
Protein expression and purification
Plasmid construction and protein purification were performed as previously described (Ni et al., 2013). Specifically in this study, the fragments of Q46-T203 and M57-T203 were used for citrate-bound and -free MCP2201LBD crystal structure determination, respectively, and the former was used for size-exclusion chromatography assay and analytical ultracentrifugation assay. For gene cloning and expression, plasmids were transformed into E. coli BL21 (DE3). E. coli strains were grown at 37 °C in LB medium containing 25 μg/ml kanamycin or 100 μg/ml penicillin. Protein expression was initiated with 0.1 mM IPTG induction at OD600 of 0.8, and cells were further cultivated for 5 h at 30 °C. Cells were harvested by centrifugation at 5,000 g for 30 min, resuspended in 50 mM Tris buffer, pH 7.5, 200 mM NaCl, and 10 mM imidazole, and were lysed by sonication. After centrifugation at 20,000 g for 30 min, the supernatant was incubated with a nickel affinity resin (Ni-NTA, Qiagen) at 4 °C for 30 min. The resin was rinsed three times with a buffer containing 20 mM Tris pH 7.5, 1 M NaCl, and 20 mM imidazole. The MCP2201LBD proteins were eluted from the affinity resin with a buffer containing 20 mM Tris pH 7.5, 200 mM NaCl, and 250 mM imidazole. The eluant was concentrated and sequentially applied to a HiTrap Desalting and a HiTrap Q HP column (GE Healthcare, Uppsala, Sweden), followed by size-exclusion chromatography (Superdex-200, GE Healthcare) in a buffer containing 20 mM Tris, pH 7.5, and 150 mM NaCl (SEC buffer). The fractions with MCP2201LBD proteins were collected and concentrated to approximately 10 mg/ml for crystallization. Se-methionine labeled MCP2201LBD was prepared using previous protocol (Doublie, 2007) and purified in the same way as the native protein in the presence of a reducing agent (1 mM TCEP or 2 mM DTT).
Crystallization, data collection and structure determination
Protein solutions with or without 5 mM citrate were used for crystal culture. All crystals were obtained using the hanging drop vapor diffusion method by mixing 1 μl protein solution and 1 μl reservoir buffer (0.1 M bis-tris propane, pH 9.4, 20% PEG5000 MME, 3% glycerol, 5% 2-propanol for ligand-free LBD, 4 °C; 0.1 M sodium citrate, pH 4.4, 13% ethylene glycol, 14% PEG400 for both citrate-bound LBD complexes, 22 °C). The diffraction data for ligand-free and citrate-bound crystals were collected at the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China) with a beamline BL17U1. All the data were processed and scaled with the iMOSFLM(Kabsch, 2010a), XDS (Kabsch, 2010b) and/or CCP4 suite (Winn et al., 2011). Parts of the models were manually built in COOT (Emsley et al., 2010). All structures were refined with PHENIX (Adams et al., 2010). The figures were prepared with PyMOL (Schrodinger, 2015). The statistics for the data collection and structure refinement are summarized in Table S1. The structure factor and coordinate files for the ligand-free and citrate-bound MCP2201LBD have been deposited in the Protein Data Bank under the accession codes numbers 5XUA, 5XUB and 6ITS, respectively.
Site-directed mutagenesis
Briefly, site-directed mutagenesis was performed via overlapping PCR and DpnI digestion. Overlapping primers that contained specific mutations at the 5’ ends were used for PCR amplification of the entire circular plasmid pBBR1MCS2-MCP2201. PCR products were treated with DpnI endonuclease (NEB) to eliminate the template plasmids. To create cysteine-containing mutants for the in TMEA crosslinking assays, site-directed mutagenesis was also performed with slight modifications. The template plasmid used was pBBR1MCS2-MCP2201His, a derivative of pBBR1MCS2-MCP2201, which contained a 6X-His-tag sequence at the C-terminal of MCP2201. The final purified products were transformed into E. coli cells for replication, and all mutations were identified with DNA sequencing. The appropriate mutation-containing plasmid was electroporated into CNB-1Δ20 strain and used in a swimming plate assay or a TMEA crosslinking assay.
TMEA crosslinking
TMEA (Tris-[2-maleimidoethyl-amide]) is a maleimide-containing cross-linker, which has three functional groups that cross-link sulfhydryls within a distance 10.3 Å. The crosslinking assay was performed as previously described (Studdert & Parkinson, 2007). The MCP-null strain CNB-1Δ20 carrying the corresponding cysteine replacement in a plasmid was grown to log-phase at 30 °C. The cells were harvested by low-speed centrifugation and washed twice with KEP buffer (10 mM potassium phosphate, pH 7.0, 0.1 mM EDTA) and subsequently resuspended to OD600=2. After incubation at 30 °C for 5 min, a ligand or a buffer was added to 100 μl cells to a final concentration of 10 mM for another 1 min incubation. TMEA was added to the cell mixture to 50 μM for 10 min, followed by quenching with 10 mM NEM. The samples were boiled in SDS-loading buffer, separated by 8% SDS-PAGE and transferred to a PVDF membrane (Amersham Biosciences Europe GmbH, Freiburg, Germany). MCP2201 was probed with a primary monoclonal antibody targeted to a His tag at a 1:2000 dilution and with a secondary antibody (goat anti-mouse HRP conjugate) at a 1:3000 dilution (Transgen, Beijing, China). Visualization was carried out with a Pro-light HRP Chemiluminescent Kit (Tiangen, Beijing, China).
Size-exclusion chromatography assay
Purified proteins were loaded on a Superdex 75 10/300 GL column (GE Healthcare, US) in the presence or absence of 5 mM citrate and were subsequently eluted by SEC buffer (20 mM Tris, pH 7.5, and 150 mM NaCl) with or without 5 mM citrate. The apparent molecular masses were calculated using protein markers in Gel Filtration Calibration Kits (GE Healthcare, US) as control.
Analytical ultracentrifugation assay
Velocity sedimentation experiments were performed on a Beckman XL-I analytical ultracentrifuge at 4 °C with a rotor speed of 60,000 rpm for 7 h. The samples contain 75 μM MCP2201LBD in SEC buffer with or without 5 mM citrate. Raw data were processed using Sednterp and the sedimentation coefficients and apparent molecular masses were calculated (Schuck, 2000).
Data sources and bioinformatics
Sequences closely related to the MCP2201 LBD were identified by BLAST searches against the NCBI RefSeq database (default parameters; limited to 500 hits) using the MCP2201 (NCBI accession number WP_041744100.1) LBD and its adjacent transmembrane regions (residues 5 to 212) as a query. Orthologous sequences were identified as mutual best BLAST hits in reciprocal searches. Incomplete sequences and paralogs were excluded from further analysis. The final dataset included sequences from 201 bacterial genomes (Dataset S1). Transmembrane regions in all sequences were identified using Phobius (Kall et al., 2007) and multiple sequence alignment of all LBDs and adjacent transmembrane regions was constructed using L-INS-I algorithm from MAFFT (Katoh & Toh, 2008).
Supplementary Material
Acknowledgements
We thank the staff members at the BL19U1/BL17U1 beamline at the National Center for Protein Sciences Shanghai and the Shanghai Synchrotron Radiation Facility, Shanghai, China, for technical assistance with the data collection. Constructive comments and discussion with G. Hazelbauer (University of Missouri) are highly appreciated. This work was supported by grants from the National Natural Science Foundation of China (31230003 to S.-J.L. and 31870037 to D.-F. L), the program Youth Innovation Promotion Association CAS (2014079 to D.-F.L) and by National Institutes of Health grant GM072295 (to I.B.Z.).
Footnotes
Conflict of Interest Statement. The authors declare no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, and Crane BR (2013) Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol 425: 886–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemann HP, and Koshland DE Jr. (1994) Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry 33: 629–634. [DOI] [PubMed] [Google Scholar]
- Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, and Crane BR (2012) Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A 109: 3766–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, and Jensen GJ (2009) Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A 106: 17181–17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, and Falke JJ (1996) Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci U S A 93: 2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S (2007) Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol Biol 363: 91–108. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M, Matilla MA, Ortega A, and Krell T (2017) Metabolic Value Chemoattractants Are Preferentially Recognized at Broad Ligand Range Chemoreceptor of Pseudomonas putida KT2440. Frontiers in microbiology 8: 990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavira JA, Ortega A, Martin-Mora D, Conejero-Muriel MT, Corral-Lugo A, Morel B, Matilla MA, and Krell T (2018) Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida. J Mol Biol 430: 1950–1963. [DOI] [PubMed] [Google Scholar]
- Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, and Guillemin K (2012) Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure 20: 1177–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gushchin I, and Gordeliy V (2018) Transmembrane Signal Transduction in Two-Component Systems: Piston, Scissoring, or Helical Rotation? Bioessays 40. [DOI] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, and Parkinson JS (2008) Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends in biochemical sciences 33: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Ni B, Jiang CY, Wu YF, He YZ, Parales RE, and Liu SJ (2016) Direct sensing and signal transduction during bacterial chemotaxis toward aromatic compounds in Comamonas testosteroni. Mol Microbiol 101: 224–237. [DOI] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, and Coles M (2006) The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126: 929–940. [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010a) Integration, scaling, space-group assignment and post-refinement. Acta crystallographica. Section D, Biological crystallography 66: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010b) XDS. Acta crystallographica. Section D, Biological crystallography 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, and Sonnhammer EL (2007) Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic acids research 35: W429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in bioinformatics 9: 286–298. [DOI] [PubMed] [Google Scholar]
- Kim KK, Yokota H, and Kim SH (1999) Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400: 787–792. [DOI] [PubMed] [Google Scholar]
- Lacal J, Alfonso C, Liu X, Parales RE, Morel B, Conejero-Lara F, Rivas G, Duque E, Ramos JL, and Krell T (2010) Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J Biol Chem 285: 23126–23136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazova MD, Butler MT, Shimizu TS, and Harshey RM (2012) Salmonella chemoreceptors McpB and McpC mediate a repellent response to L-cystine: a potential mechanism to avoid oxidative conditions. Mol Microbiol 84: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, and Hazelbauer GL (2011) Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A 108: 9390–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu B, Morado DR, Jani S, Manson MD, and Margolin W (2012) Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A 109: E1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Machuca MA, Beckham SA, Gunzburg MJ, and Roujeinikova A (2015) Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta crystallographica. Section D, Biological crystallography 71: 2127–2136. [DOI] [PubMed] [Google Scholar]
- Martin-Mora D, Ortega A, Matilla MA, Martinez-Rodriguez S, Gavira JA, and Krell T (2019) The Molecular Mechanism of Nitrate Chemotaxis via Direct Ligand Binding to the PilJ Domain of McpN. Mbio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Mora D, Ortega A, Perez-Maldonado FJ, Krell T, and Matilla MA (2018) The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Scientific reports 8: 2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Mora D, Ortega A, Reyes-Darias JA, Garcia V, Lopez-Farfan D, Matilla MA, and Krell T (2016a) Identification of a Chemoreceptor in Pseudomonas aeruginosa That Specifically Mediates Chemotaxis Toward alpha-Ketoglutarate. Frontiers in microbiology 7: 1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Mora D, Reyes-Darias JA, Ortega A, Corral-Lugo A, Matilla MA, and Krell T (2016b) McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environmental microbiology 18: 3284–3295. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE Jr., and Kim SH (1991) Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254: 1342–1347. [DOI] [PubMed] [Google Scholar]
- Milligan DL, and Koshland DE Jr. (1993) Purification and characterization of the periplasmic domain of the aspartate chemoreceptor. J Biol Chem 268: 19991–19997. [PubMed] [Google Scholar]
- Ni B, Huang Z, Fan Z, Jiang CY, and Liu SJ (2013) Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol Microbiol 90: 813–823. [DOI] [PubMed] [Google Scholar]
- Ortega A, Zhulin IB, and Krell T (2017) Sensory Repertoire of Bacterial Chemoreceptors. Microbiology and molecular biology reviews : MMBR 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega DR, and Zhulin IB (2016) Evolutionary Genomics Suggests That CheV Is an Additional Adaptor for Accommodating Specific Chemoreceptors within the Chemotaxis Signaling Complex. PLoS computational biology 12: e1004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottemann KM, Xiao W, Shin YK, and Koshland DE Jr. (1999) A piston model for transmembrane signaling of the aspartate receptor. Science 285: 1751–1754. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Hazelbauer GL, and Falke JJ (2015) Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Molina E, Reyes-Darias JA, Lacal J, Ramos JL, Garcia-Ruiz JM, Gavira JA, and Krell T (2012) Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc Natl Acad Sci U S A 109: 18926–18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Jimenez M, Munoz-Martinez F, Garcia-Fontana C, Fernandez M, Morel B, Ortega A, Ramos JL, and Krell T (2013) Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol Microbiol 88: 1230–1243. [DOI] [PubMed] [Google Scholar]
- Rico-Jimenez M, Reyes-Darias JA, Ortega A, Diez Pena AI, Morel B, and Krell T (2016) Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Scientific reports 6: 28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC (2015) The PyMOL Molecular Graphics System, Version 1.8 [Google Scholar]
- Schuck P (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophysical journal 78: 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, and Parkinson JS (2007) In vivo crosslinking methods for analyzing the assembly and architecture of chemoreceptor arrays. Method Enzymol 423: 414–431. [DOI] [PubMed] [Google Scholar]
- Tajima H, Imada K, Sakuma M, Hattori F, Nara T, Kamo N, Homma M, and Kawagishi I (2011) Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J Biol Chem 286: 42200–42210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Alexander RP, and Zhulin IB (2007) Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Method Enzymol 422: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, and Zhulin IB (2010) Origins and diversification of a complex signal transduction system in prokaryotes. Science signaling 3: ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, and Maser E (2001) Regulation of the steroid-inducible 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase gene in Comamonas testosteroni. J Biol Chem 276: 9961–9970. [DOI] [PubMed] [Google Scholar]
- Yeh JI, Biemann HP, Pandit J, Koshland DE, and Kim SH (1993) The three-dimensional structure of the ligand-binding domain of a wild-type bacterial chemotaxis receptor. Structural comparison to the cross-linked mutant forms and conformational changes upon ligand binding. J Biol Chem 268: 9787–9792. [PubMed] [Google Scholar]
- Yeh JI, Biemann HP, Prive GG, Pandit J, Koshland DE Jr., and Kim SH (1996) High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J Mol Biol 262: 186–201. [DOI] [PubMed] [Google Scholar]
- Yu D, Ma X, Tu Y, and Lai L (2015) Both piston-like and rotational motions are present in bacterial chemoreceptor signaling. Scientific reports 5: 8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


