Abstract
Interleukin 33 (IL-33) is an IL-1 family member protein that is a potent driver of inflammatory responses in both allergic and non-allergic disease. This proinflammatory effect is mediated primarily by extracellular release of IL-33 from stromal cells and binding of the C-terminal domain of IL-33 to its receptor ST2 on targets such as CD4+ Th2 cells, ILC2, and mast cells. Notably, IL-33 has a distinct N-terminal domain that mediates nuclear localization and chromatin binding. However, a defined in vivo cell-intrinsic role for IL-33 has not been established. We identified IL-33 expression in the nucleus of pro-B and large pre-B cells in the bone marrow, an expression pattern unique to B cells among developing lymphocytes. The IL-33 receptor ST2 was not expressed within the developing B cell lineage at either the transcript or protein level. RNA sequencing analysis of WT and IL-33-deficient pro-B and large pre-B cells revealed a unique, IL-33-dependent transcriptional profile wherein IL-33-deficiency led to an increase in E2F targets, cell cycle genes, and DNA replication and a decrease in the p53 pathway. Using mixed bone marrow chimeric mice, we demonstrated that IL-33-deficiency resulted in an increased frequency of developing B cells via a cell-intrinsic mechanism starting at the pro-B cell stage paralleling IL-33 expression. Finally, IL-33 was detectable during early B cell development in humans and IL33 mRNA expression was decreased in B cell chronic lymphocytic leukemia (B-CLL) samples compared to healthy controls. Collectively, these data establish a cell-intrinsic, ST2-independent role for IL-33 in early B cell development.
INTRODUCTION:
Interleukin 33 (IL-33) is an IL-1 family member protein that is a key mediator of innate and adaptive immune responses. IL-33 has been extensively studied as an extracellular signal that binds to a heterodimeric receptor complex composed of IL1RL1, also known as ST2, and the shared IL-1 receptor accessory protein (IL1RAcP) (1-4). Primary targets of IL-33 include group 2 innate lymphoid cells (ILC2), mast cells, macrophages, dendritic cells, and CD4+ Th2 cells with resultant type 2 inflammatory responses (3, 5). Additionally, a subset of CD4+ T regulatory cells expresses ST2 and expands in response to IL-33, demonstrating that extracellular IL-33 can both promote and attenuate inflammation (5).
Major sources of IL-33 include epithelial cells in the lung, skin, gastrointestinal tract, and reproductive tract (2, 6, 7). IL-33 is also highly expressed in endothelial cells in adipose tissue, liver, and secondary lymphoid organs, as well as in fibroblastic reticular cells within lymph nodes (7-12). During acute inflammation and in adipose tissue, macrophages may also be a source of IL-33 (6, 13). However, IL-33 expression has largely been observed and studied in non-hematopoietic cells.
IL-33 has also been hypothesized to have an intracellular role as a transcriptional regulator. IL-33 is a two-domain protein. The C-terminal domain (amino acids 112-270 in humans) contains IL-1 family member homology and is responsible for mediating the extracellular, ST2-dependent effects of IL-33 (2). In contrast, the N-terminal domain (amino acids 1-111 in humans) targets IL-33 to the nucleus (9, 14), has a chromatin-binding motif (15), and exhibits potent transcriptional repressor capacity in an artificial tethered gene reporter assay (14, 15). Multiple studies have attempted to define gene targets of IL-33 regulation. Several small, focused investigations have suggested single or a few putative targets including IL6 and RELA (NF-κB p65) (16-19). Yet, in two large studies, intracellular IL-33 had no influence on the global transcriptome or proteome of cultured human esophageal epithelial cells or human umbilical vein endothelial cells, respectively (20, 21). Notably, all of these studies were conducted in vitro. Thus, an intracellular regulatory role for endogenous IL-33 and its in vivo relevance has yet to be conclusively established.
B cell development in the bone marrow is characterized by somatic recombination leading to formation of the B cell receptor (BCR) and rapid expansion of the B cell lineage. This process proceeds in a stepwise fashion, with committed development starting at the progenitor B (pro-B) cell stage, where the B cell receptor (BCR) undergoes heavy-chain recombination. Hyperproliferation marks the transition to the large precursor B (large pre-B, LPB) cell stage, wherein the developing B cells rapidly expand to increase the number of cellular templates for BCR light chain rearrangement. Cessation of proliferation and initiation of light chain rearrangement denotes the transition to the small pre-B (SPB) stage. Following light chain rearrangement, developing B cells proceed to the immature B cell stage where they undergo clonal selection and functional maturation prior to exiting the bone marrow. While substantial effort and progress have been made to define the molecular cues that guide B cell development, the full complement of cell exogenous and endogenous regulators of B cell development remains undetermined.
Herein, we identified IL-33 expression during the early stages of B cell development in the bone marrow of both mice and humans. IL-33 expression restricted the capacity of B cells to repopulate the hematopoietic compartment in vivo through a cell-intrinsic, ST2-independent mechanism. Collectively, these data reveal an intracellular role for IL-33 in regulating early B cell development.
MATERIALS AND METHODS:
Mice
Animal experiments were performed with age-matched 8-14 week old male and female mice unless otherwise noted. WT BALB/c, WT BALB/c CD45.1 (CByJ.SJL(B6)-Ptprca/J; stock #006584), Il33GFP/GFP (also referred to as Il33fl/fl; B6(129S4)-Il33tm1.1Bryc/J; stock #030619), CD19-Cre (B6.129P2(C)-Cd19tm1(cre)Cgn/J, stock #006785), and WT C57BL/6 CD45.1 (B6.SJL-PtprcaPepcb/BoyJ; stock #002014) mice were obtained from The Jackson Laboratory. Il33cit/cit mice (also referred to as Il33−/− mice as the citrine reporter construct replaces endogenous Il33) and St2−/− mice were graciously provided by Dr. Andrew McKenzie. Il33Cit/Cit and St2−/− mice were on a BALB/c background. Il33GFP/GFP (Il33fl/fl) and CD19-Cre mice were on a C57BL/6 background. In indicated experiments, Il33Cit/Cit mice were bred with WT BALB/c to generate Il33Cit/+ hemizygous reporter mice with an intact copy of Il33. Il33fl/fl mice were bred with CD19-Cre mice to generate mice with a CD19-specific reduction of Il33 expression (Il33fl/fl Cd19Cre/+). WT BALB/c and WT BALB/c CD45.1 mice were bred to generate WT BALB/c mice with co-expression of CD45.1 and CD45.2 (referred to as WT BALB/c CD45.1/CD45.2). Animals were maintained in specific pathogen free housing. All animals were used in compliance with the revised 1996 Guide for the Care and Use of Laboratory Animals prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council. All animal experiments were approved by the Vanderbilt Institutional Animal Care and Use Committee.
Human Samples
Bone marrow aspirates from healthy human volunteers were collected and processed for flow cytometric analysis of IL-33 in accordance with the Vanderbilt Institutional Review Board (IRB #160060, 081012, and 160420). Briefly, bone marrow aspirates were collected in acid citrate dextrose tubes. RBC lysis was performed using RT ACK buffer and washed samples were frozen in liquid nitrogen. All processing occurred within 24 hours of sample collection; samples were thawed immediately before flow cytometric analysis. Donors averaged 34.4 (± 11.0) years of age and were 75% men/25% women and 91.7% white/8.3% Hispanic. For gene expression analysis in human B cell malignancies, we analyzed 45 publicly available microarray gene expression-profiling data sets for CLL, six different B-cell lymphomas (Burkitt, DLBCL, follicular, mantle cell, marginal zone, and Hodgkin), and normal B-cells (germinal center centroblasts and centrocytes, memory B cells, and CD19+ B cells). We collected the data from gene expression omnibus (GEO) (22) or the authors’ website (23). The GEO ids for CLL data are - GSE26725, GSE50006, GSE16455, GSE26526, GSE21029, GSE69034, GSE49896, GSE39671, GSE9250, GSE22762 and the data sets for the six lymphoma types and normal samples are previously described (24). The data were generated on the Affymetrix Human Genome U133 Plus 2.0 platform following the standard Affymetrix protocol. The data were separated into batches, normalized using the robust multi-array average (RMA) algorithm (25) in the Affy package for R (26), and batch effects corrected using ComBat (27). Probe sets were averaged to obtain a single-expression intensity measure per gene per array if multiple probe sets corresponded to the same gene ID. Expression values from replicate samples were averaged. All analyses were carried out in R, version 3.3.2. Differential expression was measured by unpaired, two-tailed t-test with Welch’s correction using Graphpad Prism v.7.
Flow Cytometry and FACS
Murine bone marrow was prepared by flushing the tibia and femur of euthanized mice with RPMI and performing RBC lysis followed by straining the cells through a 70 micron filter to remove debris. Cells were incubated with anti-CD16/CD32 for 15 minutes at 4C to block Fc-mediated binding of antibodies, followed by staining with the indicated antibodies to discriminate populations of interest (Supplemental Table I). Live/Dead discrimination was performed by staining with PI, DAPI, 7-AAD, or GhostDye UV450 (for fixed samples) prior to analysis or fixation. Spleens were initially homogenized through a 70 micron filter and processed otherwise identical to bone marrow. For BrdU analysis, animals were injected intraperitoneally with 1 mg of Bromodeoxyuridine (BrdU) at 1 hour prior to harvest and processed via manufacturer instructions. For intracellular IL-33 detection, human bone marrow was stained for surface markers and fixed for 60 minutes with the Foxp3/Transcription Factor staining kit (Tonbo, Cat. #TNB-0607-KIT). Samples were stained at 4C for 30 minutes with anti-human IL-33 or isotype control antibody in permeabilization buffer. Samples were evaluated on a BD LSR II and analyzed with FlowJo software. For FACS isolation, samples were magnetically-enriched for CD19+ cells (Miltenyi, Cat. # 130-121-301), surface stained as described for analytical flow cytometry, and separated on a BD FACS Aria III.
Quantitative PCR
RNA was isolated from FACS-purified cells using the QIAGEN RNeasy Micro Kit (Cat. #74004) per the manufacturer protocol with on-column DNase I digestion. cDNA was generated using the Invitrogen SuperScript IV First-Strand Synthesis System (Cat. #18091050) per the manufacturer protocol using Oligo(dT) priming. Quantitative PCR was performed using FAM-MGB TaqMan primer/probes from Applied Biosystems/Thermo Fisher: Il33 exon 2-3 (Mm00505399_m1), Il33 exon 3-4 (Mm01195783_m1), Il33 exon 6-7 (Mm00505403_m1) and Gapdh (Mm99999915_g1). Reactions were carried out with the TaqMan Gene Expression Master Mix (Cat. #4369016) on an Applied Biosystems QuantStudio12k Flex Real-Time PCR machine. Samples were analyzed using Applied Biosystems QuantStudio 12k Flex Software v.1.2.2. Relative gene expression was calculated using the ΔΔCt protocol with respect to the indicated reference gene.
Western Blot
Nuclear and cytoplasmic protein fractions from FACS-purified cells were isolated using the Thermo Scientific NE-PER Nuclear and Cytoplasmic Extraction Reagents (Cat. #78833) per the manufacturer protocol. Total protein was assessed for each sample using the Pierce BCA Protein Assay Kit (Cat. # 23225). Equivalent protein (2.5 μg) per sample was loaded and separated on a 4-20% gradient polyacrylamide gel. Gels were rinsed, wet transferred to a nitrocellulose membrane, and blocked with LI-COR Odyssey Blocking Buffer in TBS (Cat. #927-50000). Blots were incubated overnight at 4°C while shaking with the indicated primary antibodies: anti-IL-33 (R&D, Cat. #AF3626), anti-Lamin B1 (Abcam, Cat. #ab16048), or anti-Tubulin (Cell Signaling Technology, Cat. #3873). Blots were rinsed and incubated for 1 hour at room temperature while shaking with the appropriate secondary antibody: anti-mouse IgG 680 LT (LI-COR, Cat. #68022), anti-rabbit IgG 680 LT (LI-COR, Cat. #68023), or anti-goat IgG 800 CW (LI-COR, Cat. #32214). Blots were rinsed and imaged on an Odyssey Classic Infrared Imaging system. Image capture and densitometry were performed using Image Studio v.3.1 software.
RNA-sequencing
Pro-B and LPB cells were MACS-enriched and FACS-purified from the bone marrow of WT and Il33−/− mice. Cells were sorted into cold PBS, pelleted, and lysed with Buffer RLT (QIAGEN). RNA was isolated using the QIAGEN RNA Mini Kit (Cat. #74104) with on-column DNase I digestion following the manufacturer instructions. RNA-seq libraries were prepared using 20-50 ng of total RNA and Illumina’s TruSeq Stranded mRNA kit (Illumina, Cat #: 20020595) per manufacturer’s instructions, with mRNA enriched via poly-A-selection using oligoDT beads. The RNA was then thermally fragmented and converted to cDNA, adenylated for adapter ligation and PCR amplified. The libraries were sequenced using the HiSeq 3000 with 75 bp paired end reads with a depth of approximately 30 million reads per sample. RTA (version 2.7.6; Illumina) was used for base calling and analysis was completed using MultiQC v1.2. On paired fastq files, transcript abundance was estimated, and these read counts were imported into edgeR for differential expression analysis (28). In edgeR, data was normalized based on negative binomial distribution. The differential expression of genes between Il33−/− and WT samples was assessed by estimating an exact test P-value. Venn diagram of differential gene expression was generated with Meta-Chart (https://www.meta-chart.com/venn#/display). RNA-seq data have been deposited in the GEO database under the access code GSE132123 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132123).
GSEA
We conducted GSEA (29) using the KEGG suite (30) and the Hallmark gene set (31) embedded in GSEA to identify pathways that were differentially expressed in Il33−/− samples compared to WT. GSEA was run on pre-ranked list of the genes obtained from the edgeR analyses. Ranking was based on fold-change induction in Il33-null cells compared to control cells. Mouse genes were converted to human gene symbols before conducting GSEA pre-ranked analysis. GSEA results were assessed as being statistically significant by permutation of 1,000 samples.
Bone Marrow Chimeras
WT BALB/c CD45.1/CD45.2 mice or WT C57BL/6 CD45.1 mice were lethally irradiated with 9 Gy or 12 Gy, respectively, from a Cs-137 source. Recipient mice were transplanted with a 1:1 mixture of whole bone marrow (10 million total cells) from the indicated strains via retroorbital injection. Mice were maintained on antibiotic water (0.025% trimethoprim/0.125% sulfamethoxazole) for 2 weeks following transplant. Mice were rested for 5-8 weeks to allow for reconstitution prior to analysis. For WT BALB/c CD45.1/CD45.2 mice, residual cells from the recipient mice were gated as CD45.1+CD45.2+ by flow cytometry and excluded from analysis.
B Cell Receptor Sequencing
SPB cells were MACS-enriched and FACS-purified from the bone marrow of female 8-week old naïve adult WT, Il33−/−, and St2−/− mice. Genomic DNA (gDNA) was isolated using the QIAGEN DNeasy Blood & Tissue kit (Cat. #69504) and 0.5 μg of gDNA per sample was used for sequencing. Sequencing of the BCR hypervariable complementary determining region 3 (CDR3) was performed by the multiplex PCR-based immunoSEQ Assay (Adaptive Biotechnologies). BCR repertoire analyses were performed using the immunoSEQ Analyzer 3.0 (Adaptive Biotechnologies).
Statistics
Data was managed and analyzed using GraphPad Prism version 7. When possible, data were pooled from multiple experiments. Unpaired t test or one-way ANOVA with Tukey post-test were used to determine statistical significance, as appropriate and as indicated.
RESULTS:
IL-33 is expressed during early B cell development.
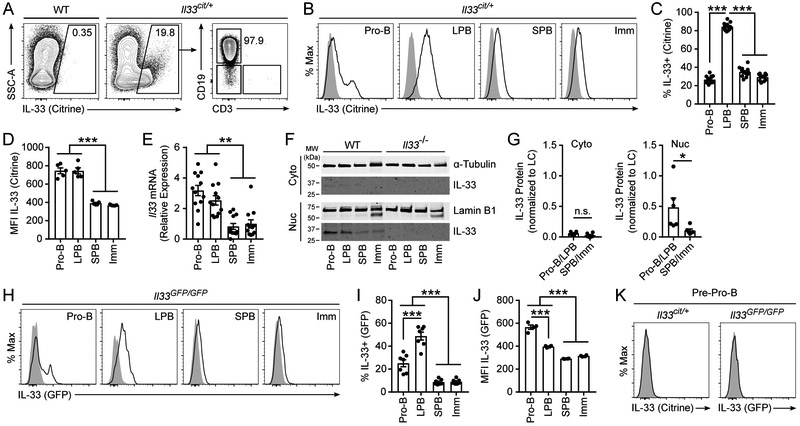
We sought to characterize IL-33 expression in the bone marrow compartment. Using a BALB/c mouse model with a citrine reporter construct under the control of the endogenous IL-33 promoter (Il33cit/+), we identified a population of IL-33-expressing cells in the bone marrow of which >95% were CD19+ B cells (Fig. 1A). Subset analysis in Il33cit/+ mice demonstrated IL-33 expression during the pro-B cell stage that persisted through the LPB cell stage and decreased in SPB and immature B cells (Fig. 1B-C, gating strategy in Supplemental Fig. 1A). Per cell expression of IL-33 was similar in pro-B and LPB cells, and higher in these early B cell populations than in SPB and immature B cells (Fig. 1D). Consistent with our flow cytometric analysis, FACS-purified pro-B and LPB cells expressed significantly higher levels of Il33 mRNA than SPB and immature B cells (Fig. 1E). Fractionation of FACS-purified B cells into nuclear and cytoplasmic protein followed by immunoblotting demonstrated directly the presence of IL-33 protein, and that this IL-33 protein was localized almost exclusively in the nucleus in pro-B and LPB cells (Fig. 1F-G).
Figure 1: IL-33 is expressed in the nucleus of early developing B cells.
Bone marrow was isolated from the tibia and femur of naïve adult WT BALB/c or IL-33 citrine reporter mice (Il33cit/+) and analyzed by flow cytometry. (A) IL-33 citrine reporter expression in bone marrow CD19+ B cells. (B) IL-33 citrine reporter expression in bone marrow B cells by developmental subset; pro-B (B220+ IgM− CD19+ CD43+), large pre-B (LPB, B220+ IgM− CD19+ CD43− FSC-Ahi SSC-Ahi), small pre-B (SPB, B220+ IgM− CD19+ CD43− FSC-Alo SSC-Alo), and immature B cells (Imm, B220+ IgM+ CD23−). Grey histogram is WT; black line is Il33cit/+. (C) Percent citrine-expressing IL-33+ B cells from B. (D) Geometric MFI of IL-33 citrine in bone marrow B cells. (E) Il33 transcript levels by quantitative PCR normalized to Gapdh and compared by the ΔΔCt method in MACS-enriched, FACS-purified bone marrow B cells. (F) IL-33 protein by western blot in MACS-enriched, FACS-purified bone marrow B cells fractionated by nuclear and cytoplasmic protein from WT and Il33−/− mice. (G) Quantification of F by densitometry from multiple blots; pro-B and LPB are aggregated together and compared to aggregated SPB and Imm. LC = loading control. (H) IL-33 GFP reporter expression in bone marrow B cells by developmental subset in WT C57BL/6 (grey histogram) and Il33GFP/GFP mice (purple line). (I) Percent GFP-expressing IL-33+ cells from H. (J) Geometric MFI of IL-33 GFP in the marrow B cells. (K) IL-33 citrine or IL-33 GFP expression in bone marrow pre-pro-B cells (B220+ IgM− CD19− CD43+). Grey histogram is WT, black line is Il33cit/+, and purple line is Il33GFP/GFP. Data are combined from 2 (I) or 3 (C, E, and G) or representative of 2 (H, J, and K) or 3 (A, B, D, and F) independent experiments. Data are displayed as the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 by one-way ANOVA with Tukey posttest or Mann-Whitney test; n.s. = not significant.
We verified these results using an IL-33-GFP reporter mouse strain (Il33GFP/GFP) independently developed on a C57BL/6 background. We identified IL-33 expression beginning in the pro-B cell stage that peaked as a percentage of the total population in the LPB cell stage and was only minimally detected in SPB and immature B cells (Fig. 1H-I). In addition, the MFIs of GFP in IL-33-expressing pro-B and LPB cells were higher than SPB and immature B cells (Fig. 1J).
Pre-pro-B cells are CD19− but represent the stage of B cell development immediately preceding pro-B cells. Flow cytometric analysis of both Il33cit/+ and Il33GFP/GFP mice demonstrated no expression of IL-33 in pre-pro-B cells, indicating that IL-33 expression first occurs during the pro-B cell stage (Fig. 1K). Peripherally, IL-33 expression was not detected in splenic B cells or peritoneal B-1 B cells in adult naïve mice (Supplemental Fig. 2A-B). IL-33 expression was rare but detectable in B cells within Peyer’s patches, representing <1% of CD19+ B220+ B cells (Supplemental Fig. 2C). Within this IL-33+ B cell population, a larger percentage were IgM− representing class-switched B cells (Supplemental Fig. 2C). Moreover, IL-33 expression among developing lymphocytes was unique to the B cell lineage, as developing T cells in the thymus universally lacked IL-33 expression (Supplemental Fig. 2D). Taken together, these data demonstrate that IL-33 is expressed within the B cell lineage primarily during the pro-B and LPB cell stages of development and this expression pattern is independent of the mouse strain.
Developing B cells do not express the IL-33 receptor ST2.
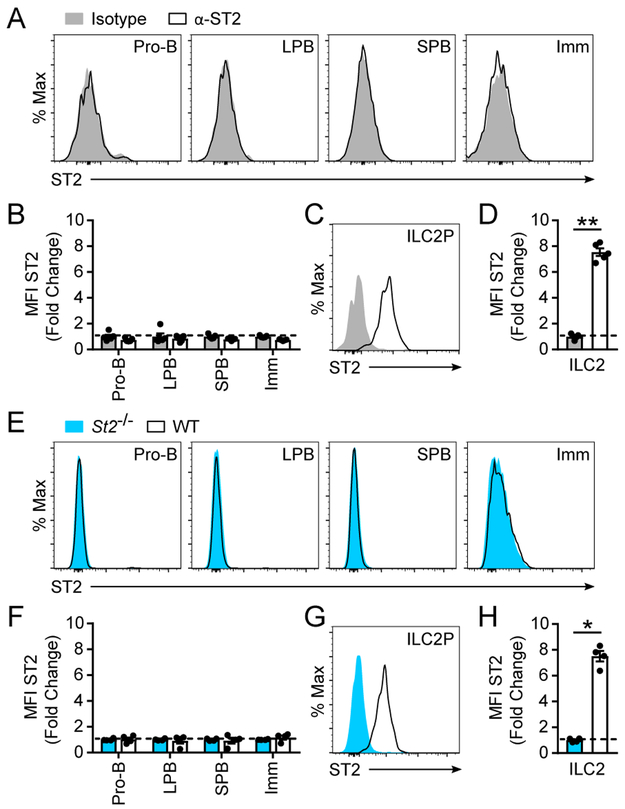
Given this expression pattern of IL-33 in pro-B and LPB cells and the known importance of IL-33 as an extracellular signal, we sought to determine whether developing B cells expressed ST2 and therefore had the capacity to respond to extracellular IL-33. ST2 expression was absent in all stages of B cell development in wild-type (WT) BALB/c mice with respect to an isotype control antibody (Fig. 2A-B). In contrast, ST2 was readily detectable on bone marrow ILC2 progenitors (ILC2P), which are highly responsive to IL-33 (Fig. 2C-D) (32). Equivalent results were obtained using St2−/− mice as a negative control (Fig. 2E-H). Collectively, these data demonstrate that developing B cells lack expression of the IL-33 receptor ST2.
Figure 2: Developing B cells do not express the IL-33 receptor ST2.
Bone marrow B cells from WT BALB/c mice were analyzed by flow cytometry for surface expression of ST2. (A) Expression of ST2 on B cell subsets. (B) Fold change of ST2 expression as measured by MFI compared to isotype control within the indicated B cell subsets. (C) Expression of ST2 on bone marrow ILC2. (D) Fold change of ST2 expression on ILC2 as measured by MFI compared to isotype control antibody. For (A) to (D), grey histograms are isotype control antibody and black lines are anti-ST2 antibody. Bone marrow B cells from WT and St2−/− mice were analyzed by flow cytometry for surface expression of ST2. (E) Expression of ST2 on WT and St2−/− B cell subsets. (F) Fold change of ST2 expression on WT B cells as measured by MFI compared to St2−/− B cells within the indicated subsets. (G) Expression of ST2 on WT and St2−/− bone marrow ILC2. (H) Fold change of ST2 expression on WT bone marrow ILC2 as measured by MFI compared to St2−/− bone marrow ILC2. For (E) to (H), blue histograms are St2−/− and black lines are WT mice, all probed with anti-ST2 antibody. Data are representative of 3 independent experiments. Data are displayed as the mean ± SEM. ***p < 0.001 by Mann-Whitney test.
IL-33-deficiency alters the transcriptome of early developing B cells.
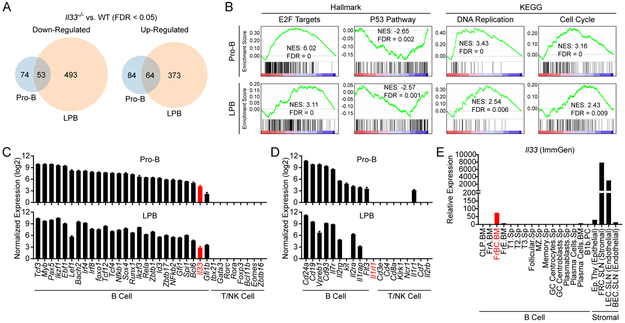
To assess for functional differences between WT and Il33−/− pro-B and LPB cells, we performed transcriptome analysis with mRNA sequencing (RNA-seq). RNA-seq of FACS-purified WT and Il33−/− pro-B and LPB cells revealed a distinct IL-33-dependent transcriptome in both pro-B and LPB cells. A total of 275 and 983 genes were differentially expressed with a false discovery rate (FDR) < 0.05 between WT and Il33−/− pro-B and LPB cells, respectively (Fig. 3A). Of these, 179 and 281 genes were differentially expressed with at least a 1.5 fold change between WT and Il33−/− pro-B and LPB cells, respectively. Among differentially expressed transcripts, 53 upregulated genes and 64 downregulated genes were shared between pro-B and LPB cells (Fig. 3A).
Figure 3: IL-33 alters the transcriptional profile of pro-B and LPB cells.
RNA was prepared from MACS-enriched and FACS-purified WT and Il33−/− pro-B and LPB cells of naïve adult mice (n = 4 mice of each genotype). RNA sequencing was performed on these samples followed by differential expression analysis by edgeR. (A) Venn diagram of shared and distinct genes between pro-B and LPB cells that were up-regulated (top) or down-regulated (bottom) with a FDR < 0.05 in Il33−/− samples compared to WT. (B) GSEA plots for select pathways with normalized enrichment score (NES) and false discovery rate (FDR). (C) Normalized expression in counts per million (CPM) for Il33 in pro-B and LPB compared with known B, T, and NK cell transcriptional regulators. (D) Normalized expression in CPM for Il1rl1, the gene encoding ST2, in pro-B and LPB compared with known B, T, and NK cell surface receptors and proteins. (E) Il33 relative expression in B cell and stromal cell populations in the Immunological Genome Project (ImmGen) ULI RNA-seq dataset. Relative expression approximates transcripts per million. Data are displayed as the mean ± SEM.
Gene set enrichment analysis (GSEA) identified numerous candidate pathways within the Molecular Signatures Database (MSigDB) Hallmark(31) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set collections(30) that were modulated in early developing B cells by IL-33 (FDR < 0.05). Notably, we identified multiple pathways related to proliferation and cell death, which were of particular interest given that rapid cellular expansion and survival are cardinal features of early B cell development at the pro-B and LPB cell stages. Specifically, Il33−/− pro-B and LPB cells were enriched for targets of E2F transcription factors and genes involved in DNA replication and the cell cycle, consistent with possible enhanced proliferation (Figure 3B).(33) Il33−/− pro-B and LPB cells also exhibited a downregulation of p53-dependent pathways, suggesting increased potential for cell survival (Figure 3B). Collectively, these data suggest that IL-33 may attenuate fitness during early B cell development by decreasing proliferation or cell survival.
We next evaluated Il33 expression in WT pro-B and LPB cells in relation to known transcriptional regulators of B cell development. Il33 expression in pro-B and LPB cells was less than hallmark early B cell transcriptional regulators including Pax5, Tcf3 (encoding E2A), and Ebf1, but was at the low end of expression of endogenous early B cell regulatory proteins, which include Gfi1, Spi1 (encoding PU.1), Bcl6, and Gfi1b (Fig. 3C). In contrast, key T cell and NK cell transcription factors were not detected (Fig. 3C). Il1rl1 encodes for the IL-33 receptor ST2 and was undetectable in both pro-B and LPB cells (Fig. 3D), consistent with our flow cytometric analysis of ST2 expression on developing B cells (Fig. 2). This is similar to other non-B cell receptors and surface proteins including Cd4, Cd8, and Ncr1, which were not detected, and in contrast to known pro-B and LPB cell receptors and surface proteins including Cd24a, Cd19, and Il7r that were highly expressed (Fig. 3D).
We sought to compare Il33 expression between early developing B cells and stromal cells, many of which are known to express and release IL-33 extracellularly for binding to ST2 and immune modulation. We compared Il33 expression in the ULI RNA-seq dataset (GSE127267) from the Immunological Genome Project (ImmGen) among B cell subsets and stromal cells (34). Within B cell subsets, Il33 expression was detected in Hardy fraction B-C cells within the bone marrow (FrBC.BM), which represent pro-B cells, and absent in mature splenic B cell populations consistent with our expression analysis (Fig. 3E). In contrast, Il33-expresing stromal cells including fibroblastic reticular cells (FRC.SLN) and lymphatic endothelial cells (LEC.SLN) of subcutaneous lymph nodes expressed Il33 at 40-100 fold higher levels than pro-B cells (Fig. 3E). Overall, expression of Il33 in early developing B cells is markedly less than stromal cells and more aligned with established intrinsic regulators of developing B cells.
IL-33-deficiency increases early developing B cell fitness in a cell-intrinsic, receptor independent manner.
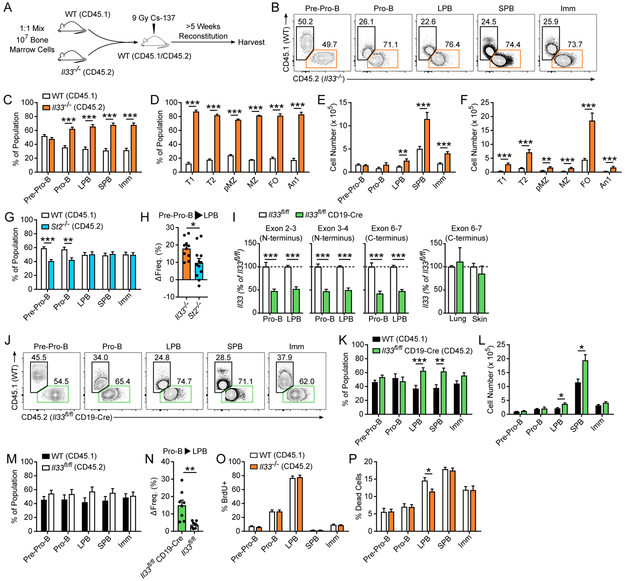
Given the identification of pro-survival gene signatures in Il33−/− pro-B and LPB cells, we assessed during in vivo B cell development for differences in B cell fitness (i.e. the ability of B cells to survive and mature). Moreover, since developing B cells lacked ST2 expression, we evaluated whether any IL-33-dependent differences in fitness were due to a cell-intrinsic or cell-extrinsic mechanism. In competitive reconstitution experiments with mixed bone marrow chimeric mice, WT and Il33−/− pre-pro-B cells were identified in approximately equal quantities (Fig. 4A-C). Alternatively, highly stringent gating for pre-pro-B cells (B220+ IgM− CD19− CD93+ Ly6D+ PDCA−) resulted in a similar frequency as our initial pre-pro-B cell analysis (Supplemental Fig. 1B). However, Il33−/− pro-B cells accumulated to a significantly higher frequency than WT pro-B cells, with a ratio of ~2:1 Il33−/− to WT (Fig. 4B-C). This enhanced fitness of the Il33−/− B cell lineage compared to WT B cells persisted throughout development, with a nearly 3:1 ratio of Il33−/− to WT cells at the immature B cell stage (Fig. 4B-C). Moreover, this increased fitness during B cell development led to alterations in peripheral B cell repopulation, with increased frequency of Il33−/− B cells in the spleen compared to WT (Fig. 4D). These frequency differences translated into significant increases in the total number of Il33−/− relative to WT B cells in the bone marrow and spleen downstream of IL-33 expression (Fig. 4E-F).
Figure 4: IL-33 is a cell-intrinsic, ST2-independent regulator of fitness during early B cell development.
(A) Recipient WT CD45.1/CD45.2 mice were irradiated with 9 Gy and transplanted with a 1:1 mixture of WT (CD45.1) and Il33−/− (CD45.2) whole bone marrow (107 cells). Transplanted mice were rested for >5 weeks and bone marrow and spleen were harvested for flow cytometric analysis. (B) Representative flow plots and (C) aggregate data of the frequency of WT and Il33−/− B cells by subset in the bone marrow (n = 10) and (D) spleen of reconstituted mice (n = 9). Total B cell number by subset in the (E) bone marrow and (F) spleen of WT: Il33−/− chimeric mice from C-D. (G) Frequency of WT and St2−/− B cells by subset in the bone marrow of WT:St2−/− bone marrow chimeric mice (n = 12). (H) Change in Il33−/− and St2−/− lineage frequency from pre-pro-B cells to LPB cells in WT:Il33−/− and WT: St2−/− bone marrow chimeric mice, respectively (n = 10-12). (I) Il33 expression measured at the junctions of exons 2-3, 3-4, and 6-7 spanning N and C terminal regions in MACS-enriched, FACS-purified pro-B and LPB cells (n = 6) as well as from whole lung and skin (n = 3) from Il33fl/fl and Il33fl/fl CD19-Cre mice. Il33 was normalized to Gapdh, quantified by the ΔΔCt method, and displayed as a percent of Il33fl/fl samples, which were standardized to 100% within each subset. (J) Representative flow plots and (K) aggregate data of the frequency and (L) total number of bone marrow B cells in WT: Il33fl/fl CD19-Cre chimeras (n = 8). (M) Frequency of bone marrow B cells in WT:Il33fl/fl chimeras (n = 8). (N) Change in the Il33fl/fl CD19-Cre and Il33fl/fl lineage frequency from pro-B to LPB cells in WT: Il33fl/fl CD19-Cre and WT:Il33fl/fl chimeras (n = 8). (O) WT:Il33−/− were exposed to 1.25 Gy radiation, treated 1 hour prior to harvest with 1 mg BrdU, and harvested 72 hours post-irradiation. Frequency of live BrdU+ bone marrow B cells (n = 9) and (P) the percentage of dead cells (Annexin V+ 7-AAD+) by B cell subset (n = 9). Data are combined from 2 (D, F, I, K-P) or 3 (C, E, G-H) or representative of 2 (J) or 3 (B) independent experiments. Orange bars = Il33−/− (CD45.2), blue bars = St2−/− (CD45.2), green bars = Il33fl/fl CD19-Cre (CD45.2), black bars = WT C57BL/6 (CD45.1), white bars (C-G, O-P) = WT BALB/c (CD45.1), and white bars (I, M-N) = Il33fl/fl (CD45.2). Data are displayed as the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 by Mann-Whitney test.
To further control for exogenous, IL-33 signaling via ST2, we evaluated developing B cell frequencies in WT:St2−/− mixed bone marrow chimeric mice. St2−/− pre-pro-B cells were recovered at a modestly, but statistically significantly, reduced frequency compared to WT pre-pro-B (Fig. 4G), possibly related to differences at early hematopoietic stem and progenitor cell stages where ST2 is expressed (35, 36). Across the span of B cell development, WT and St2−/− B cell frequencies normalized to a 1:1 ratio (Fig. 4G). In comparison, Il33−/− B cells increased in frequency significantly more across the arc of IL-33 expression from the pre-pro-B cell to LPB stage compared to St2−/− B cells within their respective bone marrow chimeras, consistent with a B cell-intrinsic role for IL-33 in regulating cellular fitness (Fig. 4H). Of note, the total number of developing B cells by subset in intact and naïve adult WT, Il33−/−, and St2−/− mice was comparable, indicating that additional regulatory mechanisms restrict pathologic overexpansion of the B cell compartment in healthy adult Il33−/− mice (data not shown).
To verify these results, we generated mice with a B cell-specific reduction of Il33. Il33fl/fl mice were crossed with mice expressing Cre recombinase under the control of the endogenous CD19 promoter/enhancer (Il33fl/fl CD19Cre/+ mice, referred to as Il33fl/fl CD19-Cre), leading to a 50-60% reduction in whole Il33 transcript in sorted pro-B and LPB cell populations as assessed by quantitative PCR of both N-terminal and C-terminal sequences (Fig. 4I). Similar expression levels of Il33 were found in the lungs and skin of Il33fl/fl and Il33fl/fl CD19-Cre mice confirming B cell lineage specificity of our Il33 knockdown (Fig. 4I). In WT:Il33fl/fl CD19-Cre chimeric mice, Il33fl/fl CD19-Cre B cells were significantly more fit than their WT counterparts as both a percentage of each B cell subset and as measured by total B cell numbers starting at the LPB cell stage (Fig. 4J-L), supporting our WT:Il33−/− chimera data using a separate, distinct mouse model. Of note, the delayed fitness advantage to the LPB cell stage in Il33fl/fl CD19-Cre chimeras compared to Il33−/− chimeras may reflect strain-specific differences (C57BL/6 vs. BALB/c), partial (50-60% in Il33fl/fl CD19-Cre) versus complete (Il33−/−) abrogation of IL-33, or progressive Cre-mediated recombination in CD19-Cre mice from pro-B to LPB cells (37, 38). In contrast, an approximately even ratio of WT and Il33fl/fl cells was detected at all stages of B cell development from pre-pro-B to immature B cells in WT: Il33fl/fl chimeric mice, confirming that IL-33 attenuation is required for this fitness advantage (Fig. 4M). Viewed another way, from the pro-B to LPB stage, the frequency of Il33fl/fl CD19-Cre B cells increased significantly more than the frequency of Il33fl/fl controls within their respective chimeras (Fig. 4N).
We next assessed for differences in proliferation and cell death that might contribute to a difference in B cell frequency. After reconstitution, WT:Il33−/− chimeras were subsequently treated with 1.25 Gy radiation to induce a hematopoietic burst.(39, 40) In regard to proliferation, no difference in BrdU+ B cells was observed at any stage of B cell development (Fig. 4O). At the LPB cell stage, there was a decrease in the frequency of Il33−/− dead cells (Annexin V+ 7-AAD+) as compared to WT dead cells, consistent with an overall increase in fitness of the Il33−/− B cell lineage (Fig. 4P). No difference in the frequency of either proliferating or dead cells was observed at the pro-B cell stage, where the dichotomy in fitness between WT and Il33−/− B cells was initiated. This may reflect insufficient sensitivity of our present assays to capture a difference in proliferation or cell death in the pro-B stage. However, the reproducible difference in cell death captured at the LPB cell stage suggests that enhanced fitness in the Il33−/− B cell lineage may at least in part be due to resistance to cell death. Collectively, these bone marrow chimeric experiments demonstrate that IL-33 is functioning as a negative regulator of fitness via a cell-intrinsic, ST2-independent mechanism during early B cell development.
Diversification of the BCR repertoire is independent of IL-33 expression.
Given that IL-33 expression spans the arc of immunoglobulin heavy chain (IgH) rearrangement, we considered whether IL-33 altered the diversity of the BCR repertoire. We assessed the IgH diversity by multiplex PCR and sequencing of FACS-sorted SPB cells. We chose to characterize the SPB cell stage as it was immediately downstream of peak IL-33 expression and occurs prior to receptor editing, which takes place during the immature B cell stage and is a mechanism of central tolerance that eliminates autoreactive B cells leading to modification of the initially-encoded BCR repertoire.(41)
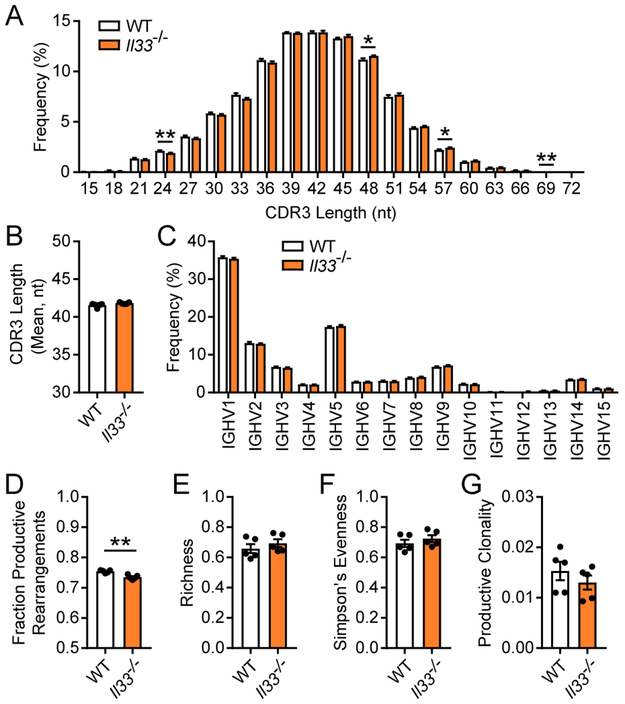
Epitope specificity is encoded in part by the complementarity determine region 3 (CDR3) region of the BCR.(42) Accordingly, we first assessed average CDR3 length between WT and Il33−/− SPB cells. The overall CDR3 length distribution was consistent between WT and Il33−/− SPB cells, with minor (<1%) but statistically significant differences at lengths of 24, 48, 57, and 69 nucleotides (Fig. 5A). The average CDR3 length assessed by Gaussian distribution analysis was not different between WT and Il33−/− SPB cells (Fig. 5B). Further, BCR diversity is achieved by the somatic recombination of V, D, and J gene segments within the IgH locus of each developing B cell clone. Our sequencing approach allowed for assessment of IgH V-family usage, which was comparable between WT and Il33−/− SPB cells (Fig. 5C). We next assessed the amino acid sequence composition of the recombined IgH. There was a small but statistically significant decrease in the fraction of IgH rearrangements in Il33−/− SPB cells that encoded a productive IgH sequence (i.e. in-frame and without early stop codons) (Fig. 5D).
Figure 5: IL-33 does not appreciably alter the heavy chain BCR repertoire in developing B cells.
Genomic DNA was isolated from SPB cells that were MACS-enriched and FACS-purified from naïve, adult WT and Il33−/− mice. IgH regions were amplified by multiplex PCR and sequenced to assess for BCR repertoire diversity. (A) CDR3 nucleotide (nt) length from productive rearrangements. (B) Mean CDR3 length in nt for WT and Il33−/− SPB cells determined by Gaussian distribution analysis. (C) IgH variable region usage by family in productive recombinations. (D) Fraction of productive rearrangements, defined as the number of IgH templates sequenced that are in-frame and without early stop codons relative to the total number of templates sequenced. (E) Richness as measured by the frequency of unique IgH sequences relative to the total number of templates sequenced. (F) Evenness was assessed by Simpson’s calculations. (G) Calculated clonality of productive templates, with values close to 0 representing comparatively polyclonal samples. n = 5 mice per group; data are displayed as the mean ± SEM. *p < 0.05 and **p < 0.01 by Mann-Whitney test.
To quantitate global diversity within the samples, we assessed for the following three parameters—richness, evenness, and clonality. Richness measures the number of unique IgH rearrangements within the sequencing pool, and is displayed as a fraction of the total number of sequenced templates. There was no difference in richness between WT and Il33−/− SPB cells (Fig. 5E). Evenness assesses for the distribution of the measured sequences within the sample. We measured evenness with Simpson’s calculation where values of one and zero indicate maximally even or skewed distribution, respectively. There was no difference in evenness between WT and Il33−/− SPB cells (Fig. 5F). Furthermore, values for evenness in WT and Il33−/− SPB cells were closer to one indicating a comparatively even distribution of each sequence within the sample. Clonality is related to evenness and assesses for the dominance of single or a few clones within a sample. Highly monoclonal or oligoclonal samples have a clonality approximating one, while infinitely polyclonal samples have a clonality of zero. There was no difference in productive clonality between WT and Il33−/− SPB cells (Fig. 5G). Moreover, both WT and Il33−/− SPB cells had productive clonality values approaching zero indicating a lack of a single or few dominate clones. Collectively, these data demonstrate that IL-33 during early B cell development does not profoundly alter the BCR repertoire.
IL-33 is expressed during early B cell development in humans and is downregulated in chronic lymphocytic leukemia.
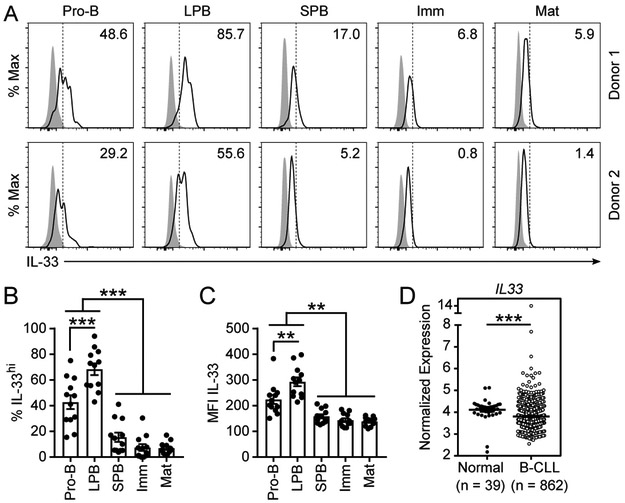
Human B cell development proceeds in a stepwise manner analogous to mice. We considered whether IL-33 was similarly expressed in early developing B cells in humans. Bone marrow was collected from healthy human donors and analyzed by intracellular flow cytometry for IL-33 expression (gating strategy in Supplemental Fig. 1C). Compared to an isotype control antibody, IL-33 expression was detected in all stages of B cell development and in mature B cells (CD19+ CD10−), which include recirculating B cells (Fig. 6A). High IL-33 expression (IL-33hi) was clustered in pro-B (>40%) and LPB (>65%) cells, whereas fewer than 20% of SPB, 10% of immature B, and 10% of mature B cells were IL-33hi (Fig. 6A-B). Moreover, the per cell expression of IL-33 as measured by the MFI of IL-33 staining in pro-B and LPB cells was significantly higher than SPB, immature, and mature B cells (Fig. 6C). Collectively, these data demonstrate that IL-33 is expressed during human B cell development, peaking at the pro-B and LPB stages, paralleling what we observed in mice.
Figure 6: IL-33 is expressed during early B cell development in human bone marrow.
Human bone marrow from healthy donors was collected and leukocytes were isolated for flow cytometric analysis with intracellular staining. (A) Representative intracellular staining for IL-33 in two donor samples by B cell subset compared to isotype control. Dashed line represents gating cutoff for IL-33hi cells. Pro-B (CD19+ CD10+ IgM− CD34+), large pre-B (LPB, CD19+ CD10+ IgM− CD34− FSC-Ahi SSC-Ahi), small pre-B (SPB, CD19+ CD10+ IgM− CD34− FSC-Alo SSC-Alo), immature B cells (Imm, CD19+ CD10+ CD34− IgM+ IgD−), and mature B cells including recirculating B cells (Mat, CD19+ CD10−). (B) Percentage of IL-33hi bone marrow B cells by subset. (C) Geometric MFI of IL-33 in bone marrow B cells by subset. (D) Normalized IL33 expression in B cell CLL (n=862) collected from peripheral blood as compared to normal peripheral blood CD19+ B cells (n=39) of healthy controls displayed as log2 fold change. Data are displayed as the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001) by one-way ANOVA with Tukey posttest or unpaired two tailed t-test with Welch’s correction.
Given that reduced or absent IL-33 expression increased developing B cell fitness in mice and that IL-33 expression was low but detectable in mature recirculating B cells in human bone marrow, we considered whether IL-33 expression was reduced in human B cell malignancies wherein enhanced cell fitness is a defining disease characteristic. First, IL33 mRNA was detectable in peripheral B cells from healthy donors and patients with B cell chronic lymphocytic leukemia (B-CLL). Compared to peripheral blood CD19+ B cells from healthy donors, expression of IL33 mRNA was significantly decreased in peripheral blood tumor cells from patients with B-CLL (Fig. 6D). In contrast, IL33 mRNA expression was increased in biopsies of all B cell lymphoma types that we evaluated, including follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), Burkitt’s lymphoma (BL), mantle cell lymphoma (MCL), marginal zone lymphoma (MZL), and Hodgkin’s lymphoma (HL) (Supplemental Fig. 3). Collectively, these data suggest that IL-33 downregulation is favored in B-CLL, possibly secondary to an adaptive fitness advantage, but not B cell lymphomas.
DISCUSSION:
Herein, our data establish an in vivo role of IL-33 during early B cell development that is both cell-intrinsic and ST2-independent. IL-33 is a two-domain protein, with the C-terminal domain primarily involved in binding to ST2 on immune cells following release of IL-33 from epithelial and endothelial cells. The N-terminal domain has a nuclear localization sequence, a putative homeodomain-like helix-turn-helix motif, and a chromatin-binding motif that interacts with histones H2A and H2B suggesting a potential cell-intrinsic role for IL-33 (9, 14, 15). Early evidence supported a gene regulatory role for IL-33, with an IL-33-Gal4-DNA-binding domain fusion protein potently repressing GAL4-responsive luciferase expression in an in vitro gene reporter assay (14, 15). However, if such a gene regulatory role exists, endogenous targets have largely remained elusive to date. Identification of targets has focused on endothelial and epithelial cells given the high level of expression of IL-33 in these lineages. Knockdown of IL-33 in primary human umbilical vein endothelial cells via multiple siRNAs did not alter the global proteome (21). Moreover, inducible overexpression of IL-33 in a human esophageal epithelial cell line lacking endogenous IL-33 and ST2 expression did not alter the transcriptome of these cells (20). While these global, unbiased approaches have failed to identify a nuclear role for IL-33 in regulating gene expression, multiple smaller in vitro studies have posited targets including IL6, IL13, CCL5, and RELA (NF-κB p65) based on gene expression differences with overexpression or knockdown of IL-33 complemented by chromatin immunoprecipitation of IL-33 (16-19). While intriguing, these studies were all conducted in vitro in cell lines with variable potential for endogenous ST2 expression and whether such interactions exist in vivo and have any phenotypic or mechanistic significance remained to be determined.
Consistent with a unique cell-intrinsic role for IL-33, the magnitude of Il33 expression in developing B cells was 40-100 fold lower than in stromal cells and instead at the low end of expression of well-established B cell-intrinsic regulators. Our present work does not definitively establish a nuclear vs. cytoplasmic role for IL-33. However, expression of IL-33 was largely restricted to the nucleus in pro-B and LPB cells indicating a high likelihood that the IL-33 is acting in the nucleus. Moreover, we determined a cell-intrinsic role of IL-33 in the context of cellular and environmental stress using competitive reconstitutions in mixed bone marrow chimeric mice. Prior studies on endothelial and epithelial cells have evaluated the role of IL-33 under optimal growth conditions or during catastrophic cellular insult (20, 21). Further investigations will be needed to clarify whether IL-33 functions in a cell-intrinsic manner in other cell types including stromal cells in the context of cellular and environmental stress. Precedent exists within the IL-1 family for cell-intrinsic functions associated with cellular fitness, with the N-terminal domain of IL-1α conferring nuclear translocation and promoting apoptosis in the context of malignancy but not in non-transformed cells (43, 44).
Increasing evidence suggests that IL-33 sequestration in the nuclei of stromal cells may serve a protective function to restrict autoinflammation (3). Transgenic mice expressing an N-terminal deletion of Il33 under the control of the endogenous Il33 promoter accumulated high serum levels of IL-33 and developed fatal autoinflammation in the first 6 months of life via an ST2-dependent mechanism (45). Similarly, skin overexpression of truncated IL-33 lacking the N-terminal domain but not full-length IL-33 induced epidermal and dermal thickening, inflammation, and hyperkeratosis (46). Recent kinetic analysis of IL-33 release demonstrated that chromatin-binding promotes a slower, phased extracellular release of IL-33 during necrosis, serving as an additional post-translational mechanism of IL-33 regulation (20). These collective data provide evidence that, particularly among high IL-33-expressing cells, the nuclear localization of IL-33 may serve a host protective function. Our data suggest that that nuclear IL-33 may serve an additional function depending on the cell type (i.e. B cells vs. stromal cells) or the environmental conditions (i.e. stress vs. homeostasis). These multiple roles of IL-33 support the high degree of sequence conservation across species (14) and may reconcile data regarding the potential of IL-33 to regulate gene expression (14, 15) versus the lack of identifiable targets or cell-intrinsic, IL-33-dependent phenotypes in high IL-33 expressing cells (20, 21).
Signals for both increased proliferation (E2F targets, cell cycle, and DNA replication) and decreased cell death (p53 pathway) in Il33−/− pro-B and LPB cells were present in our transcriptomic analysis. Using in vivo competitive reconstitution experiments with radiation-stimulated hematopoiesis, we consistently identified fewer dead cells among LPB cells of Il33−/− origin and no difference in BrdU incorporation at any B cell developmental stage. One interpretation of these data is that the increase in fitness of Il33−/− B cells is most substantially attributable to a decrease in cell death. Alternatively, the sensitivity or timing of our assays to measure proliferation and cell death may be insufficient to detect differences. Further work will be needed to assess the relative balance of proliferation and apoptosis in the context of IL-33 expression or deficiency.
Given the effect of IL-33 on modulating fitness during B cell development, we considered whether IL-33 expression levels are increased or decreased in B cell malignancies. Interestingly, B-CLL samples were associated with a decreased expression of IL33 mRNA compared to healthy controls. While B-CLL is not considered to originate from pro-B or LPB cells, cancer cells frequently employ diverse pro-survival strategies including those classically used during B cell development regardless of their initiating cell stage. Our data suggest this reduction in IL-33 expression in B-CLL samples from baseline may promote increased leukemic cell fitness. In contrast, all tested B cell lymphomas displayed significantly increased expression of IL33 mRNA. This dichotomy may be related to the differences in tumor microenvironment between B cells that form tumors (i.e. lymphomas) and those that do not (i.e. CLL) and the diverse roles of IL-33, including in the activation of T regulatory cells enhancing immunotolerance to the tumor. Such IL-33-induced immunotolerance has been observed in breast, colorectal, and certain lung cancers (47). Most notably, another group has similarly shown selection for increased IL33 expression in DLBCL, which in mice promoted Treg activation (48). Additionally, while a large proportion of B-CLL samples (79%) had a decrease in IL33 expression, there were samples that displayed an increase in IL33 mRNA compared to healthy controls, suggesting that additional factors including tumor genetics and the tumor microenvironment may influence the relative benefit of modulated IL33 expression. Collectively, the correlation in B-CLL with downregulated IL33 expression, unique among the tested B cell malignancies, suggests beneficial adaptation at the IL33 locus derived from mechanism(s) wherein decreased IL33 expression promotes CLL cell survival.
While our data establish an intracellular role for IL-33 during early B cell development, it remains unclear whether this is due to the hypothesized role of IL-33 as a transcriptional regulator bound either directly to DNA or as part of a transcriptional regulatory complex, or whether IL-33 is acting via an alternative mechanism. Moreover, peak IL-33 expression in pro-B and LPB cells is well conserved across mouse strains and in humans, but the effectors that govern IL-33 expression during early B cell development remain unidentified. Further investigation will be needed to clarify these factors. Collectively, our work defines IL-33 as a B cell-intrinsic, ST2-independent modulator of fitness during in vivo B cell development.
Supplementary Material
KEY POINTS.
IL-33 is expressed during early B cell development in both mice and humans.
Deficiency of IL-33 promotes B cell fitness via a cell-endogenous mechanism.
IL-33 expression is modulated in B cell malignancies.
ACKNOWLEDGMENTS
We appreciate the critical insights of Rachel Brown on this manuscript. We are thankful to Dr. Paul Bryce who provided us with Il33fl/fl-GFP mice prior to deposition in The Jackson Laboratory repository, and to Dr. Andrew McKenzie who provided us with Il33cit/cit and St2−/− mice. We appreciate the assistance of Kevin Weller, David Flaherty, Brittany Matlock, and Chris Warren in the Vanderbilt University Medical Center Flow Cytometry Shared Resource.
Funding: R01AI145265 (NIH) – R.S.P.
R01AI124456 (NIH) – R.S.P.
R01AI111820 (NIH) – R.S.P.
U19AI095227 (NIH) – R.S.P.
I01BX004299 (U.S. Department of VA) – R.S.P.
R21AI145397 (NIH) – R.S.P.
T32 GM007347 – Vanderbilt MSTP
F30 AI114262 – M.T.S.
R01CA226432 – C.M.E.
I01 BX 002882 (Department of VA) – P.L.K.
R01DK084246 (NIH) – P.L.K.
REFERENCES:
- 1.Chackerian AA, Oldham ER, Murphy EE, Schmitz J, Pflanz S, and Kastelein RA. 2007. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J. Immunol 179: 2551–5. [DOI] [PubMed] [Google Scholar]
- 2.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, and Kastelein RA. 2005. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 23: 479–90. [DOI] [PubMed] [Google Scholar]
- 3.Cayrol C, and Girard J-P. 2018. Interleukin-33 (IL-33): A nuclear cytokine from the IL-1 family. Immunol. Rev 281: 154–168. [DOI] [PubMed] [Google Scholar]
- 4.Ali S, Huber M, Kollewe C, Bischoff SC, Falk W, and Martin MU. 2007. IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc. Natl. Acad. Sci 104: 18660–18665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liew FY, Girard J-P, and Turnquist HR. 2016. Interleukin-33 in health and disease. Nat. Rev. Immunol 16: 676–689. [DOI] [PubMed] [Google Scholar]
- 6.Hardman CS, Panova V, and McKenzie ANJ. 2013. IL-33 citrine reporter mice reveal the temporal and spatial expression of IL-33 during allergic lung inflammation. Eur. J. Immunol 43: 488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichery M, Mirey E, Mercier P, Lefrancais E, Dujardin A, Ortega N, and Girard J-P. 2012. Endogenous IL-33 is highly expressed in mouse epithelial barrier tissues, lymphoid organs, brain, embryos, and inflamed tissues: in situ analysis using a novel Il-33-LacZ gene trap reporter strain. J. Immunol 188: 3488–95. [DOI] [PubMed] [Google Scholar]
- 8.Moussion C, Ortega N, and Girard J-P. 2008. The IL-1-Like Cytokine IL-33 Is Constitutively Expressed in the Nucleus of Endothelial Cells and Epithelial Cells In Vivo: A Novel ‘Alarmin’? PLoS One 3: e3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baekkevold ES, Roussigné M, Yamanaka T, Johansen F-E, Jahnsen FL, Amalric F, Brandtzaeg P, Erard M, Haraldsen G, and Girard J-P. 2003. Molecular Characterization of NF-HEV, a Nuclear Factor Preferentially Expressed in Human High Endothelial Venules. Am. J. Pathol 163: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Küchler AM, Pollheimer J, Balogh J, Sponheim J, Manley L, Sorensen DR, De Angelis PM, Scott H, and Haraldsen G. 2008. Nuclear Interleukin-33 Is Generally Expressed in Resting Endothelium but Rapidly Lost upon Angiogenic or Proinflammatory Activation. Am. J. Pathol 173: 1229–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlock CI, Wu J, Zhou C, Tatum K, Adams HP, Tan F, and Lou Y. 2014. Unique Temporal and Spatial Expression Patterns of IL-33 in Ovaries during Ovulation and Estrous Cycle Are Associated with Ovarian Tissue Homeostasis. J. Immunol 193: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molofsky AB, Van Gool F, Liang H-E, Van Dyken SJ, Nussbaum JC, Lee J, Bluestone JA, and Locksley RM. 2015. Interleukin-33 and Interferon-γ Counter-Regulate Group 2 Innate Lymphoid Cell Activation during Immune Perturbation. Immunity 43: 161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wills-Karp M, Rani R, Dienger K, Lewkowich I, Fox JG, Perkins C, Lewis L, Finkelman FD, Smith DE, Bryce PJ, Kurt-Jones EA, Wang TC, Sivaprasad U, Hershey GK, and Herbert DR. 2012. Trefoil factor 2 rapidly induces interleukin 33 to promote type 2 immunity during allergic asthma and hookworm infection. J. Exp. Med 209: 607–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carriere V, Roussel L, Ortega N, Lacorre D-A, Americh L, Aguilar L, Bouche G, and Girard J-P. 2007. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc. Natl. Acad. Sci. U. S. A 104: 282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roussel L, Erard M, Cayrol C, and Girard J-P. 2008. Molecular mimicry between IL-33 and KSHV for attachment to chromatin through the H2A-H2B acidic pocket. EMBO Rep. 9: 1006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao D, Perros F, Caramori G, Meng C, Dormuller P, Chou P-C, Church C, Papi A, Casolari P, Welsh D, Peacock A, Humbert M, Adcock IM, and Wort SJ. 2014. Nuclear IL-33 regulates soluble ST2 receptor and IL-6 expression in primary human arterial endothelial cells and is decreased in idiopathic pulmonary arterial hypertension. Biochem. Biophys. Res. Commun 451: 8–14. [DOI] [PubMed] [Google Scholar]
- 17.Ni Y, Tao L, Chen C, Song H, Li Z, Gao Y, Nie J, Piccioni M, Shi G, and Li B. 2015. The Deubiquitinase USP17 Regulates the Stability and Nuclear Function of IL-33. Int. J. Mol. Sci 16: 27956–27966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan J, Oshima T, Wu L, Fukui H, Watari J, and Miwa H. 2016. Interferon γ-Induced Nuclear Interleukin-33 Potentiates the Release of Esophageal Epithelial Derived Cytokines. PLoS One 11: e0151701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi Y-S, Park JA, Kim J, Rho S-S, Park H, Kim Y-M, and Kwon Y-G. 2012. Nuclear IL-33 is a transcriptional regulator of NF-κB p65 and induces endothelial cell activation. Biochem. Biophys. Res. Commun 421: 305–11. [DOI] [PubMed] [Google Scholar]
- 20.Travers J, Rochman M, Miracle CE, Habel JE, Brusilovsky M, Caldwell JM, Rymer JK, and Rothenberg ME. 2018. Chromatin regulates IL-33 release and extracellular cytokine activity. Nat. Commun 9: 3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier V, Cayrol C, Farache D, Roga S, Monsarrat B, Burlet-Schiltz O, Gonzalez de Peredo A, and Girard J-P. 2016. Extracellular IL-33 cytokine, but not endogenous nuclear IL-33, regulates protein expression in endothelial cells. Sci. Rep 6: 34255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T, and Edgar R. 2006. [19] Gene Expression Omnibus: Microarray Data Storage, Submission, Retrieval, and Analysis. In Methods in enzymology vol. 411 352–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma E-J, Greiner TC, Weisenburger DD, Rosenwald A, Ott G, Müller-Hermelink H-K, Gascoyne RD, Delabie J, Rimsza LM, Braziel RM, Grogan TM, Campo E, Jaffe ES, Dave BJ, Sanger W, Bast M, Vose JM, Armitage JO, Connors JM, Smeland EB, Kvaloy S, Holte H, Fisher RI, Miller TP, Montserrat E, Wilson WH, Bahl M, Zhao H, Yang L, Powell J, Simon R, Chan WC, Staudt LM, and Lymphoma/Leukemia Molecular Profiling Project. 2006. Molecular Diagnosis of Burkitt’s Lymphoma. N. Engl. J. Med 354: 2431–2442. [DOI] [PubMed] [Google Scholar]
- 24.Adams CM, Mitra R, Gong JZ, and Eischen CM. 2017. Non-Hodgkin and Hodgkin Lymphomas Select for Overexpression of BCLW. Clin. Cancer Res. 23: 7119–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, and Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 26.Gautier L, Cope L, Bolstad BM, and Irizarry RA. 2004. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315. [DOI] [PubMed] [Google Scholar]
- 27.Johnson WE, Li C, and Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, and Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, and Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P. 2015. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 1: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stier MT, Zhang J, Goleniewska K, Cephus JY, Rusznak M, Wu L, Van Kaer L, Zhou B, Newcomb DC, and Peebles RS. 2018. IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J. Exp. Med 215: 263–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards S, Watanabe C, Santos L, Craxton A, and Clark EA. 2008. Regulation of B-cell entry into the cell cycle. Immunol. Rev 224: 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heng TSP, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, Asinovski N, Davis S, Fassett M, Feuerer M, Gray DHD, Haxhinasto S, Hill JA, Hyatt G, Laplace C, Leatherbee K, Mathis D, Benoist C, Jianu R, Laidlaw DH, Best JA, Knell J, Goldrath AW, Jarjoura J, Sun JC, Zhu Y, Lanier LL, Ergun A, Li Z, Collins JJ, Shinton SA, Hardy RR, Friedline R, Sylvia K, and Kang J. 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol 9: 1091–1094. [DOI] [PubMed] [Google Scholar]
- 35.Le H, Kim W, Kim J, Cho HR, and Kwon B. 2013. Interleukin-33: A Mediator of Inflammation Targeting Hematopoietic Stem and Progenitor Cells and Their Progenies. Front. Immunol 4: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allakhverdi Z, Comeau MR, Smith DE, Toy D, Endam LM, Desrosiers M, Liu Y-J, Howie KJ, Denburg JA, Gauvreau GM, and Delespesse G. 2009. CD34+ hemopoietic progenitor cells are potent effectors of allergic inflammation. J. Allergy Clin. Immunol 123: 472–478.e1. [DOI] [PubMed] [Google Scholar]
- 37.Dai X, Chen Y, Di L, Podd A, Li G, Bunting KD, Hennighausen L, Wen R, and Wang D. 2007. Stat5 is essential for early B cell development but not for B cell maturation and function. J. Immunol 179: 1068–79. [DOI] [PubMed] [Google Scholar]
- 38.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, and Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. U. S. A 103: 13789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labi V, Erlacher M, Krumschnabel G, Manzl C, Tzankov A, Pinon J, Egle A, and Villunger A. 2010. Apoptosis of leukocytes triggered by acute DNA damage promotes lymphoma formation. Genes Dev. 24: 1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puccetti MV, Fischer MA, Arrate MP, Boyd KL, Duszynski RJ, Bétous R, Cortez D, and Eischen CM. 2017. Defective replication stress response inhibits lymphomagenesis and impairs lymphocyte reconstitution. Oncogene 36: 2553–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemazee D 2017. Mechanisms of central tolerance for B cells. Nat. Rev. Immunol 17: 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu JL, and Davis MM. 2000. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13: 37–45. [DOI] [PubMed] [Google Scholar]
- 43.POLLOCK AS, TURCK J, and LOVETT DH. 2003. The prodomain of interleukin 1α interacts with elements of the RNA processing apparatus and induces apoptosis in malignant cells. FASEB J. 17: 203–213. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson FT, Turck J, Locksley RM, and Lovett DH. 1997. The N-terminal propiece of interleukin 1 alpha is a transforming nuclear oncoprotein. Proc. Natl. Acad. Sci. U. S. A 94: 508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bessa J, Meyer CA, de Vera Mudry MC, Schlicht S, Smith SH, Iglesias A, and Cote-Sierra J. 2014. Altered subcellular localization of IL-33 leads to non-resolving lethal inflammation. J. Autoimmun 55: 33–41. [DOI] [PubMed] [Google Scholar]
- 46.Kurow O, Frey B, Schuster L, Schmitt V, Adam S, Hahn M, Gilchrist D, McInnes IB, Wirtz S, Gaipl US, Krönke G, Schett G, Frey S, and Hueber AJ. 2017. Full Length Interleukin 33 Aggravates Radiation-Induced Skin Reaction. Front. Immunol 8: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afferni C, Buccione C, Andreone S, Galdiero MR, Varricchi G, Marone G, Mattei F, and Schiavoni G. 2018. The Pleiotropic Immunomodulatory Functions of IL-33 and Its Implications in Tumor Immunity. Front. Immunol 9: 2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arima H, Nishikori M, Otsuka Y, Kishimoto W, Izumi K, Yasuda K, Yoshimoto T, and Takaori-Kondo A. 2018. B cells with aberrant activation of Notch1 signaling promote Treg and Th2 cell–dominant T-cell responses via IL-33. Blood Adv. 2: 2282–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.